Abstract
TP53INP1 (tumor protein 53-induced nuclear protein 1) is a tumor suppressor, whose expression is downregulated in cancers from different organs. It was described as a p53 target gene involved in cell death, cell-cycle arrest and cellular migration. In this work, we show that TP53INP1 is also able to interact with ATG8-family proteins and to induce autophagy-dependent cell death. In agreement with this finding, we observe that TP53INP1, which is mainly nuclear, relocalizes in autophagosomes during autophagy where it is eventually degraded. TP53INP1-LC3 interaction occurs via a functional LC3-interacting region (LIR). Inactivating mutations of this sequence abolish TP53INP1-LC3 interaction, relocalize TP53INP1 in autophagosomes and decrease TP53INP1 ability to trigger cell death. Interestingly, TP53INP1 binds to ATG8-family proteins with higher affinity than p62, suggesting that it could partially displace p62 from autophagosomes, modifying thereby their composition. Moreover, silencing the expression of autophagy related genes (ATG5 or Beclin-1) or inhibiting caspase activity significantly decreases cell death induced by TP53INP1. These data indicate that cell death observed after TP53INP1-LC3 interaction depends on both autophagy and caspase activity. We conclude that TP53INP1 could act as a tumor suppressor by inducing cell death by caspase-dependent autophagy.
Keywords: tumor suppressor, autophagy, p53, LC3, p62
Increased resistance to cell death participates in pancreatic cancer progression. Cells unable to undergo self elimination accumulate mutations and epigenetic modifications that in turn induce uncontrolled replication.1 Various concomitant mechanisms exist in normal cells to induce death and, as consequence, a number of known cell-death regulators are missing in cancer cells. One of the most studied is the tumor suppressor TP53, which is inactivated in >50% of pancreatic tumors.2 p53 induces cell death by both direct permeabilization of the outer mitochondrial membrane or translocation to the nucleus where it activates the transcription of several target genes. One of the p53 target genes is TP53INP1 (tumor protein 53-induced nuclear protein 1).3, 4, 5, 6 p53-dependent expression of TP53INP1 is triggered in response to several stress agents such as mutagens, ethanol, heat shock or conditions promoting reactive oxygen species formation (i.e., exposure to UV light or γ-irradiation).4, 6 TP53INP1 interacts with kinases, HIPK2 and PKCδ, which in turn phosphorylate p53 creating a positive feedback loop between p53 and TP53INP1.7, 8 Our laboratory demonstrated that TP53INP1 is a tumor suppressor on the basis of the following observations: (i) TP53INP1 deficient mice present with an increased susceptibility to tumor development; (ii) TP53INP1 is lost at very early stages of pancreatic carcinogenesis through a mechanism involving the oncogenic miR-155 microRNA and (iii) when TP53INP1 expression is restored in pancreatic cells, it suppresses xenograft growth by increasing apoptotic cell death through a caspase-dependent mechanism.3, 9, 10 More recently, in an attempt to decipher the molecular mechanism by which TP53INP1 induces cell death, we found that it interacts with a family of proteins involved in autophagy. Such interaction had already been reported for the TP53INP1 paralog TP53INP2 (also known as DOR) that shows 30% of amino-acid identity with TP53INP1. TP53INP2 interacts with the pre-autophagosomal membrane protein VMP1 allowing the recruitment of LC3 to the autophagosome.11 It was also shown that TP53INP2 regulates autophagy in Drosophila cells.12 These findings led us to hypothesize that TP53INP1 also has a role in autophagy.
Macroautophagy (termed autophagy in this manuscript) is an evolutionarily conserved process that degrades cytosolic proteins and organelles. Cellular components are engulfed into double-membraned vesicles called autophagosomes, which finally fuse to lysosomes and form autophagolysosomes.13 Autophagy is a physiological process constitutively present in cells, but it can also be induced in response to various stimuli such as nutrient starvation, endoplasmic reticulum stress, trophic factor withdrawal, genotoxic agents or cytokines.14, 15 A hallmark event in the autophagic process is the reversible conjugation of proteins of the ATG8 family to the autophagosomal membrane after an ubiquitin-like conjugation to phosphatidylethanolamine. Mammalian cells contain multiple ATG8 orthologs belonging to three subfamilies: microtubule-associated protein 1 light chain 3, γ-aminobutyric acid receptor-associated protein (GABARAP) and γ-aminobutyric acid receptor-associated protein like 2 (GABARAPL2).
Autophagy helps maintaining pools of cellular metabolites and is considered as a survival response of the cell. However, autophagosome accumulation is frequently observed within cells undergoing cell death. This observation renders controversial the link between autophagy and cell death16 as autophagy may be induced in dying cells and/or may promote cell death. In an attempt to resolve this discrepancy several studies have assessed autophagy as a potential contributor to cell death in particular when autophagy is prolonged or sustained.17, 18, 19
The implication of autophagy in oncogenesis has also been debated, some reports indicating a role in tumor persistence, whereas others suggested that it could be a cell death mechanism that, along with proliferative senescence, is a safeguard against excessive or unscheduled proliferative signals.20, 21 In support to that idea, Elgendy et al.22 recently provided an interesting example of ‘autophagic cell death' induced by oncogenic H-RasV12 in a human ovarian surface epithelial cell line.
In this paper we describe the involvement of TP53INP1 in autophagy. TP53INP1 binds to proteins of the ATG8-family and relocalizes in autophagosomes to induce autophagy-dependent cell death.
Results and Discussion
TP53INP1 interacts with GABARAP, GABARAPL2 and LC3 proteins
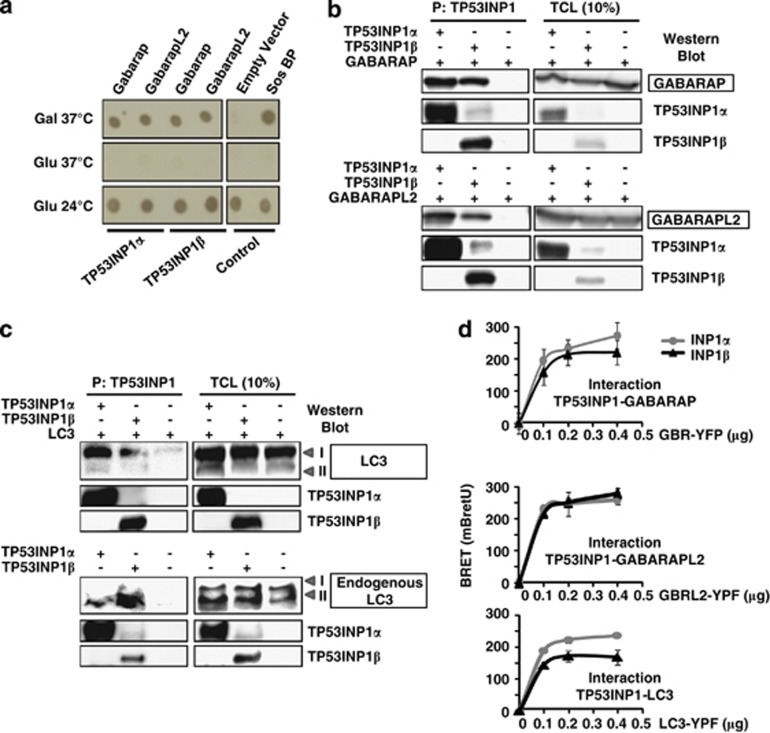
The TP53INP1 primary transcript generates by alternatively splicing two protein isoforms named TP53INP1α and TP53INP1β,4 both presenting with tumor suppressor activity. In order to identify the proteins with which they interact, both molecules were used as baits in a Sos recruitment system (SRS)-based yeast two-hybrid screening. SRS uses a temperature-sensitive Saccharomyces cerevisiae strain mutant for the Cdc25 gene (the yeast ortholog for the human Sos gene) encoding a protein that binds and activates Ras GTPase. Interaction between the prey and TP53INP1 in yeast cytoplasm leads to the translocation to the membrane of Sos protein, thereby activating Ras-signaling pathway and allowing the cdc25H yeast strain to grow at 37 °C. This system is particularly adapted to nuclear proteins because they often interact with transcriptional factors and non-specifically activate the classical Gal4 two hybrid system. We indeed observed that TP53INP1 generated such non-specific activation. Two human cDNA libraries were screened (derived from HeLa cells and from human testes). After the screening many independent clones encoding GABARAP or GABARAPL2 were identified as partners for both TP53INP1 isoforms. To confirm this result, yeasts were cotransfected with different plasmids, and growth in stringent conditions such as at 37 °C in the presence of galactose was only observed when TP53INP1α or β were cotransfected with GABARAP or GABARAPL2, indicating interaction between bait and prey proteins (Figure 1a). These results were verified in mammalian cells by coprecipitation. HEK293T cells were cotransfected with TP53INP1α or β tagged with streptavidin-binding peptide together with GABARAP-YFP or GABARAPL2-YFP. TP53INP1s were precipitated with a streptavidin-containing resin and the presence of GABARAP-YFP or GABARAPL2-YFP in the precipitate was monitored by western blot (Figure 1b). As these proteins are members of a structurally and functionally related multigenic family (mammalian homologs of ATG8), which also include LC3,23 we also assessed by the same strategy that TP53INP1α and β interact with transfected and endogenous LC3 (Figure 1c). To check whether interaction between TP53INP1s with GABARAP, GABARAPL2 and LC3 occurred in living cells we performed a Bioluminescence Resonance Energy Transfer (BRET) assay, which allows the real-time detection of protein–protein interactions. As shown in Figure 1d, the BRET signal reveals a similar level of interaction between GABARAP, GABARAPL2 and LC3 with both TP53INP1 isoforms.
Figure 1.
TP53INP1 interacts with proteins of the ATG8 family. (a) Yeast two hybrid experiments. Yeast were cotransfected with TP53INP1α-Sos or TP53INP1β-Sos and GABARAP-Myr or GABARAPL2-Myr and plated in selective medium (-leu–Ura) with galactose at 37 °C (Gal 37 °C), glucose at 37 °C (Glu 37 °C) that prevents expression of the prey and glucose at 24 °C (Glu 24 °C) for control of growth at permissive temperature. Yeast cotransfection with empty vector and Sos-binding protein (Sos BP) were used as negative and positive controls, respectively (see Materials and Methods). (b) GABARAP and GABARAPL2 coprecipitation. HEK293T cells were cotransfected with plasmids encoding GABARAP-YFP, GABARAPL2-YFP and TP53INP1α or β-NTAP. TP53INP1α or β-NTAP were precipitated with a streptavidin containing resin (P), resolved in PAGE and western blot was performed with antibodies anti-TP53INP1 (TP53INP1 precipitation control) and anti-GFP to identify presence of interactors. Western blot on total cellular extracts (TCL) served as transfection control (on the right). (c) Transfected and endogenous LC3 coprecipitation. HEK293T cells were cotransfected with plasmids encoding LC3-RFP and TP53INP1α or β-NTAP. TP53INP1α or β-NTAP were precipitated as in (b) and western blot developed with anti-LC3 antibodies. For endogenous LC3 coprecipitation, U2OS cells were transfected with plasmids encoding TP53INP1α or β-NTAP and treated with bafilomycin A1 100 nM for 6 h to stabilize endogenous LC3. TP53INP1s were precipitated as in (b) and western blot was performed with antibodies anti-TP53INP1 (TP53INP1 precipitation control) and anti-LC3. Western blot on TCL served as transfection control (on the right). (d) BRET analysis of interactions. HEK293T cells were cotransfected with 0.4 μg of TP53INP1α or β-HRluc plasmids (donor) and variable quantities (0–0.4 μg) of GABARAP-YFP GABARAPL2-YFP or LC3-YFP (acceptor) plasmids. After 1 day, the cells were harvested and distributed in 96-well microplates. On the following day, the Rluc substrate coelenterazin was added 15 min before reading. Data represent the BRET ratio in milli-BRET units (mBretU) as described in Materials and Methods and are expressed as the mean±S.D. of results from three independent experiments
TP53INP1 colocalizes with LC3 in autophagosomes
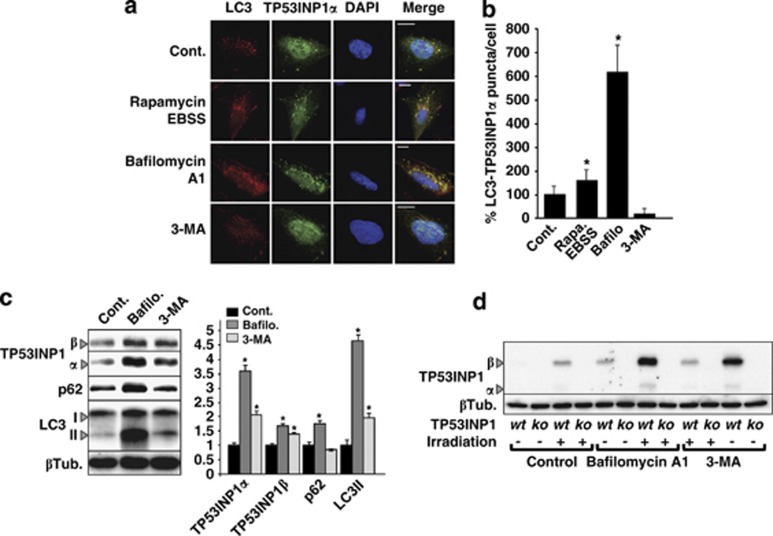
TP53INP1s are mostly nuclear proteins and LC3 is mainly cytoplasmic. In order to determine the subcellular localization of TP53INP1-LC3 interaction, the osteosarcoma U2OS cell line was modified to express a TP53INP1α-GFP fusion protein upon ponasterone A treatment.3 As shown in Figure 2a, confocal microscopic analysis reveals substantial colocalization of TP53INP1α-GFP and endogenous LC3 in cytoplasmic puncta. Because LC3 is localized on autophagosomes, we made the hypothesis that this puncta correspond to autophagosomes. To check this point, we induced autophagy by treating the cells with rapamycin and Earle's balanced salt solution (EBSS) medium or inhibited autophagy with bafilomycin A1 or 3-methyladenine (3-MA). As observed in Figures 2a and b, autophagy induction significantly increased TP53INP1-LC3 colocalization. In addition, treatment with and bafilomycin A1, which inhibits lysosome acidification that blocks the autophagic flux and enlarges autophagosomes, strongly increased the size and the percentage of colocalization puncta. By contrast, the inhibition of autophagosome formation by 3-MA completely prevented TP53INP1α-LC3 colocalization. In order to extend these results to the TP53INP1β isoform as well as to other cell lines, HeLa and MiaPaCa-2 cells were cotransfected and treated as before, with very similar results (Supplementary Figure 1) indicating that both TP53INP1 isoforms colocalize with LC3 in autophagosomes and that such colocalization is not specific to U2OS cells.
Figure 2.
TP53INP1 is localized in autophagosomes and degraded by autophagy. (a) Confocal microscopy of U2OS cells expressing TP53INP1α. Cells were induced for TP53INP1α expression with ponasterone A during 24 h and treated or not either with 5 μM rapamycin and EBSS medium, or 100 nM bafilomycin A1 (6 h) or 5 mM 3-MA (6 h). Cells where then fixed and analyzed for TP53INP1-LC3 colocalization by confocal microscopy. Red staining represents total endogenous LC3; green, TP53INP1α-GFP and blue, the cellular nucleus. On the right column, merge indicates colocalization of molecules. Scale bar represents 50 μm. (b) Quantification of LC3-TP53INP1-positive puncta per cell. Non-treated cells (Cont) were considered as 100%. Results represent counting of 50 cells in three different experiments expressed as the mean±S.D. (*P<0.05). (c) Western blot showing protein bands corresponding to TP53INP1α and β, p62, LC3-I and -II and β tubulin (loading control) in U2OS cells. Cells were transfected with TP53INP1α and β-GFP and treated with bafilomycin A1 (Bafilo, 100 nM during 6 h), 3-MA (5 mM during 6 h) or not treated (Cont). On the right, ImageJ quantification of bands indicates the relative level of proteins normalized against β tubulin (βTub). Values are expressed as the mean±S.D. relative to the control condition (Cont) considered as 1 (*P<0.05). (d) Western blot showing the bands corresponding to endogenous TP53INP1α and β and β tubulin (βTub, loading control) in primary MEFs, deficient (ko) or not (wt) for TP53INP1. Cells were irradiated (30 Gy) or not and treated with bafilomycin A1 (100 nM during 6 h) or 3-MA (5 mM during 6 h). All experiments in this figure were performed three times with comparable results
TP53INP1 is degraded during autophagy
Proteins that interact with LC3 are frequently degraded during autophagy.24 Moreover, many observations indicated that TP53INP1 is a short-lived protein that undergoes rapid degradation after synthesis (unpublished results). As autophagy is a prominent mechanism in protein degradation we assessed whether autophagy could be implicated in TP53INP1 degradation, by monitoring the levels of p62 and of the lipidated form of LC3 (LC3-II) in western blots. U2OS cells were transfected with plasmids encoding TP53INP1α or β and treated or not with bafilomycin A1 or 3-MA. Figure 2c shows that bafilomycin A1 and 3-MA treatments induced significant accumulation of TP53INP1α and β, suggesting that the TP53INP1 isoforms are indeed degraded during autophagy. As expected, levels of p62 and LC3-II increased after bafilomycin A1 treatment. Because these experiments were performed under TP53INP1s overexpression conditions, we decided to confirm whether endogenous TP53INP1s are also degraded during autophagy. Because mouse embryonic fibroblasts (MEFs) express very low levels of TP53INP1, gamma irradiation was used to induce its expression.4 As shown by western blot (Figure 2d), blocking autophagy with bafilomycin A1 or 3-MA in cells, irradiated or not, led to a significant accumulation of endogenous TP53INP1α and β. Altogether, these results show that TP53INP1 isoforms are not only relocalized in autophagosomes during autophagy but also degraded during this process.
TP53INP1 is implicated in autophagy
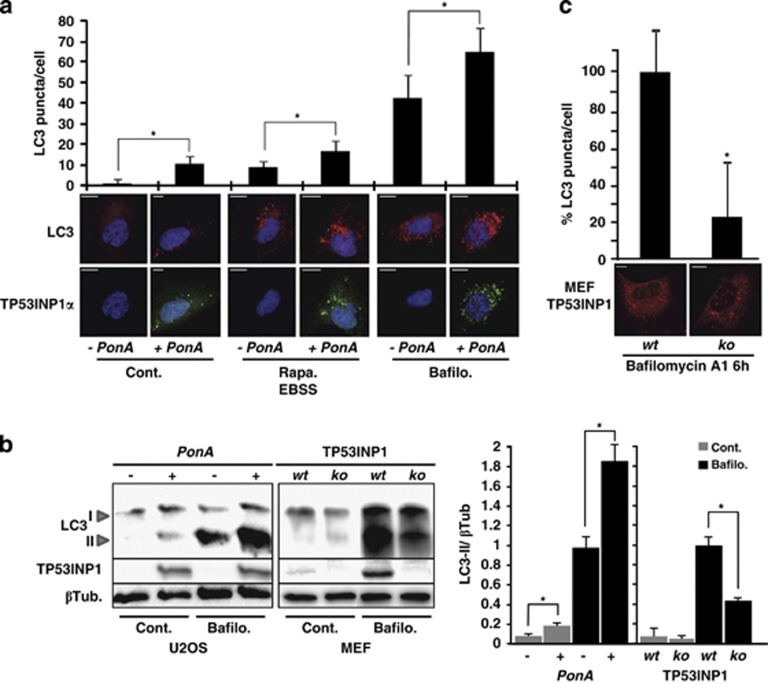
In order to analyze if TP53INP1 could also influence the autophagic process, we induced the expression of TP53INP1α-GFP by ponasterone A in the U2OS inducible model. Cells were also treated or not with rapamycin and EBSS to induce autophagy or with bafilomycin A1 to block the autophagic flux. Endogenous LC3 and TP53INP1α-GFP localizations were monitored by immunofluorescence followed by confocal microscopy. We quantified the number of LC3-positive puncta under different conditions, monitored TP53INP1 expression and assessed its presence in autophagosomes. As shown in Figure 3a, the number of autophagosomes (LC3-positive puncta) is increased in cells expressing TP53INP1, in all tested conditions. As control, U2OS cells that do not express TP53INP1 were treated or not with ponasterone A for 24 h (Supplementary Figure 2). In both conditions, the cells presented with similar levels of LC3-positive puncta, likewise upon various treatments to induce or block autophagy. These results indicate that, in our experimental conditions, ponasterone A per se does not affect autophagy. To confirm by a different approach that TP53INP1 is implicated in autophagy, LC3-II level was assessed by western blot after treatment with bafilomycin A1 of two cell lines, TP53INP1α-inducible U2OS (tumoral) and wild-type or TP53INP1-deficient MEFs (non-tumoral). As shown in Figure 3b, a significant increase in LC3-II level was observed in U2OS cells expressing TP53INP1α. In addition, TP53INP1-deficient cells presented with a significant reduction in LC3-II level upon bafilomycin A1 treatment. The densitometric quantification of the corresponding bands is shown in Figure 3b. Consistently, TP53INP1-deficient MEFs presented with a significant reduction in LC3-containing puncta compared with wild type, indicating that autophagy decreases in the absence of TP53INP1 (Figure 3c). Altogether, these results suggest that TP53INP1 facilitates autophagy.
Figure 3.
TP53INP1 is associated with autophagy. (a) TP53INP1α-inducible U2OS cells were treated or not with ponasterone A (PonA). In parallel, cells were incubated during 6 h with 10 μM rapamycin (Rapa) followed by 30 min in EBSS or with 100 nM bafilomycin A1 (Bafilo) for 6 h. Cells were then fixed and analyzed by confocal microscopy after immunocytofluorescence staining with anti-LC3 and anti-GFP antibodies. Autophagy was measured by quantification of LC3-positive dots (n=25, P<0.05). Scale bar represents 50 μm in the panels. (b) U2OS TP53INP1α inducible cells treated or not with ponasterone A and MEFs wild type (wt) or deficient (ko) for TP53INP1, were incubated or not with Bafilo for 6 h. Then the levels of LC3-I and II, TP53INP1 and β tubulin as control (βTub) were monitored by western blot in total cellular extracts. On the right column the graph represents the densitometric quantification of bands from the western blot. (c) LC3 immunostaining on MEFs wt or ko for TP53INP1 and quantification of LC3-positive puncta considering the value of wt MEFs as 100%. Results are expressed as the mean±S.D. of results from three independent experiments (*P<0.05)
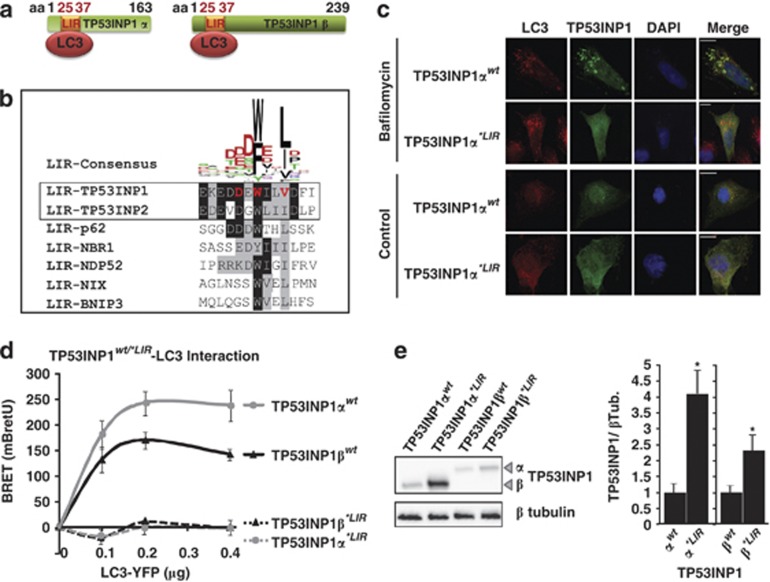
TP53INP1 interacts with LC3 via a functional LC3-interacting region (LIR) motif
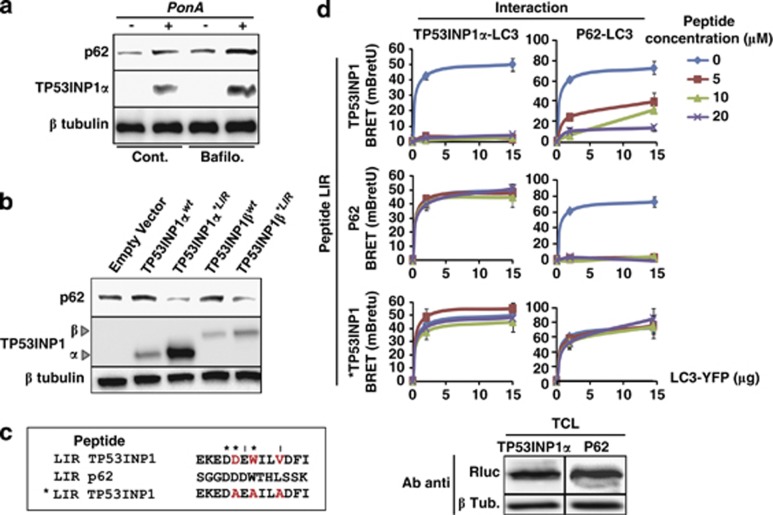
In silico analysis of TP53INP1 amino-acid sequence allowed identification of a bona fide LIR at position 25–37 on the TP53INP1α and β isoforms (Figure 4a). A very similar LIR was also identified in TP53INP2, the TP53INP1 paralog (Figure 4b). The LIR consensus sequence mediates interactions between several proteins including p62, NDP52, NBR1, Nix and BNIP3 and proteins of the LC3/GABARAP family.25 This consensus holds an aromatic (W/F) and hydrophobic (L/I) residue content required for binding the two hydrophobic pockets present on proteins of the ATG8 family (Figure 4b).26, 27 We hypothesized that interaction between TP53INP1 and LC3 could take place via this LIR motif. To check this hypothesis, we mutated three essential amino acids in the TP53INP1α and β LIR consensus to alanine (amino acids in red, Figure 4b). Resulting constructs, named TP53INP1α*LIR and TP53INP1β*LIR were tagged with GFP and their sub-cellular localization analyzed by immunofluorescence. As shown in Figure 4c for TP53INP1α and Supplementary Figure 3 for TP53INP1β, wild-type TP53INP1 isoforms but not TP53INP1α*LIR or TP53INP1β*LIR mutants colocalized with endogenous LC3 in autophagosomes. These results indicate that mutations on the LIR consensus sequence completely abolished TP53INP1 and LC3 colocalization. These results were confirmed by BRET (Figure 4d). Because TP53INP1 degradation occurs, at least in part, via autophagy, we analyzed whether or not this degradation could depend on TP53INP1-LC3 interaction. Wild-type or LIR-mutated TP53INP1α and β levels were monitored after transfection of the corresponding constructs. Western blot in Figure 4e indicates that LIR mutations stabilized TP53INP1. We conclude that TP53INP1 is degraded during autophagy after interaction with LC3 through its functional LIR.
Figure 4.
TP53INP1 interacts with LC3 via a LIR. (a) Schematic representation of LIR position in TP53INP1α and β sequences. (b) LIR sequence alignment including the human sequences for TP53INP1, TP53INP2, p62, NBR1, NDP52, NIX and BNIP3. Amino acids in red on TP53INP1 sequence were mutated to alanine in the TP53INP1α*LIR and TP53INP1β*LIR constructs. The upper sequence represents the LIR consensus established with all known sequences for LIRs in different species (adapted from Johansen and Lamark25). (c) Subcellular colocalization analysis for overexpressed wild-type and mutant TP53INP1 and endogenous LC3. U2OS cells were transfected with wild-type (TP53INP1αwt) or mutant TP53INP1α inactivated on the LIR sequence (TP53INP1α*LIR). Bafilomycin A1 was used to block the autophagic flux and enlarge specifically autophagosomes. DNA DAPI staining shows the nuclei. Scale bars represent 50 μm. (d) BRET analysis of the TP53INP1-LC3 interactions. HEK293T cells were cotransfected with 0.4 μg of wild-type (wt) or mutant (*LIR) TP53INP1α or β-HRluc (donor) and variable quantities (0–0.4 μg) of LC3-YFP (acceptor) plasmids. Data represent the BRET ratio in milli-BRET units (mBretU) as described in Materials and Methods and are expressed as the mean±S.D. of results from three independent experiments. (e) Western blot showing the TP53INP1α and β level for the wild-type and mutant LIR. HeLa cells were transfected with plasmids allowing expression of wild-type (wt) or mutant (*LIR) TP53INP1α or β. After 24 h of transfection, cell lysates were resolved in SDS-PAGE and revealed with anti-TP53INP1 antibodies or anti-β tubulin (βTub) as loading control. On the right, ImageJ quantification of bands indicates the relative level of proteins normalized against βTub. Results are expressed as the mean±S.D. of results from three independent experiments (*P<0.05)
TP53INP1 interacts with LC3 with higher affinity than p62
As shown before (Figure 3b), the expression of TP53INP1α upon ponasterone A treatment of U2OS cells results in the accumulation of LC3-II, which reflects autophagy activation. Unexpectedly, we observed accumulation of p62 in this experimental setting (Figure 5a). This observation is in apparent contradiction with the fact that p62 is known to be degraded by autophagy; p62 levels should therefore decrease. In fact, p62 acts as cargo receptor for the autophagic degradation of ubiquitinated substrates before being selectively degraded after binding LC3 through its LIR motif. We made the hypothesis that TP53INP1 interacts with LC3 with a higher affinity than p62, thus could partially displace p62 from LC3 when overexpressed, avoiding thereby p62 degradation by autophagy. To test this hypothesis we assessed whether p62 displacement by TP53INP1 is dependent on the TP53INP1 LIR sequence. The wild-type (TP53INP1α or βwt) or mutant (TP53INP1α or β*LIR) TP53INP1s isoforms were transfected and p62 level monitored by western blot (Figure 5b). Overexpression of wild-type TP53INP1s, but not of the mutants, resulted in p62 accumulation. Hence, p62 displacement depends on the integrity of the TP53INP1 LIR sequence. To confirm this observation we used synthetic peptides to perform competition assays for the TP53INP1-LC3 and p62-LC3 interactions. Competition was monitored by BRET in cellular extracts. Peptides were designed to contain the 13 most conserved amino acids of the TP53INP1 and p62 LIR consensus. As negative control we used a peptide containing the TP53INP1 LIR modified in the same three amino acids (changed to alanine) than for the TP53INP1α*LIR construct (Figures 4b and 5c). HEK293T cells were transfected with TP53INP1α-HRluc, p62-HRluc or LC3-YFP, total cellular extracts were prepared (see Materials and Methods) and the levels of fusion proteins were assayed by western blot with an anti-HRluc antibody (Figure 5d bottom). Only extracts with similar levels of TP53INP1α-HRluc, p62-HRluc were used for the competition assays. As shown in Figure 5d, TP53INP1α-LC3 interaction is completely inhibited by the LIR TP53INP1 peptide at a concentration as low as 5 μM. On the contrary, no significant inhibition is observed with the LIR p62 peptides or the mutant *LIR TP53INP1 at concentrations up to 20 μM. Interestingly, the LIR TP53INP1 peptide reduced the p62-LC3 interaction by about 50% and 15% at 10 and 20 μM, respectively. As expected, the LIR p62 completely abolished p62-LC3 interaction at 5 μM whereas the mutant peptide *LIR TP53INP1 at a concentration up to 20 μM was unable to inhibit p62-LC3 interaction. It is important to note that the LIR TP53INP1 peptide can displace the TP53INP1-LC3 interaction, whereas the LIR p62 peptide cannot. Similar results were obtained with TP53INP1β isoforms and also in similar studies addressing the interactions between the TP53INP1 isoforms and GABARAPL2 (Supplementary Figures 4 and 5). Taken together, these results shows that the TP53INP1-LC3 interaction is stronger than the p62-LC3 interaction, suggesting that TP53INP1, when present in sufficient amount, can displace p62 from LC3.
Figure 5.
TP53INP1 interacts with LC3 with higher affinity than p62. (a) Western blots showing p62 and TP53INP1 protein levels in U2OS cells expressing TP53INP1α in ponasterone A (PonA)-treated cells and (b) HeLa cells overexpressing wild-type TP53INP1 (TP53INP1α or βwt) or the mutant TP53INP1 containing a non-functional LIR (TP53INP1α or β*LIR). (c) Sequence comparison for the synthetic LIR peptides: wild-type TP53INP1 LIR (LIR TP53INP1), wild-type p62 LIR (LIR p62) and mutant TP53INP1 LIR (*LIR TP53INP1) as control. *Indicates identity and I indicates conservative changes between TP53INP1 and p62 LIRs. Amino acids in red indicate point mutations that inactivate the LIR. (d) BRET on cellular extracts. HEK293T cells were transfected with TP53INP1α-HRluc or p62-HRluc (donor) and LC3-YFP (acceptor) plasmids. After cell lysis, cellular extracts were tested by western blot with anti-Rluc antibodies for similar content in donor HRluc fusion proteins (bottom). They were then incubated with 0–20 μM of LIR TP53INP1, LIR p62 or *LIR TP53INP1 peptides and coelenterazin was added just before reading. Data represent the BRET ratio in milli-BRET units (mBretU) as described in Materials and Methods and are expressed as the mean±S.D. of results from three independent experiments
TP53INP1 promotes autophagy-dependent cell death
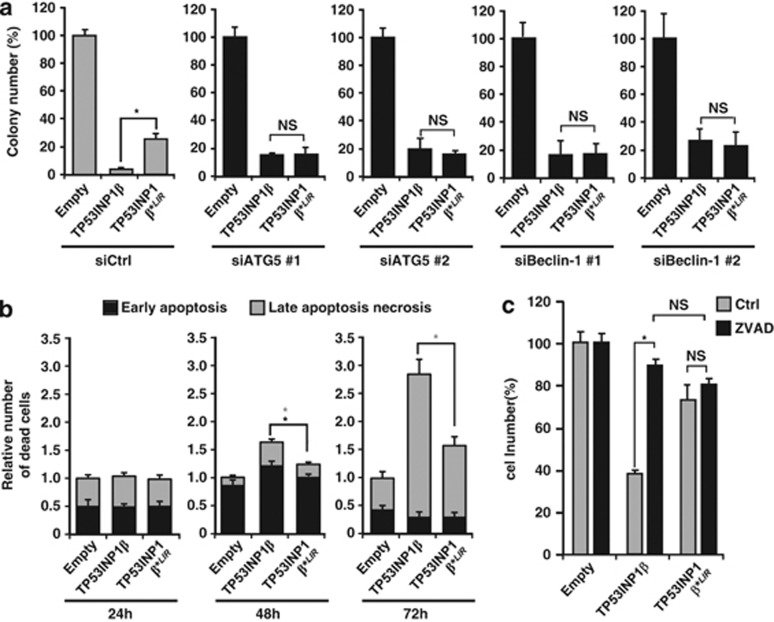
In previous works, we and others presented evidences that TP53INP1 induces cell death by apoptosis.4, 5, 6, 7 Taking into account that TP53INP1 interacts with proteins of the LC3 family and that it is involved in autophagy, we looked whether TP53INP1-associated autophagy could induce cell death. To this purpose U2OS cells were transfected with wild-type TP53INP1β, mutant TP53INP1β*LIR or an empty vector as control. All constructs were fused to YFP and epifluorescence was used to normalize transfection efficiency. The cells were grown for 12 days in selective medium and the number of clones forming colonies was counted. As shown in Figure 6a, cells transfected with wild type formed less colonies than cells transfected with the mutant TP53INP1β*LIR or with the empty vector. The lower number of colonies in cells expressing TP53INP1β suggests a significant increase in cell death, which requires the integrity of the TP53INP1 LIR. In order to assess the extent to which the observed cell death depends on autophagy, ATG5 and Beclin-1, two molecules required in the early stages of autophagosome formation, were efficiently knocked down with siRNAs before monitoring colony formation (Supplementary Figure 6). As shown in Figure 6, ATG5 or Beclin-1 knockdown abolished the difference in the number of colonies observed between the wild-type TP53INP1β and the mutant TP53INP1β*LIR. These results suggest that TP53INP1β induces cell death through its interaction with LC3 in an autophagy-dependent way. As a decreased capacity of cells to form colonies could depend on other cellular processes than cell death, we transfected U2OS cells as before and flow cytometry was performed to determine the percentage of cells showing early apoptosis, late apoptosis or necrosis (see Materials and Methods). As shown in Figure 6b, TP53INP1 expression induces a significant increase in late apoptosis and necrosis at 48 and 72 h. Transfection with the mutant TP53INP1β*LIR resulted in a significant decrease in cell death indicating that TP53INP1-LC3 interaction is required for TP53INP1-induced cell death. Interestingly early apoptosis was induced by TP53INP1 only at 48 h and a slightly but significant decrease with the mutant TP53INP1β*LIR was observed also only at 48 h. This result suggests that apoptosis could be associated with the autophagy-dependent cell death induced by TP53INP1. To further characterize this phenomenon, we evaluated the role of caspases in the TP53INP1-dependent cell death. To this end, U2OS cells were transfected with wild-type TP53INP1β, mutant TP53INP1β*LIR or with an empty vector and treated with the pan-caspase inhibitor benzyloxycarbonyl-Val-Ala-Asp (OMe) fluoromethylketone (Z-VAD-FMK) for 6 days. As shown in Figure 6c, Z-VAD-FMK suppresses the differences observed between the wild-type and the mutant TP53INP1β*LIR. All these experiments were also performed with the TP53INP1α isoform with similar results (data not shown). Altogether, these data suggest that cell death observed after TP53INP1-LC3 interaction depends on both autophagy and caspase activity suggesting that TP53INP1-associated autophagy could be a prelude to subsequent apoptosis. Interestingly, cell death induced by TP53INP1 and its degradation by autophagy actually form a negative feedback loop that in absence of a sustained stimulus would slow down the process. As consequence, TP53INP1 should be described as an inducer of autophagy-dependent cell death rather than an inducer of autophagic cell death. Further investigation is required to elucidate the relative roles of autophagy, caspases and apoptosis in this process.
Figure 6.
TP53INP1 triggers cell death by an autophagy and caspase dependent mechanism. (a) Clonogenic assays were performed in triplicate, using U2OS cells transfected with two different ATG5 or Beclin-1 siRNAs (#1 and #2) or with a scrambled siRNA (siCtrl). The cells were then transfected with TP53INP1β-YFP, TP53INP1 β*LIR-YFP or an empty vector (pEYFP-C1) as control. Transfection efficiency was normalized by YFP epifluorescence (see Materials and Methods). Results are presented as a percentage of the number of colonies observed with the empty vector. The graph represents the number of colonies present in the plate at the end of the study. ATG5 and Beclin-1 knockdown were confirmed by qPCR (see Supplementary Figure 6). (b) Flow cytometry assays with U2OS cells performed 24, 48 or 72 h after transfection with TP53INP1β, TP53INP1β*LIR or empty vector. Only YFP-positive cells were analyzed. Histograms show the relative number of dead cells compared with transfection with the empty vector for the indicated time. The black star (*) indicates a significant difference in early apoptosis (annexin V positive and 7-AAD negative, in black), the gray stars (*) indicate significant difference in late apoptosis/necrosis (annexin V and 7-AAD positive, in gray; *P<0.05) (c) U2OS cells were transfected as in (b) and treated or not for 6 days with a caspase inhibitor (Z-VAD-FMK). The graph represents the number of living cells counted at the end of the experiment as percentage of empty vector transfection. All assays were performed three times with comparable results (*P<0.05; NS, no significant difference)
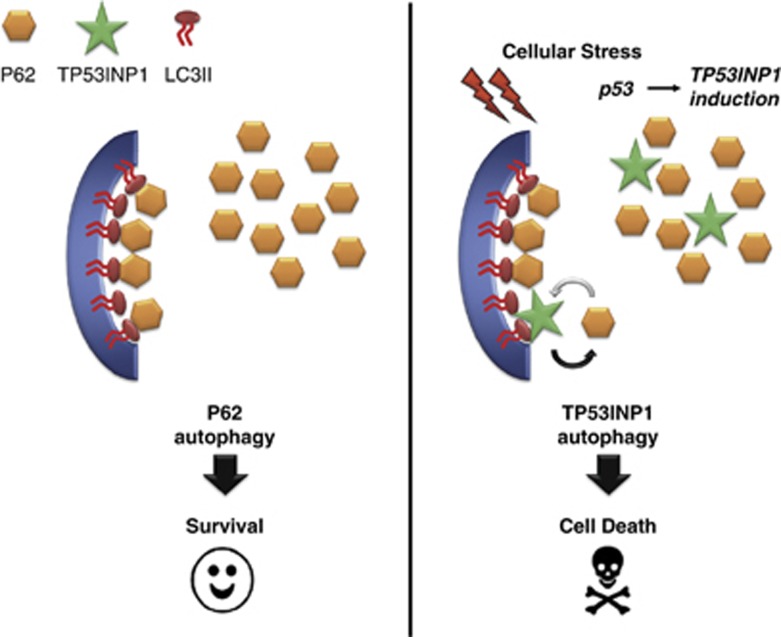
Several reports have shown that autophagy can prevent cancer development by acting as tumor suppressor.28 In this context several tumor suppressors such as p53, PTEN, TSC1, TSC2 and DAPk, were shown to be involved in autophagy29, 30, 31, 32 suggesting that autophagy modulation could be a mechanism for tumor suppression. The discovery of TP53INP1-associated autophagy reveals a novel link between p53 and this process. Interestingly, recent data associated two p53 transcriptional targets, Sestrin2 and Dram, that contribute to autophagic and apoptotic cell death.33, 34 Autophagy was supposed to have no selectivity until autophagy receptors, such as p62, NBR1 and Nix, were identified. TP53INP1 could participate in this selectivity by displacing p62 from LC3, therefore facilitating a particular type of targeted degradation. In this way, TP53INP1-mediated autophagy might induce degradation of certain proteins, for example, antiapoptotic proteins, whose depletion would lead to cell death (Figure 7). At present, the role of autophagy in cell death and cell survival remains controversial. Although several recent studies indicate that autophagy contributes to kill tumor cells,35, 36, 37 others suggest, on the contrary, that autophagy inactivation contributes to tumor cell killing.38 It is therefore critical to examine the contribution of autophagy in several tumor types, cancer stages and genetic contexts. Better characterization of autophagy actors such as TP53INP1 will also help understanding how autophagy affects cell viability or cell death in cancer.
Figure 7.
Schematic representation of the hypothetical role of TP53INP1 in autophagy-dependent cell death. Without stress, TP53INP1 is absent. In this context, p62 (in orange) acts as a cargo by binding ubiquitinated proteins and LC3-II (in red) on the membrane of the autophagosomes (in blue). This mechanism is rather considered as pro-survival. In response to stress, p53 activates TP53INP1 transcription (in green) the level of the resulting TP53INP1 protein remaining low compared with p62 protein level. Better affinity of TP53INP1 LIR sequence for LC3-II allows TP53INP1 to partially displace p62 from its interaction with LC3-II promoting autophagy-dependent cell death
Materials and Methods
Cell culture
The cells were maintained in DMEM Glutamax medium (Invitrogen, Cergy Pontoise, France) supplemented with 10% (v/v) fetal bovine serum (FBS), in a humidified atmosphere with 5% CO2 at 37 °C. The human HeLa (cervical cancer cells containing wild-type p53 inactivated by the expression of E6 protein from human papillomavirus type 18), MiaPaCa-2 (pancreatic adenocarcinoma with a mutant form of p53 R248W/R273H), HEK293T (human embryonic kidney cells containing wild-type p53) and U2OS (human osteosarcoma containing wild-type p53) cell lines were purchased from the American Type Culture Collection (Manassas, VA, USA). TP53INP1wt and TP53INP1ko primary MEFs were obtained from embryos derived from homozygous breeding at 14.5 days postcoitum (E14.5) according to standard procedure.39 MEFs were cultured in presence of penicillin (100 μ/ml) and streptomycin (100 μg/ml), and used at early passages. TP53INP1α-inducible U2OS cells were obtained by stable cotransfection with pVgRXR and pIND-TP53INP1α-EGFP vectors as indicated by Gironella et al.3 and were cultured in presence of zeocin (0.05 mg/ml) and G418 (0.2 mg/ml).
Cell treatments
All TP53INP1 transcriptional activations were done on ∼80% confluent cells, plated 24 or 48 h (for MEF) before induction. Endogenous TP53INP1 expression was induced in MEFs by 30Gy irradiation with a γ irradiation device (CisBio IBL637, Saclay, France), 6 h before cell harvesting. In TP53INP1α-inducible U2OS cells, TP53INP1α-GFP expression was induced using 10 μM ponasterone A (Invitrogen) for 24 h.
Cells were treated either with 100 nM bafilomycin A1 for 6 h (LcLabs, Woburn, MA, USA), or with 5 mM 3-MA (Sigma-Aldrich, Saint-Quentin Fallavier, France) for 6 h, or with 100 nM wortmannin for 6 h (LcLabs) or with 5 or 10 μM rapamycin for 5h30 (LcLabs) followed by 30 min in EBSS medium (Invitrogen). For caspase inhibition, cells were treated with 20 μM Z-VAD-FMK (Promega, Charbonnieres, France).
Molecular cloning and plasmid construction
Human full-length TP53INP1α and β cDNAs were subcloned into the pSos vector (Agilent, Massy, France), the pNTAP vector (Agilent), the pEYFP-C1 vector (Clontech, Saint-Germain-en-Laye, France), the pEGFP-N1 vector (Clontech) and the pHRluc-C vector (PerkinElmer Life and Analytical Sciences, Boston, MA, USA). TP53INP1*LIR mutants were obtained by PCR site directed mutagenesis on TP53INP1: D29 to A, W31 to A and V34 to A. Rat full-length LC3 cDNA was subcloned into the pERFP-C1 and the pEYFP-C1 vectors. The human full-length GABARAP and GABARAPL2 cDNAs were subcloned into the pEYFP-C1 vector (Clontech). The Human full-length p62 cDNA was ligated into the pHRluc-C.
Transfections and oligonucleotides
DNA transfection and siRNA were performed using FuGENE HD (Promega) and INTERFERin (Polyplus-transfection, Illkirch, France), respectively, according to the manufacturer's instructions. siRNA for ATG5 (#1: 5′-GCAACUCUGGAUGGGAUUG-3′ #2: 5′-GACGUUGGUAACUG ACAAA-3′), Beclin-1 (#1: 5′- CAGUUUGGCACAAUCAAUA-3′ #2: 5′-AAGAUUGAAGACACAGGAGGC-3′) and scramble (5′-UUCUCCGAACGUGUCACGU-3′) were synthesized by Eurogentec (Angers, France).
Immunoblotting
Cell were resuspended in lysis buffer (50 mM Hepes, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 10% glycerol, 1% triton X-100, 25 mM NaF, 10 μM ZnCl2, 1 mM Na3VO4, protease inhibitors cocktail (Sigma-Aldrich)). Proteins from total cell lysates were resolved by SDS-PAGE, transferred to polyvinylidene fluoride membrane, blocked in 5% non-fat milk in phosphate-buffered saline (PBS)/Tween-20, blotted and developed with antibodies anti-GFP, which cross reacts with YFP (11814460001-1 : 5000, Roche Applied Science, Meylan, France), anti-LC3 (2775-1 : 1000, Cell Signaling Technology, Saint-Quentin-en-Yvelines, France), anti-p62 (610833-1 : 500, BD Biosciences, Le Pont de Claix, France), anti-R-luc (MAB4400-1 : 1000, Millipore, Molsheim, France), anti-β tubulin (T4026-1 : 4000, Sigma-Aldrich) and anti-TP53INP1 (rat monoclonal antibody generated in our laboratory, clone F8). Secondary antibodies were purchased from Santa Cruz Biotechnology (Heidelberg, Germany): anti-rabbit horseradish peroxidase (HRP)-conjugate (sc-2004-1 : 4000), anti-mouse HRP-conjugate (sc-2005–1 : 5000) and anti-rat HRP-conjugate (sc-2006-1 : 3000). Immunoblots were developed using the Immobilon Western Chemiluminescent HRP Substrate (Millipore). Chemiluminescence was detected using Fusion FX7 device (Fisher Bioblock Scientific, Illkirch, France) and quantified by ImageJ analysis software (http://rsb.info.nih.gov/ij). The experiments for the immunoblotting were performed at least three times with comparable results.
Immunocytochemistry
Cells on glass coverslips in 12-well plates were transfected or induced for TP53INP1 expression. After 24 h, cells were treated as indicated, then fixed with 4% paraformaldehyde in PBS. Following fixation, cells were permeabilized (0.2% Triton in PBS) during 3 min, incubated for 20 min in 50 mM NH4Cl in PBS (in order to unmask epitopes) and incubated in blocking buffer (5% FBS in PBS) for 30 min before the addition of primary antibodies anti-GFP, which cross reacts with YFP (11814460001-1 : 50, Roche Applied Science) or anti-LC3 (PM036-1 : 100 for U2OS and HeLa cells and 1 : 500 for MEF, MBL, Nanterre, France) diluted in blocking buffer. Cells were incubated 1 h at room temperature and washed three times before the addition of secondary antibodies (Alexa fluor 488 anti-mouse, A-11001-1 : 500, Invitrogen, Alexa fluor 568 anti-rabbit, A-11011-1 : 500, Invitrogen). Following 1 h of incubation at room temperature, cells were stained with DAPI (Invitrogen) for 5 min and mounted in ProLong Gold antifade reagent (Invitrogen) for imaging. Quantification of LC3-positive puncta per cell was done by counting 50 cells by condition in three different experiments.
Yeast two-hybrid screening
pSos-TP53INP1 was used as bait to screen a human HeLa and Testis cDNA library (catalog no. 975212 and 975205, respectively, Agilent) constructed into the pMyr vector. The GAL1 promoter in the pMyr vector is repressed when glucose is used as a carbon source and quickly induced in the presence of galactose. After cotransfection into S. cerevisiae strain cdc25H, 5 × 106 clones were screened and several positives were identified. All clones were PCR amplified. Their sequences were identified by comparison with the GenBank repertoire. All yeast transformations were performed using the standard lithium acetate method. For CytoTrap screening, the cdc25H yeast strain was cotransformed with 40 μg of pMyr-cDNA library plasmid and 40 μg of pSos-bait recombinant plasmid. Resulting transformants were grown for 3 days at permissive temperature (24 °C) on selection medium containing glucose and additional supplements excluding leucine and uracil (SC-Leu–Ura). After replica plating onto selective minimal galactose plates, colonies that showed growth under restrictive temperature (37 °C) were considered positive clones. Colonies were patched on glucose containing medium and grown at 24 °C for 2 days. They were then replica plated onto glucose and galactose plates and grown at 37 °C. Colonies that showed growth only on galactose and not on glucose medium were selected and confirmed as true positives by a second round of screening. These clones were grown in liquid medium, yeast cell wall was disrupted by vortexing with glass beads (Sigma-Aldrich) and plasmids were isolated by theWizard Plus SV Miniprep Kit (Promega). A total of 5 μl of the plasmid preparation was used for each PCR reaction using GoTaq (Promega) and pMyr vector specific primers (forward 5′-AACCCCGGATCGGACTACTA-3′, reverse 5′-AATAAGCTCTAGAGGGCCGC-3′). Presence of a single amplicon was confirmed by agarose gel electrophoresis and sequencing was carried out by the Genome Express Sequencing Service (http://www.cogenics.com/). The interactions between TP53INP1 and GABARAP or GABARAPL2 were confirmed by transforming S. cerevisiae with the bait and prey couple and allowing the transformants to grow on synthetic dropout (SD) glucose and galactose agar plates lacking leucine and uracil (SD/glu(-LU) and SD/gal(-LU)) at the stringent temperature of 37 °C. Clones growing on SD/gal(-LU) plates but not on SD/glu(-LU) plates at 37 °C were considered as interactants. As control, no growth was observed at 37 °C in presence of glucose, which prevents the expression of prey proteins or when cells were transfected with the pMyr empty vector. The growth of yeast transfectants was verified at non-stringent conditions (glucose at 24 °C) or in presence of a positive control Sos-binding protein that interacts with the Sos tag in the N-terminal of the bait protein.
Coprecipitation assay
HEK293T or U2OS cells in six-well plates were cotransfected by TP53INP1α or β-NTAP vector and LC3-RFP, GABARAP-YFP or GABARAPL2-YFP vector. After 24 h of transfection, cells were harvested and lysed and protein complexes were purified using streptavidin containing resin according to the manufacturer's instructions of InterPlay Mammalian TAP Purification Kit (Agilent). Then, proteins were resolved by SDS-PAGE and immunobloted.
BRET assay
BRET in vivo (in living cells)
HEK293T cells in 12-well culture plates were cotransfected with 0.4 μg of BRET donor TP53INP1α or β-HRluc vectors, and 0–0.4 μg of BRET acceptor GABARAP-YFP, GABARAPL2-YFP or LC3-YFP vectors. The empty vector (pEYFP-C) was used to equalize DNA amounts to 0.8 μg in each sample. After 6 h, cells were harvested and distributed in 96-well microplate (20 000 cells per well). On the following day, the cell-permeable Rluc substrate coelentherazin-h (Promega) was added in culture medium to a final concentration of 5 μM and cells were incubated 15 min at 37 °C before reading.
BRET in vitro (with cellular extracts)
HEK293T cells were transfected separately with a BRET donor (TP53INP1α or β-HRluc or p62-HRluc vector) or a BRET acceptor (GABARAP-YFP, GABARAPL2-YFP or LC3-YFP vector). After 48 h, total cellular proteins were extracted with CelLytic NuCLEAR Extraction Kit (Sigma-Aldrich) according to manufacturer's instructions. A total of 3 μg of lysate containing a BRET donor were mixed with 0–40 μg of lysate containing a BRET acceptor and incubated during 30 min at 4 °C. Coelentherazin-h (Promega) was added to a final concentration of 5 μM in lysis buffer, just before reading. For BRET competition assays, cellular extracts containing BRET donor or acceptor were pre-incubated during 2 h at 4 °C with 0–20 μM of synthetic peptides which mimic the TP53INP1-LIR (EKEDDEWILVDFI) or the p62-LIR (SGGDDDWTHLSSK) or a control peptide (EKEDAEAILADFI), and then mixed as indicated above. Peptides were synthesized by GeneCust (Dudelange, Luxembourg).
Readings were done using a LB 941 Tristar reader (Berthold France SA, Thoiry, France), with signal detection in the 470–490 nm (donor) and 520–540 nm (acceptor). BRET signal represents the BRET ratio of the Rluc and EYFP expression constructs against the BRET ratio for the Rluc expression construct alone. To assess signal variation, the BRET values were determined by using the following equation, expressed in milli-BRET unit:
 |
where E0 corresponds to the ratio 530 nm acceptor signal per 480 nm donor signal obtained with the HRluc construct alone in the same experiment. All experiments were performed at least three times with comparable results.
Clonogenicity assay
U2OS cells in 10 cm dishes were sequentially transected with two different ATG5 or Beclin-1 siRNAs or with a scramble siRNA, then with TP53INP1wt-YFP, TP53INP1*LIR-YFP or empty pEYFP-C1 as control. Transfection efficiency was controlled by YFP fluorescence. After 24 h of transfection, cells were harvested and distributed in six-well plates (60 000 YFP fluorescent cells per well), with G418 (0.8 mg/ml) as selection pressure. The remaining cells were saved for RNA extraction and quantitative PCR to control ATG5 or Beclin-1 inhibition, as indicated below. Cells were allowed to grow during 12 days under selection pressure and stained with crystal violet to quantify surviving colonies. These experiments were performed three times with comparable results.
RNA extraction and quantitative PCR
Total RNA was obtained from U2OS cells 24 h after siRNA transfection, using TRIzol (Invitrogen). cDNA synthesis was performed using Improm-II Kit (Promega) according to the manufacturer's protocol. The ATG5 mRNA was assayed using the primers sense: 5′-TGACGTTGGTAACTGACAAAGT-3′ antisense: 5′-GTGATGTTCCAAGGAAGAGC-3′, the Beclin-1 mRNA with the primers sense: 5′-ACCGTGTCACCATCCAGGAA-3′ antisense: 5′-GAAGCTGTTGGCACTTTCTGT-3′ and the primers sense: 5′-ATGGTCAACCCCACCGTGT-3′ antisense: 5′-TTCTTGCTGTCTTTGGAACTTTGTC-3′ were used to detect cyclophilin (house-keeping gene). Real-time q-PCR was performed in a MX3005P machine (Agilent) using the SYBR Premix Ex Taq and ROX reference dye (Takara Bio, Otsu, Shiga, Japan).
Flow cytometry analysis
U2OS cells were transfected with TP53INP1-YFP, TP53INP1*LIR-YFP or empty pEYFP as control. After 24, 48 or 72 h after transfection, cells were stained with annexin V-APC (BD Biosciences) and 7-aminoactinomycin (7-AAD; Sigma-Aldrich) in binding buffer (0.1 M Hepes pH 7.4, 1.4 M NaCl and 25 mM CaCl2) during 15 min at room temperature in the dark. At the end of incubation, cells were analyzed on a FACSCalibur flow cytometer (BD Biosciences). Flow cytometry was performed to determine the percentage of dead cells discriminating cells with early apoptosis (annexin V+, 7-AAD−) and necrotic or late apoptotic cells (annexin V+, 7-AAD+).
Image acquisition and manipulation
Fluorescent images were captured using a Zeiss LSM 510 confocal microscope (Carl Zeiss, Le Pecq, France). Base images were captured and exported as TIFF files, figures were created using Adobe Photoshop 11. No manipulations other than global contrast and brightness adjustments were performed on the images obtained.
Statistical analysis
Statistical analyses were performed using Microsoft Excel. Data are given as mean±S.D., with the number of experiments indicated. Statistical significance was established by Student's t-tests.
Acknowledgments
This work was supported in part by the Institut National de la Santé et de la Recherche Médicale (INSERM), the Centre National de la Recherche Scientifique and grants from the Institut National du Cancer, Cancéropôle PACA, La Ligue Nationale Contre le Cancer, Association pour la Recherche sur le Cancer, the Agence Nationale de la Recherche and the ECOS SUD programme. MS was supported by Ministère de la Recherche et de la Technologie, SP by Association pour la Recherche sur le Cancer. We acknowledge P Spoto for technical help, F Ladelfa for critically reading the manuscript and J-C Dagorn, P Soubeyran, P Pierre and E Gatti for helpful comments.
Glossary
- TP53INP1
tumor protein 53-induced nuclear protein 1
- LIR
LC3-interacting region
- BRET
Bioluminescence Resonance Energy Transfer
- EBSS
Earle's balanced salt solution
- MEF
mouse embryonic fibroblast
- Z-VAD-FMK
benzyloxycarbonyl-Val-Ala-Asp (OMe) fluoromethylketone
- 7-AAD
7-aminoactinomycin D
- PBS
phosphate-buffered saline
- HRP
horseradish peroxidase
- FBS
fetal bovine serum
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by H-U Simon
Supplementary Material
References
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Soussi T, Beroud C. Significance of TP53 mutations in human cancer: a critical analysis of mutations at CpG dinucleotides. Hum Mutat. 2003;21:192–200. doi: 10.1002/humu.10189. [DOI] [PubMed] [Google Scholar]
- Gironella M, Seux M, Xie MJ, Cano C, Tomasini R, Gommeaux J, et al. Tumor protein 53-induced nuclear protein 1 expression is repressed by miR-155, and its restoration inhibits pancreatic tumor development. Proc Natl Acad Sci USA. 2007;104:16170–16175. doi: 10.1073/pnas.0703942104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasini R, Samir AA, Vaccaro MI, Pebusque MJ, Dagorn JC, Iovanna JL, et al. Molecular and functional characterization of the stress-induced protein (SIP) gene and its two transcripts generated by alternative splicing. SIP induced by stress and promotes cell death. J Biol Chem. 2001;276:44185–44192. doi: 10.1074/jbc.M105647200. [DOI] [PubMed] [Google Scholar]
- Tomasini R, Samir AA, Pebusque MJ, Calvo EL, Totaro S, Dagorn JC, et al. P53-dependent expression of the stress-induced protein (SIP) Eur J Cell Biol. 2002;81:294–301. doi: 10.1078/0171-9335-00248. [DOI] [PubMed] [Google Scholar]
- Okamura S, Arakawa H, Tanaka T, Nakanishi H, Ng CC, Taya Y, et al. p53DINP1, a p53-inducible gene, regulates p53-dependent apoptosis. Mol Cell. 2001;8:85–94. doi: 10.1016/s1097-2765(01)00284-2. [DOI] [PubMed] [Google Scholar]
- Tomasini R, Samir AA, Carrier A, Isnardon D, Cecchinelli B, Soddu S, et al. TP53INP1s and homeodomain-interacting protein kinase-2 (HIPK2) are partners in regulating p53 activity. J Biol Chem. 2003;278:37722–37729. doi: 10.1074/jbc.M301979200. [DOI] [PubMed] [Google Scholar]
- Yoshida K, Liu H, Miki Y. Protein kinase C delta regulates Ser46 phosphorylation of p53 tumor suppressor in the apoptotic response to DNA damage. J Biol Chem. 2006;281:5734–5740. doi: 10.1074/jbc.M512074200. [DOI] [PubMed] [Google Scholar]
- Gommeaux J, Cano C, Garcia S, Gironella M, Pietri S, Culcasi M, et al. Colitis and colitis-associated cancer are exacerbated in mice deficient for tumor protein 53-induced nuclear protein 1. Mol Cell Biol. 2007;27:2215–2228. doi: 10.1128/MCB.01454-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cano CE, Gommeaux J, Pietri S, Culcasi M, Garcia S, Seux M, et al. Tumor protein 53-induced nuclear protein 1 is a major mediator of p53 antioxidant function. Cancer Res. 2009;69:219–226. doi: 10.1158/0008-5472.CAN-08-2320. [DOI] [PubMed] [Google Scholar]
- Nowak J, Archange C, Tardivel-Lacombe J, Pontarotti P, Pebusque MJ, Vaccaro MI, et al. The TP53INP2 protein is required for autophagy in mammalian cells. Mol Biol Cell. 2009;20:870–881. doi: 10.1091/mbc.E08-07-0671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauvezin C, Orpinell M, Francis VA, Mansilla F, Duran J, Ribas V, et al. The nuclear cofactor DOR regulates autophagy in mammalian and Drosophila cells. EMBO Rep. 2010;11:37–44. doi: 10.1038/embor.2009.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer AJ, Codogno P. Regulation and role of autophagy in mammalian cells. Int J Biochem Cell Biol. 2004;36:2445–2462. doi: 10.1016/j.biocel.2004.02.002. [DOI] [PubMed] [Google Scholar]
- Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- Lum JJ, Bauer DE, Kong M, Harris MH, Li C, Lindsten T, et al. Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell. 2005;120:237–248. doi: 10.1016/j.cell.2004.11.046. [DOI] [PubMed] [Google Scholar]
- Kroemer G, Levine B. Autophagic cell death: the story of a misnomer. Nat Rev Mol Cell Biol. 2008;9:1004–1010. doi: 10.1038/nrm2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Shimizu S, Kanaseki T, Mizushima N, Mizuta T, Arakawa-Kobayashi S, Thompson CB, et al. Role of Bcl-2 family proteins in a non-apoptotic programmed cell death dependent on autophagy genes. Nat Cell Biol. 2004;6:1221–1228. doi: 10.1038/ncb1192. [DOI] [PubMed] [Google Scholar]
- Hoyer-Hansen M, Bastholm L, Szyniarowski P, Campanella M, Szabadkai G, Farkas T, et al. Control of macroautophagy by calcium, calmodulin-dependent kinase kinase-beta, and Bcl-2. Mol Cell. 2007;25:193–205. doi: 10.1016/j.molcel.2006.12.009. [DOI] [PubMed] [Google Scholar]
- Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112:1809–1820. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgendy M, Sheridan C, Brumatti G, Martin SJ. Oncogenic Ras-induced expression of Noxa and Beclin-1 promotes autophagic cell death and limits clonogenic survival. Mol Cell. 2011;42:23–35. doi: 10.1016/j.molcel.2011.02.009. [DOI] [PubMed] [Google Scholar]
- Mann SS, Hammarback JA. Gene localization and developmental expression of light chain 3: a common subunit of microtubule-associated protein 1A(MAP1A) and MAP1B. J Neurosci Res. 1996;43:535–544. doi: 10.1002/(SICI)1097-4547(19960301)43:5<535::AID-JNR3>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- Johansen T, Lamark T. Selective autophagy mediated by autophagic adapter proteins. Autophagy. 2011;7:279–296. doi: 10.4161/auto.7.3.14487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura Y, Kumanomidou T, Sou YS, Mizushima T, Ezaki J, Ueno T, et al. Structural basis for sorting mechanism of p62 in selective autophagy. J Biol Chem. 2008;283:22847–22857. doi: 10.1074/jbc.M802182200. [DOI] [PubMed] [Google Scholar]
- Noda NN, Ohsumi Y, Inagaki F. Atg8-family interacting motif crucial for selective autophagy. FEBS Lett. 2010;584:1379–1385. doi: 10.1016/j.febslet.2010.01.018. [DOI] [PubMed] [Google Scholar]
- Levine B. Cell biology: autophagy and cancer. Nature. 2007;446:745–747. doi: 10.1038/446745a. [DOI] [PubMed] [Google Scholar]
- Gozuacik D, Kimchi A. DAPk protein family and cancer. Autophagy. 2006;2:74–79. doi: 10.4161/auto.2.2.2459. [DOI] [PubMed] [Google Scholar]
- Sakamoto Y, Kato S, Takahashi M, Okada Y, Yasuda K, Watanabe G, et al. Contribution of autophagic cell death to p53-dependent cell death in human glioblastoma cell line SF126. Cancer Sci. 2011;102:799–807. doi: 10.1111/j.1349-7006.2011.01857.x. [DOI] [PubMed] [Google Scholar]
- Galluzzi L, Kepp O, Kroemer G. TP53 and MTOR crosstalk to regulate cellular senescence. Aging (Albany NY) 2010;2:535–537. doi: 10.18632/aging.100202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno T, Sato W, Horie Y, Komatsu M, Tanida I, Yoshida M, et al. Loss of Pten, a tumor suppressor, causes the strong inhibition of autophagy without affecting LC3 lipidation. Autophagy. 2008;4:692–700. doi: 10.4161/auto.6085. [DOI] [PubMed] [Google Scholar]
- Maiuri MC, Malik SA, Morselli E, Kepp O, Criollo A, Mouchel PL, et al. Stimulation of autophagy by the p53 target gene Sestrin2. Cell Cycle. 2009;8:1571–1576. doi: 10.4161/cc.8.10.8498. [DOI] [PubMed] [Google Scholar]
- Crighton D, Wilkinson S, O'Prey J, Syed N, Smith P, Harrison PR, et al. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell. 2006;126:121–134. doi: 10.1016/j.cell.2006.05.034. [DOI] [PubMed] [Google Scholar]
- Puissant A, Robert G, Fenouille N, Luciano F, Cassuto JP, Raynaud S, et al. Resveratrol promotes autophagic cell death in chronic myelogenous leukemia cells via JNK-mediated p62/SQSTM1 expression and AMPK activation. Cancer Res. 2010;70:1042–1052. doi: 10.1158/0008-5472.CAN-09-3537. [DOI] [PubMed] [Google Scholar]
- Tomic T, Botton T, Cerezo M, Robert G, Luciano F, Puissant A, et al. Metformin inhibits melanoma development through autophagy and apoptosis mechanisms. Cell Death Dis. 2011;2:e199. doi: 10.1038/cddis.2011.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar M, Carracedo A, Salanueva IJ, Hernandez-Tiedra S, Lorente M, Egia A, et al. Cannabinoid action induces autophagy-mediated cell death through stimulation of ER stress in human glioma cells. J Clin Invest. 2009;119:1359–1372. doi: 10.1172/JCI37948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Wang X, Contino G, Liesa M, Sahin E, Ying H, et al. Pancreatic cancers require autophagy for tumor growth. Genes Dev. 2011;25:717–729. doi: 10.1101/gad.2016111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey M, Sands AT, Weiss RS, Hegi ME, Wiseman RW, Pantazis P, et al. In vitro growth characteristics of embryo fibroblasts isolated from p53-deficient mice. Oncogene. 1993;8:2457–2467. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.