Abstract
A small group of human papillomaviruses (HPVs) cause almost all cervical carcinoma and a significant percentage of other anogenital tract and oral carcinoma. Another group of HPVs causes non-melanoma skin cancers in genetically predisposed or immune suppressed patients upon UV exposure. HPV genome replication requires the host cell’s DNA synthesis machinery and HPVs encode proteins that maintain differentiated epithelial cells in a replication competent state. The resulting rewiring of cellular signal transduction circuits triggers several innate cellular tumor suppressor responses that HPVs need to inactivate in order to establish persistent and/or productive infections. This review emphasizes this interplay between virus and the infected host cells and points out biological similarities and differences between different groups of HPVs.
Introduction
Human papillomaviruses (HPVs) are members of the papillomaviridae family. These epitheliotropic viruses contain ~8 kB double stranded circular DNA genomes encapsidated within 52–55 nm diameter non-enveloped particles. Peyton Rous discovered that warts caused by experimental infection with cotton tail rabbit papillomavirus (CRPV) progressed to carcinomas when they were treated with chemical carcinogens [1]. A link of HPVs to human cancers was first documented in patients with a rare hereditary skin disorder, epidermodysplasia verruciformis (EV), who develop a large number of warts, some of which progress to carcinomas particularly in sun-exposed areas of the body [2]. EV-associated HPVs are members of an extensive group of β HPVs that colonize cutaneous epithelia. Harald zur Hausen’s group discovered that some members of a distinct group of mucosatropic α HPVs are etiological agents of almost all cervical carcinomas [3]. These “high-risk” HPVs also cause other anogenital tract carcinomas including anal, vulvar, vaginal and penile cancers and more recently have been linked to oral cancer, particularly oropharyngeal carcinomas. Prophylactic vaccines targeting the most abundant high-risk and low-risk α HPVs proved highly efficacious in preventing premalignant cervical and anal lesions as well as genital warts [reviewed in 4].
Due to frequent integration of HPV sequences into a host cell chromosome during malignant progression, HPV-associated cervical carcinoma often represent non-productive infections that only express two viral proteins, E6 and E7 [reviewed in 5]. Thus, cancer formation is not part of the viral life cycle, and the transforming activities of HPV “onco”-proteins are manifestations of their activities for the productive viral life cycle.
HPV E6 and E7 protein structures
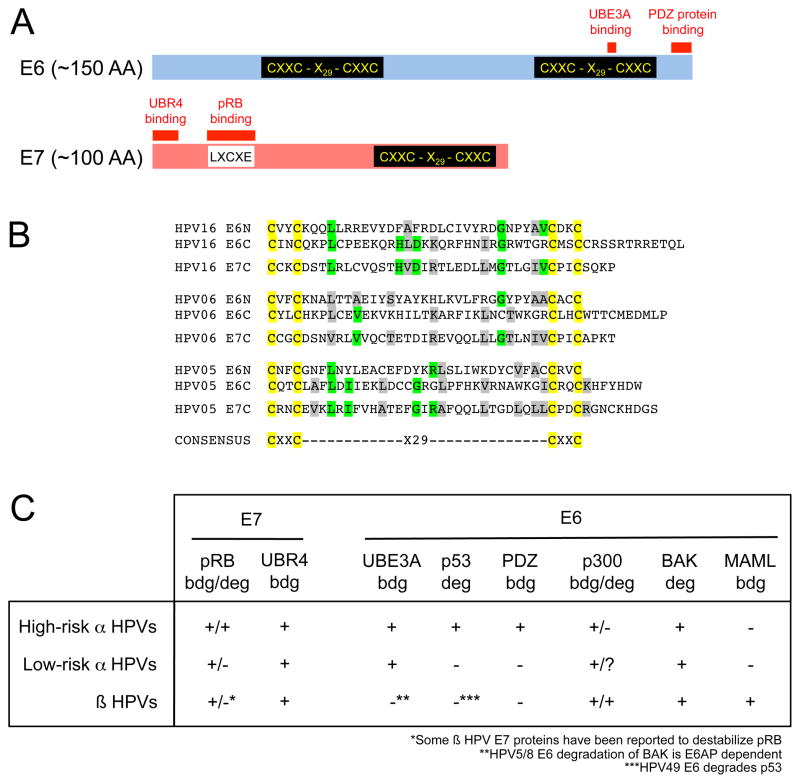
HPV E6 and E7 encode low molecular weight proteins of ~150 and ~100 amino acids, respectively (Fig 1A). They each contain zinc-binding domains consisting of two copies of CXXC separated by 29 amino acid residues (Fig 1B) and may have evolved from a common ancestor. Interestingly, some animal papillomaviruses contain only E6 and no E7 or vice versa. There is also evidence for domain shuffling as the PDZ binding motif found on high-risk HPV E6 proteins is at the carboxyl termini of the Rhesus macaque papillomavirus (RhPV) and some cynomolgus macaque papillomavirus E7 proteins [6]. The recently identified Mus musculus papillomavirus 1 (MmuPV1) E6 protein contains an LXCXE sequence similar to the core pRB-binding site in many HPV E7 proteins [7].
Figure 1.
Biochemical and biological activities of HPV E6 and E7 proteins. (A) Schematic depiction of high-risk α HPV E6 and E7 proteins. The approximate locations of the zinc-binding domains and sequences necessary for association with host cell proteins are indicated. (B) Sequence alignments of E6 and E7 zinc binding domains. HPV16 (high-risk α), HPV6 (low-risk α) and HPV5 (β) are shown as representative examples. Cysteine residues are highlighted in yellow, green denote residues identical in E7 and at least one of the E6 domains and grey are chemically similar residues in E7 and at least one of the E6 domains. (C) Association of E6 and E7 proteins encoded by high-risk αlow-riskαand β HPVs with cellular proteins. “bdg” denotes “binding” and “deg” denotes “degradation”. See text for details and references.
Structures of several E7 proteins have been reported, and a structure of the C-terminal E6 zinc-binding site has also been published. Both E6 and E7 can form dimers and, despite the conserved CXXC domain architecture (Fig 1B), the 3 dimensional structures of these domains in E6 and E7 are quite distinct [8–10]. The E7 amino terminus represents an intrinsically unstructured region, and a peptide corresponding to the HPV16 E7 core pRB-binding site assumes an extended structure when bound to pRB [11].
Mucosal α HPV E6 and E7 proteins reprogram cellular ubiquitin ligases
HPV E6 and E7 proteins lack intrinsic enzymatic activities and function by associating with and functionally reprogramming key components of host cellular signal transduction networks. The most extensively studied cellular E6 and E7 targets are the p53 and retinoblastoma (pRB) tumor suppressors, respectively (Fig 1C). High-risk HPV E6 and E7 proteins do not inactivate these tumor suppressors by stoichiometric sequestration but target them for ubiquitin-mediated proteasomal degradation.
High-risk HPV E6 proteins target p53 for ubiquitination by associating with and reprogramming the UBE3A (E6AP) ubiquitin ligase [reviewed in 5]. High-risk HPV E6 proteins also contain a type I PDZ binding motif (X-X-S/T-X-I/L/V) at their carboxyl termini, and E6 associated PDZ proteins can also be targeted for degradation through UBE3A [reviewed in 12]. PDZ protein binding is critical for E6 transforming activities. Even though the physiologically relevant cellular PDZ target(s) have not yet been identified, association of E6 with the discs large tumor suppressor homolog DLG1 causes constitutive activation of the ras homolog gene family, member G (RHOG), which contributes to the invasive properties of cervical carcinoma cells [13]. Low-risk HPV E6 proteins also form complexes with UBE3A. But since low-risk HPV E6 proteins do not associate with p53 and lack carboxyl terminal PDZ binding domains, low-risk HPV E6/UBE3A complexes do not target p53 or cellular PDZ proteins for degradation [reviewed in 14]. Ubiquitination substrates of low-risk HPV E6/UBE3A complexes remain to be determined.
High-risk HPV E6 proteins can also induce degradation of cellular proteins through UBE3A independent mechanisms [15]. HPV E6 modulates cell adhesion and focal adhesion pathways through UBE3A-independent of TAp63β thereby contributing to anchorage independent growth and cellular transformation [16].
E7 mediated pRB degradation has been mechanistically elucidated only for HPV16 E7, which associates with a ZER1 containing cullin 2 (CUL2) ubiquitin ligase complex [17]. Whereas pRB degradation is shared with other high-risk HPV E7 proteins, it does not involve the same CUL2 complex. Many papillomavirus E7 proteins associate with the UBR4 ubiquitin ligase, a 600 kDa protein that may regulate anoikis [reviewed in 5]. Association with UBR4 is independent of pRB and the potential degradation substrates of the E7/UBR4 complex remain unknown.
Mucosal α HPV E6 and E7 proteins reprogram cellular transcriptional programs
HPV E6 and E7 proteins lack specific DNA binding properties but associate with cellular transcription factor complexes and alter their activities. High-risk HPV E6 degradation of p53 blunts the p53 transcriptional response. E6 proteins also associate with other components of the p53 transcriptional response, including the coactivator p300, the adaptor protein TADA3 [reviewed in 14] and the TIP60 complex [18]. Each of these also affects p53 independent transcriptional responses. HPV16 E7 also inhibits p53 transcriptional activities and inactivates the p53 dependent G1 cell cycle inhibitor p21CIP1 [reviewed in 14]. Hence high-risk HPVs inhibit p53 signaling through multiple mechanisms.
High-risk HPV E6 proteins increase expression of the catalytic protein subunit of human telomerase, TERT. Several mechanisms have been suggested, some dependent on MYC and UBE3A, whereas others involve UBE3A mediated degradation of the TERT transcriptional repressor NFX1-91 [reviewed in 5,14].
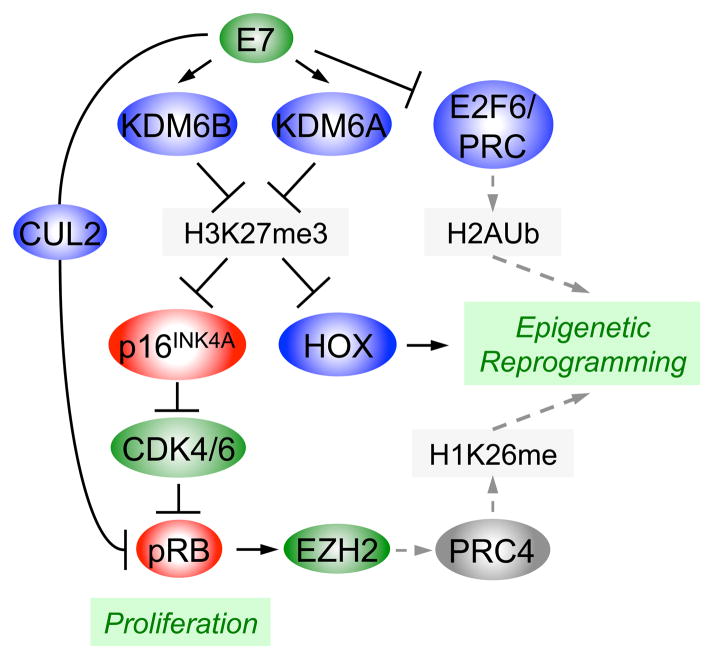
The pRB family of proteins regulates the activity of canonical E2F family members, and E7 degradation of pRB subverts this regulatory loop. E7 has also been reported to interact with canonical and non-canonical E2F family members and modulate their transcriptional activities [19,20]. The non-canonical E2F6 is a transcriptional repressor for E2F and E box containing enhancers, and E7 association abrogates E2F6 repressor activity. E2F6 is also a component of polycomb repressive complexes (PRC), and the detection of E2F6/PRCs is diminished in E7 expressing cells [20]. PRCs are transcriptional repressor complexes that play important roles in establishing and maintaining transcriptionally silenced chromatin through trimethylation of lysine 27 on histone H3 (H3K27).
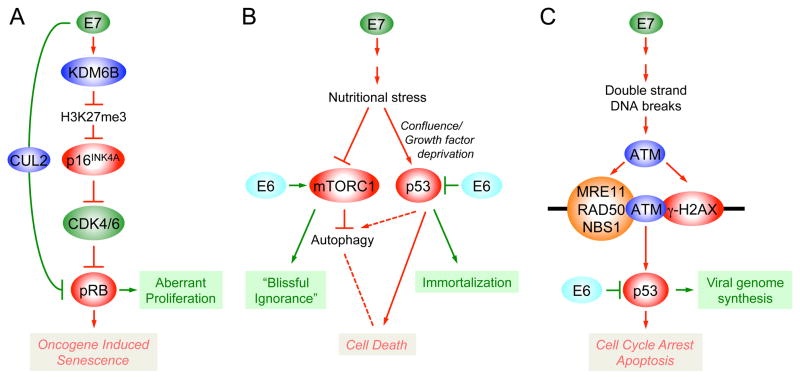
HPV16 E7 expression causes a dramatic reduction of the H3K27 mark that is essential for PRC repression. This is due to transcriptional induction of the two H3K27 specific demethylases KDM6A and KDM6B through an unknown but pRB independent mechanism [21]. H3K27 demethylation causes enhanced expression of “poised” genes, which contain both repressive (H3K27me3) and activating (H3K4me3) marks. These include homeobox genes as well as the CDK4/6 inhibitor and tumor suppressor p16INK4A, a well established biomarker for high-risk HPV infected cells (Fig 2). Induction of p16INK4A through KDM6B mediated H3K27 demethylation is also the mechanistic basis for oncogene induced senescence (OIS), a cell intrinsic innate tumor suppressive mechanism triggered by oncogenes such as RAS [22,23]. HPV16 E7 degrades pRB, the major mediator of p16INK4A induced growth arrest and senescence, thereby abrogating OIS. Hence, the ability of E7 to inactivate pRB may be driven by the necessity to avoid elimination of virally infected cells by E7 triggered OIS (Fig 3A). HPV E6/E7 expressing cells also show evidence for decreased expression of the PRC protein BMI1 [24]. As a consequence HPV E7 expression causes epigenetic reprogramming as evidenced by aberrant expression of homeobox genes, which control cell fate and identity [21,24] (Fig 2). The CaSki cervical carcinoma cell line is addicted to KDM6A and KDM6B expression suggesting that E7 induced epigenetic reprogramming may contribute to cancer formation [21].
Figure 2.
HPV16 E7 expression causes global H3K27 demethylation and potentially other changes in the histone code. Oncogenic genes are shown in green, tumor suppressors in red, components that are predicted to be altered by E7 in grey and all the other genes are highlighted in blue. Dashed grey arrows represent connections that have not been fully explored experimentally. See text for details and references.
Figure 3.
Schematic depiction of innate tumor suppressor responses triggered by high-risk α HPV E7 expression. (A) Oncogene Induced Senescence (OIS); (B) “Trophic Sentinel Response”; (C) Double strand DNA break response. Red lines represent innate tumor suppressor signaling pathways with the respective cytostatic or cytotoxic outcomes indicated in red letters. Viral subversion of these innate tumor suppressor responses is indicated by the green lines, with the respective biological outcome indicated in green letters. Autophagy can both negatively and positively affect cell death. Solid lines denote experimentally established pathways and connections, whereas dotted lines denote hypothetical connections. See text for details and references.
The H3K27 methyltransferase and PRC component EZH2 is expressed at high levels in HPV E7 expressing cells through E2F activation [25]. EZH2 over-expression enhances formation of the normally low-abundance PRC4 that methylates H1 on lysine 26 [26]. Moreover, EZH2 in HPV E6/E7 expressing cells is serine 21 phosphorylated by AKT, which reduces EZH2 enzymatic activity [24].
Unlike other DNA viruses HPVs do not encode miRNAs. However, viral infection alters the expression of cellular miRNAs such as mir203 to regulate the abundance of cellular proteins, including p63, to allow viral progeny synthesis [27]. As a consequence, HPV-positive cancers display changes in the expression of a number of cellular oncogenic and tumor suppressive miRNAs. However, a consensus as to a distinct miRNA expression profile(s) for HPV-positive lesions has not yet been reached [reviewed in 28].
Mucosal α HPV E6 and E7 proteins reprogram cellular metabolism
Similar to what has been reported for the Adenovirus E1A and CMYC proteins, HPV16 E7 expressing primary human cells are acutely dependent on exogenous growth factor and undergo non-apoptotic cell death when they reach confluence and/or are growth factor deprived [29]. Evidence of autophagy, a cellular recycling mechanism that is activated under conditions of metabolic stress, was detected in HPV16 E7 expressing primary human epithelial cells even when grown in normal culture medium [30] (Fig 3B). HPV E7 expression has been reported to induce the Warburg effect, a reprogramming of the cellular metabolism to aerobic fermentation and a general hallmark of human tumor cells [31]. Aerobic fermentation yields less ATP but provides necessary precursors for many anabolic processes including amino acid, nucleotide and lipid synthesis. HPV16 E6 expression balances E7 induced metabolic stress by short-circuiting the mTORC1 metabolic rheostat [32]. E6 activates mTORC1 through two cooperating pathways, the mTORC2 complex and PDK1, a kinase that transmits growth factor signals downstream of phosphoinositide-3-kinase (PI3K) to AKT. Activity of mTORC1 is sustained for extended periods of time when growth factors are removed from E6 expressing cells [32], suggesting that E6 induces a state of blissful ignorance of cellular nutrient supply (Fig 3B). Increased mTORC1 activity has been detected in HPV associated premalignant lesions and cancers [33].
Mucosal HPVs also encode E5 proteins, but unlike the BPV1 small single pass transmembrane E5 protein that induces constitutive, growth factor independent PDGF receptor signaling by forcing receptor dimerization [reviewed in 34], α HPV E5 proteins contain three membrane-spanning domains [35]. HPV E5 proteins are only weakly transforming and are not always expressed in cancers. Some reports indicate that HPV E5 proteins may activate EGF receptor signaling potentially by inhibiting endosomal EGF receptor trafficking, which may also lead to a sustained ligand-dependent growth factor receptor signaling [reviewed in 14].
Cancer-associated α HPV E6 and E7 proteins subvert genomic stability
HPV E6 and E7 proteins contribute to carcinogenic progression through induction of genomic instability. In addition to centrosome-associated mitotic abnormalities, HPV E6/E7 expressing cells also show evidence for anaphase bridges, which are likely caused by accumulation of double strand DNA breaks [36]. HPV episome containing primary human epithelial cells show evidence of activated DNA damage signaling including activation of the ATM kinase and downstream signaling events [37]. This is at least in part caused by E7 expression. The DNA double strand break response may represent yet another tumor suppressive cellular response to HPV E7 expression, but activation of the DNA repair machinery is harnessed for HPV genome replication in differentiated keratinocytes [37] (Fig 3C).
Cutaneous β HPVs subvert NOTCH signaling
Despite the strong association with SCCs in EV and immune compromised patients, the mechanistic role(s) of β HPVs in human cancers is not clearly defined. Unlike cancers caused by α HPVs, β HPV associated SCCs represent productive infections. Mutations of the EVER1 and EVER2 genes on 17q25 have been linked to EV development but how defects in these zinc-binding proteins mechanistically contribute to HPV-associated SCC development is unclear [reviewed in 38]. There are no sensitive cell based transformation assays for β HPVs, but E6, E7 and E2 score as tumorigenic when expressed in basal epithelial cells of transgenic mice [39,40].
While the prototype β HPV5 and HPV8 E6 and E7 proteins do not target p53 or pRB tumor suppressors for degradation, some other β HPVs have been reported to cause pRB and p53 degradation [41] (Fig 1C) and/or to inhibit p53 functions indirectly by causing ΔNp73 accumulation [42]. Moreover, HPV5 and HPV8 E6 proteins associate with and induce degradation of p300 [43,44], a critical co-activator for many transcriptional programs, including p53.
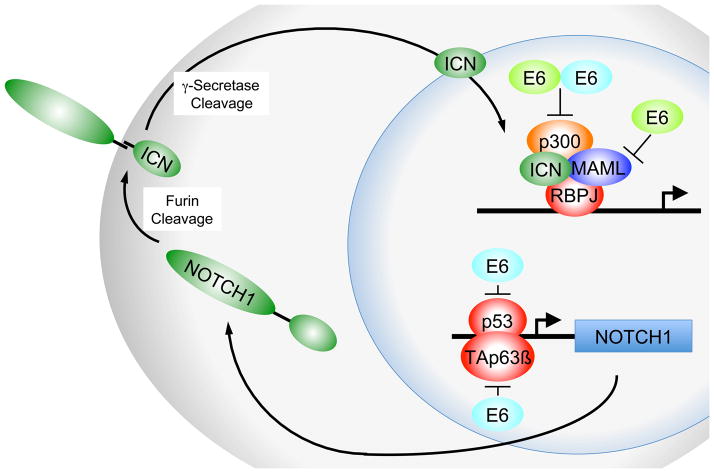
Similar to high-risk α HPVs, β HPVs disrupt epithelial differentiation through E6 and E7 expression. A recently discovered interaction between β HPV E6 proteins and mastermind-like (MAML) proteins provides a possible mechanism for the disruption of epithelial differentiation [45]. MAML family members are co-activators of Notch signaling complexes that contain p300 and the intracellular fragment of Notch bound to the DNA-binding protein RBPJ (CBF1). Notch signaling drives terminal differentiation of keratinocytes and acts as a tumor suppressor in epithelial tumors [reviewed in 46]. Whereas α HPVs do not directly target MAML, high-risk α HPV E6 proteins inhibit Notch indirectly through p53 inactivation [reviewed in 47] and degradation of TAp63β [16] (Fig 4).
Figure 4.
Inhibition of Notch signaling by β HPV E6 proteins (bright green) binding to members of the human mastermind like (MAML) coactivators. High-risk α HPV E6 proteins (bright blue) inhibit NOTCH1 signaling though inhibition of p53 and/or TAp63β mediated NOTCH1 transcription. ICN denotes the intracellular NOTCH fragment. See text for details and references.
Cutaneous β HPV E6 proteins may also contribute to carcinogenesis by targeting Bak for proteasomal degradation. This activity is shared with α HPV E6 proteins [reviewed in 14]. The pro-apoptotic Bak protein plays a critical role in UV-induced apoptosis. Since UV exposure is a critical cofactor for β HPV associated SCC development, Bak degradation may contribute to tumor formation by protecting infected cells from UV-induced apoptosis thereby promoting proliferation of cells that have accumulated DNA damage. E6 degradation of Bak has been reported to be E6AP-dependent, although no stable interaction of β HPV E6 and E6AP is observed [48].
Exploiting HPV induced cellular alterations for therapeutic purposes
Since HPVs cause tumors years or decades after the initial infection, the currently available prophylactic HPV vaccines will not have a measurable impact on HPV associated cancer rates for two or three decades [reviewed in 49]. Therapeutic vaccines may be a viable approach to therapy, and in a small study an HPV16 E6/E7 based peptide vaccine caused regression of HPV associated premalignant vulvar lesions [50]. Given that HPV associated cancers are addicted to E6/E7 expression, HPV associated cancers are prime candidates for development of RNAi based therapeutic approaches.
As pointed out above, HPV E6/E7 expression causes striking rewiring of cellular signal transduction pathways, which generates unique dependences on specific cellular enzymes. Such vulnerabilities can be discovered by RNAi based loss-of-function genetic screens. In a small proof of principle study, the cellular PAK3 and SGK2 kinases that are dispensable in normal cells were found to be essential in cervical carcinoma cell lines as a consequence of E6 mediated p53 inactivation [51]. Similarly, as mentioned above, HPV cancer cells are addicted to the KDM6A and KDM6B H3K27 demethylases [21]. Small molecule inhibitors of these enzymes may be useful therapeutics for HPV associated tumors. Since the transforming activities of α HPV E6 and E7 are at least in part based on ubiquitination and proteasomal degradation of associated cellular proteins recently developed small molecule proteasome and/or cullin based ubiquitin ligase inhibitors should be evaluated for therapy of HPV associated lesions and cancers.
HPV E7 expression triggers several cell intrinsic tumor suppressor pathways, including cell death due to accumulation of double strand DNA breaks or in response to growth factor deprivation, as well as oncogene induced senescence. These cell abortive responses are balanced by other E7 activities and/or by the co-expressed E6 oncoprotein (Fig 3). Once the mechanistic interplay of these activities has been delineated in more detail one might envision therapeutic approaches based on reactivating these cell intrinsic tumor suppressive responses.
Highlights.
HPV progeny synthesis is confined to differentiated epithelial cells
HPVs encode proteins that retain differentiated epithelial cells in a replication competent state
Rewiring of cellular signaling circuits by HPV infection triggers innate cellular tumor suppressor responses
Different groups of HPVs have evolved different strategies to subvert these cellular defenses
This interplay between HPVs and their hosts contributes to the oncogenic activity of some HPVs
Acknowledgments
We apologize to those authors whose important contributions could not be appropriately discussed and cited due to space limitations. The current work on HPV transformation in our laboratory is supported by Public Health Service grants CA081135, CA066980, CA141583, and HG004233 (KM) and CA143010 (MEM-D).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest published within the annual period of review, have been highlighted as:
* of special interest
** of outstanding interest
- 1.Rous P, Beard JW. The Progression to Carcinoma of Virus-Induced Rabbit Papillomas (Shope) J Exp Med. 1935;62:523–548. doi: 10.1084/jem.62.4.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Orth G, Jablonska S, Favre M, Croissant O, Jarzabek-Chorzelska M, Rzesa G. Characterization of two types of human papillomaviruses in lesions of epidermodysplasia verruciformis. Proc Natl Acad Sci U S A. 1978;75:1537–1541. doi: 10.1073/pnas.75.3.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dürst M, Gissmann L, Ikenberg H, zur Hausen H. A papillomavirus DNA from cervical carcinoma and its prevalence in cancer biopsy samples from different geographic regions. Proceedings of the National Academy of Sciences USA. 1983;80:3812–3815. doi: 10.1073/pnas.80.12.3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schiffman M, Castle PE, Jeronimo J, Rodriguez AC, Wacholder S. Human papillomavirus and cervical cancer. Lancet. 2007;370:890–907. doi: 10.1016/S0140-6736(07)61416-0. [DOI] [PubMed] [Google Scholar]
- 5.Moody CA, Laimins LA. Human papillomavirus oncoproteins: pathways to transformation. Nat Rev Cancer. 2010;10:550–560. doi: 10.1038/nrc2886. [DOI] [PubMed] [Google Scholar]
- 6.Tomaic V, Gardiol D, Massimi P, Ozbun M, Myers M, Banks L. Human and primate tumour viruses use PDZ binding as an evolutionarily conserved mechanism of targeting cell polarity regulators. Oncogene. 2009;28:1–8. doi: 10.1038/onc.2008.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Joh J, Jenson AB, King W, Proctor M, Ingle A, Sundberg JP, Ghim SJ. Genomic analysis of the first laboratory-mouse papillomavirus. J Gen Virol. 2011;92 :692–698. doi: 10.1099/vir.0.026138-0. [DOI] [PubMed] [Google Scholar]
- 8.Liu X, Clements A, Zhao K, Marmorstein R. Structure of the human Papillomavirus E7 oncoprotein and its mechanism for inactivation of the retinoblastoma tumor suppressor. J Biol Chem. 2006;281:578–586. doi: 10.1074/jbc.M508455200. [DOI] [PubMed] [Google Scholar]
- 9.Ohlenschlager O, Seiboth T, Zengerling H, Briese L, Marchanka A, Ramachandran R, Baum M, Korbas M, Meyer-Klaucke W, Durst M, et al. Solution structure of the partially folded high-risk human papilloma virus 45 oncoprotein E7. Oncogene. 2006 doi: 10.1038/sj.onc.1209584. [DOI] [PubMed] [Google Scholar]
- 10.Nomine Y, Masson M, Charbonnier S, Zanier K, Ristriani T, Deryckere F, Sibler AP, Desplancq D, Atkinson RA, Weiss E, et al. Structural and functional analysis of E6 oncoprotein: insights in the molecular pathways of human papillomavirus-mediated pathogenesis. Mol Cell. 2006;21:665–678. doi: 10.1016/j.molcel.2006.01.024. [DOI] [PubMed] [Google Scholar]
- 11.Lee JO, Russo AA, Pavletich NP. Structure of the retinoblastoma tumour-suppressor pocket domain bound to a peptide from HPV E7. Nature. 1998;391:859–865. doi: 10.1038/36038. [DOI] [PubMed] [Google Scholar]
- 12.Thomas M, Narayan N, Pim D, Tomaic V, Massimi P, Nagasaka K, Kranjec C, Gammoh N, Banks L. Human papillomaviruses, cervical cancer and cell polarity. Oncogene. 2008;27:7018–7030. doi: 10.1038/onc.2008.351. [DOI] [PubMed] [Google Scholar]
- 13**.Krishna Subbaiah V, Massimi P, Boon SS, Myers MP, Sharek L, Garcia-Mata R, Banks L. The Invasive Capacity of HPV Transformed Cells Requires the hDlg-Dependent Enhancement of SGEF/RhoG Activity. PLoS Pathog. 2012;8:e1002543. doi: 10.1371/journal.ppat.1002543. This publication establishes a molecular mechanism by which the human discs large protein regulates cell migration and invasion and explains how this is harnessed by the HPV E6 oncoprotein to activate these processes. The study suggests that E6 association with PDZ proteins does not necessarily cause their functional inactivation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klingelhutz AJ, Roman A. Cellular transformation by human papillomaviruses: Lessons learned by comparing high- and low-risk viruses. Virology. 2012;424:77–98. doi: 10.1016/j.virol.2011.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Massimi P, Shai A, Lambert P, Banks L. HPV E6 degradation of p53 and PDZ containing substrates in an E6AP null background. Oncogene. 2008;27 :1800–1804. doi: 10.1038/sj.onc.1210810. [DOI] [PubMed] [Google Scholar]
- 16*.Khalifa YB, Teissier S, Tan MK, Phan QT, Daynac M, Wong WQ, Thierry F. The human papillomavirus E6 oncogene represses a cell adhesion pathway and disrupts focal adhesion through degradation of TAp63beta upon transformation. PLoS Pathog. 2011;7:e1002256. doi: 10.1371/journal.ppat.1002256. This publication shows that HPV16 E6 causes degradation of the TAp63β isoform through a UBE3A independent mechanism. TAp63β activates cell adhesion and focal adhesion pathways and degradation by HPV E6 contributes to establishment of anchorage independent growth. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.White EA, Sowa ME, Tan MJ, Jeudy S, Hayes SD, Santha S, Munger K, Harper JW, Howley PM. Systematic identification of interactions between host cell proteins and E7 oncoproteins from diverse human papillomaviruses. Proc Natl Acad Sci U S A. 2012;109:E260–267. doi: 10.1073/pnas.1116776109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18*.Jha S, Vande Pol S, Banerjee NS, Dutta AB, Chow LT, Dutta A. Destabilization of TIP60 by human papillomavirus E6 results in attenuation of TIP60-dependent transcriptional regulation and apoptotic pathway. Mol Cell. 2010;38:700–711. doi: 10.1016/j.molcel.2010.05.020. This publication reports that high-risk as well as low-risk HPV E6 proteins destabilize TIP60 through a proteasome-dependent, UBE3A-independent mechanism. This causes de-repression of HPV early gene transcription and abrogates p53 induction of pro-apoptotic genes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hwang SG, Lee D, Kim J, Seo T, Choe J. Human papillomavirus type 16 E7 binds to E2F1 and activates E2F1-driven transcription in a retinoblastoma protein-independent manner. J Biol Chem. 2002;277:2923–2930. doi: 10.1074/jbc.M109113200. [DOI] [PubMed] [Google Scholar]
- 20.McLaughlin-Drubin ME, Huh KW, Munger K. Human papillomavirus type 16 E7 oncoprotein associates with E2F6. J Virol. 2008;82:8695–8705. doi: 10.1128/JVI.00579-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21**.McLaughlin-Drubin ME, Crum CP, Munger K. Human papillomavirus E7 oncoprotein induces KDM6A and KDM6B histone demethylase expression and causes epigenetic reprogramming. Proc Natl Acad Sci U S A. 2011;108:2130–2135. doi: 10.1073/pnas.1009933108. This publication demonstrates that induction of p16INK4A by HPV16 E7 is through KDM6B mediated H3K27 demethylation and may reflect E7 triggered oncogene induced senescence (OIS). Since HPV16 E7 degrades pRB, the major mediator of p16INK4A induced growth arrest and senescence, HPV E7 expressing cells escape OIS and show evidence of epigenetic reprogramming. HPV16 positive CaSki cervical carcinoma cells are addicted to KDM6B expression. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barradas M, Anderton E, Acosta JC, Li S, Banito A, Rodriguez-Niedenfuhr M, Maertens G, Banck M, Zhou MM, Walsh MJ, et al. Histone demethylase JMJD3 contributes to epigenetic control of INK4a/ARF by oncogenic RAS. Genes Dev. 2009;23:1177–1182. doi: 10.1101/gad.511109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Agger K, Cloos PA, Rudkjaer L, Williams K, Andersen G, Christensen J, Helin K. The H3K27me3 demethylase JMJD3 contributes to the activation of the INK4A-ARF locus in response to oncogene- and stress-induced senescence. Genes Dev. 2009;23:1171–1176. doi: 10.1101/gad.510809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24*.Hyland PL, McDade SS, McCloskey R, Dickson GJ, Arthur K, McCance DJ, Patel D. Evidence for alteration of EZH2, BMI1, and KDM6A and epigenetic reprogramming in human papillomavirus type 16 E6/E7-expressing keratinocytes. J Virol. 2011;85:10999–11006. doi: 10.1128/JVI.00160-11. This publication shows that HPV16 E6/E7 expressing cells express increased levels of KDM6A as well as AKT phosphorylated EZH2 histone methyltransferase, whereas expression of the polycomb repressive complex 1 component BMI1 is decreased. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25*.Holland D, Hoppe-Seyler K, Schuller B, Lohrey C, Maroldt J, Durst M, Hoppe-Seyler F. Activation of the enhancer of zeste homologue 2 gene by the human papillomavirus E7 oncoprotein. Cancer Res. 2008;68:9964–9972. doi: 10.1158/0008-5472.CAN-08-1134. This publication reports that HPV16 E7 induces transcription of the polycomb group gene enhancer of zeste homologue 2 (EZH2) through E2F. EZH2 expression is required for proliferation of HPV-positive tumor cells and may also contribute to apoptosis resistance of cervical cancer cells. [DOI] [PubMed] [Google Scholar]
- 26.Kuzmichev A, Margueron R, Vaquero A, Preissner TS, Scher M, Kirmizis A, Ouyang X, Brockdorff N, Abate-Shen C, Farnham P, et al. Composition and histone substrates of polycomb repressive group complexes change during cellular differentiation. Proc Natl Acad Sci U S A. 2005;102:1859–1864. doi: 10.1073/pnas.0409875102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Melar-New M, Laimins LA. Human papillomaviruses modulate expression of microRNA 203 upon epithelial differentiation to control levels of p63 proteins. J Virol. 2010;84:5212–5221. doi: 10.1128/JVI.00078-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gardiner AS, Wald AI, Khan SA. Alterations in Cellular miRNAs Induced by Human Papillomaviruses. In: Gaston K, editor. Small DNA Tumour Viruses. Caister Academic Press; 2012. pp. 151–173. [Google Scholar]
- 29.Jones DL, Thompson DA, Munger K. Destabilization of the RB tumor suppressor protein and stabilization of p53 contribute to HPV type 16 E7-induced apoptosis. Virology. 1997;239:97–107. doi: 10.1006/viro.1997.8851. [DOI] [PubMed] [Google Scholar]
- 30.Zhou X, Munger K. Expression of the human papillomavirus type 16 E7 oncoprotein induces an autophagy-related process and sensitizes normal human keratinocytes to cell death in response to growth factor deprivation. Virology. 2009;385:192–197. doi: 10.1016/j.virol.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zwerschke W, Mazurek S, Massimi P, Banks L, Eigenbrodt E, Jansen-Durr P. Modulation of type M2 pyruvate kinase activity by the human papillomavirus type 16 E7 oncoprotein. Proc Natl Acad Sci U S A. 1999;96 :1291–1296. doi: 10.1073/pnas.96.4.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spangle JM, Munger K. The human papillomavirus type 16 E6 oncoprotein activates mTORC1 signaling and increases protein synthesis. J Virol. 2010;84 :9398–9407. doi: 10.1128/JVI.00974-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feng W, Duan X, Liu J, Xiao J, Brown RE. Morphoproteomic evidence of constitutively activated and overexpressed mTOR pathway in cervical squamous carcinoma and high grade squamous intraepithelial lesions. Int J Clin Exp Pathol. 2009;2:249–260. [PMC free article] [PubMed] [Google Scholar]
- 34.Talbert-Slagle K, DiMaio D. The bovine papillomavirus E5 protein and the PDGF beta receptor: it takes two to tango. Virology. 2009;384:345–351. doi: 10.1016/j.virol.2008.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krawczyk E, Suprynowicz FA, Sudarshan SR, Schlegel R. Membrane orientation of the human papillomavirus type 16 E5 oncoprotein. J Virol. 2010;84:1696–1703. doi: 10.1128/JVI.01968-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Duensing S, Münger K. The human papillomavirus type 16 E6 and E7 oncoproteins independently induce numerical and structural chromosome instability. Canc Res. 2002;62:7075–7082. [PubMed] [Google Scholar]
- 37**.Moody CA, Laimins LA. Human papillomaviruses activate the ATM DNA damage pathway for viral genome amplification upon differentiation. PLoS Pathog. 2009;5:e1000605. doi: 10.1371/journal.ppat.1000605. This publication reports activation of ATM mediated double strand DNA break signaling in HPV31 episome expressing cells and shows that induction of this response is essential for HPV genome amplification in differentiating epithelial cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lazarczyk M, Cassonnet P, Pons C, Jacob Y, Favre M. The EVER proteins as a natural barrier against papillomaviruses: a new insight into the pathogenesis of human papillomavirus infections. Microbiol Mol Biol Rev. 2009;73:348–370. doi: 10.1128/MMBR.00033-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39**.Pfefferle R, Marcuzzi GP, Akgul B, Kasper HU, Schulze F, Haase I, Wickenhauser C, Pfister H. The human papillomavirus type 8 E2 protein induces skin tumors in transgenic mice. J Invest Dermatol. 2008;128:2310–2315. doi: 10.1038/jid.2008.73. This publication reports that the beta HPV8 E2 protein is tumorigenic when expressed from a basal epithelial cell specific promoter in transgenic mice. This is the first report to show tumorigenic activity by any HPV E2 protein. [DOI] [PubMed] [Google Scholar]
- 40.Marcuzzi GP, Hufbauer M, Kasper HU, Weissenborn SJ, Smola S, Pfister H. Spontaneous tumour development in human papillomavirus type 8 E6 transgenic mice and rapid induction by UV-light exposure and wounding. J Gen Virol. 2009;90:2855–2864. doi: 10.1099/vir.0.012872-0. [DOI] [PubMed] [Google Scholar]
- 41.Cornet I, Bouvard V, Campo MS, Thomas M, Banks L, Gissmann L, Lamartine J, Sylla BS, Accardi R, Tommasino M. Comparative analysis of transforming properties of E6 and E7 from different beta human papillomavirus types. J Virol. 2012;86:2366–2370. doi: 10.1128/JVI.06579-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Accardi R, Dong W, Smet A, Cui R, Hautefeuille A, Gabet AS, Sylla BS, Gissmann L, Hainaut P, Tommasino M. Skin human papillomavirus type 38 alters p53 functions by accumulation of deltaNp73. EMBO Rep. 2006;7:334–340. doi: 10.1038/sj.embor.7400615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43*.Howie HL, Koop JI, Weese J, Robinson K, Wipf G, Kim L, Galloway DA. Beta-HPV 5 and 8 E6 promote p300 degradation by blocking AKT/p300 association. PLoS Pathog. 2011;7:e1002211. doi: 10.1371/journal.ppat.1002211. This publication reports that α and β HPVE6 proteins associate with the transcriptional coactivator p300. HPV5 and 8 E6 destabilize p300 through an E6AP-independent mechanism and the decrease in cellular p300 levels affects the expression of several epithelial differentiation markers. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44*.Muench P, Probst S, Schuetz J, Leiprecht N, Busch M, Wesselborg S, Stubenrauch F, Iftner T. Cutaneous papillomavirus E6 proteins must interact with p300 and block p53-mediated apoptosis for cellular immortalization and tumorigenesis. Cancer Res. 2010;70:6913–6924. doi: 10.1158/0008-5472.CAN-10-1307. This publication reports that β HPV38 and cotton tail rabbit papillomavirus (CRPV) E6 proteins associate with p300 and inhibit p53 by abrogating p300 mediated p53 acetylation. HPV38 and CRPV p300 binding defective E6 mutants are inactive for supporting cellular immortalization and tumor formation, respectively. [DOI] [PubMed] [Google Scholar]
- 45**.Brimer N, Lyons C, Wallberg AE, Vande Pol SB. Cutaneous papillomavirus E6 oncoproteins associate with MAML1 to repress transactivation and NOTCH signaling. Oncogene. 2012 doi: 10.1038/onc.2011.589. This publication reports that MAML1, a crucial coactivator of Notch signaling, associates with β HPV and BPV E6 proteins. E6/MAML1 association causes decreased expression of several known NOTCH targets. BPV lesions show impaired tissue differentiation and a concomitantly reduced expression of NOTCH target genes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brakenhoff RH. Cancer. Another NOTCH for cancer. Science. 2011;333:1102–1103. doi: 10.1126/science.1210986. [DOI] [PubMed] [Google Scholar]
- 47.Dotto GP. Crosstalk of Notch with p53 and p63 in cancer growth control. Nat Rev Cancer. 2009;9:587–595. doi: 10.1038/nrc2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Underbrink MP, Howie HL, Bedard KM, Koop JI, Galloway DA. E6 proteins from multiple human betapapillomavirus types degrade Bak and protect keratinocytes from apoptosis after UVB irradiation. J Virol. 2008;82:10408–10417. doi: 10.1128/JVI.00902-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Frazer IH. Prevention of cervical cancer through papillomavirus vaccination. Nat Rev Immunol. 2004;4:46–54. doi: 10.1038/nri1260. [DOI] [PubMed] [Google Scholar]
- 50.Kenter GG, Welters MJ, Valentijn AR, Lowik MJ, Berends-van der Meer DM, Vloon AP, Essahsah F, Fathers LM, Offringa R, Drijfhout JW, et al. Vaccination against HPV-16 oncoproteins for vulvar intraepithelial neoplasia. N Engl J Med. 2009;361:1838–1847. doi: 10.1056/NEJMoa0810097. [DOI] [PubMed] [Google Scholar]
- 51.Baldwin A, Grueneberg DA, Hellner K, Sawyer J, Grace M, Li W, Harlow E, Munger K. Kinase requirements in human cells: V. Synthetic lethal interactions between p53 and the protein kinases SGK2 and PAK3. Proc Natl Acad Sci U S A. 2010;107:12463–12468. doi: 10.1073/pnas.1007462107. [DOI] [PMC free article] [PubMed] [Google Scholar]