Abstract
Uridine diphosphoglucuronosyltransferases (UGTs) 1A6 is the only UGT1A isoform expressed in lung tissue. It is responsible for the detoxification of carcinogens such as benezo[a]pyrene from cigarette smoke. The purpose of this study was to evaluate the association of UGT1A6 polymorphisms and haplotypes with lung cancer risk and to evaluate the functional significance of UGT1A6 polymorphisms. Genomic DNA was isolated from leukocytes. Eight UGT1A6 polymorphisms were sequenced in a test set of 72 Chinese lung cancer patients and 62 healthy controls. Potential risk modifying alleles were validated in a separate set of 95 Chinese lung cancer patients and 100 healthy controls. UGT1A6 19T>G, 541A>G and 552A>C showed significant association with increased lung cancer risk, while UGT1A6 105C>T and IVS1+130G>T were significantly associated with reduced lung cancer risk. Multivariate logistic regression analysis demonstrated a significant association of lung cancer with UGT1A6 541A>G (OR: 3.582, 95% CI: 1.27–10.04, p = 0.015), 552A>C (OR: 5.364, 95% CI: 1.92–14.96, p = 0.001) and IVS1+130G>T (OR: 0.191, 95% CI: 0.09–0.36, p<0.001). Functional test demonstrated that UGT1A6 105C>T increased mRNA stability, providing a plausible explanation of its association with reduced lung cancer risk. Thus UGT1A6 polymorphisms may be used to identify people with increased risk of developing lung cancer.
Introduction
Non-small cell lung cancer (NSCLC) accounts for more than 85% of primary lung cancers and approximately two-thirds of NSCLC patients are diagnosed at an advanced stage. Cigarette smoking has been linked to lung cancer [1]–[4], and over 60 carcinogens had been identified in cigarette smoke [5]–[7]. Of these, benzo[a]pyrene (BaP) and 4-(Methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) are the most studied. Metabolic activation of BaP and NNK by cytochrome P450 enzymes results in the formation of more reactive metabolites that can bind covalently to DNA, an important step in lung cancer induction [8]–[10].
The uridine diphosphoglucuronosyltransferases (UGTs) belong to a superfamily of metabolizing enzymes responsible for detoxifying numerous endobiotics and xenobiotics [11]. UGTs are responsible for the detoxification of BaP and NNK [12]–[15]. Lower glucuronidation activity had been implicated in lung cancer susceptibility [16]. In addition, genetic polymorphisms in UGTs have correlated with risk and incidence of different cancers including lung cancer [12], [17]–[20]. Amongst all UGT1A isoforms, UGT1A6 had been shown to be the only UGT1A isoform expressed in lung tissue [21].
We hypothesized that polymorphisms in the UGT1A6 may affect the ability to detoxify lung cancer carcinogens and modulate lung cancer risk. Here, we present the findings of a case-control study using 2 different cohorts and functionally characterized UGT1A6 105C>T polymorphism in vitro.
Materials and Methods
Study population
A total of 167 Chinese lung cancer cases and 162 Chinese healthy controls from 2 independent cohorts were analyzed. The first cohort comprised of 72 lung cancer patients and 62 healthy controls from the Cancer Center and Blood Donation Center from the National University Hospital, Singapore, respectively. A second cohort consisted of 95 lung cancer patients and 100 controls from 4 clinical studies and was used as validation set. The subjects in this cohort were lung cancer patients from two therapeutic phase II studies (n = 44 and 14 respectively) and a germline biomarkers study (n = 37) and eligible cancer-free controls from a case-control study of familial colorectal cancer in Singapore. The study protocol was approved by the institutional ethnics review board at the National University Hospital, Singapore, and all study subjects provided written informed consent.
Data collection
In the first cohort, study subjects were interviewed in person by trained medical professionals who used a structured questionnaire. Demographic information including age, gender, smoking status, pack years of smoking and histological cell type was collected for all subjects. Similar information for the second cohort was retrieved from case records, and information on the smoking status for the controls was not obtained. Smoking status was categorized into never-smoker, ex-smoker and current smoker. Histological cell types were categorized into three groups, namely adenocarcinoma, squamous cell carcinoma and poorly differentiated carcinoma.
Single Nucleotide Polymorphism (SNP) genotyping
Eight SNPs which were relatively common in the Asian population based on past literature search, were selected for this study. Of the four variants in exon 1, three were coding (cSNPs) resulting in amino acid changes, UGT1A6 19T>G (S7A), 541A>G (T181A) and 552A>C (R184S), whereas UGT1A6 105C>T was a synonymous variant. The remaining three sequence variants in the 5′ regulatory region included 2 SNPs, UGT1A6 −556C>T and −427G>C, and one deletion mutation −1310 to −1306 delete (aggag). UGT1A6 intronic variant IVS1+130G>T was also studied.
Genomic DeoxyriboNucleic Acid (DNA) was extracted from buffy coat fractions using Puregene DNA purification kit (Gentra Systems, Inc., Minneapolis, MN, US). The quality and quantity of DNA were assessed by NanoDrop (ND-1000 Spectrophotometer). All DNA has an A260/A280 ratio of 1.7–1.9. Genotyping was conducted using Pyrosequencing. For pyrosequencing assays, one positive and one negative control were included in each 96-well plate. Briefly, primers were designed based on the UGT1A6 sequence. The PCR primers and sequencing primers were designed using Pyrosequencing Assay Design Software (version 1.0.6: Biotage AB, Uppsala, Sweden). BLAST analysis was performed to confirm the specificity of the primers for each exon. Details of primer sequences and the PCR conditions are listed in Table S1. 250 ng genomic DNA at 50 ng/µl was used for each PCR reaction and PCR products were then subjected to pyrosequencing with PyroMark™ ID device (Pyrosequencing, Uppsala, Sweden). Validation steps were carried out by direct sequencing reactions using the ABI PRISM® BigDye™ terminator v 3.1 cycle sequencing chemistry.
Plasmid constructs
Functional studies on nonsynonymous polymorphisms (UGT1A6 19T>G, 541A>G and 552A>C) have been done, whereas IVS1+130G>T is an intronic SNP, therefore we selected UGT1A6 105C>T, which is a synonymous polymorphism for our functional testing. To evaluate whether UGT1A6 105C>T has any impact on UGT1A6 activity, the variant 105TT was generated. Briefly, plasmid vector encoding full-length UGT1A6*1 sequence was kindly provided by Dr. Michael H. Court (Tufts University School of Medicine, Boston, Massachusetts). The UGT1A6 coding region was subcloned into pcDNA 3.1 V5/His Topo expression plasmid vector (Invitrogen). BLAST analysis was performed to confirm the UGT1A6 sequence of the plasmid. The plasmid was then used as template to generate substitution at nucleotide 105 by using the GeneEditor in vitro site-directed mutagenesis system (Promega) with the indicated primer 105CT (5′-5Phos/ccc tca gga Tgg aag cca c-3′) and primer Bottom Strand (provided). All DNA constructs were verified by sequencing analysis. Briefly, HEK293 cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and 50 U/ml penicillin (Gibco) in a humidified atmosphere of 5% carbon dioxide at 37C. Stable transfections of plasmid into HEK293 cells were performed using Lipofectamine 2000 (Invitrogen) at a reagent to DNA ratio of 3∶1 in Opti-MEM® I Reduced Serum Medium (Gibco). All transfection experiments were carried out in triplicates.
RNA stability assay
We next examined the possible effect of the variant UGT1A6 105TT on mRNA stability. 10 µg/ml actinomycin D (Sigma) was applied to the cell cultures grown overnight and cells were harvested at selected intervals up to 24 hours. The Rneasy Plus Mini kit (Qiagen) was used to extract total RNA from transfected cells, before and after incubation with actinomycin D.
Quantification of transcripts
RNA was isolated from 5×105 cells at the indicated times (0, 2, 4, 8 and 24 hours) after treatment with ActD and were reverse transcribed to cDNA using SuperScript III (Invitrogen). The PCR reaction was then carried out comprising the following: 1 µl cDNA, 10 µM of each primers (UGT1A6: 5′-cagctgtcctcaagagagatgtgga-3′ and 5′-ccaatgaagaccatgttgggc-3′, GAPDH: 5′-tgaaggtcggagtcaacggatttggt-3′ and 5′-catgtgggccatgaggtccaccac-3′) and 2× Power SYBR Green Master Mix (which include SYBR Green dye, Taq Polymearse, ROX, and dNTP) in a final volume of 20 µl. Empty vector-transfected cells was utilized as control, with data normalized by GAPDH expression. All PCR reactions were carried out in triplicates.
Western blotting
We further examined the effect of the variant UGT1A6 105TT on protein stability. Treated cells were lysed and protein concentrations were measured. Proteins (7.5 µg/well) were resolved by 4 to 15% gradient SDS-polyacrylamide gel electrophoresis (Invitrogen) followed by transferred to a polyvinylidene fluoride (PVDF) microporous membrane (MILLIPORE, Billerica, MA, USA) blocked in PBS with 5% milk. After blocking, the membrane was probed using a rabbit anti-human UGT1A6 primary antibody diluted 1∶1000 (WB-UGT1A6; BD Gentest, Woburn, MA) and a goat anti-rabbit horseradish peroxidase-conjugated secondary antibody (1∶10,000 diluted). Blots were imaged by ECL reagents (ECL Prime, Amersham Pharmacia Biotech AB, Uppsala, Sweden) and by film processor (Konica SRX 101). Results are from two independent experiments.
Enzyme assay
Serotonin was used as substrate as it demonstrated substrate specificity to UGT1A6 [22], [23], however, a different platform methodology was used in this study. 0.01 mg of proteins was subjected to 384-well black plate (Corning, NY) with Transcreener ® HTS Fluorescent Intensity (FI) Assay kit (BellBrook Labs, Madison, WI). Each enzyme reaction was carried out comprising the following: 50 mM HEPES (pH 7.5), containing NaCl (100 mM), MgCl2 (4 mM), EGTA (2 mM), 0.01% Brij-35, dithiothreitol (DTT; 2 mM), DMSO (1%), UDPGA (5 mM) and with varying concentrations of serotonin (Sigma) 0.5–30 mM in a final volume of 25 µl. The plate was incubated at RT for 45 min and reactions were stopped by the addition of 25 µl of ice-cold methanol. The plate was then read on a Tecan Infinite M200 plate reader. All enzyme reactions were carried out in triplicates. The initial readouts were converted to the amount of UDP formed using GraphPad Prism to interpolate these values from the UDPGA/UDP standard curves. Michaelis-Menten curves were generated using GraphPad Prism. Data plotted are from two independent experiments and results are presented as the means ± S.D. of triplicates.
Statistical Analyses
Case-control differences in baseline characteristics were evaluated using t-tests (for continuous variables) and Chi-square tests (for categorical variables). Odds ratios (OR) were derived from Chi-square test, and 95% confidence intervals (CI) of the OR were given. The association between SNPs with age, gender, smoking status and TNM stages were tested in lung cancer subjects using chi-square test. All statistical analysis including univariate and multivariate logistic regression analysis were performed using SPSS version 11.5 (SPSS Inc., Chicago,Ill.,USA). Hardy-Weineberg equilibrium test, linkage disequilibrium analysis (denoted as D′) and case-control haplotype analysis (permutation test) were calculated using SNPAlyze ver.7.
Results
Characteristics of patients and controls
Baseline characteristics of lung cancer patients and controls of both cohorts are summarized in Table 1. There was a statistically significant difference in age, gender and smoking status between lung cancer patients and healthy controls, with more male (p = 0.003) and smokers (p<0.001) in the lung cancer group. Compared to healthy controls, patients with lung cancers, were generally older (p = 0.04). Adenocarcinoma was the most common histological subtype, accounting for 60% in lung cancer patients. There was no significant difference in the baseline characteristics of lung cancer patients and healthy controls between the testing and validation cohorts (data not shown).
Table 1. Baseline characteristics of Lung cancer patients and controls.
| Characteristics | Patients = 167 (%) | Controls = 162 (%) | p-values |
| Age (Range) | 63 (39–79) | 36 (18–67) | 0.04 |
| Gender | |||
| Male | 129 (77%) | 100 (62%) | 0.003 |
| Female | 38 (23%) | 62 (38%) | |
| Smoking status | |||
| Never | 55 (33%) | 120 (74%) | <0.001 |
| Ex-smoker | 55 (33%) | 26 (16%) | |
| Current smoker | 57 (34%) | 16 (10%) | |
| Pack-years of smoking (Range) | 21.6 (0–160) | 0 (0–35) | ns |
| Histological Cell Type | |||
| Adenocarcinoma | 101 (60%) | ||
| Squarmous cell carcinoma (SCC) | 25 (15%) | ||
| Poorly differentiated | 41 (25%) |
*Smoking history was not obtained for the 100 controls.
Genotype and allele frequencies of UGT1A6 polymorphisms (Table 2)
Table 2. Genotype and allele frequencies of UGT1A6 SNPs in both first and second cohorts.
| First cohort: 72 lung cancer patients and 62 controls | |||||||
| SNPs | Genotyping frequency (%) | Allele frequency (%) | p-value | OR (95%CI) | |||
| −1310del5 (rs45549435) | Aggag/Aggag | Aggag/– | –/– | Aggag | – | ||
| Patients | 50 (69) | 18 (25) | 4 (6) | 0.82 | 0.18 | 0.4903 | 0.783 (0.389 to 1.571) |
| Controls | 38 (61) | 22 (36) | 2 (3) | 0.79 | 0.21 | ||
| −556C>T (rs45568235) | CC | CT | TT | C | T | ||
| Patients | 70 (97) | 2 (3) | 0 (0) | 0.99 | 0.01 | 0.5303 | 0.562 (0.091 to 3.478) |
| Controls | 59 (95) | 3 (5) | 0 (0) | 0.98 | 0.02 | ||
| −427G>C (rs12476197) | GG | GC | CC | G | C | ||
| Patients | 49 (68) | 23 (32) | 0 (0) | 0.84 | 0.16 | 0.9691 | 0.986 (0.476 to 2.040) |
| Controls | 45 (73) | 16 (26) | 1 (2) | 0.86 | 0.15 | ||
| 19T>G (rs6759892) | TT | TG | GG | T | G | ||
| Patients | 19 (26) | 51 (71) | 2 (3) | 0.62 | 0.38 | <0.0001 | 5.579 (2.681 to 11.61) |
| Controls | 40 (65) | 22 (35) | 0 (0) | 0.83 | 0.17 | ||
| 105C>T (rs45535938) | CC | CT | TT | C | T | ||
| Patients | 55 (76) | 17 (24) | 0 (0) | 0.88 | 0.12 | 0.0376 | 0.458 (0.217 to 0.963) |
| Controls | 37 (60) | 25 (40) | 0 (0) | 0.8 | 0.2 | ||
| 541A>G (rs2070959) | AA | AG | GG | A | G | ||
| Patients | 7 (10) | 65 (90) | 0 (0) | 0.55 | 0.45 | <0.0001 | 25.07 (9.819 to 64.00) |
| Controls | 48 (77) | 14 (23) | 0 (0) | 0.89 | 0.11 | ||
| 552A>C (rs1105879) | AA | AC | CC | A | C | ||
| Patients | 5 (7) | 65 (90) | 2 (3) | 0.52 | 0.48 | <0.0001 | 22.72 (7.992 to 64.60) |
| Controls | 38 (61) | 24 (39) | 0 (0) | 0.8 | 0.2 | ||
| IVS1+130G>T (rs7592281) | GG | GT | TT | G | T | ||
| Patients | 55 (76) | 16 (23) | 1 (1) | 0.88 | 0.12 | <0.0001 | 0.241 (0.116–0.502) |
| Controls | 26 (43) | 36 (57) | 0 (0) | 0.72 | 0.28 | ||
Of the eight genotyped SNPs, five were significantly associated with lung cancer risk, when tested in the first cohort consisting of 72 lung cancer patients and 62 controls. UGT1A6 19T>G, 541A>G and 552A>C were associated with increased lung cancer risk, whereas UGT1A6 105C>T and IVS1+130G>T were inversely associated with lung cancer risk. UGT1A6 19T>G, 541A>G and 552A>C were common in lung cancer patients with minor allele frequencies of 38%, 45% and 48% respectively.
These 5 SNPs were further evaluated in the second cohort consisting of 95 Chinese lung cancer patients and 100 Chinese unmatched controls. UGT1A6 19T>G, 541A>G and 552A>C remained associated with increased lung cancer risk, and UGT1A6 105C>T and IVS1+130G>T remained significantly associated with reduced lung cancer risk.
Determination of factors associated independently with lung cancer: Univariate and multivariate analyses
To determine factors associated with lung cancer risk, we first used univariate analysis and identified the following factors: age, gender, smoking status, and five SNPs (Table 3). After adjusting for covariables identified by univariate analysis using multivariate analysis we found only smoking status (OR: 3.385, 95% CI: 1.86–6.15, p<0.001), UGT1A6 541A>G (OR: 3.582, 95% CI: 1.27–10.04, p = 0.015), 552A>C (OR: 5.364, 95% CI: 1.92–14.96, p = 0.001) and IVS1+130G>T (OR: 0.191, 95% CI: 0.09–0.36, p<0.001) were independent predictive factor for lung cancer risk.
Table 3. Univariate and multivariate models for baseline characteristics.
| Univariate analysis using chi-square test | Multivariate analysis using logistic regression | ||||||||
| Parameter | Patients = 167 (%) | Control = 162 (%) | p-value | OR | 95%CI | Adjusted p-value | OR | 95%CI | |
| Age (Median) | >63 | 78 (47%) | 1 (0.6%) | <0.001 | 141 | 19–1030 | |||
| ≤63 | 89(53%) | 161(99.4%) | |||||||
| Gender | Male | 129(77%) | 100(62%) | 0.002 | 2.105 | 1.30–3.40 | 0.134 | 1.615 | 0.86–3.02 |
| Female | 38(23%) | 62(38%) | |||||||
| Smoking | Currentsmoker/ex-smoker | 112(67%) | 66(41%) | <0.001 | 2.962 | 1.89–4.64 | <0.001 | 3.385 | 1.86–6.15 |
| Neversmoker | 55(33%) | 96(59%) | |||||||
| 19TG | TG,GG | 111(66%) | 50 (31%) | <0.001 | 4.440 | 2.79–7.05 | 0.436 | 1.331 | 0.64–2.73 |
| TT | 56(34%) | 112(69%) | |||||||
| 105CT | CT,TT | 24(14%) | 47 (29%) | 0.001 | 0.411 | 0.23–0.71 | 0.169 | 0.602 | 0.29–1.24 |
| CC | 143(86%) | 115 (71%) | |||||||
| 541AG | AG,GG | 116(69%) | 30(19%) | <0.001 | 10.008 | 5.97–16.75 | 0.015 | 3.582 | 1.27–10.04 |
| AA | 51(31%) | 132(81%) | |||||||
| 552AC | AC,CC | 125(75%) | 33(20%) | <0.001 | 11.634 | 6.93–19.53 | 0.001 | 5.364 | 1.92–14.96 |
| AA | 42(25%) | 129(80%) | |||||||
| IVS1+130 | GT,TT | 45(27%) | 85(52%) | <0.001 | 0.334 | 0.21–0.52 | <0.001 | 0.191 | 0.09–0.36 |
| GG | 122(73%) | 77(48%) | |||||||
Hardy-Weinberg Equilibrium, Linkage Disequilibrium (LD) and Haplotype analysis
Further analysis of the set of DNA polymorphisms that are inherited together, also known as haplotypes was performed with the following results.UGT1A6 19T>G and IVS1+130G>T showed significant deviation from Hardy-Weinberg equilibrium among controls. UGT1A6 552A>C was in close LD with UGT1A6 541A>G (D′ = 0.9706, r2 = 0.7795) in lung cancer patients. UGT1A6 105C allele was strongly linked to UGT1A6 541G and 552A allele in control (Table 4). Table 5 summarizes the haplotype frequencies of case-control comparisons from permutation tests. UGT1A6 541A>G were excluded from this analysis because it was in close LD with 552A>C. Ten unique haplotypes could be inferred using 4 risk modifying alleles, with data censored at frequency of 1%.
Table 4. Analysis of pairwise linkage disequilibrium (LD) coefficients and statistics (D′) for lung cancer patients (below diagonal) and control (above diagonal) among five SNPs.
| 19T>G | 105C>T | 541A>G | 552A>C | IVS1+130G>T | |
| (rs6759892) | (rs45535938) | (rs2070959) | (rs1105879) | (rs7592281) | |
| 19T>G (rs6759892) | −0.1822 | 0.5688 | 0.504 | 0.1226 | |
| 105C>T (rs45535938) | 0.2539 | −0.9999 | −1 | 0.2469 | |
| 541A>G (rs2070959) | 0.7045 | 0.3647 | 0.7759 | 0.5854 | |
| 552A>C (rs1105879) | 0.8419 | 0.2342 | 0.9706 | 0.4159 | |
| IVS1+130G>T (rs7592281) | 0.5757 | 0.0512 | 0.3512 | 0.447 |
Table 5. Haplotype frequency estimates and significant levels of case-control comparison from permutation tests.
| 19T>G | 105C>T | 552A>C | IVS1+130G>T | Frequency | permutation | OR | CI | ||
| (rs6759892) | (rs45535938) | (rs1105879) | (rs7592281) | Lung cancer | Control | p-value | |||
| 1 | T | C | A | G | 0.466 | 0.538 | ns | 0.75 | 0.55–1.01 |
| 2 | G | C | C | G | 0.231 | 0.023 | *** | 13.57 | 6.15–29.93 |
| 3 | T | C | A | T | 0.035 | 0.142 | *** | 0.23 | 0.12–0.43 |
| 4 | T | C | C | G | 0.077 | 0.031 | ** | 2.65 | 1.26–5.59 |
| 5 | T | T | A | G | 0.055 | 0.067 | ns | 0.78 | 0.70.41–1.49 |
| 6 | G | C | C | T | 0.096 | 0.043 | * | 2.35 | 1.23–4.48 |
| 7 | T | T | A | T | 0.008 | 0.066 | *** | 0.13 | 0.04–0.44 |
| 8 | G | C | A | G | 0.026 | 0.058 | ns | 0.44 | 0.20–1.00 |
| 9 | G | T | A | G | 0.004 | 0.018 | ns | 0.16 | 0.02–1.33 |
| 10 | G | C | A | T | 0.004 | 0.014 | ns | 0.19 | 0.02–1.65 |
P<0.05,
P<0.01,
P<0.001.
Among these, five haplotypes showed significant p-value in the permutation test, of which, three were associated with increased lung cancer risk. Haplotype #2 containing two risk alleles of UGT1A6 19G and 552C had a higher OR of 13.57 (95% CI: 6.15–29.93) compared to haplotype #4 (OR: 2.65, 95% CI: 1.26–5.59) containing one risk allele of UGT1A6 552A and haplotype #6 (OR: 2.35, 95% CI: 1.23–4.48) containing two risk alleles of UGT1A6 19G and 552C, and one ‘protective’ allele of UGT1A6 intron 1 +130T. Two haplotypes were associated with reduced lung cancer risk, including haplotype #3 (OR: 0.23, 95% CI: 0.12–0.43) and haplotype #7 (OR: 0.13, 95% CI: 0.04–0.44).
In vitro functional characterization of variant UGT1A6 105TT
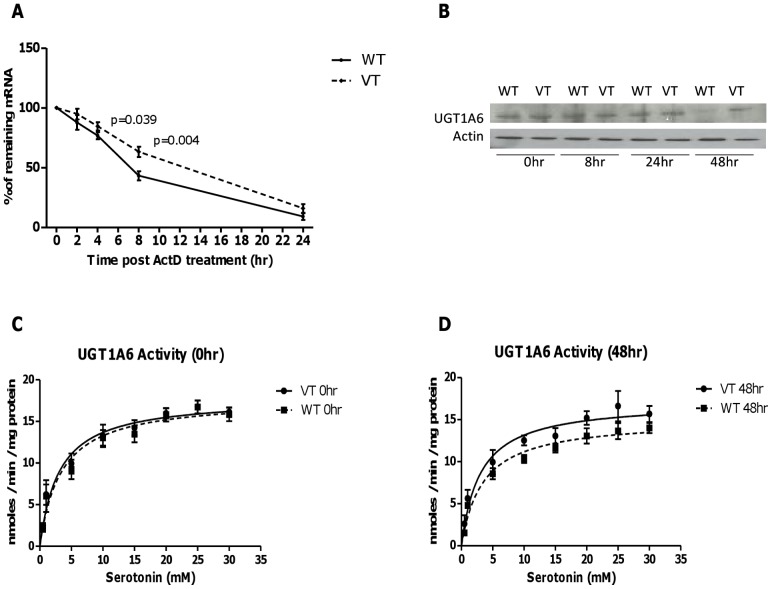
In vitro, mRNA stability was tested after blocking transcription by treating cells with actinomycin D (ActD). Variant UGT1A6 105TT mRNA was more stable than wild type with significantly higher mRNA level observed at 4 hours (p = 0.039), and 8 hours (p = 0.004) after treatment with ActD (Figure 1A). Increased stability of variant UGT1A6 105TT mRNA resulted in higher protein expression and enzyme activity compared to wild-type at 48 hours after treatment with ActD (Figure 1B, C and D).
Figure 1. In vitro functional characterization of UGT1A6 105C>T polymorphism.
UGT1A6*1 and UGT1A6 105TT constructs were stably transfected into HEK293 cells. (A) Cells were harvested upon treatment with ActD at different time point up to 24 hours. The data plotted are time course changes in the remaining amount of UGT1A6 mRNA after ActD treatment. There was significantly higher mRNA level in UGT1A6 105TT than UGT1A6*1 transfected cells at 4 hours (p = 0.039) and 8 hours (p = 0.004) after treatment with ActD. (B) Western blot analysis. Cell lysate used was from 5×105 cells at the indicated times (0, 8, 24 and 48 hours) after treatment with ActD. There was a down-regulation in protein expression in UGT1A6*1 as compared to UGT1A6 variant at 48 hours after treatment with ActD. UGT1A6 activity was assessed by evaluating the production of serotonin glucuronide using lysate from variant UGT1A6 (VT) and UGT1A6*1 (WT) at 0 hour (C) and 48 hours (D) after ActD treatment, with substrate concentrations varied from 0.5 to 30 mM serotonin. Activities are expressed as reaction velocity (nanomoles of serotonin glucuronide formed per minute per milligram of protein).
Discussion
This is the first study to describe the association between UGT1A6 polymorphisms and lung cancer. The UGT1A6 gene is responsible for the detoxification of carcinogens such as BaP from cigarette smoke [13]–[15]. Quantitative or qualitative alterations in UGT1A6 expression may affect the rate of glucuronidation, thereby modifying the risk of developing lung cancer.
Several polymorphisms have been reported in the 5′-regulatory region, exon 1 and introns of UGT1A6 [24], [25]. Among these, three non-synonymous polymorphisms at codons 7, 181 and 184 (UGT1A6 19T>G, 541A>G and 552A>C, respectively), collectively referred to as UGT1A6*2, was the most commonly studied. It is classified as a ‘low activity’ variant for several phenolic compounds, including aspirin [26] and is associated with reduced colorectal adenoma in subjects taking long-term aspirin [27].
In this study, we had demonstrated that individuals with UGT1A6*2 (haplotype #2 and 6) had increased lung cancer risk. Our finding was consistent with a report of reduced glucuronidation activity in individuals with the UGT1A6*2 allele compared with wild type [26]. Krishnaswamy.et al also demonstrated 50% lower UGT1A6 mRNA levels (p = 0.026) in carriers of UGT1A6 19T>G polymorphism compared with noncarriers but without significant effect on UGT1A6 protein content or glucuronidation activities [24]. It should be noted that there were contradictory reports from at least 2 studies, suggesting increased enzyme activity in UGT1A6*2 allozymes [28], [29]. However, the increased activity in the variant UGT1A6 allozyme did not translate into increased substrate clearance in variant human liver microsomes [29]. It is likely that apart from enzyme activity, mRNA degradation may be responsible for the variability of in UGT1A6 activity.
Two polymorphisms UGT1A6 105C>T and IVS1+130G>T were found to be associated with reduced lung cancer risk. The UGT1A6 105C>T is a synonymous SNP that is located in exon 1 of the UGT1A6 gene. Although the polymorphism does not result in qualitative modification of UGT1A6, our in vitro results have demonstrated that the T allele is associated with increased mRNA stability and high UGT1A6 activity in variant protein compared to wild-type. This may explain the protective effect of the UGT1A6 105 T allele in reducing lung cancer risk. In addition, the protective effect of UGT1A6 105C>T may be due to its inverse linkage with UGT1A6*2 (UGT1A6 541A>G and 552A>C).
UGT1A6 IVS1+130G>T was found to be the only SNP associated with decreased risk of lung cancer in multivariate analysis. It is unclear how UGT1A6 IVS1+130G>T modulates lung cancer risk as it is located in the first intronic region of the UGT1A6 gene. In-silico analysis using the Human Splicing Finder program (http://www.umd.be/HSF/) suggested that this SNP did not affect any conserved nucleotides of the branch site in the intronic region (data not shown) and would be unlikely to affect splicing activity.
Although we found UGT1A6 IVS1+130G>T to be in moderate linkage disequilibrium with UGT1A6 541A>G (denoted as UGT1A6*5), it cannot explain the protective effect observed. While UGT1A6 is the predominant UGT1A isoform expressed in lung, other UGT1A isoforms (UGT1A1 and 1A9) expressed in the liver may still affect systemic clearance of carcinogens associated with cigarette smoking [30], [31].
It is possible that IVS1+130G>T may be in linkage disequilibrium with polymorphisms in other UGT1A isoforms that play a ‘protective’ role in lung cancer risk. For example, Saeki.et al [32] demonstrated that UGT1A6 IVS1+130G>T is in close LD with UGT1A9*22, a high enzymatic activity allele which is known to increase transcription. In addition, UGT1A6 has been demonstrated to be in linkage disequilibrium with UGT1A1 and UGT1A7 [33]–[36]. Although not expressed in lung, these UGT isoforms have been demonstrated to be important in clearance of BaP [30], [31].
The main strength of this study is that our finding was validated in an independent cohort. The analysis was restricted to one ethnic group to minimize the likelihood of false-positive results due to population heterogeneity. There are, however, several limitations in our study that we acknowledge. Firstly, the lung cancer patients and healthy controls were not matched for age, gender and smoking history. Secondly, two SNPs, UGT1A6 19T>G and IVS1+130G>T deviated from Hardy Weinberg equation. Deviation from Hardy Weinberg equation may be caused by genotyping errors [37], [38]. We had taken measure to rule out genotyping error by using both pyrosequencing and direct sequencing method to identify the variant SNPs in our study subjects.
In conclusion, this study suggests that UGT1A6 polymorphisms may modulate lung cancer risk. UGT1A6 105C>T was demonstrated to increase mRNA stability, providing a plausible explanation of its association with reduced lung cancer risk. Our study suggests that UGT1A6 genotyping may be integrated into lung cancer screening strategy to help identify susceptible populations.
Supporting Information
List of PCR and sequencing primers. List of all primers used to amplify and sequence each of the eight UGT1A6 SNPs is shown, with information on SNP rsid and their localization provided. The size of PCR products and the annealing temperature for each PCR reaction are also shown. Biotinylated primers for pyrosequencing assay are indicated with §.
(DOC)
Acknowledgments
We thank Dr.Michael H.Court (Tufts University School of Medicine, Boston, Massachusetts) for providing us plasmid vector encoding full-length UGT1A6*1 sequence.
Funding Statement
This work was supported by the National Healthcare Group (Singapore) small innovative grant [NHG-SIG/06007]. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Auerbach O, Forman JB, Gere JB, Kassouny DY, Muehsam GE, et al. (1957) Changes in the bronchial epithelium in relation to smoking and cancer of the lung; a report of progress. N Engl J Med 256: 97–104. [DOI] [PubMed] [Google Scholar]
- 2. Auerbach O, Stout AP, Hammond EC, Garfinkel L (1961) Changes in bronchial epithelium in relation to cigarette smoking and in relation to lung cancer. N Engl J Med 265: 253–267. [DOI] [PubMed] [Google Scholar]
- 3. Lubin JH, Caporaso N, Wichmann HE, Schaffrath-Rosario A, Alavanja MC (2007) Cigarette smoking and lung cancer: modeling effect modification of total exposure and intensity. Epidemiology 18: 639–648. [DOI] [PubMed] [Google Scholar]
- 4. Lubin JH, Caporaso NE (2006) Cigarette smoking and lung cancer: modeling total exposure and intensity. Cancer Epidemiol Biomarkers Prev 15: 517–523. [DOI] [PubMed] [Google Scholar]
- 5. Hoffmann D, Adams JD, Lisk D, Fisenne I, Brunnemann KD (1987) Toxic and carcinogenic agents in dry and moist snuff. J Natl Cancer Inst 79: 1281–1286. [PubMed] [Google Scholar]
- 6. Hoffmann D, Djordjevic MV, Fan J, Zang E, Glynn T, et al. (1995) Five leading U.S. commercial brands of moist snuff in 1994: assessment of carcinogenic N-nitrosamines. J Natl Cancer Inst 87: 1862–1869. [DOI] [PubMed] [Google Scholar]
- 7. Swauger JE, Steichen TJ, Murphy PA, Kinsler S (2002) An analysis of the mainstream smoke chemistry of samples of the U.S. cigarette market acquired between 1995 and 2000. Regul Toxicol Pharmacol 35: 142–156. [DOI] [PubMed] [Google Scholar]
- 8. Denissenko MF, Pao A, Tang M, Pfeifer GP (1996) Preferential formation of benzo[a]pyrene adducts at lung cancer mutational hotspots in P53. Science 274: 430–432. [DOI] [PubMed] [Google Scholar]
- 9. Shields PG, Bowman ED, Harrington AM, Doan VT, Weston A (1993) Polycyclic aromatic hydrocarbon-DNA adducts in human lung and cancer susceptibility genes. Cancer Res 53: 3486–3492. [PubMed] [Google Scholar]
- 10. Hecht SS (1999) Tobacco smoke carcinogens and lung cancer. J Natl Cancer Inst 91: 1194–1210. [DOI] [PubMed] [Google Scholar]
- 11. Izukawa T, Nakajima M, Fujiwara R, Yamanaka H, Fukami T, et al. (2009) Quantitative analysis of UDP-glucuronosyltransferase (UGT) 1A and UGT2B expression levels in human livers. Drug Metab Dispos 37: 1759–1768. [DOI] [PubMed] [Google Scholar]
- 12. Zheng Z, Park JY, Guillemette C, Schantz SP, Lazarus P (2001) Tobacco carcinogen-detoxifying enzyme UGT1A7 and its association with orolaryngeal cancer risk. J Natl Cancer Inst 93: 1411–1418. [DOI] [PubMed] [Google Scholar]
- 13. Gschaidmeier H, Seidel A, Burchell B, Bock KW (1995) Formation of mono- and diglucuronides and other glycosides of benzo(a)pyrene-3,6-quinol by V79 cell-expressed human phenol UDP-glucuronosyltransferases of the UGT1 gene complex. Biochem Pharmacol 49: 1601–1606. [DOI] [PubMed] [Google Scholar]
- 14. Jin CJ, Miners JO, Burchell B, Mackenzie PI (1993) The glucuronidation of hydroxylated metabolites of benzo[a]pyrene and 2-acetylaminofluorene by cDNA-expressed human UDP-glucuronosyltransferases. Carcinogenesis 14: 2637–2639. [DOI] [PubMed] [Google Scholar]
- 15. Mackenzie PI, Rodbourn L, Iyanagi T (1993) Glucuronidation of carcinogen metabolites by complementary DNA-expressed uridine 5′-diphosphate glucuronosyltransferases. Cancer Res 53: 1529–1533. [PubMed] [Google Scholar]
- 16. Richie JP Jr, Carmella SG, Muscat JE, Scott DG, Akerkar SA, et al. (1997) Differences in the urinary metabolites of the tobacco-specific lung carcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone in black and white smokers. Cancer Epidemiol Biomarkers Prev 6: 783–790. [PubMed] [Google Scholar]
- 17. Araki J, Kobayashi Y, Iwasa M, Urawa N, Gabazza EC, et al. (2005) Polymorphism of UDP-glucuronosyltransferase 1A7 gene: a possible new risk factor for lung cancer. Eur J Cancer 41: 2360–2365. [DOI] [PubMed] [Google Scholar]
- 18. Guillemette C, Millikan RC, Newman B, Housman DE (2000) Genetic polymorphisms in uridine diphospho-glucuronosyltransferase 1A1 and association with breast cancer among African Americans. Cancer Res 60: 950–956. [PubMed] [Google Scholar]
- 19. Guillemette C, De Vivo I, Hankinson SE, Haiman CA, Spiegelman D, et al. (2001) Association of genetic polymorphisms in UGT1A1 with breast cancer and plasma hormone levels. Cancer Epidemiol Biomarkers Prev 10: 711–714. [PubMed] [Google Scholar]
- 20. Gallagher CJ, Muscat JE, Hicks AN, Zheng Y, Dyer AM, et al. (2007) The UDP-glucuronosyltransferase 2B17 gene deletion polymorphism: sex-specific association with urinary 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol glucuronidation phenotype and risk for lung cancer. Cancer Epidemiol Biomarkers Prev 16: 823–828. [DOI] [PubMed] [Google Scholar]
- 21. Zhang W, Liu W, Innocenti F, Ratain MJ (2007) Searching for tissue-specific expression pattern-linked nucleotides of UGT1A isoforms. PLoS One 2: e396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Krishnaswamy S, Duan SX, Von Moltke LL, Greenblatt DJ, Court MH (2003) Validation of serotonin (5-hydroxtryptamine) as an in vitro substrate probe for human UDP-glucuronosyltransferase (UGT) 1A6. Drug Metab Dispos 31: 133–139. [DOI] [PubMed] [Google Scholar]
- 23. Krishnaswamy S, Hao Q, Von Moltke LL, Greenblatt DJ, Court MH (2004) Evaluation of 5-hydroxytryptophol and other endogenous serotonin (5-hydroxytryptamine) analogs as substrates for UDP-glucuronosyltransferase 1A6. Drug Metab Dispos 32: 862–869. [DOI] [PubMed] [Google Scholar]
- 24. Krishnaswamy S, Hao Q, Al-Rohaimi A, Hesse LM, von Moltke LL, et al. (2005) UDP glucuronosyltransferase (UGT) 1A6 pharmacogenetics: I. Identification of polymorphisms in the 5′-regulatory and exon 1 regions, and association with human liver UGT1A6 gene expression and glucuronidation. J Pharmacol Exp Ther 313: 1331–1339. [DOI] [PubMed] [Google Scholar]
- 25. Saeki M, Saito Y, Jinno H, Sai K, Kaniwa N, et al. (2005) Genetic polymorphisms of UGT1A6 in a Japanese population. Drug Metab Pharmacokinet 20: 85–90. [DOI] [PubMed] [Google Scholar]
- 26. Ciotti M, Marrone A, Potter C, Owens IS (1997) Genetic polymorphism in the human UGT1A6 (planar phenol) UDP-glucuronosyltransferase: pharmacological implications. Pharmacogenetics 7: 485–495. [DOI] [PubMed] [Google Scholar]
- 27. Chan AT, Tranah GJ, Giovannucci EL, Hunter DJ, Fuchs CS (2005) Genetic variants in the UGT1A6 enzyme, aspirin use, and the risk of colorectal adenoma. J Natl Cancer Inst 97: 457–460. [DOI] [PubMed] [Google Scholar]
- 28. Nagar S, Zalatoris JJ, Blanchard RL (2004) Human UGT1A6 pharmacogenetics: identification of a novel SNP, characterization of allele frequencies and functional analysis of recombinant allozymes in human liver tissue and in cultured cells. Pharmacogenetics 14: 487–499. [DOI] [PubMed] [Google Scholar]
- 29. Krishnaswamy S, Hao Q, Al-Rohaimi A, Hesse LM, von Moltke LL, et al. (2005) UDP glucuronosyltransferase (UGT) 1A6 pharmacogenetics: II. Functional impact of the three most common nonsynonymous UGT1A6 polymorphisms (S7A, T181A, and R184S). J Pharmacol Exp Ther 313: 1340–1346. [DOI] [PubMed] [Google Scholar]
- 30. Fang JL, Beland FA, Doerge DR, Wiener D, Guillemette C, et al. (2002) Characterization of benzo(a)pyrene-trans-7,8-dihydrodiol glucuronidation by human tissue microsomes and overexpressed UDP-glucuronosyltransferase enzymes. Cancer Res 62: 1978–1986. [PubMed] [Google Scholar]
- 31. Fang JL, Lazarus P (2004) Correlation between the UDP-glucuronosyltransferase (UGT1A1) TATAA box polymorphism and carcinogen detoxification phenotype: significantly decreased glucuronidating activity against benzo(a)pyrene-7,8-dihydrodiol(-) in liver microsomes from subjects with the UGT1A1*28 variant. Cancer Epidemiol Biomarkers Prev 13: 102–109. [DOI] [PubMed] [Google Scholar]
- 32. Saeki M, Saito Y, Jinno H, Sai K, Ozawa S, et al. (2006) Haplotype structures of the UGT1A gene complex in a Japanese population. Pharmacogenomics J 6: 63–75. [DOI] [PubMed] [Google Scholar]
- 33. Yea SS, Lee SS, Kim WY, Liu KH, Kim H, et al. (2008) Genetic variations and haplotypes of UDP-glucuronosyltransferase 1A locus in a Korean population. Ther Drug Monit 30: 23–34. [DOI] [PubMed] [Google Scholar]
- 34. Kohle C, Mohrle B, Munzel PA, Schwab M, Wernet D, et al. (2003) Frequent co-occurrence of the TATA box mutation associated with Gilbert's syndrome (UGT1A1*28) with other polymorphisms of the UDP-glucuronosyltransferase-1 locus (UGT1A6*2 and UGT1A7*3) in Caucasians and Egyptians. Biochem Pharmacol 65: 1521–1527. [DOI] [PubMed] [Google Scholar]
- 35. Lampe JW, Bigler J, Horner NK, Potter JD (1999) UDP-glucuronosyltransferase (UGT1A1*28 and UGT1A6*2) polymorphisms in Caucasians and Asians: relationships to serum bilirubin concentrations. Pharmacogenetics 9: 341–349. [DOI] [PubMed] [Google Scholar]
- 36. Pacheco PR, Brilhante MJ, Ballart C, Sigalat F, Polena H, et al. (2009) UGT1A1, UGT1A6 and UGT1A7 genetic analysis: repercussion for irinotecan pharmacogenetics in the Sao Miguel Island Population (Azores, Portugal). Mol Diagn Ther 13: 261–268. [DOI] [PubMed] [Google Scholar]
- 37. Hosking L, Lumsden S, Lewis K, Yeo A, McCarthy L, et al. (2004) Detection of genotyping errors by Hardy-Weinberg equilibrium testing. Eur J Hum Genet 12: 395–399. [DOI] [PubMed] [Google Scholar]
- 38. Xu J, Turner A, Little J, Bleecker ER, Meyers DA (2002) Positive results in association studies are associated with departure from Hardy-Weinberg equilibrium: hint for genotyping error? Hum Genet 111: 573–574. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of PCR and sequencing primers. List of all primers used to amplify and sequence each of the eight UGT1A6 SNPs is shown, with information on SNP rsid and their localization provided. The size of PCR products and the annealing temperature for each PCR reaction are also shown. Biotinylated primers for pyrosequencing assay are indicated with §.
(DOC)