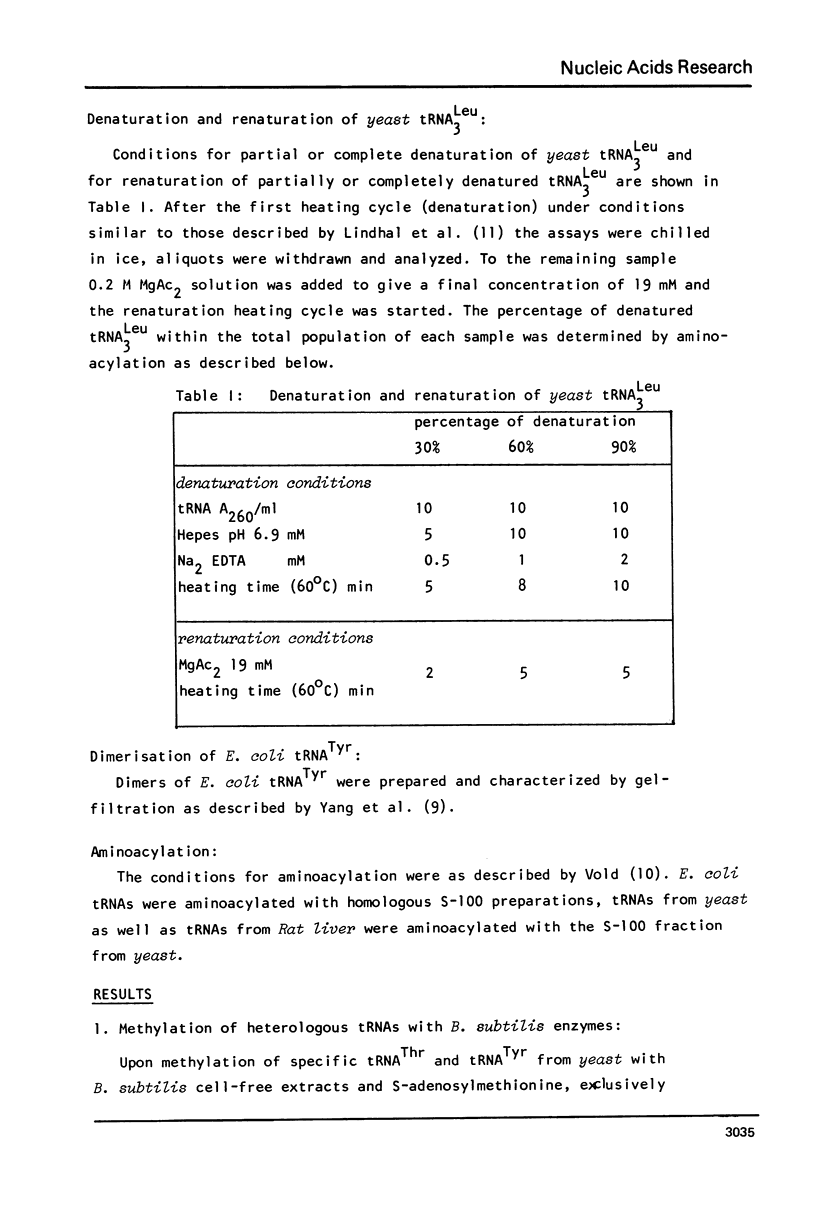
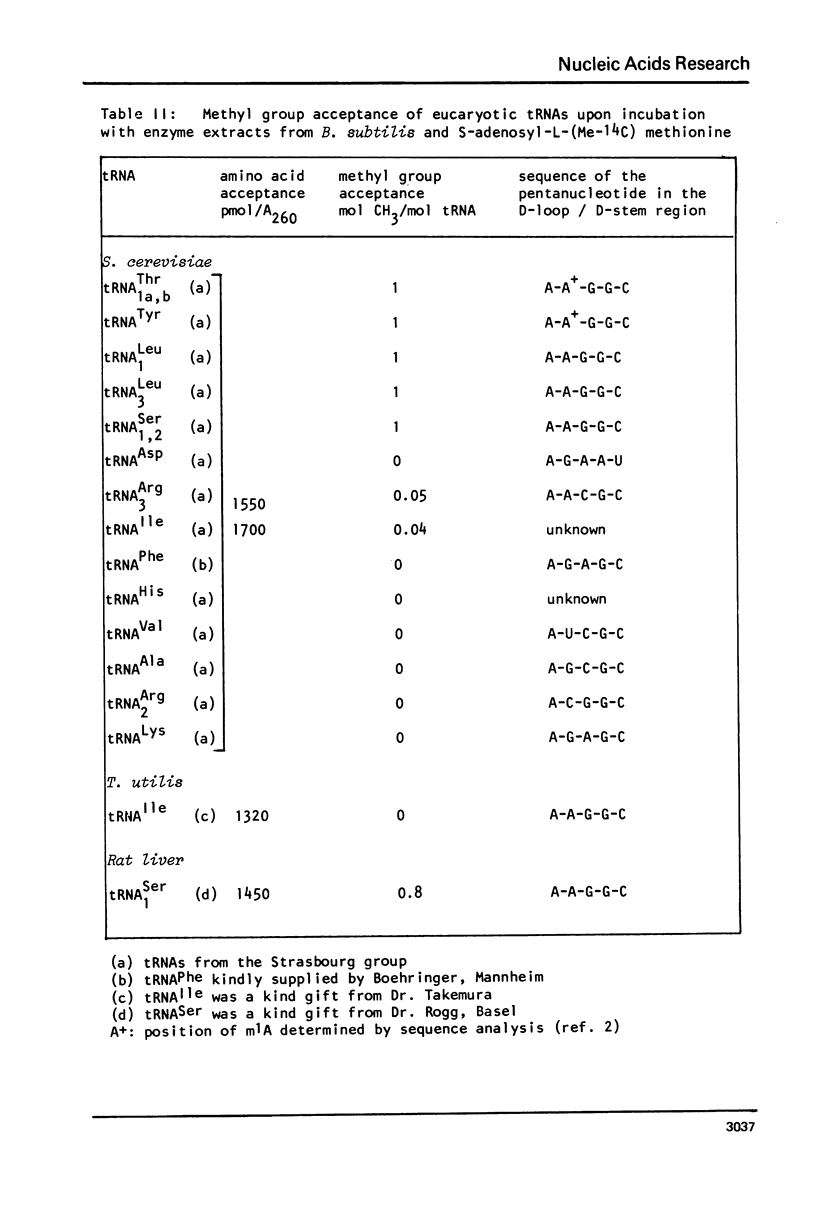
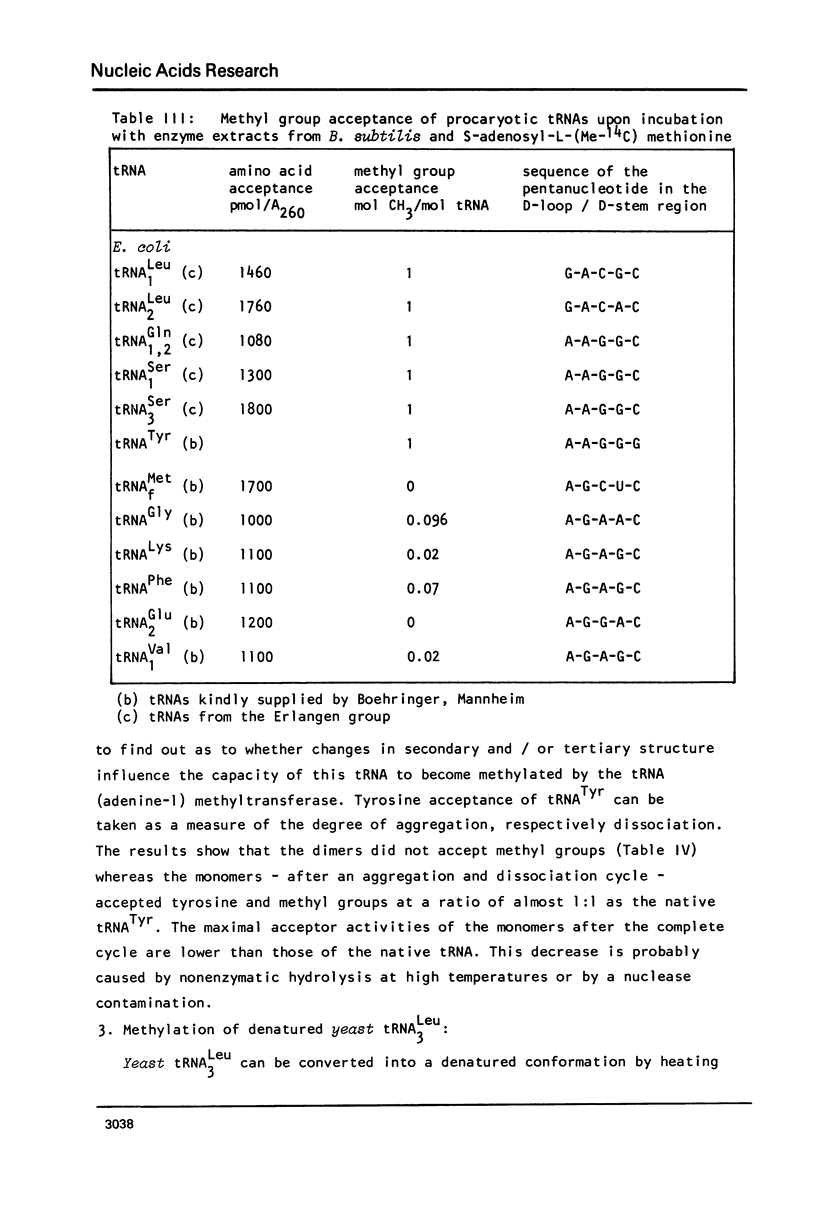
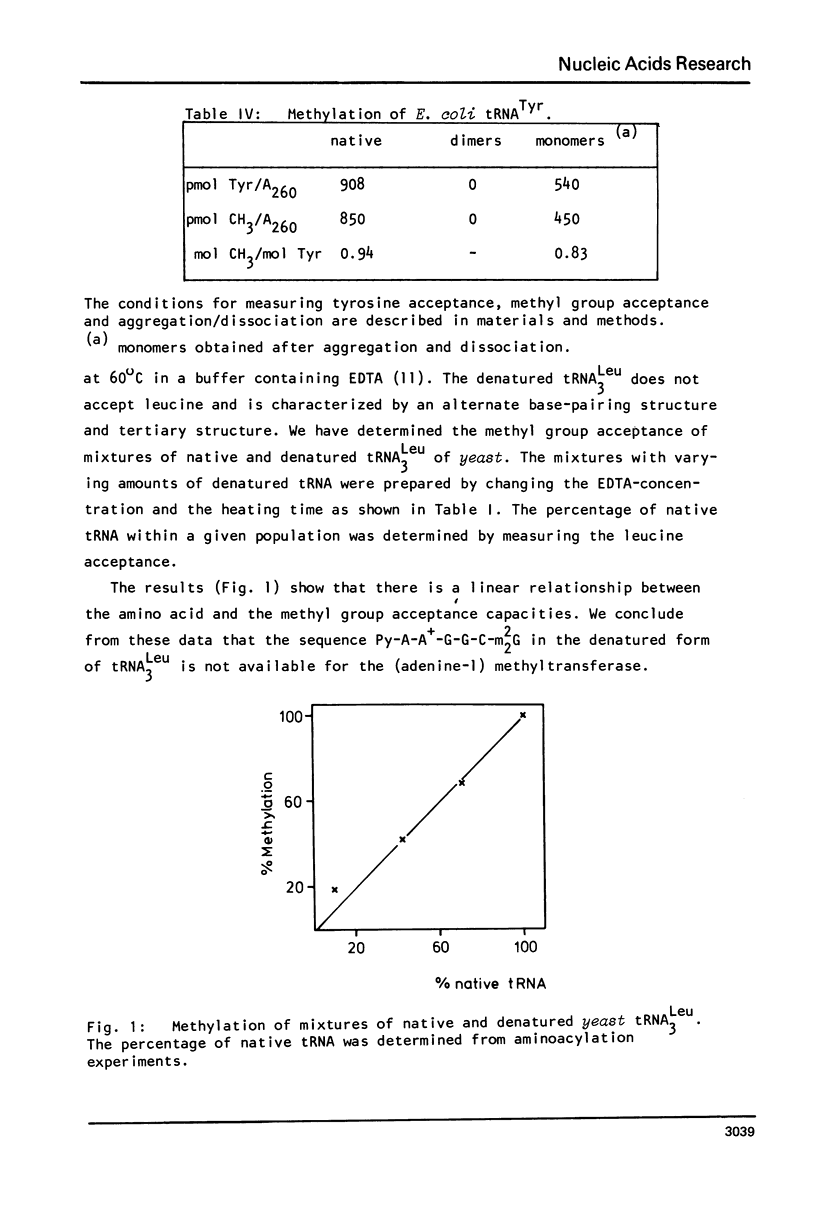
Abstract
Bulk tRNA from yeast and Rat liver can be methylated in vitro with -adenosylmethionine and B, subtilis extracts. The sole product formed is 1-methyladenosine (m1A). This tRNA (adenine-1) methyltransferase converts quantitatively the 3'-terminal adenosine-residue in the dihydrouridine-loop of tRNAThr and tRNATyr from yeast into m1A. Out of 16 eucaryotic tRNAs with known sequences 6 accepted methyl groups, all at a molar ratio of 1. These tRNAs have in common an unpaired adenosine-residue at the specific site in the sequence Py-A-A+-G-G-C-m2G. Out of 12 tRNAs from E. coli 6 served as specific substrates. These E. coli tRNAs also have an unpaired adenosine-residue at the 3'-end of the D-loop. Besides restrictions in primary structure intact secondary and tertiary structure is important for recognition of the specific tRNAs by the enzyme.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arnold H., Kersten H. The occurrence of ribothymidine, 1-methyladenosine, methylated guanosines and the corresponding methyltransferases in E. coli and Bacillus subtilis. FEBS Lett. 1973 Oct 1;36(1):34–38. doi: 10.1016/0014-5793(73)80331-x. [DOI] [PubMed] [Google Scholar]
- Dirheimer G., Ebel J. P. Fractionnement des tRNA de levure de bière par distribution en contre-courant. Bull Soc Chim Biol (Paris) 1967;49(12):1679–1687. [PubMed] [Google Scholar]
- Glick J. M., Averyhart V. M., Leboy P. S. Purification and characterization of two tRNA-(guanine)-methyltransferases from rat liver. Biochim Biophys Acta. 1978 Mar 29;518(1):158–171. doi: 10.1016/0005-2787(78)90125-9. [DOI] [PubMed] [Google Scholar]
- Kerr S. J., Borek E. The tRNA methyltransferases. Adv Enzymol Relat Areas Mol Biol. 1972;36:1–27. doi: 10.1002/9780470122815.ch1. [DOI] [PubMed] [Google Scholar]
- Kraus J. Recognition of individual Escherichia coli transfer ribonucleic acids by 1-adenine-specific methyltransferase from rat liver. Biochem J. 1978 Jan 1;169(1):247–249. doi: 10.1042/bj1690247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus J., Staehelin M. N2-guanine specific transfer RNA methyltransferase I from rat liver and leukemic rat spleen. Nucleic Acids Res. 1974 Nov;1(11):1455–1478. doi: 10.1093/nar/1.11.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchino Y., Nishimura S. Methylation of Escherichia coli transfer ribonucleic acids by adenylate residue-specific transfer ribonucleic acid methylase from rat liver. Biochemistry. 1974 Aug 27;13(18):3683–3688. doi: 10.1021/bi00715a010. [DOI] [PubMed] [Google Scholar]
- Lindahl T., Adams A., Fresco J. R. Renaturation of transfer ribonucleic acids through site binding of magnesium. Proc Natl Acad Sci U S A. 1966 Apr;55(4):941–948. doi: 10.1073/pnas.55.4.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nau F. The methylation of tRNA. Biochimie. 1976;58(6):629–645. doi: 10.1016/s0300-9084(76)80387-2. [DOI] [PubMed] [Google Scholar]
- Pegg A. E. Sites of methylation of purified transfer ribonucleic acid preparations by enzymes from normal tissues and from tumours induced by dimethylnitrosamine and 1,2-dimethylhydrazine. Biochem J. 1974 Feb;137(2):239–248. doi: 10.1042/bj1370239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope W. T., Brown A., Reeves R. H. The identification of the tRNA substrates for the supK tRNA methylase. Nucleic Acids Res. 1978 Mar;5(3):1041–1057. doi: 10.1093/nar/5.3.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raettig R., Kersten H., Weissenbach J., Dirheimer G. Methylation of an adenosine in the D-loop of specific transfer RNAs from yeast by a procaryotic tRNA (adenine-1) methyltransferase. Nucleic Acids Res. 1977 Jun;4(6):1769–1782. doi: 10.1093/nar/4.6.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raettig R., Schmidt W., Mahal G., Kersten H., Arnold H. H. Purification and characterization of tRNAMet-f, tRNAPhe and tRNATyr2 from Baccillus subtilis. Biochim Biophys Acta. 1976 Jun 18;435(2):109–118. doi: 10.1016/0005-2787(76)90241-0. [DOI] [PubMed] [Google Scholar]
- Rordorf B. F., Kearns D. R. Nuclear magnetic resonance investigation of the base-pairing structure of Escherichia coli tRNATyr monomer and dimer conformations. Biochemistry. 1976 Jul 27;15(15):3320–3330. doi: 10.1021/bi00660a024. [DOI] [PubMed] [Google Scholar]
- Simoncsits A., Brownlee G. G., Brown R. S., Rubin J. R., Guilley H. New rapid gel sequencing method for RNA. Nature. 1977 Oct 27;269(5631):833–836. doi: 10.1038/269833a0. [DOI] [PubMed] [Google Scholar]
- Vold B. S. Analysis of isoaccepting transfer ribonucleic acid species of Bacillus subtilis: chromatographic differences between transfer ribonucleic acids from spores and cells in exponential growth. J Bacteriol. 1973 Feb;113(2):825–833. doi: 10.1128/jb.113.2.825-833.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S. K., Söll D. G., Crothers D. M. Properties of a dimer of tRNA I Tyr 1 (Escherichia coli). Biochemistry. 1972 Jun 6;11(12):2311–2320. doi: 10.1021/bi00762a016. [DOI] [PubMed] [Google Scholar]



