Abstract
Merkel cell polyomavirus (MCV), discovered in 2008, is clonally integrated in ~80% Merkel cell carcinoma (MCC). MCV is a common skin flora and initiates cancer in susceptible hosts only after it acquires a precise set of mutations that render it replication incompetent. Both MCV large and small T proteins promote cancer cell survival and proliferation. Large T targets pocket proteins regulating cell cycle transit while small T activates cap-dependent translation critical for cancer cell growth. These findings already have led to new diagnostics and clinical trials to target MCV-induced survivin and to promote antitumor immunity. In four years, the cause, diagnosis and therapy for an intractable cancer has been changed due to the molecular discovery of MCV.
Introduction
Merkel cell polyomavirus (MCV or MCPyV) is the newest member of the surprisingly small group of viruses known to cause human cancer [1]. It is also one of seven new human polyomaviruses discovered in the past five years [2–10]. In a very short time, newly identified viral markers and serologic assays for MCV infection have been developed that improve Merkel cell carcinoma (MCC) diagnosis. Discovery of MCV has already led to studies on a precise molecular-targeted therapy based on rational drug testing that may alter clinical treatment for this often-intractable disease [11]. Work on immune-based therapies to complement existing cancer treatments is being explored as well [12,13]. MCV has also helped us understand a new cancer mechanism in which mutations to a typically harmless component of our skin flora—rather than the cancer cell genome itself—contributes to tumor formation [14]. Taken together, this recent research led to the WHO International Agency for Research on Cancer (IARC) to classify MCV as a group 2A carcinogen [15]. These recent advances all stem from isolation of a small piece of RNA from a Merkel cell carcinoma tumor four years ago [6].
Polyomaviruses have formed much of the basis for our understanding of the molecular biology of cancer. Animal polyomavirus tumor (T) antigens led to discoveries of p53 and PI3K as well as other oncogene/tumor suppressor signaling pathways [16,17]. They have also contributed to uncovering fundamental cellular processes such as protein nuclear localization signals and mammalian DNA replication [16,17]. MCV, which is clonally integrated into the MCC cell genome, adds new insights into the mechanisms of polyomavirus-induced cancers. In contrast to small T (sT) protein of other polyomaviruses, MCV sT is the major transforming oncogene, and exerts its tumor promoting effects at least in part through targeting of the cap-dependent translation regulator, 4E-BP1 [18]. Similar to other polyomavirus large T (LT) proteins, MCV LT targets cellular pocket proteins (pRB, p107 and p130) [14] but one critical consequence of this is the activation of survivin, an important mediator for cancer cell proliferation [11].
Discovery of MCV
Merkel cell carcinoma is an uncommon but aggressive primary cutaneous neoplasm having a poor prognosis once disseminated [19,20]. It arises from mechanoreceptor Merkel cells sparsely distributed in the basal layer of the epidermis [21,22]. Similar to other skin cancers, prolonged UV exposure is a risk factor for MCC, as is advanced age, and the risk for MCC increases dramatically in persons 50 years or older [23]. The risk for MCC is also strikingly associated with loss of immune competence; the risk of MCC is 13-fold higher in AIDS patients and 10-fold higher among organ transplant recipients than in the general population [24], an epidemiologic pattern reminiscent of Kaposi’s sarcoma and other cancers having a viral etiology [25].
Population-based studies from the United States and Europe reveal a rising MCC incidence [20,26,27–29] and the public health burden of this cancer is generally underappreciated. Approximately 1,500 MCC cases occur annually in the US with MCC being responsible for more deaths than chronic myelogenous leukemia [30]. Other cancers, such as chronic lymphocytic leukemia, basal cell carcinoma and squamous cell carcinoma [29,31–36], occur in conjunction with MCC at unexpectedly high rates. None of these secondary cancers have been robustly linked to MCV infection and reports vary as to whether MCV might also be present in these non-MCC tumors [37–44].
A focused search for oncogenic viruses in MCC was initiated by Feng et al. in 2007 [6]. This approach, called digital transcriptome subtraction (DTS), uses high-throughput complementary DNA (cDNA) sequencing and in silico subtraction of human sequences from tumor transcriptome to isolate candidate viral sequences [6,45]. Two MCV transcript sequences were found in the DTS analysis of MCC tumors, one of which had high sequence homology to a primate lymphotropic polyomavirus sequence [46].
MCV was initially found to be clonally-integrated into the human genome in tumors [6]. No preferential integration sites have so far been found [6,47–49]. In addition to disruption of the viral genome as a result of integration, viral sequences revealed a second peculiar feature for tumor-associated MCV. All tumor isolates possessed truncating mutations that deleted the origin-binding or helicase domains of the virus’ LT protein [14]. Additionally, tumor isolates have been found possessing mutations in the noncoding origin sequence [50] and VP1 structural genes [51] that prevent replication and virion formation. This suggests that there is a strong selection pressure to eliminate MCV replication within MCC and consistent with the notion that virus-induced tumors generally do not support productive (“lytic”) viral replication [1,50,51]. Active virus replication activates innate immune signaling and, in the case of MCV, unlicensed viral origin firing from the viral-human integrant will generate fragmented DNA [14], which will kill the nascent tumor cell.
MCV Virology
MCV is a non-enveloped, double-stranded DNA virus belonging to the mammalian genus Orthopolyomavirus [52]. MCV has been difficult to cultivate in the laboratory as a natural infection but several attempts have been made to produce infectious MCV molecular clones [53–55]. In each case, primary low-level virion production can be achieved (Figure 1) but secondary transmission to uninfected cells has not been successful. While early electron microscopy studies suggested that MCV virions might be seen in some MCC tumors [56], the weight of evidence now indicates that structural proteins required for encapsidation are not expressed in MCC tumors and encapidated viruses seen in tumors are likely to be coincidental [57,58].
Figure 1. Merkel cell polyomavirus virions.
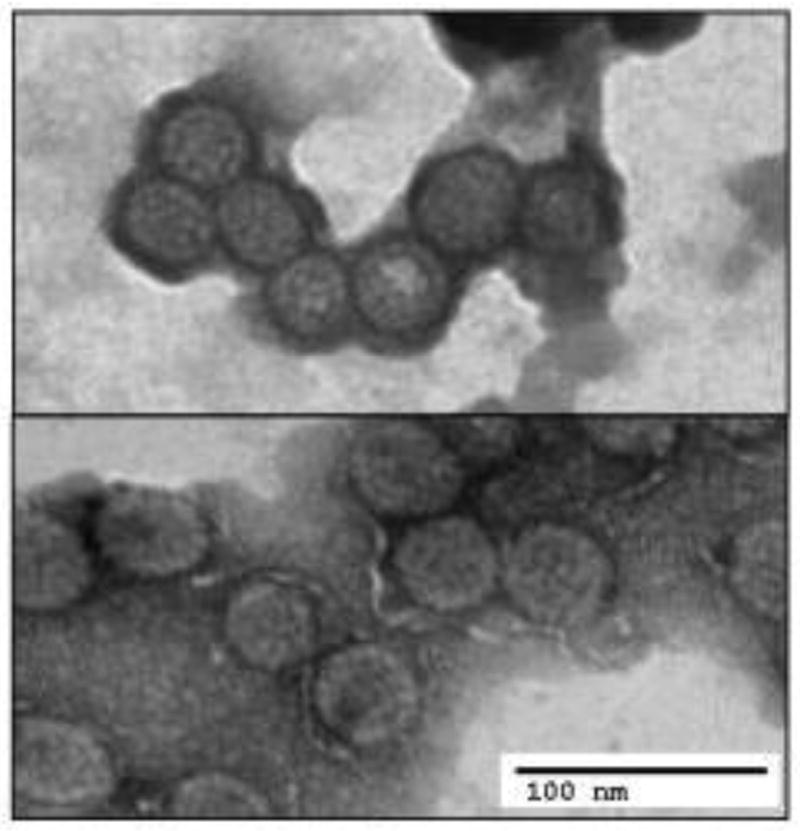
Top panel shows typical Merkel cell polyomavirus particles produced by transfection of whole genome in 293 cells. In comparison, lower panel reveals assembled MCV virus-like particles (VLP), generated by expression of VP1 and VP2 genes alone, that can be used in serologic assays (Modfied from Feng et al., PLoS one, 2011).
MCV Genes and Genome
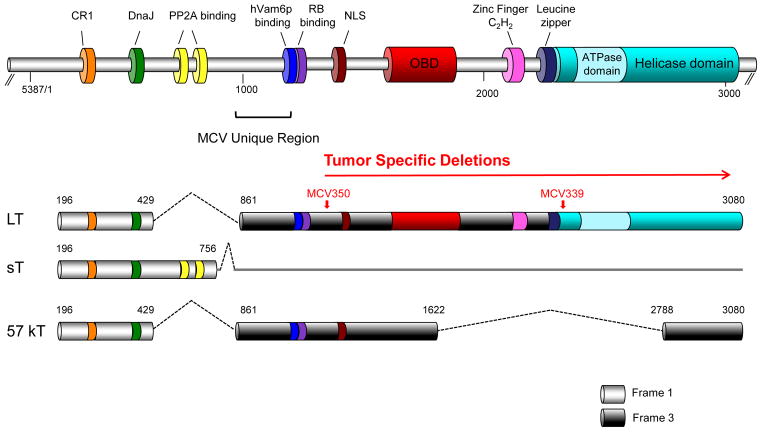
The MCV genome displays features found in other polyomaviruses. It has a ~5.4 kb genome divided into early and late gene regions by a noncoding regulatory region (NCRR). The early region encodes for alternatively spliced, overlapping RNAs that generate large T (LT), small T (sT) and 57kT antigens (analogous to the SV40 17-kT antigen [59]), and share a common 78 amino acid N terminus encoded by exon1 (Figure 2) [14]. Mutations to the T antigen region that arise in tumor-derived MCV (substitutions, frameshift, missense, insertions and deletions) [14,49] truncate LT and 57kT proteins but do not affect full length sT protein translation [14]. Despite MCV’s similarity to murine polyomavirus (MPyV), no middle T antigen has been identified.
Figure 2. MCV T antigen locus.
Three MCV T antigen isoforms are generated by alternative splicing of the T antigen gene: Large T (LT), small T (sT) and 57kT (amino acid positions, shown). Exon1 is common to all three T antigen proteins. Major conserved MCV T antigen motifs (top, base pair positions, shown) are in color (CR1, conserved region 1; DnaJ, Hsp70-binding conserved region; RB, retinoblastoma-binding; PP2A, protein phosphatase 2A-binding; NLS, nuclear localization signal; OBD, origin-binding domain). LT and 57kT encode a MCV-unique region (MUR) that includes the Vam6p/Vps39-binding motif. Tumor specific mutations (red arrow) occur C-terminal to the RB-binding domain and disrupt the helicase activity of LT but do not eliminate tumor suppressor binding domains. Locations for mutations for two MCV tumor strains (MCV350 and MCV339) are shown.
MCV LT antigen retains conserved domains that are present across different polyomaviruses, such as DnaJ and LXCXE retinoblastoma (Rb) protein binding motifs [16,60], as well as the origin binding and helicase/ATPase regions needed for viral replication [14]. Tumor-specific mutations spare the LXCXE domain (aa 212–216), indicating its importance to MCC tumorigenesis [14,47–49,61]. Similar to other polyomaviruses, MCV LT interaction with RB1 requires the LXCXE domain [14,47,48,61], which is expected to deregulate E2F-related gene transcription. Direct evidence for the requirement of this motif for cell survival, has been generated by complementing T antigen knockdown experiments in MCC cell lines by Houben and colleagues [62]. One unexpected consequence of LT targeting of pocket proteins is the specific activation of survivin transcription, a finding that has been exploited in therapeutic studies [11].
MCV LT protein contains a nuclear localization signal (NLS) at aa 277–280 (RKRK) [61], resulting in a typical nuclear LT localization pattern for most cell cultures and tumors [37] (Figure 3). Signature tumor truncation mutations can disrupt this domain resulting in diffuse nuclear and cytoplasmic distribution of LT [37,54]. A novel interaction, so far only found for MCV LT, between human Vamp6 protein (Vps39) and the MCV unique region in LT adjacent to its Rb binding motif [54], causes this cytoplasmic protein to relocalize to the nucleus. The function(s) of Vam6p relocalization in MCC tumors is unknown; evidence suggests that in non-tumor MCV infections, LT targeting of Vam6p may regulate MCV replication [54].
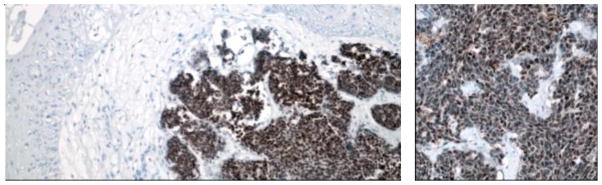
Figure 3. Merkel cell polyomavirus large T (left) and small T (right) antigen expression in MCC tumors.
MCV large T antigen usually shows distinct nuclear expression in MCC cells (dependent on an intact nuclear localization signal that can be deleted in some tumors), while MCV small T antigen displays both nuclear and cytoplasmic staining patterns. Only tumor cells show strong positivity with antibody staining, and not the surrounding non-tumor tissues.
The MCV early region mRNA also splices to produce a 57kT antigen (predicted size = 47kDa [14]) that is identical to large T protein but lacking an origin-binding domain. Similar multiply spliced T antigen isoforms occur in other polyomaviruses [14,17,59] and their functions remain poorly understood.
MCV sT is encoded by a read-through of the exon1-intron1 splice donor site [14]. In tissue sections of tumors, MCV sT is more commonly expressed than MCV LT antigen (Figure 3) [18]. Knockdown studies, however, reveal that both MCV sT and LT antigens are independently required for MCC tumor cell survival and proliferation [18,63] and both are likely to contribute to tumorigenesis.
In polyomaviruses, LT primarily target tumor suppressor pathways and sT activates Akt-mTOR signaling by binding to protein phosphatase 2A (PP2A) [64], a pathway which has been found to be critical for tumor cell survival in many types of genetic cancers [65]. For SV40, LT is a potent in vitro transforming oncoprotein while sT plays a supporting role and is not transforming alone [16,64,66]. In contrast, MCV sT is the primary transforming oncoprotein in vitro while MCV LT has no effect in focus formation and soft agar assays [18].
SV40 sT acts to inhibit Akt dephosphorylation by binding the cellular protein phosphatase 2A (PP2A) A and C subunits while displacing its B subunit [64,67,68]. MCV sT similarly binds PP2A, but this interaction is dispensable for MCV sT-induced transformation [18]. MCV sT instead promotes hyperphosphorylation of 4E-BP1, a downstream target of mTORC1 kinase through interaction with unidentified cellular partner protein(s). MCV sT thus may be a useful tool to dissect cap-dependent regulation of 4E-BP1 in cancer signaling.
The MCV late region encodes 3 capsid proteins (VP1, VP2 and VP3), expressed after the onset of viral DNA replication. These structural proteins, when expressed in uninfected cells, self-assemble into a ~55-nm diameter icosahedral viral particles that can be harvested as antigen for serological assays [57,69]. MCV does not encode an agnoprotein [70,71] or VP4 [72] found in some polyomaviruses. Formally, little is known about the kinetics and regulation of MCV late gene expression because virus replication studies have been limited. Comparison of late gene expression for the MCV-HF molecular clone to a replication defective mutant clone suggests that MCV late gene expression depends on active DNA replication of the viral genome, analogous to late gene expression among large DNA viruses (e.g., herpesviruses) [54]. MCV encodes an miRNA, MCV-mir-M-5p that is generated from long RNAs transcribed late in infection [73,74]. It is antisense to early transcripts (regions 1217–1238) and may behave similar to the SV40 miRNA in negatively regulating early gene expression during late phases of virion encapsidation [75].
The NCRR region of MCV separates early and late gene regions and contains a core 71-bp origin sufficient to initiate DNA replication. This core sequence is comprised of an AT-rich tract involved in DNA melting and a region containing 8 GAGGC pentanucleotide sequences (PS) that are bound by the MCV LT origin-binding domain at the initiation of replication [14,50,76]. Four of these PS sites are absolutely required for virus replication [50], including a core of three PS that form an interacting helicase complex seen in crystallization studies [76]. Unlike SV40, but similar to JCV, MCV origin replication is highly activated by coexpression of MCV sT proteins [50,54]. Early evidence suggested that this may be due to sT sequestration of PP2A, but this has subsequently been shown to be PP2A-independent. The NCRR also contains bidirectional transcriptional promoters and regulatory elements for early and late viral gene expression.
MCV Epidemiology
Similar to most of the human polyomaviruses, MCV is a near-ubliquitous infection of adults. MCV seroassays based on late structural capsid protein VP1 reveal MCV prevalence of 60–80% in adults [69,77–79]. Both VLP-based EIA and neutralization tests demonstrate that conformational epitopes are important for the immunodominant antibody response after infection [57,69], and that assembled particles are generally a more sensitive serologic reagent than purified VP1 recombinant protein. Seroconversion to MCV IgG positivity is generally stable and antibodies can be detected for decades after primary infection [80]. Among persons with MCC, antibody titers to MCV VLP are significantly elevated giving evidence that an episode of viremia probably precedes tumor development [57,69].
Primary MCV infection, at least among adults, is generally asymptomatic [80]. MCV antibodies are detected in children with the prevalence of infection increasing with age [69,77,78,81]. In contrast to VLP, healthy adults do not generally have antibody responses to MCV T antigens [69,82], although T antigen antibodies can also develop in a subset of MCC patients and have been used to monitor tumor recurrence or dissemination [69,82].
Serologic and molecular studies indicate that MCV is a persistent and life-long infection [7]. MCV DNA is predominantly found in skin [7,61,83–85] but can be detected in a variety of tissues including, respiratory tract samples and nasopharyngeal aspirates [86–89], saliva [84], gut [27,90], lymphoid tissue [27,37], urine [91–94] and whole blood from healthy donors [44,90,93,95,96]. For this reason, PCR-based studies identifying MCV in tumors or other diseases require confirmation-using techniques less prone to experimental false positivity than PCR (e.g., immunohistochemistry, Southern blotting). Transmission is through a form of casual contact but the precise mode is not known.
MCV – a new human carcinogen
In Feng et al.’s original description of MCV, 8 of 10 tumors harbored MCV infection [6] and this has been confirmed through multiple studies worldwide. Of 2354 MCC tumors examined in various settings, 1743 (74.2%) were positive for MCV (Supplementary Table 1). Little is known about the cause of MCV-negative MCC—although low MCV VLP antibody levels in these patients makes a hit-and-run event by MCV seem unlikely. Further, careful examination of MCV-negative MCC often reveals differences in immunophenotype (e.g., CK20) and miRNA profiles (unpublished results) from MCV-positive tumors, making it likely that MCV-positive and MCV-negative MCC have different histogeneses.
Evidence is now abundant that MCV is a component of healthy skin flora that only rarely initiates tumorigenesis. What are the factors that promote transformation of this harmless agent into a cancer virus?
Immunity
Similar to other human tumor viruses, cell-mediated immune (CMI) surveillance is critical in suppressing Merkel cell carcinoma formation and AIDS, post-transplant and other immune-deficient populations are at increased risk for MCC [19,24]. The elevated risk among the elderly is also consistent with age-related loss of immune surveillance having a critical role in promoting MCC [32]. Tumor infiltrating lymphocytes are a common feature of MCV-positive tumors [97] and virus-specific CD8+ and CD4+ T cells have been isolated from MCC [98]. The immune defect contributing to MCC may be subtle, however, Iyer et al. have shown virus-reactive T cell responses for both MCC patients and healthy volunteers [98]. Reports of spontaneous MCC remission may reflect reconstitution of CMI against tumor antigens [99] and provides hope for adoptive immunotherapies in the treatment of this cancer.
Persistence and Loss of MCV Replication
MCV, when present, is nearly uniformly integrated into MCC genomes [6,49]. Whether integration occurs spontaneously or requires exogenous mutagenesis, such as UV exposure, is unknown. One possibility is that loss of immune surveillance allows active MCV replication, leading to nonhomologous recombination of genome replication fragments that generate the integrated virus in the proto-tumor cell. This is an appealing explanation for why MCV-positive MCC patients have high capsid antibody titers, but no direct evidence for this is currently exists.
Regardless of how viral integration occurs, expression of T antigen will lead to unlicensed viral DNA replication from a viral origin fused into the human genome—a potential catastrophe for the nascent tumor cell. Precise and independent mutations eliminating the T antigen replication capacity, without disturbing oncogenic domains, are also required for MCC cell survival. Each successive step in this evolutionary process—loss of immune surveillance, virus integration and T antigen mutation—are required for MCC formation but are uncommon. Thus, rare tumors can emerge from infection with this common skin infection.
MCV Oncoprotein Expression
Knockdown experiments show that MCV LT and sT oncoproteins are needed for MCV-positive tumor cell survival and replication once MCV integrates. These experiments provide critical support for MCV being the causative agent for MCV positive MCC [18,62,63]. Research on how these proteins contribute to tumorigenesis has progressed rapidly because of the existing knowledge base gained from other polyomaviruses.
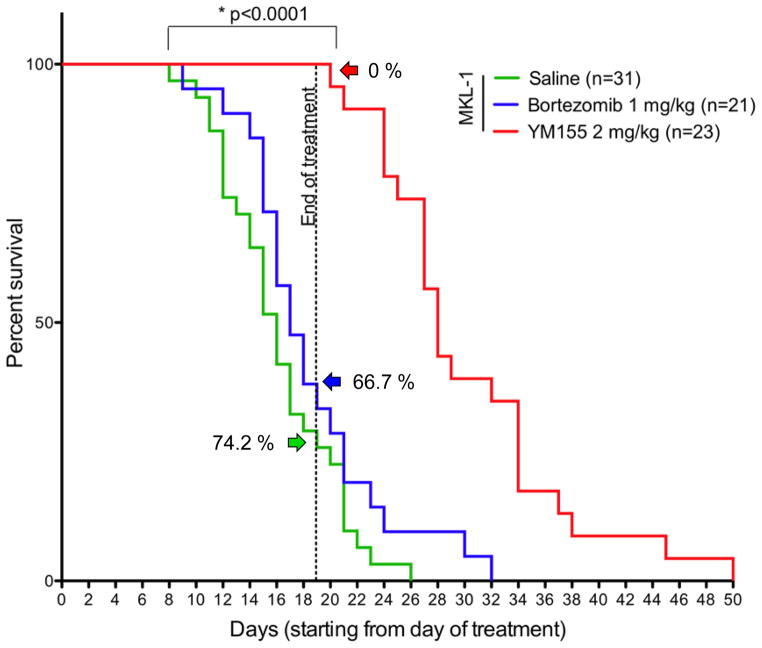
The importance of understanding the molecular causes for MCC is not limited to basic science. New MCV diagnostics help distinguish MCC from other closely related neuroendocrine cancers and may help predict the severity of the cancer when it does occur [27,37]. Even more importantly, these studies have prompted the search for fundamental changes in therapy for this difficult-to-treat tumor. Interferons are being explored to harness innate immune responses to this viral tumor [13]. Examination of cellular genes activated by MCV identified the BIRC5 gene encoding survivin oncoprotein as being highly upregulated by MCV LT sequestration of RB. This in turn led to examination of a small molecule survivin inhibitor (YM155) as a potential therapy for MCV-MCC [11]. YM155 inhibits MCV-positive MCC growth at nanomolar concentrations whereas a screen of over other 1300 drugs, including those in the NCI Oncology Drug Set, revealed only one compound (bortezomib) having similar potency. Early MCC xenograft studies (Figure 4) reveal that YM155 prolongs survival of mice bearing MCC tumors [11]. An Eastern Cooperative Oncology Group trial is slated to open in late 2012 to test efficacy of survivin inhibition in MCC. Thus, MCC has progressed from being a cancer with no known etiology and “More deaths but still no pathway to blame” [30] to having rationally-targeted molecular therapeutic trials based on its viral etiology, in just four years.
Figure 4. Survivin inhibition improves survival of mice bearing human MCC xenografts.
Survival curves are shown for mice with MCC xenografts and treated with YM155 (red line), bortezomib (blue line) or saline (green line) for three weeks. MCV positive MKL-1 cells were injected into immune deficient mice and the three-week treatment was given once tumors became palpable. Only 26–33% of bortezomib/saline-treated mice survived three weeks after appearance of tumors while 100% of YM155-treated mice survived the treatment period. Tumors resumed growth once YM155 was discontinued indicating a cytostatic rather than cytocidal effect for YM155 with short-term treatment. (Modified from Arora et al., STM, 2012)
The pace of MCV and MCC research has been rapid and is only growing faster. Speed records for research on virus discovery, viral oncogene studies, and “bench-to-bedside” research have been broken in the MCV field but it is still at a very early stage. Ever since the discovery of Epstein-Barr virus in 1964, discovery of each new human tumor virus has led to new and fundamental insights into carcinogenesis. MCV and related human polyomaviruses hold open the promise to continue this scientific tradition.
Supplementary Material
Supplementary Figure S1: Updated amino acid alignment of MCV with all known human polyomaviruses, SV40, LPyV and MPyV.
Supplementary Table S1: MCV Detection in MCC Samples
Highlights.
MCV was discovered in 2008 by digital transcriptome subtraction and is one of seven new human polyomaviruses described in the past five years.
Merkel cell polyomavirus (MCV), a new human polyomavirus, is clonally integrated in 70–80% of Merkel cell carcinoma (MCC) tumors.
MCV is part of the normal, healthy skin flora but causes cancer after viral genome mutations eliminate its replication capacity.
While similar to known polyomaviruses, MCV oncogenes act in new ways, such as activation of the survivin oncoprotein and PP2A-independent targeting of cap-dependent translation.
In four years, the diagnosis and treatment potential for an intractable and enigmatic cancer has dramatically changed through discovery of the viral cause of MCC.
Acknowledgments
We would like to thank Ezra Mirvish for the initial literature review and compiling the data on MCV and MCC association.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Paper of particular interest, published within the period of review have been highlighted as
* of special interest
**of outstanding interest
- 1*.Moore PS, Chang Y. Why do viruses cause cancer? Highlights of the first century of human tumour virology. Nat Rev Cancer. 2010;10(12):878–889. doi: 10.1038/nrc2961. This review summarizes common features for the seven current human tumor viruses. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gardner SD, Field AM, Coleman DV, Hulme B. New human papovavirus (b.K) isolated from urine after renal transplantation. Lancet. 1971;1 (7712):1253–1257. doi: 10.1016/s0140-6736(71)91776-4. [DOI] [PubMed] [Google Scholar]
- 3.Padgett BL, Walker DL, ZuRhein GM, Eckroade RJ, Dessel BH. Cultivation of papova-like virus from human brain with progressive multifocal leucoencephalopathy. Lancet. 1971;1(7712):1257–1260. doi: 10.1016/s0140-6736(71)91777-6. [DOI] [PubMed] [Google Scholar]
- 4.Allander T, Andreasson K, Gupta S, Bjerkner A, Bogdanovic G, Persson MA, Dalianis T, Ramqvist T, Andersson B. Identification of a third human polyomavirus. J Virol. 2007;81(8):4130–4136. doi: 10.1128/JVI.00028-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gaynor AM, Nissen MD, Whiley DM, Mackay IM, Lambert SB, Wu G, Brennan DC, Storch GA, Sloots TP, Wang D. Identification of a novel polyomavirus from patients with acute respiratory tract infections. PLoS Pathog. 2007;3(5):e64. doi: 10.1371/journal.ppat.0030064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6**.Feng H, Shuda M, Chang Y, Moore PS. Clonal integration of a polyomavirus in human merkel cell carcinoma. Science. 2008;319(5866):1096–1100. doi: 10.1126/science.1152586. This article describes the discovery of Merkel cell polyomavirus, found clonally integrated in 80% of Merkel cell carcinoma. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7*.Schowalter RM, Pastrana DV, Pumphrey KA, Moyer AL, Buck CB. Merkel cell polyomavirus and two previously unknown polyomaviruses are chronically shed from human skin. Cell Host Microbe. 2010;7(6):509–515. doi: 10.1016/j.chom.2010.05.006. This article describes the isolation of Merkel cell polyomavirus from skin swabs from healthy individuals and the discovery of two human polyomaviruses (HPyV6 and HPyV7), as part of the healthy human skin flora. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van der Meijden E, Janssens RW, Lauber C, Bouwes Bavinck JN, Gorbalenya AE, Feltkamp MC. Discovery of a new human polyomavirus associated with trichodysplasia spinulosa in an immunocompromized patient. PLoS Pathog. 2010;6(7):e1001024. doi: 10.1371/journal.ppat.1001024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scuda N, Hofmann J, Calvignac-Spencer S, Ruprecht K, Liman P, Kuhn J, Hengel H, Ehlers B. A novel human polyomavirus closely related to the african green monkey-derived lymphotropic polyomavirus. J Virol. 2011;85(9):4586–4590. doi: 10.1128/JVI.02602-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sauvage V, Foulongne V, Cheval J, Ar Gouilh M, Pariente K, Dereure O, Manuguerra JC, Richardson J, Lecuit M, Burguiere A, Caro V, et al. Human polyomavirus related to african green monkey lymphotropic polyomavirus. Emerg Infect Dis. 2011;17(8):1364–1370. doi: 10.3201/eid1708.110278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11**.Arora R, Shuda M, Guastafierro A, Feng H, Toptan T, Tolstov Y, Normolle D, Vollmer LL, Vogt A, Domling A, Brodsky JL, et al. Survivin is a therapeutic target in Merkel cell carcinoma. Science Translational Medicine. 2012;4(133):133ra156. doi: 10.1126/scitranslmed.3003713. This article describes LT-dependent induction of survivin and use of a survivin inhibitor, YM155, which shows selective in vitro and in vivo activity against MCV positive MCC cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bhatia S, Afanasiev O, Nghiem P. Immunobiology of Merkel cell carcinoma: Implications for immunotherapy of a polyomavirus-associated cancer. Curr Oncol Rep. 2011;13(6):488–497. doi: 10.1007/s11912-011-0197-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Willmes C, Adam C, Alb M, Volkert L, Houben R, Becker JC, Schrama D. Type i and ii IFNs inhibit Merkel cell carcinoma via modulation of the Merkel cell polyomavirus T antigens. Cancer Res. 2012;72(8):2120–2128. doi: 10.1158/0008-5472.CAN-11-2651. [DOI] [PubMed] [Google Scholar]
- 14*.Shuda M, Feng H, Kwun HJ, Rosen ST, Gjoerup O, Moore PS, Chang Y. T antigen mutations are a human tumor-specific signature for Merkel cell polyomavirus. Proc Natl Acad Sci U S A. 2008;105(42):16272–16277. doi: 10.1073/pnas.0806526105. This article describes tumor-specific Merkel cell polyomavirus LT antigen truncation mutations. These mutations renders the virus replication incompetent in tumors while N-terminal tumor suppressor targeting domains are retained. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bouvard V, Baan RA, Grosse Y, Lauby-Secretan B, El Ghissassi F, Benbrahim-Tallaa L, Guha N, Straif K. Carcinogenicity of malaria and of some polyomaviruses. The Lancet Oncology. 2012 [Google Scholar]
- 16.Ahuja D, Saenz-Robles MT, Pipas JM. SV40 large T antigen targets multiple cellular pathways to elicit cellular transformation. Oncogene. 2005;24(52):7729–7745. doi: 10.1038/sj.onc.1209046. [DOI] [PubMed] [Google Scholar]
- 17.Gjoerup O, Chang Y. Update on human polyomaviruses and cancer. Adv Cancer Res. 2010;106:1–51. doi: 10.1016/S0065-230X(10)06001-X. [DOI] [PubMed] [Google Scholar]
- 18*.Shuda M, Kwun HJ, Feng H, Chang Y, Moore PS. Human Merkel cell polyomavirus small T antigen is an oncoprotein targeting the 4E-BP1 translation regulator. J Clin Invest. 2011 doi: 10.1172/JCI46323. This article describes how MCV small T antigen, but not large T antigen transforms rodent cells by inhibiting 4E-BP1 and activating cap-dependent translation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Agelli M, Clegg LX. Epidemiology of primary Merkel cell carcinoma in the United States. J Am Acad Dermatol. 2003;49(5):832–841. doi: 10.1016/s0190-9622(03)02108-x. [DOI] [PubMed] [Google Scholar]
- 20.Hodgson NC. Merkel cell carcinoma: Changing incidence trends. J Surg Oncol. 2005;89(1):1–4. doi: 10.1002/jso.20167. [DOI] [PubMed] [Google Scholar]
- 21.Pearse AG. The neuroendocrine (apud) cells of the skin. Am J Dermatopathol. 1980;2(2):121–123. doi: 10.1097/00000372-198000220-00002. [DOI] [PubMed] [Google Scholar]
- 22.Maricich SM, Wellnitz SA, Nelson AM, Lesniak DR, Gerling GJ, Lumpkin EA, Zoghbi HY. Merkel cells are essential for light-touch responses. Science. 2009;324(5934):1580–1582. doi: 10.1126/science.1172890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller RW, Rabkin CS. Merkel cell carcinoma and melanoma: Etiological similarities and differences. Cancer Epidemiol Biomarkers Prev. 1999;8(2):153–158. [PubMed] [Google Scholar]
- 24*.Engels EA, Frisch M, Goedert JJ, Biggar RJ, Miller RW. Merkel cell carcinoma and HIV infection. Lancet. 2002;359(9305):497–498. doi: 10.1016/S0140-6736(02)07668-7. Describes 13-fold higher risk for MCC among HIV positive and AIDS patients as compared to the healthy population. [DOI] [PubMed] [Google Scholar]
- 25.Chang Y, Cesarman E, Pessin MS, Lee F, Culpepper J, Knowles DM, Moore PS. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science. 1994;265:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 26.Albores-Saavedra J, Batich K, Chable-Montero F, Sagy N, Schwartz AM, Henson DE. Merkel cell carcinoma demographics, morphology, and survival based on 3870 cases: A population based study. J Cutan Pathol. 2010;37(1):20–27. doi: 10.1111/j.1600-0560.2009.01370.x. [DOI] [PubMed] [Google Scholar]
- 27*.Sihto H, Kukko H, Koljonen V, Sankila R, Bohling T, Joensuu H. Clinical factors associated with Merkel cell polyomavirus infection in Merkel cell carcinoma. J Natl Cancer Inst. 2009;101(13):938–945. doi: 10.1093/jnci/djp139. A population-based study of Finnish patients showing improved prognosis for MCV-positive over MCV-negative MCC patients. [DOI] [PubMed] [Google Scholar]
- 28.Reichgelt BA, Visser O. Epidemiology and survival of Merkel cell carcinoma in the Netherlands. A population-based study of 808 cases in 1993–2007. Eur J Cancer. 2011;47(4):579–585. doi: 10.1016/j.ejca.2010.11.002. [DOI] [PubMed] [Google Scholar]
- 29.Kaae J, Hansen AV, Biggar RJ, Boyd HA, Moore PS, Wohlfahrt J, Melbye M. Merkel cell carcinoma: Incidence, mortality, and risk of other cancers. J Natl Cancer Inst. 2010;102(11):793–801. doi: 10.1093/jnci/djq120. [DOI] [PubMed] [Google Scholar]
- 30*.Lemos B, Nghiem P. Merkel cell carcinoma: More deaths but still no pathway to blame. J Invest Dermatol. 2007;127(9):2100–2103. doi: 10.1038/sj.jid.5700925. A survey of current knowledge on MCC pathogenesis immediately prior to discovery of MCV. [DOI] [PubMed] [Google Scholar]
- 31.Buell JF, Trofe J, Hanaway MJ, Beebe TM, Gross TG, Alloway RR, First MR, Woodle ES. Immunosuppression and Merkel cell cancer. Transplant Proc. 2002;34(5):1780–1781. doi: 10.1016/s0041-1345(02)03065-8. [DOI] [PubMed] [Google Scholar]
- 32.Heath M, Jaimes N, Lemos B, Mostaghimi A, Wang LC, Penas PF, Nghiem P. Clinical characteristics of Merkel cell carcinoma at diagnosis in 195 patients: The AEIOU features. J Am Acad Dermatol. 2008;58(3):375–381. doi: 10.1016/j.jaad.2007.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Penn I, First MR. Merkel’s cell carcinoma in organ recipients: Report of 41 cases. Transplantation. 1999;68(11):1717–1721. doi: 10.1097/00007890-199912150-00015. [DOI] [PubMed] [Google Scholar]
- 34.Takabayashi M, Sakai R, Sakamoto H, Iemoto Y, Kanamori H, Inayama Y, Ishigatsubo Y. Merkel cell carcinoma developing after antithymocyte globulin and cyclosporine therapy for aplastic anemia. Anticancer Drugs. 2003;14(3):251–253. doi: 10.1097/00001813-200303000-00010. [DOI] [PubMed] [Google Scholar]
- 35.Howard RA, Dores GM, Curtis RE, Anderson WF, Travis LB. Merkel cell carcinoma and multiple primary cancers. Cancer Epidemiol Biomarkers Prev. 2006;15(8):1545–1549. doi: 10.1158/1055-9965.EPI-05-0895. [DOI] [PubMed] [Google Scholar]
- 36.Vlad R, Woodlock TJ. Merkel cell carcinoma after chronic lymphocytic leukemia: Case report and literature review. Am J Clin Oncol. 2003;26 (6):531–534. doi: 10.1097/01.coc.0000037108.86294.5E. [DOI] [PubMed] [Google Scholar]
- 37*.Shuda M, Arora R, Kwun HJ, Feng H, Sarid R, Fernandez-Figueras MT, Tolstov Y, Gjoerup O, Mansukhani MM, Swerdlow SH, Chaudhary PM, et al. Human Merkel cell polyomavirus infection I. MCV T antigen expression in merkel cell carcinoma, lymphoid tissues and lymphoid tumors. Int J Cancer. 2009;125(6):1243–1249. doi: 10.1002/ijc.24510. This article describes the use of a MCV large T antigen specific antibody and qPCR to detect MCV T antigen expression in MCC and other tumors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tolstov YL, Arora R, Scudiere SC, Busam K, Chaudhary PM, Chang Y, Moore PS. Lack of evidence for direct involvement of Merkel cell polyomavirus (MCV) in chronic lymphocytic leukemia (CLL) Blood. 2010;115(23):4973–4974. doi: 10.1182/blood-2010-03-273177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Toracchio S, Foyle A, Sroller V, Reed JA, Wu J, Kozinetz CA, Butel JS. Lymphotropism of Merkel cell polyomavirus infection, Nova Scotia, Canada. Emerg Infect Dis. 2010;16(11):1702–1709. doi: 10.3201/eid1611.100628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Andres C, Belloni B, Puchta U, Sander CA, Flaig MJ. Prevalence of MCPyV in Merkel cell carcinoma and non-MCC tumors. J Cutan Pathol. 2010;37(1):28–34. doi: 10.1111/j.1600-0560.2009.01352.x. [DOI] [PubMed] [Google Scholar]
- 41.Murakami M, Imajoh M, Ikawa T, Nakajima H, Kamioka M, Nemoto Y, Ujihara T, Uchiyama J, Matsuzaki S, Sano S, Daibata M. Presence of Merkel cell polyomavirus in japanese cutaneous squamous cell carcinoma. J Clin Virol. 2011;50(1):37–41. doi: 10.1016/j.jcv.2010.09.013. [DOI] [PubMed] [Google Scholar]
- 42.Reisinger DM, Shiffer JD, Cognetta AB, Jr, Chang Y, Moore PS. Lack of evidence for basal or squamous cell carcinoma infection with Merkel cell polyomavirus in immunocompetent patients with Merkel cell carcinoma. J Am Acad Dermatol. 2010;63(3):400–403. doi: 10.1016/j.jaad.2009.08.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kassem A, Technau K, Kurz AK, Pantulu D, Loning M, Kayser G, Stickeler E, Weyers W, Diaz C, Werner M, Nashan D, et al. Merkel cell polyomavirus sequences are frequently detected in nonmelanoma skin cancer of immunosuppressed patients. International journal of cancer Journal international du cancer. 2009;125(2):356–361. doi: 10.1002/ijc.24323. [DOI] [PubMed] [Google Scholar]
- 44.Pantulu ND, Pallasch CP, Kurz AK, Kassem A, Frenzel L, Sodenkamp S, Kvasnicka HM, Wendtner CM, Zur Hausen A. Detection of a novel truncating Merkel cell polyomavirus large T antigen deletion in chronic lymphocytic leukemia cells. Blood. 2010;116(24):5280–5284. doi: 10.1182/blood-2010-02-269829. [DOI] [PubMed] [Google Scholar]
- 45.Feng H, Taylor JL, Benos PV, Newton R, Waddell K, Lucas SB, Chang Y, Moore PS. Human transcriptome subtraction by using short sequence tags to search for tumor viruses in conjunctival carcinoma. J Virol. 2007;81(20):11332–11340. doi: 10.1128/JVI.00875-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.zur Hausen H, Gissmann L. Lymphotropic papovaviruses isolated from african green monkey and human cells. Med Microbiol Immunol. 1979;167(3):137–153. doi: 10.1007/BF02121180. [DOI] [PubMed] [Google Scholar]
- 47.Sastre-Garau X, Peter M, Avril MF, Laude H, Couturier J, Rozenberg F, Almeida A, Boitier F, Carlotti A, Couturaud B, Dupin N. Merkel cell carcinoma of the skin: Pathological and molecular evidence for a causative role of MCV in oncogenesis. J Pathol. 2009;218(1):48–56. doi: 10.1002/path.2532. [DOI] [PubMed] [Google Scholar]
- 48*.Laude HC, Jonchere B, Maubec E, Carlotti A, Marinho E, Couturaud B, Peter M, Sastre-Garau X, Avril MF, Dupin N, Rozenberg F. Distinct Merkel cell polyomavirus molecular features in tumour and non tumour specimens from patients with Merkel cell carcinoma. PLoS Pathog. 2010;6(8):e1001076. doi: 10.1371/journal.ppat.1001076. Survey of MCV copy number, mutations and integration sites in MCC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martel-Jantin C, Filippone C, Cassar O, Peter M, Tomasic G, Vielh P, Briere J, Petrella T, Aubriot-Lorton MH, Mortier L, Jouvion G, et al. Genetic variability and integration of Merkel cell polyomavirus in Merkel cell carcinoma. Virology. 2012;426(2):134–142. doi: 10.1016/j.virol.2012.01.018. [DOI] [PubMed] [Google Scholar]
- 50.Kwun HJ, Guastafierro A, Shuda M, Meinke G, Bohm A, Moore PS, Chang Y. The minimum replication origin of Merkel cell polyomavirus has a unique large T-antigen loading architecture and requires small T-antigen expression for optimal replication. J Virol. 2009;83 (23):12118–12128. doi: 10.1128/JVI.01336-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kassem A, Schopflin A, Diaz C, Weyers W, Stickeler E, Werner M, Zur Hausen A. Frequent detection of merkel cell polyomavirus in human Merkel cell carcinomas and identification of a unique deletion in the VP1 gene. Cancer Res. 2008;68(13):5009–5013. doi: 10.1158/0008-5472.CAN-08-0949. [DOI] [PubMed] [Google Scholar]
- 52.Johne R, Buck CB, Allander T, Atwood WJ, Garcea RL, Imperiale MJ, Major EO, Ramqvist T, Norkin LC. Taxonomical developments in the family polyomaviridae. Arch Virol. 2011;156(9):1627–1634. doi: 10.1007/s00705-011-1008-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Neumann F, Borchert S, Schmidt C, Reimer R, Hohenberg H, Fischer N, Grundhoff A. Replication, gene expression and particle production by a consensus Merkel cell polyomavirus (MCPyV) genome. PLoS One. 2011;6(12):e29112. doi: 10.1371/journal.pone.0029112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Feng H, Kwun HJ, Liu X, Gjoerup O, Stolz DB, Chang Y, Moore PS. Cellular and viral factors regulating Merkel cell polyomavirus replication. PLoS One. 2011;6(7):e22468. doi: 10.1371/journal.pone.0022468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schowalter RM, Pastrana DV, Buck CB. Glycosaminoglycans and sialylated glycans sequentially facilitate Merkel cell polyomavirus infectious entry. PLoS Pathog. 2011;7(7):e1002161. doi: 10.1371/journal.ppat.1002161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wetzels CT, Hoefnagel JG, Bakkers JM, Dijkman HB, Blokx WA, Melchers WJ. Ultrastructural proof of polyomavirus in Merkel cell carcinoma tumour cells and its absence in small cell carcinoma of the lung. PLoS One. 2009;4(3):e4958. doi: 10.1371/journal.pone.0004958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pastrana DV, Tolstov YL, Becker JC, Moore PS, Chang Y, Buck CB. Quantitation of human seroresponsiveness to Merkel cell polyomavirus. PLoS Pathog. 2009;5(9):e1000578. doi: 10.1371/journal.ppat.1000578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chang Y, Moore PS. Merkel cell carcinoma: A virus-induced human cancer. Annu Rev Pathol. 2012;7:123–144. doi: 10.1146/annurev-pathol-011110-130227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zerrahn J, Knippschild U, Winkler T, Deppert W. Independent expression of the transforming amino-terminal domain of SV40 large T antigen from an alternatively spliced third SV40 early mrna. Embo J. 1993;12(12):4739–4746. doi: 10.1002/j.1460-2075.1993.tb06162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pipas JM. Common and unique features of T antigens encoded by the polyomavirus group. J Virol. 1992;66(7):3979–3985. doi: 10.1128/jvi.66.7.3979-3985.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nakamura T, Sato Y, Watanabe D, Ito H, Shimonohara N, Tsuji T, Nakajima N, Suzuki Y, Matsuo K, Nakagawa H, Sata T, et al. Nuclear localization of Merkel cell polyomavirus large T antigen in Merkel cell carcinoma. Virology. 2010;398(2):273–279. doi: 10.1016/j.virol.2009.12.024. [DOI] [PubMed] [Google Scholar]
- 62.Houben R, Adam C, Baeurle A, Hesbacher S, Grimm J, Angermeyer S, Henzel K, Hauser S, Elling R, Brocker EB, Gaubatz S, et al. An intact retinoblastoma protein-binding site in Merkel cell polyomavirus large T antigen is required for promoting growth of Merkel cell carcinoma cells. International journal of cancer Journal international du cancer. 2012;130(4):847–856. doi: 10.1002/ijc.26076. [DOI] [PubMed] [Google Scholar]
- 63*.Houben R, Shuda M, Weinkam R, Schrama D, Feng H, Chang Y, Moore PS, Becker JC. Merkel cell polyomavirus-infected Merkel cell carcinoma cells require expression of viral T antigens. J Virol. 2010;84(14):7064–7072. doi: 10.1128/JVI.02400-09. This article describes the requirement for MCV T antigen expression in MCV-positive MCC cell survival. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pallas DC, Shahrik LK, Martin BL, Jaspers S, Miller TB, Brautigan DL, Roberts TM. Polyoma small and middle T antigens and SV40 small T antigen form stable complexes with protein phosphatase 2A. Cell. 1990;60(1):167–176. doi: 10.1016/0092-8674(90)90726-u. [DOI] [PubMed] [Google Scholar]
- 65.Buchkovich NJ, Yu Y, Zampieri CA, Alwine JC. The torrid affairs of viruses: Effects of mammalian DNA viruses on the PI3K-Akt-mTOR signalling pathway. Nat Rev Microbiol. 2008;6(4):266–275. doi: 10.1038/nrmicro1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pipas JM. Sv40: Cell transformation and tumorigenesis. Virology. 2009;384(2):294–303. doi: 10.1016/j.virol.2008.11.024. [DOI] [PubMed] [Google Scholar]
- 67.Rodriguez-Viciana P, Collins C, Fried M. Polyoma and SV40 proteins differentially regulate PP2A to activate distinct cellular signaling pathways involved in growth control. Proc Natl Acad Sci U S A. 2006;103(51):19290–19295. doi: 10.1073/pnas.0609343103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hahn WC, Dessain SK, Brooks MW, King JE, Elenbaas B, Sabatini DM, DeCaprio JA, Weinberg RA. Enumeration of the simian virus 40 early region elements necessary for human cell transformation. Mol Cell Biol. 2002;22(7):2111–2123. doi: 10.1128/MCB.22.7.2111-2123.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tolstov YL, Pastrana DV, Feng H, Becker JC, Jenkins FJ, Moschos S, Chang Y, Buck CB, Moore PS. Human Merkel cell polyomavirus infection II. MCV is a common human infection that can be detected by conformational capsid epitope immunoassays. Int J Cancer. 2009;125(6):1250–1256. doi: 10.1002/ijc.24509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sariyer IK, Saribas AS, White MK, Safak M. Infection by agnoprotein-negative mutants of polyomavirus JC and SV40 results in the release of virions that are mostly deficient in DNA content. Virol J. 2011;8:255. doi: 10.1186/1743-422X-8-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jay G, Nomura S, Anderson CW, Khoury G. Identification of the SV40 agnogene product: A DNA binding protein. Nature. 1981;291(5813):346–349. doi: 10.1038/291346a0. [DOI] [PubMed] [Google Scholar]
- 72.Fischer H, Sauer G. Identification of virus-induced proteins in cells productively infected with simian virus 40. J Virol. 1972;9(1):1–9. doi: 10.1128/jvi.9.1.1-9.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73*.Seo GJ, Chen CJ, Sullivan CS. Merkel cell polyomavirus encodes a microrna with the ability to autoregulate viral gene expression. Virology. 2009;383(2):183–187. doi: 10.1016/j.virol.2008.11.001. This article is the first to identify a microRNA encoded by MCV. [DOI] [PubMed] [Google Scholar]
- 74.Lee S, Paulson KG, Murchison EP, Afanasiev OK, Alkan C, Leonard JH, Byrd DR, Hannon GJ, Nghiem P. Identification and validation of a novel mature microrna encoded by the Merkel cell polyomavirus in human Merkel cell carcinomas. J Clin Virol. 2011;52(3):272–275. doi: 10.1016/j.jcv.2011.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sullivan CS, Grundhoff AT, Tevethia S, Pipas JM, Ganem D. SV40-encoded micrornas regulate viral gene expression and reduce susceptibility to cytotoxic T cells. Nature. 2005;435(7042):682–686. doi: 10.1038/nature03576. [DOI] [PubMed] [Google Scholar]
- 76.Harrison CJ, Meinke G, Kwun HJ, Rogalin H, Phelan PJ, Bullock PA, Chang Y, Moore PS, Bohm A. Asymmetric assembly of merkel cell polyomavirus large T-antigen origin binding domains at the viral origin. J Mol Biol. 2011;409(4):529–542. doi: 10.1016/j.jmb.2011.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chen T, Hedman L, Mattila PS, Jartti T, Ruuskanen O, Soderlund-Venermo M, Hedman K. Serological evidence of Merkel cell polyomavirus primary infections in childhood. J Clin Virol. 2011;50 (2):125–129. doi: 10.1016/j.jcv.2010.10.015. [DOI] [PubMed] [Google Scholar]
- 78.Touze A, Gaitan J, Arnold F, Cazal R, Fleury MJ, Combelas N, Sizaret PY, Guyetant S, Maruani A, Baay M, Tognon M, et al. Generation of Merkel cell polyomavirus (MCV)-like particles and their application to detection of MCV antibodies. J Clin Microbiol. 2010;48(5):1767–1770. doi: 10.1128/JCM.01691-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Carter JJ, Paulson KG, Wipf GC, Miranda D, Madeleine MM, Johnson LG, Lemos BD, Lee S, Warcola AH, Iyer JG, Nghiem P, et al. Association of Merkel cell polyomavirus-specific antibodies with Merkel cell carcinoma. J Natl Cancer Inst. 2009;101(21):1510–1522. doi: 10.1093/jnci/djp332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80*.Tolstov YL, Knauer A, Chen JG, Kensler TW, Kingsley LA, Moore PS, Chang Y. Asymptomatic primary Merkel cell polyomavirus infection among adults. Emerg Infect Dis. 2011;17(8):1371–1380. doi: 10.3201/eid1708.110079. This article reveals that adult MCV antibody seroconversion on primary infection is generally asymptomatic. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kean JM, Rao S, Wang M, Garcea RL. Seroepidemiology of human polyomaviruses. PLoS Pathog. 2009;5(3):e1000363. doi: 10.1371/journal.ppat.1000363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82*.Paulson KG, Carter JJ, Johnson LG, Cahill KW, Iyer JG, Schrama D, Becker JC, Madeleine MM, Nghiem P, Galloway DA. Antibodies to Merkel cell polyomavirus T antigen oncoproteins reflect tumor burden in Merkel cell carcinoma patients. Cancer Res. 2010;70(21):8388–8397. doi: 10.1158/0008-5472.CAN-10-2128. This article describes the detection of antibodies against MCV T antigen in MCV positive MCC patients and its correlation to tumor burden and prognosis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Foulongne V, Dereure O, Kluger N, Moles JP, Guillot B, Segondy M. Merkel cell polyomavirus DNA detection in lesional and nonlesional skin from patients with merkel cell carcinoma or other skin diseases. Br J Dermatol. 2010;162(1):59–63. doi: 10.1111/j.1365-2133.2009.09381.x. [DOI] [PubMed] [Google Scholar]
- 84.Foulongne V, Kluger N, Dereure O, Mercier G, Moles JP, Guillot B, Segondy M. Merkel cell polyomavirus in cutaneous swabs. Emerg Infect Dis. 2010;16(4):685–687. doi: 10.3201/eid1604.091278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Loyo M, Guerrero-Preston R, Brait M, Hoque MO, Chuang A, Kim MS, Sharma R, Liegeois NJ, Koch WM, Califano JA, Westra WH, et al. Quantitative detection of Merkel cell virus in human tissues and possible mode of transmission. International journal of cancer Journal international du cancer. 2010;126(12):2991–2996. doi: 10.1002/ijc.24737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bialasiewicz S, Lambert SB, Whiley DM, Nissen MD, Sloots TP. Merkel cell polyomavirus DNA in respiratory specimens from children and adults. Emerg Infect Dis. 2009;15(3):492–494. doi: 10.3201/eid1503.081067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Goh S, Lindau C, Tiveljung-Lindell A, Allander T. Merkel cell polyomavirus in respiratory tract secretions. Emerg Infect Dis. 2009;15 (3):489–491. doi: 10.3201/eid1503.081206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kantola K, Sadeghi M, Lahtinen A, Koskenvuo M, Aaltonen LM, Mottonen M, Rahiala J, Saarinen-Pihkala U, Riikonen P, Jartti T, Ruuskanen O, et al. Merkel cell polyomavirus DNA in tumor-free tonsillar tissues and upper respiratory tract samples: Implications for respiratory transmission and latency. J Clin Virol. 2009;45(4):292–295. doi: 10.1016/j.jcv.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sharp CP, Norja P, Anthony I, Bell JE, Simmonds P. Reactivation and mutation of newly discovered WU, KI, and Merkel cell carcinoma polyomaviruses in immunosuppressed individuals. J Infect Dis. 2009;199(3):398–404. doi: 10.1086/596062. [DOI] [PubMed] [Google Scholar]
- 90.Campello C, Comar M, D’Agaro P, Minicozzi A, Rodella L, Poli A. A molecular case-control study of the Merkel cell polyomavirus in colon cancer. J Med Virol. 2011;83(4):721–724. doi: 10.1002/jmv.22004. [DOI] [PubMed] [Google Scholar]
- 91.Bofill-Mas S, Rodriguez-Manzano J, Calgua B, Carratala A, Girones R. Newly described human polyomaviruses Merkel cell, KI and WU are present in urban sewage and may represent potential environmental contaminants. Virol J. 2010;7:141. doi: 10.1186/1743-422X-7-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Husseiny MI, Anastasi B, Singer J, Lacey SF. A comparative study of Merkel cell, BK and JC polyomavirus infections in renal transplant recipients and healthy subjects. J Clin Virol. 2010;49(2):137–140. doi: 10.1016/j.jcv.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mertz KD, Junt T, Schmid M, Pfaltz M, Kempf W. Inflammatory monocytes are a reservoir for Merkel cell polyomavirus. J Invest Dermatol. 2010;130(4):1146–1151. doi: 10.1038/jid.2009.392. [DOI] [PubMed] [Google Scholar]
- 94.Wieland U, Mauch C, Kreuter A, Krieg T, Pfister H. Merkel cell polyomavirus DNA in persons without Merkel cell carcinoma. Emerg Infect Dis. 2009;15(9):1496–1498. doi: 10.3201/eid1509.081575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Helmbold P, Lahtz C, Enk A, Herrmann-Trost P, Marsch W, Kutzner H, Dammann RH. Frequent occurrence of RASSF1A promoter hypermethylation and merkel cell polyomavirus in Merkel cell carcinoma. Mol Carcinog. 2009;48(10):903–909. doi: 10.1002/mc.20540. [DOI] [PubMed] [Google Scholar]
- 96.Helmbold P, Lahtz C, Herpel E, Schnabel PA, Dammann RH. Frequent hypermethylation of RASSF1A tumour suppressor gene promoter and presence of Merkel cell polyomavirus in small cell lung cancer. Eur J Cancer. 2009;45(12):2207–2211. doi: 10.1016/j.ejca.2009.04.038. [DOI] [PubMed] [Google Scholar]
- 97.Paulson KG, Iyer JG, Tegeder AR, Thibodeau R, Schelter J, Koba S, Schrama D, Simonson WT, Lemos BD, Byrd DR, Koelle DM, et al. Transcriptome-wide studies of Merkel cell carcinoma and validation of intratumoral CD8+ lymphocyte invasion as an independent predictor of survival. J Clin Oncol. 2011;29(12):1539–1546. doi: 10.1200/JCO.2010.30.6308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98*.Iyer JG, Afanasiev OK, McClurkan C, Paulson K, Nagase K, Jing L, Marshak JO, Dong L, Carter J, Lai I, Farrar E, et al. Merkel cell polyomavirus-specific CD8 and CD4 T-cell responses identified in Merkel cell carcinomas and blood. Clin Cancer Res. 2011;17(21):6671–6680. doi: 10.1158/1078-0432.CCR-11-1513. This article describes MCV virus-reactive T cell responses in MCC patients. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wooff JC, Trites JR, Walsh NM, Bullock MJ. Complete spontaneous regression of metastatic Merkel cell carcinoma: A case report and review of the literature. Am J Dermatopathol. 2010;32(6):614–617. doi: 10.1097/DAD.0b013e3181cd3158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1: Updated amino acid alignment of MCV with all known human polyomaviruses, SV40, LPyV and MPyV.
Supplementary Table S1: MCV Detection in MCC Samples