Abstract
In the past decade, Clostridium difficile has emerged as an important gut pathogen. Symptoms of C. difficile infection range from mild diarrhea to pseudomembranous colitis, sometimes resulting in colectomy or death. The main virulence factors of C. difficile are toxin A and toxin B. Besides the genes encoding these toxins (tcdA and tcdB), the pathogenicity locus (PaLoc) also contains genes encoding a sigma factor (tcdR) and a putative anti-sigma factor (tcdC). The important role of TcdR as a sigma factor for toxin expression is undisputed, whereas the role of TcdC as an anti-sigma factor, inhibiting toxin expression, is currently the subject of debate. To clarify the role of TcdC in toxin expression, we generated an isogenic ClosTron-based mutant of tcdC in Clostridium difficile strain 630Δ Erm (CT::tcdC) and determined the transcription levels of the PaLoc genes and the expression levels of the toxins in the wild type strain and the tcdC mutant strain. We found only minor differences in transcription levels of the PaLoc genes between the wild type and CT::tcdC strains and total toxin levels did not significantly differ either. These results suggest that in C. difficile 630Δerm TcdC is not a major regulator of toxin expression under the conditions tested.
Introduction
Clostridium difficile is an anaerobic, Gram-positive, spore forming rod shaped bacterium that can cause disease with a wide variety of symptoms, ranging from mild diarrhea to severe forms of pseudomembranous colitis [1]–[3]. Since 2004, numerous countries have reported outbreaks in health-care facilities caused by hypervirulent C. difficile PCR Ribotype (Type) 027 [1]–[6]. Clostridium difficile infection (CDI) caused by Type 027 is associated with a more severe course of the disease and a higher mortality rate than other ribotypes [1], [3], [6]. Recently, increasing numbers of the hypervirulent Type 078 are reported [7]. C. difficile Type 078 is more frequently associated with community acquired CDI and affects a younger population than Type 027 [6]–[9]. Furthermore, CDI caused by Type 078 is associated with an increased morbidity compared to other ribotypes [8].
The main virulence factors of the enteropathogenic C. difficile are the two large clostridial toxins, toxin A (TcdA) and toxin B (TcdB). These toxins are glycosyltransferases that inactivate Rho, Rac and Cdc42, thereby disrupting the cytoskeleton and tight junctions of the cells, resulting in apoptosis [10]. This induces an inflammatory response and degradation of the intestinal epithelial cell layer. Besides the genes encoding these toxins (tcdA and tcdB), the pathogenicity locus (PaLoc) also contains genes encoding a sigma factor (tcdR) and a putative anti-sigma factor (tcdC) [11]–[13]. In between the toxin genes the tcdE gene is situated, which encodes a putative holin protein [14]. Interestingly, both hypervirulent Types 027 and 078 have been shown to contain mutations in the tcdC gene, encoding the putative negative regulator of toxin gene transcription, and this has been proposed as a possible explanation for their increased virulence [8], [15].
The exponential growth phase of C. difficile has been reported to be associated with a high transcription level of the tcdC gene and low transcription levels of tcdR and the toxin genes, whereas the stationary growth phase is associated with a low transcription level of the tcdC gene and high transcription levels of tcdR and the toxin genes in strain VPI10463 [16]. The synthesis and secretion of the toxins is increased upon entry into the stationary growth phase [16]–[19]. The decreasing transcription of tcdC correlates with diminishing TcdC protein levels in stationary growth phase [16], [20].
TcdR is an alternative sigma factor that positively regulates toxin production [11], [12]. The direct interaction of TcdR and the RNA polymerase core enzyme mediates recognition of the toxin promoters and the tcdR promoter [11], [12], [21]. TcdC has been reported to act like an anti-sigma factor for toxin production by destabilizing the TcdR-RNA polymerase core enzyme complex in a way that is not yet fully understood [12].
The reported inverse correlation between the transcription of tcdC and the toxin genes and the expression patterns of the corresponding proteins, together with the biochemical data, has led to the prevailing model that TcdC is an important repressor of toxin expression [12], [16], [17], [20]. This model seems to be supported by the finding that the absence of a functional TcdC caused by a frame shift mutation (Δ117 bp) in the tcdC gene is linked to a supposed increased toxin production in certain (hyper) virulent strains [15], [22].
Recently, some doubts were raised about the importance of TcdC for regulation of toxin expression on the basis of two findings. First, two studies have found increasing levels of tcdC transcription in time that coincide with increasing transcription of the toxin genes and increasing amounts of toxin production [18], [19]. Second, there is a great variability in toxin expression levels among (hyper) virulent strains, even though these generally carry mutations in tcdC [15], [18], [19]. Therefore, a minor (or modulatory) role for TcdC in the regulation of toxin expression was proposed [18], [19].
Here, we sought to clarify the role of TcdC in regulation of the toxin production by generating an isogenic tcdC mutant (CT::tcdC) using the ClosTron technology. We find only minor differences in transcription levels of the PaLoc genes between the wild type and CT::tcdC strains and the expressed total toxin levels did not significantly differ, suggesting that the role of TcdC in toxin regulation is not of significance under the conditions tested in C. difficile strain 630ΔErm.
Results
The importance of TcdC for regulation of toxin expression was recently challenged by two studies [18], [19]. It was proposed, based on the increasing transcription levels of the PaLoc genes in time and the variability in toxin expression levels among virulent strains, that TcdC has a minor or modulatory role on toxin expression rather than a major role as previously assumed. In this study we sought to clarify the role of TcdC for toxin expression by generating an isogenic tcdC mutant. As toxin gene expression is subject to complex regulation influenced by glucose and cysteine, we performed our experiments in a trypton-yeast (TY) based broth [17], [23]. TY broth does not contain glucose and no cysteine was added. We verified that in TY broth earlier and higher expression of toxins was achieved in comparison to the commonly used Brain Heart Infusion (BHI) broth (data not shown).
Generation and Characterization of a TcdC Mutant
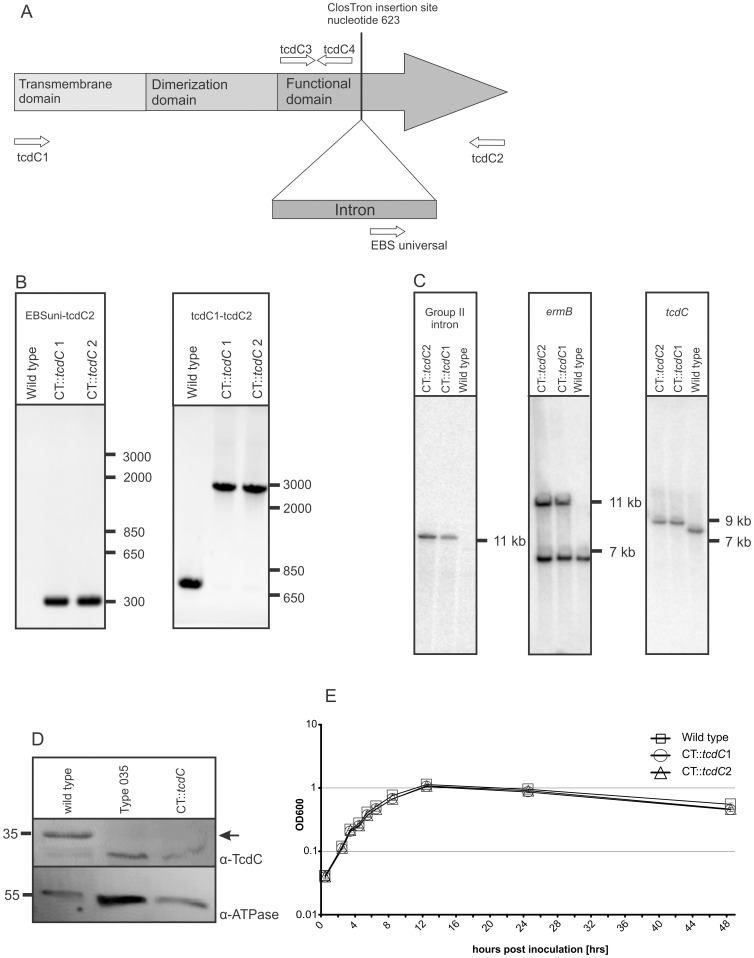
TcdC consists of three domains: a hydrophobic domain, a proposed dimerization domain and a proposed C-terminal repressor domain (Figure 1A) [12]. We successfully disrupted the tcdC gene in the region coding for the repressor domain using ClosTron technology. Disruption of genes using the ClosTron technology results in stable mutants and no or non-functional proteins [24]–[26]. The genotype of the disruption was confirmed with conventional PCRs using the tcdC2 primer and the EBS universal primer and with a primer pair (tcdC1 and tcdC2) flanking the ClosTron insertion site (Figure B). Sequence analysis confirmed that the disruption was in the proposed repressor domain of the tcdC gene at the expected site (data not shown). In addition, Southern blot analysis using intron-, ermB and tcdC- specific probes clearly confirmed a specific single insertion of the Group II intron in the genome (Figure 1C).
Figure 1. Characterization of the C. difficile tcdC mutant.
(A) Schematic representation of 3 different domains of TcdC and the intron insertion site for the inactivation of TcdC. The arrows in the putative repressor domain represent the locations and orientation of the primers used in the RT-q-PCR and conventional control PCRs. (B) PCR confirmation of the wild type strain and the CT::tcdC mutant. The primer EBS universal and tcdC2 generated a PCR product of 302 bp for the CT::tcdC strains. Primers tcdC1 and tcdC2 generated a 699 bp PCR product for the wild type and for the CT::tcdC strain a PCR product of circa 2800 bp. (C) Southern blot analysis of EcoRV digested genomic DNA of wild type and CT::tcdC strains with a Group II intron, ermB gene and tcdC specific probes. Note that probing with the ermB probe results in 2 bands for the CT::tcdC strains, since wild type already carries a copy of the ermB gene in the genome [35]. (D) Western blot analysis of TcdC production in wild type and CT::tcdC strain 8 hours post inoculation. The arrow indicates the location of TcdC protein based on MW and absence of the protein in the PaLoc negative Type 035 strain. Note that cross-reaction of TcdC antibody with a protein of similar MW was also observed in Carter et al. [30]. (E) Growth curves of C. difficile 630ΔErm and C. difficile CT::tcdC mutant strains. The absorbance (OD600) was measured over 48 hrs of growth in TY medium. The error bars indicate the standard error of the mean of six experiments.
Western blot analysis, using antibodies against TcdC, confirmed that the isogenic tcdC mutant no longer expressed TcdC (Figure 1D). A control blot using antibodies against F0F1 ATPase indicated that the lack of signal in the TcdC Western blot was not a result of lower amounts of proteins loaded in the lanes of PCR ribotype 035 (a PaLoc negative strain) and the tcdC mutant.
The growth kinetics of the wild type and CT::tcdC strains showed no significant differences in various media tested (Figure 1E and data not shown). In TY broth, which does not contain glucose or added cysteine, the wild type strain and the CT::tcdC strains showed an exponential growth phase in the first 8 hours post inoculation and after 12 hours post inoculation both strains entered into the stationary growth phase (Figure 1E). Conventional control PCRs confirmed that the disruption of the tcdC gene had remained intact during our growth curves experiments (data not shown).
Comparable Relative Transcription Levels of PaLoc Genes in Wild Type and CT::tcdC
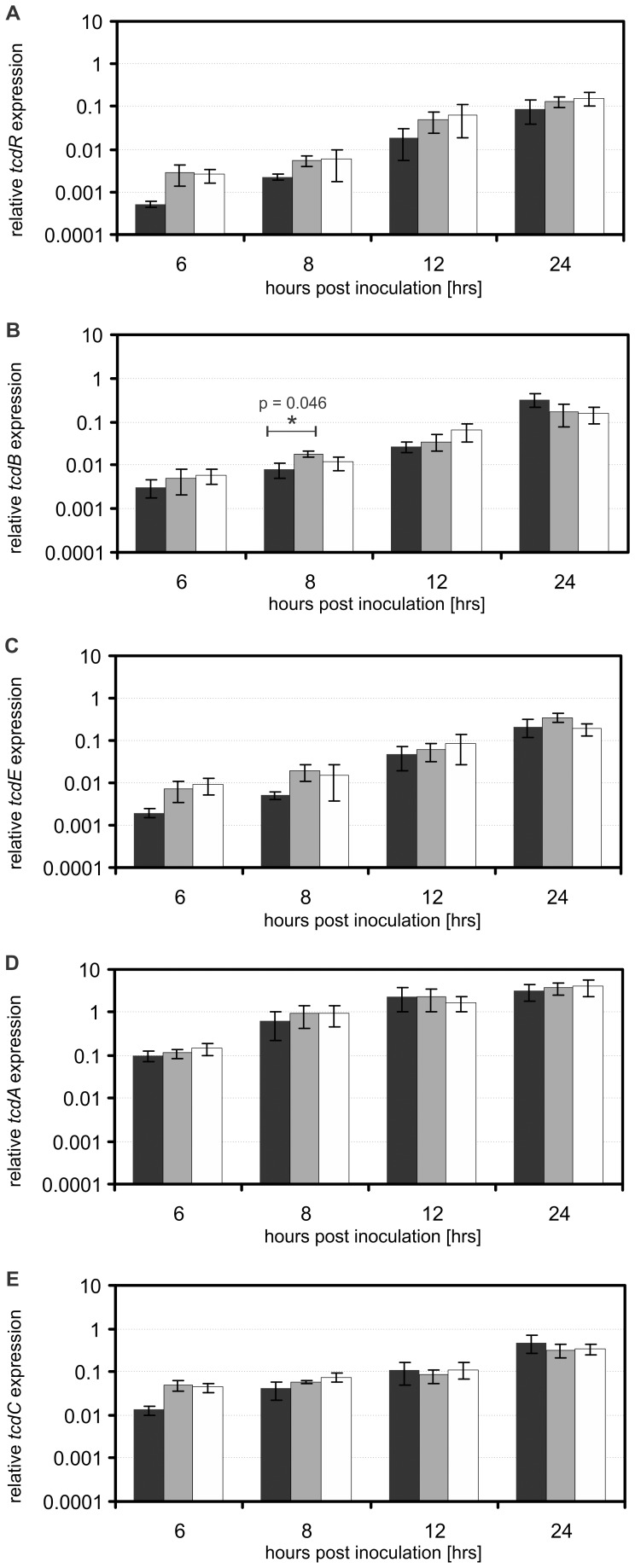
In order to determine the influence of TcdC on the transcription levels of the PaLoc genes we compared the relative transcription levels of the PaLoc genes of wild type and CT::tcdC strains by reverse transcriptase quantitative real-time PCR (RT-qPCR). We found comparable transcription levels of all PaLoc genes in wild type and CT::tcdC strains.
Overall, the logarithmic growth phase was associated with lower transcription levels of the PaLoc genes and by entering into the stationary phase increasing transcription levels of PaLoc genes were found, as previously described for tcdR, tcdE, tcdB and tcdA [16], [18], [19] and tcdC [18], [19] (Figure 2). The transcription levels of tcdR in wild type and CT::tcdC strains increased approximately 100-fold between 6 and 24 hours post inoculation (Figure 2A). Though the expression of tcdR was, on average, 3-fold higher at the various time points in the CT::tcdC strains compared to the wild type, this difference was not statistically significant (Figure 2A, all p values ≥0.088). Similarly, we observed a 10- to 100-fold increase in the transcription levels of tcdB (Figure 2B), tcdE (Figure 2C), tcdA (Figure 2D) and tcdC (Figure 2E) when comparing values from the logarithmic growth phase with those observed in the stationary growth phase. The expression levels of tcdB, tcdE, tcdA and tcdC were, on average, 1.5-fold, 2.5-fold, 1.4-fold and 1.7-fold higher, respectively, in the CT::tcdC strains compared to the wild type. With one exception, these differences were not found to be significant. The transcription level of tcdB in the CT::tcdC1 strain is significantly (P = 0.046) higher compared to wild type level at 8 hours post inoculation (Figure 2D). However, no significant differences are found between the wild type and CT::tcdC strains at any of the other time points.
Figure 2. The relative PaLoc gene expression profiles of wild type and CT::tcdC in time.
The error bars indicate the standard error of the mean (n = 6). The asterisk (*) indicate a significant difference between wild type and CT::tcdC strain. Values are normalized to rpsJ expression. Wild type corresponds to black bars, CT::tcdC1 mutant strains to gray bars, CT::tcdC2 to the white bars. (A) The relative expression of tcdR. (B) The relative expression of tcdB. (C) The relative expression of tcdE (D) The relative expression of tcdA. (E) The relative expression of tcdC.
Therefore, we conclude that the disruption of the tcdC gene does not result in a consistently and significantly increased transcription level of the PaLoc genes.
Comparable Toxin Expression in Wild Type and CT::tcdC
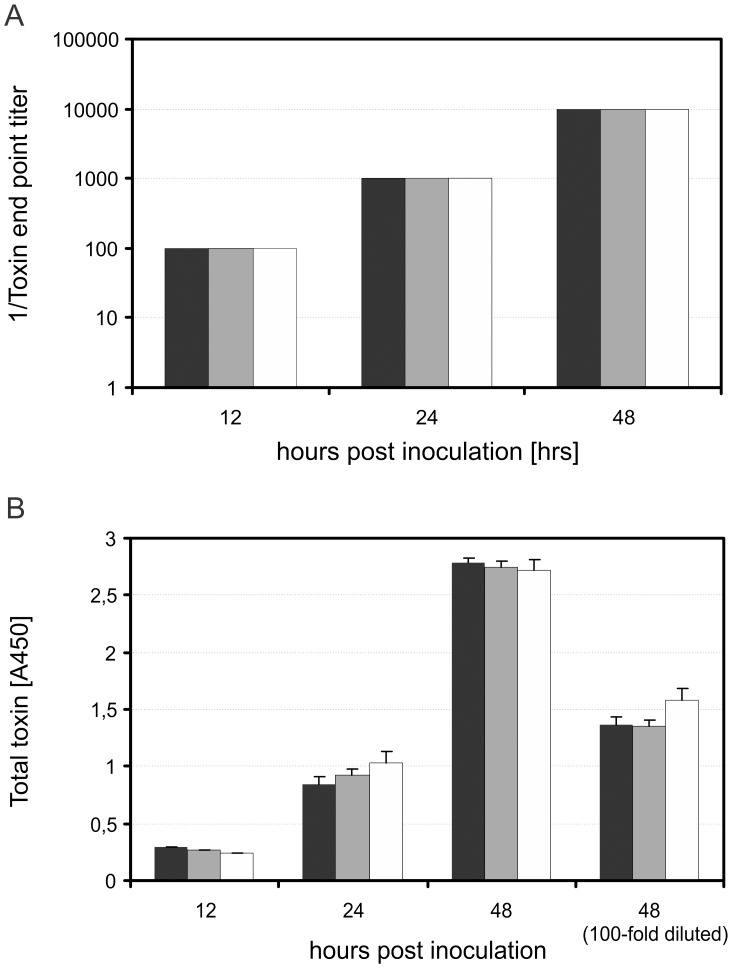
Considering the small increase in PaLoc gene expression in the CT::tcdC mutants observed in the RT-qPCR experiments, we were interested to see if this difference translated into higher toxin levels. We determined toxin levels using two independent assays, but found no consistent difference between wild type and mutant cells.
First, filter sterilized bacterial supernatants were incubated on a Vero cell (a kind gift of Dr. E.J. Snijder [27]) monolayer and cytotoxic effects were quantified after 24 hours by determining the end-point titer (Figure 3A) [26]. In the exponential growth phase (5 and 8 hours post inoculation) no cytotoxic effects were detectable (data not shown). In the stationary growth phase (12, 24 and 48 hours post inoculation) we observed increasing cytotoxic effects, indicative of the presence of toxin. Importantly, the observed cytotoxic effects were specific for C. difficile toxin A and B, as a pre-incubation of the filter sterilized bacterial supernatants with anti-toxin, a polyclonal antibody against toxin A and toxin B (Techlab), resulted in complete neutralization of cytotoxic effects on the Vero cells at all time points (data not shown). The tcdC mutant strains showed no significant differences in toxin levels compared to the wild type strain (Fig. 3A).
Figure 3. The toxin production profiles of wild type and CT::tcdC mutant strains in time.
Wild type corresponds to black bars, CT::tcdC1 mutant strains to gray bars, CT::tcdC2 to the white bars. Total toxin amounts were quantified by using two independent assays. (A) The supernatants were incubated in a ten fold dilutions series on Vero cell monolayers. After 24 hrs the cytotoxic effects were quantified by determing the toxin end point titer. Values are given as means (n = 6). (B) An enzyme immunoassay was used for direct quantification of the secreted toxins according manufacters protocol. The supernatants of 12 and 24 hours post inoculation were 10 times diluted. The supernatants of 48 hours post inoculation were diluted 10 and 100 times. Values are given as means ± standard error of the mean (n = 6).
Next, we used an enzyme immunoassay (Ridascreen, Biopharma) for the direct detection and relative quantification of the secreted toxins. In the exponential growth phase (5 and 8 hours post inoculation) no toxins were detectable (data not shown), consistent with the lack of toxicity towards Vero cells described above. In the stationary growth phase (12, 24 and 48 hours post inoculation) increasing toxin levels were detectable. When we compared the toxin levels at various time points, there were equal amounts of toxins in the wild type and tcdC mutant strains.
We conclude that the disruption of the tcdC gene does not result in consistently and significantly increased toxin levels.
Discussion
C. difficile infections caused by the (hyper-)virulent Type 027 (NAP1/REA B1) and Type 078 (NAP7/REA BK) are associated with an increased morbidity and severity of disease compared to other types [1], [3]. This increase is suggested to be linked to toxin hyper production [3], [22], [28]. A potential mechanism by which this could occur is through inactivation of a negative regulator of the toxin gene transcription. TcdC has been identified as a negative regulator of toxin production [12].
In the currently prevailing model, a major role for TcdC in the repression of toxin genes has been proposed on the basis of three lines of evidence. First, in C. difficile VPI10463 (a high toxin producing strain that also expresses high levels of TcdC [18], [29]), an inverse correlation between the transcription of tcdC and the genes encoding the toxins is found [16], [18], [29]. This correlation for TcdC is also observed in protein levels [20]. Second, elegant in vitro experiments have established that heterologously produced and purified TcdC protein can interfere with TcdR-mediated transcription of toxin genes in a way that is not yet fully understood [12]. Finally, a frame shift mutation (Δ117 bp) in tcdC, that results in a non-functional protein, is associated with increased toxin production in certain (hyper)virulent strains [15], [22].
Recently, it was reported that the introduction of a functional tcdC gene from a high toxin-producing strain that lacks any of the hyper virulence associated tcdC mutations (VPI10463, PCR ribotype 087) into an epidemic strain carrying a non-functional tcdC (M7404, PCR ribotype 027/NAP1/REA B1) can reduce toxin expression levels and moderately attenuate virulence [30]. This data seems consistent with the model discussed above. However, it is unclear how the levels of TcdC in the complemented strain relate to the physiological levels of the protein prior to the inactivation of TcdC in this strain background. The introduced tcdC gene, including its transcription signals, was derived from a different genetic background (VPI10463, Type 087) and was introduced on a multicopy plasmid. In addition, the reintroduction of TcdC in a strain lacking a functional TcdC, may affect processes that are not normally affected. Finally, the experiments were not corrected for the additional copies of the tcdC promoter that could result in the titration of regulators binding to those sequences.
In an alternative approach that addresses many of the issues above, the role of tcdC in toxin expression could be addressed by removing it from a background in which it is normally functional. To this end, we generated two independent isogenic ClosTron-based tcdC mutants strain that could be directly compared to its wild type counterpart, in which the TcdC protein was expected to be functional. Our data obtained with these mutant strains show that TcdC does not exert a major or even significant effect on the transcription of the PaLoc genes or the expression levels of the toxins under the conditions tested.
Our experiments were performed in a glucose free TY broth medium, since glucose is a known repressor of toxin production [17]. Indeed, we observed earlier and higher levels of toxin production in TY broth than in the commonly used Brain-Heat-Infusion broth (BHIS) based media, that does contain low amounts of glucose (0.2%) and to which frequently cysteine is added. However, also in BHIS we did not observe a significant effect of a tcdC deletion on toxin expression (data not shown).
We controlled critical parameters in our experiments by performing conventional PCRs which confirmed that the disruption of tcdC remained intact throughout the growth curve. Western blot analysis with antibodies raised against a TcdC epitope confirmed that the disruption of the tcdC gene resulted in the absence of TcdC protein (Figure 1D). The disruption of the tcdC gene did not affect the growth kinetics compared to the wild type strain (Figure 1E).
In the RT-qPCR experiments, sample to sample variation was corrected by normalizing to the reference gene rpsJ [31]. The rpsJ gene was selected for normalization, since rpsJ was overall the highest ranked reference gene regarding gene expression stability [31].Reverse transcription was carried out using random hexamers, to prevent gene specific biases [32]. PCR efficiency in the qPCR was determined using a standard curve for each gene, enabling post run correction [33]. To obtain objective data concerning the quantification of the secreted toxins, we used an end point titer assay and an enzyme immunoassay rather than a manual (subjective) cell scoring system [26].
The trends observed in the transcription of the PaLoc genes and the expression of the toxins generally conform to previously reported data [18], [19]. It should be noted that the up-regulation in time of tcdC transcription was not observed in earlier studies on C. difficile VPI10463 [16] but is consistent with more recent reports [18], [19]. We observed an increase in transcription of the PaLoc genes in time, and a concomitant increase in toxicity of culture supernatant in stationary phase that can be attributed to the toxins as it is fully neutralized by anti-toxin against toxin A and B.
The disruption of the tcdC gene resulted in an on average 1.7 fold higher transcription level of tcdC in time compared to the wild type strain, although this difference was not found to be statistically significant. It should be noted that we detect these differences because the real time PCR probe detects a region of the gene upstream of the ClosTron insertion site (Figure 1A). This finding might indicate some kind of feedback mechanism on TcdC expression. Similar to tcdC gene expression, the disruption of tcdC resulted in a slightly higher transcription level of the other PaLoc genes, although this was generally not significant. Moreover, the increased transcription level of the toxin genes did not result in a detectable increase in toxin levels as measured with two independent assays.
Based on the paradigm that TcdC is a major suppressor of toxin production we expected precocious and significantly elevated transcription levels of tcdA, tcdB, tcdE and tcdR in the CT::tcdC strains compared to wild type. However, our data indicate that TcdC exerts a moderate, if any, effect on the transcriptional levels of the PaLoc genes and the expression of toxins in C. difficile 630Δerm under the conditions tested.
Clostridium difficile strain 630ΔErm is a derivative of the clinical isolate 630 [34], [35], a PCR ribotype 012 strain. PCR ribotypes 012 strains constitute 4% of the clinically isolated toxinogenic isolates in Europe [7]. Clostridium difficile 630 (PCR ribotype 012)-derived strains are commonly used to investigate virulence of mutants [26], [36], [37].
An independent study, published during the preparation of this manuscript, reached a similar conclusion with respect to the role of TcdC in toxin regulation in C. difficile 630Δerm using an allelic exchange technique [38]. In that paper reintroduction of a single functional copy of tcdC at its native locus did not affect toxin production in strain R20291 either [38]. R20291 is a strain from problematic PCR ribotype 027 (NAP1/REA B1) that was isolated following an outbreak in Stoke Mandeville, UK.
Our work and that of Cartman and coworkers [38] seem at odds with the previous reports that clearly demonstrate that TcdC can act as a repressor for toxin gene expression [12], [30]. However, we cannot exclude the possibility that TcdC exerts a more profound effect under specific conditions, or in other strains of C. difficile than 630Δerm and R20291. It should be clear though that in vivo relevance of TcdC for toxin regulation in these two strains is limited.
In conclusion, we suggest that TcdC might have a modulatory role in regulating toxin expression, and that TcdC functionality is therefore not a major determinant of the (hyper)virulence of C. difficile. This is supported by the lack of correlation between virulence, toxin production and tcdC gene variants that was noted by several other studies [18], [19], [30], [39].
Materials and Methods
Bacterial Strains and Growth Conditions
The Clostridium difficile and Escherichia coli (E. coli) strains and plasmids used in this study are described in Table 1. E. coli strains were grown in Luria Bertani (LB, USB cooperation) medium supplemented with appropriate antibiotics when required. C. difficile strains were grown anaerobically in a microaerobic cabinet (Don Whitley VA1000) at 37°C in pre-reduced 3% Bacto Tryptose (Difco), 2% Yeast extract (Difco) and 0.1% thioglycolate (pH 7.4) medium (TY) or Brain Heart Infusion broth (Oxoid) supplemented with 0.5% yeast extract and 0.01% L-cysteine (Sigma) (BHIS) [40], [41]. When required, the broths were supplemented with appropriated antibiotics. For RNA extraction and toxin quantification, C. difficile 630ΔErm (wild type) and two independent isogenic tcdC mutant strains (CT::tcdC) were serially diluted and pre-cultured (overnight) in pre-reduced TY broth. Mid-logarithmic growth phase pre-cultures (OD600 0.4–0.8) were used to inoculate pre-reduced TY broth to a starting OD600 of 0.05 (±0.01). Optical density readings and samples for total toxin quantification were taken at 2, 3, 4, 5, 6 and 8 hours post inoculation in the exponential growth phase and at 12, 24 and 48 hours post inoculation in the stationary phase. Samples for RNA extraction were taken at 6, 8, 12, and 24 hours post inoculation. Samples for Western blot detection of TcdC were taken at 8 hours post inoculation. We routinely monitored the purity of the C. difficile cultures by culturing on appropriate agar plates and performed control PCRs to ensure that the insertional disruption of the tcdC gene had remained intact during our experiments. All experiments were performed six times.
Table 1. Strains and plasmids used in this study.
| Strains | Description | Origin |
| Escheria coli | ||
| DH5α | ErythromycinS, LincomycinS | Laboratory stock |
| CA434 | ErythromycinS, LincomycinS, KanamycinR, plasmid R702 | [41] |
| Clostridium difficile | ||
| 630ΔErm (wt) | Erythromycins, LincomycinS | [34] |
| Leeds_ 035 | Type 035, tcdC negative, PaLoc negative | [44] |
| CT::tcdC1 | 630ΔErmΔtcdC623as, ErythromycinR, LincomycinR | This study |
| CT::tcdC2 | 630ΔErmΔtcdC623as, ErythromycinR, LincomycinR | This study |
| Plasmids | ||
| pMTL007C-E2 | ThiamphenicolR, ErythromycinS | [25] |
| pDB001AAATTAGAAACTTGCGTTCAGTAAACE2:tcdC623as | pMTL007C- E2:tcdC623as | This study |
Generation of tcdC Mutant Strains
We generated two independent isogenic tcdC mutants by insertional inactivation of the tcdC gene in the wild type strain 630Δerm using ClosTron technology [24], [25]. Briefly, the Perutka algorithm on the ClosTron website (http://www.clostron.com) was used to design primers (Table 2) for retargeting the Group II intron (Sigma; Targetron). The retargeted intron was cloned using the restriction enzymes BsrGI and HindIII into plasmids pMTL007C-E2 and the constructs were verified by sequencing [25]. The verified plasmid (pDB001) was transformed to E. coli CA434 and transferred to the wild type strain 630Δerm via conjugation [34], [41]. The selection of C. difficile transconjugants was done by subculturing on pre-reduced BHIS agar supplemented with thiamphenicol (Sigma; 10 µg/ml) and C. difficile selective supplement (Oxoid). This was followed by several rounds of subculturing on pre-reduced BHIS agar supplemented with lincomycin (Sigma; 20 µg/ml) and C. difficile selective supplement to promote integration of the GroupII intron into the gene of interest. Chromosomal DNA isolated from the transconjugants using a QIAamp blood kit (Qiagen) was used in conventional PCRs and sequence runs to confirm the disruption of tcdC and the nucleotide position of the insertion in the tcdC gene. Primers used for cloning and sequencing are listed in Table 2.
Table 2. Primers and probes used in this study.
| Primers | ||
| IBS-tcdC623as | AAAAAAGCTTATAATTATCCTTAGTTATCGTTCCAGTGCGCCCAGATAGGGTG | This study |
| EBS2-tcdC623as | TGAACGCAAGTTTCTAATTTCGATTATAACTCGATAGAGGAAAGTGTCT | This study |
| EBS1d-tcdC623as | CAGATTGTACAAATGTGGTGATAACAGATAAGTCGTTCCAGCTAACTTACCTTTCTTTGT | This study |
| EBS universal | Intron mutagenesis/Control PCR/CGAAATTAGAAACTTGCGTTCAGTAAAC TGAACGCAAGTTTCTAATTTCGATTATAACTCGATAGAGGAAAGTGTCT | [25] |
| tcdC1 | Control PCR/ATGTTTTCTAAAAAAAATGAT | This study |
| tcdC2 | Control PCR/TTAATTAATTTTCTCTACAGCT | This study |
| tcdR Forward | Multiplex 1/ATAATGATGCCCACAAGATGATTTAG | This study |
| tcdR Reverse | Multiplex 1/AAAGAAGTGATCTATATCATCAGTTAC | This study |
| tcdR probe | Multiplex 1/TEX-TATGACCTGAACCACCTTCCATTCTCC-BHQ-2 | This study |
| tcdB forward | Multiplex 1/ATAATGATGCCCACAAGATGATTTAG | This study |
| tcdB Reverse | Multiplex 1/AAAGAAGTGATCTATATCATCAGTTAC | This study |
| tcdB probe | Multiplex 1/TEX-TATGACCTGAACCACCTTCCATTCTCC-BHQ-2 | This study |
| tcdE Forward | Multiplex 2/ATTTGATACATTATTAGGATGTTTAAG | This study |
| tcdE Reverse | Multiplex 2/AAATATACATGCTATCATTGCTAC | This study |
| tcdE probe | Multiplex 2/FAM-TGATTCCTCCATCTATTCCAAAACTAGAA-BHQ-1 | This study |
| tcdA forward | Multiplex 1/AATTCCAATACAAGCCCTGTAG | This study |
| tcdA Reverse | Multiplex 1/TATCAGCCCATTGTTTTATGTATTC | This study |
| tcdA probe | Multiplex 1/FAM-ATCACTGACTTCTCCACCTATCCATACAA-BHQ-1 | This study |
| tcdC3 | Multiplex 1/CATAATTTCCAGACACAGCTAATC | This study |
| tcdC4 | Multiplex 1/GGATATGATACTGGTATTACTTATGAC | This study |
| tcdC probe | Multiplex 1/YAK-TGCACCTCATCACCATCTTCAATAACTTG-BHQ1 | This study |
| rspJ Forward | GATCACAAGTTTCAGGACCTG | This study |
| rspJ Reverse | GTCTTAGGTGTTGGATTAGC | This study |
| tcdC5 | CATATCCTTTCTTCTCCTCTTC | This study |
| tcdC6 | AATTGTCTGATGCTGAACC | This study |
| oWKS-1131 | AAAGCGATGCCGAGAATCTG | This study |
| oWKS-1132 | TCTCGGAGTATACGGCTCTG | This study |
| Sal-R1 | ATTACTGTGACTGGTTTGCACCACCCTCTTCG | [45] |
Complementation can be a valuable control for knockout studies. However, as our tcdC mutant strains have no clearly detectable phenotype regarding toxin production, complemented mutant strains are expected to be comparable to wild type and tcdC mutant strains, as also reported recently in an independent study [38]. Therefore, a complementation study would not add to the message of this manuscript.
Southern Blots
Southern blot analysis was performed to verify a specific single integration into the genome. Genomic DNA was extracted using a Phenol-chloroform extraction [42]. Four µg of genomic DNA was digested with EcoRV enzyme and separated on a 0.8% agarose/0.5xTris-acetate-EDTA gel by electrophoresis. DNA was transferred onto a Hybond N+ filter (Amersham) in 10x saline sodium citrate (SSC) solution. The filter was washed in 2X SSC and baked at 80°C for 2 hours. Prehybridization of the filter was done for 2 hrs at 60°C in 5x SCC, 5x Denhart and 100 mg/ml of yeast tRNA. Probes specific for the group II intron (EBS2-tcdC623as/Sal-R1), ermB gene (oWKS1131/oWKS1132) and tcdC gene (tcdC5-tcdC6) were generated. Primers are listed in Table 2. The generated probes (100 ng) were radiolabeled (32P dATP) using Klenow enzyme (Roche) and overnight hybridized in 10 ml fresh pre-hybridization buffer at 60°C. The filter was washed for 30 min in 2x SCC, 0.5% SDS, 30 min in 1X SSC, 0.5% SDS and 30 min in 0.5X SSC, 0.5% SDS and analyzed using phosphorimage screen and a Typhoon 9410 scanner (GE healthcare).
Western Blots
Antibodies against TcdC were generated by immunizing rabbits with a synthetic peptide (CQLARTPDDYKYKKV) representing a specific TcdC epitope (Genscript). Note that this epitope is located before the Clostron insertion site, and would therefore also be expected to detect truncated TcdC protein, would this be produced. Western blots were performed as follows. C. difficile (2 mL) cultures were harvested by centrifugation (2 min, 11.000×g, 4°C) and washed with Phosphate Buffered Saline (PBS). The bacterial pellets were resuspended in PBS containing protease inhibitor cocktail (Complete, Roche) and lysed by sonification. The bacterial lysates were centrifuged at low speed (3 min, 1000×g, 4°C) to remove unbroken bacterial cells [20]. To separate the cytosolic proteins from the membrane associated proteins the bacterial supernatant was centrifuged at 200.000×g, 4°C for 1 hr [20]. The pelleted membrane associated proteins were resuspended in 10 mM Tris-HCl (pH 7.4), 5 mM EDTA with 2% Triton X-100 for 30 min at room temperature. Equal amounts of the resuspended membrane associated proteins were separated on 15% SDS PAGE gel and transferred onto polyvinyl difluoride (PVDF) membranes. Similarly generated membranes with the transferred membrane associated proteins of a Type 035 (PaLoc negative) strain were used for pre incubation of the TcdC antibodies. The membranes were probed with the pre incubated TcdC antibody and an antibody against the β subunit of the E. coli F0F1 ATPase that cross reacts with the homologous protein in C. difficile [20], [43]. The probed membranes were analyzed using secondary anti-mouse horse radish peroxidase conjugated antibodies (Dako), a chemiluminescence detection kit (Amersham) and a Typhoon 9410 scanner (GE Healthcare).
RNA Extraction
Five mL of the C. difficile cultures were 1∶1 diluted with ice cold methanol and stored overnight at −80°C. Bacterial pellets, obtained by centrifugation (20 min, 3000×g, 4°C), were resuspended into 200 µl lysisbuffer (100 mM EDTA, 200 mM Tris-HCl pH 7.0, 50 mg/ml lysozyme) and incubated for 1 hr at 37°C. Tri-pure reagent (Roche) was used for the extraction of RNA according to the manufacturer’s instruction with minor modifications. Briefly, 1 ml Tri-pure was added to the lysed bacterial pellets and incubated for 5 min at room temperature. Per 1 ml Tri-pure, 200 µl chloroform was added and carefully shaken by hand for 3 min, followed by an incubation of 2–5 min at room temperature. The aqueous phase was collected after centrifugation (12,000×g for 15 min at 4°C) and transferred to a fresh tube. RNA was precipitated by mixing the aqueous phase with 500 µl isopropanol, followed by an incubation of 10 min at room temperature. The precipitated RNA was collected by centrifugation (12,000×g, 10 min, 4°C) and resuspended in 100 µl DNase/RNase free water. The RNA was re-precipitated overnight at −80°C with ammonium acetate (Fluka; 10 mM) and 3 volumes of absolute ethanol. The re-precipitated RNA was washed once with 80% ethanol and dissolved in 50 µl DNase/RNase free water. The RNA was treated twice with a TurboDNase (Ambion) according to the manufacturer’s instruction followed by another Tri-pure RNA isolation. The quality and purity of the extracted RNA was assessed using a RNA nano chip on an Agilent Bioanalyzer.
Transcriptional Analysis of the PaLoc Genes
A RevertAid™ H Minus Reverse Transcriptase kit (Fermentas) was used to synthesize cDNA according to the manufacturer’s instruction. Random hexamers were used to convert 750 ng RNA into cDNA. The synthesized cDNA was treated with RNase (Qiagen) for 1 hour at 37°C and stored at −20°C. The software program Molecular Beacon (Premier Biosoft) was used to design primer pairs and probes (Table 2) for the 2 multiplex quantitative PCRs (qPCR), based on the available genome of C. difficile strain 630 [35]. All primer pairs were first tested by conventional PCR and multiplex PCR to confirm specificity and amplicon sizes. The primer pair and the probe for the amplification of the tcdC gene are in front of the insertion site in the tcdC gene (Figure 1A), allowing detection of tcdC transcription levels in wild type and CT::tcdC strains. The real-time multiplex qPCR amplification of the PaLoc genes and the reference gene encoding for a ribosomal protein (rpsJ) was performed on a CFX96 real-time PCR detection system (Biorad) [31]. The amplification efficiencies of the PaLoc and reference genes were determined using serially diluted genomic DNA (standard curve). The manually calculated efficiencies and the reference gene rpsJ were used to normalize the expression levels of the PaLoc genes. The amplification was performed in a 25 µl final volume. The first real-time multiplex qPCR (target genes: tcdA, tcdA and tcdC) contained 25 µl Hotstar mastermix (Qiagen), forward and reverse primers (80 nm each primer), 2.5 mM MgCl2, 100 nM of each probe and 2 µl synthesized cDNA. The second multiplex real-time multiplex Q-PCR (target genes: tcdR, tcdE) contained 25 µl Hotstar mastermix (Qiagen), forward and reverse primers (80 nm each primer), 3.5 mM MgCl2, 100 nM of each probe and 2 µl synthesized cDNA. The real-time qPCR to quantify the reference gene rpsJ contained 25 µl Hotstar mastermix (Qiagen), forward and reverse primers (80 nm each primer), 2.5 mM MgCl2, 0.06% SYBRgreen (Sigma) and 2 µl synthesized cDNA. The real-time qPCR protocol included an enzyme activation step for 15 min at 95°C, followed by 50 cycles of amplification; 95°C for 30 sec, 52°C for 30 sec and 72°C for 30 sec.
Relative Quantification of Toxin Expression
Total toxin amounts were quantified using 2 assays; a toxin end point titer assay and a commercial available ELISA (Ridascreen, Biopharma). The supernatants of culture samples (1 mL) were collected after centrifugation (30 min, 3000×g, 4°C), filter sterilized (0.45 µM cellulose acetate membrane) and stored at 4°C.
For the toxin end point titer assay, Vero cells were seeded into a 96 wells plate at a density of 1×104 cells per well and incubated overnight at 37°C and 5% CO2. The filter sterilized supernatants of 5, 8, 12, 24 and 48 hours post inoculation (hpi) were diluted 2, 101, 102, 103, 104 and 105 fold in cell culture medium (Dulbecco modified Eagle medium (Lonza) supplemented with penicillin 100 u/mL, streptomycin 100 U/mL, fetal calf serum(10%). Fifty µl of the dilutions were added onto the Vero cell monolayers and incubated for 1 hr at 37°C and 5% CO2. For the neutralization assay a 2-fold dilution of each tested time point (5, 8, 12, 24 and 48 hpi) was pre-incubated with a 1/100 diluted anti-toxin (Techlab) for 1 hr at 37°C and 5% CO2. After the pre-incubation, 50 µl was added onto the Vero cell monolayers. The incubated bacterial supernatants were aspirated off after one hour and replaced with 100 µl cell culture medium. After 24 hrs of incubation the end-point titer was determined of each diluted time point [26]. The end-point titer was defined as the first dilution at which the Vero Cell morphology was indistinguishable from the neutralized 2-fold diluted supernatants [26]. The enzyme immunoassay (Ridascreen, Biopharma) was performed according manufacture’s protocol.
Statistical Analysis
Statistical analysis was performed using the software package SPSS 18 (IBM). An independent sample t-test was employed to compare the strains at different time points.
Acknowledgments
We thank N. Minton and S. Cartman for providing the ClosTron system and for helpful ClosTron related discussions. We thank G. Deckers-Hebestreit for kindly providing the anti-ATPase β subunit and R. Govind for advice on anti-TcdC Western blots.
Funding Statement
W.K. Smits was supported by Veni grant 016.116.043 from The Netherlands Organization for Health, Research and Development (NWO-ZOnMW) and a Gisela Thier fellowship (LUMC). This study was supported by the European Union (HYPERDIFF-The Physiological Basis of Hypervirulence in Clostridium difficile: a Prerequisite for Effective Infection Control; Health-F3-2008-223585). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Goorhuis A, van der Kooi T, Vaessen N, Dekker FW, van den Berg R, et al. (2007) Spread and epidemiology of Clostridium difficile polymerase chain reaction ribotype 027/toxinotype III in The Netherlands. Clin Infect Dis 45: 695–703. [DOI] [PubMed] [Google Scholar]
- 2. Kuijper EJ, Coignard B, Tull P (2006) Emergence of Clostridium difficile-associated disease in North America and Europe. Clin Microbiol Infect 12 Suppl 62–18. [DOI] [PubMed] [Google Scholar]
- 3. Loo VG, Poirier L, Miller MA, Oughton M, Libman MD, et al. (2005) A predominantly clonal multi-institutional outbreak of Clostridium difficile-associated diarrhea with high morbidity and mortality. N Engl J Med 353: 2442–2449. [DOI] [PubMed] [Google Scholar]
- 4.Kuijper EJ, Barbut F, Brazier JS, Kleinkauf N, Eckmanns T, et al. (2008) Update of Clostridium difficile infection due to PCR ribotype 027 in Europe, 2008. Euro Surveill 13. [PubMed]
- 5. Paltansing S, van den Berg RJ, Guseinova RA, Visser CE, van der Vorm ER, et al. (2007) Characteristics and incidence of Clostridium difficile-associated disease in The Netherlands, 2005. Clin Microbiol Infect 13: 1058–1064. [DOI] [PubMed] [Google Scholar]
- 6. Pepin J, Valiquette L, Alary ME, Villemure P, Pelletier A, et al. (2004) Clostridium difficile-associated diarrhea in a region of Quebec from 1991 to 2003: a changing pattern of disease severity. CMAJ 171: 466–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bauer MP, Notermans DW, van Benthem BH, Brazier JS, Wilcox MH, et al. (2011) Clostridium difficile infection in Europe: a hospital-based survey. Lancet 377: 63–73. [DOI] [PubMed] [Google Scholar]
- 8. Goorhuis A, Bakker D, Corver J, Debast SB, Harmanus C, et al. (2008) Emergence of Clostridium difficile infection due to a new hypervirulent strain, polymerase chain reaction ribotype 078. Clin Infect Dis 47: 1162–1170. [DOI] [PubMed] [Google Scholar]
- 9.Wilcox MH (2009) Clostridium difficile Ribotyping Network for England and Northern Ireland.
- 10. Just I, Gerhard R (2004) Large clostridial cytotoxins. Rev Physiol Biochem Pharmacol 152: 23–47. [DOI] [PubMed] [Google Scholar]
- 11. Mani N, Dupuy B (2001) Regulation of toxin synthesis in Clostridium difficile by an alternative RNA polymerase sigma factor. Proc Natl Acad Sci U S A 98: 5844–5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Matamouros S, England P, Dupuy B (2007) Clostridium difficile toxin expression is inhibited by the novel regulator TcdC. Mol Microbiol 64: 1274–1288. [DOI] [PubMed] [Google Scholar]
- 13. Voth DE, Ballard JD (2005) Clostridium difficile toxins: mechanism of action and role in disease. Clin Microbiol Rev 18: 247–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tan KS, Wee BY, Song KP (2001) Evidence for holin function of tcdE gene in the pathogenicity of Clostridium difficile . J Med Microbiol 50: 613–619. [DOI] [PubMed] [Google Scholar]
- 15. Curry SR, Marsh JW, Muto CA, O’Leary MM, Pasculle AW, et al. (2007) tcdC genotypes associated with severe TcdC truncation in an epidemic clone and other strains of Clostridium difficile . J Clin Microbiol 45: 215–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hundsberger T, Braun V, Weidmann M, Leukel P, Sauerborn M, et al. (1997) Transcription analysis of the genes tcdA-E of the pathogenicity locus of Clostridium difficile . Eur J Biochem 244: 735–742. [DOI] [PubMed] [Google Scholar]
- 17. Dupuy B, Sonenshein AL (1998) Regulated transcription of Clostridium difficile toxin genes. Mol Microbiol 27: 107–120. [DOI] [PubMed] [Google Scholar]
- 18. Merrigan M, Venugopal A, Mallozzi M, Roxas B, Viswanathan VK, et al. (2010) Human hypervirulent Clostridium difficile strains exhibit increased sporulation as well as robust toxin production. J Bacteriol 192: 4904–4911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Vohra P, Poxton IR (2011) Comparison of toxin and spore production in clinically relevant strains of Clostridium difficile . Microbiology 157: 1343–1353. [DOI] [PubMed] [Google Scholar]
- 20. Govind R, Vediyappan G, Rolfe RD, Fralick JA (2006) Evidence that Clostridium difficile TcdC is a membrane-associated protein. J Bacteriol 188: 3716–3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mani N, Lyras D, Barroso L, Howarth P, Wilkins T, et al. (2002) Environmental response and autoregulation of Clostridium difficile TxeR, a sigma factor for toxin gene expression. J Bacteriol 184: 5971–5978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Warny M, Pepin J, Fang A, Killgore G, Thompson A, et al. (2005) Toxin production by an emerging strain of Clostridium difficile associated with outbreaks of severe disease in North America and Europe. Lancet 366: 1079–1084. [DOI] [PubMed] [Google Scholar]
- 23. Karlsson S, Lindberg A, Norin E, Burman LG, Akerlund T (2000) Toxins, butyric acid, and other short-chain fatty acids are coordinately expressed and down-regulated by cysteine in Clostridium difficile . Infect Immun 68: 5881–5888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Heap JT, Pennington OJ, Cartman ST, Carter GP, Minton NP (2007) The ClosTron: a universal gene knock-out system for the genus Clostridium . J Microbiol Methods 70: 452–464. [DOI] [PubMed] [Google Scholar]
- 25. Heap JT, Kuehne SA, Ehsaan M, Cartman ST, Cooksley CM, et al. (2010) The ClosTron: Mutagenesis in Clostridium refined and streamlined. J Microbiol Methods 80: 49–55. [DOI] [PubMed] [Google Scholar]
- 26. Kuehne SA, Cartman ST, Heap JT, Kelly ML, Cockayne A, et al. (2010) The role of toxin A and toxin B in Clostridium difficile infection. Nature 467: 711–713. [DOI] [PubMed] [Google Scholar]
- 27. Snijder EJ, Bredenbeek PJ, Dobbe JC, Thiel V, Ziebuhr J, et al. (2003) Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J Mol Biol 331: 991–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. O’Connor JR, Johnson S, Gerding DN (2009) Clostridium difficile infection caused by the epidemic BI/NAP1/027 strain. Gastroenterology 136: 1913–1924. [DOI] [PubMed] [Google Scholar]
- 29. Lyerly DM, Sullivan NM, Wilkins TD (1983) Enzyme-linked immunosorbent assay for Clostridium difficile toxin A. J Clin Microbiol. 17: 72–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Carter GP, Douce GR, Govind R, Howarth PM, Mackin KE, et al. (2011) The Anti-Sigma Factor TcdC Modulates Hypervirulence in an Epidemic BI/NAP1/027 Clinical Isolate of Clostridium difficile . PLoS Pathog 7: e1002317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Metcalf D, Sharif S, Weese JS (2010) Evaluation of candidate reference genes in Clostridium difficile for gene expression normalization. Anaerobe 16: 439–443. [DOI] [PubMed] [Google Scholar]
- 32. Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Graeber K, Linkies A, Wood AT, Leubner-Metzger G (2011) A guideline to family-wide comparative state-of-the-art quantitative RT-PCR analysis exemplified with a brassicaceae cross-species seed germination case study. Plant Cell 23: 2045–2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hussain HA, Roberts AP, Mullany P (2005) Generation of an erythromycin-sensitive derivative of Clostridium difficile strain 630 (630Deltaerm) and demonstration that the conjugative transposon Tn916DeltaE enters the genome of this strain at multiple sites. J Med Microbiol 54: 137–141. [DOI] [PubMed] [Google Scholar]
- 35. Sebaihia M, Wren BW, Mullany P, Fairweather NF, Minton N, et al. (2006) The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat Genet 38: 779–786. [DOI] [PubMed] [Google Scholar]
- 36. Ho TD, Ellermeier CD (2011) PrsW is required for colonization, resistance to antimicrobial peptides, and expression of extracytoplasmic function sigma factors in Clostridium difficile . Infect Immun 79: 3229–3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lyras D, O’Connor JR, Howarth PM, Sambol SP, Carter GP, et al. (2009) Toxin B is essential for virulence of Clostridium difficile . Nature 458: 1176–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cartman ST, Kelly ML, Heeg D, Heap JT, Minton NP (2012) Precise Manipulation of the Clostridium difficile Chromosome Reveals a Lack of Association between the tcdC Genotype and Toxin Production. Appl Environ Microbiol 78: 4683–4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Murray R, Boyd D, Levett PN, Mulvey MR, Alfa MJ (2009) Truncation in the tcdC region of the Clostridium difficile PathLoc of clinical isolates does not predict increased biological activity of Toxin B or Toxin A. BMC Infect Dis. 9: 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Garnier T, Cole ST (1986) Characterization of a bacteriocinogenic plasmid from Clostridium perfringens and molecular genetic analysis of the bacteriocin-encoding gene. J Bacteriol 168: 1189–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Purdy D, O’Keeffe TA, Elmore M, Herbert M, McLeod A, et al. (2002) Conjugative transfer of clostridial shuttle vectors from Escherichia coli to Clostridium difficile through circumvention of the restriction barrier. Mol Microbiol 46: 439–452. [DOI] [PubMed] [Google Scholar]
- 42. Wren BW, Tabaqchali S (1987) Restriction endonuclease DNA analysis of Clostridium difficile . J Clin Microbiol 25: 2402–2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Deckers-Hebestreit G, Altendorf K (1986) Accessibility of F0 subunits from Escherichia coli ATP synthase. A study with subunit specific antisera. Eur J Biochem 161: 225–231. [DOI] [PubMed] [Google Scholar]
- 44. Knetsch CW, Terveer EM, Lauber C, Gorbalenya AE, Harmanus C, et al. (2012) Comparative analysis of an expanded Clostridium difficile reference strain collection reveals genetic diversity and evolution through six lineages. Infect Genet Evol. 15 12(7): 1577–1585. [DOI] [PubMed] [Google Scholar]
- 45. Dawson LF, Donahue EH, Cartman ST, Barton RH, Bundy J, et al. (2011) The analysis of para-cresol production and tolerance in Clostridium difficile 027 and 012 strains. BMC Microbiol 11 86 86. [DOI] [PMC free article] [PubMed] [Google Scholar]