Abstract
Arabidopsis thaliana EARLY FLOWERING 3 (ELF3) as a zeitnehmer (time taker) is responsible for generation of circadian rhythm and regulation of photoperiodic flowering. There are two orthologs (OsELF3-1 and OsELF3-2) of ELF3 in rice (Oryza sativa), but their roles have not yet been fully identified. Here, we performed a functional characterization of OsELF3-1 and revealed it plays a more predominant role than OsELF3-2 in rice heading. Our results suggest OsELF3-1 can affect rice circadian systems via positive regulation of OsLHY expression and negative regulation of OsPRR1, OsPRR37, OsPRR73 and OsPRR95 expression. In addition, OsELF3-1 is involved in blue light signaling by activating EARLY HEADING DATE 1 (Ehd1) expression to promote rice flowering under short-day (SD) conditions. Moreover, OsELF3-1 suppresses a flowering repressor GRAIN NUMBER, PLANT HEIGHT AND HEADING DATE 7 (Ghd7) to indirectly accelerate flowering under long-day (LD) conditions. Taken together, our results indicate OsELF3-1 is essential for circadian regulation and photoperiodic flowering in rice.
Introduction
Flowering, referred to as heading date in rice (Oryza sativa), is an important agronomical trait for rice to adapt to specific cropping environments. Determined by photoreceptors, circadian clock systems and floral integrator genes, photoperiodic regulation is a key means in controlling plant flowering [1], [2], [3]. A sophisticated signaling network of photoperiodic flowering has been revealed initially based on the identification of key functional genes in Arabidopsis thaliana [4]. In the evolutionarily conserved GIGANTEA(GI)-CONSTANS(CO)-FLOWERING LOCUS T (FT) signaling pathways [3], clock-associated protein GI positively regulates CO expression under long-day conditions (LD) [5]. CO encodes a zinc-finger type transcriptional activator and promotes flowering under LD through up-regulation of its downstream target FT. FT as a floral activator controls flowering time by integrating input signals of various pathways [6], [7]. Interestingly, rice shares the similar pathway with Arabidopsis thaliana in photoperiodic flowering [8], [9]. Under short-day (SD) conditions, OsGI is an activator of Heading date 1 (Hd1), the rice ortholog of Arabidopsis CO [10]; Hd1 promotes flowering by activating Heading date 3α (Hd3α), the rice ortholog of FT [11], [12], [13]. Besides, RICE FLOWERING LOCUS T 1 (RFT1/FT-L3) is extensively homologous to Hd3α, and these two homologs function redundantly in the flowering [14], [15].
Recently, many studies have indicated rice possesses a distinct flowering signaling machinery from Arabidopsis. There are some rice specific genes that have no orthologs in Arabidopsis, such as EARLY HEADING DATE 1 (Ehd1), Ehd2, Ehd3 and GRAIN NUMBER, PLANT HEIGHT AND HEADING DATE 7 (Ghd7), play significant roles in photoperiodic flowering. Ehd1 is a B-type response regulator and positively regulates Hd3α and RFT independently of Hd1 [16]. Ghd7 represses Ehd1 expression to delay flowering under LD [17], [18]. Ehd2 is an ortholog of maize (Zea mays) INDETERMINATE 1 (ID1), and encodes a transcription factor for the upregulation of Ehd1 [19]. Ehd3 positively regulates Ghd7 to repress heading under LD. On the contrary, Ehd3 is independent of Ghd7 and positively regulates Ehd1 to promote flowering under SD [20]. Additionally, some MADS-box genes are crucial to rice photoperiodic flowering. OsMADS50, an ortholog of Arabidopsis SUPPRESSOR OF OVEREXPRSSION OF CONSTANS1 (SOC1), acts in a dramatically distinct manner from SOC1. SOC1 accumulates abundant mRNA and protein in the apical meristem and stimulates floral organ development downstream of FT [21], [22], [23]. However, OsMADS50, with a low expression level in the apical meristem, functions upstream of Ehd1 to indirectly induce RFT1 transcription under LD [24]. OsMADS56, another SOC1 ortholog, attenuates Ehd1 expression during LD [25]. OsMADS51, a rice-specific gene, also can positively regulate Ehd1 under SD [25].
In Arabidopsis, EARLY FLOWERING 3 (ELF3) is a zeitnehmer (time taker) to modulate resetting of the circadian clock and integrate temperature and photoperiod [26], [27], [28], [29], [30], [31]. Structurally, ELF3 exhibits no similarity to known functional factors. The C-terminal domain of ELF3 is required for its nuclear localization, while the N-terminus mediates the physical interaction of ELF3 with phytochrome B (phyB), a red light photoreceptor, and an E3 ubiquitin ligase CONSTITUTIVE PHOTOMORPHOGENIC1 (COP1) [32], [33], [34]. The ELF3-COP1 interaction enables ELF3 to associate with GI, and to further mediate GI degradation [34]. The central domain of ELF3 is necessary and sufficient to bridge EARLY FLOWERING 4 (ELF4) and a single-MYB domain-containing and SHAQYF-type GARP transcription factor LUX ARRHYTHMO (LUX) [35]. Mediated by LUX, the ELF4-ELF3-LUX complex directly binds to the promoters of PHYTOCHROME INTERACTING FACTOR 4 (PIF4) and PIF5 to regulate hypocotyl growth in the early evening, and the PSEUDO-RESPONSE REGULATOR 9 (PRR9) promoter to sustain plant circadian rhythms [33], [36]. In rice, there are at least two orthologs of Arabidopsis ELF3 [37], named as OsELF3-1 (LOC_Os06g05060) and OsELF3-2 (LOC_Os01g38530) hereafter. Late flowering was found in two mutants (NIL-Hd17 and ef7) of OsELF3-1 [38], [39] and a mutant (osef3) of OsELF3-2 [40]. However, the roles of OsELF3-1 and OsELF3-2 have not yet been fully explored. In this study, we performed a functional characterization of OsELF3-1 and found it plays a predominant role over OsELF3-2 in rice heading. Our results suggest OsELF3-1 affects rice circadian system via positive regulation of OsLHY and negative regulation on OsPRR1, OsPRR37, OsPRR73 and OsPRR95. In addition, OsELF3-1 is involved in blue light-induced activation of Ehd1 expression to promote flowering under SD. Moreover, OsELF3-1 suppresses a flowering repressor Ghd7 to indirectly accelerate flowering under LD conditions. Taken together, our results reveal OsELF3-1 is essential for circadian rhythm regulation and photoperiodic flowering in rice.
Results
The oself3-1 mutant shows late heading
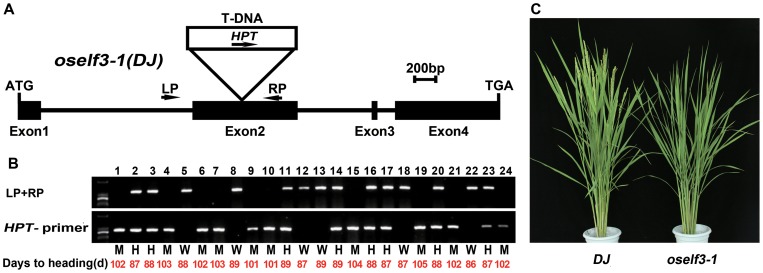
There are two putative orthologs of Arabidopsis ELF3, OsELF3-1 and OsELF3-2, in the rice genome [37], [38], [39], [40]. OsELF3-1 and OsELF3-2 respectively share 35% and 30% identity with ELF3 in their amino acid sequences (Fig. S1). To investigate whether these two genes are involved in rice flowering, we examined the heading dates of oself3-1 and oself3-2 mutant in the experimental field of Sichuan province (30°67′) (summer) and Hainan province (18°15′) (winter) in China. A natural long-day condition (day length >14h) exists in Sichuan province in the summer, while a natural short-day condition (day length <10h) exists in Hainan province in winter. As shown in Fig. S2, two T-DNA insertion lines of oself3-2 in Zhonghua11 (ZH11) background and Dongjin (DJ) background both harbor a T-DNA fragment in the 4th exon of OsELF3-2. These two lines did not show significantly different heading dates in either Sichuan or Hainan from wild-type plants (Table S1). However, oself3-1 in DJ background with a T-DNA inserted into the 2nd exon of OsELF3-1 (Fig. 1A) delayed flowering 14 days in Sichuan (LD) (Fig. 1C) and 38 days in Hainan (SD) compared with DJ (Table 1). To confirm this phenotype of oself3-1 is caused by the T-DNA insertion, we analyzed the co-segregation between the late heading defect and the existence of the T-DNA insertion. In 120 plants derived from the T2 T-DNA inserted population, two DNA markers were used to identify HPT and T-DNA insertion respectively. The correspondence between flowering phenotype and DNA markers suggested it is the T-DNA insertion that leads to a late flowering mutation (Fig. 1B).
Figure 1. Schematic diagram of OsELF3-1 gene structure, the T-DNA insertion site and oself3-1 phenotype.
(A) Schematic diagram of OsELF3-1 gene structure and the T-DNA insertion site. Filled boxes indicate exons and solid lines indicate introns. Arrows indicate the primers used for analyzing the insertion site. LP and RP respectively represent the primers around the left and right borders of the T-DNA. (B) PCR genotyping of OsELF3-1 segregants. W, wild type; M, homozygous; H, heterozygous. (C) Phenotypes of wild type and oself3-1. Photograph was taken when WT plant (Dongjin) flowered.
Table 1. Days to heading in oself3-1 and OsELF3-1 RNAi mutants.
| Genotypes | Days to Heading under Wenjiang | Days to Heading under Hainan |
| WT(DJ) | 87±0.9 | 65±1.5 |
| oself3-1WT(Ni)OsELF3-1 RNAi Line1OsELF3-1 RNAi Line2OsELF3-1 RNAi Line3 | 101±1.594±2.0139±1.0143±1.6144±1.2 | 103±1.875±2.8137±1.3146±2.7149±1.6 |
Days to heading was calculated when the first panicle appeared. WT means wild type.
OsELF3-1 RNAi plants flower late
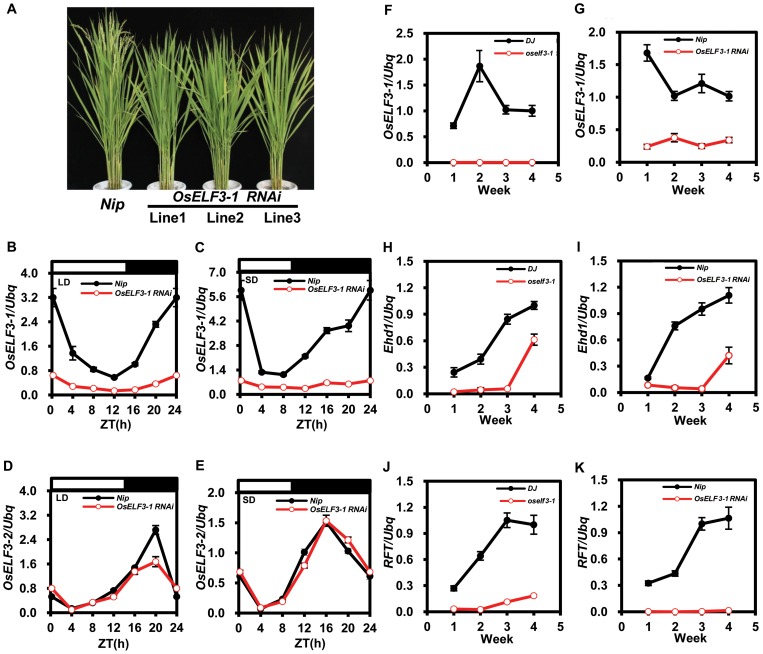
To further confirm the role of OsELF3-1 in heading date, we constructed OsELF3-1 RNAi lines in Nipponbare (Nip) background. All the homozygous and stable T4 progenies of three independent transgenic lines consistently showed delayed flowering compared with Nip in both Sichuan and Hainan provinces (Table 1 and Fig. 2A). OsELF3-1 RNAi Line 1 was employed in our further experiments. Next, gene expression during a 24-hour period of OsELF3-1 and OsELF3-2 was examined in OsELF3-1 RNAi and Nip under SD and LD. We found in OsELF3-1 RNAi, OsELF3-1 instead of OsELF3-2 was constitutively repressed regardless of the conditions (Fig. 2B–E). Then we analyzed the OsELF3-1 expression in Nip and OsELF3-1 RNAi using 60-day-old plants grown in an experimental field in Sichuan (LD) for another 4 weeks. Quantitative RT-PCR (qRT-PCR) results showed the OsELF3-1 mRNA level was continuously suppressed in OsELF3-1 RNAi (Fig. 2F, G). Therefore, the late flowering phenotype in OsELF3-1 RNAi resulted from OsELF3-1 gene repression.
Figure 2. Phenotypes of OsELF3-1 RNAi lines and expression of flowering-related genes in OsELF3-1 RNAi mutant.
(A) Flowering phenotypes of three independent OsELF3-1 RNAi lines. Photograph was taken when WT plant (Nip) flowered. (B–E) Diurnal expression of OsELF3-1 and OsELF3-2 in Nip and OsELF3-1 RNAi mutant under LD and SD. Rice leaves were harvested from 30-day old plants grown in the chambers at the indicated time points. White bars, light; black bars, darkness. ZT means Zeitgeber time. Values are means ± SD. (F–K) Expression of flowering-related genes in OsELF3-1 RNAi mutant. Rice leaves were harvested once a week at exactly the same time point for four weeks until the wild-type plants flowered. Values are means ± SD.
In Arabidopsis, elf3 mutants flower much earlier than wild-type plants under LD and SD conditions [26]. ELF3 indirectly represses FT expression through positively regulating degradation of GI under SD [34]. In rice, it was noted that RFT integrates all flowering signals and induces the development of floral organs under LD [3], so we next asked whether RFT and its regulator Ehd1 are affected by OsELF3-1. Rice leaves were harvested once a week at exactly the same time point for four weeks until the wild-type plants flowered. We observed that the transcription of Ehd1 maintained at a lower level in oself3-1 and OsELF3-1 RNAi mutants than that in their wild-type counterparts (Fig. 2H, I). There was a notable increase in RFT expression in wild-type plants, whereas RFT expression was severely attenuated in oself3-1 and OsELF3-1 RNAi mutants (Fig. 2J, K). Taken together, our results suggest that loss of OsELF3-1 function leads to delayed flowering via repressing Ehd1 and RFT under LD.
OsELF3-1 affects the expression of circadian clock components
Earlier studies demonstrated that florigen is the hypothetical floral signal that is produced in leaves [41]. Gene products of florigen, mature FT and Hd3α proteins, specifically translocate to shoot apical meristem (SAM) to induce flowering [13], [42]. We are also interested whether OsELF3-1 exhibits organ-specific expression in rice. Using rice grown under natural LD at the booting stage, qRT-PCR analyses were performed with total RNA isolated from roots, stems, leaves, leaf sheaths and young panicles (Fig. S3). The transcript levels of OsELF3-1 were highest in leaves and leaf sheaths tissues while lowest in roots and stems. OsELF3-1 shares the similar tissue-specific expression profile with Hd3α and RFT in leaves and leaf sheaths, indicating OsELF3-1 may play a vital role in flowering regulation. Previous reports have shown that Arabidopsis ELF3 expression varies in the time span of one day, with a trough in the early day and a peak in the early night. Therefore ELF3 was described as an evening-phased gene [43]. Intriguingly, OsELF3-1 mRNA showed a peak at dawn (ZT0) under LD and SD (Fig. 2B, C). Accordingly, OsELF3-1 is a morning-phased gene, suggesting a distinct role of OsELF3-1 from Arabidopsis ELF3.
The core circadian clock components, which are connected by positive–negative feedback loops, form a sophisticated network in Arabidopsis [4], [44]. The central loop identified first contains two morning-phased transcription factors CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY), and an evening-phased gene PSEUDO-RESPONSE REGULATOR 1 (PRR1 or TOC1) [4]. In the morning, CCA1 and LHY, the transcriptional repressors, can associate with the promoter of TOC1 and negatively regulate TOC1 transcription [45], [46], [47], [48], [49]. TOC1 directly represses the expression of CCA1 and LHY, through binding to the promoters of CCA1 and LHY at night [50], [51]. ELF3 physically interacts with a transcription factor LUX to associate with the PRR9 promoter and represses the transcription of PRR9 [36], [52], [53]. Because PRR9 is a repressor of CCA1 and LHY expression, ELF3 enables itself to indirectly activate CCA1 and LHY [52]. It has been well documented that OsLHY and OsPRR1 have similar circadian expression profiles with their Arabidopsis orthologs CCA1/LHY and TOC1 respectively [37]. We next studied the effects of OsELF3-1 on the expression of clock-associated genes.
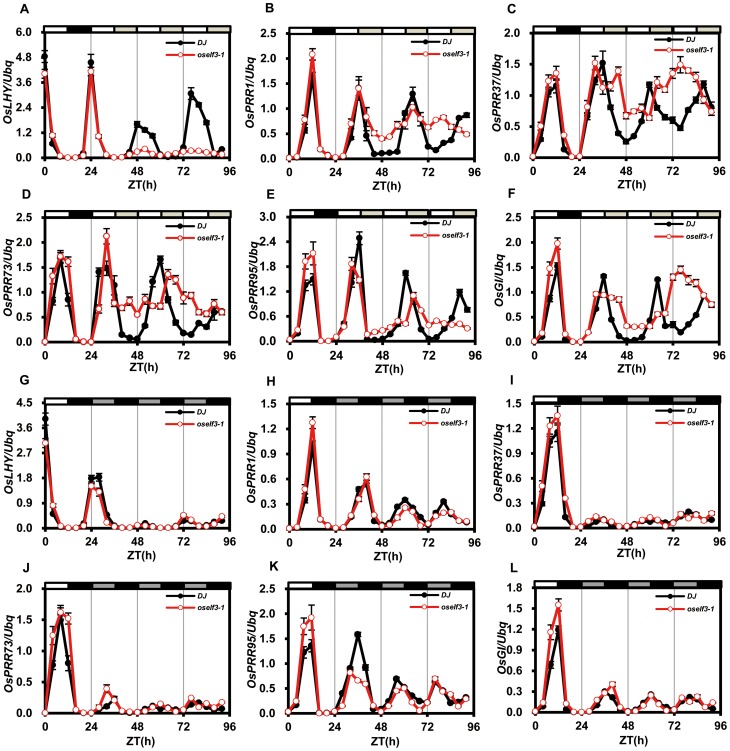
In oself3-1 mutant, we found that the maintenance of the daily expression oscillation of OsLHY and OsPRR1 was disrupted under continuous light (LL) rather than continuous darkness (DD) (Fig. 3A, B, G and H). Previously, Murakami et al. identified the rice orthologs of Arabidopsis PRR3/PRR7 and PRR5/PRR9. Since it is difficult to distinguish the exact counterparts in rice, they were designated as OsPRR37 and OsPRR73, and OsPRR59 and OsPRR95 [37], [54]. We then asked whether OsPRR37, OsPRR73, OsPRR59 and OsPRR95 transcription are regulated by OsELF3-1. The altered circadian profiles were observed in the expression of OsPRR37, OsPRR73, OsPRR95 and OsGI (Fig. 3C–F and I–L) under LL rather than DD. The similar defects in the circadian rhythm were also shown in OsELF3-1 RNAi (Fig. S4).
Figure 3. Circadian expression of clock-associated genes in oself3-1.
Circadian expression of OsLHY, OsPRR1, OsPRR37, OsPRR73, OsPRR95 and OsGI in DJ (filled circles) and oself3-1 (open circles) under LL and DD. The plants were first grown to cycles of 12-hour light/12-hour darkness for 30 days at 26°C and then transferred to the continuous light (LL) or continuous darkness (DD) at dawn. In A–F panels, white bars, light; black, darkness; gray, subjective light. In G–L panels, white bars, light; black, darkness; gray, subjective darkness. ZT means Zeitgeber time. Values are means ± SD.
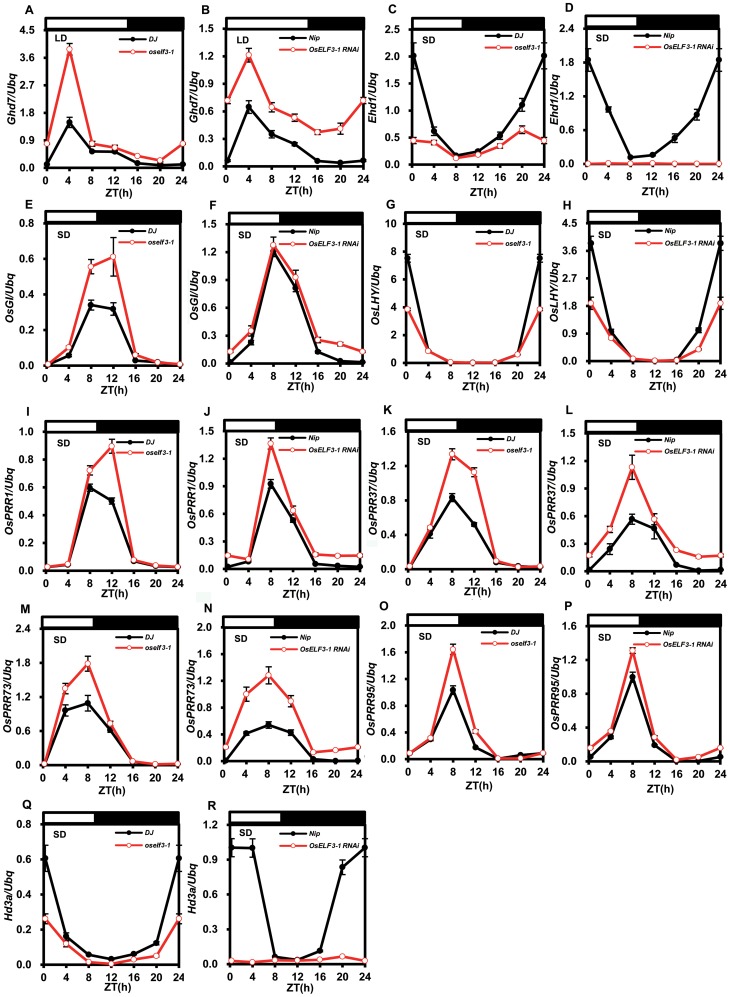
In addition, we found that the mutation of OsELF3 led to the abnormal diurnal expression of these genes. OsLHY and OsPRR1 transcription peaks appeared at dawn and dusk, respectively, in wild-type plants under SD (Fig. 4G–J). However, the expression altitude of OsLHY showed a significant reduction in oself3-1 and OsELF3-1 RNAi under SD (Fig. 4G, H). OsPRR1 transcrption level was higher at dusk in these mutants (Fig. 4I, J). Therefore, we suggested OsELF3-1 can positively regulate OsLHY expression and negatively regulate OsPRR1 expression. Meanwhile, OsPRR37, OsPRR73 and OsPRR95 showed higher expression peaks at dusk under SD in oself3-1 and OsELF3-1 RNAi mutants than that in wild-type plants (Fig. 4K–P), suggesting OsELF3-1 negatively regulates OsPRR37, OsPRR73 and OsPRR95.
Figure 4. Diurnal expression of clock-associated genes in oself3-1 and OsELF3-1 RNAi mutants.
Diurnal expression patterns of Ghd7, Ehd1, OsGI, OsLHY, OsPRR1, OsPRR37, OsPRR73, OsPRR95 and Hd3α in DJ (filled circles) and oself3-1 (open circles), and in Nip (filled circles) and OsELF3-1 RNAi (open circles) plants. Rice leaves were harvested from 30-day old plants grown in the chambers at the indicated time points. White bars, light; black bars, darkness. ZT means Zeitgeber time. Values are means ± SD.
OsELF3-1 activates flowering through a rice photoperiodic flowering pathway
It has been well documented that the mutants of core circadian clock genes in Arabidopsis, such as cca1 and lhy, suffer from severe flowering defects [46], [47]. In contrast, there is no evidence showing mutants of core circadian clock genes in rice exhibit abnormal flowering phenotypes. It is still unknown how oself3-1 and OsELF3-1 RNAi mutants delay flowering. Therefore, we analyzed whether OsELF3-1 affects Ghd7, Ehd1 and Hd1 at the transcriptional level.
Xue et al. suggested Ghd7 is a flowering repressor to negatively regulate Ehd1 under LD [17]. Ehd1, an important flowering activator, promotes RFT1 and Hd3α transcription under LD and SD respectively. The mRNA level of Ehd1 peaks at dawn [3], [16]. We observed that Ghd7 in oself3-1 and OsELF3-1 RNAi mutants accumulates more transcripts than that in wild-type plants in the subject day under LD (Fig. 4A, B), as evidence that OsELF3-1 represses Ghd7 under LD. Moreover, under SD, Ehd1 in oself3-1 and OsELF3-1 RNAi mutants accumulates less transcripts than that in wild-type plants at dawn (Fig. 4C, D); Hd3α expression in oself3-1 and OsELF3-1 RNAi mutants remained at a very low level over the 24-hour period (Fig. 4Q, R); OsGI expression level was elevated in oself3-1 (Fig. 4E). Taken together, we proposed that OsELF3-1 represses Ghd7 expression in the morning under LD and activates Ehd1 expression at dawn under SD.
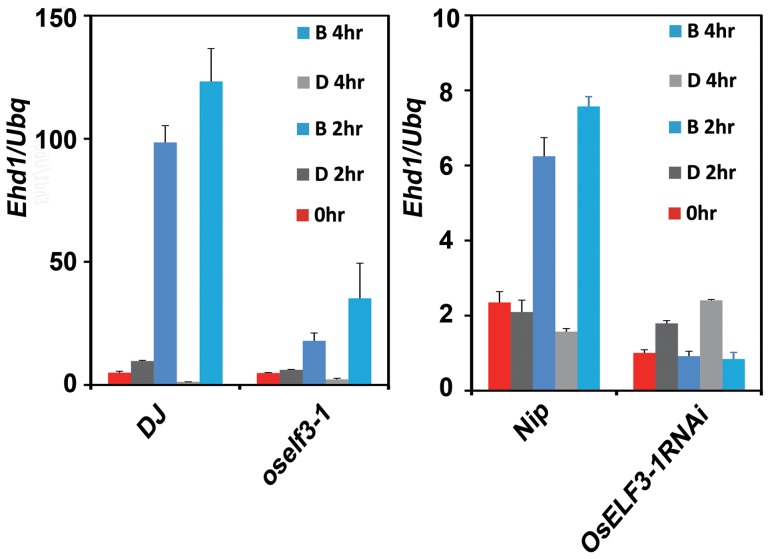
OsELF3-1 is involved in the blue light-mediated activation of Ehd1 expression
In Arabidopsis, ELF3 is an adaptor to target GI to the E3 ubiquitin ligase COP1-mediated degradation. This process may be repressed by the physical interaction of blue light receptors cryptochromes (CRY) with COP1 under blue light [34]. It was also pointed out that OsGI is a gate of circadian clocks in response to blue light in the morning to activate Ehd1 regardless of photoperiod in rice [18]. Since OsELF3 is important for Ehd1 expression (Fig. 4C, D), OsELF3-1 induces Ehd1 expression at dawn under SD, we examined whether OsELF3-1 is involved in Ehd1 activation in response to blue light in the morning. Wild type, oself3-1 and OsELF3-1 RNAi plants were entrained for 14 days under SD conditions and then transferred to blue light and dark surroundings in the following morning. In wild-type plants, activation of Ehd1 transcription was detectable within 2 hours after blue light treatment (Fig. 5). However, Ehd1 gained little induction in oself3-1 and OsELF3-1 RNAi mutants (Fig. 5). Thus, our results indicated OsELF3-1 is essential for Ehd1 activation in response to blue light in the morning under SD.
Figure 5. Blue light promotes activation of Ehd1 dependent on OsELF3-1.
Rice seedlings were first grown under SD for 14 days and then transferred to the blue light (75 μmol m−2 s−1) or darkness at dawn, and harvested at the indicated time points. Values are means ± SD.
Discussion
OsELF3-1 plays a pivotal role in rice flowering
Previous results show OsELF3-1 and OsELF3-2 (OsEF3) are involved in rice flowering [38], [39], [40]. However, their functions have not been fully explored. In our study, we obtained two T-DNA insertion lines of OsELF3-2 in ZH11 and DJ backgrounds. Each line has an insertion site in the 4th exon which might cause an abnormal C-terminus of OsELF3-2 protein (Fig. S2). In Arabidopsis, the C-terminal domain of ELF3 is responsible for its nuclear localization [32], [33]. Since OsELF3 shares intensive amino acid similarity with Arabidopsis ELF3 especially in its C-terminal region, we speculated that OsELF3-2 might be misallocated in these mutants. Surprisingly, these mutants do not exhibit flowering defects (Table S1), whereas a T-DNA insertion line oself3-1 showed a severe late flowering phenotype under both LD and SD (Fig. 1C, Table 1). Thus, we conclude that OsELF3-1 probably plays a predominant role over OsELF3-2 in rice flowering. Furthermore, OsELF3-1 exhibits difference diurnal expression pattern with Arabidopsis ELF3 [43] and OsELF3-2 (Fig. 2B–E). It raises the possibility that OsELF3-1 might play a divergent role from Arabidopsis ELF3 and OsELF3-2 through sub-functionalization in the control of rice development.
In Arabidopsis, ELF3 activity is required to restrain expression of temperature-sensitive genes like PRR7 and PRR9 in the day phase, and to establish enhanced sensitivity of these genes to warm temperature cues during the night. It is necessary for the core oscillator to progress from day to night accompanied by temperature changes [28], [29]. ELF3 is also engaged in phyB-mediated responses to ambient temperature [30]. Recently, Kolmos et al. identified a dual role of ELF3 as an integrator of phytochrome signals and a repressor of core oscillator [55]. Thus it is interesting to explore the potential functions of OsELF3-1 in coordinating various environmental stimuli and circadian regulation in our future analysis.
OsELF3-1 is important for rice circadian clock
Earlier researchers have identified ELF3 is a zeitnehmer in affecting the signaling transduction from light input to oscillators and modulates circadian clock in Arabidopsis [28], [31]. Recently, Matsubara et al. found no obvious difference in the expression of clock-associated genes between Nip and NIL-Hd17 [39]. Saito et al. reported Cab1R: LUC expression under DD conditions was not affected in the oself3-1 (ef7) mutant, while the period of free-running rhythms was slightly shortened in the oself3-1 under LL conditions [38]. However, our data clearly demonstrate OsELF3-1 is important for the circadian expression of OsLHY, OsPRR1, OsPRR37, OsPRR73 and OsPRR59 under free-running LL conditions, taking advantage of the oself3-1 and OsELF3-1 RNAi mutants (Fig. 3A–E, Fig. S4). These results showed OsELF3-1 is essential for sustaining the robust oscillations of OsLHY and OsPRRs expression. Furthermore, OsLHY, OsPRR1, OsPRR37, OsPRR73 and OsPRR59 were not affected under DD in these two mutants, indicating that OsELF3-1 might be responsible for mediating light input to the core circadian oscillator.
Recently, identification of a novel evening complex has profoundly elucidated the biochemical role of ELF3 in the maintenance of the circadian clock in Arabidopsis [33], [36]. ELF3 recruits ELF4 and LUX proteins to form a transcriptional-repressor complex that associates with the promoter of PRR9, a repressor of CCA1 and LHY expression, and then down-regulates PRR9 expression [33]. Thus, this complex indirectly activates LHY and CCA1 in a double-negative connection via PRR9 [33], [52]. Our results showed OsELF3-1 positively regulates OsLHY at dawn (Fig. 4G, H), while it negatively regulates OsPRR1, OsPRR37, OsPRR73 and OsPRR59 at dusk (Fig. 4I–P). Therefore, we deduce OsELF3-1 might associate with the promoters of OsPRRs to repress OsPRRs in the early evening and to indirectly induce OsLHY at dawn.
Functioning mode of OsELF3-1 in rice photoperiodic flowering
Lately, Ioth et al. reported that a critical day length of less than 13 hours is essential for the initiation of rice photoperiodic flowering. The reduction of day length from 13.5 to 13 hours can activate Ehd1 expression along with the observable increase in Hd3α mRNA accumulation. This process is triggered by blue light which coincides with the morning phase set by OsGI-dependent circadian clocks [18]. Our results showed blue light-induced expression of Ehd1 is also dependent on OsELF3-1 in the morning under SD (Fig. 4A, B). The observation of increased OsGI mRNA levels in oself3-1 mutants under LL and SD conditions (Fig. 3F, 4E and F) gives rise to the possibility that OsGI might feedback on the reduced Ehd1 expression to function in a compensatory way for the loss of OsELF3-1. It is also known that Arabidopsis ELF3 protein can physically interact with COP1 to mediate GI degradation in vivo [34]. Thus, further efforts will be made to elucidate the functional interaction between OsELF3-1 and OsGI in rice flowering, particularly in Ehd1 activation induced by blue light under SD.
It is worth pointing out that under LD, phytochromes play pivotal roles in controlling rice flowering. This fact can be explained mainly through two possible signaling pathways. Firstly, the light-dependent interaction between phyB and Hd1 attenuates the Hd1 activation activity on Hd3α, leading to low mRNA abundance of Hd3α in the evening under LD [56], [57]. Secondly, Ghd7 repression on Ehd1 in the morning also rests on phyB under LD [17], [58]. In our study, OsELF3-1 promotes rice flowering under LD, via repressing Ghd7 and indirectly activating Ehd1 expression (Fig. 1C, 2A, H, I, 4A–D, Table 1 and Table S1). It is promising that OsELF3-1 might be involved in phyB-mediated flowering regulation. Besides, in Arabidopsis, ELF3-phyB complex regulates early photomorphogenic development while ELF3 and phytochromes work in concert to control flowering via independent signaling pathways [32], [59], [60]. It is also of importance to explore the roles of OsELF3-1 in response to red light.
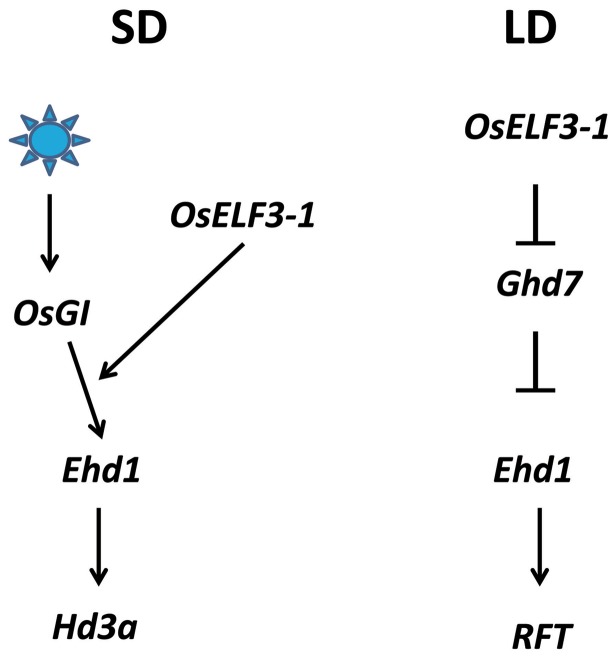
In summary, we point out OsELF3-1 functionally associates with the rice circadian clock and control of photoperiodic flowering. OsELF3-1 is essential for the expression of core oscillator genes and the maintenance of circadian clocks (Fig. 3 and Fig. S4). In photoperiodic flowering pathway, OsELF3-1 is a flowering inducer via transcriptional regulation of Ehd1 and Ghd7 (Fig. 6).
Figure 6. A working model for OsELF3-1 in rice flowering.
OsELF3-1 is a flowering inducer via transcriptional regulation on Ehd1 and Ghd7. The blue light-mediated activation of Ehd1 expression is dependent on OsELF3-1. Arrows, positive regulation. Bars, negative regulation.
Materials and Methods
Plant Materials and Growth Conditions
The T-DNA insertion mutants oself3-1 and oself3-2 (DJ) were obtained from the Salk Institute Genomic Analysis Laboratory http://signal.salk.edu/cgi-bin/RiceGE [61]. oself3-2 (ZH11) was isolated by screening a T-DNA insertion rice (Oryza sativa) var. japonica “Zhonghua11” population generated by Shigui Li's laboratory at Sichuan Agricultural university [40]. To make the OsELF3-1 RNAi construct, an OsELF3-1 cDNA fragment (406–967 bp) was amplified and cloned into the XbaI/BamHI and HindIII/SalI sites of pBluescript SK- vector in sense/antisense directions respectively. The DNA fragment containing the sense/antisense OsELF3-1 fragment was excised by BamHI/SalI and cloned in pOsActin2-1-nos binary vector [62], under the rice Actin2 promoter. Rice plants were transformed according to Agrobacterium mediated co-cultivation methods [63]. All the primers used in this study were listed in Table S2.
Screening of the flowering mutants was carried out by growing the rice plants in the experimental field of Sichuan Agricultural University in Sichuan province, China, in summer (natural LD) and in Hainan province in winter (natural SD). For diurnal expression experiments, the plants were grown in chambers at 70% humidity under LD with daily cycles of 14-hour light at 26°C, 10-hour darkness at 22°C, or under SD with 10-hour light at 26°C and 14-hour darkness at 22°C. Light was provided by fluorescent white light tubes (400 to 700 nm, 100 μmol m−2 s−1). For circadian rhythm experiments, the plants were first grown to cycles of 12-hour light/12-hour darkness for 30 days at 26°C and then transferred to the continuous light (LL) or continuous darkness (DD) at dawn. For Blue light activation experiments, rice seedlings were first grown under SD for 14 days and then transferred to the blue light (75 μmol m−2 s−1) or darkness at dawn.
RNA extraction and qRT-PCR
Total RNA was extracted using a RNeasy plant mini kit (Qiagen, USA), and treated with DNaseI (Qiagen, USA). cDNA was synthesized from 2 μg of total RNA using SuperScript III Reverse Transcriptase (Invitrogen, USA). The quantitative analysis of gene expression was performed with SsoFast EvaGreen PCR Master Mix (BIO-RAD, USA). Data were collected using the BIO-RAD CFX96 Real-Time PCR System according to the manufacturer's instructions. Each experiment was repeated with three independent samples, and Real-Time PCR was performed in three technical replicates for each sample. Changes in gene expression were calculated via the ΔΔCt method [64]. The primers were listed in Table S2.
Supporting Information
Alignment of ELF3 orthologs from the model organisms. CLC Protein Workbench 5 software was used in this analysis.
(TIF)
Schematic diagram of OsELF3-2 gene structure and the T-DNA insertion sites of oself3-2 mutants. Filled boxes indicate exons and solid lines indicate introns.
(TIF)
Tissue-specific expression of OsELF3-1. QRT-PCR analysis of OsELF3-1 expression in different tissues. Rice were grown in Sichuan natural LD field and harvested at 9∶00 am at the booting stage. Values are means ± SD.
(TIF)
Circadian expression of clock-associated genes in Nip and OsELF3-1 RNAi. Circadian expression of OsLHY, OsPRR1, OsPRR37, OsPRR73, OsPRR95 and OsGI in Nip (filled circles) and OsELF3-1 RNAi (open circles) under LL and DD. The plants were first grown to cycles of 12-hour light/12-hour darkness for 30 days at 26°C and then transferred to the continuous light (LL) or continuous darkness (DD) at dawn. In A-F panels, white bars, light; black, darkness; gray, subjective light. In G-L panels, white bars, light; black, darkness; gray, subjective darkness. ZT means Zeitgeber time. Values are means ± SD.
(TIF)
Days to heading in oself3-2 plants in 2010 and 2011. Days to heading was calculated when the first panicle appeared. WT means wild type.
(DOCX)
Primers used in this study.
(DOCX)
Acknowledgments
We thank Dr. Gynheung An for providing mutants of oself3-1 and oself3-2 in DJ background. We also thank Ray Wu and Yongsheng Liu for providing vectors. We are grateful to Haodong Chen, Peng Qin, Chongyong Fu, Yuping Wang and Bingtian Ma for their suggestions on the project, and Alex Roth and Panyu Yang for their critical comments on the manuscript.
Funding Statement
China Postdoctoral Science Foundation Grant (2012M510266) and The Postdoctoral Fellowship at Center for Life Sciences to XO (http://www.chinapostdoctor.org.cn/V3/Program/Main.aspx) (http://www.cls.edu.cn/). and National Natural Science Foundation of China (30971763 and 31025017) to SL (http://www.nsfc.gov.cn/Portal0/default152.htm). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Jiao Y, Ma L, Strickland E, Deng XW (2005) Conservation and divergence of light-regulated genome expression patterns during seedling development in rice and Arabidopsis. Plant Cell 17: 3239–3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Izawa T, Oikawa T, Sugiyama N, Tanisaka T, Yano M, et al. (2002) Phytochrome mediates the external light signal to repress FT orthologs in photoperiodic flowering of rice. Genes Dev 16: 2006–2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tsuji H, Taoka K, Shimamoto K (2011) Regulation of flowering in rice: two florigen genes, a complex gene network, and natural variation. Current Opinion in Plant Biology 14: 45–52. [DOI] [PubMed] [Google Scholar]
- 4. Harmer SL (2009) The circadian system in higher plants. Annual Review of Plant Biology 60: 357–377. [DOI] [PubMed] [Google Scholar]
- 5. Park DH, Somers DE, Kim YS, Choy YH, Lim HK, et al. (1999) Control of circadian rhythms and photoperiodic flowering by the Arabidopsis GIGANTEA gene. Science 285: 1579–1582. [DOI] [PubMed] [Google Scholar]
- 6. Suarez-Lopez P, Wheatley K, Robson F, Onouchi H, Valverde F, et al. (2001) CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410: 1116–1120. [DOI] [PubMed] [Google Scholar]
- 7. Kardailsky I, Shukla VK, Ahn JH, Dagenais N, Christensen SK, et al. (1999) Activation tagging of the floral inducer FT. Science 286: 1962–1965. [DOI] [PubMed] [Google Scholar]
- 8. Izawa T (2007) Adaptation of flowering-time by natural and artificial selection in Arabidopsis and rice. Journal of Experimental Botany 58: 3091–3097. [DOI] [PubMed] [Google Scholar]
- 9. Izawa T (2007) Daylength measurements by rice plants in photoperiodic short-day flowering. Int Rev Cytol 256: 191–222. [DOI] [PubMed] [Google Scholar]
- 10. Hayama R, Yokoi S, Tamaki S, Yano M, Shimamoto K (2003) Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature 422: 719–722. [DOI] [PubMed] [Google Scholar]
- 11. Kojima S, Takahashi Y, Kobayashi Y, Monna L, Sasaki T, et al. (2002) Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol 43: 1096–1105. [DOI] [PubMed] [Google Scholar]
- 12. Yano M, Katayose Y, Ashikari M, Yamanouchi U, Monna L, et al. (2000) Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 12: 2473–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tamaki S, Matsuo S, Wong HL, Yokoi S, Shimamoto K (2007) Hd3a protein is a mobile flowering signal in rice. Science 316: 1033–1036. [DOI] [PubMed] [Google Scholar]
- 14. Komiya R, Ikegami A, Tamaki S, Yokoi S, Shimamoto K (2008) Hd3a and RFT1 are essential for flowering in rice. Development 135: 767–774. [DOI] [PubMed] [Google Scholar]
- 15. Komiya R, Yokoi S, Shimamoto K (2009) A gene network for long-day flowering activates RFT1 encoding a mobile flowering signal in rice. Development 136: 3443–3450. [DOI] [PubMed] [Google Scholar]
- 16. Doi K, Izawa T, Fuse T, Yamanouchi U, Kubo T, et al. (2004) Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Genes Dev 18: 926–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Xue W, Xing Y, Weng X, Zhao Y, Tang W, et al. (2008) Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat Genet 40: 761–767. [DOI] [PubMed] [Google Scholar]
- 18. Itoh H, Nonoue Y, Yano M, Izawa T (2010) A pair of floral regulators sets critical day length for Hd3a florigen expression in rice. Nat Genet 42: 635–638. [DOI] [PubMed] [Google Scholar]
- 19. Matsubara K, Yamanouchi U, Wang ZX, Minobe Y, Izawa T, et al. (2008) Ehd2, a rice ortholog of the maize INDETERMINATE1 gene, promotes flowering by up-regulating Ehd1. Plant Physiol 148: 1425–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Matsubara K, Yamanouchi U, Nonoue Y, Sugimoto K, Wang ZX, et al. (2011) Ehd3, encoding a plant homeodomain finger-containing protein, is a critical promoter of rice flowering. Plant J 66: 603–612. [DOI] [PubMed] [Google Scholar]
- 21. Lee J, Lee I (2010) Regulation and function of SOC1, a flowering pathway integrator. Journal of Experimental Botany 61: 2247–2254. [DOI] [PubMed] [Google Scholar]
- 22. Samach A, Onouchi H, Gold SE, Ditta GS, Schwarz-Sommer Z, et al. (2000) Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288: 1613–1616. [DOI] [PubMed] [Google Scholar]
- 23. Seo E, Lee H, Jeon J, Park H, Kim J, et al. (2009) Crosstalk between cold response and flowering in Arabidopsis is mediated through the flowering-time gene SOC1 and its upstream negative regulator FLC. Plant Cell 21: 3185–3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lee S, Kim J, Han JJ, Han MJ, An G (2004) Functional analyses of the flowering time gene OsMADS50, the putative SUPPRESSOR OF OVEREXPRESSION OF CO 1/AGAMOUS-LIKE 20 (SOC1/AGL20) ortholog in rice. Plant J 38: 754–764. [DOI] [PubMed] [Google Scholar]
- 25. Ryu CH, Lee S, Cho LH, Kim SL, Lee YS, et al. (2009) OsMADS50 and OsMADS56 function antagonistically in regulating long day (LD)-dependent flowering in rice. Plant Cell Environ 32: 1412–1427. [DOI] [PubMed] [Google Scholar]
- 26. Zagotta MT, Hicks KA, Jacobs CI, Young JC, Hangarter RP, et al. (1996) The Arabidopsis ELF3 gene regulates vegetative photomorphogenesis and the photoperiodic induction of flowering. Plant J 10: 691–702. [DOI] [PubMed] [Google Scholar]
- 27. Hicks KA, Millar AJ, Carre IA, Somers DE, Straume M, et al. (1996) Conditional circadian dysfunction of the Arabidopsis early-flowering 3 mutant. Science 274: 790–792. [DOI] [PubMed] [Google Scholar]
- 28. McWatters HG, Bastow RM, Hall A, Millar AJ (2000) The ELF3 zeitnehmer regulates light signalling to the circadian clock. Nature 408: 716–720. [DOI] [PubMed] [Google Scholar]
- 29. Thines B, Harmon FG (2010) Ambient temperature response establishes ELF3 as a required component of the core Arabidopsis circadian clock. Proc Natl Acad Sci U S A 107: 3257–3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Strasser B, Alvarez MJ, Califano A, Cerdan PD (2009) A complementary role for ELF3 and TFL1 in the regulation of flowering time by ambient temperature. Plant J 58: 629–640. [DOI] [PubMed] [Google Scholar]
- 31. Covington MF, Panda S, Liu XL, Strayer CA, Wagner DR, et al. (2001) ELF3 modulates resetting of the circadian clock in Arabidopsis. Plant Cell 13: 1305–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liu XL, Covington MF, Fankhauser C, Chory J, Wagner DR (2001) ELF3 encodes a circadian clock-regulated nuclear protein that functions in an Arabidopsis PHYB signal transduction pathway. Plant Cell 13: 1293–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Herrero E, Kolmos E, Bujdoso N, Yuan Y, Wang M, et al.. (2012) EARLY FLOWERING4 Recruitment of EARLY FLOWERING3 in the Nucleus Sustains the Arabidopsis Circadian Clock. Plant Cell. [DOI] [PMC free article] [PubMed]
- 34. Yu JW, Rubio V, Lee NY, Bai S, Lee SY, et al. (2008) COP1 and ELF3 control circadian function and photoperiodic flowering by regulating GI stability. Mol Cell 32: 617–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hazen SP, Schultz TF, Pruneda-Paz JL, Borevitz JO, Ecker JR, et al. (2005) LUX ARRHYTHMO encodes a Myb domain protein essential for circadian rhythms. Proc Natl Acad Sci U S A 102: 10387–10392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nusinow DA, Helfer A, Hamilton EE, King JJ, Imaizumi T, et al. (2011) The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature 475: 398–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Murakami M, Tago Y, Yamashino T, Mizuno T (2007) Comparative overviews of clock-associated genes of Arabidopsis thaliana and Oryza sativa. Plant and Cell Physiology 48: 110–121. [DOI] [PubMed] [Google Scholar]
- 38. Saito H, Ogiso-Tanaka E, Okumoto Y, Yoshitake Y, Izumi H, et al. (2012) Ef7 Encodes an ELF3-like Protein and Promotes Rice Flowering by Negatively Regulating the Floral Repressor Gene Ghd7 under Both Short- and Long-Day Conditions. Plant Cell Physiol 53: 717–728. [DOI] [PubMed] [Google Scholar]
- 39. Matsubara K, Ogiso-Tanaka E, Hori K, Ebana K, Ando T, et al. (2012) Natural Variation in Hd17, a Homolog of Arabidopsis ELF3 That is Involved in Rice Photoperiodic Flowering. Plant Cell Physiol 53: 709–716. [DOI] [PubMed] [Google Scholar]
- 40. Fu C, Yang XO, Chen X, Chen W, Ma Y, et al. (2009) OsEF3, a homologous gene of Arabidopsis ELF3, has pleiotropic effects in rice. Plant Biol (Stuttg) 11: 751–757. [DOI] [PubMed] [Google Scholar]
- 41. Zeevaart JA (2008) Leaf-produced floral signals. Current Opinion in Plant Biology 11: 541–547. [DOI] [PubMed] [Google Scholar]
- 42. Corbesier L, Vincent C, Jang S, Fornara F, Fan Q, et al. (2007) FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316: 1030–1033. [DOI] [PubMed] [Google Scholar]
- 43. Hicks KA, Albertson TM, Wagner DR (2001) EARLY FLOWERING3 encodes a novel protein that regulates circadian clock function and flowering in Arabidopsis. Plant Cell 13: 1281–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Herrero E, Davis SJ (2012) Time for a Nuclear Meeting: Protein Trafficking and Chromatin Dynamics Intersect in the Plant Circadian System. Mol Plant 5: 28–39. [DOI] [PubMed] [Google Scholar]
- 45. Strayer C, Oyama T, Schultz TF, Raman R, Somers DE, et al. (2000) Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 289: 768–771. [DOI] [PubMed] [Google Scholar]
- 46. Schaffer R, Ramsay N, Samach A, Corden S, Putterill J, et al. (1998) The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell 93: 1219–1229. [DOI] [PubMed] [Google Scholar]
- 47. Wang ZY, Tobin EM (1998) Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93: 1207–1217. [DOI] [PubMed] [Google Scholar]
- 48. Somers DE, Webb AA, Pearson M, Kay SA (1998) The short-period mutant, toc1-1, alters circadian clock regulation of multiple outputs throughout development in Arabidopsis thaliana. Development 125: 485–494. [DOI] [PubMed] [Google Scholar]
- 49. Pruneda-Paz JL, Breton G, Para A, Kay SA (2009) A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science 323: 1481–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Huang W, Perez-Garcia P, Pokhilko A, Millar AJ, Antoshechkin I, et al. (2012) Mapping the core of the Arabidopsis circadian clock defines the network structure of the oscillator. Science 336: 75–79. [DOI] [PubMed] [Google Scholar]
- 51. Gendron JM, Pruneda-Paz JL, Doherty CJ, Gross AM, Kang SE, et al. (2012) Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proc Natl Acad Sci U S A 109: 3167–3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Dixon LE, Knox K, Kozma-Bognar L, Southern MM, Pokhilko A, et al. (2011) Temporal repression of core circadian genes is mediated through EARLY FLOWERING 3 in Arabidopsis. Curr Biol 21: 120–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Helfer A, Nusinow DA, Chow BY, Gehrke AR, Bulyk ML, et al. (2011) LUX ARRHYTHMO encodes a nighttime repressor of circadian gene expression in the Arabidopsis core clock. Curr Biol 21: 126–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Murakami M, Ashikari M, Miura K, Yamashino T, Mizuno T (2003) The evolutionarily conserved OsPRR quintet: rice pseudo-response regulators implicated in circadian rhythm. Plant Cell Physiol 44: 1229–1236. [DOI] [PubMed] [Google Scholar]
- 55. Kolmos E, Herrero E, Bujdoso N, Millar AJ, Toth R, et al. (2011) A reduced-function allele reveals that EARLY FLOWERING3 repressive action on the circadian clock is modulated by phytochrome signals in Arabidopsis. Plant Cell 23: 3230–3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hayama R, Coupland G (2004) The molecular basis of diversity in the photoperiodic flowering responses of Arabidopsis and rice. Plant Physiol 135: 677–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ishikawa R, Tamaki S, Yokoi S, Inagaki N, Shinomura T, et al. (2005) Suppression of the floral activator Hd3a is the principal cause of the night break effect in rice. Plant Cell 17: 3326–3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Osugi A, Itoh HD, Ikeda-Kawakatsu KD, Takano MD, Izawa T (2011) Molecular dissection of the roles of phytochrome in photoperiodic flowering in rice. Plant Physiol 157: 1128–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zust T, Joseph B, Shimizu KK, Kliebenstein DJ, Turnbull LA (2011) A Reduced-Function Allele Reveals That EARLY FLOWERING3 Repressive Action on the Circadian Clock Is Modulated by Phytochrome Signals in Arabidopsis. The plant cell 23: 3230–3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Reed JW, Nagpal P, Bastow RM, Solomon KS, Dowson-Day MJ, et al. (2000) Independent action of ELF3 and phyB to control hypocotyl elongation and flowering time. Plant Physiol 122: 1149–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Jeong DH, An S, Park S, Kang HG, Park GG, et al. (2006) Generation of a flanking sequence-tag database for activation-tagging lines in japonica rice. Plant J 45: 123–132. [DOI] [PubMed] [Google Scholar]
- 62. He C, Lin Z, McElroy D, Wu R (2009) Identification of a rice actin2 gene regulatory region for high-level expression of transgenes in monocots. Plant Biotechnology Journal 7: 227–239. [DOI] [PubMed] [Google Scholar]
- 63. Hiei Y, Ohta S, Komari T, Kumashiro T (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 6: 271–282. [DOI] [PubMed] [Google Scholar]
- 64. Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Alignment of ELF3 orthologs from the model organisms. CLC Protein Workbench 5 software was used in this analysis.
(TIF)
Schematic diagram of OsELF3-2 gene structure and the T-DNA insertion sites of oself3-2 mutants. Filled boxes indicate exons and solid lines indicate introns.
(TIF)
Tissue-specific expression of OsELF3-1. QRT-PCR analysis of OsELF3-1 expression in different tissues. Rice were grown in Sichuan natural LD field and harvested at 9∶00 am at the booting stage. Values are means ± SD.
(TIF)
Circadian expression of clock-associated genes in Nip and OsELF3-1 RNAi. Circadian expression of OsLHY, OsPRR1, OsPRR37, OsPRR73, OsPRR95 and OsGI in Nip (filled circles) and OsELF3-1 RNAi (open circles) under LL and DD. The plants were first grown to cycles of 12-hour light/12-hour darkness for 30 days at 26°C and then transferred to the continuous light (LL) or continuous darkness (DD) at dawn. In A-F panels, white bars, light; black, darkness; gray, subjective light. In G-L panels, white bars, light; black, darkness; gray, subjective darkness. ZT means Zeitgeber time. Values are means ± SD.
(TIF)
Days to heading in oself3-2 plants in 2010 and 2011. Days to heading was calculated when the first panicle appeared. WT means wild type.
(DOCX)
Primers used in this study.
(DOCX)