Abstract
Adipose tissue structure is altered during obesity, leading to deregulation of whole-body metabolism. Its function depends on its structure, in particular adipocytes number and differentiation stage. To better understand the mechanisms regulating adipogenesis, we have investigated the role of an endoribonuclease, endoribonuclease L (RNase L), using wild-type and RNase L-knockout mouse embryonic fibroblasts (RNase L−/−-MEFs). Here, we identify C/EBP homologous protein 10 (CHOP10), a dominant negative member of the CCAAT/enhancer-binding protein family, as a specific RNase L target. We show that RNase L is associated with CHOP10 mRNA and regulates its stability. CHOP10 expression is conserved in RNase L−/−-MEFs, maintaining preadipocyte state while impairing their terminal differentiation. RNase L−/−-MEFs have decreased lipids storage capacity, insulin sensitivity and glucose uptake. Expression of ectopic RNase L in RNase L−/−-MEFs triggers CHOP10 mRNA instability, allowing increased lipids storage, insulin response and glucose uptake. Similarly, downregulation of CHOP10 mRNA with CHOP10 siRNA in RNase L−/−-MEFs improves their differentiation in adipocyte. In vivo, aged RNase L−/− mice present an expanded adipose tissue, which, however, is unable to correctly store lipids, illustrated by ectopic lipids storage in the liver and in the kidney. These findings highlight RNase L as an essential regulator of adipogenesis via the regulation of CHOP10 mRNA.
Keywords: adipogenesis, CHOP10, RNase L, insulin response
In the past 50 years, the incidence of obesity and related metabolic disorders has increased markedly, reaching a worldwide epidemic scale. Obesity is associated with several health problems, including insulin resistance, type 2 diabetes, fatty liver disease, atherosclerosis, degenerative disorders, airway diseases and some cancers.
Obese individuals suffer from a higher adiposity level resulting from an increased adipocytes number (hyperplasia) and/or size (hypertrophy).1 Adipose tissue is involved in metabolic regulation and particularly lipids storage, insulin sensitivity and glucose uptake.2 It has therefore a central role in controlling whole-body glucose and lipids homeostasis. Adipose tissue functions depend on its structure defined by adipocytes number and differentiation stage; this structure is altered during obesity.3 Understanding the mechanisms implicated in the regulation of adipocyte differentiation should help elucidate adipose tissue development and its link to metabolic diseases.
After activation by various factors, such as hormones, adipogenesis occurs in successive phases leading from preadipocytes to mature adipocytes. Particularly, the expansion phase of preadipocytes precedes their differentiation to adipocytes.4, 5 Like other differentiation pathways, adipogenesis involves sequential and orderly expression of many factors triggering a whole activation/inactivation cascade of genes expression. Among the several transcription factors known to have a significant role in promoting adipogenesis, two of them have a central role by regulating several downstream target genes: the CCAAT/enhancer-binding protein α (C/EBPα) and the nuclear receptor peroxisome proliferator-activated receptor γ2 (PPARγ2). C/EBPα and PPARγ2 expression and activity are required for the adipose tissue development, insulin sensitivity and maintenance of mature adipocyte functions, such as triglycerides synthesis and storage. Their expression is closely regulated at different levels; in particular, their transcription is regulated by other members of the C/EBP family (β,δ),6 which expression precedes the induction of C/EBPα and PPARγ2. Terminal adipogenesis is also regulated by another C/EBP family member named C/EBP homologous protein 10 (CHOP10). By forming heterodimers with other C/EBPs, CHOP10 functions as a dominant negative member of the C/EBP family. In preadipocytes, it forms a heterodimer with C/EBPβ, inhibiting its DNA-binding activity. This inhibition of C/EBPβ is necessary for the expansion phase of preadipocytes before terminal differentiation.7 Thus, downregulation of CHOP10 allows C/EBPβ release and the transactivation of C/EBPα and PPARγ, which expression is essential for terminal adipocyte differentiation and the expression of most genes characterizing fat cells.7 Moreover, CHOP10 has been implicated in the production of reactive oxygen species (ROS),8 important regulators of adipocyte differentiation and insulin response.9
As now demonstrated by several studies, an essential mechanism of gene expression control is through mRNA stability regulation.10, 11 One nuclease firmly implicated in cellular mRNA stability is the endoribonuclease L (RNase L).12 RNase L is an ubiquitous endoribonuclease, which activity is strictly dependent on an oligoadenylate, the 2-5A, synthesized by the 2-5A synthetase(s).12, 13 RNase L regulates mRNAs stability during their translation and its activity is negatively regulated by RLI (RNase L inhibitor).14, 15, 16 We have previously shown that RNase L and its inhibitor RLI are induced during differentiation and control gene expression and cell fate during myogenesis.11, 17 Transcriptome analysis of myoblasts overexpressing RNase L allowed us to identify some mRNA targets of RNase L, among which CHOP10 mRNA.11 As CHOP10 is a conductor of adipogenesis,7 we were interested in studying the role of RNase L in adipogenesis and insulin sensitivity. Hence, we used wild-type (WT) and RNase L-knockout mouse embryonic fibroblasts (MEFs),18 adipogenesis of MEFs induced by hormonal treatment being a well-established model system for the study of adipocyte differentiation.19 Here, we show that RNase L directly regulates CHOP10 mRNA. Indeed, we demonstrate that RNase L is associated with CHOP10 mRNA and controls CHOP10 expression by regulating its mRNA stability. As CHOP10 expression is maintained after induction of adipocyte differentiation in RNase L−/−-MEFs, this leads to the impairment of terminal adipocyte differentiation, the decrease in insulin sensitivity and glucose uptake, and to increased ROS production. Conversely, expression of ectopic human RNase L (Hu-RNase L) or transfection of CHOP10-siRNA (siCHOP) in RNase L−/−-deficient cells triggers CHOP10 mRNA instability, decreases ROS level and allows an increase in lipids storage, insulin response and glucose uptake. Consolidating these results, RNase L−/− mice present an expansion of their adipose tissue (hyperplasia) and a defect in lipids storage. Our results highlight RNase L as an essential regulator of the adipogenesis processes and insulin response by regulating CHOP10 mRNA stability.
Results
Lack of RNase L impairs adipogenesis
MEFs are interesting cell models to study the specific role of a factor during adipogenesis, as they can differentiate into mature adipocytes after hormonal stimulation.19 To explore RNase L function during adipogenesis at a molecular level, we used WT-MEFs and RNase L-knockout MEFs (RNase L−/−-MEFs).18
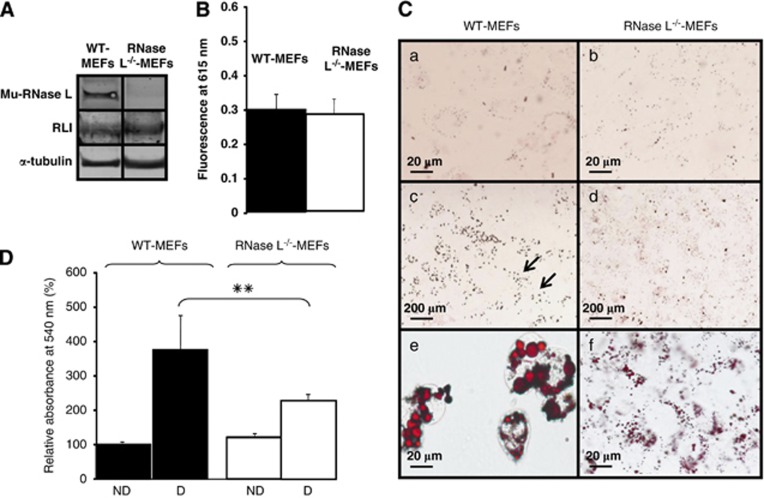
RNase L−/−-MEFs did not express mouse RNase L protein, as the first exon of RNase L gene was deleted in the RNase L−/− mouse.18 However, these cells expressed an equivalent level of RLI as WT-MEFs (Figure 1A). WT-MEFs and RNase L−/−-MEFs present the same viability under our experimental conditions, as shown in Figure 1B.
Figure 1.
RNase L and RLI expression, viability and lipids accumulation in WT-MEFs and RNase L−/−-MEFs. (A) WT-MEFs and RNase L−/−-MEFs proteins were analyzed by western blot with antibodies against mouse RNase L and mouse RLI. α-tubulin antibody was used as an indicator of proteins quantity loading. (B) Cells viability was measured in WT-MEFs and RNase L−/−-MEFs by measuring fluorescence emission at 615 nm of PrestoBlue reagent reduced by living cells. Error bars refer to the S.D. obtained in four independent experiments. (C) WT-MEFs and RNase L−/−-MEFs were induced to differentiate in ADM. Cells were fixed and stained with oil red O to reveal lipids accumulation at day 0 (a and b) and after 6 days of differentiation (c–f). Cells were observed at a magnification of × 40 (a, b, e and f) or × 5 (c and d). Arrows in (c) indicate groups of differentiated cells observed at upper magnification in e. (D) After staining with oil red O, lipids were quantified in WT-MEFs and RNase L−/−-MEFs at day 0 (ND) and day 6 after induction of adipocyte differentiation (D) by spectrophotometric analysis at 540 nm, following elution of cell retained oil red O with ethanol. The amount of lipids in WT-MEFs at day 0, was set to 100%. Error bars refer to the S.D. obtained in four independent experiments conducted in duplicate. **P=0.009 RNase L−/−-MEFs compared with WT-MEFs at day 6
First, we compared lipids accumulation in WT-MEFs and RNase L−/−-MEFs before and after adipocyte differentiation. If WT-MEFs and RNase L−/−-MEFs both increased their lipids content after differentiation (Figure 1C, compare a and e to b and f, respectively), their lipids staining with oil red O showed a very different pattern. Indeed, when observed at a lower magnification, lipids staining in WT-MEFs was not uniform, small groups of highly labeled cells being separated by unlabeled cells (Figure 1C, c). On the other hand, lipids staining in RNase L−/−-MEFs revealed a low but uniform labeling of cells, indicating that more cells accumulated lipids (Figure 1C, d). However, observation of lipids staining at a greater magnification showed that RNase L−/−-MEFs accumulated smaller lipid droplets than WT-MEFs (Figure 1C, compare e and f). This difference in lipids accumulation was confirmed by quantification: RNase L−/−-MEFs accumulated 1.7 time lesser lipids than WT-MEFs (Figure 1D). This impairment in lipids storage, a major adipocyte function, was confirmed by studying expression of two proteins specifically expressed in mature adipocytes: adipocyte protein 2 (aP2)/fatty acid-binding protein 4 (Fabp4), involved in intracellular lipids trafficking,20 and perilipin, a lipid droplet-associated protein21 (Figure 2A). As illustrated in Figure 2A, aP2/Fabp4 is less expressed in RNase L−/−-MEFs than in WT-MEFs (compare Figure 2A, a and b). Surprisingly, even if we observed an increase in lipids accumulation after differentiation of RNase L−/−-MEFs (Figures 1C and D), we were unable to detect perilipin surrounding lipid droplets in these cells (compare Figure 2A, c and d). The same result was obtained by western blot, using an anti-perilipin antibody (data not shown). These results indicated impairment in lipids accumulation and storage in RNase L−/−-MEFs.
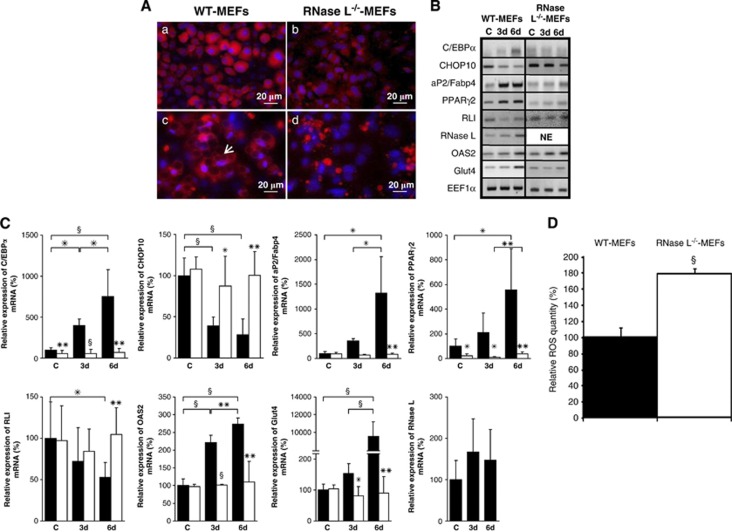
Figure 2.
RNase L deficiency impairs adipocyte functions. (A) WT-MEFs and RNase L−/−-MEFs were induced to differentiate in ADM. Cells were fixed at day 6 after induction of adipocyte differentiation and then incubated with an antibody against aP2/Fabp4 (red; WT-MEFs, (a) and RNase L−/−-MEFs, (b) and perilipin (red; WT-MEFs, (c) and RNase L−/−-MEFs, (d)). DNA was stained with DAPI (blue). Cells were observed at a magnification of × 40. Arrow in (c) indicates perilipin surrounding lipid droplets. (B) Confluent WT-MEFs and RNase L−/−-MEFs were induced to differentiate in ADM. mRNAs were extracted at confluence (C) and at days 3 (3d) and 6 (6d) after switch to ADM. Following RT and PCR amplification using specific primers (Supplementary Table 1), PCR products were analyzed on 1.2% agarose gel. Pictures of the gels are shown. NE, non expressed. (C) After RT, mRNA expression was then measured by q-PCR. Sample data were analyzed according to the comparative cycle threshold method and were normalized by stable reference expression of EEF1α gene. For each mRNA, the level of expression at confluence was set at 100% in WT-MEFs allowing on one side a comparison between WT-MEFs (black bars) and RNase L−/−-MEFs (white bars) for each phase of the differentiation and, on the other side, a comparison between the different phases of the differentiation in each cell line. Error bars refer to the S.D. obtained in four independent experiments conducted in duplicate. *P≤0.05, **P≤0.01 and §P≤0.001. (D) ROS level was measured by NBT reduction in WT-MEFs and RNase L−/−-MEFs at confluence. Data are expressed relative to ROS level in WT-MEFs, which was set at 100%. Error bars refer to the S.D. obtained in three independent experiments conducted in duplicate. §P≤0.001 ROS level in RNase L−/−-MEFs compared with WT-MEFs
Genes expression during adipocyte differentiation
Adipogenesis involves a differentiation switch that activates a new program of gene expression. As previously shown during myoblast differentiation,11 RNase L activity is induced throughout adipocyte differentiation of WT-MEFs. Indeed, OAS2 (oligoadenylate synthetase 2) mRNA is induced (Figures 2B and C); OAS2 synthesizes the 2-5A allowing RNase L activation.12, 13 In parallel, RLI mRNA expression is decreased 6 days after induction of adipocyte differentiation. To explore the molecular basis through which RNase L deficiency impairs adipogenesis, we also followed expression levels of mRNAs coding for factors involved in adipogenesis (C/EBPα, CHOP10 and PPARγ2), lipids metabolism (aP2/Fabp4) and insulin response (Glut4: glucose transporter 4; Figures 2B and C). The expression profiles of these mRNAs during the differentiation were modified in RNase L−/−-MEFs compared with WT-MEFs. Induction of C/EBPα, aP2/Fabp4 and PPARγ2 was much lower in RNase L−/−-MEFs. Importantly, Glut4 mRNA was not induced in the absence of RNase L but increased >10 times in WT-MEFs. Contrarily, CHOP10 mRNA expression was maintained at a high level during the differentiation in RNase L−/−-MEFs as it declined >70% in WT-MEFs from confluence to day 6 of differentiation (Figures 2B and C).
ROS production
In RNase L−/−-MEFs, we noted a higher CHOP10 expression than in WT-MEFs (Figures 2B and C). This constant expression of CHOP10 could favor increased ROS production.22, 23 Indeed, as shown in Figure 2D, the level of ROS was significantly higher in RNase L−/−-MEFs compared with WT-MEFs.
RNase L is associated with CHOP10 messenger-ribonucleoprotein complex (mRNP) and regulates CHOP10 mRNA stability
We were particularly interested in the differential CHOP10 mRNA expression that we noticed between WT-MEFs and RNase L−/−-MEFs, CHOP10 being, as described in the introduction, an essential regulator of adipogenesis. RNase L has no great sequence specificity and cleaves single-stranded RNA at UpNp sequences.12 Moreover, RNase L has no specific RNA recognition motif sequence allowing its direct interaction with mRNA. However, we have previously shown that RNase L could participate to mRNPs via its interaction with RNA-binding proteins.15 To analyze if RNase L is associated with mRNP containing CHOP10 mRNA, we immunoprecipitated cytoplasmic mRNPs with a specific monoclonal antibody raised against RNase L in RNase L−/−-MEFs and WT-MEFs cell extracts. By real-time quantitative PCR (q-PCR), we could specifically amplify and detect CHOP10 mRNA in the mRNPs immunoprecipitated with a specific antibody against RNase L (Figure 3a). The presence of RNase L protein associated with CHOP10 mRNA was only observed in WT-MEFs and no interaction between RNase L and CHOP10 mRNA could be identified without any antibody or using an irrelevant antibody (α-tubulin; Figure 3a). These results confirmed the specificity of the constated interaction. To confirm that RNase L regulates CHOP10 mRNA stability, we then compared CHOP10 mRNA stability in WT-MEFs and RNase L−/−-MEFs after transcription inhibition by the actinomycin D treatment. In RNase L−/−-MEFs, we clearly observed a stabilization of CHOP10 mRNA compared with WT-MEFs (Figures 3b and c).
Figure 3.
RNase L is associated with CHOP10 mRNP and specifically degrades CHOP10 mRNA. (a) CHOP10 mRNA interaction with RNase L was evaluated by RNPs immunoprecipitation in WT-MEFs and RNase L−/−-MEFs. Upper panel represents western blot analysis with an antibody against RNase L of total cell extract (TE), and RNPs immunoprecipitated with no antibody (C), anti-α-tubulin antibody (T) or anti-RNase L antibody (R). Lower panel represents CHOP10 mRNA expression after RNAs extraction from RNPs immunoprecipitated with no antibody (C), anti-α-tubulin antibody (T) or anti-RNase L antibody (R) and after RT then q-PCR amplification with specific CHOP10 mRNA primers (Supplementary Table 1). q-PCR products were analyzed on 1.2% agarose gel. Photographies of the gels are shown. RNPs immunoprecipitation in WT-MEFs and RNase L−/−-MEFs, followed by western blot analysis, RT and q-PCR was realized on two independent experiments. (b) WT-MEFs and RNase L−/−-MEFs were treated with actinomycin D, then cells were collected at the indicated time points and mRNAs were analyzed, after RT, by PCR amplification with specific primers of EEF1α and CHOP10. PCR products were analyzed on 1.2% agarose gel. Pictures of the gels are shown. (c) cDNAs obtained for analysis of mRNA stability after the actinomycin D treatment were then used for quantification by q-PCR. Sample data were analyzed according to the comparative cycle threshold method and were normalized by stable reference expression of EEF1α gene. Levels of CHOP10 mRNA expression were set at 100% at time 0 in WT-MEFs (⧫) and RNase L−/−-MEFs (□). Error bars refer to the S.D. obtained in four independent experiments conducted in duplicate. *P≤0.05
Insulin response
RNase L−/−-MEFs presented altered adipocyte properties such as lipids accumulation (Figures 1C, D and 2A), Glut4 expression (Figures 2B and C) and a higher ROS level (Figure 2D) compared with WT-MEFs. All these features are indicative of lower insulin response in RNase L−/−-MEFs. So we measured Akt phosphorylation at serine 473 (Ser473) before and after insulin treatment of WT-MEFs and RNase L−/−-MEFs. Akt phosphorylation was significantly lower in RNase L−/−-MEFs than in WT-MEFs (Figures 4a and b). Akt phosphorylation is crucial for the intracellular signal transduction of insulin leading to glucose uptake by adipocytes.24 Measurement of glucose uptake in RNase L−/−-MEFs also showed a defect in insulin response of these cells compared with WT-MEFs (Figure 4c). Indeed, no increase in glucose uptake after insulin treatment of RNase L−/−-MEFs was observed while it was increased two times in WT-MEFs after insulin treatment. Glucose uptake was equivalent in WT-MEFs and RNase L−/−-MEFs without insulin treatment (data not shown). This basal glucose uptake in the absence of insulin would be due to Glut1 activity, being that WT-MEFs and RNase L−/−-MEFs expressed similarly this glucose receptor (data not shown).
Figure 4.
Insulin response. (a) WT-MEFs and RNase L−/−-MEFs were treated (+) or not (−) 10 min at 37 °C with insulin at confluence (ND) or after 6 days of differentiation (D). Total cellular extracts were analyzed by western blot with anti-P-Akt (Ser473), anti-Akt and anti-α-tubulin antibodies. (b) Protein bands shown in (a) were quantified with the ImageJ software. The levels of P-Akt (Ser473)/Akt were corrected with corresponding α-tubulin levels, used as indicator of proteins loading. P-Akt (Ser473)/Akt rate in WT-MEFs and RNase L−/−-MEFs before insulin treatment was set at 1. Error bars refer to the S.D. obtained in four independent experiments conducted in duplicate. *P=0.024 P-Akt (Ser473)/Akt level after insulin treatment in differentiated RNase L−/−-MEFs compared with WT-MEFs. (c) Glucose uptake was measured in WT-MEFs and RNase L−/−-MEFs. Differentiated cells were incubated (+) or not (−) with insulin then medium was changed to DMEM without glucose and with 6-NBDG. After a 45-min incubation at 37 °C, fluorescence intensity of 6-NBDG was measured at 540 nm wavelength (465 nm excitation wavelength). Glucose uptake represents the ratio of fluorescence measured in basal condition (without insulin) and with insulin. Fluorescence values were normalized by cell number. Error bars refer to the S.D. obtained in three independent experiments conducted in duplicate. *P=0.011 insulin-stimulated glucose uptake level in differentiated RNase L−/−-MEFs compared with WT-MEFs
Ectopic expression of RNase L in RNase L−/−-MEFs restored terminal adipocyte differentiation
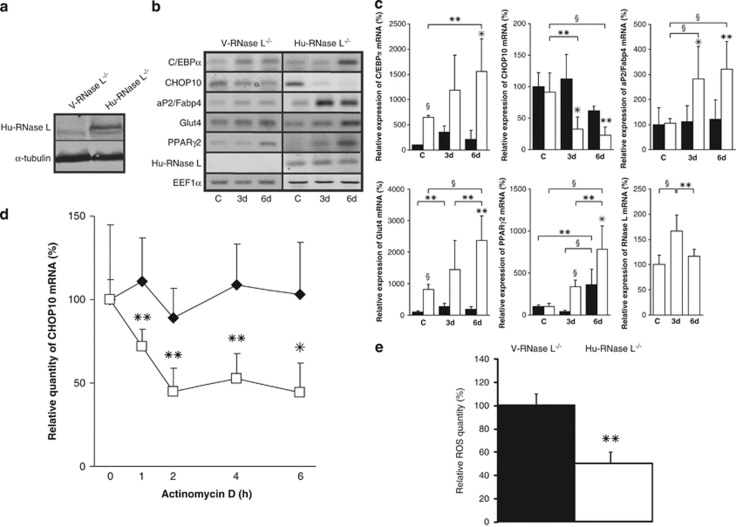
To verify that the impairment of adipocyte differentiation observed in RNase L−/−-MEFs was specifically due to the absence of RNase L, we generated RNase L−/−-MEFs-based stable cell lines by transfection with an empty pcDNA3 vector (V-RNase L−/−-MEFs) or with a vector containing Hu-RNase L complementary DNA (cDNA; Hu-RNase L−/−-MEFs; Figure 5a).
Figure 5.
Ectopic expression of Hu-RNase L in RNase L−/−-MEFs. (a) Hu-RNase L expression was measured in V-RNase L−/−-MEFs (V-RNase L−/−) and Hu-RNase L−/−-MEFs (Hu-RNase L−/−) by western blot using a specific antibody against Hu-RNase L. (b) Confluent V-RNase L−/−-MEFs and Hu-RNase L−/−-MEFs were induced to differentiate in ADM. mRNAs were extracted at confluence (C) and at days 3 (3d) and 6 (6d) after switch to ADM. After RT and PCR amplification using specific primers (Supplementary Table 1), PCR products were analyzed on 1.2% agarose gel. Pictures of the gels are shown. (c) After RT, mRNA expression was measured by q-PCR. Sample data were analyzed according to the comparative cycle threshold method and were normalized by stable reference expression of EEF1α gene. For each mRNA, the level of expression at confluence was set at 100% in V-RNase L−/−-MEFs, allowing on one side a comparison between V-RNase L−/−-MEFs (black bars) and Hu-RNase L−/−-MEFs (white bars) for each phase of the differentiation and, on the other side, a comparison between the different phases of the differentiation in each cell line. Error bars refer to the S.D. obtained in four independent experiments conducted in duplicate. *P≤0.05, **P≤0.01 and §P≤0.001. (d) At confluence, V-RNase L−/−-MEFs and Hu-RNase L−/−-MEFs were treated with actinomycin D, then cells were collected at the indicated time points and mRNA expression was quantified, after RT, by q-PCR. Sample data were analyzed according to the comparative cycle threshold method and were normalized by stable reference expression of EEF1α gene. Levels of CHOP10 mRNA expression were set at 100% at time 0 in V-RNase L−/−-MEFs (⧫) and Hu-RNase L−/−-MEFs (□). Error bars refer to the S.D. obtained in four independent experiments conducted in duplicate. *P≤0.05, **P≤0.01. (e) ROS level was measured by NBT reduction in V-RNase L−/−-MEFs and Hu-RNase L−/−-MEFs at confluence. Data are expressed relative to ROS level in V-RNase L−/−-MEFs, which was set at 100%. Error bars refer to the S.D. obtained in three independent experiments conducted in duplicate. **P=0.002 ROS level in Hu-RNase L−/−-MEFs compared with V-RNase L−/−-MEFs
Interestingly, ectopic expression of RNase L restored high expression of C/EBPα, aP2/Fabp4, PPARγ2 and Glut4 mRNA during adipocyte differentiation of Hu-RNase L−/−-MEFs. Moreover, RNase L expression led to a decreased CHOP10 mRNA level during differentiation of these cells (Figures 5b and c). Actinomycin D experiments in V-RNase L−/−-MEFs and Hu-RNase L−/−-MEFs confirmed that this downregulation of CHOP10 mRNA was due to a decrease of its stability (Figure 5d). Beside, this lower CHOP10 mRNA level was accompanied by a reduced ROS level (Figure 5e). Moreover, lipids accumulation (Figure 6A), insulin response (Figures 6B and C) and glucose uptake (Figure 6D) were also significantly improved in Hu-RNase L−/−-MEFs compared with V-RNase L−/−-MEFs in differentiated cells.
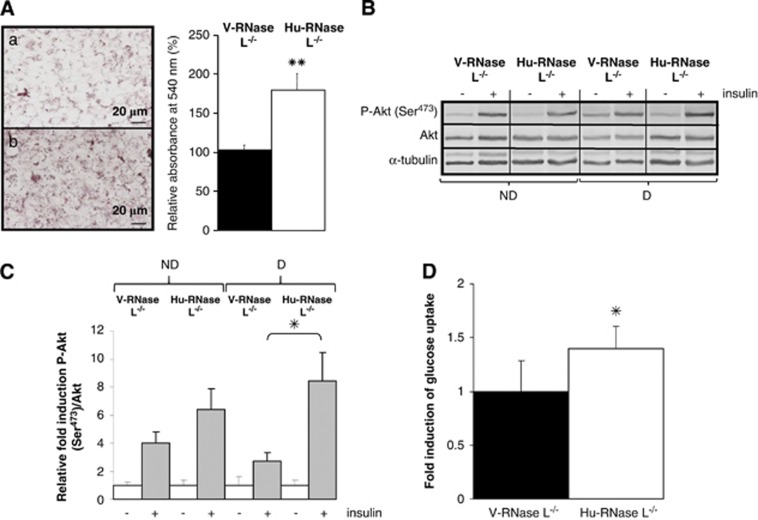
Figure 6.
Lipids accumulation and insulin response after re-introduction of RNase L expression in RNase L−/−-MEFs. (A) V-RNase L−/−-MEFs (a) and Hu-RNase L−/−-MEFs (b) were induced to differentiate in ADM. Cells were fixed and stained with oil red O to reveal lipids accumulation after 6 days of differentiation. Cells were observed at a magnification of × 40. After staining, lipids were quantified by spectrophotometric analysis at 540 nm following elution of cell retained oil red O with ethanol. The amount of lipids in V-RNase L−/−-MEFs was set at 100%. Error bars refer to the S.D. obtained in four independent experiments conducted in duplicate. **P=0.005 lipids quantity in Hu-RNase L−/−-MEFs compared with V-RNase L−/−-MEFs. (B) V-RNase L−/−-MEFs and Hu-RNase L−/−-MEFs were treated (+) or not (−) 10 min at 37 °C with insulin at confluence (ND) or after 6 days of differentiation (D). Total cellular extracts were analyzed by western blot with anti-P-Akt (Ser473), anti-Akt and anti-α-tubulin-specific antibodies. (C) Protein bands shown in (B) were quantified with the ImageJ software. The levels of P-Akt (Ser473)/Akt were corrected with corresponding α-tubulin levels, used as indicator of proteins loading. P-Akt (Ser473)/Akt rate in V-RNase L−/−-MEFs and Hu-RNase L−/−-MEFs before insulin treatment was set at 1. Error bars refer to the S.D. obtained in four independent experiments. *P=0.05 P-Akt (Ser473)/Akt level after insulin treatment in differentiated Hu-RNase L−/−-MEFs compared with differentiated V-RNase L−/−-MEFs. (D) Glucose uptake was measured in V-RNase L−/−-MEFs and Hu-RNase L−/−-MEFs after differentiation. Cells were incubated (+) or not (−) with insulin, then medium was changed to DMEM without glucose and 6-NBDG. After a 45-min incubation at 37 °C, fluorescence intensity of 6-NBDG was measured at 540 nm wavelength (465 nm excitation wavelength). Glucose uptake represents the ratio of fluorescence measured in basal condition (without insulin) and in the presence of insulin. Fluorescence values were normalized by cell number. Error bars refer to the S.D. obtained in three independent experiments conducted in duplicate. *P=0.028 insulin-stimulated glucose uptake level in differentiated Hu-RNase L−/−-MEFs compared with V-RNase L−/−-MEFs
Downregulation of CHOP10 mRNA in RNase L−/−-MEFs improves their adipocyte differentiation
To confirm that regulation of CHOP10 expression by RNase L is key for adipocyte differentiation, we then downregulated expression of CHOP10 mRNA by siCHOP in RNase L−/−-MEFs. RNase L−/−-MEFs were transfected with siCHOP or a control siRNA (siCT) before induction of differentiation. Only a certain percentage of the cells population was transfected by the siCHOP, as checked by using a fluorescent siRNA (data not shown). However, we could observe a downregulation of CHOP10 mRNA in RNase L−/−-MEFs transfected with siCHOP compared with RNase L−/−-MEFs transfected with siCT (Figure 7A). This allowed a better terminal adipocyte differentiation of RNase L−/−-MEFs, as demonstrated by an increase in lipids accumulation and the detection of perilipin surrounding lipid droplets in RNase L−/−-MEFs transfected with siCHOP compared with RNase L−/−-MEFs transfected with siCT (Figures 7B and C). We also observed a slight improvement in insulin response, as shown by a small increase of Akt phosphorylation at Ser473 in RNase L−/−-MEFs transfected with siCHOP compared with RNase L−/−-MEFs transfected with siCT (Figures 7D and E). However, this result was not statistically significant. The latter could be because of the difficulty to obtain sufficient cells transfected with siCHOP. Nevertheless, these results clearly showed that CHOP10 mRNA downregulation is necessary to allow complete adipocyte differentiation of MEFs.
Figure 7.
Downregulation of CHOP10 mRNA with CHOP10 siRNA allows a better adipocyte differentiation of RNase L−/−-MEFs. (A) RNase L−/−-MEFs were transfected with a siCHOP or a siCT, then cells were collected and mRNAs were analyzed, after RT, by q-PCR with specific primers for CHOP10 or EEF1α (Supplementary Table 1). PCR products were analyzed on 1.2% agarose gel. Photographies of the gels are shown. (B) Cells were stained with oil red O to reveal lipids accumulation after 6 days of differentiation. RNase L−/−-MEFs transfected with siCT (a and c) and RNase L−/−-MEFs transfected with siCHOP (b and d) were observed at a magnification of × 40. After staining with oil red O, lipids were quantified by spectrophotometric analysis at 540 nm following elution of cell retained oil red O with ethanol. (e) Quantification of lipids in RNase L−/−-MEFs transfected with siCT or siCHOP, 6 days after induction of differentiation in ADM. The amount of lipids in RNase L−/−-MEFs transfected with siCT was set at 100%. Error bars refer to the S.D. obtained in four independent experiments conducted in duplicate. **P=0.003 RNase L−/−-MEFs transfected with siCHOP compared with RNase L−/−-MEFs transfected with siCT. (C) RNase L−/−-MEFs transfected with siCT or siCHOP were induced to differentiate during 6 days in ADM. Cells were fixed and then incubated with an antibody against perilipin (red; RNase L−/−-MEFs transfected with siCT (a) and RNase L−/−-MEFs transfected with siCHOP (b). DNA was stained with DAPI (blue). (D) RNase L−/−-MEFs transfected with siCT or siCHOP were treated (+) or not (−) 10 min at 37 °C with insulin after 6 days of differentiation. Total cellular extracts were analyzed by western blot with anti-P-Akt (Ser473), anti-Akt and anti-α-tubulin-specific antibodies. (E) Protein bands shown in (D) were quantified with the ImageJ software. Levels of P-Akt (Ser473)/Akt were corrected with corresponding α-tubulin levels, used as indicator of proteins loading. P-Akt (Ser473)/Akt rate in RNase L−/−-MEFs transfected with siCT or siCHOP before insulin treatment was set at 1. Error bars refer to the S.D. obtained in four independent experiments. NS, non statistically significant P-Akt (Ser473)/Akt level after insulin treatment in differentiated RNase L−/−-MEFs transfected with siCHOP compared with differentiated RNase L−/−-MEFs transfected with siCT
Discussion
Adipose tissue has a central role in metabolic regulations, partly due to its capacity to store lipids. Adipogenesis is controlled by several functionally coherent mechanisms, which are intricately coordinated and regulated in time to ensure the activation/inactivation cascade of genes expression. The transition from preadipocyte to mature adipocyte involves successive stages: growth arrest, clonal expansion and differentiation. The clonal expansion phase is necessary for terminal differentiation. These different steps are predominantly regulated by C/EBP family members and PPARγ.
Regulation of mRNA stability is an essential mechanism for controlling genes expression.10, 11 Cytoplasmic mRNAs are engaged in mRNA/proteins complexes called mRNPs, which have a major role in mRNA translation and stability.25 Here, we show that an endoribonuclease, RNase L, directly regulates mRNA stability of CHOP10, a member of the C/EBP family and a fundamental regulator of adipogenesis. RNase L has no great sequence specificity and cleaves single-stranded RNA at UpNp sequences.12 RNase L specificity is above all due to its interaction with transacting factors, such as eukaryotic releasing factor 315 and mitochondrial initiation factor 2.16 In this study, we show for the first time that RNase L interacts with CHOP10 mRNA in mRNPs. Indeed, a specific monoclonal antibody directed against RNase L immunoprecipitated RNase L protein associated with CHOP10 mRNA only in WT-MEFs, while no RNase L-CHOP10 mRNA interaction was detected in RNase L−/−-MEFs. Association of RNase L with CHOP10 mRNA in mRNP enables CHOP10 mRNA regulation by RNase L, as illustrated by the preservation of CHOP10 mRNA expression in RNase L−/−-MEFs when it strongly decreases in WT-MEFs during adipocyte differentiation.
CHOP10 interacts with another member of the C/EBP family, C/EBPβ, by inhibiting its transcriptional activity. This inhibition is necessary for clonal expansion of preadipocytes, a prerequisite to adipocyte differentiation.7 As CHOP10 expression decreases, C/EBPβ is released, allowing transactivation of C/EBPα and terminal adipocyte differentiation. Maintaining CHOP10 expression could lead to an excessive clonal expansion (hyperplasia), at the expense of late differentiation and lipids storage capacity of mature adipocytes. Here, we show that terminal adipocyte differentiation is impaired in RNase L−/−-MEFs. In fact, these cells accumulated fewer lipids than WT-MEFs. Moreover, RNase L−/−-MEFs expressed a lower level of aP2/Fabp4, indicating a dysfunction of intracellular lipids trafficking.20 Beside, we failed to detect perilipin at the surface of lipid droplets of RNase L−/−-MEFs. In RNase L−/−-MEFs, CHOP10 expression was maintained even after induction of differentiation, leading to lower induction of C/EBPα and PPARγ2 expression. Therefore, as terminal adipocyte differentiation could not correctly proceed, we observed more adipocytes with small lipid droplets while lipids accumulation, Glut4 mRNA expression and insulin response were lower in these cells than in WT-MEFs. On the other hand, it has been shown in different cell systems that overexpression of CHOP10 leads to an increased ROS production.26 ROS, in turn, could induce CHOP10 expression.27 Actually, in RNase L−/−-MEFs cells, we noticed an increase in ROS level. Several studies relating to the consequences of ROS production have been performed and contradicting effects have been reported. Some studies describe ROS as positive regulators of adipocyte differentiation by accelerating mitotic clonal expansion28 and enhancing insulin sensitivity.29 Conflictingly, other studies present ROS as inhibitors of adipocyte differentiation by regulating CHOP10 expression27 and inhibiting insulin response22 and Glut4 expression.30 The apparent discrepancies between these different studies reflect how the level and kinetics of ROS expression have vital effects on cell fate.9 Here, we showed higher ROS level in RNase L−/−-MEFs with a decrease in insulin signaling, which participated in insulin-stimulated glucose uptake inhibition. The lower level of Glut4 expression in RNase L−/−-MEFs could also favor the restricted glucose uptake.
To confirm the essential role of RNase L in CHOP10 mRNA regulation and adipogenesis, we re-introduced RNase L expression in RNase L−/−-MEFs. In these cells, which express Hu-RNase L (Hu-RNase L−/−-MEFs), CHOP10 mRNA stability was highly reduced compared with RNase L−/−-MEFs transfected with a control vector (V-RNase L−/−-MEFs). This also led to a decreased CHOP10 mRNA expression after induction of adipocyte differentiation and to higher levels of C/EBPα, PPARγ2, aP2/Fabp4 and Glut4 mRNAs expression. Moreover, in Hu-RNase L−/−-MEFs, ROS level were decreased by 50%, while lipids accumulation was increased by 75%. Finally, insulin response and glucose uptake were also significantly improved after re-expression of RNase L in RNase L−/−-MEFs. However, we could not completely reverse RNase L−/−-MEFs phenotype to WT-MEFs with Hu-RNase L transfection. One reason could be that transfected RNase L mRNA was not regulated as was the endogenous RNase L mRNA in WT-MEFs. RNase L expression is constitutive in Hu-RNase L−/−-MEFs, and RNase L activity could only be regulated by OAS2 expression, 2-5A synthesis and RLI.
To confirm the effects observed following the overexpression of CHOP10 on RNase L−/−-MEFs differentiation inhibition, we downregulated CHOP10 mRNA in RNase L−/−-MEFs by CHOP10 siRNA (siCHOP) transfection. Even if not all cells were transfected with siCHOP, we notified a significant downregulation of CHOP10 mRNA expression, which was accompanied by improved adipocyte differentiation. Our results clearly identify RNase L as an essential effector of terminal adipocyte differentiation by regulating CHOP10 level.
Maintaining a high level of CHOP10 expression in vivo could lead to an excessive clonal expansion (hyperplasia) of developing adipocytes at the expense of late differentiation and lipids storage capacity of mature adipocytes.5 Actually, RNase L−/− mice possess a higher number of smaller adipocytes than WT mice (Figure 8A), that is characteristic of hyperplasia.31 This number of adipocytes increases with aging, as we observed an adipose tissue expansion in aged RNase L−/− mice. The average body weight of aged RNase L−/− mice was 21% higher than that of aged WT mice (Figure 8B). Despite this excessive adipose tissue development, RNase L−/− mice present a defect in lipids storage, as indicated by ectopic fat depots in other organs, such as the liver and the kidneys. Indeed, RNase L−/− mice present hepatic steatosis and lipid droplets within renal tubular epithelial cell cytoplasms (Figure 8C). Interestingly, adipose tissue expansion is observed in aged RNase L−/− mice and not in young mice. At present, it is known that CHOP10 is more expressed in old individuals preadipocytes than from young ones.5 Change in adipogenesis process is due to the incapacity of preadipocytes to express appropriate levels of key adipogenic regulators: CHOP10, C/EBPα and PPARγ.4, 32 We assume that overexpression of CHOP10 in RNase L−/− mice added to the physiological overexpression of CHOP10 in old age would lead to even higher levels of CHOP10. In the aged RNase L−/− mice, this cumulative effect would increase hyperplasia of adipose tissue at the expense of its terminal differentiation and efficient lipids storage.
Figure 8.
RNase L−/− mice present impairments of adipogenesis. (A) Adipose tissue sections from male WT and RNase L−/− mice (a and b, respectively) and female WT and RNase L−/− mice (c and d, respectively) were stained with H&E. (e) Relative number of adipocytes from male and female WT (black bars) and RNase L−/− mice (white bars). Number of adipocytes from WT male mouse was set at 100%. §P≤0.001 number of adipocytes in male and female RNase L−/− mice compared with adipocytes number in male and female WT mice, respectively. (f) Average size of adipocytes from abdominal adipose tissue from male and female WT (black bars) and RNase L−/− mice (white bars). §P≤0.001 average size of adipocytes in male and female RNase L−/− mice compared with adipocytes size in male and female WT mice, respectively. (B) Eight pairs of age- and gender-matched WT and RNase L−/− mice were maintained for 18 months and fed with a chew diet. The average body weight was 65.8±5.9 g for RNase L−/− mice and 54.4±3.3 g for WT mice (P=0.056). (C) H&E staining of the liver (a and b) and kidney (c and d) sections from WT and RNase L−/− mice. (D) Scheme of potential role of RNase L during adipogenesis. In RNase L−/− mouse, CHOP10 expression is maintained after induction of differentiation. Consequently, clonal expansion of preadipocytes is more important in RNase L−/− mice that leads to an important adipose tissue development but with less differentiated adipocytes. These adipocytes are not able to stock correctly lipids as do WT mouse adipocytes
Interestingly, Tomaru et al.33 recently produced a transgenic mouse with decreased proteasomal chymotrypsin-like activity. These mice overexpressed RNase L and presented a loss of adipose tissue.
Here, we identify RNase L as a regulator of adipogenesis by regulating CHOP10 mRNA stability. As cited above and demonstrated in vivo by others, CHOP10 has a main role in the regulation of adipose tissue expansion by controlling the proliferation phase of preadipocytes. The decrease of RNase L activity, through excessive CHOP10 expression could lead to high preadipocytes proliferation (hyperplasia) and impairment of their terminal differentiation. RNase L activity could be crucial in the control of adipose tissue development and function during obesity and aging; based on our results we propose here a scheme of the role of RNase L in adipogenesis (Figure 8D).
Materials and Methods
MEFs isolation – cell viability
Five embryos from each mouse (WT and RNase L−/− mice, C57BL/6 strain) were collected aseptically at the stage of 16 days (the day of plug being the first day) in phosphate-buffered saline (PBS: 140 mM NaCl, 2 mM KCl, 8 mM Na2HPO4, 1.5 mM KH2PO4 (pH 7.4)). The limbs were cut and chipped to very small pieces. After digestion with 0.05% trypsin–EDTA solution, the cells were dissociated in two volumes of Dulbecco's modified eagle medium (DMEM, BioWhittaker, Lonza, Basel, Switzerland) containing 10% (V/V) of fetal calf serum (FCS, PAN Biotech, Dutscher, Brumath, France) by vigorous shaking. Then the cells were plated in tissue culture dishes with DMEM 10% (V/V) FCS. The cell line was immortalized by letting the cells pass the crisis with a total of 37 passages.34
Cell viability was controlled for each cell line with the PrestoBlue Cell Viability Reagent kit (Life Technologies, Fisher Scientific, Illkirch, France) following the manufacturer instructions. PrestoBlue reagent was added in the cell culture medium, and the cells were incubated at 37 °C for 30 min. Fluorescence intensity of the PrestoBlue reagent reduced by living cells was measured at 615 nm wavelength (535 nm excitation wavelength) with an Infinite 200PRO TECAN (Lyon, France). Fluorescence level was normalized by cell number.
Cell culture – induction of adipocyte differentiation
WT-MEFs and RNase L−/−-MEFs were cultured, as described previously, in the growth medium (GM): DMEM with 10% (V/V) FCS, 100 U/ml penicillin and 100 μg/ml streptomycin at 37°C.18
At 80% confluence, cells were switched to adipocyte differentiating medium (ADM): GM supplemented with 0.5 mM 3-isobutyl-1-methylxanthine, 1 μM dexamethasone and 10 μM insulin (all from Sigma-Aldrich, Saint-Quentin Fallavier, France) for 2 days. Cells were then incubated in fresh ADM for 2 supplementary days and then the medium was changed to GM for 2 days.19
For insulin response, cells were treated or not with insulin (1 nmol) for 10 min at 37 °C. Cells were then washed twice with PBS and lysed in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer (300 mM Tris (pH 8.9), 5% (W/V) SDS, 750 mM β-mercaptoethanol, 20% (V/V) glycerol and bromophenol blue) for proteins analysis by western blot.
Oil red O labeling – immunofluorescence
Adipogenic cell phenotype was characterized by specific oil red O staining of lipids.35 Cells were observed with a Nikon AZ100 and TIFF images were captured with a Nikon Digital Camera (DXM 1200c; Champigny sur Marne, France) with ACT-1C program for the control of DXM 1200C platform. Lipids accumulation was quantified by spectrophotometric analysis at 540 nm after eluting the oil red O retained in the cells with ethanol. Optical density was measured with an Infinite 200PRO TECAN.
For immunofluorescence analysis, cells were fixed in 3.7% (V/V) formalin for 10 min and permeabilized with PBS-Triton X100 0.5% (V/V) at room temperature for 5 min. After blocking with 10% (V/V) FCS in PBS, cells were incubated with an anti-perilipin or an anti-aP2/Fabp4 antibody (both from Cell Signaling, Ozyme, Saint-Quentin Yvelines, France) for 1 h at room temperature. Cells were then washed and incubated with a donkey anti-rabbit secondary antibody conjugated to tetramethylrhodamine-5,6-isothiocyanate (Santa Cruz Biotechnology, Santa Cruz, CA, USA) for 1 h at room temperature. Cells were observed at room temperature with an Imager MI Zeiss (Le Pecq, France), and TIFF images were captured with an Axiocam MRm Zeiss and the Axiovision 4.7 (05-2008) program.
Reverse transcription (RT), semi-quantitative PCR and q-PCR
Total RNAs were isolated using TRIzol (Invitrogen, Fisher Scientific). To avoid genomic DNA contamination and amplification during PCR, RNAs were treated with DNase-RNase free (Euromedex, Souffelweyersheim, France) before RT. cDNAs were generated by RT with oligo-(dT) primers and the Moloney murine leukemia virus (M-MLV) Reverse Transcriptase (Jena Bioscience, Euromedex). Briefly, 5 μg of total RNAs were denatured at 70 °C and then reverse transcribed at 42 °C for 120 min following the manufacturer instructions.
Semi-quantitative PCR of the cDNAs was performed using a Biometra thermocycler (LABGENE, Archamps, France) in a total volume of 25 μl. Cycle number was 32. Amounts of cDNA were adjusted for each primer pair to be in the linear range amplification and to give the same quantity of amplified cDNA with eukaryotic elongation factor 1α (EEF1α) primers. PCR products were visualized in 1.2% agarose gel and stained with ethidium bromide. Photographies of the gels were captured as TIFF files with a Vilber Lourmat camera and Ecapt program (both from VWR, Fontenay-sous-bois, France).
To quantify genes expression allowing a comparison between WT-MEFs and RNaseL−/−-MEFs or V-RNaseL−/−-MEFs and Hu-RNaseL−/−-MEFs, cDNAs were used as templates in SYBR Green I Master real-time q-PCR assays on a LightCycler 480 (Roche Diagnostics, Meylan, France), in a total volume of 10 μl. Cycle number was 45. Sample data were analyzed according to the comparative cycle threshold method and were normalized by stable reference gene of EEF1α.
Gene sequences for primer design were obtained from the NCBI Reference Sequences database. Primers were chosen using the Primer3 and the LightCycler Probe Design (Roche) softwares. When possible, forward and reverse primers were designed on different exon sequences. Primers sequences are provided in Supplementary Table 1.
mRNPs immunoprecipitation
We used the protocol described by Sanchez et al.36 with some modifications. Briefly, confluent WT-MEFs and RNase L−/−-MEFs were lysed with IPP50 buffer (10 mM Tris-HCl (pH 8), 150 mM NaCl,) without NP40 and supplemented with 2 mM EDTA and 10 μg/ml aprotinin, 150 μg/ml leupeptin, 1 mM phenylmethanesulfonyl fluoride and 80U RNaseOut (Invitrogen). Cell extracts were centrifuged at 10 000 × g. Supernatants (2 mg of proteins) were incubated with protein A-sepharose equilibrated in the same modified IPP50 buffer, and no antibody, mouse monoclonal anti-α-tubulin (Sigma-Aldrich) or mouse monoclonal anti-RNase L antibody (Abcam, Paris, France). After an overnight incubation at 4 °C with gentle shaking, protein A-sepharose beads were centrifuged and washed three times with ice-cold-modified IPP50 buffer. The pellet was separated in two fractions. One fraction was analyzed for RNase L protein content by western blot as described below. RNAs were extracted from the other fraction by adding TRIzol (1 ml) to the beads and following the manufacturer instructions (Invitrogen). Glycogen (10 μg) was added to ease RNA precipitation with isopropanol. cDNA was generated from total extracted RNA by RT with oligo-(dT) primers and the M-MLV Reverse Transcriptase (Jena Bioscience) as described above. 2.5 μl of RT product was amplified with specific CHOP10 primers (Supplementary Table 1) by q-PCR following the manufacturer instructions (Roche) with a LightCycler 480 as described previously.
mRNA stability
WT-MEFs and RNase L−/−-MEFs were plated in GM and, at confluence, cells were treated with actinomycin D (5 μg/ml) for 0, 1, 2, 4 or 6 h. Then RNAs were extracted and analyzed after RT by semi-quantitative PCR as described above. Following agarose gel electrophoresis, pictures of the gels were saved as TIFF files with a Vilber Lourmat camera and Ecapt program. cDNAs were also analyzed by q-PCR following the manufacturer instructions (Roche) with a LightCycler 480 as described above.
Western blot assay
At confluence, cells were switched to differentiation medium as indicated above. Cells were then collected in SDS-PAGE buffer at confluence or after 6 days in ADM. Protein extracts were incubated with benzonase (Sigma-Aldrich) 15 min at 37 °C before western blot analysis. Nitrocellulose membranes were then incubated in blocking buffer for fluorescent western blotting (Odyssey Infrared Imaging System LI-COR Biosciences, ScienceTec, Courtabœuf, France) for 1 h and incubated with a rabbit polyclonal anti-RLI antibody (Novus Biological, Interchim, Montluçon, France), or a mouse monoclonal anti-Hu-RNase L (Abcam) or a rabbit polyclonal anti-mouse RNase L (Santa Cruz), or a rabbit polyclonal anti-phospho-Akt (Ser473) or a rabbit polyclonal anti-Akt (both from Cell Signaling), or a mouse monoclonal anti-α-tubulin (Sigma-Aldrich) in the same buffer overnight at 4 °C. Membranes were washed in PBS supplemented with 0.05% (V/V) Tween 20 then with PBS alone and incubated for 45 min at room temperature with a donkey anti-rabbit or anti-mouse conjugated to IRDye 700DX or IRDye800 (Rockland, Tebu-Bio). Specific proteins were visualized with a LI-COR Odyssey Biosciences Imaging System. Protein bands were quantified by image analysis with the ImageJ software (http://rsbweb.nih.gov/ij/index.html).
Glucose uptake
A fluorescent glucose analog, 6-(N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl) amino)-2-deoxy-glucose (6-NBDG, Molecular Probes, Fisher Scientific), was used to measure glucose uptake in WT-MEFs, RNase L−/−-MEFs, V-RNase L−/−-MEFs and Hu-RNase L−/−-MEFs.37 After differentiation, cells were incubated 10 min with or without insulin (1 nmol) then medium was changed to glucose-free DMEM and 6-NBDG (300 μM) was then added to all cells for 45 min at 37 °C. Cells were thoroughly washed to remove all exogenous fluorescent glucose analog, and fluorescence intensity of 6-NBDG was measured at 540 nm wavelength (465 nm excitation wavelength) with an Infinite 200PRO TECAN. Fluorescence level was normalized with cell number.
ROS production in WT-MEFs and RNase L−/−-MEFs
ROS level was measured using the nitroblue tetrazolium (NBT) assay (Sigma-Aldrich).38 Cells were incubated for 90 min in DMEM 0.2% (W/V) NBT. Produced formazan was then dissolved in 50% (V/V) acetic acid and the absorbance was determined at 560 nm with an Infinite 200PRO TECAN. Optical density values were normalized by protein levels.
Ectopic expression of Hu-RNase L in RNase L−/−-MEFs
The coding sequence of Hu-RNase L cDNA was subcloned in pcDNA3neo (Invitrogen) by the standard procedures.39 RNase L-pcDNA3neo or the empty vector pcDNA3neo were transfected into RNase L−/−-MEFs with JetPEI (Qbiogene, Illkirch, France) following the manufacturer instructions. As observed previously, the yield of transfection was low,40 so transfectants were selected by culturing cells in the presence of 1 mg of G418 (Gibco-BRL, Fisher Scientific) per ml of cell culture medium. The polyclonal cell population expressing the transfected Hu-RNase L cDNA was named Hu-RNase L−/−-MEFs; the polyclonal cell population expressing the transfected empty vector was named V-RNase L−/−-MEFs.
siCHOP and siCT transfection
The siRNA duplexes have been designed and manufactured by Qiagen (Courtabœuf, France).
siCHOP sense: (5′-GGAAGUGCAUCUUCAUACA-3′)dTdT; siCHOP antisense: (5′-UGUAUGAAGAUGCACUUCC-3′)dTdT; siCT: sense (5′-UUCUCCGAACGUGUCACGU-3′)dTdT; siCT antisense: (5′-ACGUGACACGUUCGGAGA-3′)dTdT. siRNAs were transfected into RNase L−/−-MEFs with HiPerFect (Qiagen) following the manufacturer instructions.
Statistical analysis
Means±S.D.'s were calculated and statistical analyses using the Holm–Sidak method were performed with the SigmaStat software. Significant differences between two groups are considered for *P≤0.05, **P≤0.01 and §P≤0.001.
Histological analysis of white adipose tissue from WT and RNase L−/− mice
Abdominal fat pad was removed from each mouse (two females and two males, 21 weeks aged) and fixed in 10% formalin. Fixed specimens were embedded in paraffin. Sections were then stained with hematoxylin and eosin (H&E). Adipocytes were counted in four different sections for each mouse.
Phenotype of mouse RNase L−/−
Eight pairs of age-matched WT and RNase L−/− male mice (C57BL/6 strain) were maintained for 18 months and fed with chew diet. Food intake was similar for both types of mice. The average body weight was measured in WT mice (five survived) and in RNase L−/− (six survived). For histological analyses of the liver and kidney, tissues were fixed in formalin and then embedded in paraffin. Sections were deparaffinized, rehydrated and stained with H&E.
Acknowledgments
This work was supported by grants from the Institut National de la Santé et de la Recherche Médicale (INSERM) and the Université de Montpellier 1 (UM1). OF was recipient of a UM1 fellowship. We thank M Hokayem for her insight on this manuscript.
Glossary
- aP2
adipocyte protein 2
- C/EBP
CCAAT/enhancer-binding protein
- CHOP10
C/EBP homologous protein 10
- DMEM
Dulbecco's modified eagle medium
- EEF1α
eukaryotic elongation factor 1α
- Fabp4
fatty acid-binding protein 4
- FCS
fetal calf serum
- Glut4
glucose transporter 4
- H&E
hematoxylin and eosin
- MEFs
mouse embryonic fibroblasts
- M-MLV
Moloney murine leukemia virus
- 6-NBDG
6-(N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl) amino)-2-deoxy-glucose
- NBT
nitroblue tetrazolium
- OAS2
oligoadenylate synthetase 2
- PPARγ
peroxisome proliferator-activated receptor γ
- q-PCR
real-time quantitative PCR
- RLI
RNase L inhibitor
- RNase L
endoribonuclease L
- ROS
reactive oxygen species
- RT
reverse transcription
- SDS-PAGE
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by H-U Simon
Supplementary Material
References
- Jo J, Gavrilova O, Pack S, Jou W, Mullen S, Sumner AE, et al. Hypertrophy and/or hyperplasia: dynamics of adipose tissue growth. PLoS Comput Biol. 2009;5:e1000324. doi: 10.1371/journal.pcbi.1000324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulos SP, Hausman DB, Hausman GJ. The development and endocrine functions of adipose tissue. Mol Cell Endocrinol. 2010;323:20–34. doi: 10.1016/j.mce.2009.12.011. [DOI] [PubMed] [Google Scholar]
- Bluher M. Adipose tissue dysfunction in obesity. Exp Clin Endocrinol Diabetes. 2009;117:241–250. doi: 10.1055/s-0029-1192044. [DOI] [PubMed] [Google Scholar]
- Cartwright MJ, Tchkonia T, Kirkland JL. Aging in adipocytes: potential impact of inherent, depot-specific mechanisms. Exp Gerontol. 2007;42:463–471. doi: 10.1016/j.exger.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchkonia T, Pirtskhalava T, Thomou T, Cartwright MJ, Wise B, Karagiannides I, et al. Increased TNFalpha and CCAAT/enhancer-binding protein homologous protein with aging predispose preadipocytes to resist adipogenesis. Am J Physiol Endocrinol Metab. 2007;293:E1810–E1819. doi: 10.1152/ajpendo.00295.2007. [DOI] [PubMed] [Google Scholar]
- Tontonoz P, Spiegelman BM. Fat and beyond: the diverse biology of PPARgamma. Annu Rev Biochem. 2008;77:289–312. doi: 10.1146/annurev.biochem.77.061307.091829. [DOI] [PubMed] [Google Scholar]
- Tang QQ, Lane MD. Role of C/EBP homologous protein (CHOP-10) in the programmed activation of CCAAT/enhancer-binding protein-beta during adipogenesis. Proc Natl Acad Sci USA. 2000;97:12446–12450. doi: 10.1073/pnas.220425597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song B, Scheuner D, Ron D, Pennathur S, Kaufman RJ. Chop deletion reduces oxidative stress, improves beta cell function, and promotes cell survival in multiple mouse models of diabetes. J Clin Invest. 2008;118:3378–3389. doi: 10.1172/JCI34587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szypowska AA, Burgering BM. The peroxide dilemma: opposing and mediating insulin action. Antioxid Redox Signal. 2011;15:219–232. doi: 10.1089/ars.2010.3794. [DOI] [PubMed] [Google Scholar]
- Hao S, Baltimore D. The stability of mRNA influences the temporal order of the induction of genes encoding inflammatory molecules. Nat Immunol. 2009;10:281–288. doi: 10.1038/ni.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salehzada T, Cambier L, Vu Thi N, Manchon L, Regnier L, Bisbal C. Endoribonuclease L (RNase L) regulates the myogenic and adipogenic potential of myogenic cells. PLoS One. 2009;4:e7563. doi: 10.1371/journal.pone.0007563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisbal C, Silverman RH. Diverse functions of RNase L and implications in pathology. Biochimie. 2007;89:789–798. doi: 10.1016/j.biochi.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovanessian AG, Justesen J. The human 2′–5′oligoadenylate synthetase family: unique interferon-inducible enzymes catalyzing 2′–5′ instead of 3′–5′ phosphodiester bond formation. Biochimie. 2007;89:779–788. doi: 10.1016/j.biochi.2007.02.003. [DOI] [PubMed] [Google Scholar]
- Bisbal C, Martinand C, Silhol M, Lebleu B, Salehzada T. Cloning and characterization of a RNAse L inhibitor. A new component of the interferon-regulated 2–5A pathway. J Biol Chem. 1995;270:13308–13317. doi: 10.1074/jbc.270.22.13308. [DOI] [PubMed] [Google Scholar]
- Le Roy F, Salehzada T, Bisbal C, Dougherty JP, Peltz SW. A newly discovered function for RNase L in regulating translation termination. Nat Struct Mol Biol. 2005;12:505–512. doi: 10.1038/nsmb944. [DOI] [PubMed] [Google Scholar]
- Le Roy F, Silhol M, Salehzada T, Bisbal C. Regulation of mitochondrial mRNA stability by RNase L is translation-dependent and controls IFNalpha-induced apoptosis. Cell Death Differ. 2007;14:1406–1413. doi: 10.1038/sj.cdd.4402130. [DOI] [PubMed] [Google Scholar]
- Bisbal C, Silhol M, Laubenthal H, Kaluza T, Carnac G, Milligan L, et al. The 2′–5′ oligoadenylate/RNase L/RNase L inhibitor pathway regulates both MyoD mRNA stability and muscle cell differentiation. Mol Cell Biol. 2000;20:4959–4969. doi: 10.1128/mcb.20.14.4959-4969.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou A, Paranjape J, Brown TL, Nie H, Naik S, Dong B, et al. Interferon action and apoptosis are defective in mice devoid of 2′,5′-oligoadenylate-dependent RNase L. EMBO J. 1997;16:6355–6363. doi: 10.1093/emboj/16.21.6355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KA, Kim JH, Wang Y, Sul HS. Pref-1 (preadipocyte factor 1) activates the MEK/extracellular signal-regulated kinase pathway to inhibit adipocyte differentiation. Mol Cell Biol. 2007;27:2294–2308. doi: 10.1128/MCB.02207-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertzel AV, Bernlohr DA. The mammalian fatty acid-binding protein multigene family: molecular and genetic insights into function. Trends Endocrinol Metab. 2000;11:175–180. doi: 10.1016/s1043-2760(00)00257-5. [DOI] [PubMed] [Google Scholar]
- Bickel PE, Tansey JT, Welte MA. PAT proteins, an ancient family of lipid droplet proteins that regulate cellular lipid stores. Biochim Biophys Acta. 2009;1791:419–440. doi: 10.1016/j.bbalip.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houstis N, Rosen ED, Lander ES. Reactive oxygen species have a causal role in multiple forms of insulin resistance. Nature. 2006;440:944–948. doi: 10.1038/nature04634. [DOI] [PubMed] [Google Scholar]
- Gregor MG, Hotamisligil GS. Adipocyte stress: the endoplasmic reticulum and metabolic disease. J Lipid Res. 2007;48:1905–1914. doi: 10.1194/jlr.R700007-JLR200. [DOI] [PubMed] [Google Scholar]
- Tanti JF, Grillo S, Gremeaux T, Coffer PJ, Van Obberghen E, Le Marchand-Brustel Y. Potential role of protein kinase B in glucose transporter 4 translocation in adipocytes. Endocrinology. 1997;138:2005–2010. doi: 10.1210/endo.138.5.5136. [DOI] [PubMed] [Google Scholar]
- Balagopal V, Parker R. Polysomes, P bodies and stress granules: states and fates of eukaryotic mRNAs. Curr Opin Cell Biol. 2009;21:403–408. doi: 10.1016/j.ceb.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namba T, Tanaka K, Ito Y, Ishihara T, Hoshino T, Gotoh T, et al. Positive role of CCAAT/enhancer-binding protein homologous protein, a transcription factor involved in the endoplasmic reticulum stress response in the development of colitis. Am J Pathol. 2009;174:1786–1798. doi: 10.2353/ajpath.2009.080864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carriere A, Carmona MC, Fernandez Y, Rigoulet M, Wenger RH, Penicaud L, et al. Mitochondrial reactive oxygen species control the transcription factor CHOP-10/GADD153 and adipocyte differentiation: a mechanism for hypoxia-dependent effect. J Biol Chem. 2004;279:40462–40469. doi: 10.1074/jbc.M407258200. [DOI] [PubMed] [Google Scholar]
- Lee H, Lee YJ, Choi H, Ko EH, Kim JW. Reactive oxygen species facilitate adipocyte differentiation by accelerating mitotic clonal expansion. J Biol Chem. 2009;284:10601–10609. doi: 10.1074/jbc.M808742200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh K, Deng H, Fukushima A, Cai X, Boivin B, Galic S, et al. Reactive oxygen species enhance insulin sensitivity. Cell Metab. 2009;10:260–272. doi: 10.1016/j.cmet.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pessler-Cohen D, Pekala PH, Kovsan J, Bloch-Damti A, Rudich A, Bashan N. GLUT4 repression in response to oxidative stress is associated with reciprocal alterations in C/EBP alpha and delta isoforms in 3T3-L1 adipocytes. Arch Physiol Biochem. 2006;112:3–12. doi: 10.1080/13813450500500399. [DOI] [PubMed] [Google Scholar]
- Johmura Y, Watanabe K, Kishimoto K, Ueda T, Shimada S, Osada S, et al. Fad24 causes hyperplasia in adipose tissue and improves glucose metabolism. Biol Pharm Bull. 2009;32:1656–1664. doi: 10.1248/bpb.32.1656. [DOI] [PubMed] [Google Scholar]
- Karagiannides I, Tchkonia T, Dobson DE, Steppan CM, Cummins P, Chan G, et al. Altered expression of C/EBP family members results in decreased adipogenesis with aging. Am J Physiol Regul Integr Comp Physiol. 2001;280:R1772–R1780. doi: 10.1152/ajpregu.2001.280.6.R1772. [DOI] [PubMed] [Google Scholar]
- Tomaru U, Takahashi S, Ishizu A, Miyatake Y, Gohda A, Suzuki S, et al. Decreased proteasomal activity causes age-related phenotypes and promotes the development of metabolic abnormalities. Am J Pathol. 2012;180:963–972. doi: 10.1016/j.ajpath.2011.11.012. [DOI] [PubMed] [Google Scholar]
- Todaro GJ, Green H. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J Cell Biol. 1963;17:299–313. doi: 10.1083/jcb.17.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green H, Kehinde O. Sublines of mouse 3T3 cells that accumulate lipids. Cell. 1974;1:113–116. [Google Scholar]
- Sanchez M, Galy B, Hentze MW, Muckenthaler MU. Identification of target mRNAs of regulatory RNA-binding proteins using mRNP immunopurification and microarrays. Nat Protoc. 2007;2:2033–2042. doi: 10.1038/nprot.2007.293. [DOI] [PubMed] [Google Scholar]
- Le TT, Cheng JX. Single-cell profiling reveals the origin of phenotypic variability in adipogenesis. PLoS One. 2009;4:e5189. doi: 10.1371/journal.pone.0005189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa S, Fujita T, Shimabukuro M, Iwaki M, Yamada Y, Nakajima Y, et al. Increased oxidative stress in obesity and its impact on metabolic syndrome. J Clin Invest. 2004;114:1752–1761. doi: 10.1172/JCI21625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory: Cold Spring Harbor, NY; 1982. [Google Scholar]
- Khabar KS, Siddiqui YM, al-Zoghaibi F, al-Haj L, Dhalla M, Zhou A, et al. RNase L mediates transient control of the interferon response through modulation of the double-stranded RNA-dependent protein kinase PKR. J Biol Chem. 2003;278:20124–20132. doi: 10.1074/jbc.M208766200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.