Abstract
Impaired regulation of mitochondrial dynamics, which shifts the balance towards fission, is associated with neuronal death in age-related neurodegenerative diseases, such as Alzheimer's disease or Parkinson's disease. A role for mitochondrial dynamics in acute brain injury, however, has not been elucidated to date. Here, we investigated the role of dynamin-related protein 1 (Drp1), one of the key regulators of mitochondrial fission, in neuronal cell death induced by glutamate toxicity or oxygen–glucose deprivation (OGD) in vitro, and after ischemic brain damage in vivo. Drp1 siRNA and small molecule inhibitors of Drp1 prevented mitochondrial fission, loss of mitochondrial membrane potential (MMP), and cell death induced by glutamate or tBid overexpression in immortalized hippocampal HT-22 neuronal cells. Further, Drp1 inhibitors protected primary neurons against glutamate excitotoxicity and OGD, and reduced the infarct volume in a mouse model of transient focal ischemia. Our data indicate that Drp1 translocation and associated mitochondrial fission are key features preceding the loss of MMP and neuronal cell death. Thus, inhibition of Drp1 is proposed as an efficient strategy of neuroprotection against glutamate toxicity and OGD in vitro and ischemic brain damage in vivo.
Keywords: neuronal cell death, oxidative stress, Drp1, mitochondrial fusion and fission, cerebral ischemia
Mitochondria play crucial roles in energy metabolism, regulation of free radical formation and calcium storage, thereby determining essential metabolic functions and cell survival.1 Further, mitochondria are highly dynamic organelles that undergo constant fission and fusion and these morphological changes are required for efficient ATP production, calcium buffering, regulation of signal transduction and apoptosis.2 In neurons, mitochondrial fission is also essential for axonal transport of the organelles into areas of high metabolic demand,3 whereas mitochondrial fusion supports substitution and regeneration of mitochondrial proteins, mtDNA repair and functional recovery.2, 4
Consistent with the critical roles of mitochondrial dynamics in neurons, defects in mitochondrial fission and fusion proteins are associated with a wide array of inherited or acquired neurodegenerative diseases such as Charcot–Marie–Tooth disease or Alzheimer's disease, respectively.2 Fission and fusion defects may limit mitochondrial motility, decrease energy production, promote oxidative stress and lead to accumulating of mtDNA defects, thereby promoting neuronal dysfunction and cell death.1 Recently, enhanced mitochondrial fragmentation was associated with induction of neuronal death triggered by oxidative stress.5 These data imply that during neuronal cell death, the tubular mitochondrial network is fragmented into smaller and functionally impaired organelles.5 It is, however, a matter of ongoing controversy, whether mitochondrial fragmentation is cause or consequence in programmed cell death.
Current knowledge of the mechanisms regulating mitochondrial dynamics indicates that fission and fusion of mitochondria are under control of highly conserved dynamin-related GTPases.6 In mammalian cells, mitochondrial membrane fusion is mediated by mitofusin-1 and -2 (Mfn-1, -2) and optical atrophy 1 (OPA1), respectively.6 Mitochondrial fission is mediated by dynamin-related protein 1 (Drp1).7, 8, 9 It has been suggested that the outer membrane proteins Fis1 and Mff and the proapoptotic Bcl-2 family protein Bax10, 11 function directly or indirectly as Drp1 receptors to promote mitochondrial fission. The underlying mechanisms that promote and regulate the recruitment of cytosolic Drp1 to mitochondria are largely unknown.
The observation that enhanced mitochondrial fission is linked to neurodegeneration exposed Drp1 as a potential therapeutic target. Drp1 is highly conserved and contains many critical functional features that correspond to specific target structures within the enzyme, such as GTP binding, GTP hydrolysis, self-assembly and protein interactions with key functions in mitochondrial division.12, 13
In the present study, the mechanisms of Drp1-dependent mitochondrial death pathways were investigated in a model of glutamate toxicity in immortalized mouse hippocampal HT-22 neurons (HT-22 cells). This cell line lacks ionotropic glutamate receptors and glutamate-induced death is mediated by inhibition of the cellular cystine import, subsequent glutathione depletion and enhanced lipoxygenation, inducing a form of programmed cell death called oxytosis.14, 15, 16 Further, we examined small molecule inhibitors of Drp117 to address whether Drp1 played a major role in neuronal cell death after glutamate toxicity and oxygen–glucose deprivation (OGD) in primary neurons in vitro and ischemic brain damage in vivo.
Results
siRNA silencing of Drp1 attenuates glutamate-induced mitochondrial fragmentation and cell death in HT-22 cells
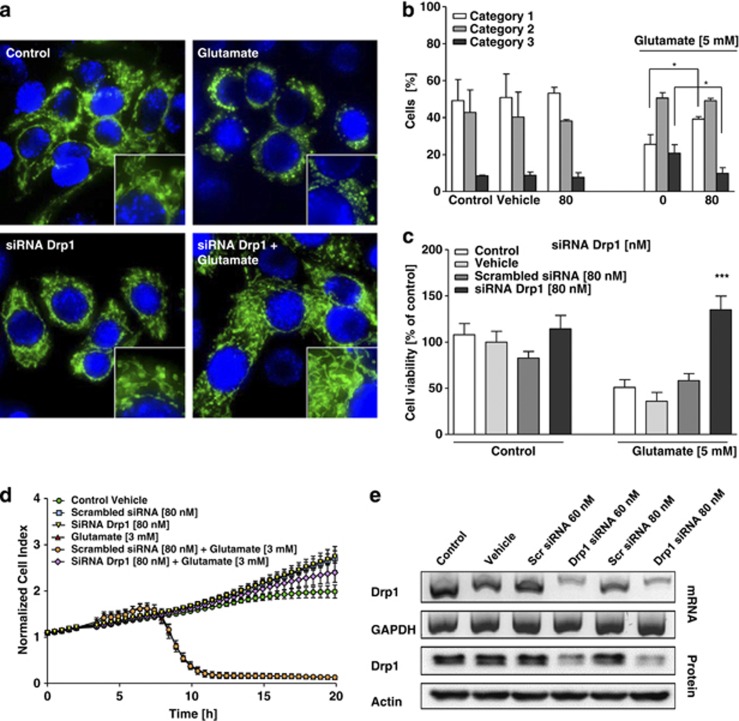
Similar to our previous findings, glutamate toxicity in HT-22 cells was associated with mitochondrial fragmentation.5 Under control conditions, mitochondria were primarily organized in an evenly distributed tubular network, indicating a balance between mitochondrial fusion and fission events (Figures 1a and b). After exposure to glutamate, mitochondria appeared as short tubules and round fragments (Figures 1a and b). Drp1 siRNA significantly reduced Drp1 mRNA and protein levels >60% in the HT-22 cells (Figure 1e). Drp1 siRNA, but not control scrambled siRNA also attenuated mitochondrial fragmentation in glutamate-treated cultures (Figures 1a and b; Supplementary Figures 1a and b). In addition, silencing of Drp1 enhanced cell viability in glutamate-treated HT-22 cells as detected by the MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay (Figure 1c) and real-time impedance measurements (Figure 1d). FACS analysis of apoptotic cells after FITC-Annexin-V-staining confirmed the protective effect of Drp1 siRNA pool (Supplementary Figure 1c). These experiments were repeated with two additional different single siRNA sequences targeting Drp1 at concentrations of 40 nM, confirming the specificity of the protective effect against glutamate toxicity by Drp1 silencing (Supplementary Figures 2a–f). Overall, these results indicate that Drp1 is a suitable target of therapeutic strategies for neuroprotection against glutamate toxicity.
Figure 1.
siRNA silencing of Drp1 attenuates glutamate-induced mitochondrial fragmentation and cell death in HT-22 cells. (a) Fluorescence photomicrographs ( × 63 objective) of mGFP-transfected and DAPI-stained HT-22 cells show that pooled Drp1 siRNA (80 nM, 48 h) prevents fission of mitochondria in glutamate-exposed (5 mM, 18 h) HT-22 cells. (b) Quantification of mitochondrial morphology of ∼500 cells per condition: category 1 (elongated tubulin-like structure), category 2 (intermediate length) and category 3 (fragmented mitochondria). *P<0.05 compared with glutamate 5 mM-treated cells (n=3 independent experiments, ANOVA, Bonferroni-test). (c) Silencing of Drp1 attenuates glutamate-induced cell death as determined by MTT assay. The presented data are normalized to vehicle control Lipofectamine 2000 (100%). ***P<0.001 compared with glutamate-treated cells (n=8, ANOVA, Scheffé's-test). For statistical analysis, the experiments were independently repeated at least three times with an n=8 per treatment condition and the results are presented as mean±S.D. (d) Representative graph of real-time detection of cellular impedance by xCELLigence System (Roche) is shown. After 24 h, Drp1 siRNA-transfected HT-22 cells were seeded out for additional 24 h in 96-well E-plates with 4500 cells/well and treated with 3 mM glutamate for 20 h (n=8 per treatment condition). Glutamate-induced cell death does not differ in vehicle- or scrambled siRNA-treated cells, whereas Drp1 siRNA exerts a persistent protective effect. The cellular impedance of glutamate alone (red triangles) is obscured by the overlapping curve of scrambled siRNA+glutamate (orange dots). (e) RT-PCR analysis of Drp1 mRNA (upper panels) and western blot analysis of Drp1 protein (lower panels) in HT-22 cells pretreated with 60 and 80 nM pooled Drp1 siRNA for 48 h. RT-PCR with primers specific for glyceraldehyde-3-phosphate-dehydrogenase (GAPDH) and anti-β-actin antibodies used as controls for respective analyses is shown. Quantification of the relative protein expression shows a reduction of 60 and 70%
Novel pharmacological inhibitors of Drp1 provide protective effects in HT-22 cells
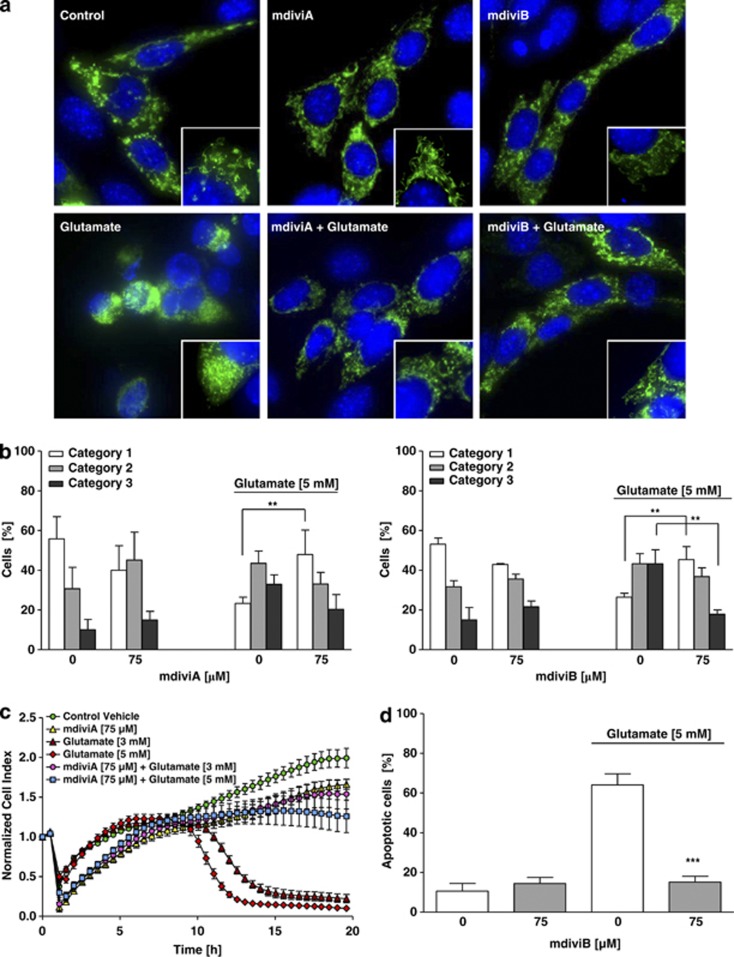
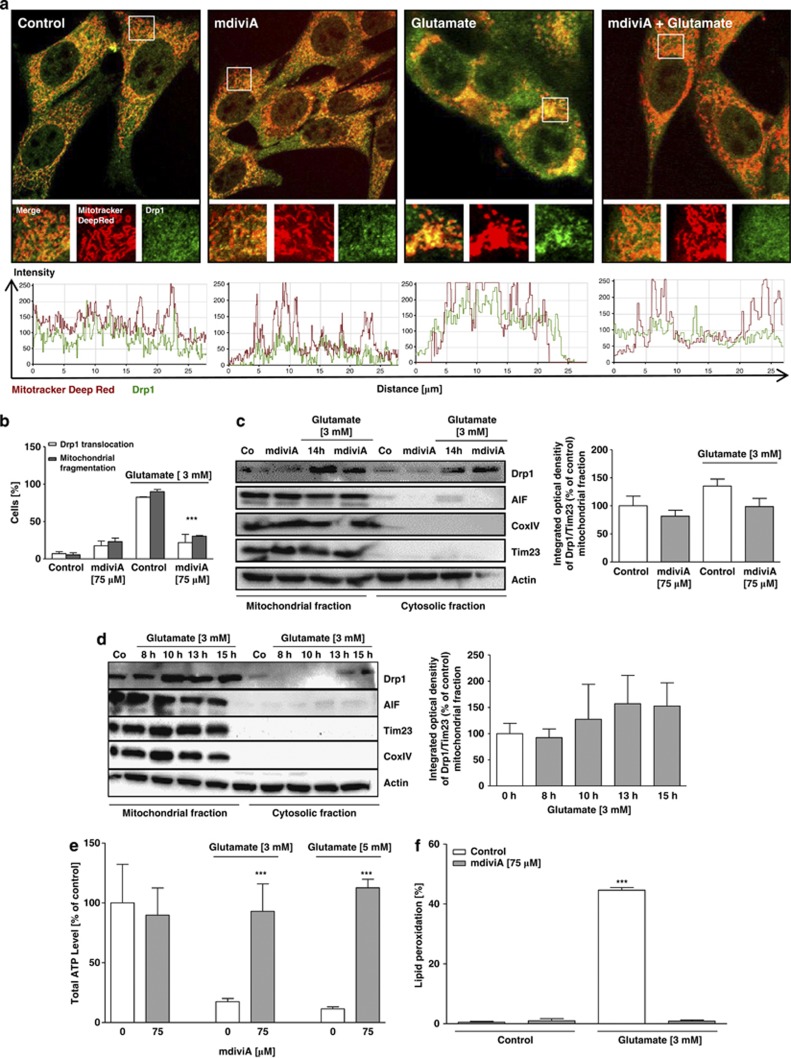
To further test the therapeutic potential of Drp1 inhibition, we applied the small molecule compounds mdiviA and mdiviB, which were previously established as Drp1 inhibitors in vitro.17 MdiviA and mdiviB blocked glutamate-induced mitochondrial fragmentation (Figure 2a). The Drp1 inhibitors increased the overall mitochondrial length and thus shifted the majority of cells into category 1, suggesting reduced mitochondrial fragmentation and increased fusion events (Figure 2b; Supplementary Figures 3c and e). It has been observed that Drp1 assembly and recruitment to mitochondria is stimulated during apoptosis.9, 12 Consistently, glutamate caused an increase in Drp1 clusters associated with mitochondria (Figure 3a). In contrast, the majority of Drp1 remained diffusely distributed in the cytoplasm in cells treated with mdiviA or mdiviB despite the glutamate challenge. The mitochondria appeared as a tubular network in cells treated with mdiviA or mdiviB, contrasting to increased mitochondrial fragmentation in cells undergoing glutamate-induced death. In mitochondrial fractions obtained up to 10 h after onset of glutamate exposure, Drp1 immunoreactivity increased compared with controls in a time-dependent manner (Figure 3d; Supplementary Figure 4c). MdiviA blocked mitochondrial translocation of Drp1 (Figures 3b and c). These results suggested that mdiviA and mdiviB attenuated mitochondrial division during glutamate-induced oxytosis by blocking Drp1 recruitment to mitochondria, consistent with previously published work on STS-induced cell death in mammalian COS-cells.12, 17 In addition, mdiviA and mdiviB enhanced cell viability of glutamate-treated HT-22 cells as detected by impedance measurements (Figure 2c) and the MTT assay (Supplementary Figures 3a and d). It is interesting to note that, once initiated, the complete loss of cellular impedance was observed within a relatively small time window of 2–3 h, suggesting that cell death occurred in a highly synchronized manner, and this was persistently blocked by the Drp1 inhibitors (Figure 2). These results were also confirmed by FACS analysis after FITC-Annexin-V-staining (Figure 2d; Supplementary Figure 3b). In addition, mdiviA also prevented ATP depletion (Figure 3e; Supplementary Figures 3f and g) and significantly reduced lipid peroxidation after glutamate exposure (Figure 3f). In this model system, the increase in early ROS formation after 6–8 h through increased lipid peroxidation is followed by a second more pronounced accumulation of ROS caused by damage of mitochondria.16 Here, lipid peroxidation was detected at the later stages of cell damage, when mitochondrial ROS formation occurred in glutamate-damaged cells. This late increase in ROS formation was blocked by the Drp1 inhibitor mdiviA, indicating protection against mitochondrial ROS formation (Figure 3f). Analyses of mitochondrial protein extracts at various time points after onset of the glutamate treatment revealed increased Drp1 translocation to the organelles at 8–10 h of glutamate exposure (Figure 3d). The Drp1 translocation to mitochondria was confirmed by immunostainings of Drp1 staining (Supplementary Figure 4a) and matches the time course of cellular death that also became detectable around 8 h by impedance measurements or MTT assays (Figure 1d). These observations indicated a continuous increase in Drp1 translocation to mitochondria while the organelles were fragmented and accumulated around the nucleus in damaged cells. Quantification of cells with Drp1 translocation to the mitochondria (Supplementary Figure 4b) and additional western blot analysis with protein quantifications determined Drp1 translocation to the mitochondria after 10 h of glutamate toxicity in HT-22 cells, which was blocked by mdiviA (Figures 3b–d). Overall, these results reveal a major upstream role for Drp1 in mitochondrial damage, energy depletion and subsequent execution of glutamate-induced cell death in HT-22 cells.
Figure 2.
The novel pharmacological inhibitor of Drp1, mdiviA, protects HT-22 cells against glutamate toxicity. (a) Fluorescence photomicrographs ( × 63 objective) of mGFP-transfected and DAPI-stained HT-22 cells show that Drp1 inhibitor mdiviA (75 μM) prevents fission of mitochondria in glutamate-exposed (5 mM, 18 h) HT-22 cells. (b) Quantification of mitochondrial morphology of ∼500 cells per condition: category 1 (elongated tubulin-like structure), category 2 (intermediate length) and category 3 (fragmented mitochondria). **P<0.01 category 1+2 mdiviA, mdiviB+glutamate compared with glutamate 5 mM-treated cells (n=3 independent experiments, ANOVA, Bonferroni-test). (c) HT-22 cells were cultured for 48 h in 96-well E-plates with 4500 cells/well and treated 24 h after seeding with 3 and 5 mM glutamate and mdiviA (75 μM) for 18 h (n=8). Representative graph of real-time detection of cellular impedance by xCELLigence System (Roche) is shown. (d) FACS analysis of HT-22 cells with n=10 000 cells per treatment condition after FITC-Annexin-V labeling to detect apoptotic cells. Exposure to glutamate (5 mM, 14 h) results in enhanced Annexin-V binding of apoptotic HT-22 cells compared with DMSO control. MdiviB (75 μM) significantly reduce glutamate-induced apoptosis. For statistical analysis, the experiments were repeated at least three times with an n=3 per treatment condition. ***P<0.001 compared with glutamate-treated cells (n=3, ANOVA, Scheffé's-test)
Figure 3.
MdiviA inhibits glutamate-induced translocation of Drp1 to the mitochondria, and prevents energy depletion and lipid peroxidation. (a) HT-22 cells were cultured for 24 h in eight-well-ibidi-plates and treated with 3 mM glutamate and mdiviA (75 μM) for 14 h. Immunostaining with specific antibody of Drp1 reveal that Drp1 (green) translocation to the mitochondria (Mitotracker Deep Red) occur after glutamate stimulus. In the magnification pictures by overlapping of the fluorescence (yellow) is shown that Drp1 interacts with the mitochondria. MdiviA inhibits the translocation of Drp1 to the mitochondria. Analysis of fluorescence intensities by Zeiss image software revealed the co-localization of Drp1 and mitochondria by overlapping of green and red fluorescence. (b) Quantification of cells with localization of Drp1 at mitochondria. Drp1 translocation (white bar) correlates with increased mitochondrial fragmentation (grey bar) after glutamate (3 mM) treatment. MdiviA inhibits the translocation of Drp1 to the mitochondria. The quantification is based on three independent experiments with an n=60–70 cells per treatment condition, which were analyzed without knowledge of the treatment history. ***P<0.001 compared with glutamate (3 and 5 mM)-treated cells (n=3, ANOVA, Scheffé's-test). (c) Representative western blot analysis of mitochondrial and cytosolic fractions of HT-22 cells show that glutamate (3 mM) induces an increase of endogenous Drp1 protein in the mitochondrial fraction after 14 h. Treatment with mdiviA inhibit Drp1 translocation to the mitochondria. Quantitative analysis of protein amount was performed by intensity measurements of Drp1 in contrast to mitochondrial loading control Tim23. (d) Representative western blot analysis of mitochondrial and cytosolic fractions of HT-22 cells to detect the translocation of Drp1 to mitochondria after glutamate treatment in a time-dependent manner (3 mM, 8–15 h). Glutamate-treated cells in comparison with the control cells (Tim23 for mitochondria protein loading; actin for cytosolic extracts). These experiments were independently repeated three times with similar results. The representative quantification of total Drp1 in relation to the respective loading controls in the cytosolic and mitochondrial fractions confirmed that Drp1 accumulates in the glutamate-treated cells in comparison with the control cells (Tim23 for mitochondria protein loading; actin for cytosolic extracts). These experiments were independently repeated three times with similar results. (e) HT-22 cells were cultured in white 96-well plates with 7000 cells/well and treated with 3 and 5 mM glutamate and mdiviA (75 μM) at 14 h. ATP-luminescence measurements were performed according to the manufacturer's protocol. The presented data are normalized to DMSO control. For statistical analysis, the experiments were repeated at least three times with an n=8 per treatment condition. ***P<0.001 compared with glutamate (3 and 5 mM)-treated cells (ANOVA, Scheffé's-test). (f) Fluorescence detection of lipid peroxidation by Bodipy assay, quantified by FACS analysis of three independent experiments with n=20 000 cells per treatment condition. ***P<0.001 compared with glutamate (3 and 5 mM)-treated cells (n=3, ANOVA, Scheffé's-test)
It is important to note that the non-functional mdiviA derivate mdiviG17 also failed to inhibit glutamate-induced mitochondrial fragmentation and cell death in the HT-22 cells (Supplementary Figures 5a–c). In contrast, the active mdiviA derivatives mdiviC and mdiviE preserved mitochondrial morphology and cell viability with similar effectivity as mdiviA (Supplementary Figures 5d and e). To further verify the proposed specificity of mdivi inhibitors, we combined treatment with Drp1 siRNA and mdiviA. The combination of Drp1 siRNA and mdiviA did not exceed the protective effects of the respective treatments alone (Supplementary Figure 6a). Notably, the combination of both approaches appeared to decrease the therapeutic effect rather than exerting additional protection, consistent with toxic effects of mdivi compounds at high concentrations and cytotoxic effects of dominant negative mutants of Drp1.18 Next, we determined the protective time window and added the Drp1 inhibitors mdiviA, B and G at different time points between 2 and 12 h after onset of the glutamate treatment. HT-22 cells were protected against glutamate toxicity even when mdiviA or mdiviB were added up to 8 h after the glutamate challenge (Supplementary Figure 6b). MdiviG, the non-functional mdivi compound failed to protect the cells in the post-treatment experiments (Supplementary Figure 6b).
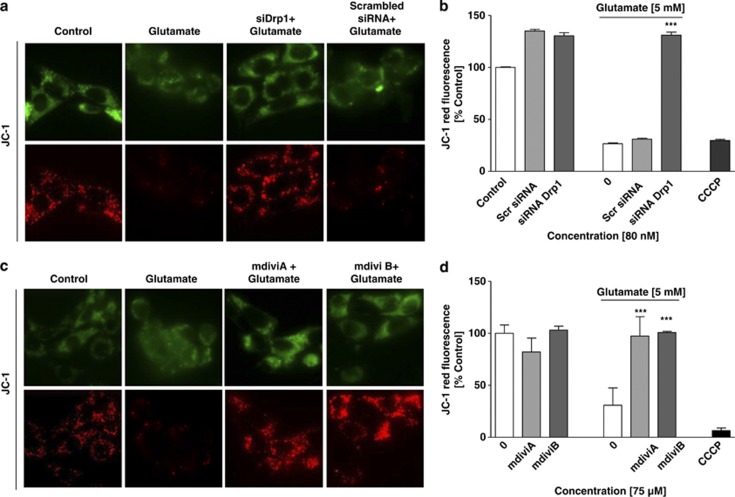
Drp1 gene silencing and pharmacological Drp1 inhibition prevent mitochondrial depolarization and Drp1 translocation to the mitochondria
Loss of mitochondrial membrane potential (MMP) is a major feature of glutamate-induced cell death that is associated with mitochondrial fragmentation and precedes the fatal release of AIF and execution of caspase-independent cell death.5, 19 In line with our previous experiments in HT-22 cells, JC-1 fluorescence indicated that there was a decrease in MMP within 12 h after glutamate treatment, similar to that observed after treatment with the uncoupler carbonylcyanide-3-chlorophenylhydrazone (CCCP) (Figures 4a and c). Three different Drp1 siRNAs significantly attenuated the glutamate-induced breakdown of the MMP, and similar effects were detected after using the Drp1 inhibitors mdiviA and mdiviB (Figures 4b and d; Supplementary Figure 7).
Figure 4.
Inhibition of Drp1 prevents glutamate-induced mitochondrial depolarization. (a and c) MMP was analyzed by JC-1 fluorescence: upper panels show epifluorescence photomicrographs indicating equal cellular uptake of JC-1 by green fluorescence; lower panels depict intact mitochondria exposing red fluorescence. Glutamate-treated (5 mM, 12 h) HT-22 cells show significantly reduce red fluorescence compared with controls whereas mdiviA and B (75 μM) and siRNA of Drp1 (80 nM) prevents the breakdown of the MMP as indicated by preservation of the red JC-1 fluorescence. (b and d) FACS analyses of n=3 independent experiments with 10 000 cells per treatment condition reveal a decrease of the red JC-1 fluorescence to 30% of control levels 12 h after glutamate treatment (5 mM), which has prevented mdiviA, B and siRNA of Drp1 (80 nM). Glutamate treatment was as effective as the positive damage-control CCCP, which causes a fast breakdown of the MMP. The presented data are normalized to DMSO control for mdiviA and mdiviB and vehicle control Lipofectamine 2000 for siRNA approach (100%). ***P<0.001 compared with glutamate-treated controls (n=3, ANOVA, Scheffé's-test)
Inhibition of Drp1 prevents tBid-induced mitochondrial fragmentation and neuronal cell death
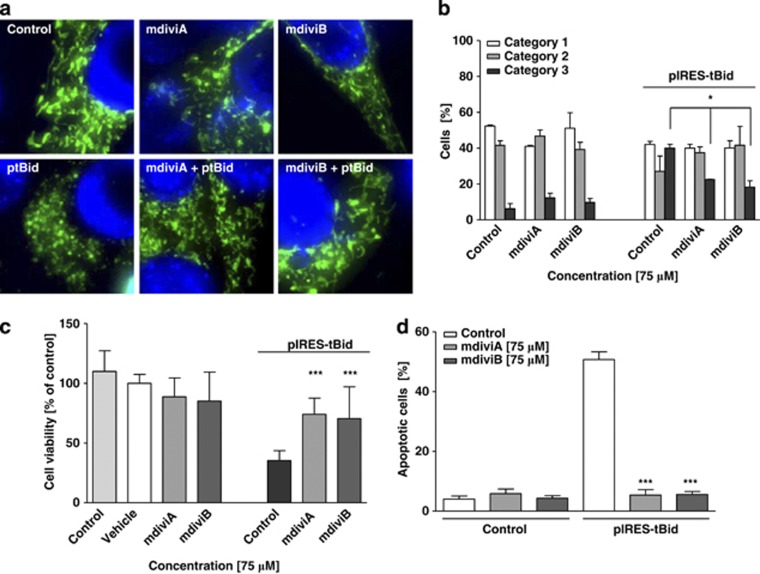
Our recent data demonstrated a role for the proapoptotic BH3-only protein Bid for enhanced mitochondrial fragmentation in the present model systems of HT-22 cell oxytosis and primary neuron cell death induced by glutamate or OGD.5, 19 We documented that Bid is involved in the loss of MMP and release of mitochondrial AIF, which mediates delayed neuronal death after cerebral ischemia.19, 20, 21 Here, we examined the role of Drp1 in Bid-mediated neuronal cell death and observed that treatment of cells with mdiviA or mdiviB also prevented tBid-induced mitochondrial fission and preserved cell viability (Figure 5). Vector-induced expression of tBid reduced cell viability to 30% of control levels, whereas cell viability was restored to control levels when the Drp1 inhibitors were applied to tBid expressing cells (Figure 5c). This protective effect of mdiviA and B against tBid-induced cell death was confirmed in FACS analysis of apoptotic cells after FITC-Annexin-V-staining (Figure 5d).
Figure 5.
Inhibition of Drp1 prevents tBid-induced mitochondrial fragmentation and neuronal cell death. (a) Fluorescence photomicrographs ( × 63 objective) showed that the Drp1 inhibitors mdiviA and mdiviB (75 μM) prevent fission of mitochondria in tBid-induced toxicity after 12 h in HT-22 cells. (b) Quantification of mitochondrial morphology of ∼500 cells per condition: category 1 (elongated tubulin-like structure), category 2 (intermediate length) and category 3 (fragmented mitochondria). *P<0.05 compared with glutamate (5 mM)-treated cells (n=3 independent experiments, ANOVA, Bonferroni-test). (c) HT-22 cells were cultured in 24-well plates with 70 000 cells/well and tBid-induced cell death determined by MTT assay after 12 h. Treatment with mdiviA and mdiviB (75 μM) was performed 1 h before transfection. The presented data are normalized to vehicle control Lipofectamine 2000. ***P<0.001, compared with tBid-treated cells (n=4, ANOVA, Scheffé's-test). For statistical analysis, the experiments were independently repeated at least three times with an n=4 per treatment condition and the results are presented as mean±S.D. (d) FACS analysis of HT-22 cells in n=3 independent experiments with 10 000 cells per treatment condition after FITC-Annexin-V labeling to detect apoptotic cells. Transfection of pIRES-tBid results in enhanced Annexin-V binding of apoptotic HT-22 cells compared with vehicle control Lipofectamine 2000. MdiviA and B (75 μM) significantly reduce tBid-induced cell death. ***P<0.001 compared with tBid toxicity (n=3, ANOVA, Scheffé's-test)
It is known that full-length Bid as well as cleaved Bid translocating to mitochondria in dying cells contributes to mitochondrial demise in HT-22 cells and may explain why overexpression of tBid triggers very similar Drp1-dependent mitochondrial death pathways.22, 23 Additional experiments using Bid siRNA to further substantiated the role of Bid in Drp1-mediated mitochondrial fission (Supplementary Figures 8a–e). As shown in Supplementary Figure 8, silencing of Bid prevents mitochondrial fragmentation and the loss of cell viability. Further, silencing of Bid inhibits the translocation of Drp1 caused by glutamate and clearly demonstrates that Bid plays a significant role in Drp1-dependent mitochondrial fission. Since Bid siRNA inhibit both, full-length Bid and tBid toxicity, similar results are detected in native cells and in cells overexpressing tBid. A direct interaction between Bid/tBid and Drp1, however, could not be detected in immunoprecipitation experiments (Supplementary Figure 9).
Mdivi compounds protect primary neurons from glutamate toxicity and OGD in vitro and attenuate ischemic brain damage in vivo
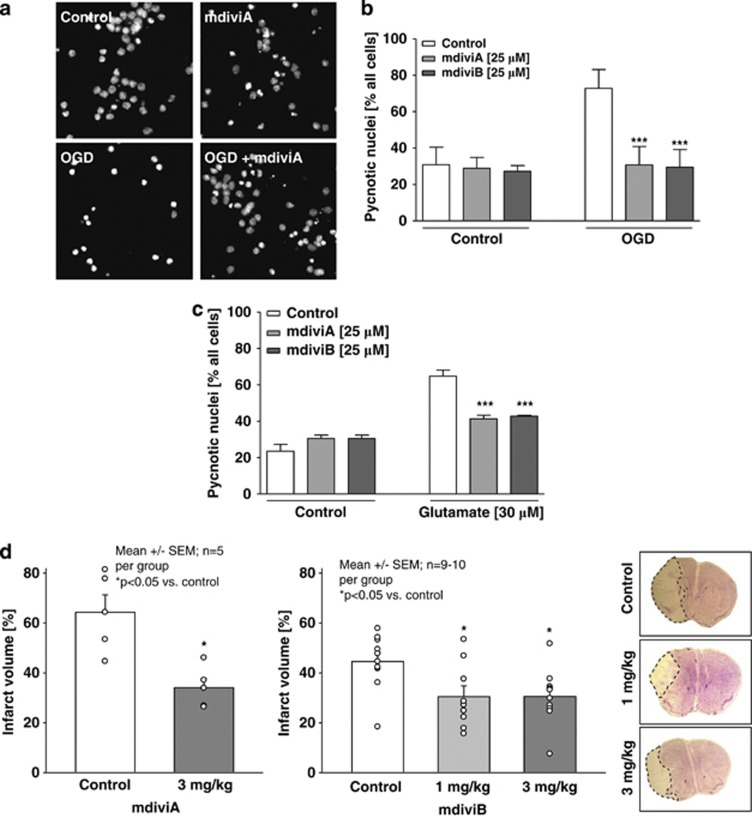
Because HT-22 cells lack i receptors, the HT-22 model of oxytosis only partly reflects the mechanisms of neuronal death after acute brain injury. Thus, we next addressed the effects of the mdivi compounds in a model of glutamate-induced excitotoxicity in primary cortical neurons, which express these ionotropic glutamate receptors. We found that mdivi A or B significantly attenuated OGD-induced damage in the primary neurons (Figures 6a and b), and similar protective effects were detected in primary neurons exposed to glutamate (Figure 6c). Control neurons displayed condensed, pycnotic nuclei 18 h after OGD, whereas the neurons receiving mdivi compounds maintained healthy nuclear morphology (Figure 6a). Quantification of four independent experiments revealed that mdivi compounds A or B reduced OGD-induced cell death by ∼55% (Figure 6b).
Figure 6.
mdivi compounds protect primary neurons against glutamate-induced excitotoxicity and OGD in vitro and attenuate ischemic brain damage in vivo. Primary rat cortical neurons (DIV 9) were exposed 4 h to OGD. After 4 h of OGD, the 1 × EBSS±glucose was replaced by NB+-medium containing glucose to all dishes and the cells were incubated for another 18–24 h in the regular oxygen-containing incubator. (a) Fluorescence microscope images ( × 40 objective) of DAPI-stained embryonic cortical neurons were obtained after 18 h of exposure to OGD. (b) Counting of ∼500 DAPI-stained neurons with a total n=5 per group displaying pycnotic nuclei after 4 h of OGD revealed that 25 μM Drp1 inhibitor mdiviA and B significantly protect neurons from hypoxic-hypoglycemic cell death. ***P<0.001 versus untreated neurons subjected to OGD (n=5, ANOVA, Scheffé's-test). Cultures treated with 25 μM mdiviA or B contained >60% healthy nuclei, whereas all other OGD-treated groups show 75% pycnotic and/or fragmented nuclei, indicating apoptotic damage (OGD, right white column). The respective control cultures with 1 × EBSS medium + glucose contained only very few apoptotic nuclei (Control, left white column). The experiments were repeated three times and the results are presented as mean±S.D. (c) Quantification of pycnotic nuclei in percentage of four independent experiments with a total n=5 per group. Primary rat cortical neurons (DIV 9) were treated for 18 h with glutamate (30 μM) and mdiviA and B (25 μM). After 18 h, cells were fixed in 4% PFA and stained with DAPI and ∼500 nuclei were counted per treatment condition. MdiviA and B significantly reduced glutamate-induced cell death. ***P<0.001 versus glutamate-treated neurons (n=5, ANOVA, Scheffé's-test). (d) Transient focal ischemia was induced for 45 min by a silicon-coated nylon filament that was introduced into the internal carotid artery to occlude the middle cerebral artery (MCA). Surgery was performed in deep isoflurane/N2O anesthesia with controlled ventilation, and ischemia and reperfusion were verified by Laser-Doppler flowmetry. The infarct volume was calculated on the basis of the histomorphometric data from 12 to 15 consecutive sections (10 μM, 500 μM apart) obtained 24 h after onset of ischemia. MdiviA (3 mg/kg) and mdiviB (1 and 3 mg/kg) reduced the mean infarct volume by 34 and 30% compared with vehicle (DMSO) controls, respectively. The photomicrographs show representative pictures of cresyl-violet-stained brain slices from animals, which received vehicle or mdiviB (1 and 3 mg/kg). The infarct area remained unstained and is highlighted by dashed lines
Findings on the efficacy of mdivi compounds in the in vitro models encouraged further investigations in vivo, in a model of ischemic stroke (Figure 6d). Remarkably, mdiviA (3 mg/kg) or mdiviB (1 and 3 mg/kg) reduced the mean infarct volume by 34% and 30% compared with vehicle (dimethylsulfoxide (DMSO)) controls, respectively. These data demonstrate a significant protective efficiency of the mdivi compounds in vivo and suggested that the mechanisms of Drp1-mediated mitochondrial pathways of neuronal cell death also play a major role in ischemic brain damage. Notably, analysis of physiological parameters did not reveal any differences between mdiviA-treated animals and vehicle controls. This observation was also confirmed in a toxicity study over 7 days, where temperature and body weight, blood gases, pH, Na+ and K+-concentrations, and cerebral blood flow did not differ between the animals of the different groups (Supplementary Figure 10).
Discussion
The present study demonstrates that Drp1-mediated mitochondrial fission plays a major role in neuronal cell death associated with acute ischemic brain damage. This conclusion is based on effects of Drp1 siRNA or the small molecule inhibitors, which significantly preserved mitochondrial morphology and MMP, and reduced glutamate toxicity in the neuronal HT-22 cell line. Further, Drp1 inhibitors prevented glutamate excitotoxicity and OGD-induced death in primary cultured neurons, and reduced the infarct size in a model of cerebral ischemia in vivo. Therefore, the current findings extend previous reports on the role of Drp1 in chronic neurodegenerative diseases, since we demonstrate that Drp1 mediates neuronal death in models of acute brain injury. In fact, Drp1-mediated detrimental fission of mitochondria appears occurring independent of the initiating trigger mechanism. The model systems applied here differ regarding the initiation of cell damage, that is, glutamate-induced ROS formation in HT-22 cells, glutamate-induced excitotoxicity in primary neurons, OGD, glutamate excitotoxicity with a burst of ROS formation upon re-oxygenation in the model of OGD in primary neurons and after ischemic brain damage and reperfusion in vivo. All these models were suitable to reveal protective effects of Drp1 inhibition. Thus, regardless of the different upstream stress stimulus, Drp1 translocation and Drp1-mediated mitochondrial fragmentation and downstream mitochondrial death pathways play a major role for the observed cell death.
Our present findings demonstrating protective effects of Drp1 inhibitors in models of OGD in vitro and cerebral ischemia in vivo are in line with recent reports in experimental models of ischemia in the retina, the heart or the kidney.24, 25, 26 In addition, mdiviA was efficacious in rodent models of cisplatin-induced renal damage,24 suggesting a therapeutic potential for Drp1 inhibitors in tissue damage caused by different insults. Earlier studies using dominant negative mutant Drp1K38A validated Drp1 as a potential therapeutic target in neurodegenerative diseases.7 In addition, a very recent study showed that enhanced Drp1 activity caused detrimental mitochondrial fission in Huntington's disease.27 This study also applied mdiviA, and together with our current findings it is suggested that Drp1 inhibition is a promising approach to prevent mitochondrial fragmentation in different models of delayed neuronal cell death relevant for acute and chronic neurological diseases. In fact, these studies show that Drp1 inhibitors are applicable to neurons in vitro and in vivo, thereby overcoming obstacles of siRNA applications or gene therapy.13
The trigger for Drp1 activation in delayed neuronal cell death has not yet been revealed, but there is growing evidence that Bcl-2 family proteins like Bax, Bak and Bid affect mitochondrial morphology and localization in healthy cells.10, 28, 29 For example, Bax can promote Drp1-dependent mitochondrial fragmentation by co-localization of Bax with Drp1 and Mfn-2 at sites of the mitochondria outer membrane (MOM), which then become fission sites. These protein interactions stabilize the association of Drp1 with the mitochondrial membrane and are prerequisite for inducing fragmentation of the organelles.28 In turn, Drp1 stimulates Bax oligomerization by accelerating formation of hemifusion/fission structures of the mitochondrial membrane.11 Such Drp1 facilitated Bax oligomerization is likely an essential step towards enhanced membrane permeability, and the Bax-dependent release of proapoptotic factors from the mitochondria into the cytosol. Further, Bax translocation to the mitochondria correlates with a reduction of mitochondrial fusion since Bax inhibits mitofusins thereby decreasing mitochondrial fusion,11 and this Bax activity may be facilitated by activated Bid or tBid.20
Our recent data demonstrate a pivotal role for the proapoptotic BH3-only protein Bid in the glutamate-induced oxidative stress model system. Further, depolarization of the MOM and mitochondrial fragmentation are both important key events of apoptosis induced by oxidative stress, and both events were apparently Bid dependent (Supplementary Figures 8a–d).5, 19 The pivotal role for Bid upstream of such mitochondrial dysfunction is substantiated by experiments demonstrating pronounced neuroprotective effects of the Bid inhibitor BI-6c9 in models of glutamate-induced excitotoxicity and OGD in primary cultured neurons, and similar studies in HT-22 neurons using Bid siRNA.19, 20, 30, 31 Further, reduced Bid expression attenuated neuronal death in a model of OGD in vitro and reduced brain damage in models of cerebral ischemia and brain trauma in vivo.21, 31
Recent studies showed that tBid and full-length Bid exert similar effects on mitochondria and AIF-dependent cell death in neurons.19, 32 Mitochondrial translocation of full-length Bid was sufficient to induce Bax oligomerization and integration into the outer mitochondrial membrane in Hela cells33 and to induce mitochondrial dysfunction in neurons.32 Activated tBid induces rapid mitochondrial demise in caspase-dependent death receptor mediated pathways, while full-length Bid-mediated caspase-independent cell death pathways, however with slower kinetics compared with tBid.23 In the present model system, activated full-length Bid and tBid may both contribute to the mitochondrial death pathways that are observed in HT-22 cells with a delay of 6–8 h after onset of glutamate exposure.
Our data suggest an essential interplay of proapoptotic Bid with Drp1, the main regulator of mitochondrial fission. We found that tBid-induced loss of MOMP and mitochondrial fragmentation in a Drp1-dependent manner, since Drp1 inhibition and Drp1 siRNA prevented tBid-induced mitochondrial fission and cell death (Figure 5). Further, Bid siRNA prevented mitochondrial translocation of Drp1 to the mitochondria after glutamate exposure, suggesting an interaction between these proteins in mitochondrial damage and associated death pathways (Supplementary Figure 8e). A direct interaction between Bid/tBid and Drp1, however, could not be detected by immunoprecipitation (Supplementary Figure 9) and the collaborative activity of Drp1 and Bid in mitochondrial death pathways may thus be mediated through more complex mechanisms. For example, the interplay between Bid and Drp1 may involve other proapoptotic Bcl-2 proteins such as Bax. It was previously reported, that in addition to full-length Bid and truncated Bid additional proteins are required to trigger Bax oligomerization at the MOM sites.33, 34, 35, 36 Consistent with previous reports,28 we found that Bax co-immunoprecipitated with Drp1 after glutamate toxicity, suggesting that in our model system, Bax and Drp1 cooperate to promote MOMP and mitochondrial fission (Supplementary Figure 9).
This conclusion is confirmed by results showing that Bid induces the oligomerization and insertion of Bax in the MOM,33 and Bax was also found to co-localize with Drp1 at fission sites.28, 37 Further, very recent findings demonstrated Drp1 recruitment to cardiolipin-rich membrane sites as a prerequisite for the formation of membrane hemifusion/fission structures and Bax oligomerization.11, 28 Notably, Drp1-induced Bax oligomerization at mitochondrial fission sites also required the presence of tBid, which further supports the conclusion that both, Drp1 and Bid concomitantly contribute to mitochondrial death pathways.11, 29 Altogether, these findings and our data in neuronal cells suggest that Bid and oligomerized Bax accumulate at Drp1 recruiting sites in the MOM, which then become mitochondrial fission sites.
In damaged neurons, inhibition of Drp1 may serve to shift the observed mitochondrial fission back to a balance of fusion and fission thereby preventing neuronal dysfunction and death. Alternatively, the role of Drp1 in neuronal cell death may be independent of its role in mitochondrial fission. Bax oligomerization and subsequent release of proapoptotic factors to the cytosol were significantly delayed in cells expressing Drp1 mutants lacking membrane recruitment and hemifusion activity.11 This observation is in line with our present findings of Drp1 recruitment to mitochondria before demise of these organelles. Thus, in dying neurons, Bid- and Drp1-dependent mitochondrial fission may be regarded as a morphological hallmark initiating the process of MMP and mitochondrial death pathways.
In summary, the inhibition of Drp1 using the mdivi compounds represents a potential therapeutic strategy for both acute and chronic neurological diseases associated with glutamate toxicity and oxidative stress. This approach may prevent neuronal death pathways upstream of mitochondrial damage thereby providing persistent neuroprotective effects in a wide variety of different neurological diseases where programmed cell death causes dysfunction and death of neurons.
Materials and Methods
Cell culture and induction of neuronal cell death
HT-22 cells derived from immortalized hippocampal neurons were cultured in Dulbecco's modified Eagle medium (DMEM; Invitrogen, Karlsruhe, Germany) supplemented with 10% heat-inactivated fetal calf serum, 100 U/ml penicillin, 100 μg/ml streptomycin and 2 mM glutamine (PAA Laboratories GmbH, Cölbe, Germany). Induction of apoptosis was performed 24 h after seeding of the cells. Growth medium was removed and replaced by medium containing inhibitors and glutamate (3 and 5 mM) for induction of apoptosis. Between 2 and 18 h later, cells were analyzed following standard procedures for flow cytometry, epifluorescence microscopy, protein or RNA analysis. It is important to note that sensitivity to glutamate toxicity varies in HT-22 neurons depending on their density and passage number. Therefore, experiments were always performed at different glutamate concentrations with 3 and 5 mM, and representative results are mostly shown from at least three pooled independent experiments where the respective glutamate concentration induced at least 50% cell death as detected with the MTT assay.
Primary rat embryonic cortical neurons were cultured in neurobasal medium with 2% (v/v) B27 Supplementary, 2 mM glutamine and 100 U/ml penicillin/streptomycin (Invitrogen, San Diego, CA, USA). Cultures contain >95% neurons as routinely controlled by neuronal-specific nuclear protein (NeuN) immunostaining.31, 38 In 6- to 7-day-old primary neurons, the culture medium was replaced immediately before glutamate treatment (25 μM, 24 and 48 h). Since cultures of primary neurons develop functional glutamate receptors after 6–8 days in culture, experimental treatments were performed with 8- to 10-day-old cultures in Earl's balanced salt solution (1 × EBSS) medium (in mg/l: 6800 NaCl, 400 KCl, 264 CaCl2 × 2 × H2O, 200 MgCl2 × 7 × H2O, 2200 NaHCO3, 140 NaH2PO4 × H2O and 1000 glucose, pH 7.2). Apoptosis was induced by glutamate (30 μM) in 1 × EBSS and quantified 18–24 h later. Cells were treated with mdiviA or mdiviB 25 μM in 1 × EBSS. After 18–24 h, the neurons were fixed with 4% paraformaldehyde (PFA) and stained with the fluorescent DNA-binding dye DAPI (4′,6-diamidino-2-phenylindole dihydrochloride) (Sigma-Aldrich, Taufkirchen, Germany). Living and pyknotic cells were counted using a fluorescence microscope (DMI6000B, Leica, Wetzlar, Germany) without knowledge of the treatment history. For the quantification of apoptotic cells, 500 cells from five areas per cell culture dish were counted in total and experiments were performed at least three times with n=5 per treatment condition.
For OGD
Primary cortical neurons were seeded onto 35 mm culture dishes with 5 × 104/cm2 cells. At day 8 or 9 in culture, the dishes were separated into two groups, one for the OGD chamber and one as control group. First, all dishes were washed twice with glucose-free 1 × EBSS supplemented with gentamycin (5 mg/l). For OGD, the cells were treated with mdiviA or mdiviB 25 μM in 1 × EBSS without glucose and transferred to the chamber containing 5% carbon dioxide (CO2) and 95% N2 for 4 h. The analog treatment in 1 × EBSS containing glucose (10 mM) or neurobasal was applied to the control cells in the regular oxygen-containing incubator. After 4 h of OGD, the 1 × EBSS±glucose was replaced by NB+-medium containing glucose to all dishes and the cells were incubated for another 18–24 h in the regular oxygen-containing incubator. For counting analysis of cell death, cells were fixed with 4% PFA and stained with the fluorescent DNA-binding dye DAPI (Sigma-Aldrich). Living and pycnotic cells were counted using a fluorescence microscope (DMI6000B, Leica) without knowledge of the treatment history. For the quantification of apoptotic cells, 500 cells from five areas per cell culture dish were counted in total and experiments were performed at least three times with n=5 per treatment condition.
Plasmids and gene transfer
The mGFP vector, the IRES-tBid vector and control vectors were generated as described previously.5, 19 For plasmid and siRNA transfections 7 × 104 HT-22 cells were seeded in 24-well plates. Standard growth medium was replaced with 900 μl antibiotic-free growth medium. Lipofectamine 2000 (Invitrogen, Germany), IRES-tBid, mGFP vector or empty vector pcDNA 3.1+ were dissolved separately in Optimem I (Invitrogen, Germany). After 10 min of equilibration at room temperature, each DNA solution was combined with the respective volume of the Lipofectamine 2000 solution, mixed gently, and allowed to form plasmid liposomes for further 20 min at room temperature. The transfection mixture was added to the antibiotic-free cell culture medium to a final concentration of 1 μg DNA and 1.5 μl/ml Lipofectamine 2000 in HT-22 cells. Controls were treated with100 μl/ml Optimem only, and vehicle controls with 1.5 μl/ml Lipofectamine 2000.
Inhibition of Drp1
Drp1 inhibition was achieved by using highly specific mdivi compounds A, B, C, E and G in HT-22 cells at concentrations of 50 and 75 μM and at concentrations of 25 μM in primary neurons.17 Notably, higher concentrations of the mdivi compounds exerted toxic effects in the respective cell cultures.
Drp1 silencing
Lipofectamine 2000 (Invitrogen, Germany) and Drp1 siRNA (On-target Plus siRNA Pool (four different sequences), Dharmacon, Thermo Scientific, Bonn, Germany) or non-functional mutant RNA (5′-AAG AGA AAA AGC GAA GAG CCA-3′ Dharmacon, Thermo Scientific) were dissolved separately in Optimem I (Invitrogen, Germany). After 10 min of equilibration at room temperature, each RNA solution was combined with the respective volume of the Lipofectamine 2000 solution, mixed gently and allowed to form siRNA liposomes for 20 min at room temperature. The transfection mixture was added to the antibiotic-free cell culture medium to a final concentration of 60 and 80 nM RNA and 2 μl/ml Lipofectamine in HT-22 cells. Controls were treated with 100 μl/ml Optimem only and vehicle controls with 2 μl/ml Lipofectamine 2000.
Immunostaining and confocal laser scanning fluorescence microscopy
For detection of mitochondrial morphology changes and Drp1 translocation during cell death, HT-22 cells were transfected with the mGFP plasmid with Lipofectamine 2000. Twenty-four hours after transfection, HT-22 cells were seeded in ibidi μ-slide eight-well plates (Ibidi GmbH, Munich, Germany) at a density of 1.7 × 104 cells/well for end point analysis on a fluorescence microscope (DMI6000B, Leica). Alternatively, mitochondria were visualized by MitoTracker Green/DeepRed according to the manufacturer's protocol (Invitrogen, Germany). Microscopic pictures were taken after fixation of the cells with 4% PFA 4–18 h after onset of treatment, followed by Drp1 immunostaining using a specific Drp1 antibody (BD Transduction Laboratories, Heidelberg, Germany) and DAPI counterstaining of the nuclei. Images were acquired using a fluorescent confocal laser scanning microscope (LSM 510, Carl Zeiss, Jena, Germany) equipped with an UV, an argon, and a Helium/Neon laser delivering light at 364, 488 and 543 nm, respectively. Light was collected × 63 1.4 NA, oil immersion objective. DAPI fluorescence was excited at 364 nm and emission was achieved by using the 385-nm long pass filter. Mitotracker Green and mGFP were excited at 488 and 543 nm and emissions were detected using 505–530 nm band pass filter (green), and Mitotracker DeepRed was excited at 620 nm band pass filter and emissions were detected using 690 nm long pass filter (red). Fluorescence of oregon green was excited at a wavelength of 488 nm and emission was detected using a 505-nm long pass filter. For digital imaging, the software LSM Image Browser 4.2.0 (Carl Zeiss) was used.
Evaluation of mitochondrial morphology
HT-22 cells were transfected with mGFP as described before or alternatively, mitochondria were visualized by MitoTracker Green/DeepRed according to the manufacturer's protocol (Invitrogen, Germany). After 24 h, the cells were reseeded on eight-well-ibidi-slides (ibidi) and treated for 18 h. Microscopic pictures were taken after fixation with 4% PFA and DAPI counterstaining of the nuclei 18 h after onset of treatment. Counting of four different types of mitochondrial morphology states was performed for quantification in at least four independent experiments with 500 cells per condition in n=2 wells by an investigator blinded towards the treatment of the cells. Three categories of mitochondrial morphology states were defined as follows: healthy cells display category 1, mitochondria form elongated tubular network. These mitochondria are equally distributed throughout the cytosol. Cells containing large round mitochondria distributed throughout the cytosol are defined as category 2. These cells do not show apoptotic features, for example, shrunken nuclei, and are not detached. In contrast, damaged and dying cells contain smaller round heterogeneous mitochondria located close to the nucleus. These mitochondria are defined as category 3. For statistical analysis, the experiments were repeated at least three times. For automated analyses of mitochondrial morphology changes, mitochondrial length was also calculated by ImageJ software (NIH, Bethesda, MD, USA) from at least six independent pictures per experimental condition.5 Imaging of mitochondrial morphology was performed using a DMI6000B fluorescence microscope, equipped with a DCF360FX-camera (Leica). Mitotracker Green and mGFP were excited at 488 and 543 nm and emissions were detected using 505–530 nm band pass filter (green), and Mitotracker DeepRed was excited at 620 nm band pass filter and emissions were detected using 690 nm long pass filter (red).
RT-PCR
Total RNA was extracted (Nucleospin RNAII kit, Macherey-Nagel, Düren, Germany) and One-step RT-PCR was performed with SuperscriptIII One Step RT-PCR (Invitrogen, Germany). Primers for Drp1 and GAPDH were used as follows: Drp1 forward primer, 5′-ACAGGAGAAGAAAATGGAGTTTGAAGCAG-3′, Drp1 reverse primer, 5′-AACAAATCCTAGCACCACGCAT-3′, GAPDH forward primer 5′-CGTCTTCACCACCATGGAGAAGGC-3′, and GAPDH reverse primer, 5′-AAGGCCATGCCAGTGAGCTTCCC-3′. PCR for GAPDH and Drp1 was performed after cDNA-synthesis at 60°C for 30 min and a denaturation at 95°C for 2 min. PCR amplification was performed by 26 cycles, each with a denaturation for 30 s at 95°C, annealing for 1 min at 57°C and elongation for 2 min at 70°C. The final extension of the PCR products was performed at 70°C for 10 min. RT-PCR products were visualized by a Bio-Rad gel detection system (ChemiDoc XRS Bio-Rad, Karlsruhe, Germany) under UV illumination after electrophoresis on a 1.5% agarose gel containing SybrGold (Invitrogen, Germany).
Evaluation of cell viability and apoptosis
For morphological analysis of cell viability, transmission light microscopy of living HT-22 cells growing as monolayers was performed using an Axiovert 200 microscope (Carl Zeiss) equipped with a Lumenera Infinity 2 digital camera (Lumenera Corporation, Ottawa, Canada). Light was collected through a × 10 0.25 NA objective (Carl Zeiss), and images were captured using phase contrast. Digital image recording and image analysis were performed with the INFINITY ANALYZE software (Lumenera Corporation). Quantification of cell viability in HT-22 cells was performed in 96-well plates with 7000 cells/well by MTT reduction at 0.25 mg/ml for 1 h. The reaction was terminated by adding DMSO after freezing the plate without media at −80°C for at least 1 h and absorbance was then determined at 590 nm versus 630 nm (Fluostar OPTIMA, BMG Labtech, Offenburg, Germany). The data are normalized to DMSO control when mdivi compounds were used in the experiment. In the case of siRNA applications, the presented cell viability data are normalized to the vehicle control Lipofectamine 2000 or Lipofectamine RNAiMax (Invitrogen, Germany). The controls were set to 100% cell viability, since absolute numbers may vary between experiments depending on cell density and MTT signal variations between independent experiments. For statistical analysis, the experiments were repeated at least three times with an n=8 per treatment condition.
For end point analysis of total ATP levels, the ViaLight HS Kit (Lonza, Cologne, Germany) was used following the manufacturer's instructions. HT-22 cells were seeded in white 96-well plates (Greiner, Frickenhausen, Germany) for luminescence measurements. Twenty-four hours after seeding, the cells were damaged with glutamate 3 and 5 mM and treated with Drp1 inhibitor mdiviA (75 μM). ATP levels were detected 0–14 h after the onset of glutamate exposure by detection of luminescence. The ATP release levels were expressed as percentage of luminescence levels in treated control cells (100% ATP release). For statistical analysis, the experiments were repeated at least three times with an n=8 per treatment condition.
Apoptotic cell death was detected by Annexin-V/propidium iodide staining and subsequent flow cytometry analysis. Cells from three-wells per condition were harvested 14 h after glutamate treatment by using Trypsin/EDTA (1 × TE; Sigma-Aldrich), washed once in 1 × PBS and stained according to the manufacturer's protocol (Annexin-V-FITC Detection Kit, PromoKine, Promocell, Heidelberg, Germany). Apoptotic and necrotic cells were determined using FACScan (BD Bioscience, Heidelberg, Germany). Annexin-V-FITC was excited at 488 nm and emission was detected through a 530±40 nm band pass filter. Propidium iodide was excited at 488 nm and fluorescence emission was detected using a 680±30 nm band pass filter. To exclude cell debris and doublets, cells were appropriately gated by forward versus side scatter and pulse width, and 1 × 104 gated events per sample were collected. Surviving cells did not show any staining whereas Annexin-V staining indicated apoptosis and cells positive for both Annexin-V and propidium iodide were regarded necrotic. For statistical analysis, the experiments were repeated at least three times.
DAPI staining
At different time points after the onset of the different treatment conditions, cultured primary neurons were fixed for 15 min in 1 ml of a 1 × PBS solution containing 4% PFA. The fixed primary neurons were stained for 15 min in 35 mm dishes with the fluorescent DNA-binding dye DAPI or Hoechst 33342 (1 μg/ml in 1 × PBS). After removing the fluorescent dyes, cells were washed twice with 1 × PBS and kept in 1 ml 1 × PBS. Stained nuclei were visualized under epifluorescence illumination (DMI6000B, Leica) with A4 (340 nm excitation and 510 nm barrier filter) using a × 40 objective. Neurons with condensed and fragmented nuclei were considered apoptotic. For the quantification of apoptotic cells, 500 cells from five areas per cell culture dish were counted in total and experiments were performed at least three times with n=5 per treatment condition without knowledge of the treatment history. For statistical analysis, the experiments were repeated at least three times.
Detection of cellular lipid peroxidation
For detection of cellular lipid peroxidation, HT-22 cells were seeded out in 24-well plates with 7 × 105 cells/well. After respective treatments, the cells were loaded with 2 μM BODIPY 581/591 C11 for 60 min in 1 ml DMEM per well. Cells were then harvested with standard 1 × TE in 1 × PBS with 100 μl/well. Trypsin incubation was stopped by adding 1 ml serum containing medium to each well. Cells were centrifuged at 1.000 × g for 5 min at room temperature, washed with 1 × PBS and resuspended in 1 ml 1 × PBS. Detection of lipid peroxidation was performed by flow cytometry on a FACScan (BD Bioscience) by using 488 nm UV line argon laser for excitation and lipid peroxidation emission was recorded on channels FL1 at 530 nm (green) and FL2 at 585 nm (red). Data were collected from at least 20 000 cells. To exclude cell debris and doublets, cells were appropriately gated by forward versus side scatter and pulse width, and 2 × 104 gated events per sample were collected from three to four independent samples per treatment condition.
Analysis of MMP
MMP of HT-22 neurons was determined by 5, 5′, 6, 6′-tetrachloro-1, 1′, 3, 3′-tetraethylbenzimidazolylcarbocyanine iodide (JC-1) reduction. HT-22 neurons were stained with JC-1 (Mitoprobe, Invitrogen, Germany) according to the manufacturer's protocol and analyzed by subsequent flow cytometry or epifluorescence microscopy. After glutamate treatment (12 h), JC-1 was added to each well of the different treatment conditions to a final concentration of 2 μM for 30 min. Living-control cells received vehicle (DMSO, 0.1% final concentration) and damage-control cells were treated with carbonyl cyanide m-chlorophenylhydrazone (CCCP) 15 min before staining to induce mitochondrial membrane depolarization. Detached cells in the medium were collected and combined with the attached cells harvested with standard 1 × TE in 1 × PBS. Trypsin incubation was stopped by adding 1 ml serum containing medium to each well. Cells were centrifuged at 1000 × g for 5 min at room temperature, washed once with 1 × PBS, and kept in 0.5 ml 1 × PBS on ice until analysis of JC-1 fluorescence using a FACScan (BD Bioscience). JC-1 green fluorescence indicating mitochondrial uptake of the dye was excited at 488 nm and emission was detected using a 530±40 nm band pass filter. JC-1 red fluorescence indicating intact MMP was excited at 488 nm and emission was detected using a 613±20 nm band pass filter. To exclude cell debris and doublets, cells were appropriately gated by forward versus side scatter and pulse width, and 1 × 104 gated events per sample were collected from three to four independent samples per treatment condition and the experiments were repeated at least three times.
Real-time measurements with xCelligence
The xCelligence System (Roche, Penzberg, Germany) monitors cellular events in real time by measuring the electrical impedance between micro-electrodes integrated into the bottom of custom made tissue culture plate (E-Plates). Since cells have a very high electrical resistance, the more cells are attached to the bottom of the E-Plate, the higher the electrical impedance will be. Thus, the electrical impedance, which is displayed as cell index (CI), can be used to monitor cell viability, number, morphology and adhesion in a large number of tissue culture wells simultaneously, at any given frequency, and over any desired period of time without removing the plate from the incubator. HT-22 cells were seeded in a density of 4000–5000 cells/well in 96-well E-plates (Roche). Twenty-four hours after seeding, cells were incubated with vehicle, mdivi compound A (75 μM), glutamate (3 and 5 mM) or glutamate + mdivi compound A (75 μM).
Cerebral ischemia
Male C57BL/6 mice (body weight, 18–22 g; Charles River Laboratories, Wilmington, MA, USA) were subjected to transient MCAO as described previously.21, 39 The Drp1 inhibitors mdiviA and B were injected i.p. at doses of 1 and 3 mg/kg body weight before the induction of MCAo. Surgery was performed in isoflurane/N2O anesthesia (1.5% isoflurane, 68.5% N2O, 30% O2). A silicone-coated nylon monofilament was inserted into the internal carotid artery and gently pushed forward until blood flow in the MCA territory decreased to <20% of baseline as determined by Laser-Doppler fluxmetry. Twenty-four hours after MCAo, animals were perfused and fixed in 4% PFA for 24 h. Thereafter, the brains were removed, post-fixed in 4% PFA overnight, dehydrated and embedded in paraffin. In all, 12–15 sections (10 μM, 500 μM apart), including the middle cerebral artery territory, were stained with cresyl-violet for quantification of infarct area or evaluation of ischemic cell death, respectively. For each experimental group, five animals were used in the experiments with mdiviA, and 9–10 animals per group were used in the experiments with mdiviB. Statistical analysis was performed using the Mann–Whitney U-test. Physiological parameters and animal temperature were obtained as described previously.40 The left external carotid artery was cannulated for blood sampling, monitoring of blood gases, pH, electrolyte status and mean arterial blood pressure (MABP) during isoflurane/N2O anesthesia. Laser-Doppler flowmetry (LDF; MBF3D, Moor Instruments) was carried out to monitor local cortical blood flow (LCBF) of cortical areas supplied by the MCA. For the placement of the LDF probe, a burr hole (1 mm diameter) was drilled 5 mm lateral and 1 mm posterior to the bregma without injury of the dura mater. LCBF was continuously measured (2 Hz sampling rate) starting before the onset of ischemia and lasting as long as 1 h after reperfusion.
Statistical analysis
All data are given as mean values±standard deviation (S.D.). For statistical comparison between two groups, Mann–Whitney U-test was used. Multiple comparisons were performed by analysis of variance (ANOVA) followed by Scheffé's post hoc test or Bonferroni-test, as indicated. Calculations were performed with the Winstat standard statistical software package (R Fitch Software, Bad Krozingen, Germany).
Acknowledgments
We thank Emma Esser for careful editing of the manuscript. Parts of this project were supported by a grant of the Michael J Fox Foundation to Carsten Culmsee.
Glossary
- AIF
apoptosis-inducing factor
- AD
Alzheimer disease
- ATP
adenosinetriphosphate
- Bcl-2
B-cell lymphom-2
- Bak
Bcl-2 antagonist/killer
- Bax
Bcl-2-associated protein X
- BCA
bicinchoninic acid
- Bcl-xl
Bcl-2-related form log isoform
- Bid
Bcl-2 interacting domain death antagonist
- Bodipy
4,4-diflouro-5-(4-phenyl1,3-butadienyl)-4-bora-3a,4a-diaza-s-indacene-3-undecanoic acid
- CCCP
carbonylcyanide-3-chlorophenylhydrazone
- CI
cell index
- DAPI
4′,6-diamidino-2-phenylindole dihydrochloride
- DMSO
dimethylsulfoxide
- DMEM
Dulbecco's modified Eagle medium
- DNA
deoxyribonucleic acid
- Drp1
dynamin-related protein 1
- EBSS
Earl's balanced salt solution
- FACS
fluorescence activated cell sorting
- FCS
fetal calf seum
- GAPDH
glyceraldehyde-3-phosphate-dehydrogenase
- GSH
gluthatione
- h
hour
- H2O2
hydrogen peroxide
- HBSS
Hank's balanced medium
- HCl
hydrochloric acid
- HEPES
4-(2-hydroxyethyl)piperazine-1-ethanesulfonic acid
- HRP
horseradish peroxidase
- JC-1
5, 5′, 6, 6′-tetrachloro-1, 1′, 3,3′-tetraethylbenzimid-azolylcarbocyanine iodide
- LDF
Laser-Doppler flowmetry
- LCBF
local cortical blood flow
- MABP
mean arterial blood pressure
- MCAO
middle carotid artery occlusion
- mdivi
mitochondrial division inhibitor
- mGFP
mitochondrial targeting green fluorescent protein
- MEM+
Eagle's minimum essential medium
- Mfn-1, -2
mitofusin-1, -2
- MMP
mitochondrial membrane potential
- MTT
3-(4,5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide
- MMP
mitochondrial membrane permeability
- MOM
mitochondrial outer membrane
- MOMP
mitochondrial outer membrane permeabilization
- NMDA
N-methyl-D-aspartic acid
- NCI
normalized cell index
- OGD
oxygen–glucose deprivation
- OPA1
optic atrophy 1
- PBS
phosphate buffered saline
- PCD
programmed cell death
- PD
Parkinson's disease
- PEI
polyethylenimine
- PFA
paraformaldeyde
- pH
potentia hydrogenii
- PI
propidium iodide
- PLL
poly-L-ornithine
- RT
room temperature
- ROS
reactive oxygen species
- SDS
sodium dodecyl sulfate
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- S.D.
standard deviation
- S.E.M.
standard deviation of the mean
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by N Chandel
Supplementary Material
References
- Chan DC. Mitochondria: dynamic organelles in disease, aging, and development. Cell. 2006;125:1241–1252. doi: 10.1016/j.cell.2006.06.010. [DOI] [PubMed] [Google Scholar]
- Benard G, Bellance N, James D, Parrone P, Fernandez H, Letellier T, et al. Mitochondrial bioenergetics and structural network organization. J Cell Sci. 2007;120:838–848. doi: 10.1242/jcs.03381. [DOI] [PubMed] [Google Scholar]
- Chen H, McCaffery JM, Chan DC. Mitochondrial fusion protects against neurodegeneration in the cerebellum. Cell. 2007;130:548–562. doi: 10.1016/j.cell.2007.06.026. [DOI] [PubMed] [Google Scholar]
- Jezek P, Plecita-Hlavata L. Mitochondrial reticulum network dynamics in relation to oxidative stress, redox regulation, and hypoxia. Int J Biochem Cell Biol. 2009;41:1790–1804. doi: 10.1016/j.biocel.2009.02.014. [DOI] [PubMed] [Google Scholar]
- Grohm J, Plesnila N, Culmsee C. Bid mediates fission, membrane permeabilization and peri-nuclear accumulation of mitochondria as a prerequisite for oxidative neuronal cell death. Brain Behav Immun. 2009;24:831–838. doi: 10.1016/j.bbi.2009.11.015. [DOI] [PubMed] [Google Scholar]
- Westermann B. Mitochondrial fusion and fission in cell life and death. Nat Rev Mol Cell Biol. 2010;11:872–884. doi: 10.1038/nrm3013. [DOI] [PubMed] [Google Scholar]
- Reddy PH, Reddy TP, Manczak M, Calkins MJ, Shirendeb U, Mao P. Dynamin-related protein 1 and mitochondrial fragmentation in neurodegenerative diseases. Brain Res Rev. 2010;67:103–118. doi: 10.1016/j.brainresrev.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knott AB, Perkins G, Schwarzenbacher R, Bossy-Wetzel E. Mitochondrial fragmentation in neurodegeneration. Nat Rev Neurosci. 2008;9:505–518. doi: 10.1038/nrn2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackner LL, Nunnari JM. The molecular mechanism and cellular functions of mitochondrial division. Biochim Biophys Acta. 2008;1792:1138–1144. doi: 10.1016/j.bbadis.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnoult D, Rismanchi N, Grodet A, Roberts RG, Seeburg DP, Estaquier J, et al. Bax/Bak-dependent release of DDP/TIMM8a promotes Drp1-mediated mitochondrial fission and mitoptosis during programmed cell death. Curr Biol. 2005;15:2112–2118. doi: 10.1016/j.cub.2005.10.041. [DOI] [PubMed] [Google Scholar]
- Montessuit S, Somasekharan SP, Terrones O, Lucken-Ardjomande S, Herzig S, Schwarzenbacher R, et al. Membrane remodeling induced by the dynamin-related protein Drp1 stimulates Bax oligomerization. Cell. 2010;142:889–901. doi: 10.1016/j.cell.2010.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank S, Gaume B, Bergmann-Leitner ES, Leitner WW, Robert EG, Catez F, et al. The role of dynamin-related protein 1, a mediator of mitochondrial fission, in apoptosis. Dev Cell. 2001;1:515–525. doi: 10.1016/s1534-5807(01)00055-7. [DOI] [PubMed] [Google Scholar]
- Lackner LL, Nunnari J. Small molecule inhibitors of mitochondrial division: tools that translate basic biological research into medicine. Chem Biol. 2010;17:578–583. doi: 10.1016/j.chembiol.2010.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagara Y, Dargusch R, Chambers D, Davis J, Schubert D, Maher P. Cellular mechanisms of resistance to chronic oxidative stress. Free Radic Biol Med. 1998;24:1375–1389. doi: 10.1016/s0891-5849(97)00457-7. [DOI] [PubMed] [Google Scholar]
- Burdo J, Dargusch R, Schubert D. Distribution of the cystine/glutamate antiporter system xc- in the brain, kidney, and duodenum. J Histochem Cytochem. 2006;54:549–557. doi: 10.1369/jhc.5A6840.2006. [DOI] [PubMed] [Google Scholar]
- Tobaben S, Grohm J, Seiler A, Conrad M, Plesnila N, Culmsee C. Bid-mediated mitochondrial damage is a key mechanism in glutamate-induced oxidative stress and AIF-dependent cell death in immortalized HT-22 hippocampal neurons. Cell Death Differ. 2011;18:282–292. doi: 10.1038/cdd.2010.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassidy-Stone A, Chipuk JE, Ingerman E, Song C, Yoo C, Kuwana T, et al. Chemical inhibition of the mitochondrial division dynamin reveals its role in Bax/Bak-dependent mitochondrial outer membrane permeabilization. Dev Cell. 2008;14:193–204. doi: 10.1016/j.devcel.2007.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uo T, Dworzak J, Kinoshita C, Inman DM, Kinoshita Y, Horner PJ, et al. Drp1 levels constitutively regulate mitochondrial dynamics and cell survival in cortical neurons. Exp Neurol. 2009;218:274–285. doi: 10.1016/j.expneurol.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landshamer S, Hoehn M, Barth N, Duvezin-Caubet S, Schwake G, Tobaben S, et al. Bid-induced release of AIF from mitochondria causes immediate neuronal cell death. Cell Death Differ. 2008;15:1553–1563. doi: 10.1038/cdd.2008.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culmsee C, Plesnila N. Targeting Bid to prevent programmed cell death in neurons. Biochem Soc Trans. 2006;34:1334–1340. doi: 10.1042/BST0341334. [DOI] [PubMed] [Google Scholar]
- Plesnila N, Zinkel S, Le DA, min-Hanjani S, Wu Y, Qiu J, et al. BID mediates neuronal cell death after oxygen/glucose deprivation and focal cerebral ischemia. Proc Natl Acad Sci USA. 2001;98:15318–15323. doi: 10.1073/pnas.261323298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song G, Chen GG, Hu T, Lai PB. Bid stands at the crossroad of stress-response pathways. Curr Cancer Drug Targets. 2010;10:584–592. doi: 10.2174/156800910791859515. [DOI] [PubMed] [Google Scholar]
- Ward MW, Rehm M, Duessmann H, Kacmar S, Concannon CG, Prehn JH. Real time single cell analysis of Bid cleavage and Bid translocation during caspase-dependent and neuronal caspase-independent apoptosis. J Biol Chem. 2006;281:5837–5844. doi: 10.1074/jbc.M511562200. [DOI] [PubMed] [Google Scholar]
- Brooks C, Wei Q, Cho SG, Dong Z. Regulation of mitochondrial dynamics in acute kidney injury in cell culture and rodent models. J Clin Invest. 2009;119:1275–1285. doi: 10.1172/JCI37829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong SB, Subrayan S, Lim SY, Yellon DM, Davidson SM, Hausenloy DJ. Inhibiting mitochondrial fission protects the heart against ischemia/reperfusion injury. Circulation. 2010;121:2012–2022. doi: 10.1161/CIRCULATIONAHA.109.906610. [DOI] [PubMed] [Google Scholar]
- Park SW, Kim KY, Lindsey JD, Dai Y, Heo H, Nguyen DH, et al. A selective inhibitor of drp1, mdivi-1, increases retinal ganglion cell survival in acute ischemic mouse retina. Invest Ophthalmol Vis Sci. 2011;52:2837–2843. doi: 10.1167/iovs.09-5010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W, Chen J, Petrilli A, Liot G, Klinglmayr E, Zhou Y, et al. Mutant huntingtin binds the mitochondrial fission GTPase dynamin-related protein-1 and increases its enzymatic activity. Nat Med. 2011;17:377–382. doi: 10.1038/nm.2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karbowski M, Norris KL, Cleland MM, Jeong SY, Youle RJ. Role of Bax and Bak in mitochondrial morphogenesis. Nature. 2006;443:658–662. doi: 10.1038/nature05111. [DOI] [PubMed] [Google Scholar]
- Landes T, Martinou JC. Mitochondrial outer membrane permeabilization during apoptosis: the role of mitochondrial fission. Biochim Biophys Acta. 2011;1813:540–545. doi: 10.1016/j.bbamcr.2011.01.021. [DOI] [PubMed] [Google Scholar]
- Becattini B, Culmsee C, Leone M, Zhai D, Zhang X, Crowell KJ, et al. Structure-activity relationships by interligand NOE-based design and synthesis of antiapoptotic compounds targeting Bid. Proc Natl Acad Sci USA. 2006;103:12602–12606. doi: 10.1073/pnas.0603460103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culmsee C, Zhu C, Landshamer S, Becattini B, Wagner E, Pellecchia M, et al. Apoptosis-inducing factor triggered by poly(ADP-ribose) polymerase and Bid mediates neuronal cell death after oxygen--glucose deprivation and focal cerebral ischemia. J Neurosci. 2005;25:10262–10272. doi: 10.1523/JNEUROSCI.2818-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konig HG, Rehm M, Gudorf D, Krajewski S, Gross A, Ward MW, et al. Full length Bid is sufficient to induce apoptosis of cultured rat hippocampal neurons. BMC Cell Biol. 2007;8:7. doi: 10.1186/1471-2121-8-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskes R, Desagher S, Antonsson B, Martinou JC. Bid induces the oligomerization and insertion of Bax into the outer mitochondrial membrane. Mol Cell Biol. 2000;20:929–935. doi: 10.1128/mcb.20.3.929-935.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovell JF, Billen LP, Bindner S, Shamas-Din A, Fradin C, Leber B, et al. Membrane binding by tBid initiates an ordered series of events culminating in membrane permeabilization by Bax. Cell. 2008;135:1074–1084. doi: 10.1016/j.cell.2008.11.010. [DOI] [PubMed] [Google Scholar]
- Lucken-Ardjomande S, Montessuit S, Martinou JC. Contributions to Bax insertion and oligomerization of lipids of the mitochondrial outer membrane. Cell Death Differ. 2008;15:929–937. doi: 10.1038/cdd.2008.9. [DOI] [PubMed] [Google Scholar]
- Terrones O, Etxebarria A, Landajuela A, Landeta O, Antonsson B, Basanez G. BIM and tBID are not mechanistically equivalent when assisting BAX to permeabilize bilayer membranes. J Biol Chem. 2008;283:7790–7803. doi: 10.1074/jbc.M708814200. [DOI] [PubMed] [Google Scholar]
- Wasiak S, Zunino R, McBride HM. Bax/Bak promote sumoylation of DRP1 and its stable association with mitochondria during apoptotic cell death. J Cell Biol. 2007;177:439–450. doi: 10.1083/jcb.200610042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culmsee C, Gerling N, Lehmann M, Nikolova-Karakashian M, Prehn JH, Mattson MP, et al. Nerve growth factor survival signaling in cultured hippocampal neurons is mediated through TrkA and requires the common neurotrophin receptor P75. Neuroscience. 2002;115:1089–1108. doi: 10.1016/s0306-4522(02)00539-0. [DOI] [PubMed] [Google Scholar]
- Plesnila N, Zhu C, Culmsee C, Groger M, Moskowitz MA, Blomgren K. Nuclear translocation of apoptosis-inducing factor after focal cerebral ischemia. J Cereb Blood Flow Metab. 2004;24:458–466. doi: 10.1097/00004647-200404000-00011. [DOI] [PubMed] [Google Scholar]
- Culmsee C, Junker V, Kremers W, Thal S, Plesnila N, Krieglstein J. Combination therapy in ischemic stroke: synergistic neuroprotective effects of memantine and clenbuterol. Stroke. 2004;35:1197–1202. doi: 10.1161/01.STR.0000125855.17686.6d. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.