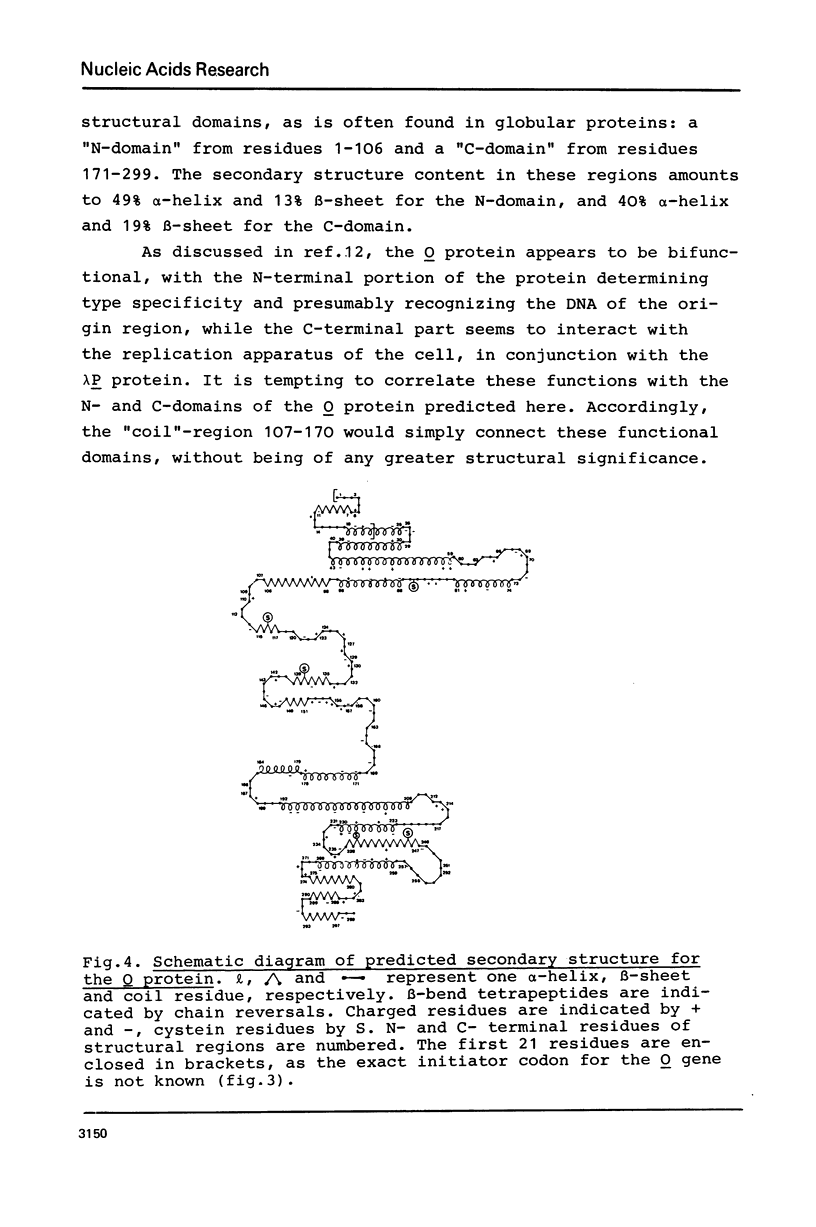
Abstract
The nucleotide sequence of the O gene in bacteriophage lambda DNA is presented. According to two possible initiator codons, the primary structure of the O protein deduced from the DNA sequence consists of 278 or 299 amino acid residues. Structure and function of the O protein--one of the two phage initiator proteins for lambda DNA replication--are discussed in the light of a secondary structure model for the O protein. The central part of the O gene contains a cluster of symmetrical sequences extending over 160 base pairs. The point mutation of the cis-dominant replication mutant ti12 is located in this region.
Full text
PDF














Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bickle T. A., Pirrotta V., Imber R. A simple, general procedure for purifying restriction endonucleases. Nucleic Acids Res. 1977 Aug;4(8):2561–2572. doi: 10.1093/nar/4.8.2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou P. Y., Adler A. J., Fasman G. D. Conformational prediction and circular dichroism studies on the lac repressor. J Mol Biol. 1975 Jul 25;96(1):29–45. doi: 10.1016/0022-2836(75)90180-1. [DOI] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Prediction of protein conformation. Biochemistry. 1974 Jan 15;13(2):222–245. doi: 10.1021/bi00699a002. [DOI] [PubMed] [Google Scholar]
- Denniston-Thompson K., Moore D. D., Kruger K. E., Furth M. E., Blattner F. R. Physical structure of the replication origin of bacteriophage lambda. Science. 1977 Dec 9;198(4321):1051–1056. doi: 10.1126/science.929187. [DOI] [PubMed] [Google Scholar]
- Eisen H. A., Fuerst C. R., Siminovitch L., Thomas R., Lambert L., Pereira da Silva L., Jacob F. Genetics and physiology of defective lysogeny in K12 (lambda): studies of early mutants. Virology. 1966 Oct;30(2):224–241. doi: 10.1016/0042-6822(66)90098-5. [DOI] [PubMed] [Google Scholar]
- Fiddes J. C., Barrell B. G., Godson G. N. Nucleotide sequences of the separate origins of synthesis of bacteriophage G4 viral and complementary DNA strands. Proc Natl Acad Sci U S A. 1978 Mar;75(3):1081–1085. doi: 10.1073/pnas.75.3.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furth M. E., Blattner F. R., McLeester C., Dove W. F. Genetic structure of the replication origin of bacteriophage lambda. Science. 1977 Dec 9;198(4321):1046–1051. doi: 10.1126/science.929186. [DOI] [PubMed] [Google Scholar]
- Hughes S. G. The sensitivity of bacteriophage lambda DNA to restriction endonuclease RII. J Mol Biol. 1975 Nov 5;98(3):645–647. doi: 10.1016/s0022-2836(75)80093-3. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmori H., Tomizawa J. I., Maxam A. M. Detection of 5-methylcytosine in DNA sequences. Nucleic Acids Res. 1978 May;5(5):1479–1485. doi: 10.1093/nar/5.5.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirrotta V. The lambda repressor and its action. Curr Top Microbiol Immunol. 1976;74:21–54. doi: 10.1007/978-3-642-66336-9_2. [DOI] [PubMed] [Google Scholar]
- Pirrotta V. Two restriction endonucleases from Bacillus globiggi. Nucleic Acids Res. 1976 Jul;3(7):1747–1760. doi: 10.1093/nar/3.7.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polisky B., Greene P., Garfin D. E., McCarthy B. J., Goodman H. M., Boyer H. W. Specificity of substrate recognition by the EcoRI restriction endonuclease. Proc Natl Acad Sci U S A. 1975 Sep;72(9):3310–3314. doi: 10.1073/pnas.72.9.3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab C., Klein A., Kluding H., Hirth P., Fuchs E. Cell free synthesis of bacteriophage lambda replication proteins. FEBS Lett. 1977 Aug 15;80(2):275–278. doi: 10.1016/0014-5793(77)80456-0. [DOI] [PubMed] [Google Scholar]
- Rambach A. Replicator mutants of bacteriophage lambda: characterization of two subclasses. Virology. 1973 Jul;54(1):270–277. doi: 10.1016/0042-6822(73)90136-0. [DOI] [PubMed] [Google Scholar]
- Scherer G., Hobom G., Kössel H. DNA base sequence of the po promoter region of phage lamdba. Nature. 1977 Jan 13;265(5590):117–121. doi: 10.1038/265117a0. [DOI] [PubMed] [Google Scholar]
- Schnös M., Inman R. B. Position of branch points in replicating lambda DNA. J Mol Biol. 1970 Jul 14;51(1):61–73. doi: 10.1016/0022-2836(70)90270-6. [DOI] [PubMed] [Google Scholar]
- Schwarz E., Scherer G., Hobom G., Kössel H. Nucleotide sequence of cro, cII and part of the O gene in phage lambda DNA. Nature. 1978 Mar 30;272(5652):410–414. doi: 10.1038/272410a0. [DOI] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalka A. M. DNA replication--bacteriophage lambda. Curr Top Microbiol Immunol. 1977;78:201–237. [PubMed] [Google Scholar]
- Smith H. O., Wilcox K. W. A restriction enzyme from Hemophilus influenzae. I. Purification and general properties. J Mol Biol. 1970 Jul 28;51(2):379–391. doi: 10.1016/0022-2836(70)90149-x. [DOI] [PubMed] [Google Scholar]
- Steitz J. A., Jakes K. How ribosomes select initiator regions in mRNA: base pair formation between the 3' terminus of 16S rRNA and the mRNA during initiation of protein synthesis in Escherichia coli. Proc Natl Acad Sci U S A. 1975 Dec;72(12):4734–4738. doi: 10.1073/pnas.72.12.4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streeck R. E., Hobom G. Mapping of cleavage sites for restriction endonucleases in lambdadv plasmids. Eur J Biochem. 1975 Sep 15;57(2):595–606. doi: 10.1111/j.1432-1033.1975.tb02335.x. [DOI] [PubMed] [Google Scholar]
- THOMAS R., BERTANI L. E. ON THE CONTROL OF THE REPLICATION OF TEMPERATE BACTERIOPHAGES SUPERINFECTING IMMUNE HOSTS. Virology. 1964 Nov;24:241–253. doi: 10.1016/0042-6822(64)90163-1. [DOI] [PubMed] [Google Scholar]
- Thomas M., Davis R. W. Studies on the cleavage of bacteriophage lambda DNA with EcoRI Restriction endonuclease. J Mol Biol. 1975 Jan 25;91(3):315–328. doi: 10.1016/0022-2836(75)90383-6. [DOI] [PubMed] [Google Scholar]
- Wyatt W. M., Inokuchi H. Stability of lambda O and P replication functions. Virology. 1974 Mar;58(1):313–315. doi: 10.1016/0042-6822(74)90168-8. [DOI] [PubMed] [Google Scholar]
- Yates J. L., Gette W. R., Furth M. E., Nomura M. Effects of ribosomal mutations on the read-through of a chain termination signal: studies on the synthesis of bacteriophage lambda O gene protein in vitro. Proc Natl Acad Sci U S A. 1977 Feb;74(2):689–693. doi: 10.1073/pnas.74.2.689. [DOI] [PMC free article] [PubMed] [Google Scholar]