Abstract
Ubiquitin-dependent protein degradation within malarial parasites is a burgeoning field of interest due to several encouraging reports of proteasome inhibitors that were able to confer antimalarial activity. Despite the growing interest in the Plasmodium proteasome system, relatively little investigation has been done to actually characterize the parasite degradation machinery. In this report, we provide an initial biological investigation of the ubiquitylating components of the endoplasmic reticulum-associated degradation (ERAD) system, which is a major pathway in targeting misfolded proteins from the ER to the cytosol for proteasome degradation. We are able to show that the ERAD system is essential for parasite survival and that the putative Plasmodium HRD1 (E3 ubiquitin ligase), UBC (E2 ubiquitin conjugating enzyme) and UBA1 (E1 ubiquitin activating enzyme) are able to mediate in vitro ubiquitylation. Furthermore, by using immunofluorescence, we report that Plasmodium HRD1 localizes to the ER membranes, while the Plasmodium UBC and UBA1 localize to the cytosol. In addition, our gene disruption experiments indicate that the Plasmodium HRD1 is likely essential. We have conducted an initial characterization of the ubiquitylating components of the Plasmodium ERAD system, a major pathway for protein degradation and parasite maintenance. In conjunction with promising proteasome inhibitor studies, we explore the possibility of targeting the Plasmodium ERAD system for future bottom-up drug development approaches.
Introduction
Malaria is one of the deadliest infectious diseases of the world, infecting up to half a billion and killing up to one million people each year [1]. Though progresses were made in combating malaria with multi-drug therapies [2], the high-cost treatment and the rise of drug resistances beckon the need for novel and cheap antimalarials. The causative agents of malaria are protozoan parasites that belong to the genus Plasmodium. Among them, Plasmodium falciparum is the deadliest human-infecting species. With the urgent need of developing new anti-malarial therapies, there has been much effort in understanding how the Plasmodium regulates its life cycle to subsequently identify potential new targets for drug development and therapeutic intervention.
In recent years, several studies showed that proteasome inhibitors have significant antimalarial properties [3]–[7] (see [8] for a review). Previous studies characterized some of P. falciparum’s 26S proteasome components [9], [10] and a bacteria-like proteasome [11]–[13], both involved in protein degradation. Though the protein degradation via the 26S proteasome closely involves ubiquitylation of target substrates, very little work has been done regarding the characterization of the parasite’s ubiquitylating machinery acting upstream of the proteasome, possibly leading to the discovery of parasite-specific divergences that can be exploited for drug targeting. In eukaryotes, the endoplasmic reticulum-associated degradation (ERAD) pathway mediates protein degradation during quality control. Aberrant proteins are recognized by ER luminal chaperone proteins and protein disulfide isomerases to help discriminate properly folded proteins from misfolded proteins [14]. Misfolded proteins are shuttled to the DER1 translocon complex, which forms a hydrophobic pore to allow the retro-translocation of proteins through the ER membrane. Within this translocon complex, the HRD1 E3 ubiquitin ligase interacts with membrane-bound proteins needed for retro-translocation and helps form the hydrophobic pore complex [15]. The HRD1 E3 enzyme also catalyzes, with the intervention of other ubiquitylating enzymes, the ubiquitylation of the target misfolded protein that is the prerequisite for subsequent retro-translocation to the cytosol and destruction by the 26S proteasome [16]–[18]. Typically, ubiquitylation involves the covalent attachment of a ubiquitin moiety to lysine residues of protein substrates via the hierarchical intervention of an E1 ubiquitin-activating enzyme, an E2 ubiquitin-conjugating enzyme, and an E3 ubiquitin ligase that is usually involved in specific substrate recognition [19], [20]. Up until now, no functional study has investigated the ubiquitylating components that compose the malaria parasite ERAD pathway. While ubiquitin E1 and E2 enzymes seem to be well conserved across eukaryotic phyla, Plasmodium E3 ubiquitin ligases have been shown to have high levels of divergences that can be utilized for the development of new antimalarials [21].
Here, we present the identification and biochemical characterization of the ubiquitylating components of the P. falciparum ERAD system: one ubiquitin-activating E1 enzyme, an ubiquitin-conjugating E2 enzyme, and an ubiquitin E3 ligase. Our results show that the Plasmodium ERAD system is vital to the parasite and that recombinant E1, E2, and E3 enzymes promote ubiquitylation in vitro. Furthermore, immunofluorescence microscopy experiments also reveal that E1 and E2 localize to the cytosol whereas the E3 ubiquitin ligase is found within the ER membrane, consistent with their respective functions in the ERAD pathway. Finally, gene disruption experiments suggest that the ubiquitylating components of the Plasmodium ERAD system are essential to the parasite survival. This analysis contributes to our understanding of the mechanisms leading to protein degradation in the human malaria parasite and proposes the components of the ERAD pathway as potential targets for the ‘bottom-up’ development of new drugs.
Results
ERAD-specific Inhibitor Eeyarestatin I Validates the Essentiality of an Ubiquitin-dependent ERAD System in Plasmodium
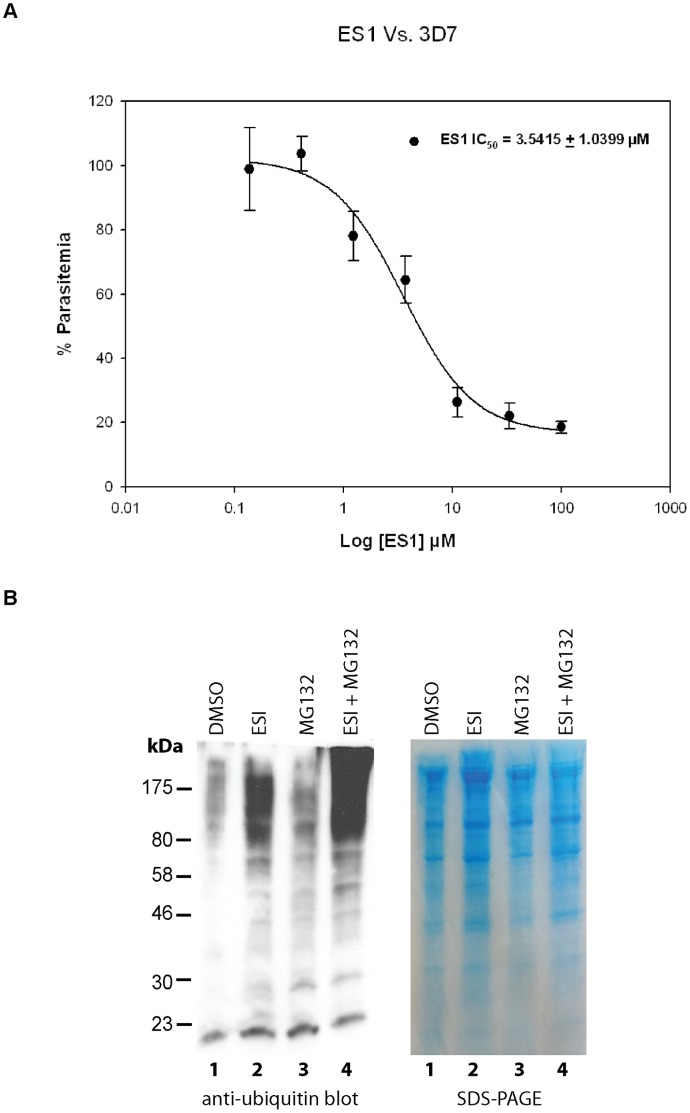
To test the essentiality of the ERAD pathway in Plasmodium, we decided to test the inhibitory effects on P. falciparum cultures with Eeyarestatin I (ESI). ESI a known ERAD inhibitor which acts to block the release of ubiquitylated misfolded proteins from the Sec61 translocon at the ER membrane [22], [23] thus preventing misfolded proteins from reaching the proteasome. P. falciparum parasites were found to be highly sensitive to ESI, with an IC50 of 3.5415±1.0399 µM (Figure 1A). In addition, we validated ESI’s ability to block the Plasmodium ERAD system by analyzing changes in the levels of ubiquitylated substrates between untreated and treated cultures. Parasites that were treated with ESI were found to have heightened abundances of ubiquitylated proteins (Figure 1B). The observed accumulation of ubiquitylated proteins is likely the result of ESI preventing the translocation of ubiquitylated misfolded proteins across the ER membrane for degradation as seen in other organisms [23]. These ESI studies give preliminary indication that Plasmodium does indeed contain an ubiquitin-dependent ERAD system that is integral to parasite survival.
Figure 1. ERAD inhibitor Eeyarestatin I inhibits P. falciparum in vitro and increases the amount of ubiquitylated Plasmodium proteins.
(A) Eeyarestatin I (ES1) inhibits P. falciparum 3D7 strain with an IC50 value of 3.5415±1.0399 µM (mean value ± standard error). Dose-response curve is plotted with increasing concentrations of ES1 (B) ES1 treatment leads to an accumulation of ubiquitylated proteins in P. falciparum. Anti-ubiquitin immunoblots show higher levels of ubiquitylated protein levels in P. falciparum parasites when treated with ES1 (lane 2) compared to the control (DMSO, lane 1). As a comparison, parasite were also treated with proteasome inhibitor MG132 [48] (lane 3) and exhibited slightly higher amounts of ubiquitylated proteins. When parasites were treated with both ES1 and MG132 (lane 4), significant increases in ubiquitylated products were detected.
In silico Analysis of the Plasmodium ERAD Ubiquitylating Components
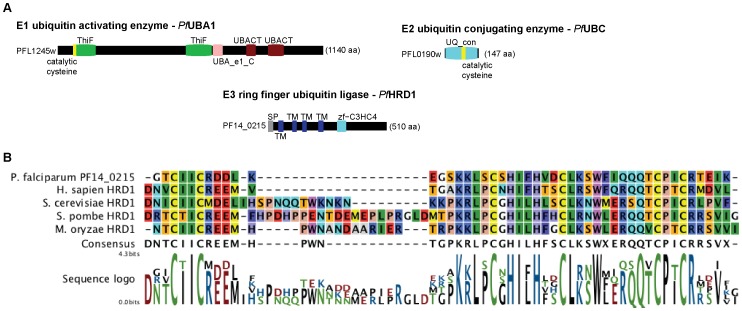
The P. falciparum genome was scanned to identify candidates for the ubiquitylating components of the ERAD system based on sequence homology and domain architecture using a previously published approach [21]. We identified PFL1245w as the putative PfUba1 E1 ubiquitin enzyme, PFL0190w, as the putative PfUbc7 E2 ubiquitin conjugating enzyme, and PF14_0215 as a putative E3 ubiquitin ligase with distant protein homology with the human and yeast HRD1.
Similar to yeast and human UBA1, the 1140 amino acid (aa) long Plasmodium protein encoded by PFL1245w contains a ubiquitin-activating enzyme active site, two ubiquitin-like activating enzyme catalytic domains, two ThiF repeats, and a catalytic cysteine at the N-terminal end (Figure 2A). The short protein (147 aa in length) encoded by PFL0190w has strong homology to the human and yeast UBC7 and contains an ubiquitin conjugating enzyme domain taking up almost its whole length (Figure 2A). Finally, the protein encoded by PF14_0215 presents some sequence similarities with the canonical HRD1 of the ERAD pathway in eukaryotic models [21], [24], [25]. Similar to HRD1, PF14_0215 has multiple transmembrane domains, an E3 RING zinc finger (zf-C3HC4) domain on its C-terminal half, and a predicted signal peptide consistent with ER targeting (Figure 2A). The presence of four transmembrane domains is compatible with an ability to form pores to participate in the recognition and translocation of misfolded proteins across the ER membrane. Based on protein alignments, we show that the PF14_0215 protein contains a highly conserved zf-C3HC4 domain (289–336 aa) similar to that of other characterized HRD1 proteins from various eukaryotic model systems (Figure 2B).
Figure 2. Domain architecture of the ubiquitylating components of the Plasmodium ERAD system and cf-C3HC4 domain alignment of PF14_0215.
(A) Proteins are represented as black boxes of length proportional to their sizes in amino acids (aa). SP = signal peptide; TM = transmembrane domain; ThiF/UBACT = repeats found in E1 ubiquitin-activating enzymes; UBA_e1_C = E1 ubiquitin-activating enzyme catalytic domain; UQ_con = E2 ubiquitin-conjugating enzyme domain; zf-C3HC4 = E3 ubiquitin ligase RING finger domain. Catalytic cysteines are marked in yellow. (B) Using MUSCLE [43], we performed protein alignments on PF14_0215 and found that its zf-C3HC4 domain had high homology to other previously characterized HRD1 of various model systems.
Based on our in silico predictions, it is likely that these proteins have the necessary components to promote ubiquitylation and have enough homology to the human and yeast ERAD components to be part of the parasite’s ERAD system. In the following sections, we conduct some initial biological characterizations to validate our in silico data analyses.
PF14_0215 is an Essential ER Membrane-bound E3 Ubiquitin Ligase
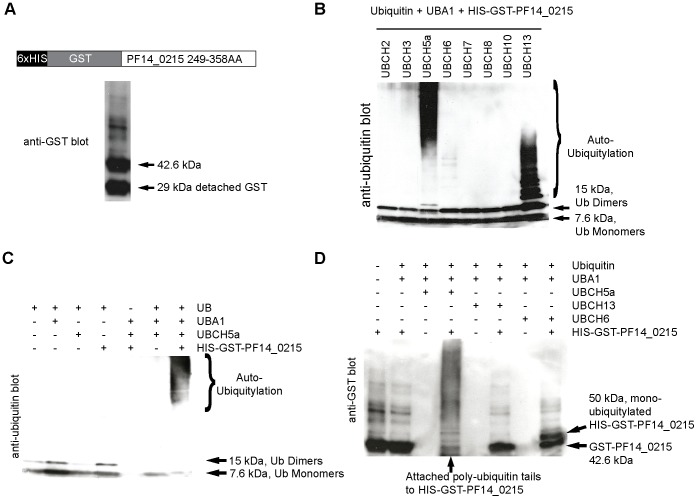
Initiating our biological validation, we investigated the biochemical activity and the cellular localization of the protein encoded PF14_0215. First, the predicted RING domain of PF14_0215 was cloned into E. coli, fused to tandem GST and 6xHis tags, expressed, and purified (Figure 3A and Figure S1). In vitro ubiquitylation assays were performed by incubating the purified recombinant PF14_0215 RING domain with commercially available human E1 (UBA1), various human E2 (UBC) enzymes, and commercial purified ubiquitin (Figure 3B). Anti-ubiquitin immunoblots show that different patterns of ubiquitylation were obtained when our recombinant E3 and the commercial E1 were mixed with UBCH5a, UBCH6, or UBCH13complex. With UBCH5a, recombinant PF14_0215 RING domain elicited a laddering effect that mostly began from the 50 kDa range, while with UBCH13, the ubiquitin laddering began around the 15 kDa mark. Ubiquitylation was no longer detected when our recombinant PF14_0215 was removed from the assay (Figure 3C), which demonstrates that PF14_0215 promotes ubiquitylation. We investigated further the differences in ubiquitylating patterns obtained when different E2 enzymes are used. Anti-GST immunoblots of our in vitro ubiquitylation products were performed to distinguish auto-ubiquitylation events, a characteristic of RING E3 ligases, from the production of free poly-ubiquitin tails (Figure 3D). The high molecular weight banding pattern that was obtained in the presence of UBCH5a indicates the auto poly-ubiquitylation of our epitope-tagged PF14_0215. On the contrary, the single 8 kDa molecular weight shift of recombinant PF14_0215 obtained in the presence of UBCH6 indicates that only mono-ubiquitylation occurs. Finally, no auto-ubiquitylation of PF14_0215 was detected in the presence of UBCH13complex (pattern not different from what obtained with our E3 alone). The previously observed pattern of ubiquitylation consisted of free poly-ubiquitin tails only (Figure 3B). Taken together, our results demonstrate that PF14_0215 is a genuine E3 ubiquitin ligase that catalyzes both the formation of poly-ubiquitin tails and its auto-ubiquitylation.
Figure 3. In vitro ubiquitylation assay of PfHRD1 (PF14_0215).
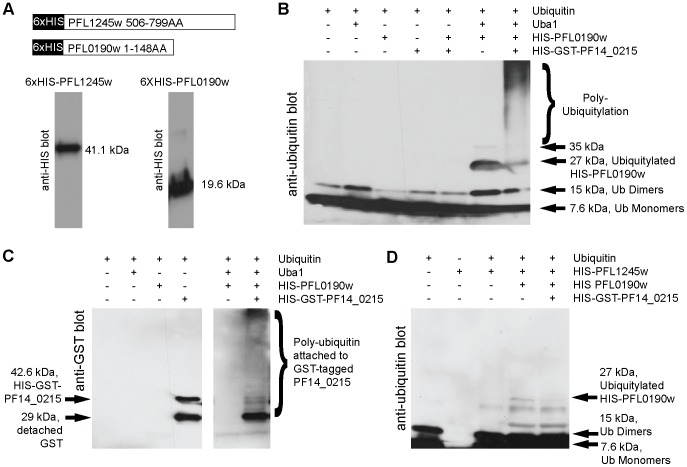
(A) Epitope-tagged recombinant PfHRD1 was expressed in E. coli and purified. Anti-GST immunoblot on purified protein extracts reveals the presence of detached free GST tags (MW = 29 kDa) and of PfHRD1-GST (MW = 42.6 kDa). (B) In vitro ubiquitylation assays were performed with commercial ubiquitin (Ub), human E1 UBA1, varying human E2 UBC enzymes (i.e., UBCH2, UBCH3, UBCH5a, UBCH6, UBCH7, UBCH8, UBCH10, and UBCH13), and our purified extract of epitope-tagged recombinant PfHRD1. Ubiquitylation patterns were revealed on anti-ubiquitin immunoblots. Different patterns of auto-ubiquitylation are observed with UBCH5a, UBCH13, and UBCH6. (C) In vitro ubiquitylation assays were performed in the presence (+) or absence (-) of commercial ubiquitin (UB), human E1 UBA1, human E2 UBCH5a, and our purified extract of epitope-tagged recombinant PfHRD. Ubiquitylation patterns were revealed on anti-ubiquitin immunoblots. Ubiquitylation is specifically observed only when all components are mixed together, including PfHRD1. (D) In vitro ubiquitylation assays were performed with commercial ubiquitin (Ub), human E1 UBA1, the human E2 enzymes UBCH5a, UBCH6, and UBCH13, and our purified extract of epitope-tagged recombinant PfHRD1. The attachment of ubiquitin on the recombinant PfHRD1 itself was detected by anti-GST immuno-detection. PfHRD1 is poly-ubiquitylated in the presence of UBCH5a and mono-ubiquitylated in the presence of UBCH6 whereas UBCH13 does not permit PfHRD1 auto-ubiquitylation.
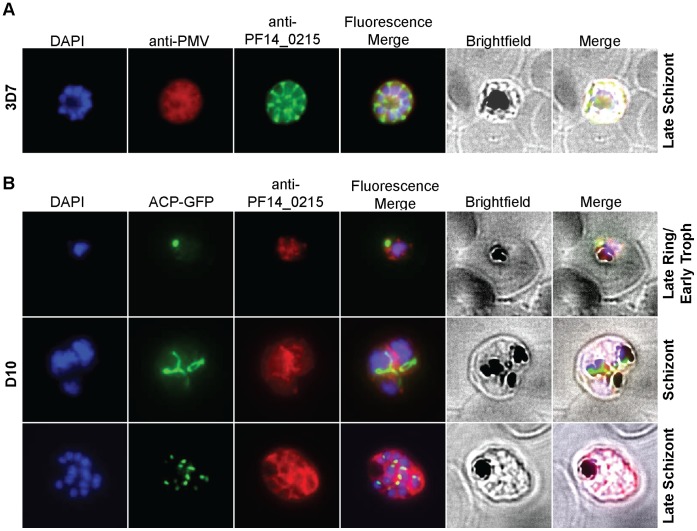
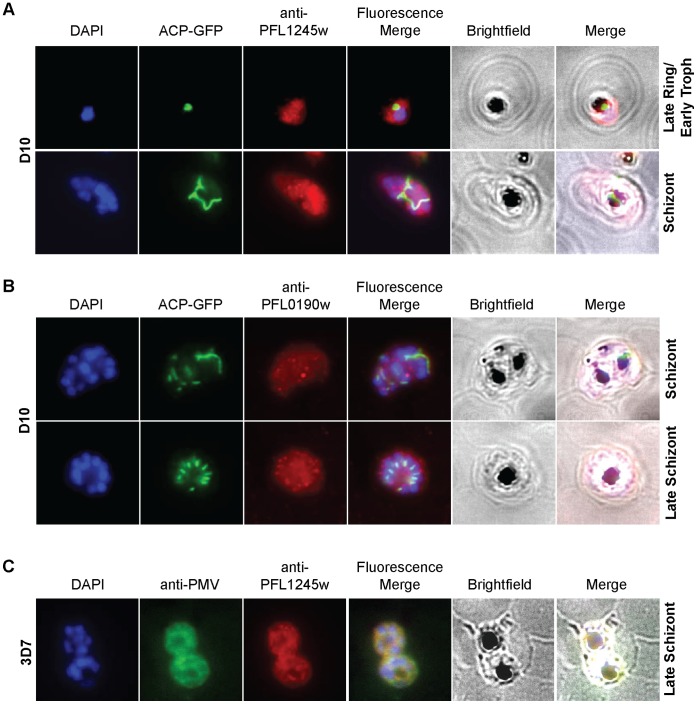
We further investigated the possible role of PF14_0215 E3 ligase and determined its localization by immunofluorescence microscopy using custom antibodies. We first verified the specificity of the custom antibody against PF14_0215 (see figure S2). Then we used the custom antibody to immuno-stain PF14_0215, which was found to co-localize with Plasmepsin V, a Plasmodium protein known to reside in the ER membrane [26] (Figure 4A). Co-imaged with DAPI-stained nuclei and ACP-GFP, an apicoplast marker, for comparative purposes, PF14_0215 was observed at various stages of the parasite life cycle and was found within reticular structures outside the nuclear regions at the trophozoite and schizont stages of the parasite, similar to the physical attributes of the ER (Figure 4B). In the late schizont stage PF14_0215 proteins reside within globular structures surrounding each budding merozoite in a pattern typical of the ER (Figure 4B). These observations support that PF14_0215 E3 ubiquitin ligase resides in the ER membrane, consistent with a putative role in the ERAD pathway. To further confirm our findings, we analyzed PF14_0215 function by reverse genetics using a double recombination gene knock out approach (Figure S3). Despite multiple attempts, we were only able to obtain crossover events at the 3′ UTR of PF14_0215 and failed to produce viable parasites with a disrupted PF14_0215 gene suggesting that PF14_0215 is essential for parasite survival. This result evokes the observed essentiality of HRD1 in other eukaryotic model organisms. Evidences showing in vitro ubiquitylation activity, ER membrane localization, and gene essentiality strongly suggest that PF14_0215 is the E3 ligase HRD1 Plasmodium homologue of the classical ERAD system.
Figure 4. Cellular localization of PfHRD1.
(A) PfHRD1 and plasmepsin V (PMV), an ER membrane marker, co-localize in P. falciparum. (B) At various stages of the parasite life cycle, PfHRD1 was co-stained with the nuclei (DAPI) and the apicoplast (ACP-GFP) in the P. falciparum strain D10.
PFL1245w and PFL0190w are Functional Cytosolic Ubiquitin Activating and Conjugating Enzymes
PFL1245w and PFL0190w are predicted to be the E1 ubiquitin-activating enzyme and the E2 ubiquitin-conjugating enzyme, respectively. In order to validate the in silico predictions, we have recombinantly cloned, expressed and purified PFL1245w and PFL1090w (Figure 5A and Figure S1) to test their in vitro activities. Since we found that PF14_0215 is likely the Plasmodium homologue of HRD1 of the ERAD system, we also tested the interaction of PFL1245w and PFL1090w with PF14_0215. Our results show that recombinant PFL0190w (19.6 kDa) is capable of in vitro mono (27 kDa band) and di-auto-ubiquitylation (35 kDa band) activity when incubated with commercially available human UBA1 (Figure 5B, second lane from the right). When the recombinant PF14_0215 RING domain was added to the reaction, a large laddering effect (poly-ubiquitin chains) was observed (Figure 5B, far right lane) similar to the effect observed using the commercial human UBCH5a (Figure 3B, third lane from the left). Anti-GST blots further revealed that these poly-ubiquitin chains were attached to the recombinant GST-tagged RING domain of PF14_0215 (Figure 5D). In addition, we immunoprecipitated the endogenous PF14_0215 E3 protein using our custom antibodies and were able to detect elevated amounts of ubiquitylated products when incubated in vitro with ubiquitylating reactions, which included our recombinant full-length PFL0190w E2 protein (Figure S4). These results show that PFL0190w is a functional E2 ubiquitin-conjugating enzyme that is compatible in vitro with endogenous PF14_0215.
Figure 5. In vitro ubiquitylation assay of PfUBA1 and PfUBC.
(A) 6xHIS-tagged recombinant PfUBA1 (E1) and PfUBC (E2) are depicted and anti-HIS blots reveal purification. (B) In vitro ubiquitylation assays were performed with recombinant PfUBC (E2), which revealed an attachment of a single ubiquitin when incubated with human UBA1 (E1) (second lane from the right) and an increased number of ubiquitylation products when both UBA1 and recombinant PfHRD1(E3) was added (far right lane). (C) When incubated with human UBA1 (E1), recombinant PfUBC (E2) (along with the other necessary reagents) attaches polymers of ubiquitin to recombinant PfHRD1 (E3) (far right lane). (D) Recombinant PfUBA1 (E1) is capable of attaching a single ubiquitin to recombinant PfUBC (E2), which is depicted by the appearance of a 27 kDa band (second lane from the right). Extra banding was not detected with the addition of recombinant PfHRD1 (E3) (far right lane).
In vitro ubiquitylation assays were repeated using recombinant parasite E1 PFL1245w together with E2 PFL0190w and E3 PF14_0215 (Figure 5D). PFL1245w was able to ubiquitylate PFL0190w as shown by the presence of a band at 27 kDa. When recombinant Plasmodium E3 ligase PF14_0215 was added to the reaction, no increase of ubiquitylating activity could be detected. Considering the mode of action of the classical ERAD-system, it is possible that the full-length of recombinant PF14_0215 as well as additional accessory proteins are required for a full in vitro activity of all three Plasmodium proteins. On the whole, these results validate (i) PFL1245w as a functional E1 ubiquitin-activating enzyme that can work in vitro with the E2 PFL0190w, and (ii) PFL0190w as a functional E2 ubiquitin-conjugating enzyme that can work in vitro with the E3 ligase PF14_0215.
To further validate PFL1245w and PFL0190w as the likely Plasmodium ERAD E1 and E2 enzymes, respectively, we investigated their localization using custom-made antibodies; antibody-specificity was verified by immunoblotting (see Figure S2). In other model organisms, the classical ERAD E1 UBA1 and E2 UBC7 proteins are reported to reside in the cytoplasm until they are needed and are recruited to the outer membrane of the ER. Using immunofluorescence microscopy, we found that both PFL0190w and PFL1245w are mainly dispersed throughout the cytoplasm with small aggregations scattered throughout the parasite (Figures 6A,B). In addition, when PFL1245w was co-immunostained with PMV, an ER membrane marker, there was noticeable overlap between the two proteins, which suggests that PFL1245w is likely recruited to the ER membrane as expected (Figure 6C).
Figure 6. Localization studies of PfUba and Pfubc7.
(A,B) IFA experiments, using different parasite stages, show that PfUBA1 and PfUBC localize mainly to the cytosol. (C) When co-immunostained with plasmepsin V (PMV), a Plasmodium ER membrane protein marker, there was noticeable overlap between the two proteins, suggesting PfUBA1 recruitment to the ER as well.
The localization pattern of PFL1245w and PFL0190w indicates cytoplasmic residence with possible transient recruitment to the ER that is consistent the classical ERAD model. Given their in vitro activities in auto-ubiquitylating assays and their pattern of localization, PFL1245w and PFL0190w are likely the Plasmodium homologues for E1 UBA1 and E2 UBC7 involved in the parasite’s ERAD system.
Discussion
Proteasome inhibitor studies have shown promising antimalarial results and have ignited interest in studying how protein degradation in Plasmodium can be targeted for effective drug discovery and synthesis. Though there have been several reports on the efficacy of various proteasome inhibitors on malaria parasites, there have been relatively few studies done on functionally characterizing the malarial protein degradation system upstream of the proteasome. An integral part of protein degradation within eukaryotic cells is the ERAD system, which relies on ubiquitylation in order to shuttle misfolded proteins across the ER membrane and label them for degradation by the 26S proteasome. Here, we show that ESI, a known ERAD inhibitor, is toxic to the Plasmodium parasite within low µM ranges and causes increased levels of ubiquitylated products. These results provide the first indications that an ubiquitin-dependent ERAD system does exist within the parasite and also gives a proof of concept that the ERAD system could be a point of antimalarial drug targeting.
According to our bioinformatics analysis, in vitro ubiquitylation assays and localization studies, it seems that the Plasmodium ERAD system may function similarly to that of other eukaryotic model systems. The domain architecture of these putative ubiquitylating Plasmodium ERAD proteins are homologues to that of their identified counterparts in other eukaryotes [21], while localization studies show them localizing to their expected destinations of either the ER membrane or cytosol. The in vitro ubiquitylation assays performed on recombinant versions of PFL1245w, PFL09190w and PF14_0215 revealed that these proteins are capable of facilitating ubiquitylation and that they can work together. In many cases, E3 ligases with RING domains are known to mediate the transfer of ubiquitin between E2 conjugating enzymes and target substrates without actually binding to ubiquitin itself [27]. And though recent studies are reporting novel RING E3 ligase complexes (e.g. RBR ubiquitin ligases) are capable of binding to ubiquitin [28], autoubiquitylation of E3 ligases have been more associated with HECT E3 ligases rather than RING E3 ligases. However, when we experimentally studied the E3 RING domain of PF14_0215, we found that PF14_0215 was capable of autoubiquitylation, which is reminiscent to the yeast HRD1p E3 ligase, which has also been reported to autoubiquitylate as a mechanism to regulate itself [29], [30]. All together, our data indicates that PFL1245w, PFL0190w, and PF14_0215 are the Plasmodium falciparum UBA1, UBC and HRD1 (PfUBA1, PfUBC and PfHRD1) respectively and serve to ubiquitylate misfolded ER proteins for translocation from the ER lumen to the cytosol for proteasome degradation (see figure 7 for a graphical model).
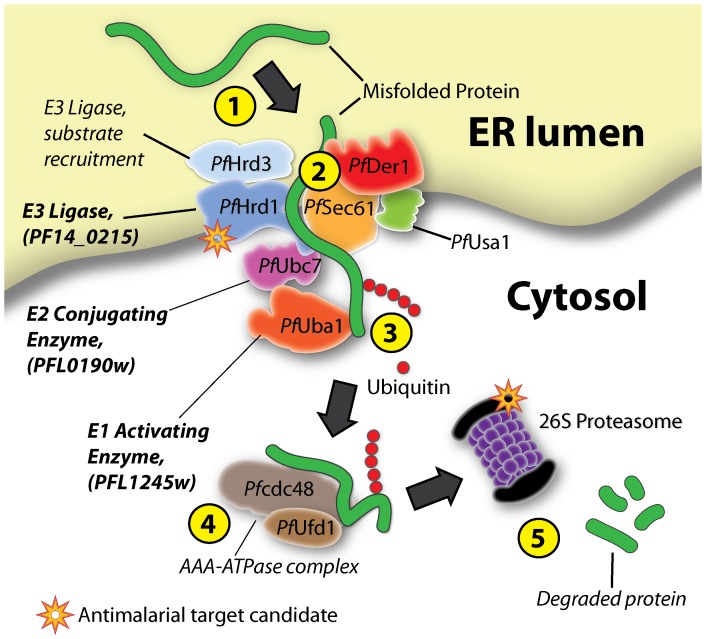
Figure 7. Graphical depiction of the Plasmodium ERAD pathway in protein quality control and its potential for antimalarial strategies.
Similar to their human and yeast counterparts, the components of the Plasmodium ERAD system likely serve to recognize and translocate misfolded ER luminal proteins to the cytosol for degradation by the 26S proteasome. A simplified flowchart is presented: (1) misfolded proteins are recognized and recruited to the ERAD complex, (2) where they are inserted across the ER membrane through a pore formed by a complex of ERAD proteins. (3) During translocation, the aberrant proteins are poly-ubiquitylated by the concerted action of the E1 ubiquitin-activating enzyme PfUBA1 (PFL1245w), the E2 ubiquitin-conjugating enzyme PfUBC (PFL0190w), and the E3 ligase PfHRD1 (PF14_0215). (4) The ubiquitylated aberrant proteins are extracted by the CDC48-UFD1-NPL4 AAA-ATPase complex and shuttled to the 26S proteasome (5) for degradation. Plasmodium ERAD drug target candidates are highlighted with an orange star.
The results from the knock out attempts of PF14_0215 suggest that PF14_0215 is an essential Plasmodium gene. As part of the ERAD system, the probable essentiality of PF14_0215 is not surprising since the ERAD system, and protein degradation in general, is known to be an essential part of eukaryotic biology. That is why there is burgeoning interest in proteasome degradation and inhibitors in regards to antimalarial drug discovery. If the protein degradation at the proteasome level can be effectively exploited as a potential antimalarial target, it is reasonable to believe that upstream pathways, such as the ubiquitylating components within the ERAD system, may also serve as viable antimalarial drug targets. Currently, a wide range of ubiquitylating enzymes, from E1 activating enzymes to deubiquitylating enzymes (DUB)s are being screened for inhibitors that may confer anti-cancer properties. In fact, there are already several inhibitors of ubiquitin or ubiquitin-like enzymes that have shown effective results against cancer (Sun, 2003; Yang et al, 2007), with some already in clinical trials (for a review, see [31]).
Some may argue that targeting a highly conserved system such as the ERAD pathway in the Plasmodium is likely to have cross-reactivity with the very similar human host ERAD counterpart and is unlikely to produce a tight parasite-specific drug. However, inhibitors of ubiquitin E1 enzymes have already been demonstrated to reduce leukemia and multiple myeloma, while maintaining limited toxicity in mouse models [32]. The limited toxicity of E1 ubiquitin inhibitors observed in normal host cells is likely explained by the notion that inhibition of proteasome degradation has more of a profound effect on highly proliferative cells, such as malignant tumors and protozoan parasites, due to their higher metabolic needs. Furthermore, since it has been shown that E3 ligases are the most various and divergent within eukaryotes [21], higher parasite-specificity can be achieved by targeting essential parasite-specific E3 ligases. Currently, there are specific inhibitors of E3 ubiquitin ligases that are in clinical trials. For example, inhibitors of MDM2, the E3 ligase regulator of the tumor suppressor protein p53, has shown promising anti-myeloma activity [33]. In theory, components of the Plasmodium ERAD degradation pathway, particularly its E3 ligases (i.e. SEL1L/HRD3 [34], gp78 [35], RNF5/RMA1 [36], TEB4/March6 [37] and TRC8/RNF139 [38]), could produce an effective new strategy against Plasmodium as they are both specific and likely essential to the parasite life cycle. Interestingly, there have also been recent identifications of a duplicated ERAD-like system within the plastids of some apicomplexans, cryptomonads, and diatoms [24], [25], with P. falciparum included. However, instead of functioning in protein degradation, the plastid ERAD-like system is hypothesized to have roles in plastid protein import [39], which may also turn out to be an effective drug target.
The biology of the Plasmodium ERAD system and overall degradation pathways are worthy themes to investigate. It will be interesting to see whether or not these core ubiquitylating enzymes do collaborate in conjugating ubiquitin to misfolded proteins in vivo and work together with other known ERAD proteins (i.e. Der1, Cdc48, Ufd1) for targeted protein degradation. Also, whether or not protein degradation in protozoan parasites has unique features compared to human and yeast models. However, this can only be uncovered with more targeted and vigilant examination. One aspect of parasite biology that we can be certain of is that protein degradation is necessary for parasite survival and has already been shown to be a good candidate for antimalarial targeting with reports of proteasome inhibitors conferring Plasmodium IC50 values within the µM to nM ranges [8]. All together our results show that the Plasmodium ERAD system offers at least one parasite-specific drug target candidate, the ERAD E3 ubiquitin ligases, which have been characterized as being divergent from their human host counterparts. Though we have just begun to explore the machinery that is responsible for protein degradation in the human malaria parasite, we believe that the Plasmodium ERAD system may provide great drug target candidates that are both effective and parasite-specific.
Materials and Methods
Half Maximal Inhibitory Concentration (IC50) Assays and Ubiquitylated Protein Abundance Experiments with Eeryarestatin I
Eeryarestatin I (ESI) (EMD Millipore Corporation) was reconstituted at 10 mM in DMSO. 3D7 parasites were introduced to a 3-fold serial dilution of ESI in complete culture medium in 96-well plates and incubated for 72 hours in a humid modular incubation chamber (Billups-rothenberg) after gassing at 1% O2/3% CO2/96% N2. Plates were frozen overnight then thawed and evaluated for relative parasite growth with SYBR Green dye and a Molecular Devices Gemini EM spectrofluorometer.
3D7 parasites were synchronized with sorbitol treatment. During the early trophozoite stages, parasites were drugged with either of four different conditions: ESI (10 µM), MG132 (0.1 µM), ESI (10 µM) + MG132 (0.1 µM) or DMSO only (equivalent volumes to the drug conditions). After 8 hours incubation, parasites were isolated with 0.15% saponin and crude protein lysates were extracted. 0.5 µg of protein lysates from each condition were loaded in each well for SDS-PAGE. Immunoblots were probed with rabbit anti-ubiquitin antibodies (Millipore; 1∶2500) and goat anti-rabbit antibodies conjugated to HRP (1∶5000; Pierce).
In silico Discovery of Candidate Ubiquitylating Proteins and Protein Alignments
The translated genome of P. falciparum v8.0 was obtained from PlasmoDB (www.plasmodb.org) and was bioinformatically searched to identify ubiquitylating proteins using a previously published approach [21]. Domain architecture analysis was performed using InterProScanSequence [40]. Transmembrane domains were predicted with TMHMM [41]. Signal peptides were predicted with signal 3.0 [42]. Pictures were generated with Matlab®. Using MUSCLE [43], we performed protein alignments on PF14_0215, comparing it to the HRD1 of H. sapien (BAC24801.1), S. cerevisiae (NP_014630.1), S. pombe (NP_596376.1) and M. oryzae (XP_003709820.1).
Cloning and Purification of Recombinant Proteins
The plasmid pGS-21a (GenScript) and a modified version of it (designed as pGS-21aHis) were used as cloning vectors. The region coding for a GST was removed so that only the two 6xHIS tags subsist. The GST tag, along with the tandem 6xHIS tag, was removed by cutting pGS-21a with ClaI and NcoI. A new 6xHis-coding fragment was then amplified by PCR from purified pGS-21a using the primers GATCGAGATCGATCTCGATC and ATCCATGGCCTTACCGCTGCTATGATGATGAT and inserted to replace the GST using ClaI and NcoI. The sequence coding for the RING domain in PF14_0215 was PCR-amplified from 3D7 strain cDNA using the primers AGGGGATCCCTTAAGCCGCGGCCTTTACATATGACAGCAGAT and AGGAAGCTTTTAACTAGTGCTAGCTTACTTTTGTGTTGTATCATTTTCTG. A sequence segment that contains the E1 ubiquitin-activating domain of PFL1245w was PCR-amplified using the primers AGGGGATCCCTTAAGCCGCGGGTGGTGAATATTTTTGGGTTGG and AGGAAGCTTTTAACTAGTGCTAGCCCAACCCAAAAATATTCACCAC. The entire gene of PFL0190w was amplified from 3D7 strain cDNA with the primers AGGGGATCCCTTAAGCCGCGGATGGCCCTTAAAAGAATAACAAAA and AGGAAGCTTTTAACTAGTGCTAGCTTATTGTGCATATTTTTGTGTCC. The amplified fragments were digested with SacII and SpeI and ligated into pGS-21a (6xHis-GST) or pGS-21aHis (6xHis only) where the gene was N-terminally tagged. Constructs were inserted into E. coli for expression.
Transformed E. coli cells were grown in LB at 25°C until OD600>0.5. IPTG 1 mM was added to the cultures, which were then incubated overnight at 12°C. Cells were collected by centrifugation at 14,000 g and 4°C and resuspended in lysis buffer (Tris-HCl pH 7.5 25 mM, NaCl 500 mM, Triton X-100 1% (v/v), Mini EDTA-free Protease Inhibitor Cocktail (Roche), and AEBSF 1 mM). Cells were sonicated and lysates were cleared by centrifugation at 14,000 g and 4°C. Purifications of GST-tagged proteins were performed on glutathione agarose beads (Sigma-Aldrich). Glutathione-bound proteins were washed three times with GST wash buffer (Tris-HCl pH 7.5 25 mM, NaCl 300 mM, Triton X-100 1% v/v) and eluted with GST elution buffer (Tris-HCl pH 7.5 25 mM, NaCl 150 mM, reduced glutathione 15 mM, Triton X-100 0.01% v/v, glycerol 40% v/v). Purifications of His-tagged proteins were performed on Ni-NTA beads (Qiagen). Beads-bound proteins were washed several times with wash buffer (Tris-HCl pH 7.5 25 mM, NaCl 500 mM, imidazole 30 mM, glycerol 5% v/v) and subsequently eluted with elution buffer (Tris-HCl pH 7.5 25 mM, NaCl 500 mM, imidazole 250 mM, glycerol 5%). Purified protein extracts were analyzed by immunoblots. GST-tagged proteins were detected using a goat anti-GST antibody (1∶5000; GE Healthcare) and a donkey anti-goat antibody conjugated to horseradish peroxidase (1∶20,000; Jackson Immuno research). His-tagged proteins were detected using a mouse anti-His antibody (1∶2500; Millipore) and a goat anti-mouse antibody conjugated to horseradish peroxidase (1∶10,000; BioRad).
Custom Primary Antibodies
The following peptide sequences were used as antigens for the production of affinity-purified antibodies (Fisher Scientific or Genscript) in rabbits: RFKSFQKYRELTKNIETK for PF14_0215, KTDRTKYHQTAKAWTQKYAQ for PFL0190w, CSDQDLVDVLIPSIQFIYK for PFL1245w.
Parasite Culture and Transfection
The P. falciparum 3D7 (MRA-102, MR4) and D10 ACP(transit)-GFP (MRA-568, MR4) strains were grown in human O+ red blood cells according to a previously described protocol [44]. Transfections were carried out by electroporation of parasite-infected red blood cells as described in [45]. Successful transfectants were obtained by positive selection with WR99210 [46], and, when relevant, negative selection with 5-fluorocytosine [47].
Immunofluorescence Assay
Immunofluorescence assays were performed using the protocol described in [25]. Briefly, cells were washed with PBS and fixed for 1 h at 37°C in PBS containing 4% of paraformaldehyde and 0.0075% glutaraldehyde. Fixed cells were then pelleted by centrifugation at 700 g for 2 min and washed once for 10 minutes with glycine 1.25 M in PBS. After another centrifugation at 700 g for 2 min, the pelleted cells were permeabilized for 10 min with TritonX-100 0.1% (v/v) in PBS. Permeabilized cells were then washed once for 10 minutes with glycine 1.25 M in PBS and pelleted by centrifugation (700 g for x min). After a minimum of a 1 h-long incubation in BSA 3% (w/v) in PBS, the desired target-specific custom primary antibody was added for at least 2 hours at room temperature or overnight at 4°C at a dilution of 1∶200. Cells were subsequently extensively washed with PBS. Secondary goat anti-rabbit IgG Alexa Fluor® 488 or donkey anti-rabbit IgG Alexa Fluor® 568 were then added according to their respective primary antibodies at a dilution of 1∶100 for two hours at room temperature. Cells were washed 3 times with PBS and DAPI stain was finally added at 100 ng/mL final concentration for nuclei visualization. Anti-plasmepsin V antibodies (MRA-815A, MR4) were used to visualize the endoplasmic reticulum membrane [26].
Microscopy slides were mounted with slow-fade mounting medium (Fluoromount-G; Southern Biotech) and viewed with an Olympus BX40 fluorescence microscope using a 100X objective lens (UPlanFI). Image capture was done with a CoolSNAP cf camera and processed by the Metavue software (Photometrics). Images were finally merged and background was reduced using ImageJ software.
In vitro Ubiquitylation Assay
A mix containing free ubiquitin 50–200 µM (Boston Biochem), Human recombinant UBE1 E1 enzyme 0.05-0.2 µM (Boston Biochem), Human recombinant UBC E2 enzyme 1–5 µM (Boston Biochem), and purified Plasmodium E3 ligase 1–12.5 µM was incubated in reaction buffer (Tris-HCl pH 7.4 50 mM, DTT 1 mM, re-energizing buffer from Boston Biochem) for 2 h at 37°C. Alternatively, Human E1 and/or E2 were replaced with our purified Plasmodium proteins. Reactions were then analyzed by SDS-PAGE and anti-ubiquitin or anti-GST immunoblotting. Anti-ubiquitin immunoblots were probed with rabbit anti-ubiquitin antibodies (1∶2500; Millipore) and goat anti-rabbit antibodies conjugated to HRP (1∶5000; Pierce). Anti-GST immunoblots were probed with goat anti-GST antibodies (1∶2500; GE Healthcare) and donkey anti-goat antibodies conjugated to HRP (1∶5000; Jackson Immunoresearch).
Gene Disruption Experiments
Double recombination-based gene disruption plasmids were constructed with the 5′ UTR and 3′UTR of PF14_0215 flanking a human dihydrofolate reductase (hDHFR) resistance cassette (for positive selection with WR99210) within the pCC1 vector that also contains a Saccharomyces cerevisiae cytosine deaminase (ScCD) cassette for subsequent negative selection [42]. Gene disruption vectors were transfected into Pf3D7. Successful integration was verified by PCR.
Immunoprecipitation and in vitro Ubiquitylation Assay of Pulled-down Proteins
Isolated 3D7 parasites were resuspended in 20 mM HEPES pH7.9, 10 mM KCl, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, 0.5 mM AEBSF (Fisher Scientific), 0.65% Igepal v/v and cocktail protease inhibitor (Roche) (lysis buffer 1). Parasites were allowed to lyse on ice for 10 minutes and then were spun down. Supernatant was collected. The pellet was then subsequently resuspended in 20 mM HEPES pH 7.9, 0.1 M NaCl, 0.1 mM EDTA, 0.1 mM EGTA, 1.5 mM MgCl2, 1 mM DTT, 1 mM AEBSF and cocktail protease inhibitor (Roche) (lysis buffer 2), and then incubated at 4°C with vigorous shaking for 20 minutes. After spinning, the supernatant was collected. The remaining pellet was then resuspended in lysis buffer 1 and then sonicated and spun down. Supernatant was collected and pooled with the previous two collected supernatants to create a mixture of extracted proteins. Extracted proteins were precleared with magnetic Protein A beads (Millipore) and then incubated with anti-PfHRD1 (PF14_0215) antibodies for 2 hours at 4°C with gentle shaking. Pre-washed magnetic Protein A beads were added to the samples and allowed to incubate overnight at 4°C with gentle shaking. Samples were washed several times with lysis buffer 1. Pulled-down PF14_0215 proteins were left bound to the beads.
Human recombinant UBE1 (Boston Biochem), Plasmodium recombinant PfUBC (PFL0190w) and pulled-down PfHRD1 were added to a solution containing 0.2 M HEPES pH7.9, 50 mM DTT, re-energizing buffer (Boston Biochem), 0.5 µg/µL final concentration of biotin conjugated ubiquitin (Boston Biochem), 0.5 µg/µL final concentration of ubiquitin (Boston Biochem), 5 mM AEBSF (Fisher Scientific) and cocktail protease inhibitor (Roche). The reactions were allowed to incubate at 37°C with gentle agitation for two hours. After incubation, reactions were terminated with Laemmli buffer and incubated at 95°C for 5 mins. Activity was visualized with affinity blots. Biotin affinity blots were probed with streptavidin conjugated with peroxidase (1∶10,000; Jackson Immunoresearch). Immunoblot intensities were measured with ImageJ64 software.
Supporting Information
Coomassie-stained SDS-PAGE gels of purified recombinant proteins. Coomassie-stained SDS-PAGE gels show the step-by step purification of recombinant (A) PF14_0215, (B) PFL1245w, and (C) PFL0190w that have been either tagged with HIS or with GST and HIS both. In each elution, a distinguishable band that represents the respective purified recombinant protein can be found at their expected molecular sizes: GST-HIS-PF14_0215(249–358 AA), 42.6 kDa; HIS-PFL1245w (506–799 AA), 41.4 kDa; HIS-PFL0190w (1–148 AA), 19.6 kDa.
(TIF)
Immunoblots showing custom antibody specificity. (A) The specificity of the custom-made antibody raised against PF14_0215 protein (59.8 kDa) was verified by western blot on crude parasite extract. (B) Using anti-PFL1245w custom antibodies on crude parasite protein extracts, a band at 131.8 kDa was detected on an immunoblot, which is the expected size for the PFL1245w protein. (C) Immunoblots were made using anti-PFL1245w custom antibodies on crude parasite protein extracts. A band at 131.8 kDa was detected, which is the expected size for the PFL1245w protein.
(TIF)
Knock-Out Attempt for PF14_0215. A PF14_0215 knockout vector was constructed with a human dihydrofolate reductase (hDHFR) selection cassette that is flanked by the 5′ UTR and 3′ UTR of the PF14_0215. A Saccharomyces cerevisiae cytosine deaminase (ScCD) cassette (not shown) was placed outside the PF14_0215 5′ UTR and 3′ UTR sections to be used for negative selection. Primers (indicated by red arrows) were designed to only amplify crossover events at either the 5′ or 3′ UTR of PF14_0215. PCR experiments show that only parasites that underwent a 3′ UTR crossover event, that still leaves the PF14_0215 intact, could be recovered. Double recombination that would excise the endogenous PF14_0215 gene was never recovered.
(TIF)
In vitro ubiquitylation activity of immunoprecipitated Pf HRD1. Using anti-PfHRD1 (PF14_0215) antibodies, endogenous PfHRD1 was immunoprecipitated from 3D7 strains. In vitro ubiquitylation assays were performed using biotinylated-ubiquitin, thus only newly ubiquitylated products can be detected with streptavidin blots. Human recombinant UBE1 (lane 1) by itself was able to produce a single ubiquitylated product when incubated with biotin-ubiquitin. Aggregates of biotinylated ubiquitin (depicted by the arrow) were seen in all lanes (even without the addition of any ubiquitylating enzymes, not shown) and are considered as background. Recombinant PfUBC (PFL0190w) (lane 2) and immunoprecitated PfHRD1 (lane 3) by themselves were unable to produce any ubiquitylated products. UBE1 and Plasmodium PfUBC together (lane 4) were able to generate significant levels of newly ubiquitylated products. With the addition of immunoprecipitated PfHRD1 (lane 5), we were able to detect around a 44% increased level of ubiquitylation when compared to lane 4.
(TIF)
Funding Statement
This study was funded by the startup funds provided by the University of California, Riverside. Dr. Rodrigues was supported by a postdoctoral fellowship from the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES, contract no. 374808-1/2009). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Garcia LS (2010) Malaria. Clinics in Laboratory Medicine 30: 93–129 doi:10.1016/j.cll.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 2. van Vugt M, van Beest A, Sicuri E, van Tulder M, Grobusch MP (2011) Malaria treatment and prophylaxis in endemic and nonendemic countries: evidence on strategies and their cost-effectiveness. Future Microbiol 6: 1485–1500 doi:10.2217/fmb.11.138. [DOI] [PubMed] [Google Scholar]
- 3. Lindenthal C, Weich N, Chia YS, Heussler V, Klinkert MQ (2005) The proteasome inhibitor MLN-273 blocks exoerythrocytic and erythrocytic development of Plasmodium parasites. Parasitology 131: 37–44. [DOI] [PubMed] [Google Scholar]
- 4. Reynolds JM, El Bissati K, Brandenburg J, Günzl A, Mamoun CB (n.d.) Antimalarial activity of the anticancer and proteasome inhibitor bortezomib and its analog ZL3B. BMC Clin Pharmacol 7: 13–13 doi:10.1186/1472-6904-7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dick LR, Cruikshank AA, Grenier L, Melandri FD, Nunes SL, et al. (1996) Mechanistic studies on the inactivation of the proteasome by lactacystin: a central role for clasto-lactacystin beta-lactone. J Biol Chem 271: 7273–7276. [DOI] [PubMed] [Google Scholar]
- 6. Kreidenweiss A, Kremsner PG, Mordmüller B (n.d.) Comprehensive study of proteasome inhibitors against Plasmodium falciparum laboratory strains and field isolates from Gabon. Malar J 7: 187–187 doi:10.1186/1475-2875-7-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prudhomme J, McDaniel E, Ponts N, Bertani S, Fenical W, et al. (2008) Marine Actinomycetes: A New Source of Compounds against the Human Malaria Parasite. PLoS ONE 3. doi:10.1371/journal.pone.0002335. [DOI] [PMC free article] [PubMed]
- 8. Chung D-WD, Le Roch KG (2010) Targeting the Plasmodium ubiquitin/proteasome system with anti-malarial compounds: promises for the future. Infect Disord Drug Targets 10: 158–164. [DOI] [PubMed] [Google Scholar]
- 9. Li GD, Li JL, Mugthin M, Ward SA (2000) Molecular cloning of a gene encoding a 20S proteasome beta subunit from Plasmodium falciparum. Int J Parasitol 30: 729–733. [DOI] [PubMed] [Google Scholar]
- 10. Certad G, Abrahem A, Georges E (1999) Cloning and partial characterization of the proteasome S4 ATPase from Plasmodium falciparum. Exp Parasitol 93: 123–131 doi:10.1006/expr.1999.4442. [DOI] [PubMed] [Google Scholar]
- 11. Ramasamy G, Gupta D, Mohmmed A, Chauhan VS (2007) Characterization and localization of Plasmodium falciparum homolog of prokaryotic ClpQ/HslV protease. Mol Biochem Parasitol 152: 139–148 doi:10.1016/j.molbiopara.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 12. Yoo SJ, Seol JH, Shin DH, Rohrwild M, Kang MS, et al. (1996) Purification and characterization of the heat shock proteins HslV and HslU that form a new ATP-dependent protease in Escherichia coli. J Biol Chem 271: 14035–14040. [DOI] [PubMed] [Google Scholar]
- 13. Gardner MJ, Hall N, Fung E, White O, Berriman M, et al. (2002) Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 419: 498–511 doi:10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Xie W, Ng DTW (2010) ERAD substrate recognition in budding yeast. Seminars in Cell & Developmental Biology 21: 533–539 doi:16/j.semcdb.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 15. Sato BK, Schulz D, Do PH, Hampton RY (2009) Misfolded Membrane Proteins Are Specifically Recognized by the Transmembrane Domain of the Hrd1p Ubiquitin Ligase. Molecular Cell 34: 212–222 doi:16/j.molcel.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bagola K, Mehnert M, Jarosch E, Sommer T (2011) Protein dislocation from the ER. Biochim Biophys Acta 1808: 925–936 doi:10.1016/j.bbamem.2010.06.025. [DOI] [PubMed] [Google Scholar]
- 17. Carvalho P, Goder V, Rapoport TA (2006) Distinct Ubiquitin-Ligase Complexes Define Convergent Pathways for the Degradation of ER Proteins. Cell 126: 361–373 doi:10.1016/j.cell.2006.05.043. [DOI] [PubMed] [Google Scholar]
- 18. Hershko A, Ciechanover A (1998) THE UBIQUITIN SYSTEM. Annu Rev Biochem 67: 425–479 doi:10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 19. Pickart CM (2004) Back to the Future with Ubiquitin. Cell 116: 181–190 doi:10.1016/S0092-8674(03)01074-2. [DOI] [PubMed] [Google Scholar]
- 20. Laney JD, Hochstrasser M (1999) Substrate Targeting in the Ubiquitin System. Cell 97: 427–430 doi:10.1016/S0092-8674(00)80752-7. [DOI] [PubMed] [Google Scholar]
- 21. Ponts N, Yang J, Chung D-WD, Prudhomme J, Girke T, et al. (2008) Deciphering the Ubiquitin-Mediated Pathway in Apicomplexan Parasites: A Potential Strategy to Interfere with Parasite Virulence. PLoS ONE 3: e2386 doi:10.1371/journal.pone.0002386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cross BCS, McKibbin C, Callan AC, Roboti P, Piacenti M, et al. (2009) Eeyarestatin I inhibits Sec61-mediated protein translocation at the endoplasmic reticulum. J Cell Sci 122: 4393–4400 doi:10.1242/jcs.054494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. McKibbin C, Mares A, Piacenti M, Williams H, Roboti P, et al. (2012) Inhibition of protein translocation at the endoplasmic reticulum promotes activation of the unfolded protein response. Biochem J 442: 639–648 doi:10.1042/BJ20111220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sommer MS, Gould SB, Lehmann P, Gruber A, Przyborski JM, et al. (2007) Der1-mediated Preprotein Import into the Periplastid Compartment of Chromalveolates? Mol Biol Evol 24: 918–928 doi:10.1093/molbev/msm008. [DOI] [PubMed] [Google Scholar]
- 25. Spork S, Hiss JA, Mandel K, Sommer M, Kooij TWA, et al. (2009) An Unusual ERAD-Like Complex Is Targeted to the Apicoplast of Plasmodium falciparum. Eukaryotic Cell 8: 1134–1145 doi:10.1128/EC.00083-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Klemba M, Goldberg DE (2005) Characterization of plasmepsin V, a membrane-bound aspartic protease homolog in the endoplasmic reticulum of Plasmodium falciparum. Mol Biochem Parasitol 143: 183–191 doi:10.1016/j.molbiopara.2005.05.015. [DOI] [PubMed] [Google Scholar]
- 27. Deshaies RJ, Joazeiro CAP (2009) RING domain E3 ubiquitin ligases. Annu Rev Biochem 78: 399–434 doi:10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 28. Wenzel DM, Klevit RE (2012) Following Ariadne’s thread: a new perspective on RBR ubiquitin ligases. BMC Biol 10: 24 doi:10.1186/1741-7007-10-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Smith MH, Ploegh HL, Weissman JS (2011) Road to Ruin: Targeting Proteins for Degradation in the Endoplasmic Reticulum. Science 334: 1086–1090 doi:10.1126/science.1209235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gardner RG, Swarbrick GM, Bays NW, Cronin SR, Wilhovsky S, et al. (2000) Endoplasmic Reticulum Degradation Requires Lumen to Cytosol Signaling Transmembrane Control of Hrd1p by Hrd3p. J Cell Biol 151: 69–82 doi:10.1083/jcb.151.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Edelmann MJ, Nicholson B, Kessler BM (2011) Pharmacological targets in the ubiquitin system offer new ways of treating cancer, neurodegenerative disorders and infectious diseases. Expert Rev Mol Med 13: e35 doi:10.1017/S1462399411002031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Xu GW, Ali M, Wood TE, Wong D, Maclean N, et al. (2010) The ubiquitin-activating enzyme E1 as a therapeutic target for the treatment of leukemia and multiple myeloma. Blood 115: 2251–2259 doi:10.1182/blood-2009-07-231191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Saha MN, Jiang H, Jayakar J, Reece D, Branch DR, et al. (2010) MDM2 antagonist nutlin plus proteasome inhibitor velcade combination displays a synergistic anti-myeloma activity. Cancer Biol Ther 9: 936–944. [DOI] [PubMed] [Google Scholar]
- 34. Mueller B, Lilley BN, Ploegh HL (2006) SEL1L, the homologue of yeast Hrd3p, is involved in protein dislocation from the mammalian ER. J Cell Biol 175: 261–270 doi:10.1083/jcb.200605196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.St-Pierre P, Dang T, Joshi B, Nabi IR (2012) Peripheral Endoplasmic Reticulum Localization of Gp78 Ubiquitin Ligase Activity. Journal of Cell Science. Available: http://www.ncbi.nlm.nih.gov/pubmed/22328510. Accessed 6 April 2012. [DOI] [PubMed]
- 36. Tcherpakov M, Delaunay A, Toth J, Kadoya T, Petroski MD, et al. (2009) Regulation of endoplasmic reticulum-associated degradation by RNF5-dependent ubiquitination of JNK-associated membrane protein (JAMP). J Biol Chem 284: 12099–12109 doi:10.1074/jbc.M808222200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hassink G, Kikkert M, van Voorden S, Lee S-J, Spaapen R, et al. (2005) TEB4 is a C4HC3 RING finger-containing ubiquitin ligase of the endoplasmic reticulum. Biochem J 388: 647–655 doi:10.1042/BJ20041241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Stagg HR, Thomas M, van den Boomen D, Wiertz EJHJ, Drabkin HA, et al. (2009) The TRC8 E3 ligase ubiquitinates MHC class I molecules before dislocation from the ER. J Cell Biol 186: 685–692 doi:10.1083/jcb.200906110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hempel F, Felsner G, Maier UG (2010) New mechanistic insights into pre-protein transport across the second outermost plastid membrane of diatoms. Molecular Microbiology 76: 793–801 doi:10.1111/j.1365-2958.2010.07142.x. [DOI] [PubMed] [Google Scholar]
- 40. Hunter S, Apweiler R, Attwood TK, Bairoch A, Bateman A, et al. (2009) InterPro: the integrative protein signature database. Nucleic Acids Res 37: D211–215 doi:10.1093/nar/gkn785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Melén K, Krogh A, von Heijne G (2003) Reliability measures for membrane protein topology prediction algorithms. J Mol Biol 327: 735–744. [DOI] [PubMed] [Google Scholar]
- 42. Bendtsen JD, Nielsen H, von Heijne G, Brunak S (2004) Improved prediction of signal peptides: SignalP 3.0. J Mol Biol 340: 783–795 doi:10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 43. Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797 doi:10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Le Roch KG, Zhou Y, Blair PL, Grainger M, Moch JK, et al. (2003) Discovery of gene function by expression profiling of the malaria parasite life cycle. Science 301: 1503–1508 doi:10.1126/science.1087025. [DOI] [PubMed] [Google Scholar]
- 45. Deitsch K, Driskill C, Wellems T (2001) Transformation of malaria parasites by the spontaneous uptake and expression of DNA from human erythrocytes. Nucleic Acids Res 29: 850–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fidock DA, Wellems TE (1997) Transformation with human dihydrofolate reductase renders malaria parasites insensitive to WR99210 but does not affect the intrinsic activity of proguanil. Proc Natl Acad Sci USA 94: 10931–10936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Maier AG, Braks JAM, Waters AP, Cowman AF (2006) Negative selection using yeast cytosine deaminase/uracil phosphoribosyl transferase in Plasmodium falciparum for targeted gene deletion by double crossover recombination. Molecular and Biochemical Parasitology 150: 118–121 doi:10.1016/j.molbiopara.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 48.Melikova M, Kondratov K, Kornilova E (2005) Two different stages of epidermal growth factor (EGF) receptor endocytosis are sensitive to free ubiquitin depletion produced by proteasome inhibitor MG132. Cell Biology International. Available: http://www.cellbiolint.org/cbi/030/0031/cbi0300031.htm. Accessed 10 July 2012. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Coomassie-stained SDS-PAGE gels of purified recombinant proteins. Coomassie-stained SDS-PAGE gels show the step-by step purification of recombinant (A) PF14_0215, (B) PFL1245w, and (C) PFL0190w that have been either tagged with HIS or with GST and HIS both. In each elution, a distinguishable band that represents the respective purified recombinant protein can be found at their expected molecular sizes: GST-HIS-PF14_0215(249–358 AA), 42.6 kDa; HIS-PFL1245w (506–799 AA), 41.4 kDa; HIS-PFL0190w (1–148 AA), 19.6 kDa.
(TIF)
Immunoblots showing custom antibody specificity. (A) The specificity of the custom-made antibody raised against PF14_0215 protein (59.8 kDa) was verified by western blot on crude parasite extract. (B) Using anti-PFL1245w custom antibodies on crude parasite protein extracts, a band at 131.8 kDa was detected on an immunoblot, which is the expected size for the PFL1245w protein. (C) Immunoblots were made using anti-PFL1245w custom antibodies on crude parasite protein extracts. A band at 131.8 kDa was detected, which is the expected size for the PFL1245w protein.
(TIF)
Knock-Out Attempt for PF14_0215. A PF14_0215 knockout vector was constructed with a human dihydrofolate reductase (hDHFR) selection cassette that is flanked by the 5′ UTR and 3′ UTR of the PF14_0215. A Saccharomyces cerevisiae cytosine deaminase (ScCD) cassette (not shown) was placed outside the PF14_0215 5′ UTR and 3′ UTR sections to be used for negative selection. Primers (indicated by red arrows) were designed to only amplify crossover events at either the 5′ or 3′ UTR of PF14_0215. PCR experiments show that only parasites that underwent a 3′ UTR crossover event, that still leaves the PF14_0215 intact, could be recovered. Double recombination that would excise the endogenous PF14_0215 gene was never recovered.
(TIF)
In vitro ubiquitylation activity of immunoprecipitated Pf HRD1. Using anti-PfHRD1 (PF14_0215) antibodies, endogenous PfHRD1 was immunoprecipitated from 3D7 strains. In vitro ubiquitylation assays were performed using biotinylated-ubiquitin, thus only newly ubiquitylated products can be detected with streptavidin blots. Human recombinant UBE1 (lane 1) by itself was able to produce a single ubiquitylated product when incubated with biotin-ubiquitin. Aggregates of biotinylated ubiquitin (depicted by the arrow) were seen in all lanes (even without the addition of any ubiquitylating enzymes, not shown) and are considered as background. Recombinant PfUBC (PFL0190w) (lane 2) and immunoprecitated PfHRD1 (lane 3) by themselves were unable to produce any ubiquitylated products. UBE1 and Plasmodium PfUBC together (lane 4) were able to generate significant levels of newly ubiquitylated products. With the addition of immunoprecipitated PfHRD1 (lane 5), we were able to detect around a 44% increased level of ubiquitylation when compared to lane 4.
(TIF)