Abstract
During development, progenitors that are committed to differentiate into oligodendrocytes, the myelinating cells of the central nervous system (CNS), are generated within discrete regions of the neuroepithelium. More specifically, within the developing spinal cord and hindbrain ventrally located progenitor cells that are characterized by the expression of the transcription factor olig2 give temporally rise to first motor neurons and then oligodendrocyte progenitors. The regulation of this temporal neuron-glial switch has been found complex and little is known about the extrinsic factors regulating it. Our studies described here identified a zebrafish ortholog to mammalian atx, which displays evolutionarily conserved expression pattern characteristics. Most interestingly, atx was found to be expressed by cells of the cephalic floor plate during a time period when ventrally-derived oligodendrocyte progenitors arise in the developing hindbrain of the zebrafish. Knock-down of atx expression resulted in a delay and/or inhibition of the timely appearance of oligodendrocyte progenitors and subsequent developmental stages of the oligodendrocyte lineage. This effect of atx knock-down was not accompanied by changes in the number of olig2-positive progenitor cells, the overall morphology of the axonal network or the number of somatic abducens motor neurons. Thus, our studies identified Atx as an extrinsic factor that is likely secreted by cells from the floor plate and that is involved in regulating specifically the progression of olig2-positive progenitor cells into lineage committed oligodendrocyte progenitors.
Keywords: myelination, glia differentiation, CNS development, floor plate, zebrafish
INTRODUCTION
During development, oligodendrocytes, the myelinating cells of the vertebrate central nervous systems (CNS), are generated from neural progenitor cells that are located within distinct regions of the neuroepithelium (Miller, 2005; Richardson et al., 2006; Rowitch and Kriegstein, 2010). The most direct known domain of progenitor cells giving rise to ventrally-derived oligodendrocytes in the developing spinal cord and hindbrain is composed of progenitor cells that sequentially generate first motor neurons and then oligodendrocytes (Richardson et al., 2000; Rowitch et al., 2002; Wu et al., 2006) and that can be identified via the expression of the transcription factor olig2 (Lu et al., 2002; Mukouyama et al., 2006; Park et al., 2002; Takebayashi et al., 2002; Zannino and Appel, 2009; Zhou and Anderson, 2002). Despite extensive research, however, the exact mechanisms and factors determining the timely appearance of oligodendrocyte progenitors from olig2-positive progenitor cells are currently not fully understood.
A well characterized factor regulating early development of both motor neurons and ventrally-derived oligodendrocyte progenitors is sonic hedgehog (Shh), which is expressed and secreted by cells of the floor plate (Jessell, 2000; Orentas and Miller, 1996; Park et al., 2004; Poncet et al., 1996; Pringle et al., 1996). The floor plate is, however, known to release extracellular factors other than Shh (Placzek and Briscoe, 2005), suggesting that it may contribute to additional developmental mechanisms, possibly including the regulation of oligodendrocyte development via a yet uncharacterized floor plate-derived signal.
In our previous studies we identified autotaxin (Atx), also known as Enpp2, phosphodiesterase-Iα/Atx or lysoPLD, as an extracellular factor that promotes differentiation of particularly the later stages of the oligodendrocytes lineage (Dennis et al., 2008; Fox et al., 2004; Fox et al., 2003; Yuelling and Fuss, 2008). More specifically, Atx was found to promote morphological maturation of differentiating oligodendrocytes via its C-terminal domain, referred to as the modulator of oligodendrocyte remodeling and focal adhesion organization (MORFO) domain. As such regulator of oligodendrocyte maturation, Atx is thought to act as an autocrine signal since it is expressed and secreted by differentiating oligodendrocytes. It is worth mentioning that while Atx has been originally thought to be a proteolytically cleavable transmembrane protein, it is now well established to represent a bona fide secretory protein (Jansen et al., 2005; Koike et al., 2006). During embryonic development, particularly during the developmental time period when ventrally-derived oligodendrocyte progenitors are generated (Pringle and Richardson, 1993; Richardson et al., 2000; Sussman et al., 2000), atx was found in the mouse to be expressed by cells of the floor plate (Bachner et al., 1999). This spatio-temporal expression raises the possibility that Atx may play an additional paracrine role in regulating the initial stages of oligodendrocyte development. Knock-out mice for atx have been generated. However, they are characterized by an early embryonic lethal phenotype due to severe vascular defects, thus rendering them uninformative with regard to a potential in vivo role of atx in regulating oligodendrocyte development (Ferry et al., 2007; Fotopoulou et al., 2010; Koike et al., 2009; Tanaka et al., 2006; van Meeteren et al., 2006).
Here, the in vivo role of atx in oligodendrocyte development was assessed using the zebrafish as a model system. The choice of the model system was based on the notion that zebrafish embryos can develop independent from a fully functional vascular system for up to seven days post fertilization (Jin et al., 2007; Ny et al., 2006; Stainier, 2001), a time frame that is sufficient for investigating oligodendrocyte development (Brosamle and Halpern, 2002; Buckley et al., 2010; Kirby et al., 2006; Park et al., 2002; Schebesta and Serluca, 2009). First, a zebrafish ortholog to mammalian atx was identified and found to display evolutionarily conserved expression pattern characteristics. Then, using morpholino antisense oligonucleotide-mediated gene silencing, our studies revealed that in the developing zebrafish hindbrain atx promotes the differentiation of oligodendrocyte progenitors from olig2-positive progenitor cells. Furthermore, our findings suggest that this novel functional role of atx occurs by a paracrine mechanism with cells of the floor plate as the most likely source for secreted Atx.
MATERIALS AND METHODS
Zebrafish Strains and Care
Wildtype embryos of the AB strain were obtained through natural matings, raised at 28.5°C and staged according to morphological criteria and hours post fertilization (hpf) (Kimmel et al., 1995).
Sequence Analysis and cDNA Cloning
Sequence data for zebrafish atx (ZDB-GENE-040426-1156, NM_200603.1) were obtained using NCBI’s Basic Local Alignment Search Tool (BLAST; Sayers et al., 2011). Amino acid sequence alignments and phylogenic guide trees were generated using the Vector NTI software package (Invitrogen, Carlsbad, CA). The authors would like to note that while zebrafish atx is recorded under the gene symbol enpp2, the symbol atx will be used throughout the manuscript since this is currently the most commonly used designation.
For the generation of gene-specific cRNA probes the following plasmid constructs were used: atx: a 1843 bp ClaI-KpnI fragment containing the 3’ 1812 bps of the 2552 bp atx coding region was excised from the I.M.A.G.E. clone 3816628 (Open Biosystems, Huntsville, AL) and inserted into pBluescript (Agilent Technologies/Stratagene, La Jolla, CA). olig1 (ZDB-GENE-050107-2): a 674 bp fragment covering most of the 708 bp olig1 coding region was amplified from zebrafish embryo RNA (3 dpf). The resulting amplification product was cloned into pBluescript. shhb (ZDB-GENE-980526-41): a 1990 bp fragment containing the entire shhb coding region was amplified from I.M.A.G.E. clone 7149635 (ATCC, Manassas, VA) and a T3 RNA polymerase binding site was introduced at the 3’ end. In addition, previously described plasmid constructs were used for the following genes: foxa1 (ZDB-GENE-990415-78): Odenthal and Nusslein-Volhard, 1998, foxa2 (ZDB-GENE-980526-404): Strahle et al., 1993, mbp (ZDB-GENE-030128-2): Brosamle and Halpern, 2002, olig2 (ZDB-GENE-030131-4013): Park et al., 2002, sox10 (ZDB-GENE-011207-1): Dutton et al., 2001.
Whole-Mount In Situ Hybridization and Immunohistochemistry
Embryos were fixed in 4% paraformaldehyde in PBS overnight at 4°C and stored in methanol at −20°C for at least 1 day. Colorimetric in situ hybridizations using digoxigenin-labeled antisense cRNA probes were performed by standard methods (Thisse and Thisse, 2008). Fluorescent immunostainings using the anti-neurofilament M antibody RMO44 (Invitrogen, Carlsbad, CA) and the anti-Neurolin/DM-GRASP antibody Zn-8 (Developmental Studies Hybridoma Bank, Iowa City, Iowa) were performed in principle as described by Waskiewicz et al. (2001) and Zannino and Appel (2009), respectively.
In situ hybridized and immunostained embryos were imaged either as whole-mounts or cryoprotected (30% sucrose/PBS), embedded in Tissue-Tek O.C.T. Compound (Sakura Finetek, Torrance, CA) and cryosectioned (20 µm; Cryotome Cryostat; Shandon, Inc., Pittsburgh, PA). Embryos and sections of colorimetric in situ hybridizations were mounted in 90% glycerol/PBS. Images were acquired using either the extended focus module of the axiovision software package in combination with an Axio Observer Z.1 or SteREO Discovery.V20 microscope equipped with an AxioCam MRc digital camera (Carl Zeiss MicroImaging, Inc., Thornwood, NY) or an Olympus SZX12 stereomicroscope equipped with a DP70 digital camera (Olympus, Center Valley, PA). Fluorescently immunostained embryos were mounted in Vectashield mounting medium (Vector laboratories, Burlingame, CA) and images were acquired using a Zeiss LSM 510 META NLO laser scanning microscope (Carl Zeiss MicroImaging, Thornwood, NY). Once captured, images were imported into Adobe Photoshop and adjustments were limited to contrast, levels, color matching settings, and cropping. For quantification of RMO44-immuno-positive pixels 2D maximum projections of confocal Z stacks of 5.66 µm optical sections were analyzed without prior image adjustments using IPLab imaging software (BioVision Technologies, Exton, PA).
Morpholino Oligonucleotide and mRNA Injections
Two antisense morpholino oligonucleotides (MOs) targeting atx were designed and synthesized by GeneTools (Philomath, OR) in accordance with the published zebrafish genome sequence (Ensembl entry ENSDART00000047920): atx TL MO (5’-TGCGTCTGGTGGCTCTCTTCCACAC-3’) was designed to target the atx translation start site, while atx E2I2 MO (5’-AAGAAGCATCCTACTTTTTGAGAGC-3’) was designed to target the exon 2-intron 2 splice site of atx. As controls, 5 base pair mismatch MOs were used: atx TL control MO (5’-TGCGTGTGGTGCCTGTCTTGCAGAC-3’) and atx E2I2 control MO (5’-AACAAGGATCGTACTTTTTGACACC-3’). MOs were reconstituted in H2O and diluted in 1× Danieau buffer (58 mM NaCl, 0.7 mM KCl,. 0.4 mM MgSO4, 0.6 mM Ca(NO3)2, 5.0 mM HEPES pH 7.6). 1 nl was injected into the yolk of one- to four-cell-stage embryos. atx TL MO and atx TL control MO were injected at a concentration of 1 mg/ml, while atx E2I2 MO and atx E2I2 control MO were used at a concentration of 10 mg/ml. To confirm that phenotypes observed upon MO injections were not due to Tp53-mediated activation of cell death pathways, an often observed MO off-target effect (Gerety and Wilkinson, 2011; Robu et al., 2007), atx TL MO injected and uninjected embryos were immunostained at 24 hpf with an anti-cleaved caspase-3 antibody. No significant difference in the number of cleaved caspase-3-positive cells was observed (data not shown).
For mRNA rescue experiments 5′ capped and 3’ polyadenylated atx sense RNA was synthesized from a full-length rat-derived cDNA cloned into the vector pEF/V5-His (Dennis et al., 2008) using mMessage mMachine T7 and Poly(A) Tailing kits (Life Technologies/Ambion, Grand Island, NY). 1 nl of mRNA (20 ng/µl) was injected into the yolk of one- to four-cell-stage embryos directly following the injection of the atx TL MO.
Western Blot Analysis
Embryos were deyolked and homogenized in lysis buffer (40 mM NaCl, 40 mM HEPES (pH 7.4), 10 mM EDTA, 0.1% SDS, 1% Triton) including the cOmplete mini protease inhibitor cocktail (Roche Diagnostics Corp., Indianapolis, IN). Lysates were centrifuged at 10,000×g for 5 min and supernatants were collected. Proteins were resolved by sodium dodecyl sulfate polyacrylamide gel electrophoresis and transferred to Immobilon-P PDVF membranes (Millipore, Billerica, MA) in principle as previously described (Fuss et al., 2000). Polyclonal anti-Atx antibodies were generated against a peptide representing amino acids 827–840 (RRTSRTYEEILALK) of the zebrafish Atx protein sequence (EZBiolab, Inc., Westfield, IN) and used at a dilution of 1:10,000. These zebrafish-specific antibodies recognized in whole zebrafish lysates protein forms with apparent molecular weights of 100 kD and 125/135 kD (Fig. 3A), which is consistent with the molecular weights described for unmodified and post-translationally modified Atx protein forms, respectively (Jansen et al., 2007; Pradere et al., 2007). In contrast, none of the alternatively spliced exon sequences found in higher vertebrates (Giganti et al., 2008) were found present in the published zebrafish genomic atx sequences. The binding to all of the above protein forms was inhibited by pre-incubation with the peptide used for antibody production. In addition, polyconal anti-Atx antibodies raised against a peptide representing amino acids 573–588 (KNKLEELNKRLHTKGS) of the rat Atx protein sequence (Cayman Chemical Company, Ann Arbor, MI) were used at a dilution of 1:1,000. These antibodies recognized, similar to the above anti-zebrafish Atx generated antibodies, proteins with apparent molecular weights of 100 kD and 125/135 kD (Fig. 6A). Anti-β-tubulin antibodies (1:1000; Sigma-Aldrich, St. Louis, MO) were used for normalization. Bound primary antibodies were detected using HRP-conjugated secondary antibodies (1:10,000; Vector Laboratories, Burlingame, CA) in combination with ECL Plus Western blot detection reagents (GE Healthcare Life Sciences, Piscataway, NJ). Chemiluminescent signals were detected by exposure of photographic film (Kodak BioMax MR, Eastman Kodak Company, Rochester, NY) and quantified by densitometry using the ImageJ software package (Abramoff et al., 2004).
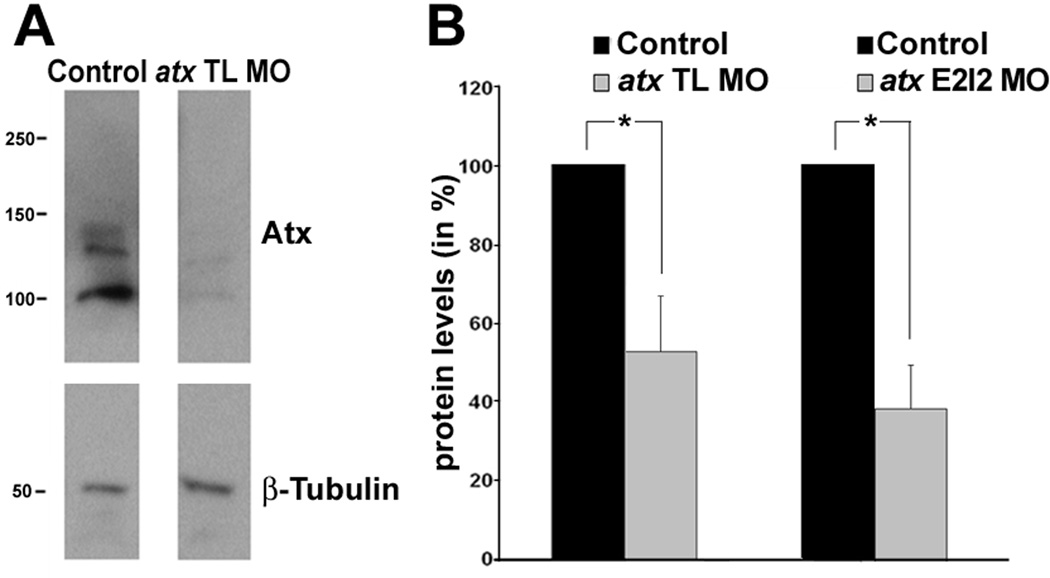
Fig. 3.
Injection of anti-atx morpholino oligonucleotides leads to a significant reduction in Atx protein levels. Embryos were injected with either an anti-atx translation blocking (atx TL MO) or an anti-atx splice site targeted (atx E2I2 MO) morpholino oligonucleotide and analyzed at 48 hpf. As controls, 5 base pair mismatch morpholino oligonucleotides were used. A: Representative Western blot depicting Atx protein levels. β-tubulin was used for normalization. Numbers on the left indicate molecular weights in kD. Protein forms with apparent molecular weights of 100 kD and 125/135 kD were detected and are most likely a result of differences in posttranslational modifications (Jansen et al., 2007; Pradere et al., 2007). B: Bar graphs depicting Atx protein levels as % of control (control = 100%). The graphs depict three independent experiments. Means and standard errors are shown. Stars indicate an overall significance level of p<0.05 (one-sample t-test).
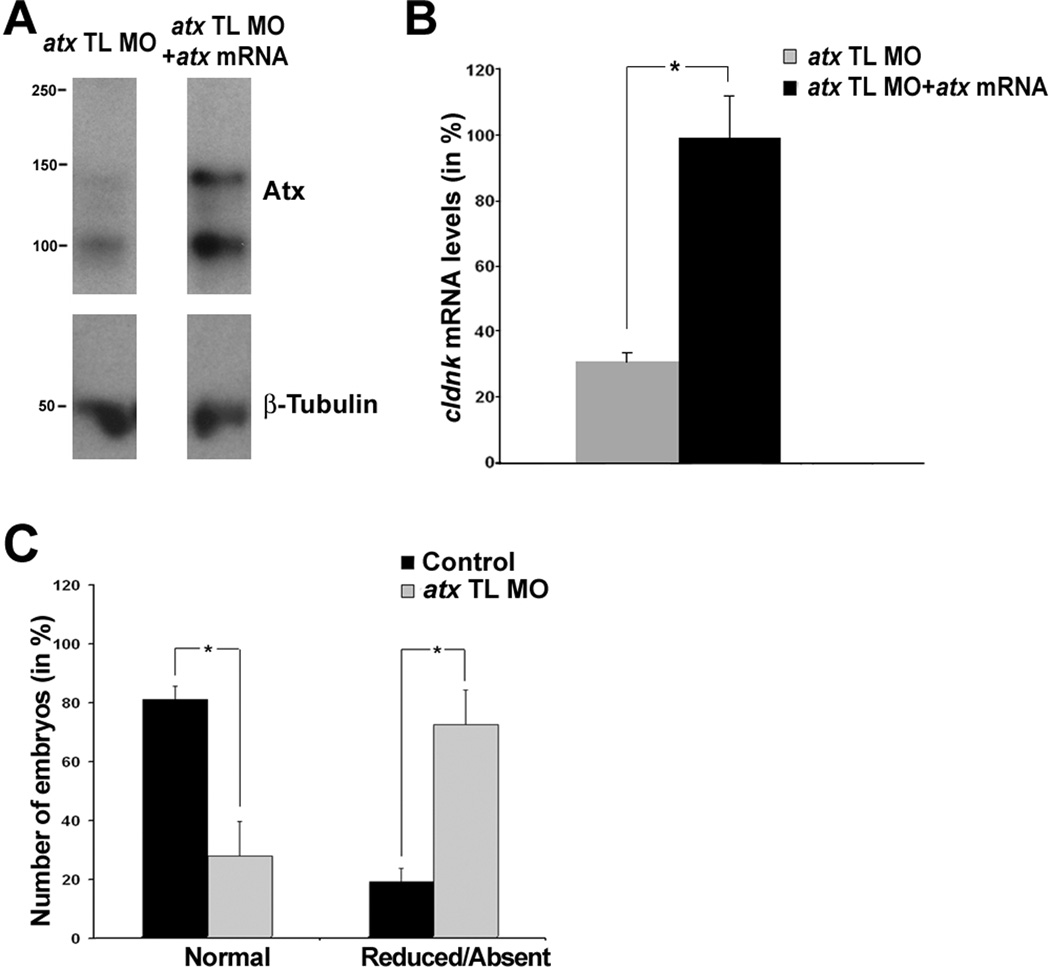
Fig. 6.
Co-injection of a synthetic atx mRNA leads to a rescue of the atx knock-down phenotype, and there is no apparent recovery from the atx knock-down-induced phenotype up to 72 hpf. A–B: Embryos were co-injected with atx TL MO and a synthetic atx mRNA and then analyzed at 48 hpf. As control, a 5 base pair mismatch morpholino oligonucleotide was used. A: Representative Western blot depicting Atx protein levels. β-tubulin was used for normalization. Numbers on the left indicate molecular weights in kD. B: Bar graph depicting cldnk mRNA levels in % (control = 100%) as determined by quantitative RT-PCR. Means and standard errors of four independent experiments are shown. The star indicates an overall two-tailed significance level of p<0.05 (Student’s t-test). No statistically significant difference in cldnk mRNA level was found between embryos injected with control MO versus atx TL MO plus atx mRNA (not shown). C: Embryos were treated as described in Fig. 3 and analyzed at 72 hpf. The bar graph depicts the number of embryos with normal or reduced/absent mbp mRNA expression in % (total number of embryos per condition = 100%). Means and standard errors of four independent experiments are shown. Stars indicate an overall two-tailed significance level of p<0.05 (Student’s t-test).
Quantitative RT-PCR
Total RNA samples were isolated from embryos using Trizol (Invitrogen, Carlsbad, CA) and treated with DNase using the DNA-Free kit (Applied Biosystems/Ambion, Austin, TX). Oligo(dT)-primed cDNAs were synthesized using the Superscript II RT kit (Invitrogen, Carlsbad, CA). Quantitative RT-PCR was performed on a Chromo4 (MJ Research, Inc., Waltham, MA) or CFX96 (BioRad, Hercules, CA) real-time PCR detection system using the iQ SYBR Green Supermix (BioRad, Hercules, CA). The following primer pairs were used at an annealing temperature of 58°C: mbp primer pair as described by Buckley et al. (2010); plp1b primer pair: Forward: 5’-TGCCATGCCAGGGGTTGTTTGTGGA-3’ and Reverse: 5’-GGCGACCATGTAAACGAACAGGGC-3’; cldnk primer pair: Forward: 5’-TGGCATTTCGGCTCAAGCTCTGGA-3’ and Reverse: 5’-GGTACAGACTGGGCAATGGACCTGA-3’; olig1 primer pair: Forward: 5’-CCGGTGTAGGGGGAGCACTGCA-3’ and Reverse 5-‘TCCGAGCCAGCACCAGTGTCGAG-3’. β-actin (Buckley et al., 2010) was used as reference gene and relative expression levels were determined using the ΔΔCT method (Livak and Schmittgen, 2001). Statistical significance was determined using the one-sample t-test (Dalgaard, 2008; Skokal and Rohlf, 1995).
RESULTS
An Ortholog of Mammalian atx is Present in the Zebrafish
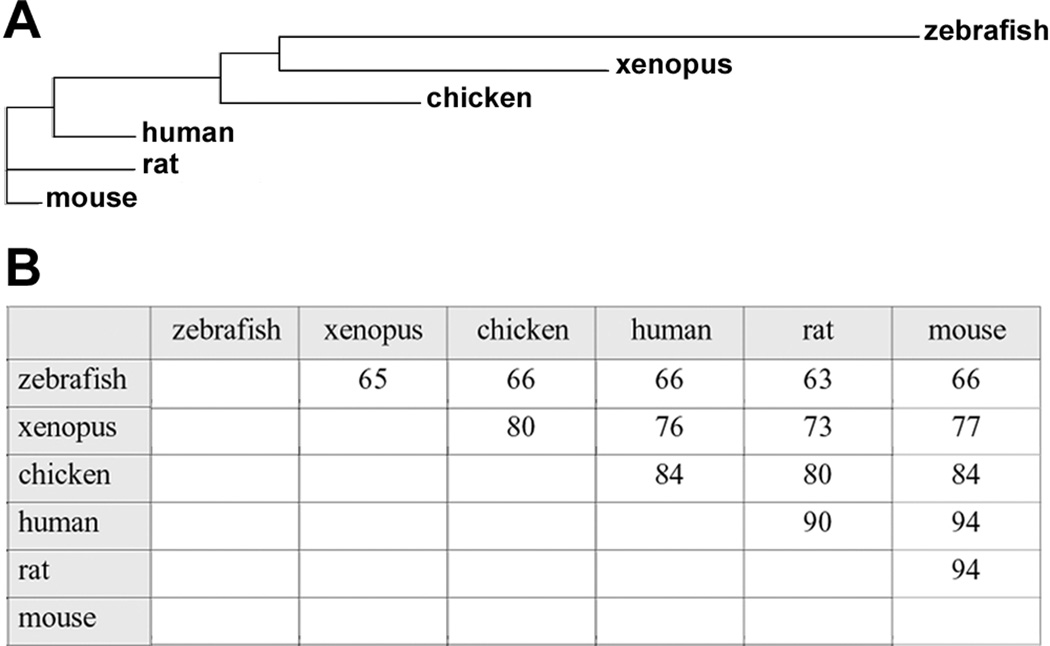
A zebrafish ortholog (ZDB-GENE-040426-1156, NM_200603.1) of the known rat and human atx mRNA sequences was identified within the ZFIN RNA/cDNA database (Bradford et al., 2011) using BLAST searches. This zebrafish ortholog is identical to the one that has been recently characterized to regulate early vascular development in the zebrafish (Yukiura et al., 2011). Translation of its open reading frame revealed an amino acid sequence with high sequence conservation compared to orthologs of other vertebrate mRNA sequences (Fig. 1; Brosamle and Halpern, 2002). In addition, amino acid sequence alignments confirmed the presence of conserved structure-function domains within zebrafish Atx, namely two somatomedin B-like domains, a catalytic lysoPLD domain and a nuclease-like domain entailing a single EF hand-like motif (Supplemental Fig. S1; Gijsbers et al., 2003; Masse et al., 2010; Moolenaar, 2002; Murata et al., 1994; Narita et al., 1994; Tokumura et al., 2002; Umezu-Goto et al., 2002; Yuelling and Fuss, 2008).
Fig. 1.
A conserved ortholog to mammalian atx exists in the zebrafish. A: Guide tree depicting evolutionary sequence relationships between known vertebrate Atx proteins. B: Identity table depicting amino acid sequence identities between known vertebrate Atx proteins. For Ensembl transcript IDs see Fig. S1.
The human and mouse atx genes span 27 exons and give rise to at least three protein isoforms, Atxα, Atxβ and Atxγ, due to alternative splicing of exons 12 and 21 (Giganti et al., 2008). The physiological relevance for these different isoforms of atx, however, is currently unknown. Out of these Atx protein isoforms, Atxβ, which lacks exons 12 and 21, is the only protein isoform represented by the zebrafish atx gene identified and appears, therefore, to be evolutionarily the oldest of the known Atx isoforms.
In the teleost fish lineage, whole-genome duplications occurred subsequent to its divergence from mammals, thus generating co-orthologs to many single mammalian genes (Ohno et al., 1968; Postlethwait et al., 2004; Postlethwait, 2007; Taylor et al., 2003). Using the Ensembl genome browser (Flicek et al., 2011), two zebrafish atx genes with a coding region of 99.6% amino acid sequence identity were identified in the most recent assembly (Zv9; http://www.sanger.ac.uk/Projects/D_rerio/). These two atx genes are not located on different linkage groups but next to each other on chromosome 16. Thus, they are likely either a result of a tandem rather than a large scale genome duplication event, or merely represent an inaccuracy in the assembly. Most importantly, based on the sequence information available and the high sequence identity between the two atx genes, all probes used in the present study recognize and/or affect the products of both genes and are referred to here as the zebrafish ortholog to mammalian atx.
In the Developing Zebrafish atx Displays Expression Pattern Characteristics that are Conserved between Different Vertebrate Species
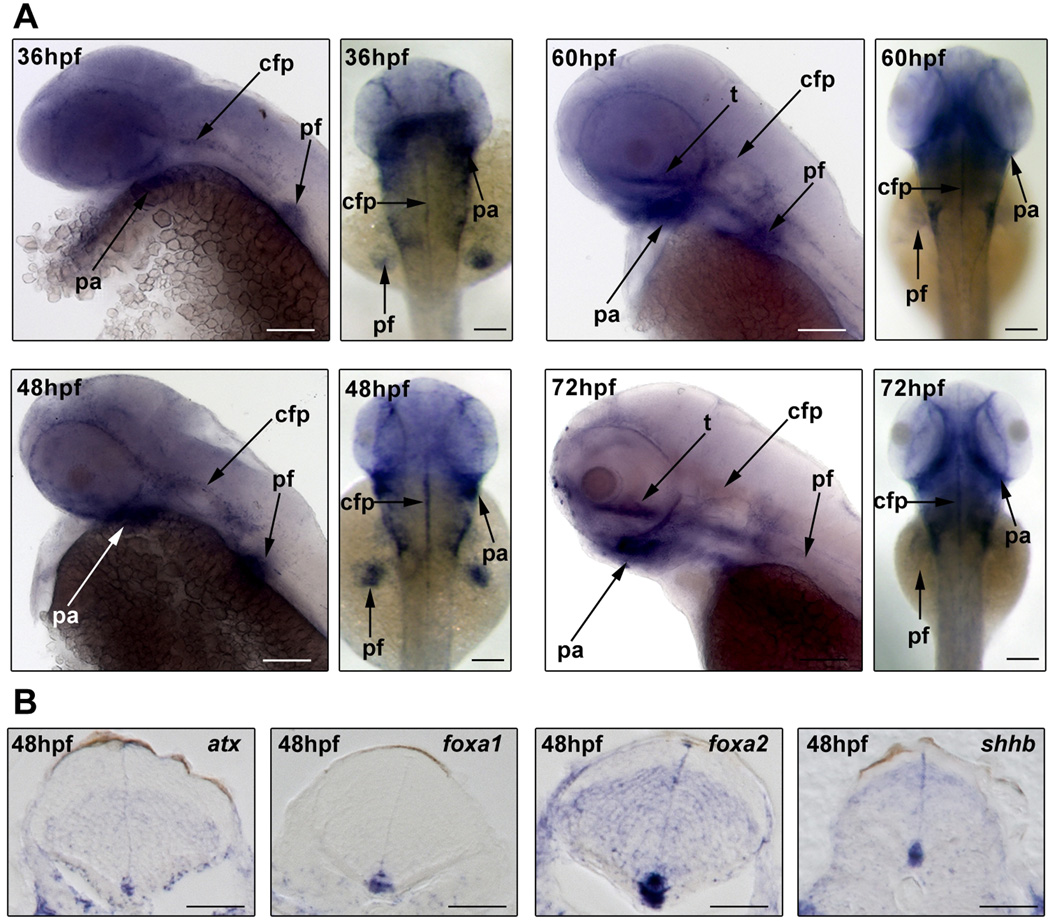
Detailed developmental atx expression profiles have so far been described in the mouse, chicken and frog (Bachner et al., 1999; Masse et al., 2010; Ohuchi et al., 2007). While species-specific expression variations exist, evolutionarily conserved atx expression does occur for example in the developing extremities (limb or fin buds) and jaw (branchial and pharyngeal arches). To assess the extent to which atx may be expressed in homologous structures located within the anterior part of the developing zebrafish, whole-mount in situ hybridizations were performed. In these studies, atx mRNA was first detected at 36 hpf. At all developmental ages analyzed (36–72 hpf), atx mRNA was found to be present in the pectoral fin buds and the pharyngeal arches (Fig. 2A). From lateral views, atx mRNA was at 60 and 72 hpf also detectable in a structure resembling the trabeculae cranii, a component of the developing neurocranium and thus part of the head mesenchyme (Fig. 2A). In support of an evolutionarily conserved expression and/or function, cavities in the head mesenchyme have been noted as a characteristic phenotype in atx knock-out mice (Koike et al., 2010). In addition, atx mRNA was detected in the developing head vasculature (Fig. 2A, not marked), which is consistent with recently published data (Yukiura et al., 2011) and the expression of atx in at least some types of blood vessels in other vertebrate species (Hoelzinger et al., 2005; Kanda et al. 2008; Masse et al. 2010; Ohuchi et al. 2007). Most interestingly, prominent expression of atx in the CNS was noted in the cephalic floor plate (Fig. 2B). A similar expression of atx in the floor plate and/or ventral spinal cord/hindbrain has been observed in all species so far analyzed with the exception of the chicken.
Fig. 2.
During zebrafish development atx is expressed in a pattern that includes a prominent expression in the ventral CNS. Embryos were collected at different developmental ages and analyzed by whole-mount in situ hybridization. A: Representative images of whole-mount embryos in situ hybridized for atx. Developmental ages are noted in hours post fertilization (hpf). Left panels for each developmental age depict lateral views, anterior is to the left. Right panels depict dorsal views, anterior is to the top. pharyngeal arches (pa), pectoral fin buds (pf), cephalic floor plate (cfp) and trabeculae cranii (t). Scale bars: 100 µm. B: Representative images of transverse sections through the hindbrain of 48 hpf embryos. Dorsal is to the top. cephalic floor plate markers (foxa1, foxa2, shhb). Scale bars: 50 µm
Taken together, the above data demonstrate that evolutionarily conserved features can be identified in atx’s developmental expression pattern in the zebrafish. These include an expression of atx by cells of the cephalic floor plate. Intriguingly, this expression coincides temporally with the appearance of differentiating oligodendrocytes in the developing hindbrain (Brosamle and Halpern, 2002; Buckley et al., 2010; Zannino and Appel, 2009).
Knock-down of atx Expression Delays and/or Inhibits the Appearance of Differentiating Oligodendrocytes in the Developing Zebrafish Hindbrain
The above described prominent cephalic floor plate expression of atx during a time period critical for oligodendrocyte development, prompted us to investigate whether knock-down of atx expression affects oligodendrocyte differentiation in the developing zebrafish hindbrain. In light of the known defects in vascular development upon knock-down of atx expression (Yukiura et al., 2011), it is of note that the vasculature is dispensable for the formation and maintenance of neuronal structures within the developing hindbrain for up to 72 hpf (Ulrich et al., 2011). However, complete knock-down of atx expression has been shown to cause edema in the head region, which could potentially affect CNS development (Yukiura et al., 2011). Thus, we felt it important to titrate antisense morpholino oligonucleotides to a concentration at which Atx protein levels were significantly reduced, while embryos lacked a gross morphological phenotype including head edema. Based on these criteria, we opted for conditions under which both an anti-atx translation blocking (atx TL MO) and an anti-atx splice site targeted (atx E2I2 MO) morpholino oligonucleotide yielded Atx protein levels of about 40–50% of control levels (Fig. 3).
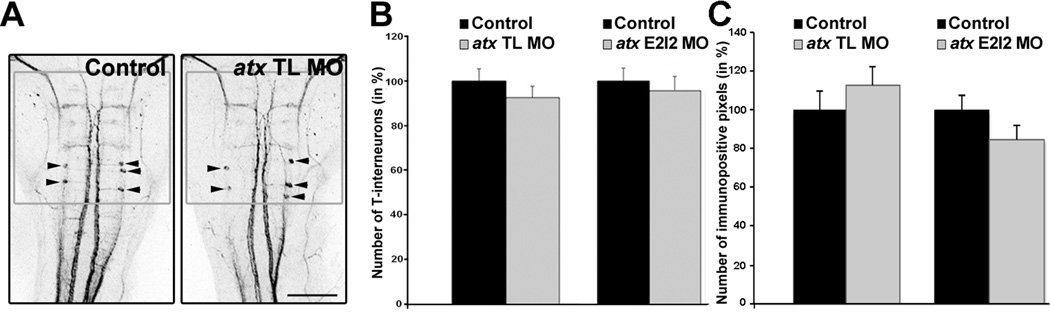
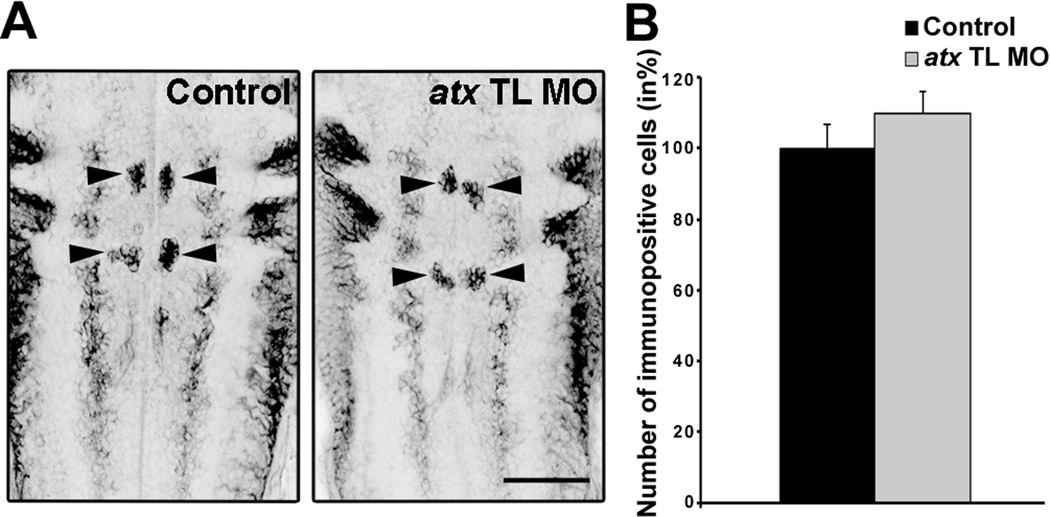
In atx knock-out mice, severe cranial neural tube defects have been described to occur in addition to vascular defects (Fotopoulou et al., 2010; Koike et al., 2011; van Meeteren et al., 2006). These neural tube defects may have developed at least in part independently from any vascular defects. Thus, to ensure that our atx knock-down conditions did not significantly affect the establishment of the axonal network within the developing hindbrain, whole-mount immunostainings were performed using the RMO44 antibody. This antibody recognizes a neurofilament protein strongly expressed in zebrafish reticulospinal neurons (Feng et al., 2010; Kimmel et al., 1985; Waskiewicz et al., 2001). As shown in Fig. 4, no effect on the overall morphology of the RMO44-immuno-positive axonal network was observed upon knock-down of atx expression.
Fig. 4.
In the developing hindbrain knock-down of atx expression does not affect neuronal/axonal organization. Embryos were treated as described in Fig. 3 and analyzed at 48 hpf. A: Representative images of embryos after whole-mount immnuostaining with the RMO44 antibody. Images represent 2D maximum projections of stacks of 5.66 µm optical sections. Arrowheads point toward T-interneurons. The gray boxes indicate the area used to determine the number of RMO44-immuno-positive pixels. Scale bar: 100 µm. B–C: Bar graphs depicting the number of RMO44-immuno-positive T-interneurons (B) and pixels (C) in % (control = 100%). Means and standard errors of three independent experiments are shown. Student’s t-test revealed no statistically significant differences.
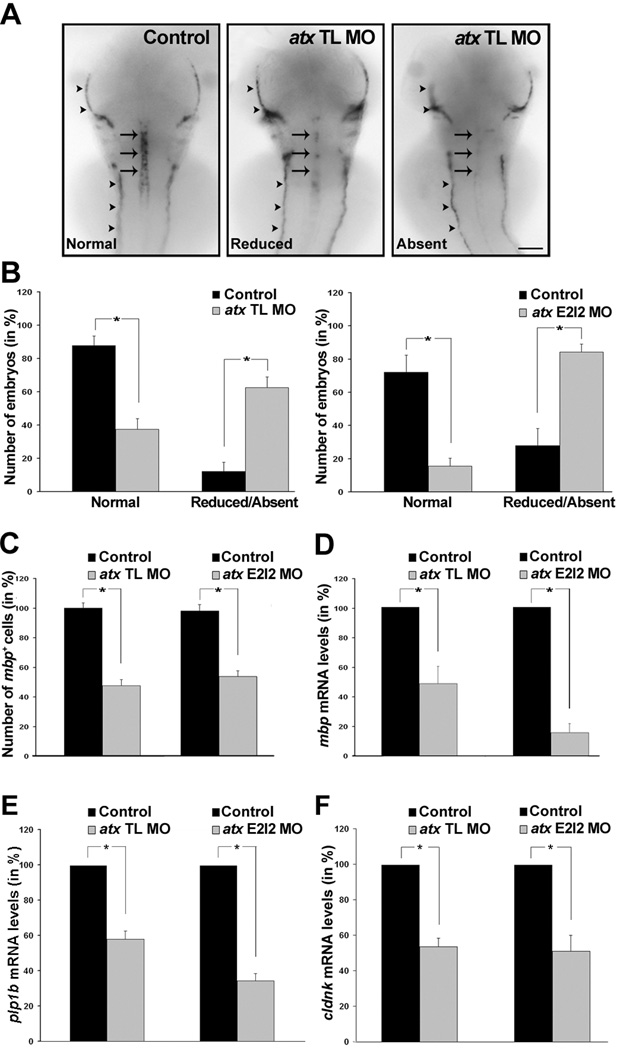
To assess the effect of knock-down of atx expression on the timely appearance of differentiating oligodendrocytes, whole-mount in-situ hybridizations were performed. In these studies embryos were analyzed at 66 hpf, a time point at which atx expression appeared prominent in the cephalic floor plate (Fig. 2) but was not detectable in differentiating oligodendrocytes (data not shown). First, the expression of myelin basic protein (mbp), a gene that is expressed by differentiating and myelinating oligodendrocytes and whose mRNA is not restricted to the cytoplasm but transported into cellular processes (Brosamle and Halpern, 2002), was investigated. As shown in Fig. 5A–C, there was a significant reduction in the number of mbp-positive oligodendrocytes in the developing hindbrain upon knock-down of atx expression. A similar reduction was observed for both anti-atx morpholino oligonucleotides (compare Fig. 5B, C left graphs with 5B, C right graphs). This effect on mbp expression in the CNS was much more dramatic than any effect noticed in the peripheral nervous system (see lateral line in Fig. 5A). The reduction in the number of mbp-positive oligodendrocytes was also associated with a significant reduction in mbp mRNA levels (Fig. 5D), suggesting that mRNA levels of oligodendrocyte-enriched genes provide a reliable measure for assessing the appearance of differentiating oligodendrocytes. This idea is consistent with previous findings (Buckley et al., 2010). To assess the extent to which the effect of atx knock-down may be specific for mbp expression, the mRNA levels for two additional genes enriched in differentiating oligodendrocytes were determined, namely proteolipid protein (plp1b) and claudin K (cldnk) (Brosamle and Halpern, 2002; Munzel et al., 2012; Takada and Appel, 2010). The mRNA levels for both genes were found to be significantly reduced (Fig. 5E, F).
Fig. 5.
In the developing hindbrain knock-down of atx expression leads to a reduction in the number of differentiating oligodendrocytes and in the mRNA levels of oligodendrocyte-enriched genes. Embryos were treated as described in Fig. 3 and analyzed at 66 hpf. A: Representative images of embryos after whole-mount in situ hybridization with a probe specific for myelin basic protein (mbp). Expression of mbp in oligodendrocytes (arrows) was classified into three categories (normal, reduced and absent). Expression of mbp in the anterior and posterior lateral line is marked by arrowheads. Dorsal views are shown, anterior is to the top. Scale bar: 100 µm. B: Bar graphs depicting the number of embryos with normal or reduced/absent mbp mRNA expression in % (total number of embryos per condition = 100%). Means and standard errors of five (left graph) and four (right graph) independent experiments are shown. Stars indicate an overall two-tailed significance level of p<0.05 (Student’s t-test). C: Bar graphs depicting the number of mbp-positive oligodendrocytes in % (control = 100%). Means and standard errors of four independent experiments are shown. Stars indicate an overall two-tailed significance level of p<0.05 (Student’s t-test). D–F: Bar graphs depicting mRNA levels in % (control = 100%) as determined by quantitative RT-PCR for mbp (D), proteolipid protein (plp1b) (E) and claudin K (cldnk) (F). Means and standard errors of three independent experiments are shown. Stars indicate an overall significance level of p<0.05 (one sample t-test).
To further confirm the specificity of the phenotype seen upon knock-down of atx expression, mRNA rescue experiments were performed. In these experiments, a rat-derived synthetic atx mRNA was used since translation from this mRNA is not affected by the anti-atx translation blocking morpholino oligonucleotide. As shown in Fig. 6A, Atx protein levels were noticeably increased at 48 hpf after co-injection of the synthetic atx mRNA with the atx TL MO. Importantly, cldnk mRNA levels, as a measure for the appearance of differentiating oligodendrocytes, were found to be similar to control levels (Fig. 6B), thus demonstrating a restoration of the wild-type phenotype. No such restoration was observed when a beta-galactosidase encoding synthetic mRNA was co-injected with the atx TL MO (data not shown).
For a better evaluation of the persistence of the phenotype seen upon knock-down of atx expression, the number of embryos with normal and reduced/absent mbp expression in the hindbrain was determined at 72 hpf. As shown in Fig. 6C, there was a significant decrease in the number of embryos with normal mbp expression and a concomitant significant increase in the number of embryos with reduced/absent mbp expression. The extent of decrease/increase was comparable to the one seen at 66 hpf (compare Fig. 6C with Fig. 5B).
Taken together, the above data demonstrate that a reduction in atx expression significantly delays and/or inhibits the appearance of differentiating oligodendrocytes in the developing hindbrain, and they suggest that this delay and/or inhibition is not due to gross morphological defects in the establishment of the axonal network.
Knock-down of atx Expression Delays and/or Inhibits the Appearance of Oligodendrocyte Progenitors in the Developing Zebrafish Hindbrain
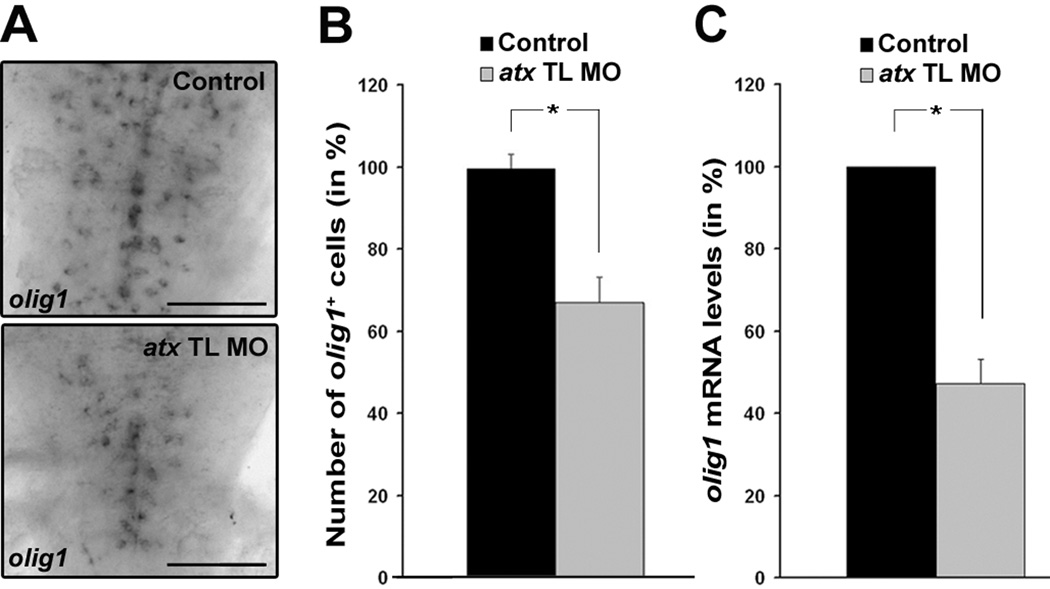
The above data raised the possibility that the effects observed on differentiating oligodendrocytes may have been a result of an effect on the developmental lineage progression of early oligodendrocyte progenitors into differentiating oligodendrocytes that is at much earlier stages than previously observed. To investigate this possibility, whole-mount in situ hybridizations were performed using a probe specific for the transcription factor olig1, which in the developing zebrafish is expressed by oligodendrocyte progenitors. As shown in Fig. 7, knock-down of atx expression led to a significant reduction in the number of olig1-positive cells and in the level of olig1 mRNA. The reduction in the number of olig1-positive cells was not found associated with a significant increase in cell death as assessed by immunostaining using an antibody specific for activated caspase-3 (data not shown). Furthermore, olig1 expression has been described to only partially overlap with the expression of the myelin genes mbp and plp1b at a developmental age similar to the one used here (Li et al., 2007; Schebesta and Serluca 2009). Thus, if the effect of atx knock-down were specific to only the later stages of the oligodendrocyte lineage, a much less pronounced effect on olig1 mRNA levels would be expected. In fact, the reduction in olig1 mRNA levels seen upon knock-down of atx expression (43.31 ± 4.19 %; Fig. 7) was comparable to the reduction in mbp (48.31 ± 11.88%, Fig. 5), plp1b (58.07 ± 4.57%, Fig. 5) and cldnk (52.34 ± 3.86%, Fig. 5) mRNA levels. Thus, the above data demonstrate that knock-down of atx expression delays and/or inhibits not only the appearance of differentiating oligodendrocytes but also oligodendrocyte progenitors.
Fig. 7.
In the developing hindbrain knock-down of atx expression leads to a reduction in the number of olig1-positive cells and the levels of olig1 mRNA. Embryos were treated as described in Fig. 3 and analyzed at 66 hpf. A: Representative images of embryos after whole-mount in situ hybridization with a probe specific for olig1. Scale bar: 100 µm. B: Bar graph depicting the number of olig1-positive oligodendrocytes in % (control = 100%). Means and standard errors of three independent experiments are shown. Stars indicate an overall two-tailed significance level of p<0.05 (Student’s t-test). C: Bar graph depicting olig1 mRNA levels in % (control = 100%) as determined by quantitative RT-PCR. Means and standard errors of three independent experiments are shown. Stars indicate an overall significance level of p<0.05 (one sample t-test).
Knock-down of atx Expression Delays and/or Inhibits the Differentiation of Oligodendrocyte Progenitors from olig2-positive Progenitor Cells in the Developing Zebrafish Hindbrain
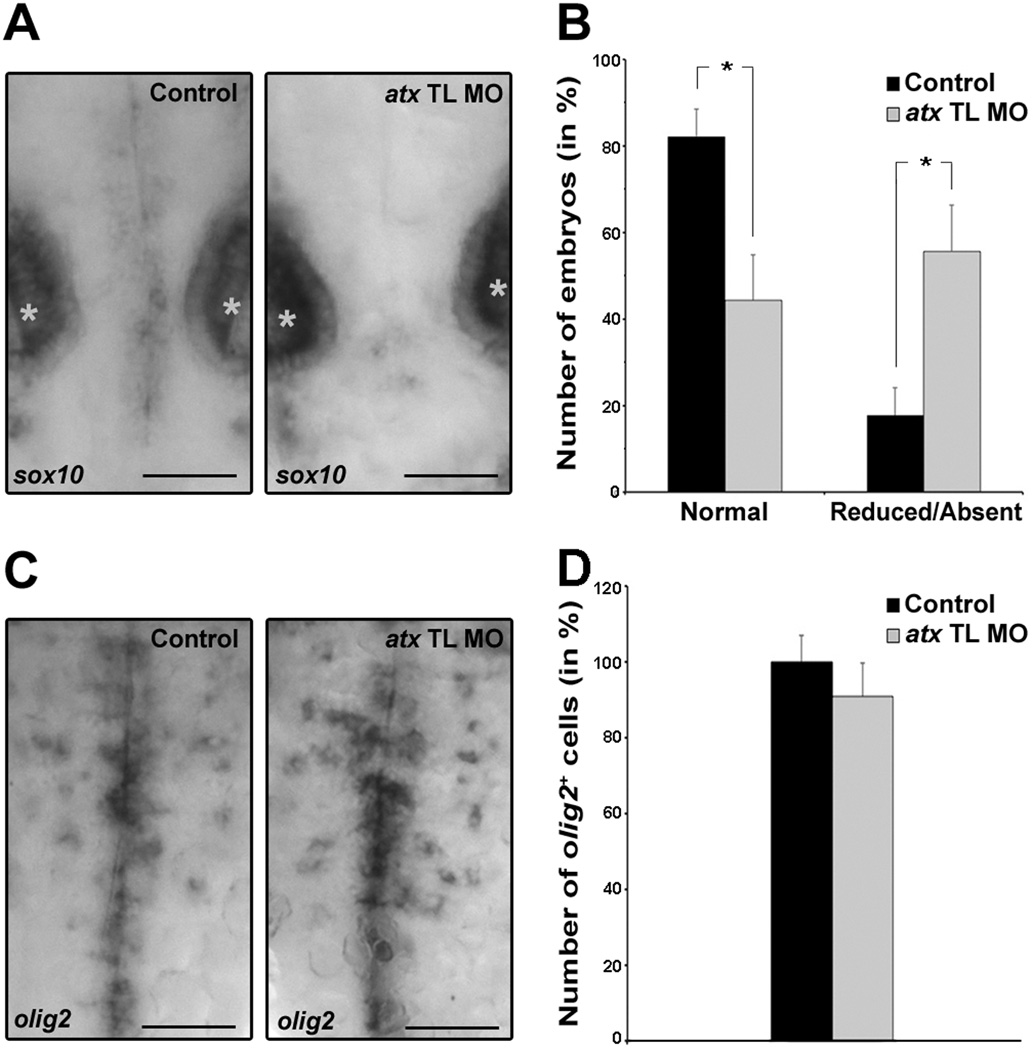
To further define the developmental stage at which knock-down of atx expression affects the developmental progression of cells of the oligodendrocyte lineage, zebrafish embryos were analyzed at 48 hpf. At this developmental time point oligodendrocyte progenitors expressing the transcription factor sox10 begin to differentiate in the developing hindbrain from olig2-positive progenitor cells (Zannino and Appel, 2009). As shown in Fig. 8A,B, knock-down of atx expression resulted in a reduction in the number of embryos with normal expression of sox10 in the developing hindbrain. Conversely, the number of embryos with reduced or absent expression of sox10 in the developing hindbrain was increased. No change in the expression of sox10 was noted in the otic vesicles. In contrast to the effects seen on sox10-positve oligodendrocyte progenitors, the number of olig2-positive cells remained unchanged (Fig. 8C,D). Thus, knock-down of atx expression affects in the developing hindbrain the differentiation of olig2-positive progenitor cells into cells of the oligodendrocyte lineage.
Fig. 8.
In the developing hindbrain knock-down of atx expression leads to a reduction in the number of sox10-positive but not olig2-positive cells. Embryos were treated as described in Fig. 3 and analyzed at 48 hpf. A: Representative images of embryos after whole-mount in situ hybridization with a probe specific for sox10. Stars indicate otic vesicles. Scale bar: 50 µm. B: Bar graph depicting the number of embryos with normal (>2 sox10-positive cells) or reduced/absent (≤2 sox10-positive cells) sox10 expression in % (total number of embryos per condition = 100%). Means and standard errors of six independent experiments are shown. Stars indicate an overall two-tailed significance level of p<0.05 (Student’s t-test). C: Representative images of embryos after whole-mount in situ hybridization with a probe specific for olig2. Scale bar: 50 µm. D: Bar graph depicting the number of olig2-positive oligodendrocytes in % (control = 100%). Means and standard errors of three independent experiments are shown. Student’s t-test revealed no statistically significant difference.
It has been shown previously that in the developing zebrafish hindbrain olig2-positive progenitor cells that are located within rhombomere r5 and r6 cell clusters can give rise to not only hindbrain oligodendrocyte progenitors but also somatic abducens motor neurons (Zannino and Appel, 2009). Thus, atx may be regulating not only the differentiation of oligodendrocyte progenitors from olig2-positive progenitor cells but also the differentiation of somatic abducens motor neurons. To test this idea, we used the Zn-8 antibody, which recognizes Neurolin/DM-Grasp and thus somatic abducens motor neurons (Chandrasekhar et al., 1997; Kanki et al., 1994; Trevarrow et al., 1990). As shown in Fig. 9, no significant change in the number of Zn-8-immuno-positive somatic abducens motor neurons was detected upon knock-down of atx expression.
Fig. 9.
In the developing hindbrain knock-down of atx expression does not affect the number of somatic abducens motor neurons. Embryos were treated as described in Fig. 3 and analyzed at 48 hpf. A: Representative images of embryos after whole-mount immunostaining with the Zn-8 antibody. Images represent 2D maximum projections of stacks of 0.87 µm optical sections. Arrows point toward somatic abducens motor neurons. Zn-8-immuno-positive hindbrain commissural axons are only partially captured. Scale bar: 50 µm. B: Bar graph depicting the number of Zn-8-immuno-positive somatic abducens motor neurons in % (control = 100%). Means and standard errors of three independent experiments are shown. Student’s t-test revealed no statistically significant difference.
Taken together, the above data demonstrate that knock-down of atx expression delays and/or inhibits specifically the differentiation of oligodendrocyte progenitors from olig2-positive progenitor cells while not significantly affecting the differentiation of somatic abducens motor neurons.
DISCUSSION
The data presented here demonstrate that the zebrafish ortholog to mammalian atx displays evolutionarily conserved expression pattern characteristics. These include an expression of atx in the developing extremities and jaw. Most prominent expression in the CNS of the anterior embryo was noted in the cephalic floor plate coinciding temporally with the time period during which cells of the oligodendrocyte lineage are generated. Antisense morpholino oligonucleotide-mediated knock-down of atx expression revealed a functional role of atx in regulating early development of cells of the oligodendrocyte lineage in the developing hindbrain, without affecting the number of olig2-positive progenitor cells, the overall morphology of the axonal network or the differentiation of somatic abducens motor neurons. This novel functional property of atx is likely mediated by a paracrine mechanism and via the expression and secretion of Atx by cells of the cephalic floor plate.
In our previous studies using rodent oligodendrocytes and cell culture systems, the effects of Atx were characterized for later stages of the oligodendrocyte lineage and found to be most likely mediated by an autocrine mechanism (Dennis et al., 2008; Fox et al., 2004). Antisense morpholino oligonucleotide-mediated effects in the zebrafish have in some cases been found penetrant for up to seven days (Rinner et al., 2005; Seiler et al., 2005; van der Sar et al., 2002). Initial analysis, however, suggests that in the case of the titrated anti-atx morpholino oligonucleotides, Atx protein levels are almost normal at about 3 dpf, thus precluding a meaningful analysis at later developmental stages. More sophisticated experimental paradigms will thus be necessary to determine whether the effects of Atx seen in the rodent system also apply to the zebrafish.
Two functionally active sites have been described for Atx, namely the MORFO domain and the catalytic lysoPLD domain. In our previous studies we demonstrated that it is the MORFO domain that mediates morphological maturation of differentiating oligodendrocytes (Dennis et al., 2008; Fox et al., 2004; Yuelling and Fuss, 2008). Atx is, however, better known for its catalytic lysoPLD activity and the generation of the lipid signaling molecule lysophosphatidic acid (LPA) (Liu et al., 2009; Moolenaar, 2002; Nakanaga et al., 2010; Samadi et al., 2011; Tokumura et al., 2002; Umezu-Goto et al., 2002; van Meeteren and Moolenaar, 2007). As an extracellular lysophospholipid, LPA exerts its functions through interactions with a family of G protein-coupled receptors, the family of LPA receptors (Chun et al., 2010; Lin et al., 2009; Yanagida and Ishii, 2011). Orthologs for at least some of the known mammalian LPA receptors have been identified in the zebrafish (Lee et al., 2008; Yukiura et al., 2011), and cells of the oligodendrocyte lineage have long been known to express a variety of LPA receptors (Dawson et al., 2003; Nogaroli et al., 2009; Stankoff et al., 2002; Weiner et al., 1998; Yu et al., 2004). In addition, LPA receptors have been functionally implicated in the regulation of early CNS development (Choi et al., 2008; Estivill-Torrus et al., 2008; Fukushima et al., 2007; Hecht et al., 1996; Kingsbury et al., 2003; Kingsbury et al., 2004). However, little is currently known about the role of LPA signaling for early glial and in particular oligodendroglial development. In support of a potential role of LPA in gliogenesis, LPA has been shown to promote the differentiation of oligodendrocytes from neural stem/progenitor cells, at least under certain cell culture conditions (Cui and Qiao, 2007; Pitson and Pebay, 2009; Svetlov et al., 2004). Such effects of LPA may be mediated by autocrine as well as paracrine mechanisms. Atx has traditionally been regarded as an autocrine factor. However, there is increasing evidence for paracrine roles, all of which have so far been attributed to the synthesis of LPA (Ferry et al., 2003; Hoelzinger et al., 2008; Kanda et al., 2008). Such paracrine roles of Atx are further facilitated by slow catalytic kinetics and a predicted mobility of Atx-lipid complexes up to a distance of approximately 65 µm (Saunders et al., 2011). With regard to our findings, all the above data point toward a primary role of Atx’s catalytic lysoPLD activity, rather than its MORFO domain, in stimulating the progression of olig2-positive progenitor cells into oligodendrocyte progenitors in a paracrine, rather than autocrine, fashion.
It has been well established in the spinal cord that there are ventral as well as dorsal origins for cells of the oligodendrocyte lineage (Miller, 2005; Richardson et al., 2006). While less well characterized, the same appears to be true for hindbrain oligodendrocytes (Davies and Miller, 2001; Vallstedt et al., 2005; Zannino and Appel, 2009). The molecular mechanisms regulating the development of ventrally- and dorsally-derived oligodendrocytes have been shown to involve different extracellular signals (Bilican et al., 2008; Cai et al., 2005; Fogarty et al., 2005; Langseth et al., 2010; Vallstedt et al., 2005). Based on our findings, one would predict that at least in the developing zebrafish, Atx’s role in regulating early oligodendrocyte development may be restricted to ventrally-derived oligodendrocytes. In homology, the same would apply to the developing mouse, where atx has been described expressed by cells of the floor plate (Bachner et al., 1999; Ohuchi et al., 2007). In contrast, atx was found expressed in the alar plate in the chicken (Ohuchi et al., 2007), and it is thus tempting to speculate that in the chicken atx may play a functional role in regulating the development of dorsally-derived oligodendrocytes.
Ventrally-derived oligodendrocytes arise in the developing spinal cord and hindbrain from olig2-positive progenitor cells that are located within a progenitor niche that gives rise first to motor neurons and then to oligodendrocytes. The regulation of this neuron-glial switch has been found complex involving cell intrinsic factors as well as extracellular signaling factors (for reviews see: Richardson et al., 2000; Rowitch, 2004; Rowitch et al., 2002). In this regard, our data identify atx as an attractive novel candidate for an extracellular signaling factor involved in promoting the generation of cells of the oligodendrocyte lineage from olig2-positive progenitor cells.
Supplementary Material
Amino acid sequence alignment. The degree of sequence similarity is color coded. Black letters on white background represent non-similar amino acid residues, red letters on yellow background represent identical amino acid residues, blue letters on turquoise background represent conserved amino acid residues, black letters on green background represent similar amino acid residues and green letters on white background represent weakly similar residues. The dashed lines at the bottom of the sequence delineate the known structure-function domains. The MORFO domain entails the nuclease-like domain but extends amino-terminal by 69 amino acid residues. The numbers on the top of the sequence indicate the position of amino acids within the sequence. The catalytic site residue is marked by a star. The EF hand-like motif is double underlined. Ensembl transcript IDs used for the alignment were as follows: zebrafish: ENSDART00000104089 and ENSDART00000047920, xenopus: ENSXETT00000006797, chicken: ENSGALT00000026501, human: ENST00000075322, rat: ENSRNOT00000005561, mouse: ENSMUST00000041591.
ACKNOWLEDGEMENTS
The authors would like to thank Bruce Appel and Marnie Halpern for generously providing reagents and invaluable discussions. The authors also thank Gregory Walsh for helpful advice. Microscopy was performed at the VCU Department of Anatomy and Neurobiology Microscopy Facility, supported, in part, with funding from NIH-NINDS Center Core grant (5P30NS047463). The monoclonal antibody Zn-8, developed by Bill Trevarrow, was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by the Department of Biology at The University of Iowa. This work was supported by grants from the NIH-NINDS (B.F.) and the National Multiple Sclerosis Society (B.F.).
REFERENCES
- Abramoff M, Magelhaes P, Ram S. Image Processing with image. Journal of Biophotonics Int. 2004;11:36–42. [Google Scholar]
- Bachner D, Ahrens M, Betat N, Schroder D, Gross G. Developmental expression analysis of murine autotaxin (ATX) Mech Dev. 1999;84:121–125. doi: 10.1016/s0925-4773(99)00048-9. [DOI] [PubMed] [Google Scholar]
- Bilican B, Fiore-Heriche C, Compston A, Allen ND, Chandran S. Induction of Olig2 precursors by FGF involves BMP signalling blockade at the Smad level. PLoS One. 2008;3:e2863. doi: 10.1371/journal.pone.0002863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford Y, Conlin T, Dunn N, Fashena D, Frazer K, Howe DG, Knight J, Mani P, Martin R, Moxon SA, et al. ZFIN: enhancements and updates to the Zebrafish Model Organism Database. Nucleic Acids Res. 2011;39(Database issue):D822–D829. doi: 10.1093/nar/gkq1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosamle C, Halpern ME. Characterization of myelination in the developing zebrafish. Glia. 2002;39:47–57. doi: 10.1002/glia.10088. [DOI] [PubMed] [Google Scholar]
- Buckley CE, Marguerie A, Alderton WK, Franklin RJM. Temporal dynamics of myelination in the zebrafish spinal cord. Glia. 2010;58:802–812. doi: 10.1002/glia.20964. [DOI] [PubMed] [Google Scholar]
- Cai J, Qi Y, Hu X, Tan M, Liu Z, Zhang J, Li Q, Sander M, Qiu M. Generation of oligodendrocyte precursor cells from mouse dorsal spinal cord independent of Nkx6 regulation and Shh signaling. Neuron. 2005;45:41–53. doi: 10.1016/j.neuron.2004.12.028. [DOI] [PubMed] [Google Scholar]
- Chandrasekhar A, Moens CB, Warren JT, Jr, Kimmel CB, Kuwada JY. Development of branchiomotor neurons in zebrafish. Development. 1997;124:2633–2644. doi: 10.1242/dev.124.13.2633. [DOI] [PubMed] [Google Scholar]
- Choi JW, Lee CW, Chun J. Biological roles of lysophospholipid receptors revealed by genetic null mice: an update. Biochim Biophys Acta. 2008;1781:531–539. doi: 10.1016/j.bbalip.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun J, Hla T, Lynch KR, Spiegel S, Moolenaar WH. International Union of Basic and Clinical Pharmacology. LXXVIII. Lysophospholipid receptor nomenclature. Pharmacol. 2010;62:579–587. doi: 10.1124/pr.110.003111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui HL, Qiao JT. Effect of lysophosphatidic acid on differentiation of embryonic neural stem cells into neuroglial cells in rats in vitro. Sheng Li Xue Bao. 2007;59(6):759–764. [PubMed] [Google Scholar]
- Dalgaard P, editor. Introductory Statistics with R. 2nd ed. New York: Springer; 2008. [Google Scholar]
- Davies JE, Miller RH. Local sonic hedgehog signaling regulates oligodendrocyte precursor appearance in multiple ventricular zone domains in the chick metencephalon. Dev Biol. 2001;233:513–525. doi: 10.1006/dbio.2001.0224. [DOI] [PubMed] [Google Scholar]
- Dawson J, Hotchin N, Lax S, Rumsby M. Lysophosphatidic acid induces process retraction in CG-4 line oligodendrocytes and oligodendrocyte precursor cells but not in differentiated oligodendrocytes. J Neurochem. 2003;87:947–957. doi: 10.1046/j.1471-4159.2003.02056.x. [DOI] [PubMed] [Google Scholar]
- Dennis J, White MA, Forrest AD, Yuelling LM, Nogaroli L, Afshari FS, Fox MA, Fuss B. Phosphodiesterase-Ialpha/autotaxin's MORFO domain regulates oligodendroglial process network formation and focal adhesion organization. Mol Cell Neurosci. 2008;37:412–424. doi: 10.1016/j.mcn.2007.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutton KA, Pauliny A, Lopes SS, Elworthy S, Carney TJ, Rauch J, Geisler R, Haffter P, Kelsh RN. Zebrafish colourless encodes sox10 and specifies non-ectomesenchymal neural crest fates. Development. 2001;128:4113–4125. doi: 10.1242/dev.128.21.4113. [DOI] [PubMed] [Google Scholar]
- Estivill-Torrus G, Llebrez-Zayas P, Matas-Rico E, Santin L, Pedraza C, De Diego I, Del Arco I, Fernandez-Llebrez P, Chun J, De Fonseca FR. Absence of LPA1 signaling results in defective cortical development. Cereb Cortex. 2008;18:938–950. doi: 10.1093/cercor/bhm132. [DOI] [PubMed] [Google Scholar]
- Feng L, Hernandez RE, Waxman JS, Yelon D, Moens CB. Dhrs3a regulates retinoic acid biosynthesis through a feedback inhibition mechanism. Dev Biol. 2010;338:1–14. doi: 10.1016/j.ydbio.2009.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferry G, Giganti A, Coge F, Bertaux F, Thiam K, Boutin JA. Functional invalidation of the autotaxin gene by a single amino acid mutation in mouse is lethal. FEBS Lett. 2007;581:3572–3578. doi: 10.1016/j.febslet.2007.06.064. [DOI] [PubMed] [Google Scholar]
- Ferry G, Tellier E, Try A, Gres S, Naime I, Simon MF, Rodriguez M, Boucher J, Tack I, Gesta S, et al. Autotaxin is released from adipocytes, catalyzes lysophosphatidic acid synthesis, and activates preadipocyte proliferation. Up-regulated expression with adipocyte differentiation and obesity. J Biol Chem. 2003;278:18162–18169. doi: 10.1074/jbc.M301158200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flicek P, Amode MR, Barrell D, Beal K, Brent S, Chen Y, Clapham P, Coates G, Fairley S, Fitzgerald S, et al. Ensembl 2011. Nucleic. 2011;39(Database issue):D800–D806. doi: 10.1093/nar/gkq1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogarty M, Richardson WD, Kessaris N. A subset of oligodendrocytes generated from radial glia in the dorsal spinal cord. Development. 2005;132:1951–1959. doi: 10.1242/dev.01777. [DOI] [PubMed] [Google Scholar]
- Fotopoulou S, Oikonomou N, Grigorieva E, Nikitopoulou I, Paparountas T, Thanassopoulou A, Zhao Z, Xu Y, Kontoyiannis DL, Remboutsika E, et al. ATX expression and LPA signalling are vital for the development of the nervous system. Dev Biol. 2010;339:451–464. doi: 10.1016/j.ydbio.2010.01.007. [DOI] [PubMed] [Google Scholar]
- Fox MA, Alexander JK, Afshari FS, Colello RJ, Fuss B. Phosphodiesterase-I alpha/autotaxin controls cytoskeletal organization and FAK phosphorylation during myelination. Mol Cell Neurosci. 2004;27:140–150. doi: 10.1016/j.mcn.2004.06.002. [DOI] [PubMed] [Google Scholar]
- Fox MA, Colello RJ, Macklin WB, Fuss B. Phosphodiesterase-Ialpha/autotaxin: a counteradhesive protein expressed by oligodendrocytes during onset of myelination. Mol Cell Neurosci. 2003;23:507–519. doi: 10.1016/s1044-7431(03)00073-3. [DOI] [PubMed] [Google Scholar]
- Fukushima N, Shano S, Moriyama R, Chun J. Lysophosphatidic acid stimulates neuronal differentiation of cortical neuroblasts through the LPA1-G(i/o) pathway. Neurochem Int. 2007;50:302–307. doi: 10.1016/j.neuint.2006.09.008. [DOI] [PubMed] [Google Scholar]
- Fuss B, Mallon B, Phan T, Ohlemeyer C, Kirchhoff F, Nishiyama A, Macklin WB. Purification and analysis of in vivo-differentiated oligodendrocytes expressing the green fluorescent protein. Dev Biol. 2000;218:259–274. doi: 10.1006/dbio.1999.9574. [DOI] [PubMed] [Google Scholar]
- Gerety SS, Wilkinson DG. Morpholino artifacts provide pitfalls and reveal a novel role for pro-apoptotic genes in hindbrain boundary development. Dev. 2011;350:279–289. doi: 10.1016/j.ydbio.2010.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giganti A, Rodriguez M, Fould B, Moulharat N, Coge F, Chomarat P, Galizzi JP, Valet P, Saulnier-Blache JS, Boutin JA, et al. Murine and human autotaxin alpha, beta, and gamma isoforms: gene organization, tissue distribution, and biochemical characterization. J Biol Chem. 2008;283:7776–7789. doi: 10.1074/jbc.M708705200. [DOI] [PubMed] [Google Scholar]
- Gijsbers R, Aoki J, Arai H, Bollen M. The hydrolysis of lysophospholipids and nucleotides by autotaxin (NPP2) involves a single catalytic site. FEBS Lett. 2003;5383:60–64. doi: 10.1016/s0014-5793(03)00133-9. [DOI] [PubMed] [Google Scholar]
- Hecht JH, Weiner JA, Post SR, Chun J. Ventricular zone gene-1 (vzg-1) encodes a lysophosphosphatidic acid receptor expressed in neurogenic regions of the developing cerebral cortex. J Cell Biol. 1996;135:1071–1083. doi: 10.1083/jcb.135.4.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoelzinger DB, Mariani L, Weis J, Woyke T, Berens TJ, McDonough WS, Sloan A, Coons SW, Berens ME. Gene expression profile of glioblastoma multiforme invasive phenotype points to new therapeutic targets. Neoplasia. 2005;7:7–16. doi: 10.1593/neo.04535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoelzinger DB, Nakada M, Demuth T, Rosensteel T, Reavie LB, Berens ME. Autotaxin: a secreted autocrine/paracrine factor that promotes glioma invasion. J Neurooncol. 2008;86:297–309. doi: 10.1007/s11060-007-9480-6. [DOI] [PubMed] [Google Scholar]
- Jansen S, Stefan C, Creemers JW, Waelkens E, Van Eynde A, Stalmans W, Bollen M. Proteolytic maturation and activation of autotaxin (NPP2), a secreted metastasis-enhancing lysophospholipase D. J Cell Sci. 2005;118:3081–3089. doi: 10.1242/jcs.02438. [DOI] [PubMed] [Google Scholar]
- Jansen S, Callewaert N, Dewerte I, Andries M, Ceulemans H, Bollen M. An essential oligomannosidic glycan chain in the catalytic domain of autotaxin, a secreted lysophospholipase-D. J Biol Chem. 2007;282:11084–11091. doi: 10.1074/jbc.M611503200. [DOI] [PubMed] [Google Scholar]
- Jessell TM. Neuronal specification in the spinal cord: inductive signals and transcriptional codes. Nat Rev Genet. 2000;1:20–29. doi: 10.1038/35049541. [DOI] [PubMed] [Google Scholar]
- Jin SW, Herzog W, Santoro MM, Mitchell TS, Frantsve J, Jungblut B, Beis D, Scott IC, D'Amico LA, Ober EA, et al. A transgene-assisted genetic screen identifies essential regulators of vascular development in vertebrate embryos. Dev Biol. 2007;307:29–42. doi: 10.1016/j.ydbio.2007.03.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanda H, Newton R, Klein R, Morita Y, Gunn MD, Rosen SD. Autotaxin, an ectoenzyme that produces lysophosphatidic acid, promotes the entry of lymphocytes into secondary lymphoid organs. Nat Immunol. 2008;9:415–423. doi: 10.1038/ni1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki JP, Chang S, Kuwada JY. The molecular cloning and characterization of potential chick DM-GRASP homologs in zebrafish and mouse. J Neurobiol. 1994;25:831–845. doi: 10.1002/neu.480250708. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Metcalfe WK, Schabtach E. T reticular interneurons: a class of serially repeating cells in the zebrafish hindbrain. J Comp Neurol. 1985;233:365–376. doi: 10.1002/cne.902330306. [DOI] [PubMed] [Google Scholar]
- Kingsbury MA, Rehen SK, Contos JJ, Higgins CM, Chun J. Non-proliferative effects of lysophosphatidic acid enhance cortical growth and folding. Nat Neurosci. 2003;6:1292–1299. doi: 10.1038/nn1157. [DOI] [PubMed] [Google Scholar]
- Kingsbury MA, Rehen SK, Ye X, Chun J. Genetics and cell biology of lysophosphatidic acid receptor-mediated signaling during cortical neurogenesis. J Cell Biochem. 2004;92:1004–1012. doi: 10.1002/jcb.20061. [DOI] [PubMed] [Google Scholar]
- Kirby BB, Takada N, Latimer AJ, Shin J, Carney TJ, Kelsh RN, Appel B. In vivo time-lapse imaging shows dynamic oligodendrocyte progenitor behavior during zebrafish development. Nat Neurosci. 2006;9:1506–1511. doi: 10.1038/nn1803. [DOI] [PubMed] [Google Scholar]
- Koike S, Keino-Masu K, Masu M. Deficiency of autotaxin/lysophospholipase D results in head cavity formation in mouse embryos through the LPA receptor-Rho-ROCK pathway. Biochem Biophys Res Commun. 2010;400:66–71. doi: 10.1016/j.bbrc.2010.08.008. [DOI] [PubMed] [Google Scholar]
- Koike S, Keino-Masu K, Ohto T, Masu M. The N-terminal hydrophobic sequence of autotaxin (ENPP2) functions as a signal peptide. Genes Cells. 2006;11:133–142. doi: 10.1111/j.1365-2443.2006.00924.x. [DOI] [PubMed] [Google Scholar]
- Koike S, Keino-Masu K, Ohto T, Sugiyama F, Takahashi S, Masu M. Autotaxin/lysophospholipase D-mediated lysophosphatidic acid signaling is required to form distinctive large lysosomes in the visceral endoderm cells of the mouse yolk sac. J Biol Chem. 2009;284:33561–33570. doi: 10.1074/jbc.M109.012716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike S, Yutoh Y, Keino-Masu K, Noji S, Masu M, Ohuchi H. Autotaxin is required for the cranial neural tube closure and establishment of the midbrain-hindbrain boundary during mouse development. Dev Dyn. 2011;240:413–421. doi: 10.1002/dvdy.22543. [DOI] [PubMed] [Google Scholar]
- Langseth AJ, Munji RN, Choe Y, Huynh T, Pozniak CD, Pleasure SJ. Wnts influence the timing and efficiency of oligodendrocyte precursor cell generation in the telencephalon. J Neurosci. 2010;6:13367–13372. doi: 10.1523/JNEUROSCI.1934-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SJ, Chan TH, Chen TC, Liao BK, Hwang PP, Lee H. LPA1 is essential for lymphatic vessel development in zebrafish. FASEB J. 2008;22:3706–3715. doi: 10.1096/fj.08-106088. [DOI] [PubMed] [Google Scholar]
- Li H, Lu Y, Smith HK, Richardson WD. Olig1 and Sox10 interact synergistically to drive myelin basic protein transcription in oligodendrocytes. J Neurosci. 2007;27:14375–14382. doi: 10.1523/JNEUROSCI.4456-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin ME, Herr DR, Chun J. Lysophosphatidic acid (LPA) receptors: signaling properties and disease relevance. Prostaglandins Other Lipid Mediat. 2009;91:130–138. doi: 10.1016/j.prostaglandins.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Murph M, Panupinthu N, Mills GB. ATX-LPA receptor axis in inflammation and cancer. Cell Cycle. 2009;8:3695–3701. doi: 10.4161/cc.8.22.9937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lu QR, Sun T, Zhu Z, Ma N, Garcia M, Stiles CD, Rowitch DH. Common developmental requirement for Olig function indicates a motor neuron/oligodendrocyte connection. Cell. 2002;109:75–86. doi: 10.1016/s0092-8674(02)00678-5. [DOI] [PubMed] [Google Scholar]
- Masse K, Bhamra S, Allsop G, Dale N, Jones EA. Ectophosphodiesterase/nucleotide phosphohydrolase (Enpp) nucleotidases: cloning, conservation and developmental restriction. Int J Dev Biol. 2010;54:181–193. doi: 10.1387/ijdb.092879km. [DOI] [PubMed] [Google Scholar]
- Miller RH. Dorsally derived oligodendrocytes come of age. Neuron. 2005;45:1–3. doi: 10.1016/j.neuron.2004.12.032. [DOI] [PubMed] [Google Scholar]
- Moolenaar WH. Lysophospholipids in the limelight: autotaxin takes center stage. J Cell Biol. 2002;158:197–199. doi: 10.1083/jcb.200206094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukouyama YS, Deneen B, Lukaszewicz A, Novitch BG, Wichterle H, Jessell TM, Anderson DJ. Olig2+ neuroepithelial motoneuron progenitors are not multipotent stem cells in vivo. Proc Natl Acad Sci U S A. 2006;103:1551–1556. doi: 10.1073/pnas.0510658103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munzel EJ, Schaefer K, Obirei B, Kremmer E, Burton EA, Kuscha V, Becker CG, Brosamle C, Williams A, Becker T. Claudin k is specifically expressed in cells that form myelin during development of the nervous system and regeneration of the optic nerve in adult zebrafish. Glia. 2012;60:253–270. doi: 10.1002/glia.21260. [DOI] [PubMed] [Google Scholar]
- Murata J, Lee HY, Clair T, Krutzsch HC, Arestad AA, Sobel ME, Liotta LA, Stracke ML. cDNA cloning of the human tumor motility-stimulating protein, autotaxin, reveals a homology with phosphodiesterases. J Biol Chem. 1994;269:30479–30484. [PubMed] [Google Scholar]
- Nakanaga K, Hama K, Aoki J. Autotaxin--an LPA producing enzyme with diverse functions. J Biochem. 2010;148:13–24. doi: 10.1093/jb/mvq052. [DOI] [PubMed] [Google Scholar]
- Narita M, Goji J, Nakamura H, Sano K. Molecular cloning, expression, and localization of a brain-specific phosphodiesterase I/nucleotide pyrophosphatase (PD-I alpha) from rat brain. J Biol Chem. 1994;269:28235–28242. [PubMed] [Google Scholar]
- Nogaroli L, Yuelling LM, Dennis J, Gorse K, Payne SG, Fuss B. Lysophosphatidic acid can support the formation of membranous structures and an increase in MBP mRNA levels in differentiating oligodendrocytes. Neurochem Res. 2009;34:182–193. doi: 10.1007/s11064-008-9772-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ny A, Autiero M, Carmeliet P. Zebrafish and Xenopus tadpoles: small animal models to study angiogenesis and lymphangiogenesis. Exp Cell Res. 2006;312:684–693. doi: 10.1016/j.yexcr.2005.10.018. [DOI] [PubMed] [Google Scholar]
- Odenthal J, Nusslein-Volhard C. fork head domain genes in zebrafish. Dev Genes Evol. 1998;208:245–258. doi: 10.1007/s004270050179. [DOI] [PubMed] [Google Scholar]
- Ohno S, Wolf U, Atkins NB. Evolution from fish to mammals by gene duplication. Hereditas. 1968;59:169–187. doi: 10.1111/j.1601-5223.1968.tb02169.x. [DOI] [PubMed] [Google Scholar]
- Ohuchi H, Hayashibara Y, Matsuda H, Onoi M, Mitsumori M, Tanaka M, Aoki J, Arai H, Noji S. Diversified expression patterns of autotaxin, a gene for phospholipid-generating enzyme during mouse and chicken development. Dev Dyn. 2007;236:1134–1143. doi: 10.1002/dvdy.21119. [DOI] [PubMed] [Google Scholar]
- Orentas DM, Miller RH. The origin of spinal cord oligodendrocytes is dependent on local influences from the notochord. Dev Biol. 1996;177:43–53. doi: 10.1006/dbio.1996.0143. [DOI] [PubMed] [Google Scholar]
- Park HC, Mehta A, Richardson JS, Appel B. olig2 is required for zebrafish primary motor neuron and oligodendrocyte development. Dev Biol. 2002;248:356–368. doi: 10.1006/dbio.2002.0738. [DOI] [PubMed] [Google Scholar]
- Park HC, Shin J, Appel B. Spatial and temporal regulation of ventral spinal cord precursor specification by Hedgehog signaling. Development. 2004;131:5959–5969. doi: 10.1242/dev.01456. [DOI] [PubMed] [Google Scholar]
- Pitson SM, Pebay A. Regulation of stem cell pluripotency and neural differentiation by lysophospholipids. Neurosignals. 2009;17:242–254. doi: 10.1159/000231891. [DOI] [PubMed] [Google Scholar]
- Placzek M, Briscoe J. The floor plate: multiple cells, multiple signals. Nat Rev Neurosci. 2005;6:230–240. doi: 10.1038/nrn1628. [DOI] [PubMed] [Google Scholar]
- Poncet C, Soula C, Trousse F, Kan P, Hirsinger E, Pourquie O, Duprat AM, Cochard P. Induction of oligodendrocyte progenitors in the trunk neural tube by ventralizing signals: effects of notochord and floor plate grafts, and of sonic hedgehog. Mech Dev. 1996;60:13–32. doi: 10.1016/s0925-4773(96)00595-3. [DOI] [PubMed] [Google Scholar]
- Postlethwait J, Amores A, Cresko W, Singer A, Yan YL. Subfunction partitioning, the teleost radiation and the annotation of the human genome. Trends Genet. 2004;20:481–490. doi: 10.1016/j.tig.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Postlethwait JH. The zebrafish genome in context: ohnologs gone missing. J Exp Zool B Mol Dev Evol. 2007;308:563–577. doi: 10.1002/jez.b.21137. [DOI] [PubMed] [Google Scholar]
- Pradere JP, Tarnus E, Gres S, Valet P, Saulnier-Blache JS. Secretion and lysophospholipase D activity of autotaxin by adipocytes are controlled by N-glycosylation and signal peptidase. Biochim Biophys Acta. 2007;1771:93–102. doi: 10.1016/j.bbalip.2006.11.010. [DOI] [PubMed] [Google Scholar]
- Pringle NP, Richardson WD. A singularity of PDGF alpha-receptor expression in the dorsoventral axis of the neural tube may define the origin of the oligodendrocyte lineage. Development. 1993;117:525–533. doi: 10.1242/dev.117.2.525. [DOI] [PubMed] [Google Scholar]
- Pringle NP, Yu WP, Guthrie S, Roelink H, Lumsden A, Peterson AC, Richardson WD. Determination of neuroepithelial cell fate: induction of the oligodendrocyte lineage by ventral midline cells and sonic hedgehog. Dev Biol. 1996;177:30–42. doi: 10.1006/dbio.1996.0142. [DOI] [PubMed] [Google Scholar]
- Richardson WD, Kessaris N, Pringle N. Oligodendrocyte wars. Nat Rev Neurosci. 2006;7:11–18. doi: 10.1038/nrn1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson WD, Smith HK, Sun T, Pringle NP, Hall A, Woodruff R. Oligodendrocyte lineage and the motor neuron connection. Glia. 2000;29:136–142. doi: 10.1002/(sici)1098-1136(20000115)29:2<136::aid-glia6>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- Rinner O, Makhankov YV, Biehlmaier O, Neuhauss SC. Knockdown of cone-specific kinase GRK7 in larval zebrafish leads to impaired cone response recovery and delayed dark adaptation. Neuron. 2005;47:231–242. doi: 10.1016/j.neuron.2005.06.010. [DOI] [PubMed] [Google Scholar]
- Robu ME, Larson JD, Nasevicius A, Beiraghi S, Brenner C, Farber SA, Ekker SC. p53 activation by knockdown technologies. PLoS Genet. 2007;3:e78. doi: 10.1371/journal.pgen.0030078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowitch DH. Glial specification in the vertebrate neural tube. Nat Rev Neurosci. 2004;5:409–419. doi: 10.1038/nrn1389. [DOI] [PubMed] [Google Scholar]
- Rowitch DH, Kriegstein AR. Developmental genetics of vertebrate glial-cell specification. Nature. 2010;468:214–222. doi: 10.1038/nature09611. [DOI] [PubMed] [Google Scholar]
- Rowitch DH, Lu QR, Kessaris N, Richardson WD. An 'oligarchy' rules neural development. Trends Neurosci. 2002;25:417–422. doi: 10.1016/s0166-2236(02)02201-4. [DOI] [PubMed] [Google Scholar]
- Samadi N, Bekele R, Capatos D, Venkatraman G, Sariahmetoglu M, Brindley DN. Regulation of lysophosphatidate signaling by autotaxin and lipid phosphate phosphatases with respect to tumor progression, angiogenesis, metastasis and chemo-resistance. Biochimie. 2011;93:61–70. doi: 10.1016/j.biochi.2010.08.002. [DOI] [PubMed] [Google Scholar]
- Saunders LP, Cao W, Chang WC, Albright RA, Braddock DT, De La Cruz EM. Kinetic analysis of Autotaxin reveals substrate-specific catalytic pathways and a mechanism for lysophosphatidic acid distribution. J Biol Chem. 2011;286:30130–30141. doi: 10.1074/jbc.M111.246884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayers EW, Barrett T, Benson DA, Bolton E, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2011;39(Database issue):D38–D51. doi: 10.1093/nar/gkq1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schebesta M, Serluca FC. olig1 expression identifies developing oligodendrocytes in zebrafish and requires hedgehog and notch signaling. Dev Dyn. 2009;238:887–898. doi: 10.1002/dvdy.21909. [DOI] [PubMed] [Google Scholar]
- Seiler C, Finger-Baier KC, Rinner O, Makhankov YV, Schwarz H, Neuhauss SC, Nicolson T. Duplicated genes with split functions: independent roles of protocadherin15 orthologues in zebrafish hearing and vision. Development. 2005;132:615–623. doi: 10.1242/dev.01591. [DOI] [PubMed] [Google Scholar]
- Skokal RR, Rohlf FJ, editors. Biometry: the principle and practice in biological research. 3d ed. New York: W. H. Freeman and Company; 1995. [Google Scholar]
- Stainier DY. Zebrafish genetics and vertebrate heart formation. Nat Rev Genet. 2001;2:39–48. doi: 10.1038/35047564. [DOI] [PubMed] [Google Scholar]
- Stankoff B, Barron S, Allard J, Barbin G, Noel F, Aigrot MS, Premont J, Sokoloff P, Zalc B, Lubetzki C. Oligodendroglial expression of Edg-2 receptor: developmental analysis and pharmacological responses to lysophosphatidic acid. Mol Cell Neurosci. 2002;20:415–428. doi: 10.1006/mcne.2002.1129. [DOI] [PubMed] [Google Scholar]
- Strahle U, Blader P, Henrique D, Ingham PW. Axial, a zebrafish gene expressed along the developing body axis, shows altered expression in cyclops mutant embryos. Genes Dev. 1993;7:1436–1446. doi: 10.1101/gad.7.7b.1436. [DOI] [PubMed] [Google Scholar]
- Sussman CR, Dyer KL, Marchionni M, Miller RH. Local control of oligodendrocyte development in isolated dorsal mouse spinal cord. J Neurosci Res. 2000;59:413–420. doi: 10.1002/(SICI)1097-4547(20000201)59:3<413::AID-JNR16>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Svetlov SI, Ignatova TN, Wang KK, Hayes RL, English D, Kukekov VG. Lysophosphatidic acid induces clonal generation of mouse neurospheres via proliferation of Sca-1- and AC133-positive neural progenitors. Stem Cells Dev. 2004;13:685–693. doi: 10.1089/scd.2004.13.685. [DOI] [PubMed] [Google Scholar]
- Takada N, Appel B. Identification of genes expressed by zebrafish oligodendrocytes using a differential microarray screen. Dev Dyn. 2010;239:2041–2047. doi: 10.1002/dvdy.22338. [DOI] [PubMed] [Google Scholar]
- Takebayashi H, Nabeshima Y, Yoshida S, Chisaka O, Ikenaka K. The basic helix-loop-helix factor olig2 is essential for the development of motoneuron and oligodendrocyte lineages. Curr Biol. 2002;12:1157–1163. doi: 10.1016/s0960-9822(02)00926-0. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Okudaira S, Kishi Y, Ohkawa R, Iseki S, Ota M, Noji S, Yatomi Y, Aoki J, Arai H. Autotaxin stabilizes blood vessels and is required for embryonic vasculature by producing lysophosphatidic acid. J Biol Chem. 2006;281:25822–25830. doi: 10.1074/jbc.M605142200. [DOI] [PubMed] [Google Scholar]
- Taylor JS, Braasch I, Frickey T, Meyer A, Van de Peer Y. Genome duplication, a trait shared by 22000 species of ray-finned fish. Genome Res. 2003;13:382–390. doi: 10.1101/gr.640303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse C, Thisse B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat Protoc. 2008;3:59–69. doi: 10.1038/nprot.2007.514. [DOI] [PubMed] [Google Scholar]
- Tokumura A, Majima E, Kariya Y, Tominaga K, Kogure K, Yasuda K, Fukuzawa K. Identification of human plasma lysophospholipase D, a lysophosphatidic acid-producing enzyme, as autotaxin, a multifunctional phosphodiesterase. J Biol Chem. 2002;277:39436–39442. doi: 10.1074/jbc.M205623200. [DOI] [PubMed] [Google Scholar]
- Trevarrow B, Marks DL, Kimmel CB. Organization of hindbrain segments in the zebrafish embryo. Neuron. 1990;4:669–679. doi: 10.1016/0896-6273(90)90194-k. [DOI] [PubMed] [Google Scholar]
- Ulrich F, Ma LH, Baker RG, Torres-Vazquez J. Neurovascular development in the embryonic zebrafish hindbrain. Dev Biol. 2011;357:134–151. doi: 10.1016/j.ydbio.2011.06.037. [DOI] [PubMed] [Google Scholar]
- Umezu-Goto M, Kishi Y, Taira A, Hama K, Dohmae N, Takio K, Yamori T, Mills GB, Inoue K, Aoki J, et al. Autotaxin has lysophospholipase D activity leading to tumor cell growth and motility by lysophosphatidic acid production. J Cell Biol. 2002;158:227–233. doi: 10.1083/jcb.200204026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallstedt A, Klos JM, Ericson J. Multiple dorsoventral origins of oligodendrocyte generation in the spinal cord and hindbrain. Neuron. 2005;45:55–67. doi: 10.1016/j.neuron.2004.12.026. [DOI] [PubMed] [Google Scholar]
- van der Sar AM, Zivkovic D, den Hertog J. Eye defects in receptor protein-tyrosine phosphatase alpha knock-down zebrafish. Dev Dyn. 2002;223:292–297. doi: 10.1002/dvdy.10059. [DOI] [PubMed] [Google Scholar]
- van Meeteren LA, Moolenaar WH. Regulation and biological activities of the autotaxin-LPA axis. Prog Lipid Res. 2007;46:145–160. doi: 10.1016/j.plipres.2007.02.001. [DOI] [PubMed] [Google Scholar]
- van Meeteren LA, Ruurs P, Stortelers C, Bouwman P, van Rooijen MA, Pradere JP, Pettit TR, Wakelam MJ, Saulnier-Blache JS, Mummery CL, et al. Autotaxin, a secreted lysophospholipase D, is essential for blood vessel formation during development. Mol Cell Biol. 2006;26:5015–5022. doi: 10.1128/MCB.02419-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waskiewicz AJ, Rikhof HA, Hernandez RE, Moens CB. Zebrafish Meis functions to stabilize Pbx proteins and regulate hindbrain patterning. Development. 2001;128:4139–4151. doi: 10.1242/dev.128.21.4139. [DOI] [PubMed] [Google Scholar]
- Weiner JA, Hecht JH, Chun J. Lysophosphatidic acid receptor gene vzg-1/lpA1/edg-2 is expressed by mature oligodendrocytes during myelination in the postnatal murine brain. J Comp Neurol. 1998;398:587–598. [PubMed] [Google Scholar]
- Wu S, Wu Y, Capecchi MR. Motoneurons and oligodendrocytes are sequentially generated from neural stem cells but do not appear to share common lineage-restricted progenitors in vivo. Development. 2006;133:581–590. doi: 10.1242/dev.02236. [DOI] [PubMed] [Google Scholar]
- Yanagida K, Ishii S. Non-Edg family LPA receptors: the cutting edge of LPA research. J Biochem. 2011;150:223–232. doi: 10.1093/jb/mvr087. [DOI] [PubMed] [Google Scholar]
- Yu N, Lariosa-Willingham KD, Lin FF, Webb M, Rao TS. Characterization of lysophosphatidic acid and sphingosine-1-phosphate-mediated signal transduction in rat cortical oligodendrocytes. Glia. 2004;45:17–27. doi: 10.1002/glia.10297. [DOI] [PubMed] [Google Scholar]
- Yuelling LM, Fuss B. Autotaxin (ATX): a multi-functional and multi-modular protein possessing enzymatic lysoPLD activity and matricellular properties. Biochim Biophys Acta. 2008;1781:525–530. doi: 10.1016/j.bbalip.2008.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yukiura H, Hama K, Nakanaga K, Tanaka M, Asaoka Y, Okudaira S, Arima N, Inoue A, Hashimoto T, Arai H, et al. Autotaxin regulates vascular development via multiple lysophosphatidic acid (LPA) receptors in zebrafish. J Biol Chem. 2011;286:43972–43983. doi: 10.1074/jbc.M111.301093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zannino DA, Appel B. Olig2+ precursors produce abducens motor neurons and oligodendrocytes in the zebrafish hindbrain. J Neurosci. 2009;29:2322–2333. doi: 10.1523/JNEUROSCI.3755-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Anderson DJ. The bHLH transcription factors OLIG2 and OLIG1 couple neuronal and glial subtype specification. Cell. 2002;109:61–73. doi: 10.1016/s0092-8674(02)00677-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Amino acid sequence alignment. The degree of sequence similarity is color coded. Black letters on white background represent non-similar amino acid residues, red letters on yellow background represent identical amino acid residues, blue letters on turquoise background represent conserved amino acid residues, black letters on green background represent similar amino acid residues and green letters on white background represent weakly similar residues. The dashed lines at the bottom of the sequence delineate the known structure-function domains. The MORFO domain entails the nuclease-like domain but extends amino-terminal by 69 amino acid residues. The numbers on the top of the sequence indicate the position of amino acids within the sequence. The catalytic site residue is marked by a star. The EF hand-like motif is double underlined. Ensembl transcript IDs used for the alignment were as follows: zebrafish: ENSDART00000104089 and ENSDART00000047920, xenopus: ENSXETT00000006797, chicken: ENSGALT00000026501, human: ENST00000075322, rat: ENSRNOT00000005561, mouse: ENSMUST00000041591.