Abstract
Hedgehog (Hh) signaling pathway is activated in diffuse large B-cell lymphoma (DLBCL). Genetic abnormalities that explain activation of Hh signaling in DLBCL are unknown. We investigate the presence of amplifications of Hh genes that might result in activation of this pathway in DLBCL. Our data showed few extra copies of GLI1 and SMO due to chromosomal aneuploidies in a subset of DLBCL cell lines. We also showed that pharmacologic inhibition of PI3K/AKT and NF-KB pathways resulted in decreased expression of GLI1 and Hh ligands. In conclusion, our data support the hypothesis that aberrant activation of Hh signaling in DLBCL mainly results from integration of deregulated oncogenic signaling inputs converging into Hh signaling.
Keywords: Gene copy number, Hedgehog pathway, GLI, diffuse large B-cell lymphoma
INTRODUCTION
The hedgehog (Hh) signaling pathway is a highly regulated signaling pathway which is not only important in tissue patterning, organogenesis and embryonic development but also in tissue repair as well as in the maintenance of stem cells in adult tissues and has been implicated in the development of various tumors [1]. The Hh family of proteins consists of three distinct ligands, sonic hedgehog (Shh), indian hedgehog (Ihh), and desert hedgehog (Dhh), which use the same receptors and signal transduction pathway [2]. Hh ligands interact with a receptor complex composed of two proteins, the 12 transmembrane protein patched (PTCH1), ligand-binding subunit, and the 7 transmembrane protein smoothened (SMO), signal transduction subunit. In the absence of Hh ligands, PTCH1 inhibits SMO, which results in phosphorylation-dependent ubiquitination of glioma-associated oncogene homolog (GLI) transcription factors, leading to degradation and clearance of GLI1 and GLI2 or partial proteolysis of GLI3 that represses transcription of Hh target genes. However, binding of Hh ligands leads to internalization of PTCH1 allowing the activation of SMO. SMO activation results in increased stability and nuclear accumulation of GLI transcription factors and transcription of Hh target genes [3]. GLI1 and SMO are positive regulators of Hh transcriptional targets and GLI1 is, itself, a transcriptional target of Hh signaling and the best reliable indicator of the activation of the pathway [4].
Inappropriate activation of the Hh signaling pathway has been shown in several cancers including hematopoetic malignancies [5–13]. Recently, we found that Hh signaling is active and functional in diffuse large B-cell lymphoma (DLBCL), the most frequent aggressive subtype of B-cell lymphoma [8]. High expression of SMO and the transcription factor GLI1 have been detected in DLBCL tumor samples and in DLBCL cell lines [8, 14]. Importantly, inhibition of Hh signaling in DLBCL cell lines results in increased apoptosis and/or cell cycle arrest, in particular in those DLBCL cell lines with high expression of SMO [8]. However, genetic abnormalities (gene amplifications) that might explain the inappropriate activation of Hh signaling in DLBCL are unknown.
To uncover potential factors contributing to Hh activation in DLBCL, we explored if gene amplifications involving Hh-related genes may contribute to explain the inappropriate Hh signaling activation in DLBCL. GLI1 is located at 12q13 and gains of this locus and/or extra copies of chromosome 12 have been reported in B-cell lymphomas, including splenic and nodal marginal zone lymphoma, DLBCL and mantle cell lymphoma [15–18].
Our data indicate the presence of few extra copies of SMO and GLI1 in a subset of DLBCL which are caused by chromosomal aneuploidy rather than true gene amplification. The presence of these extra copies of Hh related genes do not explain per se the inappropriate level of activation of Hh pathway. Recent evidence suggests that activation of Hh signaling in cancers with Hh ligand dependent activation may result, at least in part, from integration of multiple deregulated oncogenic signaling inputs in the Hh pathway [19]. For this reason, we also explored the effect of inhibition of two relevant oncogenic pathways in DLBCL, PI3K/AKT and NF-kB signaling, in the activation level of Hh signaling.
MATERIAL AND METHODS
DLBCL Cell Lines and Patient Samples
Seven DLBCL cell lines were used for this study; 3 germinal center type (BJAB, DOHH2, and SUDHL4), 3 activated cell type (OCl-LY3, SUDHL2, and HBL-1), and one primary mediastinal large B-cell lymphoma (U2940). DOHH2, SUDHL4, and U2490 were purchased from DSMZ (Braunschweig, Germany) and HBL-1 from ATCC (Manassas, VA, USA). OCI-LY3 were kindly provided by Michael G Rosenblum (Department of Experimental Therapeutics, MDACC), and SUDHL2 and BJAB cells by Felipe Samaniego (Department of Lymphoma/Myeloma, MDACC). All cell lines were maintained at 37°C in RPMI 1640 (ATCC) supplemented with 10% heat-inactivated fetal bovine serum (FBS; Sigma, St Louis, MO, USA) in a humidified atmosphere containing 5% CO2.
Gene copy number was also studied in frozen lymph node samples collected from 9 patients with DLBCL. Four DLBCL tumors were relapses collected from patients previously treated with R-CHOP and 5 DLBCL tumors were de novo lymphomas from untreated patients. The DLBCL tumors were provided by the Hematopathology Tissue Bank of the UT MD Anderson Cancer Center.
Real-Time Quantitative (q) PCR for copy number estimation
DNA was extracted from the cell lines using the DNeasy tissue kit (Qiagen, Valencia, CA, USA). Gene copy number by real-time qPCR was also assessed in DNA extracted from paraffin embedded tissue sections collected from 9 patients with DLBCL involving lymph nodes. The ABI Prism 7900 HT Sequence Detection System and Taqman probes (PE Applied Biosystems, Foster City, CA, USA) were used for performing real-time qPCR. Primers and probes were added at a final concentration of 0.3 and 0.1 µmol/L per reaction respectively. 500 ng of DNA was used for each reaction and PCR was performed in a total reaction volume of 25 µL. For each copy number estimation each target gene was amplified in duplicate. The copy number was calculated based on the difference between amplification of each gene and RNaseP and comparison with gene copy number in normal female DNA using ΔΔCT method. These experiments were performed in quadruplicate for the DLBCL cell lines and in triplicate for the tumor samples. Gene copy number was estimated as follows: a 2−ΔΔCT ratio higher than 1 was considered as a potential indication of presence of gene extra copies. Importantly, qRT-PCR assay for copy number estimation is a screening method and the results of this assay need to be confirmed by additional methods such as FISH.
G-band Karyotyping and Spectral Karyotyping (SKY)
G-band karyotyping analysis was performed using conventional cytogenetic techniques and the Applied Imaging system (Genetix, San Jose, CA). The cells were grown, arrested in metaphase by colcemid, harvested, dropped on slides, G-banded with trypsin, and stained with 4% Giemsa. Complicated chromosome abnormalities or discrepancies between chromosome number by G-banding and FISH were further investigated in one cell line (BJAB) using Spectral Karyotyping (SKY) and analyzed using SKYPaint probes (Applied Spectral Imaging).
Fluorescent In Situ Hybridization (FISH) analysis
To verify the presence of extra copies/amplifications of GLI1 and SMO, two Hh genes which might contribute in the activation status of Hh pathway, we designed FISH assays using bacterial artificial chromosome (BAC) clones which include GLI1 and SMO genes. The BAC clones used were RP11-258J5 (from 57740420 to 57928991) for GLI1 and RP11-152K21 (from 12877.4 to 128920.82) for SMO. BAC clones were selected using mapping information from the National Center of Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov) and obtained from BAC Resources PAC (Oakland, CA, USA). BAC clones were grown in TB media with chloramphenicol. DNA was isolated using a standard alkaline lysis kit (Eppendorf Plasmid Mini Prep, Fischer Scientific, Pittsburg, PA, USA). Centromere probes were also used as controls. DNA was labeled directly with spectrum Orange-dUTP or Spectrum Green-dUTP using Nick Translation Kit (Vysis, Downer’s Grove, Il, USA) following the manufacturer’s instructions.
We verified that fluorescently labeled BAC clones hybridized to the correct chromosome locations using metaphase chromosome spreads obtained using standard procedures. Following hybridization with the FISH probes, the slides were analyzed using a Zeiss Axiphot fluorescent microscope including single- and triple-band pass filters. Up to 1000 cells were counted for the FISH studies. Digital FISH images were captured by a Power Macintosh G3 system and MacProbe version 4.4 (Applied Imaging, San Jose, CA, USA).
Western blots analysis and drug treatments
Western blotting was done as described previously.[8] Antibodies used were Ser536p-P65, total P65, GLI1 (Santa Cruz Biotechnology, Santa Cruz, CA, USA), SHH monoclonal antibody (Clone C9C5), Ser473pAKT (Cell Signaling Technology, Beverly, MA, USA), and β-actin (Sigma). Reactions were visualized with suitable secondary antibodies conjugated with horseradish peroxidase using enhanced chemiluminescence reagents (Amershan, Piscataway, NJ, USA). Drug treatments were done using the PI3K inhibitor, 2-(4-morpholinyl)-8-phenyl-4H-1-benzopyran-4-one (LY294002), (Calbiochem, San Diego, CA, USA) and the NF-Kβ inhibitor BAY11-7082 (Calbiochem) as previously described [20].
RESULTS
Estimation of copy number of Hh-related genes in DLBCL cell lines and tumor samples
The results of the real-time qPCR assays for gene copy number estimation in DLBCL cell lines and in tumor samples are summarized in figures 1 and 2, respectively.
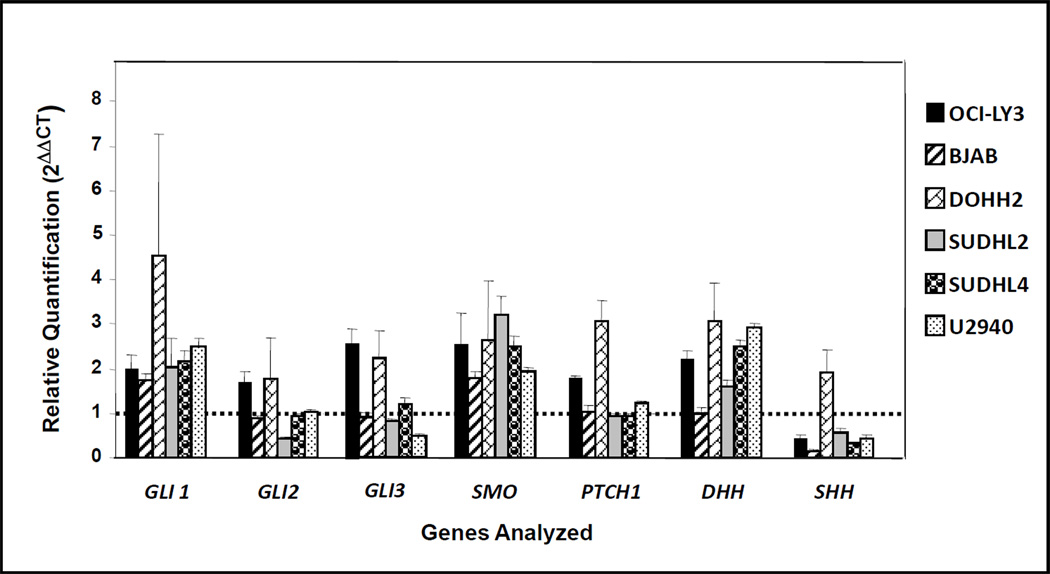
Figure 1. Real-time quantitative (q) PCR for Hh-related genes copy number estimation in DLBCL cell lines.
These experiments were performed in quadruplicate. In each experiment, Hh related genes were amplified in duplicate. The diploid status is indicated with a dotted line.
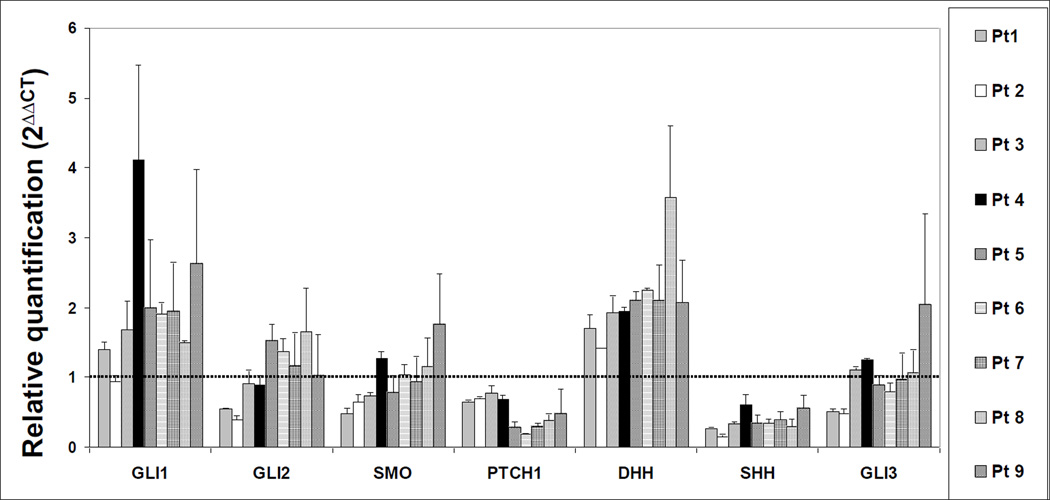
Figure 2. Real-time quantitative (q) PCR for Hh-related genes copy number estimation in DLBCL tumor samples.
These experiments were performed in triplicate. In each experiment, Hh related genes were amplified in duplicate. The diploid status is indicated with a dotted line.
The real-time qPCR data suggested the presence of extra copies of Hh-related genes in most of the cell lines analyzed. Extra copies of GLI1, SMO and DHH were detected in almost all the cell lines analyzed. The number of extra copies of GLI1 was higher in DOHH2 cells than in the other cell lines. The real-time qPCR data also suggested the presence of extra copies of GLI2, GLI3, and PTCH1 in a subset of the cell lines. Losses involving SHH gene were also suggested in 5 DLBCL cell lines except in DOHH2 cells (Figure 1). Extra copies of SHH were detected in DOHH2 (Figure 1).
In DLBCL tumor samples, extra copies of GLI1 were suggested by the real-time qPCR assay in most of the tumor samples (Figure 2). Extra copies of DHH were seen in all patient samples. SHH and PTCH1 genes seems to be frequently lost in DLBCL tumor samples.
Cytogenetic findings including G-band karyotyping, FISH and SKY and correlation with the gene copy number PCR assay
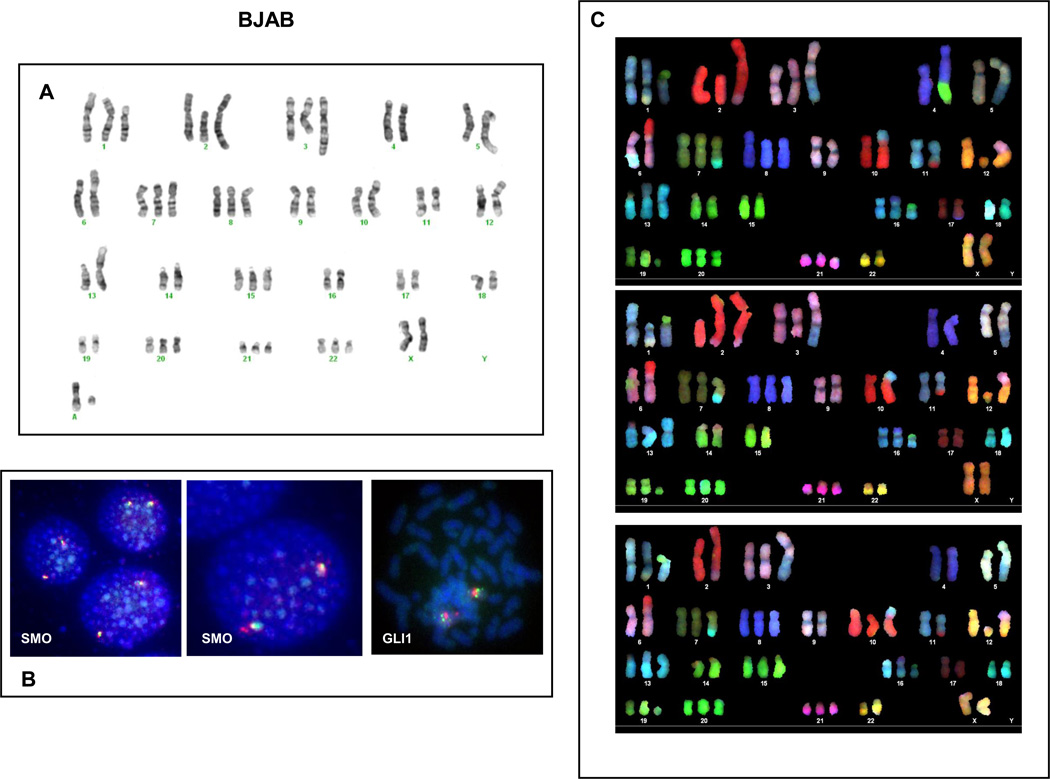
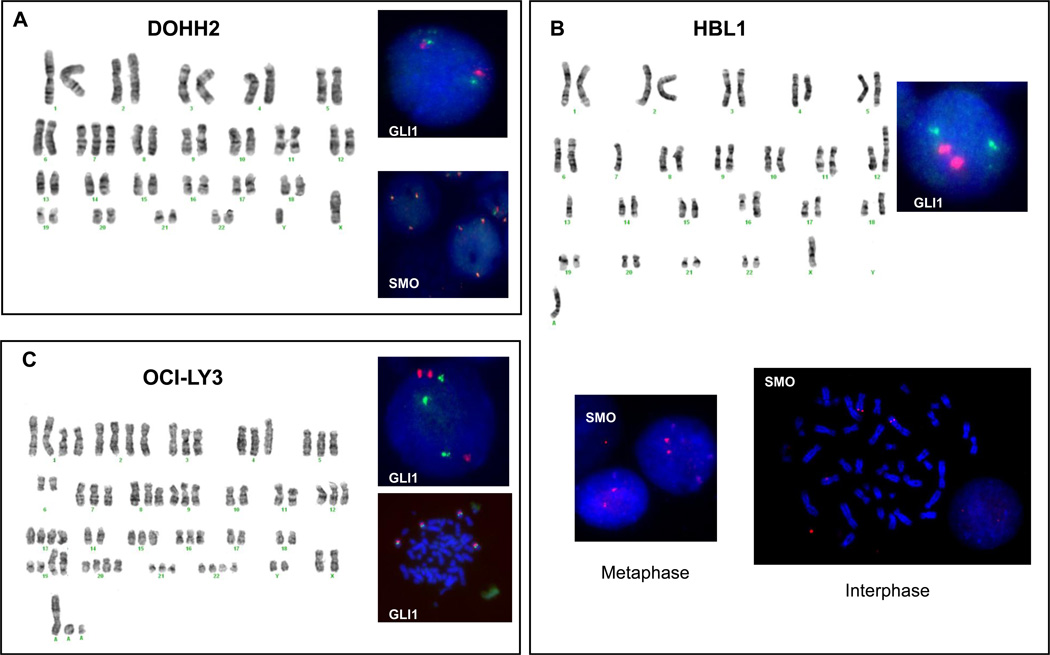
G-band karyotyping was performed on 4 DLBCL cell lines: BJAB, DOHH2, HBL1, and OCI-LY3. The results of the karyotyping analysis of these DLBCL cell lines is shown in table 1. The chromosomes of interest regarding the major Hh-related genes are the long arm of chromosome 12 (for GLI1 and DHH), the long arm of chromosome 2 (for GLI2), the short arm of chromosome 7 (for GLI3), the long arm of chromosome 7 (for SHH, and SMO) and the long arm of chromosome 9 (for PTCH1). BJAB cells showed a very complex karyotype with multiple structural abnormalities, chromosomal additions and deletions. DOHH2 cells showed trisomy 7 and the t(8;14;18)(q24.1;q32;q21.3), HBL-1 cells showed monosomy 7 and a derivative chromosome 12 with the t(7;12)(q11.2;p13) among other structural and numeric abnormalities and OCI-LY3 cells showed multiple structural and numeric abnormalities.
Table 1.
G-banding karyotyping for the DLBCL cell lines analyzed.
| Cell lines | Karyotype |
|---|---|
| BJAB | 57,XX,+1,+add(1)(p13),del(1)(p13),+2,add(2)(q37),del(2)(p12),+3,del(3)(q25q26.3),psudic(3;1)(q25;p12), der(5)t(1;5)(p13;q31), der(6)t(2;6)(?;p23),+7,+8,add(10)(p11.2),del(11)(q23q24),der(12)t(6;12)(p12;q11.2),add(13)(p11.2),+15,add(16)(p11.2),+20,+21,+22,+2 mar |
| DOHH2 | 47,XY,+7,t(8;14;18)(q24.1;q32;q21.3) |
| HBL-1 | 44,X,−Y,del(4)(q21q25),add(6)(p21),−7,der(12)t(7;12)(q11.2;p13),−13,add(16)(q24),add(17)(p11.2),add(18)(p11.2),+mar |
| OCI-Ly3 | 72~75,XXY,+Y,+1,add(1)(p13)x2,+2,add(4)(q35),−6,del(6)(q13)x2,−10,−11,+13,−14,−17,−18,+19,add(19)(p11.3)x2,+20,add(20)(q13.3),+21,+r,+1~5mar[cp4] |
Interphase and metaphase FISH studies to detect extra copies of GLI1 and SMO were also performed. In DOHH2, HBL1 and OCI-LY3, the number of signals for GLI1 and SMO detected by FISH studies correlated with the number of the respective centromeric signals (centromere 12 for GLI1 and centromere 7 for SMO). In BJAB cells there were some discrepancies between the results obtained by G-banding and FISH which were clarified using SKY. In BJAB cells, G-band karyotyping showed trisomy 7 (Figure 3A) and interphase FISH showed that most of the cells (90.1%) have 2 green signals for SMO and 2 red signals for centromere 7 indicating the presence of 2 chromosome 7 and 2 copies of SMO. Few cells (3.5%) showed 2 SMO signals and 3 centromeric signals indicating that a subset of cells with trisomy 7 has a potential deletion/translocation involving the long arm (q arm) on chromosome 7 (Figure 3B). SKY analysis confirmed the presence of trisomy 7, but with a derivative 7 having the long arm replaced with DNA material from another chromosome (Figure 3C). DOHH2 cells showed trisomy 7 by G-banding and FISH studies showed that 91% of the cells had 3 green and red signals confirming the presence of trisomy 7 and 3 copies of SMO (Figure 4A). HBL1 cells showed that one entire chromosome 7 moved to a derivative chromosome 12. Metaphase and interphase FISH studies confirmed that SMO was not lost as a consequence of this chromosomal translocation as virtually 100% of the cells had 2 red signals for SMO (Figure 4B). FISH studies also showed that 88% of the OCI-LY3 cells displayed 3 green signals (GLI1) and 7% of the cells had 4 green signals, that were associated with identical number (1:1 ratio) of centromeric (red) signals (Figure 4C).
Figure 3. G-banding karyotype, FISH and SKY for BJAB cells.
G-banding showed 3 chromosomes 7 and 2 chromosomes 12 in BJAB cells. However, dualcolor interphase FISH showed that 90% of the cells have 2 green signals for SMO and 2 red signals for centromere 7 (ratio 1:1) indicating the presence of 2 chromosome 7 and 2 copies of SMO. Few cells showed 2 SMO signals and 3 centromeric signals indicating that a subset of cells has trisomy 7 and a deletion involving the long arm on chromosome 7 (not shown). SKY analysis confirmed the presence of trisomy 7 with one derivative 7 having its long arm replaced with DNA from another chromosome.
Figure 4. G-banding karyotype and FISH assays for DOHH2, HBL1 and OCI-LY3 cells.
In DOHH2 cells, G-banding showed trisomy 7 and 2 chromosomes 12. Dual-color interphase FISH confirmed the presence of 3 copies of SMO (ratio 1:1 with the centromere probe) and 2 copies of GLI1 in this cell line. In HBL1 cells, G-banding showed one intact chromosome 7 and a der(12) having the other chromosome 7. The presence of 2 copies of SMO gene was confirmed by interphase and metaphase FISH assays. In OCI-Ly3, G-banding show trisomy 7 and 12 and dual-color interphase and metaphase FISH studies confirmed that the presence of extra copies of SMO and GLI1 genes were associated with chromosomal aneuploidies (ratio 1:1 with centromere probes).
Altogether, these results suggest that DLBCL cell lines carry extra copies GLI1 and SMO genes, and that these extra copies are due to chromosomal aneuploidy rather than true gene amplification.
Evidence of synergistic cross talk between HH signaling with PI3K/AKT and NF-kB
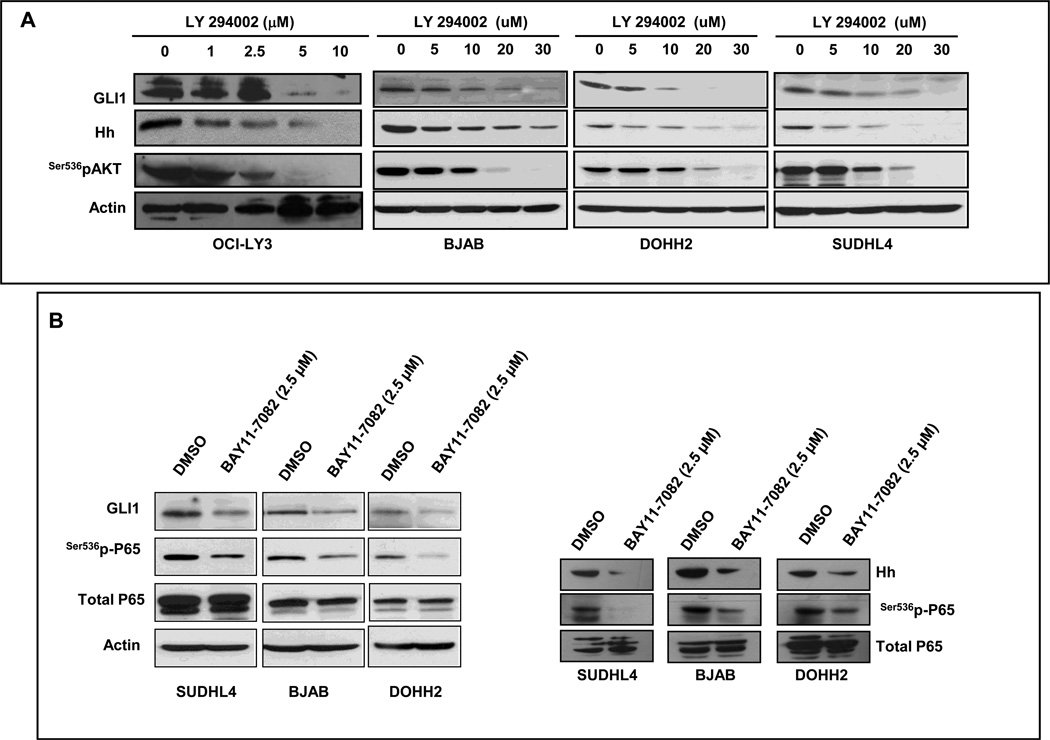
Recent evidence suggest that activation of Hh signaling in some cancers may result, at least in part, from integration of multiple deregulated oncogenic signaling inputs in the final signaling effectors of the Hh pathway [19]. To investigate if this may also apply to DLBCL, we inhibited 2 relevant oncogenic pathways known to be activated in DLBCL and assessed the effect of this inhibition on the expression levels of Hh-related genes. We measured the protein levels of GLI1, which reflects the activation status of the pathway, as well as the levels of Hh ligands. For these experiments we used 2 drugs, LY294002 and BAY11-7082, which are well-established inhibitors of PI3K and NF-KB signaling pathways, respectively. BAY11-7082 selectively and irreversibly inhibits the phosphorylation of IκBα, resulting in inhibition of NF-κB activation.
Treatments with the PI3K inhibitor, LY294002, led to a concentration dependent decrease of GLI1 and Hh protein levels in OCI-LY3, BJAB, DOHH2, and SUDHL4 cells (Figure 5A) as well as decreased Hh ligands secreted into the culture medium (Supplementary Figure 1). Similarly, inhibition of NF-kB signaling using BAY11-7082 resulted in decrease of GLI1 and Hh protein levels in SuDHL4, BJAB and DOHH2 associated with a decreased in the phosphorylation levels of P65 at ser536 (Figure 5B, left panel). It is accepted that phosphorylation of ser536 of P65 represents an active mark for NF-kB activation (mainly canonical, but also non-canonical). These data support a crosstalk between PI3K/AKT and NF-kB pathways with Hh signaling and that the activation level of Hh signaling may be modulated by the activation status of these two oncogenic pathways.
Figure 5. The activation status of PI3K and NF-kB modulates GLI1 and Hh levels in DLBCL cell lines.
(A) OCI-LY3, BJAB, DOHH2 and SUDHL4 cells were treated with increasing concentrations of the PI3K inhibitor LY294002. Western blot analysis showed a concentration dependent decrease in the expression levels of GLI1 and Hh proteins (left panel). (B) Three DLBCL cell lines (SUDHL4, BJAB and DOHH2) cells were treated with 2.5 µM of BAY11-7082. In all the cell lines, treatments with BAY11-7082 resulted in decrease of expression levels of GLI1 associated with a decreased in the phosphorylated levels of p65 (ser536) but not total p65. In addition, treatments with 2.5 µM of BAY11-7082 also resulted in decrease in the levels of Hh protein in the three cell lines.
DISCUSSION
One mechanism that explains the aberrant activation of Hh signaling in some cancers is the excessive expression of Hh ligands (ligand-dependent activation of Hh signaling), resulting in autocrine and/or paracrine stimulation of cancer cells. Recently, we have found that Hh ligands are synthesized, processed and secreted by DLBCL cells and that the canonical Hh ligand-PTCH1-SMO-GLI axis is activated and functional in DLBCL cells providing evidence of the existence of an autocrine/paracrine Hh signaling loop in DLBCL [8, 14, 21] Moreover, we also found evidence that a significant degree of Hh activation in DLBCL cell lines seem to be independent of the stimulus provided by Hh ligands [8]. Silencing the expression of various Hh ligands using siRNA approaches as well as blocking Hh ligands using 5E1 resulted only in a modest decrease of mRNA and proteins levels of GLI1 in DLBCL cell lines [8].
However, causative factors that might explain the activation of Hh signaling, independently of Hh ligands (Shh, Dhh and Ihh), in DLBCL are unknown. One possibility is the presence of gene amplifications involving Hh-related genes. Another mechanism that may explain Hh activation in DLBCL are activating or inactivating mutations involving Hh related genes. However, two recent studies analyzing the coding genome of DLBCL did not find mutations involving Hh signaling genes [22, 23].
Hedge et al, have also reported Hh activation in a subset of mantle cell lymphoma cell lines that seem to be independent of the stimulus provided by Hh ligands and/or SMO and suggested the possibility of other mechanisms responsible for Hh activation such as gene mutations of Hh-related genes or cross talking between Hh signaling with other activated oncogenic signaling pathways [11].
Here, we explored if genetic amplifications of Hh-related genes, in particular those genes responsible for fostering endogenous pathway activity, are found in DLBCL cells. Our data indicate the presence of few extra copies of GLI1 and SMO in DLBCL cell lines and GLI1 and DHH in DLBCL tumors. However, these extra copies seem to be caused by chromosomal aneuploidies but not by true gene amplification.
Activation of canonical Hh signaling pathway results in increased expression of GLI1 and gene expression profile studies have shown that GLI1 is mainly a transcriptional activator involved in the regulation of genes involved in cell cycle control including cyclin D1, D2 and E1 among others [4, 24]. Although we did not detect significant gene amplifications per se, it is likely that the presence of small number of extra copies of GLI1 may represent a biological advantage for the tumor cells. It is known that oncogene amplifications have importance for the biological behavior, prognosis, and response to treatment in cancers. For example, amplifications of MYC and ERBB2 genes in breast carcinoma have been reported to be significantly associated with high tumor grade and poor prognosis [25]. It has been also found that the presence of few extra copies of some oncogenes, due to chromosomal aneuploidy, also have consequences in the tumor biology and prognosis of DLBCL. For example, in DLBCL the presence of few extra copies of MYC (e.g. due to trisomy 8) or BCL2 (e.g. due to trisomy 18), when associated to other genetic aberrancies, such as the t(14;18) or MYC gene rearrangements, respectively, result in tumors with aggressive clinical course and poor prognosis [26]. B-cell lymphomas with extra copies of MYC associated with the t(14;18) or MYC gene rearrangement associated with extra copies of BCL2 or with just an extra chromosome 18 have been reported [26]. These patients were described to have aggressive clinical or laboratory findings, including high International Prognostic Index, high serum lactate dehydrogenase levels and poor prognosis. Snuderl and colls [27] described one patient with histologically aggressive B-cell lymphoma associated with MYC translocation and eight copies of BCL2 who also showed an aggressive clinical behavior.
DLBCL, mantle cell lymphoma and anaplastic large cell lymphoma express some of the main components of Hh signaling such as SMO, GLI1 and GLI2 [9,11,14]. Other lymphomas such as classical Hodgkin lymphoma consistently express GLI3 but not GLI1 or GLI2 [12]. Inappropriate activation of Hh signaling appears to have an important role in cell proliferation, survival and enhances tolerance to chemotherapeutic agents in malignant lymphomas as inhibition of Hh signaling results in cell cycle arrest, apoptosis and decrease of chemotolerance (reviewed in [10] and [13]). Effective inhibitors of SMO are currently available and inhibitors of GLI1 are being developed, both inhibitors could be the basis for novel therapeutic strategies for malignant lymphomas as well as for DLBCL. However, it would be important to understand the contribution of SMO and GLI1 and the contribution of Hh-ligand dependent and independent mechanisms in the pathobiology of these neoplasms.
In this study, we provide evidence of crosstalk between PI3K/AKT and NF-kB pathways with Hh signaling and that these cross talks (as well as others) may contribute to the inappropriate activation of Hh signaling pathway in DLBCL.
A possible link between PI3K/AKT and Hh signaling has been previously suggested by others as well as by us [9, 28]. Several growth factors including platelet-derived growth factor, epidermal growth factor and insulin-like growth factor 1 has been described that increase activation of Hh signaling that is dependent of activation of the PI3K/AKT pathway [24]. Previously, we have shown that adenoviral delivery of activated AKT up-regulate the expression of Hh ligand and vice versa inhibition of PI3K/AKT results in inhibition of Hh signaling in ALK-positive anaplastic large cell lymphoma (ALCL) [9]. In addition, genetic studies in mice have revealed that the PI3K/AKT pathway provides a synergistic signal for Hh pathway-induced tumor formation [29]. However, the precise mechanism by which PI3K/AKT regulates Hh signaling remains unclear.
Several lines of experimental evidence also support cross talk between NF-kB and Hh signaling. For example, during avian embryonic limb formation, inhibition of NF-kB activity in limb mesenchyma led to reduced expression of SHH [30]. Nakashima and colleagues [31] reported that blockage of NF-kB signaling suppressed constitutive expression of SHH RNA in pancreatic cancer cells. Moreover, Kasperczyk and colleagues [32] have provided evidence that SHH is a NF-kB target gene (under direct control of NF-kB) and promotes NF-kB mediated apoptosis resistance and tumor growth.
In summary, we showed that Hh signaling activation in DLBCL is not due to amplifications of the main genes involved in this signaling pathway. Our data support the hypothesis that activation of Hh signaling in DLBCL and in cancer in general may result, at least in part, from integration/crosstalk of multiple deregulated oncogenic signaling inputs in the Hh signaling pathway [19]. This view of interconnectivity between oncogenic pathways has important implications for understanding how the major oncogenic pathways interact in malignant tumors in general, and in DLBCL in particular, and also for the development of more effective and focused anticancer therapies.
Supplementary Material
Acknowledgements
This work was supported by funds from The Translational Grant of The Leukemia & Lymphoma Society (to RS and FV), and K08 Physician-Scientist Award 1 K08 CA143151-01 (NIH) (to FV). The primary tumor samples were provided by the Hematopathology Tissue Bank of the UT MD Anderson Cancer Center (supported by the NCI/NIH Grant CA16672). The cytogenetic studies were performed with the colaboration of the Genetic Core laboratory supported by the Cancer Center Core grant CA016672 by the NCI.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing Interests: The authors have declared that no competing interest exist.
Contributions: ER was involved in data collection and writing the manuscript. KK, YL, CQ and JJ performed the experiments. RRS and PCH were involved in the research plan design. AM, PAL, MH, SD, JG and SJ were involved in data analysis. FV was involved in plan design, data analysis and discussion of the manuscript.
REFERENCES
- 1.McMahon AP, Ingham PW, Tabin CJ. Developmental roles and clinical significance of hedgehog signaling. Curr Top Dev Biol. 2003;53:1–114. doi: 10.1016/s0070-2153(03)53002-2. [DOI] [PubMed] [Google Scholar]
- 2.Johnson RL, Scott MP. New players and puzzles in the Hedgehog signaling pathway. Curr Opin Genet Dev. 1998;8:450–6. doi: 10.1016/s0959-437x(98)80117-2. [DOI] [PubMed] [Google Scholar]
- 3.Murone M, Rosenthal A, de Sauvage FJ. Hedgehog signal transduction: from flies to vertebrates. Exp Cell Res. 1999;253:25–33. doi: 10.1006/excr.1999.4676. [DOI] [PubMed] [Google Scholar]
- 4.Eichberger T, Sander V, Schnidar H, Regl G, Kasper M, Schmid C, et al. Overlapping and distinct transcriptional regulator properties of the GLI1 and GLI2 oncogenes. Genomics. 2006;87:616–632. doi: 10.1016/j.ygeno.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 5.Guessous F, Li Y, Abounader R. Signaling pathways in medulloblastoma. J Cell Physiol. 2008;217:577–583. doi: 10.1002/jcp.21542. [DOI] [PubMed] [Google Scholar]
- 6.Pasca di Magliano M, Hebrok M. Hedgehog signalling in cancer formation and maintenance. Nat Rev Cancer. 2003;3:903–911. doi: 10.1038/nrc1229. [DOI] [PubMed] [Google Scholar]
- 7.Karhadkar SS, Bova GS, Abdallah N, Dhara S, Gardner D, Maitra A, et al. Hedgehog signalling in prostate regeneration, neoplasia and metastasis. Nature. 2004;431:707–712. doi: 10.1038/nature02962. [DOI] [PubMed] [Google Scholar]
- 8.Singh RR, Kim JE, Davuluri Y, Drakos E, Cho-Vega JH, Amin HM, et al. Hedgehog signaling pathway is activated in diffuse large B-cell lymphoma and contributes to tumor cell survival and proliferation. Leukemia. 2010;24:1025–1036. doi: 10.1038/leu.2010.35. [DOI] [PubMed] [Google Scholar]
- 9.Singh RR, Cho-Vega JH, Davuluri Y, Ma S, Kasbidi F, Milito C, et al. Sonic hedgehog signaling pathway is activated in ALK-positive anaplastic large cell lymphoma. Cancer Res. 2009;69:2550–258. doi: 10.1158/0008-5472.CAN-08-1808. [DOI] [PubMed] [Google Scholar]
- 10.Irvine DA, Copland M. Targeting hedgehog in hematologic malignancy. Blood. 2012;119:2196–2204. doi: 10.1182/blood-2011-10-383752. [DOI] [PubMed] [Google Scholar]
- 11.Hegde GV, Munger CM, Emanuel K, Joshi AD, Greiner TC, Weisenburger DD, et al. Targeting of sonic hedgehog-GLI signaling: a potential strategy to improve therapy for mantle cell lymphoma. Mol Cancer Ther. 2008;7:1450–1460. doi: 10.1158/1535-7163.MCT-07-2118. [DOI] [PubMed] [Google Scholar]
- 12.Greaves WO, Kim JE, Singh RR, Drakos E, Kunkalla K, Sanchez-Espiridion B, et al. Glioma-associated oncogene homologue 3, a hedgehog transcription factor, is highly expressed in Hodgkin and Reed-Sternberg cells of classical Hodgkin lymphoma. Hum Pathol. 2011;42:1643–1652. doi: 10.1016/j.humpath.2010.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ok CY, Singh RR, Vega F. Aberrant activation of the hedgehog signaling pathway in malignant hematological neoplasms. Am J Pathol. 2012;180:2–11. doi: 10.1016/j.ajpath.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim JE, Singh RR, Cho-Vega JH, Drakos E, Davuluri Y, Khokhar FA, et al. Sonic hedgehog signaling proteins and ATP-binding cassette G2 are aberrantly expressed in diffuse large B-cell lymphoma. Mod Pathol. 2009;22:1312–1320. doi: 10.1038/modpathol.2009.98. [DOI] [PubMed] [Google Scholar]
- 15.Vega F, Cho-Vega JH, Lennon PA, Luthra MG, Bailey J, Breeden M, et al. Splenic marginal zone lymphomas are characterized by loss of interstitial regions of chromosome 7q, 7q31.32 and 7q36.2 that include the protection of telomere 1 (POT1) and sonic hedgehog (SHH) genes. Br J Haematol. 2008;142:216–226. doi: 10.1111/j.1365-2141.2008.07176.x. [DOI] [PubMed] [Google Scholar]
- 16.Rao PH, Houldsworth J, Dyomina K, Parsa NZ, Cigudosa JC, Louie DC, et al. Chromosomal and gene amplification in diffuse large B-cell lymphoma. Blood. 1998;92:234–240. [PubMed] [Google Scholar]
- 17.Bea S, Colomo L, Lopez-Guillermo A, Salaverria I, Puig X, Pinyol M, et al. Clinicopathologic significance and prognostic value of chromosomal imbalances in diffuse large B-cell lymphomas. J Clin Oncol. 2004;22:3498–3506. doi: 10.1200/JCO.2004.11.025. [DOI] [PubMed] [Google Scholar]
- 18.Sander S, Bullinger L, Leupolt E, Benner A, Kienle D, Katzenberger T, et al. Genomic aberrations in mantle cell lymphoma detected by interphase fluorescence in situ hybridization. Incidence and clinicopathological correlations. Haematologica. 2008;93:680–687. doi: 10.3324/haematol.12330. [DOI] [PubMed] [Google Scholar]
- 19.Stecca B, Ruiz IAA. Context-dependent regulation of the GLI code in cancer by HEDGEHOG and non-HEDGEHOG signals. J Mol Cell Biol. 2010;2:84–95. doi: 10.1093/jmcb/mjp052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vega F, Medeiros LJ, Leventaki V, Atwell C, Cho-Vega JH, Tian L, et al. Activation of mammalian target of rapamycin signaling pathway contributes to tumor cell survival in anaplastic lymphoma kinase-positive anaplastic large cell lymphoma. Cancer Res. 2006;66:6589–6597. doi: 10.1158/0008-5472.CAN-05-3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Singh RR, Kunkalla K, Qu C, Schlette E, Neelapu SS, Samaniego F, et al. ABCG2 is a direct transcriptional target of hedgehog signaling and involved in stroma-induced drug tolerance in diffuse large B-cell lymphoma. Oncogene. 2011;30:4874–4886. doi: 10.1038/onc.2011.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pasqualucci L, Trifonov V, Fabbri G, Ma J, Rossi D, Chiarenza A, et al. Analysis of the coding genome of diffuse large B-cell lymphoma. Nat Genet. 2011;43:830–837. doi: 10.1038/ng.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morin RD, Mendez-Lago M, Mungall AJ, Goya R, Mungall KL, Corbett RD, et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011;476:298–303. doi: 10.1038/nature10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vestergaard J, Lind-Thomsen A, Pedersen MW, Jarmer HO, Bak M, Hasholt L, et al. GLI1 is involved in cell cycle regulation and proliferation of NT2 embryonal carcinoma stem cells. DNA Cell Biol. 2008;27:251–256. doi: 10.1089/dna.2007.0625. [DOI] [PubMed] [Google Scholar]
- 25.Al-Kuraya K, Schraml P, Torhorst J, Tapia C, Zaharieva B, Novotny H, et al. Prognostic relevance of gene amplifications and coamplifications in breast cancer. Cancer Res. 2004;64:8534–8540. doi: 10.1158/0008-5472.CAN-04-1945. [DOI] [PubMed] [Google Scholar]
- 26.Li S, Lin P, Fayad LE, Lennon PA, Miranda RN, Yin CC, et al. B-cell lymphomas with MYC/8q24 rearrangements and IGH@BCL2/t(14;18)(q32;q21): an aggressive disease with heterogeneous histology, germinal center B-cell immunophenotype and poor outcome. Mod Pathol. 2012;25:146–156. doi: 10.1038/modpathol.2011.147. [DOI] [PubMed] [Google Scholar]
- 27.Snuderl M, Kolman OK, Chen YB, Hsu JJ, Ackerman AM, Dal Cin P, et al. B-cell lymphomas with concurrent IGH-BCL2 and MYC rearrangements are aggressive neoplasms with clinical and pathologic features distinct from Burkitt lymphoma and diffuse large B-cell lymphoma. Am J Surg Pathol. 2010;34:327–340. doi: 10.1097/PAS.0b013e3181cd3aeb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang L, Wang Y, Mao H, Fleig S, Omenetti A, Brown KD, et al. Sonic hedgehog is an autocrine viability factor for myofibroblastic hepatic stellate cells. J Hepatol. 2008;48:98–106. doi: 10.1016/j.jhep.2007.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rao G, Pedone CA, Del Valle L, Reiss K, Holland EC, Fults DW. Sonic hedgehog and insulin-like growth factor signaling synergize to induce medulloblastoma formation from nestin-expressing neural progenitors in mice. Oncogene. 2004;23:6156–6162. doi: 10.1038/sj.onc.1207818. [DOI] [PubMed] [Google Scholar]
- 30.Bushdid PB, Brantley DM, Yull FE, Blaeuer GL, Hoffman LH, Niswander L, et al. Inhibition of NF-kappaB activity results in disruption of the apical ectodermal ridge and aberrant limb morphogenesis. Nature. 1998;392:615–618. doi: 10.1038/33435. [DOI] [PubMed] [Google Scholar]
- 31.Nakashima H, Nakamura M, Yamaguchi H, Yamanaka N, Akiyoshi T, Koga K, et al. Nuclear factor-kappaB contributes to hedgehog signaling pathway activation through sonic hedgehog induction in pancreatic cancer. Cancer Res. 2006;66:7041–7049. doi: 10.1158/0008-5472.CAN-05-4588. [DOI] [PubMed] [Google Scholar]
- 32.Kasperczyk H, Baumann B, Debatin KM, Fulda S. Characterization of sonic hedgehog as a novel NF-kappaB target gene that promotes NF-kappaB-mediated apoptosis resistance and tumor growth in vivo. FASEB J. 2009;23:21–33. doi: 10.1096/fj.08-111096. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.