Abstract
Ubiquitin (Ub)-mediated proteasome-dependent proteolysis is critical in regulating multiple biological processes including apoptosis. We show that the unstructured BH3-only protein, NOXA, is degraded by an Ub-independent mechanism requiring 19S regulatory particle (RP) subunits of the 26S proteasome, highlighting the possibility that other unstructured proteins reported to be degraded by 20S proteasomes in vitro may be bona fide 26S proteasome substrates in vivo. A lysine-less NOXA (NOXA-LL) mutant, which is not ubiquitinated, is degraded at a similar rate to wild-type NOXA. Myeloid cell leukemia 1, but not other anti-apoptotic BCL-2 family proteins, stabilizes NOXA by interaction with the NOXA BH3 domain. Depletion of 19S RP subunits, but not alternate proteasome activator REG subunits, increases NOXA half-life in vivo. A NOXA-LL mutant, which is not ubiquitinated, also requires an intact 26S proteasome for degradation. Depletion of the 19S non-ATPase subunit, PSMD1 induces NOXA-dependent apoptosis. Thus, disruption of 26S proteasome function by various mechanisms triggers the rapid accumulation of NOXA and subsequent cell death strongly implicating NOXA as a sensor of 26S proteasome integrity.
Keywords: apoptosis, MCL-1, NOXA, proteasome, ubiquitin
Apoptosis has an essential role in the development and maintenance of tissue homeostasis in mammals. BCL-2 family proteins are key regulators of the intrinsic or mitochondrial apoptotic pathway, and can be functionally classified into pro- and anti-apoptotic members.1 Pro-survival BCL-2 homologs include BCL-2, BCL-XL, myeloid cell leukemia 1 (MCL-1) and BCL2A1/BFL-1. Pro-apoptotic BCL-2 family members can be further divided into the multidomain effector proteins BAX and BAK, and the BH3-only proteins, which serve as sentinels for various cellular stresses.1 The BH3-only proteins include BID, BAD, BIK, BIM, BMF, HRK, NOXA and PUMA, and can be subdivided on the basis of their capacity to interact with either anti-apoptotic BCL-2 proteins only (‘sensitizers') or both anti-apoptotic and multidomain effector BCL-2 family members (‘direct activators').1, 2 NOXA, a sensitizer BH3-only protein, binds with high affinity to MCL-1, and also interacts with BCL2A1 and BCL-XL.2, 3
The expression of BH3-only proteins is controlled by multiple mechanisms including those at both transcriptional and post-translational levels. NOXA is regulated in a stimulus-dependent manner by various transcription factors including p53, HIF1α and c-MYC.4, 5, 6 NOXA is also ubiquitinated and its levels are regulated by proteasome-dependent degradation.7, 8, 9 The requirement for NOXA in proteasome inhibitor-induced apoptosis suggests that the degradation of NOXA is a key mechanism by which its pro-apoptotic activity is regulated.3, 8 However, whether ubiquitination of NOXA targets it for proteasomal degradation is unknown.
The major route of proteasome-dependent protein degradation in mammalian cells is by ubiquitin (Ub)-dependent targeting to 26S proteasomes, in which assembly of polyubiquitin chains containing at least four Ub moieties on the protein targeted for degradation are recognized by 26S proteasomes. Ub is initially activated by an Ub-activating enzyme (E1), transferred to one of a limited number of Ub-conjugating enzymes (E2) and subsequently covalently attached, in most cases, to lysine residues of target proteins by one of many E3 Ub protein ligases.10, 11 Alternatively, a subset of proteins containing unstructured regions, including BIM and MCL-1, may be degraded, at least in vitro, in a proteasome-dependent but Ub-independent manner by 20S proteasomes.12, 13, 14, 15 However, the physiological significance of this pathway remains controversial.13, 16
We now show that NOXA is degraded by a 26S proteasome-dependent but Ub-independent pathway in cells. NOXA is stabilized by MCL-1. Depletion of 19S regulatory particle (RP) subunits leads to loss of 26S proteasome function and NOXA-dependent apoptosis, supporting the hypothesis that NOXA is a sensor of 26S proteasome function.
Results
Proteasome-dependent degradation of NOXA is lysine-independent
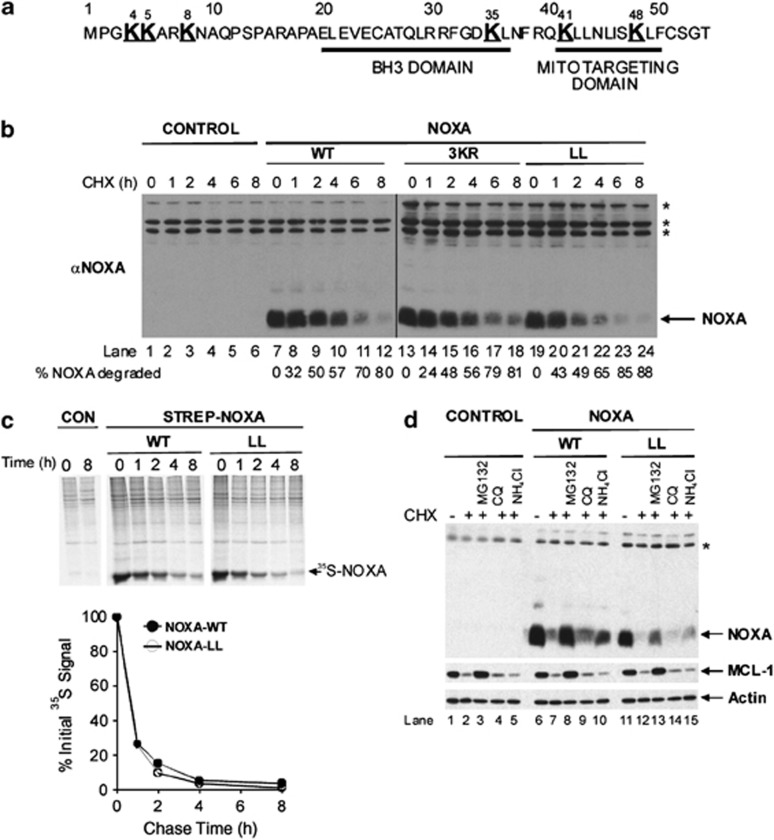
To investigate the mechanism of NOXA degradation, we mutated either the three amino terminal or all six lysine-to-arginine residues, which are unable to form isopeptide bonds with Ub, to generate either NOXA-K4/5/8R (NOXA-3KR) or lysine-less NOXA (NOXA-LL) (Figure 1a). Following transfection of these constructs, cells were incubated with cycloheximide and NOXA levels assessed. NOXA-WT, NOXA-3KR and NOXA-LL were all rapidly degraded (half life (t1/2) ∼2 h, Figure 1b), similar to t1/2 of endogenous NOXA.8 Similar results were observed using NOXA-WT and -LL constructs with a single N-terminal Strep Tag (Supplementary Figure S1A). As a control for an Ub-dependent substrate, MyoD was overexpressed and degradation of wild-type and lysine-less mutants assessed.17 Degradation of lysine-less full-length and an N-terminal deletion mutant were reduced or completely blocked compared with wild-type MyoD, respectively, in contrast to our results with NOXA-LL (Supplementary Figure S1B,17). As cycloheximide may also inhibit the synthesis of proteins involved in regulating NOXA turnover, 35S pulse-chase labelling was used followed by streptactin bead pulldown assays to compare the basal turnover of Strep Tag-NOXA-WT with -LL. Under basal conditions, Strep Tag-NOXA-WT and -LL were both rapidly turned over with similar t1/2 of ∼30 min (Figure 1c). Taken together, our results using two independent methods show that both NOXA-WT and -LL are rapidly degraded with a similar t1/2. Next, we assessed whether degradation of NOXA-WT and -LL was proteasomal- or lysosomal-dependent. Cells were incubated with the proteasome inhibitor, MG132, or two lysosomal inhibitors, chloroquine or NH4Cl, followed by incubation with cycloheximide. Only MG132 significantly blocked the degradation of untagged or Strep tagged NOXA-WT or -LL (Figure 1d and Supplementary Figure S1C). Degradation of MCL-1, a known proteasomal substrate,18 was also inhibited by MG132 but not by chloroquine or NH4Cl (Figure 1d and Supplementary Figure S1C). Together, these results suggest that NOXA is degraded by a proteasome-dependent but lysine-independent mechanism in vivo. Furthermore, addition of a single N-terminal Strep Tag to NOXA does not block its degradation via a proteasomal-dependent pathway.
Figure 1.
NOXA is degraded by proteasomes in a lysine-independent manner. (a) Primary sequence of human NOXA. Lysine residues are shown as large bold text. BH3 and mitochondrial-targeting domains are underlined. (b) NOXA is degraded via a lysine-independent pathway. Cells were transfected as indicated, and NOXA degradation assessed by immunoblotting and quantified using Odyssey Image analysis (LI-COR, Cambridge, UK). (c) NOXA-WT and -LL have similar basal turnover rates. Cells transiently expressing Strep-tagged NOXA-WT or -LL were labeled with a 35S-Cys/Met mixture for 1 h, and chased with unlabeled Cys and Met for the indicated times. Strep-tagged proteins were captured on streptactin beads. Gels were subjected to autoradiography and 35S-NOXA quantified using a phosphoimager. Results shown are from one experiment typical of two. (d) Degradation of NOXA-WT and -LL is proteasomal-dependent. Cells were transfected for 48 h and treated for the final 12 h with DMSO, MG132 (25 μM), chloroquine (200 μM) or NH4Cl (20 mM), and NOXA degradation assessed. Asterisks denotes non-specific bands
NOXA-LL is not ubiquitinated but functions like wild-type NOXA
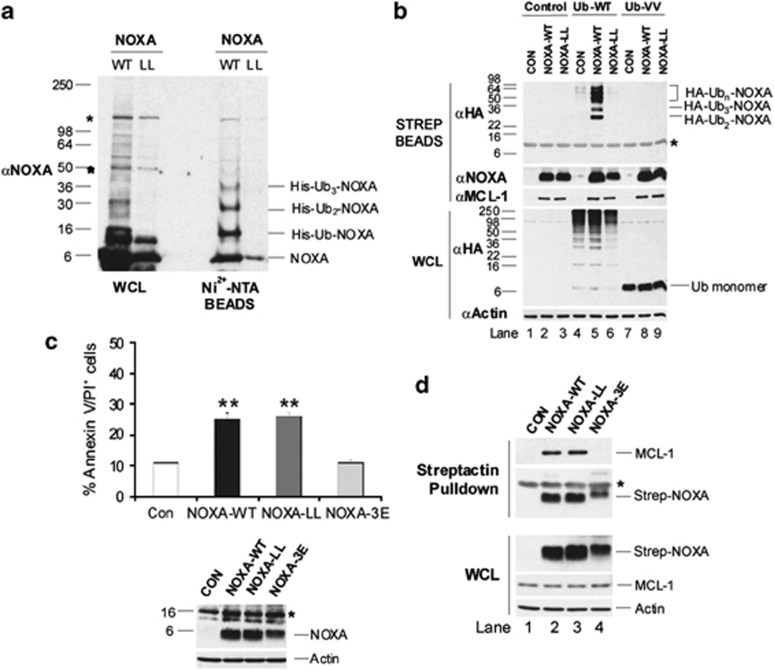
To examine whether NOXA-LL was ubiquitinated, cells were co-transfected with NOXA-WT or -LL constructs and a His6-tagged Ub vector. His6-Ub conjugates were eluted from Ni2+-nitriloacetic acid (NTA) beads in the absence of a reducing agent and analyzed by immunoblotting. NOXA-WT, but not NOXA-LL, was ubiquitinated and eluted as a ladder of mono- and polyubiquitinated species (Figure 2a). Cells were also co-transfected with either Strep Tag-NOXA-WT or -LL and either HA-tagged Ub-WT, which was incorporated into protein substrates (Figure 2b, lanes 4–6, lower panel) or Ub-VV, in which the two C-terminal glycine residues were mutated to valine. This Ub mutant cannot form isopeptide bonds with target proteins, and is thus expressed only as a monomer (Figure 2b, lanes 7–9, lower panel).19 Whereas Strep Tag-NOXA-WT was strongly ubiquitinated, Strep Tag-NOXA-LL was not ubiquitinated (Figure 2b, upper panel). The absence of a higher molecular mass species in cells co-transfected with NOXA-WT and Ub-VV indicates their formation is dependent upon Ub conjugation (Figure 2b, upper panel, compare lanes 5 with 8).
Figure 2.
NOXA is ubiquitinated on lysine residues. (a) Cells were co-transfected with either NOXA-WT or NOXA-LL and pMT107, lysed in guanidium HCl and His-tagged proteins purified on Ni2+-NTA beads or lysed in RIPA buffer (WCL). (b) Cells were transfected as indicated, and Strep Tag-NOXA and associated proteins collected on streptactin beads. (c) NOXA-WT and -LL induce similar levels of apoptosis. HeLa cells were transfected with the indicated NOXA constructs or a control vector and apoptosis quantified after 48 h (top panel). Results are the mean ±S.E.M. (n=3). **P < 0.01 from control using a Student's t test. Proteins were assessed by western blotting (lower panel). (d) NOXA-WT and -LL, but not -3E interact with MCL-1. Cells were transfected with Strep Tag-NOXA-WT, -LL or -3E or a control vector. Strep Tag-NOXA and associated proteins were collected on streptactin beads. Asterisks denotes non-specific bands
N-terminal ubiquitination on non-lysine residues promotes proteasome-dependent degradation of proteins, and addition of a large tag at the N- but not the C-terminus stabilizes N-terminal ubiquitinated proteins.17, 20 Some N-terminally ubiquitinated proteins are also stabilized by short N-terminal truncations.20 Although an N-terminal Myc6 tag stabilized NOXA-WT and -LL (Supplementary Figure S2A), fusion of a C-terminal Myc6-tag to either NOXA-WT or -LL also blocked degradation of both proteins (Supplementary Figure S2A). In contrast, deletion of the N-terminal eight amino acids from NOXA-WT, which included three potential ubiquitination sites, did not stabilize either NOXA-WT or -LL (Supplementary Figure S2B). These data strongly suggest that NOXA is not N-terminally ubiquitinated and that a NOXA-LL mutant is rapidly degraded in the absence of ubiquitination (Figures 1 and 2).
NOXA-WT and -LL induced similar, albeit moderate, levels of apoptosis and both associated with MCL-1, in contrast to the BH3 domain mutant, NOXA-L29E/F32E/L36E (NOXA-3E), which neither induced apoptosis nor interacted with MCL-1 (Figures 2c and d).21 Taken together, our results show that a NOXA-LL mutant has comparable functional activity to NOXA-WT.
NOXA is selectively stabilized by MCL-1 and requires a functional BH3 domain for its stabilization
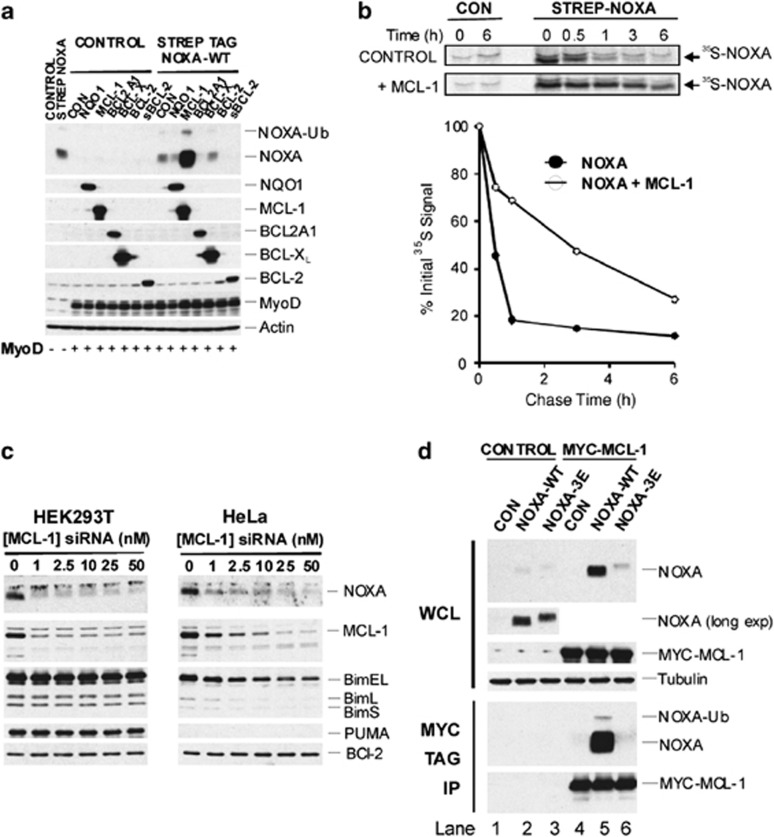
As NOXA is rapidly degraded and may interact with MCL-1, BCL2A1 and BCL-XL,2, 3 we examined their effects on NOXA levels, as well as that of NAD(P)H Quinone Oxidoreductase 1 (NQO1), which stabilizes several proteasomal substrates.12 Strikingly, co-expression of NOXA with MCL-1 strongly increased NOXA levels compared with expression of NOXA alone (Figure 3a, upper panel). Co-expression of NOXA with BCL2A1 or BCL-2 led to a decrease in NOXA levels relative to expression of NOXA only (Figure 3a, upper panel). Analysis using 35S pulse-chase labelling showed that MCL-1 decreased the turnover of Strep Tag-NOXA ∼fivefold from ∼30 to 160 min (Figure 3b). To test whether MCL-1 was required for stabilization of NOXA, MCL-1 was depleted by RNA interference (RNAi). NOXA disappeared whereas BIM isoforms or PUMA, which can also associate with MCL-1, were largely maintained with no change in BCL-2 levels (Figure 3c), supporting the idea that NOXA was selectively stabilized by MCL-1.
Figure 3.
MCL-1 stabilizes NOXA via a mechanism that requires NOXA–MCL-1 interactions. (a) MCL-1 selectively increases NOXA levels. Cells were co-transfected with plasmids encoding either Strep-tagged NOXA or a control and constructs encoding the indicated BCL-2 family members. A MyoD construct was also included as a control for equal transfection efficiency. Cell lysates were prepared and levels of the indicated proteins assessed by immunoblotting. (b) MCL-1 reduces NOXA turnover. Cells transfected with plasmids encoding Strep-tagged NOXA-WT in the presence or absence of MCL-1 were labeled with a 35S-Cys/Met mixture for 1 h and chased with unlabeled Cys and Met. Strep-tagged proteins were captured on streptactin beads. Gels were subjected to autoradiography and 35S-NOXA quantified using a phosphoimager. Results shown are from one of two experiments. (c) MCL-1 depletion lowers endogenous NOXA levels. HEK293T or HeLa cells were transfected with a pool of MCL-1 siRNAs and the indicated proteins assessed by immunoblotting. (d) MCL-1-dependent increases in NOXA levels require the NOXA BH3 domain. Cells were co-transfected with the indicated NOXA constructs or control vector and either a plasmid encoding MYC-MCL-1 or a control vector. After 24 h, lysates (WCL) were prepared and MYC-tagged MCL-1 and associated proteins precipitated with a MYC Tag mAb, and proteins assessed by immunoblotting
To examine whether interaction between NOXA and MCL-1 was necessary to maintain NOXA levels, cells were transfected with NOXA-WT or a NOXA-3E mutant, which does not bind MCL-1 (Figures 2d and 3d, lower panel21). Interaction of NOXA-WT with MCL-1 was essential for stabilization of NOXA, as no increase in NOXA levels was observed with NOXA-3E, following co-expression with MCL-1 (Figure 3d, top panel). Similar results were observed using a reciprocal pulldown assay in which Strep Tag-NOXA-WT, but not a -3E mutant, was stabilized by MCL-1 (Supplementary Figure S3D). To further understand the mechanism by which MCL-1 selectively stabilizes NOXA, we used a NOXA structure in complex with BCL2A1 (PDB ID: 3MQP) to model the complexes of NOXA with MCL-1 and BCL-XL. We asked which features in the modelled NOXA:MCL-1 complex may explain why this may be more stable than the complexes with BCL2A1 and BCL-XL. Three favourable potential electrostatic interactions are present in the NOXA:MCL-1 complex but absent from the other complexes. These involve the side chains of Glu20, Glu22 and Glu24 in NOXA interacting with Lys234 and Arg248 on the surface of MCL-1 on either side of the non-polar interaction groove present in BH3-only proteins (Supplementary Figure S3A). In BCL2A1 and BCL-XL, these basic residues are replaced with Cys/Gln or Gln/Gln, respectively (Supplementary Figure S3B). To investigate the involvement of this cluster of glutamate residues in NOXA stabilization, these residues were mutated to generate a NOXA-E20R/E22R/E24R mutant (NOXA-3R). To exclude possible differences in epitope recognition by the NOXA antibody being responsible for observed differences in NOXA levels in vivo, a coupled in vitro transcription/translation assay was used to initially express NOXA-WT, -3E and -3R. Similar levels of NOXA-WT, -3E and -3R were observed using the NOXA antibody, indicating the antibody recognizes both NOXA-WT and mutant proteins similarly (Supplementary Figure S3C). The NOXA-3R mutant weakly interacted with MCL-1 compared with NOXA-WT (Supplementary Figures S3D and E, lower panels). Levels of the NOXA-3R mutant protein were also moderately increased following its co-expression with MCL-1, albeit weakly compared with NOXA-WT (Supplementary Figures S3D–F). Moreover, cycloheximide chase studies indicated that while MCL-1 overexpression strongly blocked degradation of NOXA-WT, turnover of NOXA-3R was also partially inhibited by MCL-1, particularly compared with NOXA-3E (Supplementary Figure S3F). In summary, our results show that a cluster of acidic residues in NOXA contribute to MCL-1-mediated stabilization of NOXA, but are less critical than the BH3 domain.
NOXA contains unstructured regions
Various proteins, which can be degraded by an Ub-independent pathway, possess unstructured regions that may act as initiation sites for their subsequent degradation.12, 16 Multiple predictive tools suggested that the N-terminal region of NOXA has a high probability to be disordered (Supplementary Figure S4). Moreover, the thermal denaturation curve for synthetic NOXA (s-NOXA) showed no evidence of cooperative unfolding, which suggested that the protein was intrinsically unfolded, whereas ΔN-MCL-1-WT showed co-operative unfolding (Supplementary Figure S5A). The circular dichroism (CD) spectrum for full-length s-NOXA was characteristic of a largely unstructured protein, whereas that of ΔN-MCL-1-WT showed characteristics of an α-helical protein (Supplementary Figure S5B). Addition of NOXA to a constant concentration of ΔN-MCL-1-WT showed a characteristic increase in α-helical content, which was not observed with ΔN-MCL-1 G262A, a non-binding BH1 domain mutant of MCL-1 (Supplementary Figure S5B), suggesting that the NOXA:MCL-1 complex has an increased helical structure.
19S RP subunits of the 26S proteasome are required for NOXA degradation
Our results suggested that NOXA is an unstructured protein that can be efficiently degraded in an Ub-independent manner (Figures 1 and 2, Supplementary Figures S4 and S5). Although several proteins containing unstructured regions, including p53, p73α, p21Cip1 IκBα, MCL-1 and BIM, can be degraded in an Ub-independent manner by 20S proteasomes in vitro,12, 14, 15, 22 it remains to be demonstrated whether 20S proteasomes are required for their degradation in vivo. The CDK inhibitors p21Cip1 and p19Arf and the lysine-less protein p16Ink4a can be degraded in an Ub-independent manner in vivo by proteasomes capped with alternate REGγ activator subunits.23 Other proteins, such as ornithine decarboxylase, are degraded by 26S proteasomes by an Ub-independent pathway.24
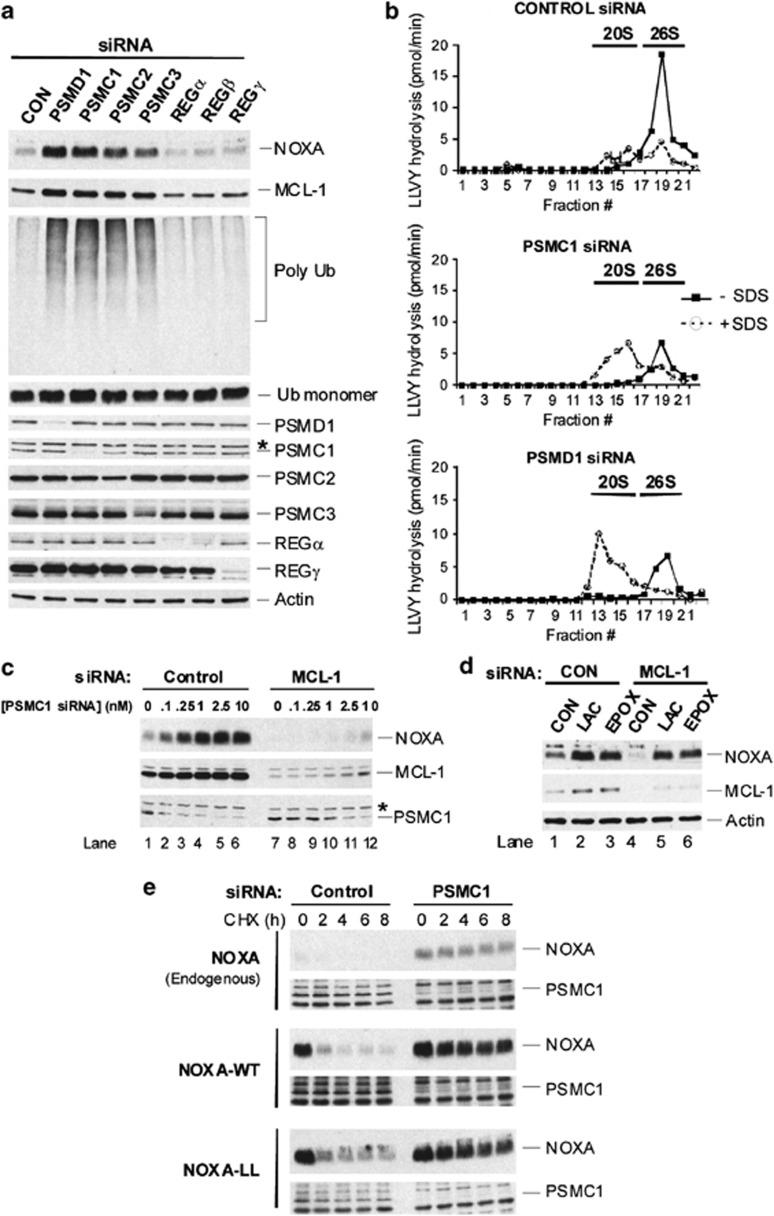
siRNA-mediated depletion of either 19S RP or REG activator subunits was used to investigate the possible involvement of 26S or REG-capped proteasomes, respectively, in NOXA degradation in vivo. Depletion of the 19S RP non-ATPase subunit PSMD1/Rpn2 or the AAA+-ATPase subunits PSMC1/Rpt2, PSMC2/Rpt1 or PSMC3/Rpt5, but not the alternate activators REGα, β or γ, increased levels of endogenous NOXA that were accompanied by moderate increases in MCL-1, concomitant with accumulation of polyubiquitinated proteins in the absence of changes in levels of monomeric Ub (Figure 4a). Increased NOXA levels were also observed, following depletion of different 19S RP subunits in HCT-116 colorectal carcinoma and H1299 lung carcinoma cells (Supplementary Figures S6A and B). The levels of BIMEL, another BH3-only protein that can be targeted to the proteasome,14 were also moderately increased (Supplementary Figure S6A). In contrast, the levels of PUMA, which may also be a proteasomal target,25, 26 were not significantly changed (Supplementary Figure S6A). Depletion of 19S RP AAA+-ATPase subunits or PSMD1, but not REG subunits, also extended the t1/2 of endogenous NOXA and MCL-1 (Supplementary Figure S6C). 35S pulse-chase labelling studies also showed that depletion of PSMC1 decreased the t1/2 of Strep-tagged NOXA more than six-fold (Supplementary Figure S6D).
Figure 4.
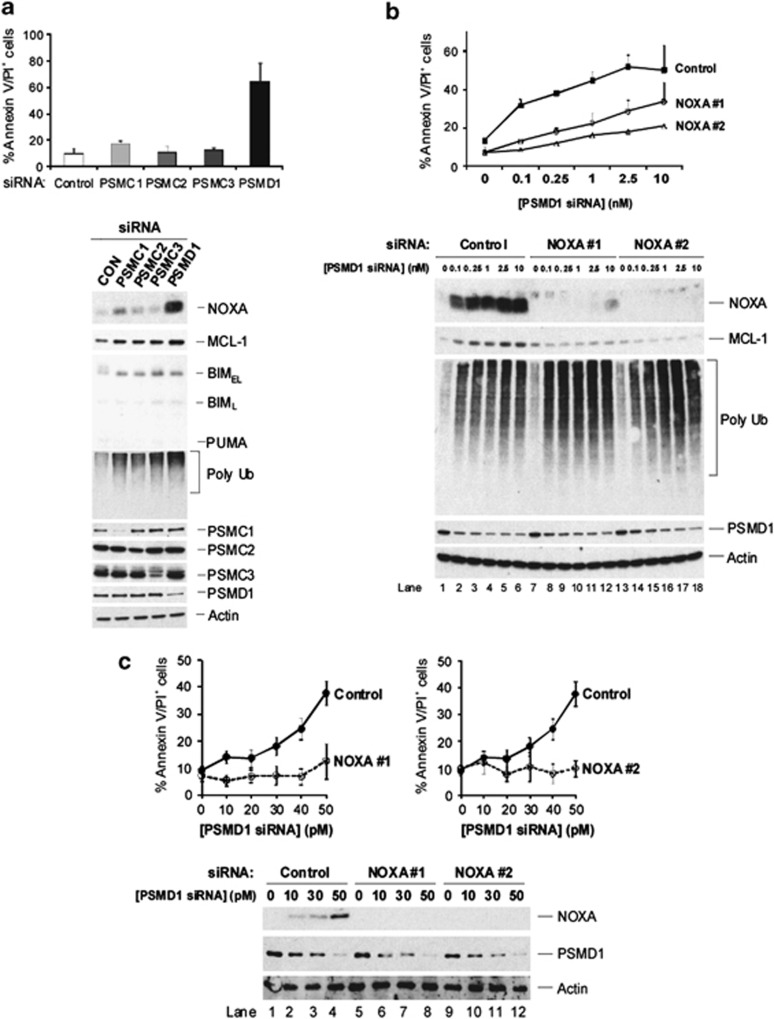
19S RP but not REG subunits are required for NOXA degradation. (a) Depletion of 19S RP subunits increases endogenous NOXA levels. HEK293T cells were transfected with siRNA pools (10 nM) against the indicated 19S RP or REG subunits for 48 h, and the indicated proteins were assessed by immunoblotting. REGβ was undetectable in these cells, although REGβ siRNA reduced REGα levels. (b) Depletion of 19S RP subunits inhibits 26S proteasome but increases 20S proteasome activity. HEK293T cells were transfected with the indicated siRNA pools (1 nM). Soluble extracts were subjected to 10–40% glycerol density gradient centrifugation and fractions assayed for Suc-LLVY-AMC hydrolyzing activity ± (0.02%). Fractions 1 and 22 correspond to the top (10%) and bottom (40%) of the gradient, respectively. Sedimentation positions of 20S and 26S proteasomes were assessed based upon co-fractionation with 20S CP subunits in the absence of 19S RP subunits or 20S CP subunits in the presence of 19S RP subunits, respectively. (c) MCL-1 is required to maintain endogenous NOXA levels, following depletion of the 19S RP subunit PSMC1. Cells were co-transfected with PSMC1 siRNA in the presence of a control or MCL-1 siRNA (10 nM). Indicated proteins were assessed by immunoblotting. (d) Proteasome inhibitor-induced accumulation of endogenous NOXA does not require MCL-1. Cells were transfected with either control or MCL-1 siRNA (1 nM) and immediately incubated with either DMSO, lactacystin (10 μM) or epoxomicin (2.5 μM) for 24 h. Indicated proteins were assessed by immunoblotting. (e) Degradation of NOXA-LL is blocked upon PSMC1 depletion. HEK293T cells were transfected for 24 h with control or PSMC1 siRNA (10 nM) and re-transfected for a further 24 h with empty vector, NOXA-WT or -LL. Degradation of NOXA was assessed by immunoblotting, following cycloheximide addition
Next, we examined the effect of siRNA-mediated depletion of both AAA+-ATPase subunit, PSMC1, and a non-ATPase 19S RP subunit, PSMD1, on the assembly and activity of 26S proteasomes using glycerol density gradient centrifugation. In control siRNA-treated cells, a major faster sedimenting peak of proteasome activity (fractions 18–21) characteristic of 26S proteasomes was observed, as it co-migrated with 19S RP subunits and half of the 20S core particle (CP) subunits (Figure 4b and Supplementary Figure S7A). A minor, slower sedimenting peak of proteasome activity (fractions 12–16) characteristic of uncapped 20S proteasomes, as it was activated by low SDS concentrations and co-migrated with the remaining half of the 20S CP subunits, was also observed (Figure 4b and Supplementary Figure S7A,27, 28). Importantly, depletion of PSMC1 or PSMD1 resulted in substantial loss of 26S proteasome activity accompanied by a 2–3-fold increase in 20S proteasome activity, together with an increased proportion of CP subunits in 20S compared with 26S proteasomal fractions and the displacement of 19S RP subunits to lower density fractions (Figure 4b, Supplementary Figures S7A and B). These results are consistent with the disruption of 26S proteasome structure and increased accumulation of 20S proteasomes.28
The increase in NOXA levels following PSMC1 depletion, but not following lactacystin or epoxomicin, was dependent upon maintenance of MCL-1 levels (Figures 4c and d). These results were consistent with the observation that PSMC1 knockdown was incomplete, allowing residual 26S proteasome activity to subsequently degrade NOXA, which was enhanced in the absence of MCL-1 (Figures 4b and c, Supplementary Figures S7B and S7C), whereas proteasome inhibition by MG132 and lactacystin was more complete (Figure 4d). Importantly, following depletion of PSMC1, NOXA-LL degradation was blocked to a similar extent as both endogenous NOXA and transfected NOXA-WT (Figure 4e), demonstrating that 19S RP subunits were also required for degradation of a NOXA-LL mutant.
NOXA is required for apoptosis mediated by 26S proteasome inhibition
Next, we examined whether depletion of individual 19S RP subunits induced apoptosis. Depletion of PSMD1 using a pool of four individual siRNAs induced significant levels of apoptosis in HeLa cells, which correlated with the highest levels of NOXA (Figure 5a). The broad-spectrum caspase inhibitor, z-VAD.fmk, completely inhibited apoptosis following suppression of PSMD1, indicating that cell death was caspase-dependent (Supplementary Figure S8). Loss of PSMD1 caused a concentration-dependent increase in apoptosis, accompanied by an accumulation of polyubiquitinated proteins, which was consistent with disruption of 26S proteasome activity (Figures 5b and c). Importantly, apoptosis triggered by depletion of PSMD1 was strongly decreased using two independent pools of NOXA siRNA in both HeLa and H1299 cells (Figures 5b and c). In summary, these results show that the degradation of NOXA in cells requires 19S RP of the 26S proteasome and demonstrate that disruption of 26S proteasomes triggers NOXA-dependent apoptosis. Although the siRNA for NOXA largely protected against apoptosis, we cannot exclude the possibility that other BH3-only proteins may also have some additional or complimentary role in apoptosis induced, following loss of 26S proteasome integrity.
Figure 5.
Inhibition of 26S proteasomes by depletion of the 19S RP subunit PSMD1 triggers NOXA-dependent apoptosis. (a) Apoptosis induced by depletion of different 19S RP subunits correlates with high endogenous NOXA levels. HeLa cells were transfected with siRNA pools of the indicated 19S RP subunits or a control siRNA (10 nM each) for 48 h and apoptosis quantified (top panel). Results are the means ±S.E.M. (n=3). Indicated endogenous proteins were assessed by immunoblotting (lower panel). (b) HeLa or (c) H1299 cells were transfected with a pool of PSMD1 siRNAs, and either a control or pool of NOXA siRNAs (10 nM) from Dharmacon (NOXA #1) or Ambion (NOXA #2). After 48 h, apoptosis was quantified (top panels). Results are the means ±S.E.M. (n=3). The levels of indicated proteins were assessed by immunoblotting (lower panels)
Discussion
Our data shows that NOXA, which possesses an unstructured region, can be degraded by 26S proteasomes in an Ub-independent manner in vivo (Figures 1, 2 and 4 and Supplementary Figure S4). This is the first demonstration that a BCL-2 family member can be degraded by 26S proteasomes in an Ub-independent manner, and raises the possibility that other BH3-only proteins containing unstructured regions may be degraded in a similar manner in vivo,29 In more general terms, there is an expanding list of unstructured proteins including p53, p73α, p21Cip1 IκB, MCL-1 and BIM that are proposed 20S proteasome substrates based on in vitro studies.12, 14, 15, 22 Our studies with NOXA suggest that some of these proteins may be bona fide 26S proteasome substrates in vivo. For example, although MCL-1 can be degraded by 20S proteasomes in vitro and a non-ubiquitinated MCL-1 mutant is degraded at a similar rate compared with wild-type MCL-1,15 other studies and our data suggest that 26S proteasomes have an important role in MCL-1 turnover in cells, possibly by the E3 ligase, MULE30 (Figure 4 and Supplementary Figure S6). Depletion of 19S RP subunits in cells showed that NOXA accumulated under conditions where 26S proteasome activity was inhibited and 20S proteasome activity was increased (Figure 4b). These results clearly support the hypothesis that NOXA degradation in cells occurs by 26S as opposed to 20S proteasomes. We suggest that this approach of using siRNA-mediated selective inhibition of 26S proteasomes without inhibiting 20S proteasome activity may be a useful tool to assess the relative contributions of 26S and 20S proteasomes to the degradation of other proteins containing unstructured regions in vivo (Figure 4).28
Previous studies have demonstrated a requirement for NOXA in cell death triggered by proteasome inhibitors that act by inhibiting the 20S CP catalytic activity.7, 8, 9 We now show that depletion of the 19S RP non-ATPase PSMD1 subunit by RNAi resulted in NOXA-dependent apoptosis (Figure 5). Concurrent increases in MCL-1 following blockade of 26S proteasomes potentiate NOXA accumulation by promoting stabilization of NOXA (Figures 3, 4 and 5), while also perturbing NOXA-mediated apoptosis. Further studies are therefore warranted to establish how these apparently opposing roles of MCL-1 during proteasome inhibitor-induced apoptosis are regulated. Our results combined with others demonstrate that disruption of proteasomal function by various mechanisms triggers the rapid accumulation of NOXA and subsequent cell death, strongly implicating NOXA as a sensor of 26S proteasome function. Thus, functional targeting of key components of the 19S RP including its AAA+-ATPase activity or disruption of critical molecular interactions involving, for example, PSMD1, could represent a more specific and targeted alternative approach to treat various tumors than existing proteasome inhibitors, which primarily inhibit the chymotrypsin-like activity of 20S proteasomes. Our observation that suppression of different 19S RP subunits resulted in similar moderate increases in MCL-1 yet varying increases in NOXA levels suggests distinct pathways may exist for controlling degradation of 19S RP substrates, possibly by utilization of distinct proteasome entry portals.31 Our findings do not exclude the possibility that NOXA turnover may occur by a trans inititation mechanism in vivo.16 Consistent with this two-part degron model,16 NOXA may associate with an uncharacterized polyubiquitinated adaptor protein that facilitates its targeting to the 19S RP and subsequently, degradation of NOXA is initiated from the unstructured region of NOXA. Further studies are necessary to distinguish this possibility from an alternative model that NOXA binds directly with sufficiently high affinity to the 19S RP, followed by degradation commencing from its unstructured region.
MCL-1 caused a dramatic stabilization of endogenous NOXA in vivo (Figures 3 and 4). A similar finding was also observed in HeLa cells.32 The NOXA BH3 domain was required for MCL-1-mediated stabilization of NOXA (Figure 3d and Supplementary Figure S3D). In addition, a cluster of acidic residues (Glu 20, Glu 22 and Glu 24) in NOXA that form potential electrostatic interactions with two exposed basic residues in MCL-1 (Supplementary Figures S3A and B) appeared to contribute to the selective stabilization of NOXA by MCL-1 (Supplementary Figure S3). These results are in apparent contrast to previous studies in MEFs, which have shown that proteasomal degradation of MCL-1 is triggered by its interaction with NOXA.21, 33, 34, 35 Recently, it has been shown that MULE, an E3 ligase for MCL-1, and USP9X, a deubiquitinase for MCL-1, regulate NOXA-mediated MCL-1 proteasomal degradation.30, 35, 36 We did not observe a loss of MCL-1 protein either following NOXA overexpression in HEK293T cells (Figure 1) or during NOXA-induced apoptosis in HeLa cells (Figures 2 and 5). The basis for these apparently conflicting observations is unclear but may relate to cell type differences, multiplicity of other E3 ligases and deubiquitinases for MCL-1, or MULE may not be present or have access to the NOXA/MCL-1 complex. Two other BH3-only proteins, BIM and PUMA, stabilize MCL-1 via a mechanism that requires their BH3 domains,21, 37 and interaction of BIMEL with BCL-2 also reduces its turnover.38 Although further studies are required to reconcile these differing results, together they highlight the critical role that interactions between BH3-only proteins and their pro-survival partners have in regulating the stability of multiple BCL-2 family proteins.
The ability of MCL-1 to stabilize NOXA may have important therapeutic implications. Tumor cells containing anti-apoptotic proteins that have sequestered significant amounts of BH3-only proteins have been described as ‘primed for death', and release of these sequestered BH3-only proteins can lead to high levels of cell death.39 The ability of MCL-1 to bind and increase the stability of NOXA could represent a mechanism by which cancer cells expressing high MCL-1 levels, such as MYC-induced acute myeloid leukemic cells in mice, are ‘primed' with high levels of NOXA.40 Thus, the development of potent specific MCL-1 inhibitors may fill an important therapeutic need to treat tumors addicted to MCL-1.
In summary, our studies show that NOXA, an unstructured BH3-only protein, can be degraded in an Ub-independent manner by 26S proteasomes in vivo. Interaction of NOXA with MCL-1 selectively increases NOXA stability. Disruption of 26S proteasomes triggers rapid accumulation of NOXA and subsequent apoptosis, strongly implicating NOXA as a sensor of 26S proteasome function.
Materials and methods
Materials
HEK293T and HeLa cells were cultured in DMEM supplemented with 10% FBS. H1299 cells were cultured in RPMI 1640 containing 10% FBS. HCT-116 cells were grown in McCoys medium supplemented with 10% FBS. Full-length NOXA, synthesized as a peptide (>95% purity), was obtained from Peptide Protein Research (Wickham, Hampshire, UK). A NOXA mAb (Clone 114C307) and a mouse tubulin mAb (Cat. No. CP06) were obtained from Merck Biosciences (Nottingham, UK). Rabbit polyclonal antibodies to MCL-1 (Cat. No. sc-819), MyoD (Cat. No. sc-304) and PSMD2/Rpn1 (Cat. No. sc-68352), a HA mAb (Cat. No. sc-7392) and a goat polyclonal antibody to NOXA (sc-26917) were obtained from Santa Cruz Biotechnology (Heidelberg, Germany). 26S proteasomes, lactacystin, epoxomicin and antibodies to Ub, REGα, β and γ, PSMC1/Rpt2, PSMC2/Rpt1, PSMC3/Rpt5, PSMC5/Rpt6, PSMC6/Rpt4, PSMD1/Rpn2, PSMB2, PSMB5 and PSMA2 were obtained from Enzo Life Sciences (Exeter, UK). Purified 26S proteasomes were also kindly provided by Dr. W. Dubiel (Charite Univ. Med Berlin., Berlin, Germany). Recombinant p53 was purchased from R&D Systems Europe Ltd (Abingdon, UK). Rabbit polyclonal antiserum to PUMA (Cat. No. 4976) and a Myc Tag (9B11) mAb (Cat. No. 2276) were obtained from Cell Signalling Technology (New England Biolabs, Hitchin, UK). Rabbit polyclonal antiserum to BCL-XL was obtained from BD Biosciences (Cat. No. 610212, Cambridge, UK). Rabbit polyclonal antiserum to BCL2A1 was a generous gift from Dr. Jannie Borst (Netherlands Cancer Institute, Amsterdam, Holland). A TNT- T7 Coupled Wheat Germ Extract System kit was obtained from Promega (Southampton, UK). The following SMARTpool siRNAs (human) were purchased from Dharmacon (Lafayette, CO, USA): PSMC1/Rpt2 (target sequences; 5′-GGCCCAAACUCGUACGGGA-3′, 5′-AGAGAGAAAUUCAGCGAAC-3′, 5′-CCGAAUAGAAACUUUGGAU-3′, 5′-CGUCCAUCGUGUUUAUUGA-3′), PSMC2/Rpt1, PSMC3/Rpt5, PSMD1/Rpn2, PSME1/REGα (target sequences; 5′-GGAAUGAUCUAUUGAGAGC-3′, 5′-GAAGACCUCUGUACCAAGA-3′, 5′-GCAGAUACCUCGGAUUGAG-3′, 5′-GUGGUGAUGCAGUGACUAA-3′), PSME2/REGβ (target sequences; 5′-CCUUGGUGCAUGAGCGAGA-3′, 5′-CGUCAAGACCAAAGUGGAA-3′, 5′-AGAAGAAAGAAGUCCAUAA-3′, 5′-GAUUGAAGAUGGAAAUGAU-3′), PSME3/REGγ (target sequences; 5′-GAAUCAAUAUGUCACUCUA-3′, 5′-UCUGAAGGAACCAAUCUUA-3′, 5′-GCUAAGAACUGUUGAGAGU-3′, 5′-GACCAGAUUUCUAGAUAUU-3′) and NOXA (target sequences; 5′-AAACUGAACUUCCGGCAGA-3′, 5′-GAACCUGACUGCAUCAAAA-3′, 5′-AAUCUGAUAUCCAAACUCU-3′, 5′-GCAAGAACGCUCAACCGAG-3′). Pools of MCL-1 and NOXA (target sequences; 5′-AGAUAUGAAUGUUUCUAAA-3′, 5′-AGUCGAGUGUGCUACUCAA-3′, 5′-GAAACUUAGUAGAACAAAU-3′) siRNAs were also obtained from Ambion (Warrington, UK). Cycloheximide was obtained from Sigma-Aldrich (Poole, UK). MG132 was from Merck Biosciences. Streptactin-sepharose was obtained from IBA GmbH (Göttingen, Germany). Ni2+-NTA agarose was obtained from Qiagen (Crawley, UK).
Plasmid constructs and site-directed mutagenesis
p3xFLAG-CMV7.1 was obtained from Sigma. pEGFP-Myc6-NFAT5 was from Addgene (Cambridge, MA, USA). pcDNA3-Myc-MCL-1 was kindly provided by Dr. Eric Eldering (Academic Medical Center, Amsterdam, The Netherlands). pEYZ-BCL2A1 was generously provided by Dr. Ingolf Berberich (University of Würzburg, Wurzburg, Germany). Full-length BCL2A1 was subcloned into either pcDNA3 or pmax to obtain pcDNA3-BCL2A1 or pmax-BCL2A1, respectively. Human BCL-2 cDNA (transcript α, IMAGE clone 4511027) was obtained from Geneservice (Cambridge, UK). The BCL-2 coding sequence was amplified by proofreading PCR then subcloned into the EcoRI and XbaI sites of pcDNA3.1 to generate pcDNA3.1-BCL-2. Human Bcl-XL cDNA was prepared from isolated chronic lymphocytic leukemia patient peripheral blood cells and subcloned with a 5′-Strep Tag II into the BamHI and XhoI sites of pcDNA6 to generate pcDNA6-Strep Tag II-Bcl-XL. pcDNA3-NQO1 was a generous gift from Dr. David Ross (University of Colorado, Boulder, CO, USA). pMT107 encoding octa-His6Ub under control of a cytomegalovirus promoter was obtained from Dr. Dirk Bohmann (University of Rochester, Rochester, NY, USA). pcDNA3.1-HA-Ub-WT and -VV were generated by proofreading PCR using pcDNA3-Ub-WT as a template, and forward and reverse primers containing flanking EcoRI and XbaI restriction sites including a 5′-HA tag, digested and ligated into pcDNA3.1. pcDNA6-NOXA-WT and Strep Tag II-NOXA-WT were prepared by amplifying NOXA-WT from pMT25M-NOXA-WT using 5′- and 3′- primers containing BamHI and XhoI sites, and ligating into corresponding restriction sites of pcDNA6.
A NOXA-LL construct was generated by two sequential site-directed mutagenesis reactions using a Quikchange Multisite-directed mutagenesis kit (Stratagene, Amsterdam, The Netherlands). In the first reaction, NOXA-WT was used as a template with three mutagenic primers: NOXA-K4R/K5R/K8R, NOXA-K35R and NOXA-K48R to generate a library of NOXA mutants including NOXA-K4R/K5R/K8R/K35R/K48R, which was subsequently used as a template with a NOXA-K41R mutagenic primer to generate NOXA-LL. To construct N-terminal Myc6-NOXA-WT and -LL, the Myc6 tag from pEGFP- Myc6-NFAT5 was amplified by proofreading PCR using Pfx DNA polymerase with primers containing flanking BamHI restriction sites, digested and ligated into the BamHI site of pcDNA6-NOXA-WT/-LL. To generate C-terminal Myc6-tagged NOXA-WT and -LL, the stop codon of NOXA was first changed into an Ala residue. Then, the Myc6 tag from pEGFP-Myc6-NFAT5 was amplified by proofreading PCR using Pfx DNA polymerase with primers flanked with XbaI restriction sites, digested and ligated into the XbaI site of pcDNA6-NOXA-WT/-LL. All DNA primers were purchased from Invitrogen (Paisley, UK). Sequences of all plasmid constructs were verified by automated DNA sequencing. Sequences of primers used for site-directed mutagenesis of human NOXA are available upon request.
Transient transfection and RNAi of cells
HEK293T and HeLa cells were transfected using JetPEI (Autogen Bioclear, Calne, UK) or TransIT-LT1 (Mirus-Cambridge Bioscience, Cambridge, UK), respectively, according to the manufacturer's instructions. Pools of four individual siRNA duplexes were transfected into cells using Lipofectamine RNAiMAX, following the manufacturer's instructions (Invitrogen).
Assessment of protein stability using cycloheximide chase
Cells were transiently transfected with the indicated constructs. After 48 h, cycloheximide (20 μM) was added for 0–8 h. Cells were washed with ice-cold PBS and lysed for 15 min with ice-cold RIPA buffer (50 mm Tris, pH 7.4, 150 mm NaCl, 10 mm Na4P2O7, 25 mm sodium β-glycerophosphate, 1 mm EDTA, 1% Igepal CA-630, 0.5% sodium deoxycholate, 0.1% SDS, 1 mM Na3VO4, 10 mM NaF, 5 mM N-ethylmaleimide, 1 mM 1,10-phenanthroline and 50 μM MG132 supplemented with a complete Protease inhibitor cocktail (Roche, Mannheim, Germany)).
Pulse-chase labelling assay to measure protein turnover
Following transient transfection with the constructs or siRNAs indicated, cells were depleted of cysteine and methionine by washing twice at 4 °C in methionine/cysteine-free medium and subsequently incubating cells for 1 h in methionine/cysteine-free media supplemented with dialyzed serum. Fresh methionine/cysteine-free media with dialyzed serum and 35S-labeled methionine/cysteine (25 μCi/ml for each amino acid) (Perkin Elmer, Buckinghamshire, UK) was added and incubated for 1 h. After the pulse, the labelling media was removed and the cells washed twice in ice-cold complete medium containing 2 mM unlabeled cysteine and methionine. Complete media with 2 mM cysteine and methionine at 37 °C was added and samples withdrawn at the indicated times. Cells were lysed in 1 ml ice-cold 20 mM TrisHCl pH 7.4, 150 mM NaCl, 10% glycerol, 1% Igepal CA630 containing a complete Protease inhibitor cocktail, MG132, protein phosphatase and deubiquitinase inhibitors. After centrifugation, washed streptactin–sepharose beads were added and the supernatant/bead mixture rotated at 4 °C overnight. The beads were washed in lysis buffer and turnover of 35S-labeled NOXA followed by SDS-PAGE and Phosphorimage analysis.
Assay for ubiquitination of NOXA in cells
Cells were plated in 10 cm dishes 24 h prior to transfection. Cells were transfected with equal quantities of a NOXA-WT or -LL construct and pMT107. After 48 h, cells were lysed in 10 mM TrisHCl pH 8, 0.1 M sodium phosphate, 5 mM imidazole containing 6 M guanidine HCl then briefly sonicated and incubated for 30 min at 4 °C. After centrifugation, Ni2+–NTA agarose beads were added and mixtures rotated for 3 h at 4 °C. Beads were washed with the following buffers: (i) 10 mM TrisHCl pH 8, 0.1 M sodium phosphate, 5 mM imidazole containing 6 M guanidine HCl (ii) 10 mM TrisHCl pH 8, 0.1 M sodium phosphate containing 8 M urea and (iii) 10 mM TrisHCl pH 6.3, 0.1 M sodium phosphate, 0.2% (w/v) Triton X-100 containing 8 M urea. His-Ub-tagged proteins were eluted with 0.15 M TrisHCl, 30% glycerol, 5% SDS, 0.5 M imidazole and resolved by SDS-PAGE.
Pulldown of NOXA and associated proteins using streptactin–sepharose beads
Cells were washed twice with ice-cold PBS and lysed for 30 min with 1 ml ice-cold 20 mM TrisHCl pH 7.4, 150 mM NaCl, 10% glycerol, 1% Igepal CA630 containing a complete Protease inhibitor cocktail, MG132, protein phosphatase and deubiquitinase inhibitors. Following centrifugation to remove insoluble material, lysates were added to pre-washed streptactin-sepharose beads and rotated for 3–4 h or overnight at 4 °C. Beads were washed five times with 1 ml ice-cold lysis buffer and Strep Tag-NOXA, and associated proteins resolved by SDS-PAGE.
Glycerol density gradient centrifugation
Cells (3 × 10 cm dishes) were collected and washed twice in PBS. Pellets (300 mg wet weight/ml) were resuspended in Proteasome Buffer (50 mM TrisHCl pH 8, 5 mM MgCl2, 0.5 mM EDTA, 1 mM DTT, 4 mM ATP) prior to disruption by vortexing for 5 min at 4 °C using washed glass beads. After incubation for 30 min at 4 °C, cell homogenates were centrifuged at 17 000 × g for 30 min at 4 °C. Supernatants (0.25 ml) were layered on top of 4.5 ml glycerol gradients (10–40% glycerol in 25 mM TrisHCl pH 7.5, 1 mM DTT, 4 mM ATP) and centrifuged at 125 000 × g for 18 h at 4 °C using a Beckman Optima L-90K centrifuge (Beckman Coulter (UK), High Wycombe, UK) with a SW60 Ti rotor. Fractions (200 μl) were collected and proteasome activity quantified in the presence or absence of 0.02% SDS, using N-succinyl-Leu-Leu-Val-Tyr 7-amido-4-methylcoumarin (Suc-LLVY-AMC; Bachem, Weil am Rhein, Germany). Fractions (10 μl) were mixed with 25 μM LLVY-amc in 100 μl Proteasome Buffer, incubated at 37 °C, and the rate of 7-amido-4-methylcoumarin liberated over 20 min measured with an excitation wavelength of 380 nm and an emission wavelength of 440 nm. In addition, fractions (10 μl) were resolved by SDS-PAGE on 10–20% Criterion Tris-Tricine gradient gels (Bio-Rad Laboratories, Hemel Hempstead, UK) and immunoblot analysis were performed.
In vitro transcription/translation of NOXA
NOXA proteins were translated in vitro using a TNT T7 Coupled Wheat Germ Extract transcription-translation kit according to the manufacturer's instructions (Promega).
Bacterial expression of MCL-1
A human MCL-1 deletion mutant (ΔN-MCL-1 WT, amino acids 152–308) lacking the N-terminal region and C-terminal transmembrane domain was expressed in E. Coli BL21 (DE3) as a His6-S Tag fusion protein. In addition, a BH1 domain mutant of ΔN-MCL-1 (ΔN-MCL-1 G162A) was generated. The protein was purified using Ni-NTA resin and following elution with imidazole, extensively dialyzed against Proteasome buffer (50 mM TrisHCl, 5% glycerol, 1.1 mM DTT, 5 mM MgCl2, pH 8.2).
UV CD measurements
CD spectra were recorded at 5 °C using a Jasco J-715 spectrapolarimeter (Jasco Spectroscopic Co. Ltd., Hachioji City, Japan) equipped with a water bath, using a 1-mm path length cell. Data were collected continuously at 50 nm/min from 200 to 250 nm, with a bandwidth of 1 nm, and results were averaged from five scans. Preparations of NOXA and MCL-1 were analyzed in 50 mM HEPES, pH 8.2 and 5% glycerol. Thermal stability measurements were performed by heating samples at a constant rate of 1 °C/min, while monitoring the ellipticity at 222 nm.
Assessment of apoptosis
Apoptosis was quantified using Annexin V-FITC, made in our laboratory, in the presence of PI to detect phosphatidylserine externalization as described.8
Acknowledgments
These studies were supported by the Medical Research Council (London, UK). We thank Dr. X-M. Sun for ΔN-MCL-1 G262A protein and Dr. David Ross (University of Colorado, Boulder) for pcDNA3-NQO1.
Glossary
- CHX
cycloheximide
- MCL-1
myeloid cell leukemia 1
- WT
wild-type
- LL
lysine-less
- BH
Bcl-2 homology
- CP
core particle
- RP
regulatory particle
- t1/2
half life
- HA
hemagglutinin
- NTA
nitriloacetic acid
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by G Melino
Supplementary Material
References
- Youle RJ, Strasser A. The BCL-2 protein family: opposing activities that mediate cell death. Nat Rev Mol Cell Biol. 2008;9:47–59. doi: 10.1038/nrm2308. [DOI] [PubMed] [Google Scholar]
- Chen L, Willis SN, Wei A, Smith BJ, Fletcher JI, Hinds MG, et al. Differential targeting of prosurvival Bcl-2 proteins by their BH3-only ligands allows complementary apoptotic function. Mol Cell. 2005;17:393–403. doi: 10.1016/j.molcel.2004.12.030. [DOI] [PubMed] [Google Scholar]
- Hagenbuchner J, Ausserlechner MJ, Porto V, David R, Meister B, Bodner M, et al. The anti-apoptotic protein BCL2L1/Bcl-xL is neutralized by pro-apoptotic PMAIP1/Noxa in neuroblastoma, thereby determining bortezomib sensitivity independent of prosurvival MCL1 expression. J Biol Chem. 2010;285:6904–6912. doi: 10.1074/jbc.M109.038331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JY, Ahn HJ, Ryu JH, Suk K, Park JH. BH3-only protein Noxa is a mediator of hypoxic cell death induced by hypoxia-inducible factor 1 alpha. J Exp Med. 2004;199:113–123. doi: 10.1084/jem.20030613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikiforov MA, Riblett M, Tang WH, Gratchouck V, Zhuang D, Fernandez Y, et al. Tumor cell-selective regulation of NOXA by c-MYC in response to proteasome inhibition. Proc Natl Acad Sci USA. 2007;104:19488–19493. doi: 10.1073/pnas.0708380104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda E, Ohki R, Murasawa H, Nemoto J, Shibue T, Yamashita T, et al. Noxa, a BH3-only member of the Bcl-2 family and candidate mediator of p53-induced apoptosis. Science. 2000;288:1053–1058. doi: 10.1126/science.288.5468.1053. [DOI] [PubMed] [Google Scholar]
- Qin JZ, Ziffra J, Stennett L, Bodner B, Bonish BK, Chaturvedi V, et al. Proteasome inhibitors trigger NOXA-mediated apoptosis in melanoma and myeloma cells. Cancer Res. 2005;65:6282–6293. doi: 10.1158/0008-5472.CAN-05-0676. [DOI] [PubMed] [Google Scholar]
- Baou M, Kohlhaas SL, Butterworth M, Vogler M, Dinsdale D, Walewska R, et al. Role of NOXA and its ubiquitination in proteasome inhibitor-induced apoptosis in chronic lymphocytic leukemia cells. Haematologica. 2010;95:1510–1518. doi: 10.3324/haematol.2010.022368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez Y, Verhaegen M, Miller TP, Rush JL, Steiner P, Opipari AW, et al. Differential regulation of Noxa in normal melanocytes and melanoma cells by proteasome inhibition: Therapeutic implications. Cancer Res. 2005;65:6294–6304. doi: 10.1158/0008-5472.CAN-05-0686. [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. The ubiquitin system. Ann Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Vucic D, Dixit VM, Wertz IE. Ubiquitylation in apoptosis: a post-translational modification at the edge of life and death. Nat Rev Mol Cell Biol. 2011;12:439–452. doi: 10.1038/nrm3143. [DOI] [PubMed] [Google Scholar]
- Tsvetkov P, Reuven N, Shaul Y. The nanny model for IDPs. Nat Chem Biol. 2009;5:778–781. doi: 10.1038/nchembio.233. [DOI] [PubMed] [Google Scholar]
- Jariel-Encontre I, Bossis G, Piechaczyk M. Ubiquitin-independent degradation of proteins by the proteasome. Biochim Biophys Acta. 2008;1786:153–177. doi: 10.1016/j.bbcan.2008.05.004. [DOI] [PubMed] [Google Scholar]
- Wiggins CM, Tsvetkov P, Johnson M, Joyce CL, Lamb CA, Bryant NJ, et al. BimEL, an intrinsically disordered protein, is degraded by 20S proteasomes in the absence of poly-ubiquitylation. J Cell Sci. 2011;124:969–977. doi: 10.1242/jcs.058438. [DOI] [PubMed] [Google Scholar]
- Stewart DP, Koss B, Bathina M, Perciavalle RM, Bisanz K, Opferman JT. Ubiquitin-independent degradation of antiapoptotic MCL-1. Mol Cell Biol. 2010;30:3099–3110. doi: 10.1128/MCB.01266-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader EK, Harstad KG, Matouschek A. Targeting proteins for degradation. Nat Chem Biol. 2009;5:815–822. doi: 10.1038/nchembio.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitschopf K, Bengal E, Ziv T, Admon A, Ciechanover A. A novel site for ubiquitination: the N-terminal residue, and not internal lysines of MyoD, is essential for conjugation and degradation of the protein. EMBO J. 1998;17:5964–5973. doi: 10.1093/emboj/17.20.5964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijhawan D, Fang M, Traer E, Zhong Q, Gao WH, Du FH, et al. Elimination of Mcl-1 is required for the initiation of apoptosis following ultraviolet irradiation. Genes Dev. 2003;17:1475–1486. doi: 10.1101/gad.1093903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabek N, Herman-Bachinsky Y, Ciechanover A. Ubiquitin degradation with its substrate, or as a monomer in a ubiquitination-independent mode, provides clues to proteasome regulation. Proc Natl Acad Sci USA. 2009;106:11907–11912. doi: 10.1073/pnas.0905746106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover A, Ben-Saadon R. N-terminal ubiquitination: more protein substrates join in. Trends Cell Biol. 2004;14:103–106. doi: 10.1016/j.tcb.2004.01.004. [DOI] [PubMed] [Google Scholar]
- Czabotar PE, Lee EF, van Delft MF, Day CL, Smith BJ, Huang DCS, et al. Structural insights into the degradation of Mcl-1 induced by BH3 domains. Proc Natl Acad Sci USA. 2007;104:6217–6222. doi: 10.1073/pnas.0701297104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asher G, Tsvetkov P, Kahana C, Shaul Y. A mechanism of ubiquitin-independent proteasomal degradation of the tumor suppressors p53 and p73. Genes Dev. 2005;19:316–321. doi: 10.1101/gad.319905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XY, Barton LF, Chi Y, Clurman BE, Roberts JM. Ubiquitin-independent degradation of cell-cycle inhibitors by the REG gamma proteasome. Mol Cell. 2007;26:843–852. doi: 10.1016/j.molcel.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami Y, Matsufuji S, Kameji T, Hayashi S, Igarashi K, Tamura T, et al. Ornithine decarboxylase is degraded by the 26S proteasome without ubiquitination. Nature. 1992;360:597–599. doi: 10.1038/360597a0. [DOI] [PubMed] [Google Scholar]
- Fricker M, O'Prey J, Tolkovsky AM, Ryan KM. Phosphorylation of puma modulates its apoptotic function by regulating protein stability. Cell Death Disease. 2010;1:e59. doi: 10.1038/cddis.2010.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandow JJ, Jabbour AM, Condina MR, Daunt CP, Stomski FC, Green BD, et al. Cytokine receptor signaling activates an IKK-dependent phosphorylation of PUMA to prevent cell death Cell Death Differ 2011. e-pub ahead of print 14 October 2011. [DOI] [PMC free article] [PubMed]
- Shibatani T, Ward WF. Sodium dodecyl sulfate (SDS) activation of the 20S proteasome in rat liver. Arch Biochem Biophys. 1995;321:160–166. doi: 10.1006/abbi.1995.1381. [DOI] [PubMed] [Google Scholar]
- Wojcik C, DeMartino GN. Analysis of Drosophila 26 S proteasome using RNA interference. J Biol Chem. 2002;277:6188–6197. doi: 10.1074/jbc.M109996200. [DOI] [PubMed] [Google Scholar]
- Hinds MG, Smits C, Fredericks-Short R, Risk JM, Bailey M, Huang DCS, et al. Bim, Bad and Bmf: intrinsically unstructured BH3-only proteins that undergo a localized conformational change upon binding to prosurvival Bcl-2 targets. Cell Death Differ. 2007;14:128–136. doi: 10.1038/sj.cdd.4401934. [DOI] [PubMed] [Google Scholar]
- Zhong Q, Gao WH, Du FH, Wang XD. Mule/ARF-BP1, a BH3-only E3 ubiquitin ligase, catalyzes the polyubiquitination of Mcl-1 and regulates apoptosis. Cell. 2005;121:1085–1095. doi: 10.1016/j.cell.2005.06.009. [DOI] [PubMed] [Google Scholar]
- Foerster F, Lasker K, Nickell S, Sali A, Baumeister W. Toward an integrated structural model of the 26S proteasome. Mol Cell Proteomics. 2010;9:1666–1677. doi: 10.1074/mcp.R000002-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez H, Zhang L, George NM, Liu X, Pang X, Evans JJ, et al. Perturbation of the Bcl-2 network and an induced Noxa/Bcl-xL interaction trigger mitochondrial dysfunction after DNA damage. J Biol Chem. 2010;285:15016–15026. doi: 10.1074/jbc.M109.086231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis SN, Chen L, Dewson G, Wei A, Naik E, Fletcher JI, et al. Proapoptotic Bak is sequestered by Mcl-1 and Bcl-x(L), but not Bcl-2, until displaced by BH3-only proteins. Genes Dev. 2005;19:1294–1305. doi: 10.1101/gad.1304105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EF, Czabotar PE, Van Delft MF, Michalak EM, Boyle MJ, Willis SN, et al. A novel BH3 ligand that selectively targets Mcl-1 reveals that apoptosis can proceed without Mcl-1 degradation. J Cell Biol. 2008;180:341–355. doi: 10.1083/jcb.200708096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Bougie P, Menoret E, Juin P, Dousset C, Pellat-Deceunynck C, Amiot M. Noxa controls Mule-dependent Mcl-1 ubiquitination through the regulation of the Mcl-1/USP9X interaction. Biochem Biophys Res Commun. 2011;413:460–464. doi: 10.1016/j.bbrc.2011.08.118. [DOI] [PubMed] [Google Scholar]
- Schwickart M, Huang X, Lill JR, Liu J, Ferrando R, French DM, et al. Deubiquitinase USP9X stabilizes MCL1 and promotes tumour cell survival. Nature. 2010;463:103–107. doi: 10.1038/nature08646. [DOI] [PubMed] [Google Scholar]
- Mei YD, Du WJ, Yang YH, Wu MA. Puma* Mcl-1 interaction is not sufficient to prevent rapid degradation of Mcl-1. Oncogene. 2005;24:7224–7237. doi: 10.1038/sj.onc.1208873. [DOI] [PubMed] [Google Scholar]
- Ewings KE, Hadfield-Moorhouse K, Wiggins CM, Wickenden JA, Balmanno K, Gilley R, et al. ERK1/2-dependent phosphorylation of Bim(EL) promotes its rapid dissociation from Mcl-1 and Bcl-x(L) EMBO J. 2007;26:2856–2867. doi: 10.1038/sj.emboj.7601723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letai AG. Diagnosing and exploiting cancer's addiction to blocks in apoptosis. Nat Rev Cancer. 2008;8:121–132. doi: 10.1038/nrc2297. [DOI] [PubMed] [Google Scholar]
- Xiang Z, Luo H, Payton JE, Cain J, Ley TJ, Opferman JT, et al. Mcl1 haploinsufficiency protects mice from Myc-induced acute myeloid leukemia. J Clin Invest. 2010;120:2109–2118. doi: 10.1172/JCI39964. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.