Abstract
U12 intron-specific spliceosomes contain U11 and U12 small nuclear ribonucleoproteins and mediate the removal of U12 introns from precursor-mRNAs. Among the several proteins unique to the U12-type spliceosomes, an Arabidopsis thaliana AtU11/U12-31K protein has been shown to be indispensible for proper U12 intron splicing and for normal growth and development of Arabidopsis plants. Here, we assessed the functional roles of the rice (Oryza sativa) OsU11/U12-31K protein in U12 intron splicing and development of plants. The U11/U12-31K transcripts were abundantly expressed in the shoot apical meristems (SAMs) of Arabidopsis and rice. Ectopic expression of OsU11/U12-31K in AtU11/U12-31K-defecient Arabidopsis mutant complemented the incorrect U12 intron splicing and abnormal development phenotypes of the Arabidopsis mutant plants. Impaired cell division activity in the SAMs and inflorescence stems observed in the AtU11/U12-31K-deficient mutant was completely recovered to normal by the expression of OsU11/U12-31K. Similar to Arabidopsis AtU11/U12-31K, rice OsU11/U12-31K was determined to harbor RNA chaperone activity. Collectively, the present findings provide evidence for the emerging idea that the U11/U12-31K protein is an indispensible RNA chaperone that functions in U12 intron splicing and is necessary for normal development of monocotyledonous plants as well as dicotyledonous plants.
Introduction
Splicing of both the major class of U2-dependent introns and minor class of U12-dependent introns is an indispensible step in the regulation of gene expression in eukaryotes. Although U12-type minor introns, which constitute less than 0.5% of all introns in animals and plants [1]–[4], are significantly less frequent than U2-type major introns, their importance is substantial in the growth and development of animals and plants. The roles of U12 intron-specific small nuclear RNAs (snRNAs) in U12 intron splicing and the development of Drosophila, zebra fish, and human have been demonstrated [5]–[9], and the roles of U11/U12-specific proteins in cell viability and nonsense-mediated mRNA decay have also been demonstrated [2], [10], [11].
Splicing of the minor class of U12-type introns and major class of U2-type introns is catalyzed by the minor spliceosome and major spliceosome, respectively [2], [12]–[14]. During minor spliceosome formation, the U11 and U12 small nuclear ribonucleoproteins (snRNPs) form a stable U11/U12 di-snRNP complex [15], [16], and seven unique proteins, denoted 65K, 59K, 48K, 35K, 31K, 25K and 20K, specifically associate with the U11/U12 di-snRNPs [11], [17]. These minor spliceosome-associated proteins were found to be well conserved in animals and plants, and in plants they are conserved in both dicotyledonous and monocotyledonous plants [11], [18], [19].
Despite the well-conserved sequences of the minor spliceosome-associated proteins in animals and plants, the significance and biological roles of many minor spliceosomal proteins have not yet been experimentally demonstrated. It has been determined that U11/U12-65K, U11/U12-59K, and U11/U12-48K proteins are integral proteins in animals necessary for 5′ splice site selection and branchpoint site selection [20], [21]. Despite our increasing understanding of the roles of minor spliceosomal proteins in animals, reports demonstrating the functional roles of minor spliceosomal proteins in plants are severely limited. Arabidopsis AtU11/U12-35K was shown to facilitate recognition of the 5′ splice site [22]. In a previous study, we reported that Arabidopsis AtU11/U12-31K was essential for U12-type intron splicing by functioning as an RNA chaperone, and the AtU11/U12-31K-dependent U12 splicing was shown to be critical for normal plant development [23]. U11/U12-31K proteins are highly conserved in animals and plants, and in plants, the sequence of the U11/U12-31K proteins are well conserved in both dicotyledonous and monocotyledonous plants [18], [23], suggesting that they perform important cellular functions in plants. Considering our limited understanding of the roles of the minor spliceosome-associated proteins including U11/U12-31K protein, it would be of interest to determine whether U11/U12-31K proteins perform a similar function in monocotyledonous plants as well as in dicotyledonous plants. Here, we assessed the functional roles of the rice (Oryza sativa) OsU11/U12-31K protein in U12 intron splicing and the development of plants. By functional complementation of OsU11/U12-31K in the AtU11/U12-31K-defecient Arabidopsis mutant plants, we provide evidence demonstrating that U11/U12-31K proteins are functionally conserved RNA chaperones in Arabidopsis thaliana and rice, which are crucial for correct U12 intron splicing and normal development of monocotyledonous plants as well as dicotyledonous plants.
Results
Expression of U11/U12-31K in Arabidopsis and rice
Plant U11/U12-31K proteins are well conserved between dicotyledonous plants and monocotyledonous plants, with over 60 to 80% amino acid sequence identity [23]. In our previous analysis, AtU11/U12-31K was determined to be ubiquitously expressed in all Arabidopsis organs, including rosette leaves, cauline leaves, inflorescence stem, floral buds, and flowers [23]. Since AtU11/U12-31K affected the formation of primary inflorescence stems and meristem activity, we investigated in more detail the expression patterns of the U11/U12-31K gene in Arabidopsis and rice by in situ hybridization analysis. Results showed that U11/U12-31K was abundantly expressed in the shoot apical meristem (SAM) of Arabidopsis and rice (Figure 1). In addition, U11/U12-31K was abundantly expressed in elongating young leaves surrounding the SAM, but weakly expressed in mature leaves.
Figure 1. Expression patterns of U11/U12–31K in Arabidopsis and rice.
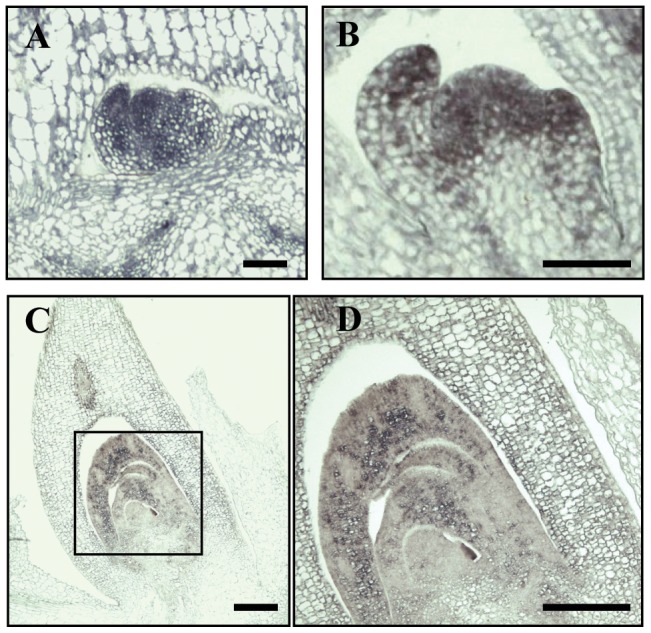
Expression patterns of U11/U12-31K in (A, B) Arabidopsis 20 days after germination and (C, D) rice 3 days after germination as analyzed by in situ hybridization. Scale bar = 50 µm.
U11/U12-31K is crucial for normal plant development
Based on our previous observation that Arabidopsis AtU11/U12-31K plays an essential role in plant development [23], we wanted to determine whether rice OsU11/U12-31K is also crucial for plant development. To facilitate the functional analysis of OsU11/U12-31K, we used an artificial microRNA (amiRNA)-mediated AtU11/U12-31K knockdown plant. It has been demonstrated that amiR1-4, the hypermorphic allele of amiRNA-mediated AtU11/U12-31K knockdown plant, produces severely arrested primary inflorescence stems [23]. Therefore, we investigated the role of OsU11/U12-31K via functional complementation in the amiR1-4 mutant. Several independent transgenic lines expressing OsU11/U12-31K in an amiR1-4 background were generated, and their phenotypes were analyzed. The amiR1-4 mutant expressing OsU11/U12-31K showed wild type phenotypes (Figure 2). It is noteworthy that the degree of recovery was dependent on the levels of the OsU11/U12-31K transcript in the amiR1-4 mutant. The findings that OsU11/U12-31K successfully complemented the development-defect phenotypes of amiR1-4 mutant clearly demonstrated that the function of U11/U12-31K is conserved in Arabidopsis and rice. Although the amiR1-4 mutant exhibited severe defects in the formation of primary inflorescence stems, the knockdown plants survived beyond the death of the wild-type and complementation lines expressing OsU11/U12-31K gene (Figure 3). These results indicate that prolonged cell division activity is maintained in AtU11/U12-31K knockdown plants when compared to the wild-type plants.
Figure 2. Recovery of normal phenotype of the mutant via OsU11/U12-31K complementation.

The phenotypes of wild type (WT), AtU11/U12-31K knockdown mutant (amiR1-4), and complementation line expressing OsU11/U12-31K (amiR1-4/Os31K) were observed at different growth stages.
Figure 3. Maintenance of the prolonged cell division activity of AtU11/U12-31K knockdown plants.

The knockdown plants (amiR1-4) survived beyond the death of the complementation lines expressing OsU11/U12-31K gene (Os31K).
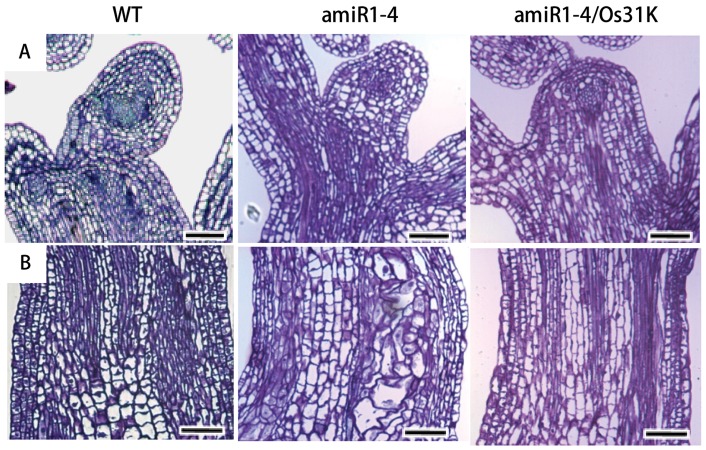
To better understand the cellular role of U11/U12-31K in regulating plant development, the morphologies of the vegetative SAM and the inflorescence stems were examined in 4-week-old wild-type, miR1-4 mutant, and miR1-4/Os31K complementation plants. The typical regular structure and organization of the SAM were maintained in miR1-4 mutant plants as well as in the wild-type and complementation plants, but the cell division activity in the SAM was diminished in the mutant plants (Figure 4A). However, the structure and shape of the cells in the inflorescence stems was significantly altered in the miR1-4 mutant, while the typical regular structure of cells was restored in the miR1-4/Os31K complementation plants (Figure 4B). The structures of the inflorescence and floral bud were also altered in the miR1-4 mutant plants compared with the wild-type plants (Figure S1). These abnormal morphologies of the miR1-4 mutant plants were completely recovered to normal structures in the miR1-4/Os31K complementation plants (Figure S1). All of these results indicate that U11/U12-31K plays a role in regulating cell division in the inflorescence stems, which is crucial for the normal development of monocot and dicot plants.
Figure 4. Images of the shoot apical meristem and the stem regions of the Arabidopsis plants.
(A) Longitudinal sections of the SAMs of the 4-week-old wild type (WT), AtU11/U12-31K knockdown mutant (amiR1-4), and complementation line expressing OsU11/U12-31K (amiR1-4/Os31K). Bar = 50 µm. (B) Longitudinal sections through the stems of 4-week-old WT, amIR1-4, and amiR1-4/Os31K plants. Bar = 50 µm.
Splicing defects in amiR1-4 mutant are rescued by the expression of OsU11/U12-31K
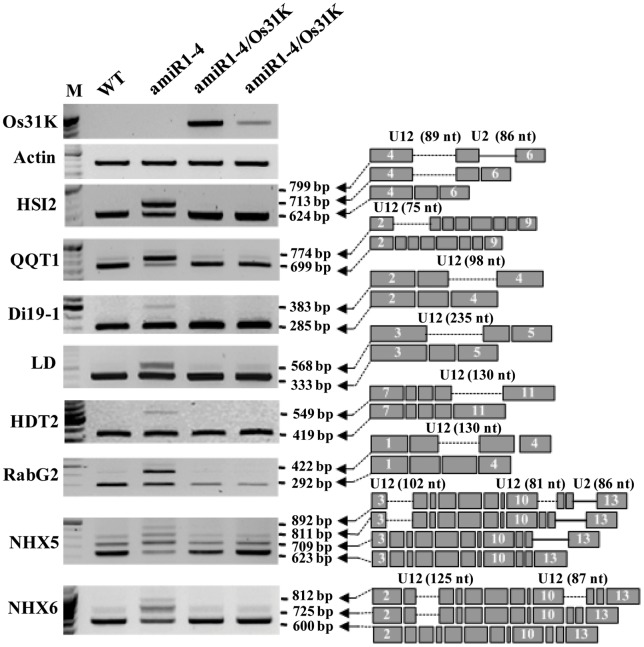
To ascertain whether the recovery of normal phenotypes observed in the complementation lines was correlated with the maintenance of normal splicing of U12 introns by the expression of OsU11/U12-31K, we determined the splicing patterns of U12-type introns in the complementation plants as well as in wild type and mutant plants. Thirty U12 intron-containing genes out of the 165 U12-type intron-containing genes in Arabidopsis [3] were randomly selected and their splicing patterns analyzed (Table S1), as previously described [23]. The splicing of most U12 introns investigated in this study was defective in the amiR1-4 mutant plants, whereas the splicing of these U12 introns was normal in the complementation lines (Figure 5 and S2). These results indicate that the occurrence of normal splicing of U12 introns is correlated with the wild-type phenotypes of the complementation lines, and confirm that the function of U11/U12-31K in the process of U12 intron splicing is conserved in dicot and monocot plants.
Figure 5. Recovery of normal splicing activity in the mutant via OsU11/U12-31K complementation.
The splicing patterns of U12 intron-containing transcripts were analyzed by RT-PCR in wild type (WT), AtU11/U12-31K knockdown mutant (amiR1-4), and complementation lines expressing OsU11/U12-31K (amiR1-4/Os31K). Identical results were obtained from three independent experiments, one of which is shown. Expression of OsU11/U12-31K in each complementation line (amiR1-4/Os31K) was confirmed by RT-PCR.
Since miR1-4 mutant exhibited normal growth until bolting and showed severe defect in the formation of primary inflorescence stems, we investigated in more detail whether the splicing of U12 intron-containing genes is regulated by a developmental stage-dependent manner. The splicing patterns of U12 intron-containing transcripts in wild type and miR1-4 mutant plants were analyzed at 3, 7, and 30 days after germination (DAG) (Figure 6). Several transcripts, including HSI2, QQT1, RabG2, NHX6, and Di19-2, were expressed at low levels at 3 DAG but the expression of all transcripts at 7 and 30 DAG was similar to each other. Importantly, the splicing patterns of U12 intron transcripts in miR1-4 mutant at 7 DAG were similar to those at 30 DAG (Figure 6 and data not shown). These results indicate that the defect in U12 intron splicing does not affect the growth of Arabidopsis until bolting, which strengthen the notion that U11/U12-31K-directed splicing of U12 intron-containing genes influences the development of Arabidopsis specifically after bolting stage.
Figure 6. Development-dependent splicing of U12 intron-containing transcripts.
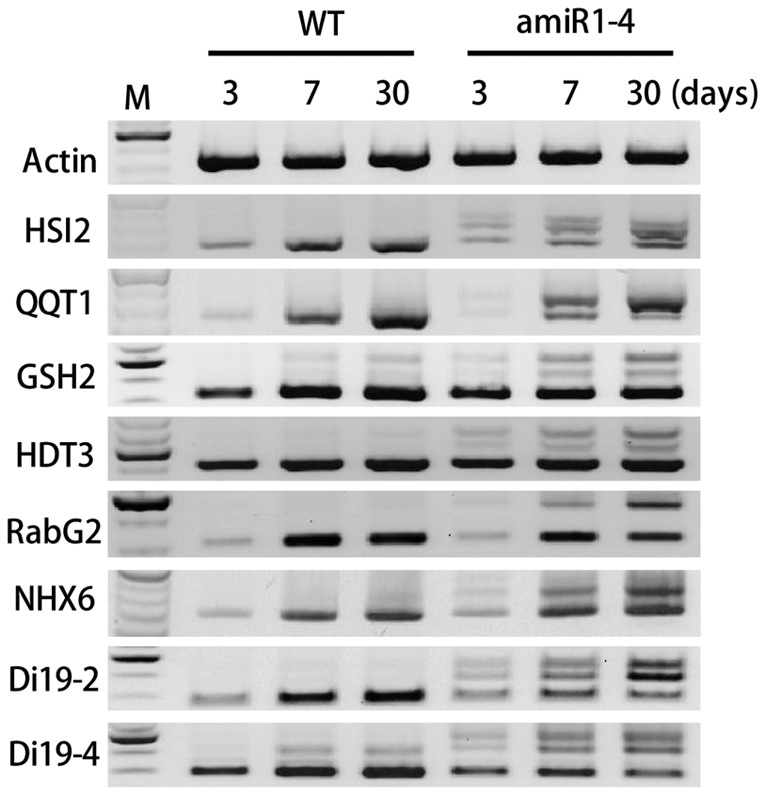
The splicing patterns of U12 intron-containing transcripts were analyzed by RT-PCR in wild type (WT) and AtU11/U12-31K knockdown mutant (amiR1-4) at 3, 7, and 30 days after germination. Identical results were obtained from three independent experiments, one of which is shown.
Both Arabidopsis and rice U11/U12-31K proteins harbor an RNA chaperone activity
The findings that U11/U12-31K is involved in the splicing of most U12 intron-containing genes indicate that it may function as an RNA chaperone, which interact with diverse RNA substrates with low sequence specificity [24], [25]. In our previous report, Arabidopsis AtU11/U12-31K was shown to harbor RNA chaperone activity [23]. To determine whether rice OsU11/U12-31K also harbors RNA chaperone activity, several well-established methods were employed. First, the complementation ability of the OsU11/U12-31K protein was evaluated in cold-sensitive E. coli BX04 mutant cells, which lack four cold shock protein (CSP) genes that are bacterial RNA chaperones [26]–[28]. The BX04 cells expressing either OsU11/U12-31K, CspA (positive control), or pINIII vector (negative control) grew well under normal growth conditions (37°C) with no noticeable differences (Figure 7A). However, when subjected to low temperature (20°C), only the BX04 cells expressing either OsU11/U12-31K or CspA grew well, whereas the BX04 cells harboring the pINIII vector did not (Figure 7A). This complementation ability of OsU11/U12-31K in the cold sensitive BX04 cells suggests that it functions as an RNA chaperone. Secondly, the nucleic acid-melting ability of OsU11/U12-31K was evaluated to further demonstrate its RNA chaperone activity. Recombinant glutathione S-transferase (GST)-31K protein and GST-CspA protein were purified from E. coli (Figure S3) and utilized for nucleic acid-melting analysis. The DNA-melting ability of OsU11/U12-31K was evaluated by using a partially double-stranded DNA molecule that produces fluorescence signals upon melting. Strong fluorescent signals were observed upon the addition of recombinant GST-31K or GST-CspA (positive control), but not the addition of GST protein (negative control), to the reaction mixture (Figure 7B), indicating that OsU11/U12-31K possesses in vitro DNA-melting ability. We next performed the RNase T1 cleavage assay to determine whether OsU11/U12-31K is capable of melting RNA secondary structures. The addition of RNase T1 to the RNA substrates transcribed from the pET-22b(+) plasmid produced several RNase T1-resistant bands (Figure 7C, indicated by arrows). These RNase-resistant bands disappeared when recombinant GST-31K was added before RNase T1. However, cleavage of RNA substrates by RNase T1 was not affected by the addition of GST. These results indicate that OsU11/U12-31K destabilizes the base pairings in the RNA substrates, allowing RNase T1 to further digest RNAs into smaller fragments. All of these results clearly demonstrate that OsU11/U12-31K is capable of melting nucleic acids and harbors RNA chaperone activity.
Figure 7. RNA chaperone activity of OsU11/U12-31K protein.
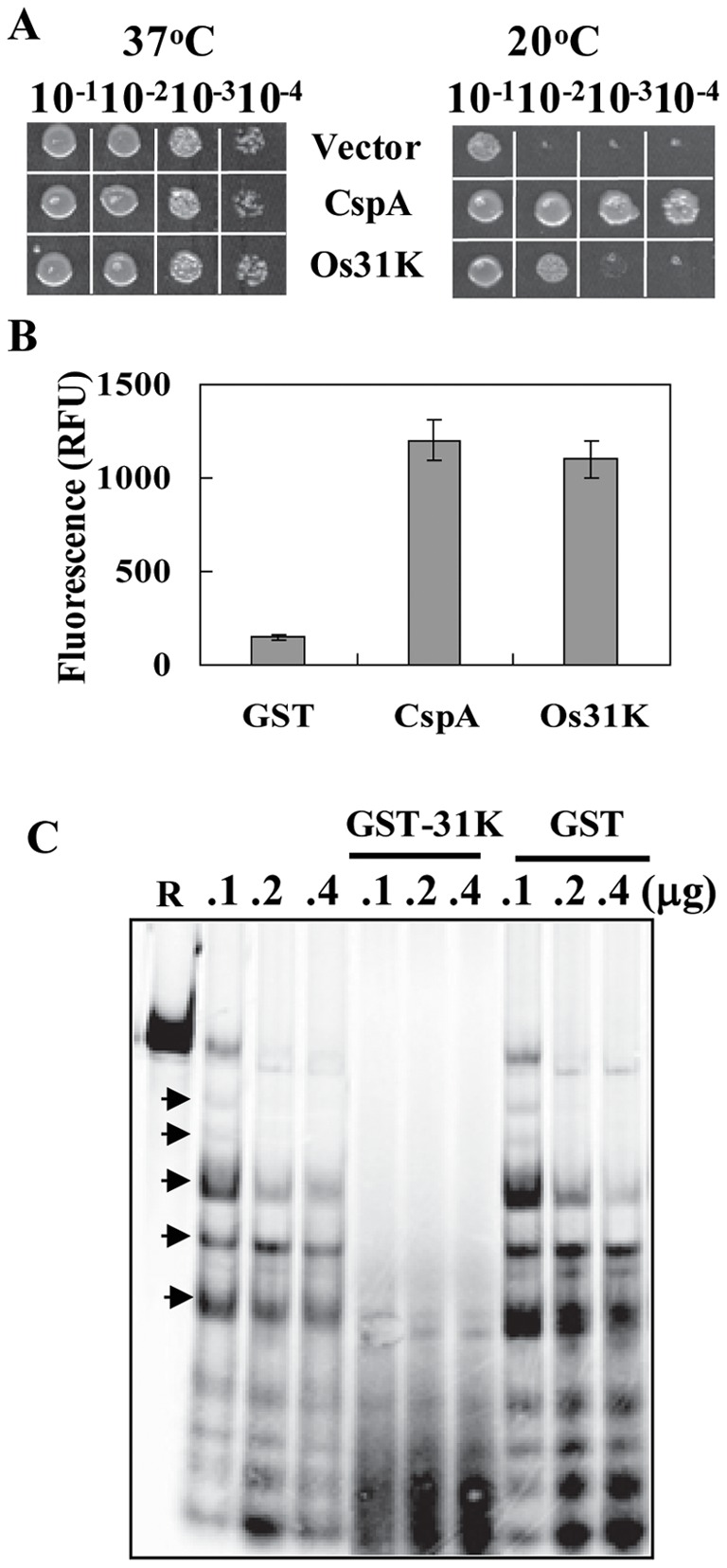
(A) The colony-forming abilities of diluted BX04 E. coli cells expressing either OsU11/U12-31K (Os31K), CspA (positive control), or pINIII (negative control) were examined under normal (37°C) and cold stress (20°C) conditions. (B) The DNA-melting activity of OsU11/U12-31K protein was examined by monitoring the fluorescence of a 78-nucleotide-long molecular beacon with the addition of GST-Os31K (10 µg), GST-CspA (5 µg), or GST (5 µg). (C) Enhanced RNase T1 cleavage of the substrate RNA (R) was measured after the addition of OsU11/U12-31K protein (GST-31K). The RNase T1-resistant bands, as indicated by arrows, disappeared in the presence of recombinant GST-31K fusion protein. The numbers above each column represent the amount of T1 added to the reaction.
Discussion
The results of our study clearly indicate that the U11/U12-31K protein, one of the seven unique minor spliceosomal proteins, is a functionally conserved RNA chaperone, which are crucial for correct U12 intron splicing and normal development of monocotyledonous plants as well as dicotyledonous plants. The presence of a single gene encoding U11/U12-31K in plant genomes and the highly similar amino acid sequences of U11/U12-31K proteins in dicots and monocots have indicated that their function is conserved in plants. The functional complementation of OsU11/U12-31K in the Arabidopsis mutant diminishing the expression of AtU11/U12-31K clearly demonstrated that U11/U12-31K plays an essential role in the development of monocot as well as dicot plants. Future research in rice is needed to fully confirm that U11/U12-31K protein is necessary for the development of monocot plants. We observed a much higher expression of U11/U12-31K in the SAM of Arabidopsis and rice (Figure 1), which correlated with the observed phenotypic defects in knockdown plants exhibiting stunted inflorescence stems. In addition, the structure and shape of the cells in inflorescence stems of U11/U12-31K knockdown mutant plants were abnormal (Figure 4). These results indicate that U11/U12-31K affects plant development by regulating meristem activity and cell division activity.
U12 introns are often found in genes that perform a function in the regulation of cell proliferation, such as DNA replication/repair, RNA processing, and translation [12], [29]. In our previous analysis, AtU11/U12-31K was shown to be responsible for the correct splicing of most of the U12 intron-containing genes [23], including QQT1(encoding ATP/GTP binding protein), HSI2 (encoding B3 DNA-binding transcription factor), Di19 (encoding gene family implicated in stress and light signaling pathways), and NHX (encoding Na+/H+ antiporter), all of which are closely related to cell division, seed maturation, and seedling growth [30]–[33]. The role for U11/U12-31K in U12-type intron splicing in dicots and monocots was confirmed in the present analysis. Defects in the correct splicing of all U12 introns in Arabidopsis AtU11/U12-31K mutants were completely recovered in the complementation lines expressing rice OsU11/U12-31K (Figure 5). These results indicate that the activity of the U11/U12-31K protein in the splicing of most U12-type introns on minor spliceosomal complexes is absolutely conserved in dicots and monocots, which is crucial for the normal development of plants.
The proposition that U11/U12-31K protein functions as an RNA chaperone during U12 intron splicing was firmly supported by the current and previous analyses. Both OsU11/U12-31K and AtU11/U12-31K proteins have been determined to harbor RNA chaperone activity, demonstrated by the functional complementation of RNA chaperone-deficient E. coli mutant and nucleic acid-melting activity (Figure 7) [23]. The observation that U11/U12-31K influences the splicing of most U12-type introns is another indication that U11/U12-31K functions as an RNA chaperone, since RNA chaperones usually adopt highly disordered structures and bind their RNA substrates with low sequence specificity [24], [25], [34], [35]. It is likely that, as an RNA chaperone, U11/U12-31K maintains the pre-RNA substrates in splicing-competent conformations or facilitates the rearrangement of spliceosomal RNAs and/or mRNAs during the splicing process. The U11/U12-31K protein is not a splicing factor, and it remains uncertain whether U11/U12-31K directly binds to the RNA substrates or if additional protein factors are required. The presence of an RNA recognition motif and CCHC-type zinc knuckle domain in U11/U12-31K strongly suggests that it directly binds to the RNA substrates. All of these results led us to hypothesize that U11/U12-31K is an RNA chaperone involved in U12 intron splicing.
In conclusion, the present findings provide evidence for the emerging idea that U11/U12-31K is an indispensible RNA chaperone that functions in U12-type intron splicing and is necessary for the normal growth and development of dicot as well as monocot plants. Considering that the biological functions of most minor spliceosomal proteins have not yet been determined in plants and animals, our findings open new opportunities to further investigate the roles of other highly conserved minor spliceosomal proteins in U12 intron splicing and for the growth and development of plants and animals.
Materials and Methods
Plant materials and growth conditions
A. thaliana Columbia-0 ecotype was grown at 23°C under long day conditions (16-hr-light/8-hr-dark cycle). Plants were also grown in half-strength Murashige and Skoog (MS) medium containing 1% sucrose. The AtU11/U12-31K knockdown Arabidopsis mutant plants (amiR1-4), in which the expression of AtU11/U12-31K is downregulated by an artificial microRNA-mediated knockdown strategy (Web MicroRNA Designer; http://wmd3.weigelworld.org/), were described in our previous report [23]. To generate complementation lines, the pCambia3301 vector carrying the rice OsU11/U12-31K gene under the control of the cauliflower mosaic virus 35S promoter was introduced into the Arabidopsis amiR1-4 mutant by vacuum infiltration [36] using Agrobacterium tumefaciens GV3101. The T3 homozygous lines were selected and used for phenotype analysis.
In situ hybridization and microscopy
To determine the expression patterns of U11/U12-31K in different organs and cells, in situ hybridization analysis was conducted using different tissues of Arabidopsis and rice. Plant samples were fixed with 4% paraformaldehyde in 50 mM sodium phosphate buffer (pH 7.0), embedded in paraffin, and sliced into thin sections. The RNA probes were synthesized in vitro using a digoxigenin RNA labeling kit (Roche Molecular Biochemicals, Mannheim, Germany). The probes were hybridized to the samples, and the signals were detected by chemiluminescence. For scanning electron microscopy (SEM) observation, the samples were fixed with a mixture of 2% glutaraldehyde and 2% paraformaldehyde, and were post-fixed with 1% osmium tetroxide in 50 mM cacodylate buffer (pH 7.2). After dehydrating the samples with a series of alcohols, the samples were dried with a HCP-2 critical point dryer (Hitach, Tokyo, Japan), coated with gold in a Emitec K550 ion sputter, and observed with a Hitachi S-2400 scanning electron microscope (Hitachi, Tokyo, Japan). For light microscope observation, the samples which were embedded in LR White resin were sectioned into thin sections, stained with 0.1% toluidine blue, and examined with a light microscope (Zeiss, Axiolba, Germany).
Analysis of the splicing of U12 intron-containing genes
For the analysis of splicing patterns of U12 intron-containing genes, total RNAs were extracted from 20-day-old wild-type, AtU11/U12-31K knockdown Arabidopsis mutant (amiR1-4), and OsU11/U12-31K-expressing amiR1-4 plants. Five to ten micrograms of total RNAs were treated with RQ1 DNase (Promega, Madison, WI, USA) and further purified using an RNeasy clean-up kit (Qiagen, Valencia, CA, USA). RT-PCR analysis of the splicing patterns was conducted as described previously [23]. Briefly, two hundred nanograms of RNAs were reverse-transcribed with gene-specific primers [23] and amplified using a one-step RT-PCR kit (Qiagen). The PCR products were separated on 1% agarose gel and visualized under UV light.
Analysis of RNA chaperone activity
In the cold shock assay using the E. coli BX04 mutant cells, OsU11/U12-31K cDNA was cloned into the pINIII vector. The cold shock test for E. coli was conducted as described previously [23], [35]. The pINIII expression vector was transformed into E. coli BX04 mutant cells, and the growth of cells was monitored in Luria-Bertani (LB) medium at low temperatures. In the nucleic acid-melting assay, the 78-nucleotide-long hairpin-shaped DNA molecule labeled with a fluorophore (tetramethyl rhodamine) and quencher (dabcyl) was synthesized as described previously [28], and nucleic acid-melting assay was conducted according to our previous studies [23], [35]. The fluorescence emitted from the molecular beacon after reaction with recombinant GST-31K fusion proteins was measured using a Spectra Max GeminiXS spectrofluorometer (Molecular Devices, Sunnyvale, CA, USA) at an excitation and emission wavelengths of 555 and 575 nm, respectively. In ribonuclease cleavage assay, the 32P-labeled RNA substrates were prepared by transcription of the pET-22b(+) plasmid using T7 RNA polymerse (Promega). The RNA substrates were incubated with recombinant GST-31K fusion proteins, and the reaction products were separated on an 8% acrylamide gel. All experimental conditions were maintained as described previously [23], [35].
Supporting Information
Morphology of inflorescence stems of the plants. (A) Light micrographs of floral bud regions of 7-week-old wild-type (WT), knockdown mutant (amiR1-4), and complementation line expressing OsU11/U12-31K gene (amiR1-4/Os31K). Scale bar = 1mm. (B) SEM of inflorescence stems of 7-week-old wild-type, mutant, and complementation line.
(TIF)
Abnormal splicing patterns of U12-type introns in the amiR1-4 mutant and complementation lines. The splicing patterns of several U12 intron-containing transcripts were analyzed by RT-PCR in wild-type (WT), knockdown plants (amiR1-4), and complementation lines expressing OsU11/U12-31K gene (amiR1-4/Os31K). The experiment was repeated three times using different batches of RNA samples, and similar results were obtained. The gray boxes with numbers represent exons, and the dashed and solid lines represent U12 and U2 introns, respectively.
(TIF)
Purification of GST-31K fusion proteins. The recombinant GST fusion proteins were purified in E. coli and the purified GST, GST-CspA and GST-Os31K fusion proteins were analyzed by SDS-PAGE.
(TIF)
List of U12 intron-containing genes investigated in this study and their splicing patterns in the amiR1-4 mutant plant.
(RTF)
Acknowledgments
We thank Drs M. Inouye and S. Phadtare for the BX04 mutant cells and pINIII vector and Dr Detlef Weigel for pRS300 vector.
Funding Statement
This work was supported in part by the Mid-career Researcher Program (2011-0017357) through the National Research Foundation (NRF) of Korea grant funded by the Ministry of Education, Science and Technology (MEST), by a grant from the Next-Generation BioGreen 21 Program (SSAC, PJ00803701), Rural Development Administration, Republic of Korea, and by Priority Research Centers Program (2011-0018393) through the NRF of Korea funded by the MEST. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Levine A, Durbin R (2001) A computational scan for U12-dependent introns in the human genome sequence. Nucleic Acids Res 29: 4006–4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhu W, Brendel V (2003) Identification, characterisation and molecular phylogeny of U12-dependent introns in the Arabidopsis thaliana genome. Nucleic Acids Res 31: 4561–4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Alioto TS (2007) U12DB: a database of orthologous U12-type spliceosomal introns. Nucleic Acids Res 35: D110–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lin CF, Mount SM, Jarmołowski A, Makałowski W (2010) Evolutionary dynamics of U12-type spliceosomal introns. BMC Evol Biol 10: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Otake LR, Scamborova P, Hashimoto C, Steitz JA (2002) The divergent U12-type spliceosome is required for pre-mRNA splicing and is essential for development in Drosophila . Mol Cell 9: 439–446. [DOI] [PubMed] [Google Scholar]
- 6. König H, Matter N, Bader R, Thiele W, Müller F (2007) Splicing segregation: The minor spliceosome acts outside the nucleus and controls cell proliferation. Cell 131: 718–729. [DOI] [PubMed] [Google Scholar]
- 7. Edery P, Marcaillou C, Sahbatou M, Labalme A, Chastang J, et al. (2011) Association of TALS developmental disorder with defect in minor splicing component U4atac snRNA. Science 332: 240–243. [DOI] [PubMed] [Google Scholar]
- 8. He H, Liyanarachchi S, Akagi K, Nagy R, Li J, et al. (2011) Mutations in U4atac snRNA, a component of the minor spliceosome, in the developmental disorder MOPD I. Science. 332: 238–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sikand K, Shukla GC (2011) Functionally important structural elements of U12 snRNA. Nucleic Acids Res 39: 8531–8543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hirose T, Shu M-D, Steitz JA (2004) Splicing of U12-type introns deposits an exon junction complex competent to induce nonsense-mediated mRNA decay. Proc Natl Acad Sci USA 101: 17976–17981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Will CL, Schneider C, Hossbach M, Urlaub H, Rauhut R, et al. (2004) The human 18S U11/U12 snRNP contains a set of novel proteins not found in the U2-dependent spliceosome. RNA 10: 929–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Burge CB, Padgett RA, Sharp PA (1998) Evolutionary fates and origins of U12-type introns. Mol Cell 2: 773–785. [DOI] [PubMed] [Google Scholar]
- 13. Will CL, Lührmann R (2005) Splicing of a rare class of introns by the U12-dependent spliceosome. Biol Chem 386: 713–724. [DOI] [PubMed] [Google Scholar]
- 14. López MD, Rosenblad MA, Samuelsson T (2008) Computational screen for spliceosomal RNA genes aids in defining the phylogenetic distribution of major and minor spliceosomal components. Nucleic Acids Res 36: 3001–3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wassarman KM, Steitz JA (1992) The low-abundance U11 and U12 small nuclear ribonucleoproteins (snRNPs) interact to form a two-snRNP complex. Mol Cell Biol 12: 1276–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Frilander MJ, Steitz JA (1999) Initial recognition of U12-dependent introns requires both U11/5′splice-site and U12/branchpoint interactions. Genes Dev 13: 851–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schneider C, Will CL, Makarova OV, Makarov EM, Lührmann R (2002) Human U4/U6 U5 and U4atac/U6atac U5 tri-snRNPs exhibit similar protein compositions. Mol Cell Biol 22: 3219–3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lorković ZJ, Lehner R, Forstner R, Barta A (2005) Evolutionary conservation of minor U12-type spliceosome between plants and humans. RNA 11: 1095–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Russell AG, Charette JM, Spencer DF, Gray MW (2006) An early evolutionary origin for the minor spliceosome. Nature 443: 863–866. [DOI] [PubMed] [Google Scholar]
- 20. Benecke H, Lührmann R, Will CL (2005) The U11/U12 snRNP 65K protein acts as a molecular bridge, binding the U12 snRNA and U11-59K protein. EMBO J 24: 3057–3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Turunen JJ, Will CL, Grote M, Lührmann R, Frilander MJ (2008) The U11-48K protein contacts the 5′ splice site of U12-type introns and the U11-59K protein. Mol Cell Biol 28: 3548–3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lorković ZJ, Lopato S, Pexa M, Lehner R, Barta A (2004) Interactions of Arabidopsis RS domain containing cyclophilins with SR proteins and U1 and U11 snRNP-specific proteins suggest their involvement in pre-mRNA splicing. J Biol Chem 279: 33890–33898. [DOI] [PubMed] [Google Scholar]
- 23. Kim WY, Jung HJ, Kwak KJ, Kim MK, Oh SH, et al. (2010) The Arabidopsis U12-type spliceosomal protein U11/U12-31K is involved in U12 intron splicing via RNA chaperone activity and affects plant development. Plant Cell 22: 3951–3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rajkowitsch L, Chen D, Stampfl S, Semrad K, Waldsich C, et al. (2007) RNA chaperones, RNA annealers and RNA helicases. RNA Biol 4: 118–130. [DOI] [PubMed] [Google Scholar]
- 25. Tompa P, Kovacs D (2010) Intrinsically disordered chaperones in plants and animals. Biochem Cell Biol 88: 167–174. [DOI] [PubMed] [Google Scholar]
- 26. Bae W, Xia B, Inouye M, Severinov K (2000) Escherichia coli CspA-family RNA chaperones are transcription antiterminators. Proc Natl Acad Sci USA 97: 7784–7789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Xia B, Ke H, Inouye M (2001) Acquirement of cold sensitivity by quadruple deletion of the cspA family and its suppression by PNPase S1 domain in Escherichia coli . Mol Microbiol 40: 179–188. [DOI] [PubMed] [Google Scholar]
- 28. Phadtare S, Inouye M, Severinov K (2002) The nucleic acid melting activity of Escherichia coli CspE is critical for transcription antitiermination and cold acclimation of cells. J Biol Chem 277: 7239–7245. [DOI] [PubMed] [Google Scholar]
- 29. Chang W-C, Chen Y-C, Lee K-M, Tarn W-Y (2007) Alternative splicing and bioinformatic analysis of human U12-type introns. Nucleic Acids Res 35: 1833–1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Milla MAR, Townsend J, Chang IF, Cushman JC (2006) The Arabidopsis AtDi19 gene family encodes a novel type of Cys2/His2 zinc-finger protein implicated in ABA-independent dehydration, high-salinity stress and light signaling pathways. Plant Mol Biol 61: 13–30. [DOI] [PubMed] [Google Scholar]
- 31. Lahmy S, Guilleminot J, Schmit AC, Pelletier G, Chaboute ME, et al. (2007) QQT proteins colocalize with microtubules and are essential for early embryo development in Arabidopsis. Plant J 50: 615–626. [DOI] [PubMed] [Google Scholar]
- 32. Tsukagoshi H, Morikami A, Nakamura K (2007) Two B3 domain transcriptional repressors prevent sugar-inducible expression of seed maturation genes in Arabidopsis seedlings. Proc Natl Acad Sci USA 104: 2543–2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bassil E, Ohto MA, Esumi T, Tajima H, Zhu Z, et al. (2011) The Arabidopsis intracellular Na+/H+ antiporters NHX5 and NHX6 are endosome associated and necessary for plant growth and development. Plant Cell 23: 224–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jiang W, Hon Y, Inouye M (1997) CspA, the major cold-shock protein of Escherichia coli, is an RNA chaperone. J Biol Chem 272: 196–202. [DOI] [PubMed] [Google Scholar]
- 35. Kim JS, Park SJ, Kwak KJ, Kim Y-O, Kim JY, et al. (2007) Cold shock domain proteins and glycine-rich RNA-binding proteins from Arabidopsis thaliana can promote the cold adaptation process in E. coli. . Nucleic Acids Res 35: 506–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bechtold N, Pelletier G (1998) In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol Biol 82: 259–266. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Morphology of inflorescence stems of the plants. (A) Light micrographs of floral bud regions of 7-week-old wild-type (WT), knockdown mutant (amiR1-4), and complementation line expressing OsU11/U12-31K gene (amiR1-4/Os31K). Scale bar = 1mm. (B) SEM of inflorescence stems of 7-week-old wild-type, mutant, and complementation line.
(TIF)
Abnormal splicing patterns of U12-type introns in the amiR1-4 mutant and complementation lines. The splicing patterns of several U12 intron-containing transcripts were analyzed by RT-PCR in wild-type (WT), knockdown plants (amiR1-4), and complementation lines expressing OsU11/U12-31K gene (amiR1-4/Os31K). The experiment was repeated three times using different batches of RNA samples, and similar results were obtained. The gray boxes with numbers represent exons, and the dashed and solid lines represent U12 and U2 introns, respectively.
(TIF)
Purification of GST-31K fusion proteins. The recombinant GST fusion proteins were purified in E. coli and the purified GST, GST-CspA and GST-Os31K fusion proteins were analyzed by SDS-PAGE.
(TIF)
List of U12 intron-containing genes investigated in this study and their splicing patterns in the amiR1-4 mutant plant.
(RTF)