Abstract
Our aim was to elucidate the physiological role of calpains (CAPN) in mammary gland involution. Both CAPN-1 and -2 were induced after weaning and its activity increased in isolated mitochondria and lysosomes. CAPN activation within the mitochondria could trigger the release of cytochrome c and other pro-apoptotic factors, whereas in lysosomes it might be essential for tissue remodeling by releasing cathepsins into the cytosol. Immunohistochemical analysis localized CAPNs mainly at the luminal side of alveoli. During weaning, CAPNs translocate to the lysosomes processing membrane proteins. To identify these substrates, lysosomal fractions were treated with recombinant CAPN and cleaved products were identified by 2D-DIGE. The subunit b2 of the v-type H+ ATPase is proteolyzed and so is the lysosomal-associated membrane protein 2a (LAMP2a). Both proteins are also cleaved in vivo. Furthermore, LAMP2a cleavage was confirmed in vitro by addition of CAPNs to isolated lysosomes and several CAPN inhibitors prevented it. Finally, in vivo inhibition of CAPN1 in 72-h-weaned mice decreased LAMP2a cleavage. Indeed, calpeptin-treated mice showed a substantial delay in tissue remodeling and involution of the mammary gland. These results suggest that CAPNs are responsible for mitochondrial and lysosomal membrane permeabilization, supporting the idea that lysosomal-mediated cell death is a new hallmark of mammary gland involution.
Keywords: apoptosis, lysosomes, mammary gland involution, calpains, Lamp2
The mammary gland is a highly dynamic tissue that undergoes several developmental stages. During pregnancy and lactation the epithelial cells proliferate and differentiate to produce milk. At the end of lactation there is a decrease in milk macronutrient synthesis, loss of secretory epithelial cells, collapse of alveolar structures and extensive tissue remodeling with re-establishment of the fat pad. This post-lactational regression, termed involution, provides an excellent ‘in vivo' model to study the molecular mechanisms underlying cell death and tissue remodeling.
Over the last decade, genomic approaches have shed light into the key regulators of the involution phase,1, 2, 3 with nuclear factor kappaB (NF-κB) playing a key role in both mammary gland development and involution.4, 5, 6 Interestingly, calpain (CAPN)-1 and -2 were found among the 268 NF-κB target genes identified by chromatin immunoprecipitation (ChIP)-on-chip experiments in 48-h-weaned glands.7
CAPNs are a family of calcium-dependent cysteine proteases. The ubiquitously expressed CAPN1 and 2 are the best-characterized isoforms. Both enzymes are heterodimers, consisting of a small regulatory subunit (30 kDa) encoded by Capn4, and a large catalytic subunit (80 kDa) encoded by the Capn1 or Capn2 genes.8 These two main isoforms differ in the calcium concentrations required for their activation ‘in vitro' (μM or mM, respectively).8 Both proteases co-exist with the endogenous specific inhibitor, calpastatin; however, upon calcium increase CAPNs become activated and destroy their inhibitor.9 More than 100 proteins have been shown to be CAPN targets ‘in vitro'. Regardless of this wide spectrum of substrates, CAPN-mediated protein cleavage is rather specific and occurs at a limited number of sites.10 Therefore, under physiological conditions, CAPNs most probably regulate the protein's biological functions rather than protein degradation per se.11
CAPNs have been implicated in apoptosis,12, 13 being involved in the mitochondrial pathway. In this sense, Bcl-2 and Bid are cleaved by activated CAPNs, promoting cytochrome c release and induction of the intrinsic apoptotic pathway.12 Besides, although CAPNs are considered cytosolic proteases, CAPN1 is enriched in the mitochondrial fraction.14, 15 Upon calcium activation, CAPN1 cleaves substrates in the inner mitochondrial membrane, such as the Na+/Ca+2 exchanger NCX3, causing mitochondrial calcium overload16 and releasing cytochrome c and apoptosis-inducing factor.17
On the other hand, CAPNs have also been involved in lysosomal rupture and the resultant release of lysosomal cathepsins into the cytoplasm during neuronal cell death.18, 19 Although the molecular mechanisms underlying this process are not well understood, the ‘CAPN–cathepsin cascade' begins with CAPN1-mediated cleavage of heat shock protein 70.1 at the luminal side and disruption of the lysosomal membrane. Consequently, the hydrolytic enzymes typically sequestered within lysosomes, primarily the cathepsins, are released into the cytosol.20
Focusing on mammary gland involution, we should point out that calcium concentration increases in mammary tissue during weaning21 and this could have a pivotal role in apoptosis13 and mammary gland remodeling. Therefore, it is likely that CAPNs become activated during post-lactational regression. In the current study, we examined the expression pattern and activity of ubiquitous CAPNs in the involuting mammary gland. The aim of this work was to elucidate the biological importance of CAPN-mediated cleavage of new substrates from lysosomal membranes, reinforcing the new theory that lysosomal membrane permeabilization is crucial for mammary gland involution.22
Results
NF-κB binds the promoter of CAPN1 and 2 genes during post-lactational mammary gland involution
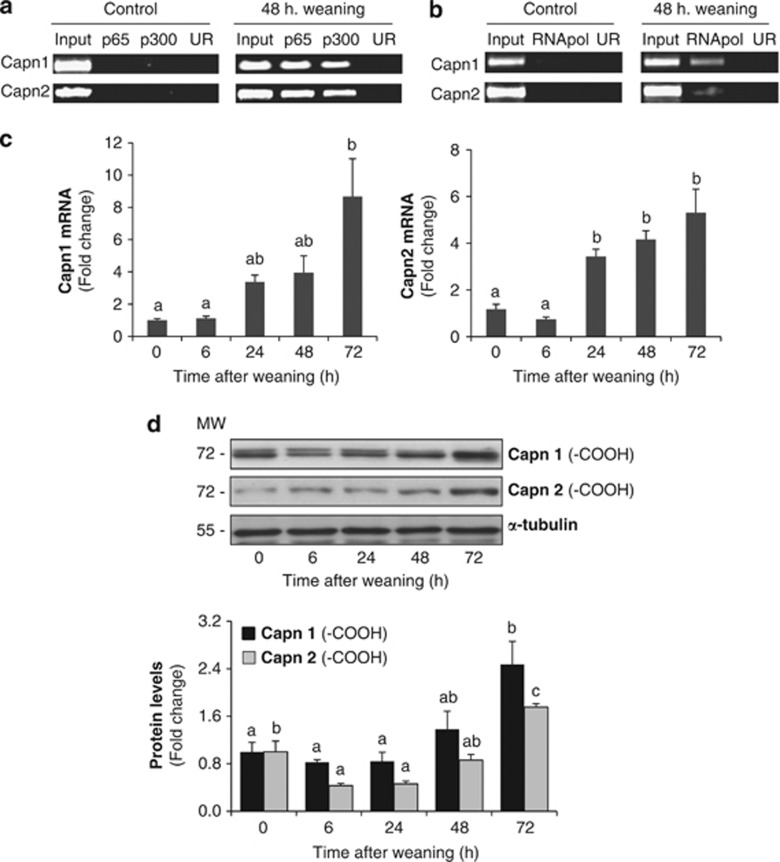
NF-κB is a key transcription factor that becomes activated early during involution and remains activated thereafter.5, 6 To identify new NF-κB target genes, our group performed ChIP-on-chip experiments during weaning; among those genes calpain-1 and calpain-2 were identified.7 ChIP showed binding of NF-κB (p65 subunit) and its coactivator p300 to capn1 and capn2 promoters after 48 h of weaning, while no NF-κB DNA binding was detectable at the peak of lactation (Figure 1a). To determine if NF-κB binding activity was involved in the current transcription of these genes, the same cross-linked chromatin samples were used for RNA polymerase II (RNApol II) ChIP.3 As shown in Figure 1b, no signal was noted in lactation; however, after weaning, the RNApol II was bound to the coding region of both genes.
Figure 1.
NF-κB activation regulates calpain expression during weaning. (a) Representative ChIP assay from control lactating and 48-h-weaned mammary glands showing the in vivo association of NF-κB (subunit p65) and its transcriptional coactivator p300 to the Capn1 and Capn2 promoters during weaning. (b) RNA pol II ChIP assay showing the presence of the RNApol II in the coding region of both Capn1 and Capn2. Chromatin was immunoprecipitated with anti-p65, anti-p300 or anti-RNApol II antibodies. An aliquot of the input chromatin and an unrelated antibody (UR) as negative control are also shown. Primers specific to the promoter or coding region for Capn1 and Capn2 genes were used to amplify the DNA isolated from the ChIP assay. (c) Real-time PCR was performed on both genes, Capn1 and Capn2, at different times after weaning (0, 6, 24, 48 and 72 h). Each PCR was normalized against the housekeeping gene 18S. Results are shown as mean±S.E.M. (n=3). (d) Western blot analysis for CAPN1 and CAPN2 was performed in protein mammary extracts at day 10 of lactation (0 h) and 6, 24, 48 and 72 h of involution. In CAPN1, besides the bands at 80 kDa, bands at smaller molecular weights were also detected. Three separate mice were used per developmental stage. Equal loading was confirmed by reprobing the blot against α-tubulin. The intensity of the bands was measured by densitometry using ImageJ, and quantified against α-tubulin. In (c) and (d) ANOVA was performed for the statistical analysis, where different superscript letters indicate significant differences, P<0.05; the letter ‘a' always represents the lowest value within the group
Quantitative PCR (qPCR) was performed to determine mRNA steady-state levels of capn1 and capn2 during weaning (Figure 1c), demonstrating that their expression was increased after 24 h of weaning, reaching the highest levels after 72 h of involution. We also found an increased expression of capn4, the gene encoding the small regulatory subunit of CAPNs, at different times after weaning (Supplementary Figure S1). These results were in agreement with the increased protein levels observed for both CAPN1 and 2 during involution (Figure 1d). It is noteworthy that for CAPN1, besides the expected 80-kDa band, a band with smaller molecular weight also appeared, indicating the aminoprocessing of this CAPN.
CAPN activation in involuting mammary gland
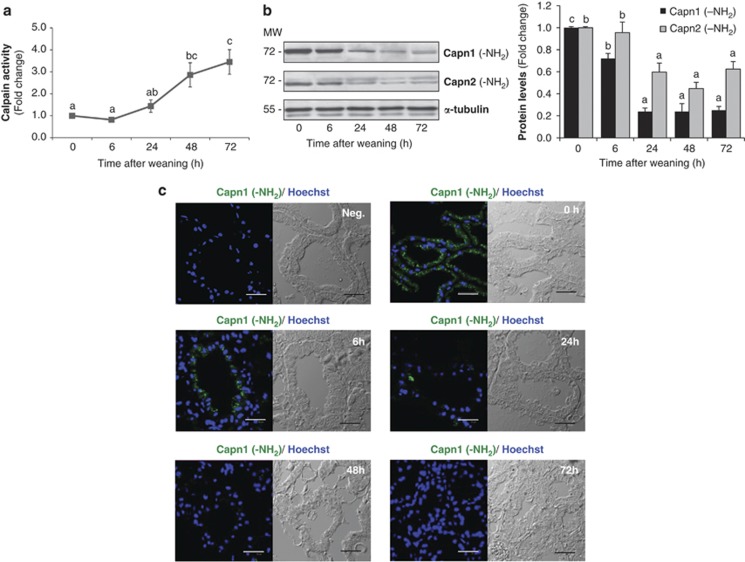
As a first step to investigate the involvement of CAPNs in mammary gland involution, we evaluated CAPN activity at different times after weaning (Figure 2a). Cleavage of the CAPN substrate Suc-LLVY-AMC was statistically significant after 48 h of weaning, reaching a fourfold induction after 72 h of weaning compared with mice at the peak of lactation. CAPNs require Ca2+ for their activation and intracellular calcium levels rise within the involuting mammary gland as early as 6 h after forced weaning (Supplementary Figure S2a). Our PCR and microarray experiments performed in lactating rats (Supplementary Figure S2b) or mice (Supplementary Figure S2c and Supplementary Table 1) confirmed the abrogation of the plasma membrane calcium transporter Atp2b2 (plasma membrane calcium ATPase (PMCA2)) after 24 h of weaning and the downregulation of the secretory pathway Ca2+–ATPases 1 and 2 (Atp2c1 and Atp2c2) at the mRNA levels. All these results are in agreement with previous studies in which loss of PMCA2 expression raised intracellular calcium levels and sensitized mammary epithelial cells to apoptosis.23
Figure 2.
Calpain activation in mammary gland involution. (a) Protease activity was measured in mammary gland samples from control lactating (0 h) and involuting mice (6, 24, 48 and 72 h) using a fluorogenic assay based on the cleavage of the calpain substrate Suc-LLY-AFC. Results are shown as mean±S.E.M. (n=3). (b) Representative immunoblot for the amino-terminal domain of both CAPN1 and CAPN2 during involution of the mammary gland. α-Tubulin was used for normalization. In (a) and (b) ANOVA was performed for the statistical analysis, where different superscript letters indicate significant differences, P<0.05; the letter ‘a' always represents the lowest value within the group. (c) Representative immunofluorescence staining for calpain 1 amino-terminal epitope (green) in the mammary gland of control (0 h) and weaned mice (0, 6, 24, 48 and 72 h). Autofluorescence and nonspecific binding was reduced using samples incubated with dilution buffer or secondary antibody (negative control). Cell nuclei are shown in blue (Hoeschst-33342). Bright-field images were included simultaneously to visualize the exact localization of immunofluorescence in mammary gland structure. Scale bar: 30 μm
In the presence of Ca2+, autoproteolysis of CAPN1, 2 and 4 occurs at the amino-terminal domain, generating cleaved activated forms.24 To show CAPN activation, western blots for CAPN1 and 2 were performed, using antibodies raised against the NH2-terminus epitope. As seen in Figure 2b, the immunoreactive band of 80 kDa disappeared after 24–48 h of weaning, for both CAPN1 and 2, indicating that autolysis was produced. Besides, immunostaining for NH2-CAPN1 showed no signal in histological samples from 24, 48 and 72 h of weaning (Figure 2c). Interestingly, we observed that CAPN1 was mainly confined to the apical side of epithelial cells during lactation (Supplementary Figure S2d), likely where the main transporter PMCA2 is localized, pumping calcium into milk in the alveolar lumen.23
CAPNs and mitochondrial cell death
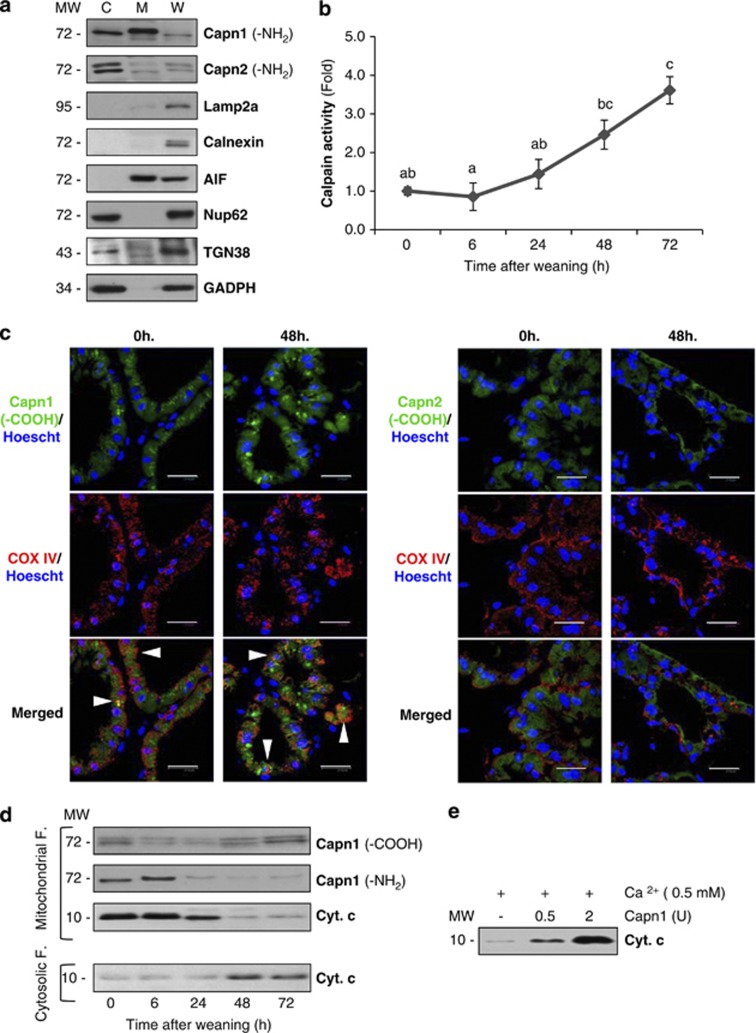
Different studies have demonstrated that CAPN1 also appears within the mitochondria.14 Therefore, one could hypothesize that CAPN activation could mediate mitochondrial programmed cell death during mammary gland involution. As can be seen in Figure 3a, CAPN1 is present not only in the cytosol but also in the mitochondria, whereas CAPN2 is only detectable in cytosolic extracts. Besides, CAPN activity was measured in mitochondrial extracts from control lactating and weaned mammary glands (Figure 3b), showing a CAPN activation pattern similar to that observed in total extracts (Figure 2a). Although CAPN2 is not detectable in the mitochondria, it could translocate upon activation and therefore be responsible together with CAPN1 for the changes observed. To elucidate this fact, immunofluorescence analysis of control and 48-h-weaned tissue was performed with antibodies against CAPN1, CAPN2 and cytochrome c oxidase IV (COX IV), used as a mitochondrial marker (Figure 3c). Co-immunostaining of both CAPN1 and COX IV during weaning confirmed the presence of this protease within the mitochondria (arrowheads) in control and weaned mice. However, no co-localization was detected for CAPN2 and COXIV, neither in lactating nor in weaned mice. In agreement with this, western blot analysis showed that CAPN1 was present in mitochondrial fractions in both control and weaned mice (Figure 3d) while CAPN2 did not translocate to the mitochondria upon activation (data not shown), supporting the idea that CAPN1 is the major player in mitochondrial destabilization. Indeed, concomitant with the increased activity, there is a loss of the CAPN1 amino-terminal domain and cytochrome c release from mitochondrial fractions to the cytosolic compartment (Figure 3d).
Figure 3.
Mitochondrial effects of calpain activation. (a) Immunoblot of the cytoplasmic (C), mitochondrial (M) and whole tissue (W) extracts with antibodies against calpain-1 and -2 in control lactating glands. To assess the purity of mitochondrial fractions, western blot analysis with antibodies against other organelle markers was performed: Lamp2a (lysosomal marker), Calnexin (endoplasmic reticulum marker), AIF (mitochondrial marker), TGN38 (Golgi apparatus), nucleoporin p62 (nuclei) and GAPDH (cytosolic marker). (b) Determination of calpain activity in mitochondrial extracts from control and weaned glands at different times of involution. All values are shown as means±S.E.M. ANOVA was performed for the statistical analysis, and different superscript letters indicate significant differences, P<0.05; the letter ‘a' always represents the lowest value within the group. (c) Mammary tissue sections from control lactating (0 h) or 48-h-weaned mice were fixed and immunostained with anti-COX IV (red) and calpain-1 or -2 (green) antibodies. Nuclei were visualized by Hoeschst-33342 (blue). Scale bar: 20 μm. (d) Western blot analysis of mitochondrial or cytosolic fractions at different times of weaning (0, 6, 24, 48 and 72 h), showing CAPN1 cleavage of the amino-terminal residues and subsequent cytochrome c release from the mitochondria to the cytosol. (e) In vitro treatment of isolated mitochondria from lactating mice with different units of recombinant calpain-1 in the presence of calcium. After 15-min incubation at 37 °C, mitochondria were pelleted and the amounts of cytochrome c released to the supernatant were assessed by western blot
To assess the role of CAPN1 in this pathway, intact mitochondria from control lactating mice were isolated and incubated with purified human CAPN1. Figure 3e shows that addition of CAPN1 triggers cytochrome c release from the mitochondria in a concentration-dependent manner. Interestingly, incubation of isolated mitochondria merely with calcium also induces a slight cytochrome c release, probably due to the intrinsic CAPN1 already detected on these organelles.
CAPN activity in lysosomes from weaned mammary glands
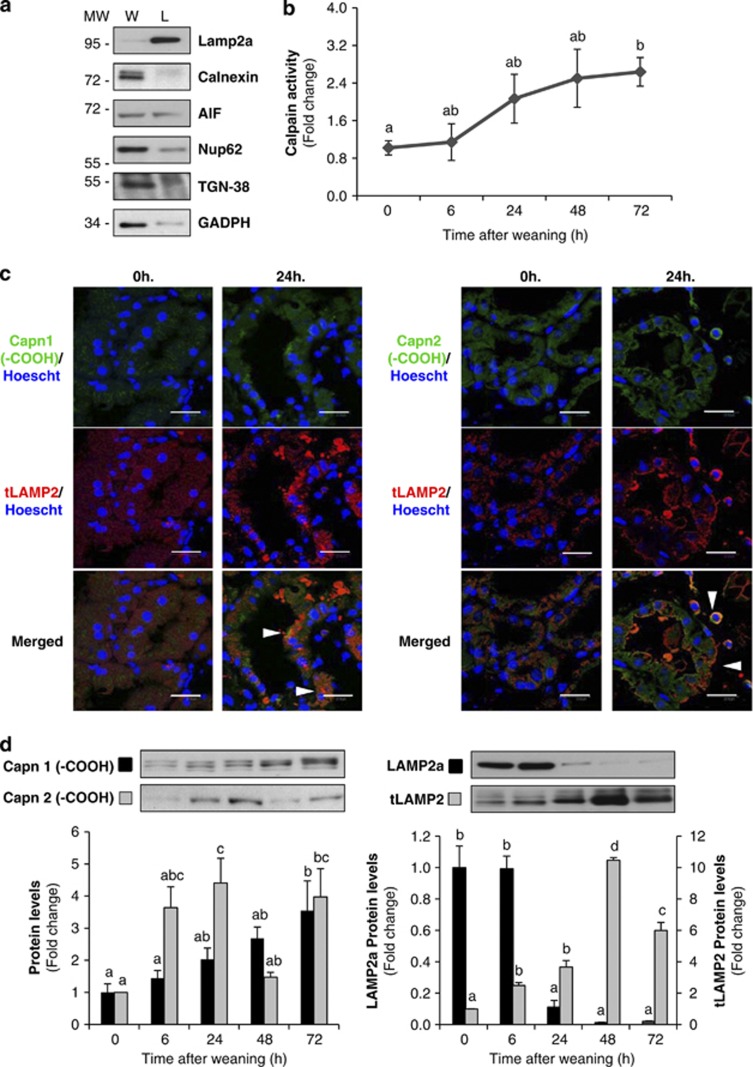
The CAPN system has recently emerged as an important player in regulating cell death, not only in the mitochondria, but also in lysosomes, where it causes apparent release of cathepsins B and D.25 To corroborate this hypothesis, we isolated lysosomal enriched fractions from control and weaned mammary glands. The purity of the lysosomal fraction was assessed by western blot, using antibodies against different organelle markers (Figure 4a). CAPN activity within lysosomes increased by twofold after 24 h of weaning, remaining elevated thereafter (Figure 4b).
Figure 4.
Calpain activity is increased in lysosomes during involution inducing Lamp2a cleavage. (a) Western blot in whole tissue (W) or lysosomal (L) extracts from control lactating mammary glands showing enrichment of the lysosomal fractions by using different organelle markers: Lamp2a (lysosomal marker), Calnexin (endoplasmic reticulum marker), AIF (mitochondrial marker), TGN38 (Golgi apparatus), nucleoporin p62 (nuclei) and GAPDH (cytosolic marker). (b) Quantification of calpain enzymatic activity on lysosomal-enriched fractions from control lactating (0 h) and weaned (6, 24, 48 and 72 h) mammary glands. (c) Intracellular co-localization of calpain-1 or -2 (green) and the lysosomal marker Lamp2 (red) is shown (arrowheads) in merged images from 24-h-weaned mammary glands. (d) Western blot analysis shows increased CAPN1 and CAPN2 protein levels in isolated lysosomes from involuting mammary glands compared with lysosomal extracts from day 10 of lactation (0 h) (left panel). Lysosomes isolated from lactating or weaned mammary glands were immunoblotted with an antibody that recognizes the cytosolic tail of Lamp2a or all Lamp2 isoforms (right panel). A representative immunoblot of four different experiments is shown. All values are shown as means±S.E.M. ANOVA was performed for the statistical analysis, and different superscript letters indicate significant differences, P<0.05; the letter ‘a' always represents the lowest value within the group
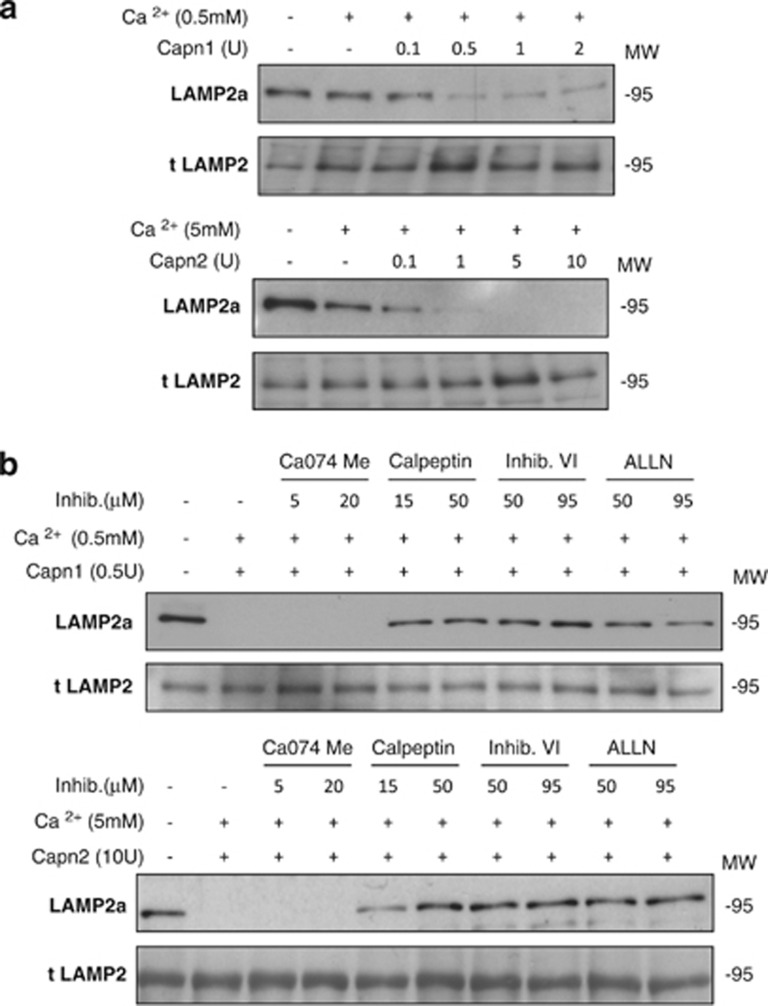
Tissue sections from control lactating or 24-h-weaned mammary glands were double stained for the lysosomal-specific protein LAMP2 (lysosomal-associated membrane protein 2) and CAPN1 or 2. As shown in Figure 4c, both CAPNs co-localize with LAMP2 in weaning samples, indicating that upon activation both proteases translocate to the lysosomes. By western blot analysis of the lysosomal fractions, both CAPN1 and 2 were detectable, increasing their levels upon activation (Figure 4d, left panel). LAMP2a has been used as a molecular marker for lysosome stability. We found that, concomitant with the increase in CAPN levels, LAMP2a almost disappeared after 24 h of weaning (Figure 4d, right panel). The reduction in LAMP2a levels was observed using an antibody that recognizes the cytosolic tail of the protein, whereas a western blot against LAMP2 (transmembrane domain) indicated that the levels of this lysosomal protein increase during involution, as has already been published.22
To test if LAMP2a was indeed a proteolytic substrate for CAPNs, isolated lysosomes from control lactating mammary glands were exposed to recombinant CAPNs. Figure 5a shows that LAMP2a disappeared when lysosomal enriched fractions were incubated with increasing concentrations of either CAPN1 or CAPN2 in the presence of calcium. In the absence of CAPN and calcium, no LAMP2a degradation was observed; however, a slight decrease in LAMP2a levels was observed upon addition of calcium, most likely due to activation of intrinsic CAPNs. LAMP2a cleavage was also time-dependent (Supplementary Figure S3). Nevertheless, it has been published that LAMP degradation is mediated by lysosomal cathepsins.26 To discard this possibility, lysosomes were incubated with either CAPN1 or 2 in the presence of calcium and a set of protease inhibitors. The addition of the specific cathepsin B inhibitor Ca-074Me did not block LAMP2a cleavage (Figure 5b, lanes 3–4). However, CAPN inhibitors, such as calpeptin (inactivates CAPN1 and 2 and papains) and CAPN inhibitor VI or ALLN (both inhibit CAPNs but also cathepsins B and L), inhibited CAPN activation, thus preventing LAMP2a degradation in vitro (Figure 5b, lanes 5–10). These results provide conclusive evidence that CAPNs are responsible for LAMP2a cleavage during mammary gland involution.
Figure 5.
Calpain-mediated proteolytic cleavage of Lamp2a at the lysosomal membrane. (a) Isolated lysosomal-enriched fractions obtained from control lactating mammary glands were incubated at 37 °C for 15 min with increasing concentrations of either recombinant calpain 1 (upper panel) or calpain 2 (lower panel) in the presence of calcium. At the end of the incubation samples were subjected to SDS-PAGE and immunoblotted with Lamp2a (anti-cytosolic) or total Lamp2. (b) Blockage of LAMP2a cleavage by different calpain inhibitors. Lysosomal fractions were incubated as described in (a), with calpain 1 (upper panel) or calpain 2 (lower panel) in the presence of calcium with a battery of inhibitors specific for cathepsin B (Ca074Me), calpains (calpeptin), or both calpains and cathepsins B and L (calpain inhibitor VI and ALLN)
Identifying new CAPN substrates in lysosomal membranes
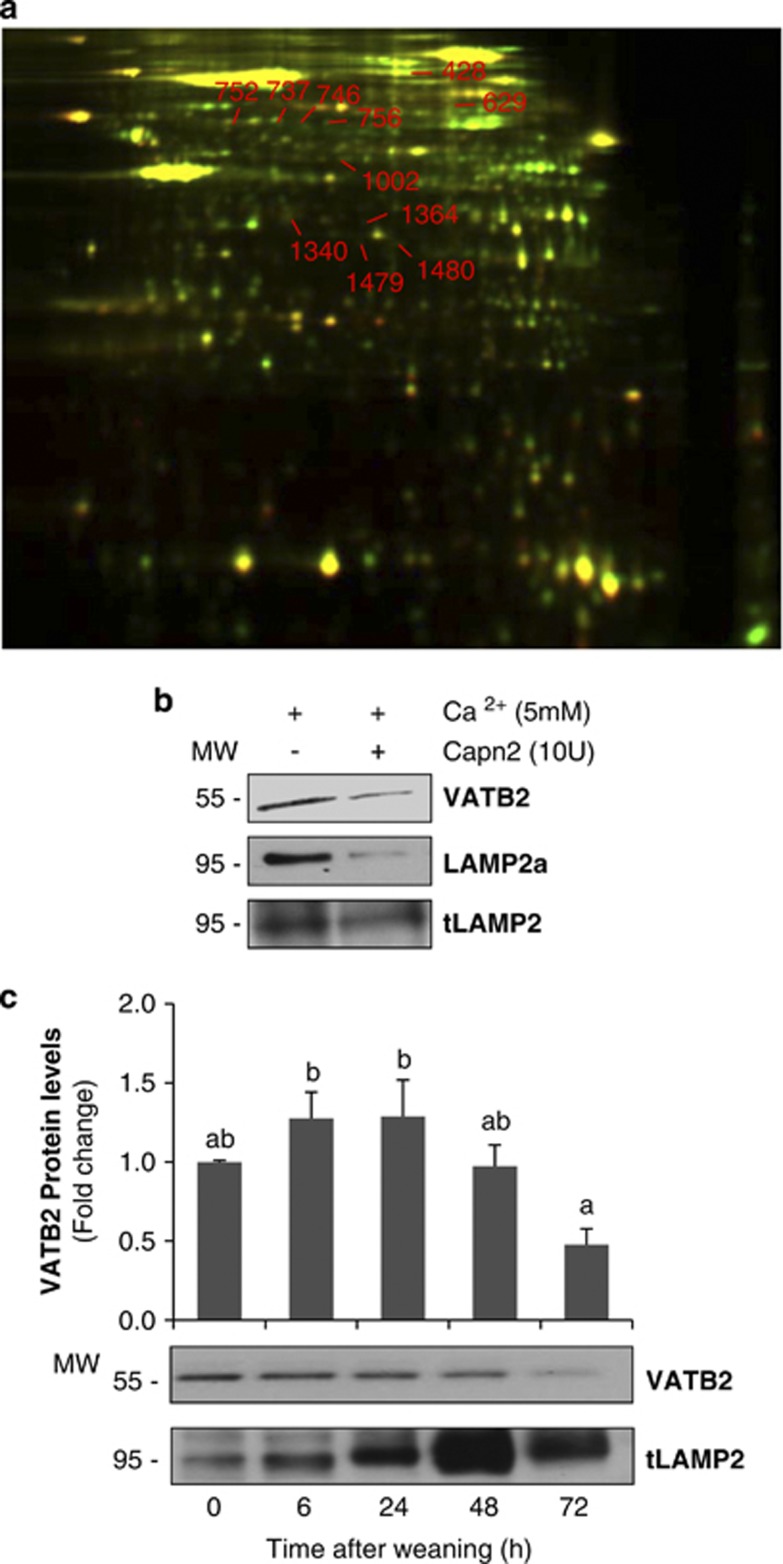
Proteomic studies were carried out in lysosomal enriched fractions from control lactating mammary gland incubated in the presence/absence of recombinant CAPN (Figure 6a). Treated and untreated samples were combined and analyzed by 2D-DIGE and mass spectrometry; 11 spots were excised and identified as potential CAPN subtrates. By using a recently published CAPN-cleavage prediction method based on Multiple Kernel Learning algorithms,27 we could predict the cleavage sites of all the proteins identified, with a high score range (Table 1). Some of these proteins are located in endosomes, such as the long-chain fatty acid CoA ligase or the β2-glycoprotein 1. As our subcellular fractionation gives us an enriched lysosomal fraction but not purified, we cannot rule out the possibility that these proteins are indeed located in endosomes and not lysosomes. The presence of the vacuolar-type proton ATPase subunit B (VATB2) was remarkably interesting, not only for its cellular function but also because the same protein was identified in several different spots. This protein of 511 amino acids is a subunit of the vacuolar H+-ATPase (v-ATPase), a membrane-anchored, multi-subunit proton pump responsible for acidification of intracellular compartments.28 CAPN breakdown of VATB2 was confirmed in vitro by immunoblot with a specific antibody (Figure 6b). Besides, it was also demonstrated that during weaning this subunit from the v-ATPase disappeared from the lysosomal fraction (Figure 6c), giving support to the idea that lysosomes from involuting mammary gland are more likely destabilized.
Figure 6.
2D DIGE analysis of calpain-mediated proteolysis in lysosomal extracts. (a) The lysosome fraction from control lactating mammary gland was treated with (green) or without (red) recombinant calpain 2 (10 units) for 1 h at 37 °C. Dye-labeled samples were combined, and the protein mixture was separated subsequently in two dimensions. Proteins were reduced in abundance following calpain treatment (i.e. calpain substrates) appear green. Protein spots resistant to calpain digestion appear yellow. The numbered spots were excised and identified by mass spectrometry. (b) Immunoblot for v-type H+ ATPase subunit b2 (VATB2), Lamp2a (anti-cytosolic) and total Lamp2 in samples used for proteomic analysis. (c) Western blot for VATB2 in mammary gland lysosomal extracts at different times of involution (0, 6, 24, 48 and 72 h). Total Lamp2 was used as a lysosomal marker. Immunoblot is representative of three different experiments. For quantification, all values are means±S.E.M. ANOVA was performed for the statistical analysis, and different superscript letters indicate significant differences, P<0.05; the letter ‘a' always represents the lowest value within the group
Table 1. Calpain-cleaved spots identified by 2D DIGE and mass spectrometry in lysosomal fraction from lactating mammary gland.
| Accession number | Protein name | NP | Sequence coverage (SC%) | MASCOT score | Gene ontology classification | Predicted cleavage site | Score range | |
|---|---|---|---|---|---|---|---|---|
| 428 | NP_032007.2 | Long-chain-fatty-acid – CoA ligase 1 (LACS 1) | 17 | 25 | 122.9 | ATP binding, long-chain fatty acid–CoA ligase activity | 40, 44, 105, 255, 280, 403, 415, 435, 448, 607 | 0.11–0.017 |
| NP_598738.1 | Serotransferrin (Transferrin) | 8 | 13 | 47.1 | Ferric iron transmembrane transporter activity | 26, 92, 273, 268, 455, 459, 462, 478, 525, 610 | 0.11–0.16 | |
| 629 | NP_032007.2 | LACS 1 | 10 | 17 | 75.4 | |||
| NP_598738.1 | Serotransferrin (Transferrin) | 7 | 12 | 47.3 | ||||
| NP_038503.4 | Beta-2-glycoprotein 1 (Apo-H) (B2GPI) (Beta(2)GPI) | 3 | 9 | 17.5 | Regulation of blood coagulation, triglyceride metabolic process | 49, 69, 75, 104, 108, 154, 214, 249, 338 | 0.09–0.13 | |
| 737 | NP_031535.2 | V-type proton ATPase subunit B, brain isoform (V-ATPase subunit B2) | 16 | 36 | 108.0 | Proton-transporting ATPase activity, rotational mechanism | 4, 41, 46, 73, 308, 337, 349, 462, 501, 503 | 0.11–0.14 |
| 746 | NP_031535.2 | V-ATPase subunit B2 | 12 | 20 | 77.2 | |||
| 752 | NP_031535.2 | V-ATPase subunit B2 | 35 | 48 | 203.1 | |||
| 756 | NP_031535.2 | V-ATPase subunit B2 | 11 | 20 | 76.1 | |||
| NP_058662.2 | 𝒟-3-phosphoglycerate dehydrogenase (3-PGDH) | 3 | 7 | 18.9 | Oxidoreductase activity | 60, 194, 262, 312, 343, 389, 405, 446, 461, 527 | 0.1–0.15 | |
| 1002 | NP_082253.1 | Adipocyte plasma membrane-associated protein | 8 | 20 | 52.2 | Strictosidine synthase activity | 9, 21, 85, 203, 256, 289, 315, 323, 354, 365 | 0.1–0.14 |
| 1340 | NP_081269.1 | N(G), N(G)-dimethylarginine dimethylaminohydrolase 1 (DDAH-1) (dimethylarginine dimethylaminohydrolase 1) | 9 | 35 | 60.5 | Hydrolase activity | 4, 18, 19, 50, 98, 142, 211, 218, 236, 256 | 0.1–0.13 |
| NP_038664.2 | Serine/threonine-protein phosphatase PP1-gamma catalytic subunit (PP-1G) | 3 | 9 | 19.2 | Protein serine/threonine phosphatase activity | 20, 121, 183, 267, 301, 305, 306, 314, 315, 318 | 0.09–0.24 | |
| 1364 | NP_031501.1 | 60S acidic ribosomal protein P0 | 4 | 15 | 34 | Structural constituent of ribosome | 55, 70, 120, 141, 239, 275, 285, 286, 291, 292 | 0.1–0.2 |
| 1479 | NP_666245.2 | Golgi phosphoprotein 3 | 3 | 13 | 19.8 | 38, 59, 62, 65, 72, 75, 88, 162, 212, 315 | 0.11–0.18 | |
| 1480 | NP_666245.2 | Golgi phosphoprotein 3 | 5 | 19 | 42.9 |
Bold numbers indicate the best predicted score for the cleavage site of each protein
Inhibition of CAPN1 attenuates LAMP2a degradation and delays mammary gland involution
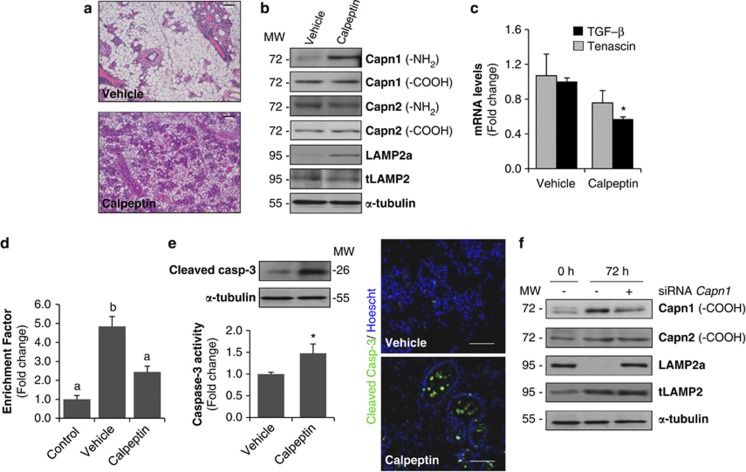
To study whether activated CAPN is involved in epithelial-cell death, we used calpeptin, a pharmacological inhibitor of CAPN,29 in lactating mice once the litter was removed, during 3 days. Histological analysis of tissue sections from CAPN-treated mice demonstrated a substantial delay in mammary tissue regression compared with vehicle-treated controls (Figure 7a). Furthermore, CAPN1 activation, as determined by NH2-terminal domain autolysis and LAMP2a cleavage, was completely prevented by calpeptin treatment (Figure 7b). Surprisingly, CAPN2 levels did not change, suggesting that Lamp2a cleavage is mainly associated with CAPN1 activation.
Figure 7.
In vivo inhibition of calpain delays involution and LAMP2a cleavage. (a) Females at the peak of lactation were forced weaned and received calpeptin (40 mg/kg) by intraperitoneal injection every 12 h during 3 days. Representative hematoxylin and eosin (H&E)-stained sections of the inguinal mammary gland of a 72-h-weaned female injected with vehicle (control) or calpeptin-treated. Scale bars: 100 μm. (b) Western blot analysis showing levels of calpain-1 and -2 (using antibodies against the amino-terminal and carboxy-terminal domains), LAMP2a and total LAMP2 in extracts from calpeptin-treated or untreated (vehicle) mammary glands. Equal loading was confirmed by reprobing the blot against α-tubulin. (c) Real-time quantitative PCR of TGF-β1 and Tenascin-C in 72-h-weaned mice treated with vehicle or calpeptin. t-test was performed for the statistical analysis, where *P<0.05 for calpeptin versus vehicle. (d) Apoptosis was assessed by measuring specific enrichment of mono- and oligonucleosomes released into the cytoplasm (enrichment factor) and was calculated as the ratio between the absorbance values obtained in control lactating mice (control) and 72-h-involuting mammary glands treated with vehicle or calpeptin. Data are shown as means±S.E.M. of three independent experiments performed in triplicate. ANOVA was performed for the statistical analysis, and different superscript letters indicate significant differences, P<0.05; the letter ‘a' always represents the lowest value within the group. (e) Right panel, caspase-3 activity in 72-h-weaned mammary tissue treated with calpeptin. Values are shown as means±S.E.M. of three different experiments performed in triplicate. *P<0.05 between calpeptin and vehicle-injected mice. Representative western blot analysis showing cleaved caspase-3 in those samples. Left panel, immunohistochemical analysis of cleaved executioner caspase-3 in vehicle and calpeptin-treated samples from 72 h of involution. (f) Calpain-1 silencing reduces Lamp2a cleavage in vivo. Representative western blot analysis of at least three independent biological samples showing calpain-1 and -2, Lamp2a and total Lamp2 (tLamp) in control lactating (0 h), 72-h-weaned mice and 72-h-weaned mice in which calpain-1 was knocked down by specific siRNA treatment (siRNA calpain-1). α-Tubulin was used as loading control
Focusing on the changes observed in tissue regression and remodeling, mRNA levels of tenascin-C, a glycoprotein known to be upregulated during involution,30 were measured. The expression of this extracellular matrix protein was lower in calpeptin samples compared to vehicle controls (Figure 7c). Besides, we also measured mRNA levels of transforming growth factor beta 1 (TGF-β1), a key local regulator of mammary tissue involution. Indeed, mice overexpressing TGF-β1 demonstrated accelerated remodeling associated with involution.31 In our calpeptin-treated mice, mRNA levels of TGF-β1 were significantly lower than in vehicle controls (Figure 7c). Thus, the decrease in both tenascin-C and TGF-β1 mRNA levels suggests that inhibition of CAPN activity is delaying (but not abrogating) the remodeling process of involution.
Tissue apoptosis was assessed by the presence of oligonucleosomal fragments in cytoplasmic extracts from control lactating mice and 72-h-weaned mice injected with calpeptin or vehicle (Figure 7d). Calpeptin treatment dramatically prevented the formation of oligonucleosomal fragments, which suggests that calpeptin administration inhibits the extent of apoptosis in involuting mammary tissue. Surprisingly, when measuring caspase-3 activation we found higher levels of cleaved caspase-3 and activity in calpeptin-treated mice. Immunostaining of cleaved-caspase 3 nicely showed epithelial cells undergoing cell death, either shed into the lumen or still within the glandular structures, in calpeptin-treated glands (Figure 7e). The absence of positive cells for active caspase-3 in vehicle-treated mice might be explained by the collapse of mammary gland architecture and apoptosis of epithelial cells that had already taken place earlier during the weaning process.
From the results presented herein, one could infer that CAPN1 has a major role in lysosomal destabilization. Thus, we separately knocked down CAPN1 with specific small interfering RNA (siRNA) in 72-h-weaned mammary glands in vivo. Transfected mammary tissue exhibited a significant decrease in CAPN1 protein levels compared with 72-h-weaned mice. However, levels of CAPN2 were similar to those of non-silenced weaned glands. Cleavage of LAMP2a was blocked by siRNA silencing of CAPN1 (Figure 7f).
Discussion
Mammary gland involution is a complex process that involves epithelial-cell death and tissue remodeling. Among the signaling pathways involved, the NF-κB pathway becomes activated early during involution.3, 4, 5, 6 NF-κB regulates the expression of a large number of genes involved in multiple biological processes, including apoptosis. We have shown that both Capn1 and 2 are regulated by NF-κB during mammary gland involution (Figure 1a). Besides, there is evidence suggesting that NF-κB is involved in the activation of Capn4 in hepatoma cells.32 The CAPN system is a key mediator of programmed cell death in several experimental models.19, 29, 33 The results presented herein demonstrate that (a) CAPN activity increases during involution and (b) the CAPN system seems to be involved not only in mitochondrial cell death but also in lysosomal destabilization.
At the end of lactation there is a substantial increase in calcium levels, which might be not only due to milk stasis within the alveolar lumen but also due to a marked decrease in the main calcium transporter, PMCA2 (Supplementary Figure 2). This Ca2+ increase21, 23 is enough to activate CAPNs, as autolysis is produced and both CAPN1 and 2 lose their amino-terminal tail after 24 h of weaning (Figure 2b). It is likely that the localization of CAPN1 near the apical side of the epithelial cells (Supplementary Figure 2d) will favor this activation, as this is where PMCA2 pumps Ca2+ within the lumen. Accordingly, when this transporter disappears, calcium would mainly accumulate on that side of the cell.
Activation of CAPNs results in the cleavage of several substrate proteins, mainly located in the plasma membrane but also in other organelles such as the mitochondria.14, 15, 16, 17, 33, 34 Upon activation, CAPNs cleave apoptosis-related proteins, triggering cytochrome c release from mitochondria and inducing caspase activation.12 Our data demonstrate that CAPN1 is already localized in the mitochondria during lactation (Figure 3a) and becomes activated during involution, releasing cytochrome c (Figure 3d). CAPN-dependent cytochrome c translocation was further demonstrated by incubation of isolated mitochondria with CAPN1 in the presence of calcium (Figure 3e). Overall, CAPN activation seems to have a key role in the mitochondrial apoptotic pathway that has already been described in mammary gland involution.
An additional finding was that CAPNs were also active within lysosomes (Figure 4b). These results provide new evidence for lysosomal-mediated cell death in mammary gland involution, supporting the idea that this event takes place even earlier than the mitochondrial intrinsic pathway and caspase-3 activation. With regard to the possible substrates for CAPN cleavage, LAMP2 was considered a good candidate. LAMP2 is a lysosomal-associated membrane protein that is highly glycosylated, the function of which remains to be elucidated. It has been proposed to protect the lysosomal membrane from attack by lysosomal proteases; besides, it is a receptor for selective degradation of cytosolic proteins.35 Lamp2 mRNA and protein levels are known to increase during involution;22 however, when using an antibody against the cytosolic tail of LAMP2a, degradation of this protein occurs after litter removal (Figure 4d). Furthermore, the data presented above demonstrated that LAMP2a turnover in the mammary tissue is CAPN-mediated. Lysosomes incubated with CAPNs in the presence of calcium and several CAPN inhibitors showed reduced rates of LAMP2a degradation, while the cathepsin B inhibitor, Ca-074, had no effect on LAMP2a cleavage (Figure 5b). This truncated LAMP2a might have a direct effect on the permeability of the lysosomal membrane35 that has been shown to increase leakiness during involution.22
To gain a deeper knowledge in CAPN function on lysosomes and discover new substrates we used 2D-DIGE in lysosomes after CAPN treatment. VATB2 was identified as a CAPN target (Figure 6b). Several papers in the literature emphasize the importance of this proton pump in lysosomes. In fact, inhibition of the v-ATPase neutralizes the pH of lysosomes, causes lysosomal dysfunction and induces apoptosis in several cancer cell lines.36, 37 Taken together, our results shed further light on the lysosomal-mediated cell death that occurs during involution. Activated CAPNs would translocate to the lysosomal membrane, cleaving substrate proteins such as the cytosolic tail of LAMP2a or the subunit b2 of the v-ATPase. As a result of this selective proteolysis, lysosomal membrane would be destabilized, inducing membrane permeabilization, which would finally lead to cathepsin release.
Finally, experiments in vivo with the CAPN inhibitor calpeptin showed a prominent role for CAPNs in mammary gland involution, as there is a substantial delay of this process in calpeptin-treated mice. Surprisingly, calpeptin treatment prevented autolysis of CAPN1, but not 2; however, LAMP2a degradation was abolished with the administration of this inhibitor. In accordance with these results, silencing calpain-1 with siRNA during weaning also blocked LAMP2a cleavage (Figure 7f). These data suggest that the lysosomal destabilization observed in weaned glands occurs mainly through CAPN1 activation. However, we do not rule out a synergistic role for CAPN2 in other lysosomal targets or compartments during mammary gland involution. Therefore, inhibiting CAPNs during involution could result in the disturbance of several signaling pathways and as a consequence apoptosis is delayed, as demonstrated by the decreased nuclear fragmentation (Figure 7c).
Physiological implications
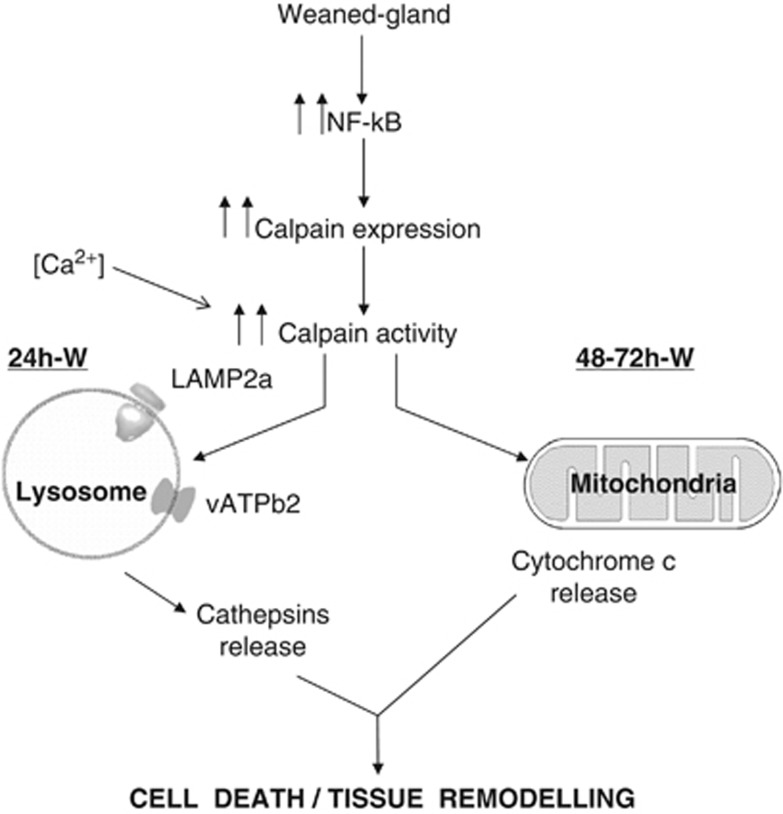
Programmed cell death is a key event of mammary gland involution by which epithelial cells are eliminated. Conventionally this process was supposed to be caspase-mediated apoptosis; however, accumulating evidence indicates that necrotic cell death can also be a regulated process occurring during mammary gland involution. We postulate that CAPNs might be involved in both forms of cell death. During mammary gland involution ubiquitous CAPNs are activated; these proteases first cleave substrates on the lysosomal membrane after 24 h of weaning and, in a second phase of involution, CAPNs induce the intrinsic mitochondrial apoptotic pathway. Taking into account these data, it seems that lysosomal dysfunction has an important role in the early events that occur during involution. Both pathways, lysosomal and mitochondrial, are involved in the epithelial-cell death and tissue remodeling by caspase-3 activation and cathepsin release (Figure 8). Finally, we have demonstrated a role for CAPNs in this programmed cell death in vivo, as inhibiting CAPN activation, either pharmacologically or by siRNA, reduces LAMP2a lysosomal cleavage and apoptosis, thus giving a direct connection between NF-κB, CAPNs and cell death.
Figure 8.
Hypothetical model of the findings described in this paper. Calpain activation during weaning is involved in both lysosomal permeabilization and mitochondrial destabilization, triggering mammary gland involution. Forced weaning induces NF-κB activation that translocates to the nucleus and promotes expression of downstream target genes such as calpains. Besides, calcium accumulates within the cells and results in the activation of calpains. During the first stage of mammary gland involution, calpain-1 is traslocated to the lysosomal membrane, where it cleaves the cytosolic tail of Lamp2 and other membrane proteins such as VATB2, contributing to destabilizing lysosomes and releasing cathepsin D into the cytosol. This cleavage is prevented by calpeptin and other calpain inhibitors. At the second phase of involution, mitochondrial calpain-1 facilitates mitochondrial permeabilization, thus inducing cytochrome c release and triggering caspase-dependent apoptosis
Materials and Methods
Animals and tissue extraction
C57BL/6 mice were obtained from Taconic (Ejby, Denmark). Ten-week-old virgin female mice were mated and males were subsequently removed at mid-gestation. Following parturition, litters were maintained with at least seven pups. Then, at the peak of lactation (days 9–11), mice were divided into different groups: control lactating mice and weaned mice, whose pups were removed 10 days after delivery to initiate involution. The weaning took place for 6, 24, 48 and 72 h before death. Mice were given intraperitoneally a single dose of sodium pentobarbital (60 mg/kg of body weight) freshly dissolved in 0.9% NaCl (Braun Medical, Rubí, Barcelona, Spain). Then, the inguinal mammary glands were removed and quickly freeze-clamped in liquid nitrogen or fixed in formaldehyde for histological studies. All the animals were given food and water ad libitum and housed in a controlled environment (12 h light/12 h dark cycle). Experimental protocols were approved by the Research Committee of the School of Medicine (University of Valencia, Valencia, Spain). The mice were cared for and handled in conformance with the National Institutes of Health guidelines and the Guiding Principles for Research Involving Animals and Humans approved by the Council of the American Physiological Society.
Histological studies
Inguinal mammary glands from lactating and weaned mice were fixed in 4% p-formaldehyde and embedded in paraffin after removal. Tissue sections of 5 μm size were cut and stained with hematoxylin and eosin for histological studies. Pictures were taken with Leica DMD 108 (Leica, Wetzlar, Germany).
Immunofluorescence analysis
For immunohistochemistry, 5-μm sections were de-paraffinized in xylene, rehydrated through a graded series of alcohol and washed with PBS. After antigen retrieval (10 min boiling in 10 mM tri-sodium citrate buffer, pH 6.0), sections were blocked in 5% normal goat serum (Dako, Glostrup, Denmark) for 1 h at room temperature and incubated overnight at 4 °C with one of the following primary antibodies: CAPN1 (both -NH2 and -COOH terminus domain), LAMP2, COX IV and cleaved caspase-3 from Abcam (Cambridge, UK) and CAPN2 from Cell Signaling (Danvers, MA, USA). Alexa Fluor 488 anti-rabbit IgG, Cy3 anti-rat IgG (both from Invitrogen, Carlsbad, CA, USA) or Cy3 anti-mouse IgG (Sigma Aldrich, St. Louis, MO, USA) were used for detection. Non-specific binding was reduced by incubating the sections in secondary antibodies. Nuclei were counterstained with Hoechst 33342 (Invitrogen). The pictures were acquired on a Leica TCS-SP 2 confocal microscope.
ChIP assays
Chromatin from mammary tissue was fixed and immunoprecipitated as described previously.3, 38 The following antibodies from Santa Cruz Biotechnology (Santa Cruz, CA, USA) were used: p65 (sc-109), p300 (sc-585) and RNA pol II (sc-899). After DNA purification, Input, Immunoprecipitated and Unrelated Ab fractions were analyzed by PCR with specific primers for either Capn1 or 2 promoter region (Capn 1 forward primer: 5′-TACCGTTTGTCTAGCGTCCG-3′ and reverse: 5′-GGGGTATTTTTAACTAGGGCC-3′ Capn2 forward primer: 5′-AGATGGACACACCAGTAGCC-3′ and reverse: 5′-ACGACAATCGCAAGTTGGG-3′) or coding regions (Capn1 forward primer: 5′-TAGTGTTGAGAGTCACACCC-3′ and reverse: 5′-ACTGAAGGATTTGGGAAGCG-3′ Capn2 forward primer: 5′-CTAGGACAGAACTCAAAGGG-3′ and reverse: 5′-CGTTTCATAGTCAGACCTCC-3′). PCR fragments were size-fractionated by 2% agarose gel electrophoresis and stained with ethidium bromide.38
Real-time quantitative PCR (RT-qPCR) analysis and reverse transcriptase (RT)-PCR
Total RNA from mammary tissue was extracted by TRIzol (Invitrogen) followed by additional column purification (RNeasy, Qiagen, Hilden, Germany). RNA quantity and purity were determined using NanoDrop ND-2000 (NanoDrop Technologies, Wilmington, DE, USA), and RNA integrity was assessed by determining the RNA 28S/18S ratio using RNA 6000 Nano Labchips in an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA).
RNA (500 ng) was reverse-transcribed into cDNA at 37 °C for 60 min and 95 °C for 5 min using a high-capacity RNA-to-cDNA kit (Applied Biosystems, Foster City, CA, USA). The cDNA products were amplified by qPCR using the GeneAmp Fast PCR Master Mix (Applied Biosystems). All reactions were carried out in triplicate. RT-q PCR was run in the 7900HT Fast Real-Time PCR System (Applied Biosystems). Pre-developed Taqman primers specific for Capn1, Capn2, Capn4, PMCA2, TGF-β, Tenascin-C and 18S were purchased from Applied Biosystems. Results were normalized according to 18S quantification in the same sample reaction. The threshold cycle (Ct) was determined, and the relative gene expression was expressed as follows:
 |
where ΔCt=Ct (target)−Ct (18S), and Δ(ΔCt)=ΔCt (weaned)−ΔCt (control).
Total protein extraction and immunoblotting
Total protein was extracted from snap-frozen mammary glands (0.1 g) by homogenization in a buffer containing detergents and protease inhibitors as described previously.5 Equal amounts of protein (15 μg) were separated by SDS–PAGE and transferred onto nitrocellulose membranes (Protran, Whatman, Dassel, Germany). After addition of the corresponding primary antibody, immunocomplexes were revealed by using a HRP-conjugated secondary antibody (Dako), as appropriate, and subsequent peroxidase-induced enhanced chemiluminescence reaction (ECL Detection Kit, GE Healthcare, Uppsala, Sweden). The intensity of the bands was measured by densitometry using ImageJ (National Institute of Health, Bethesda, MD, USA). Equal loading was confirmed by reprobing the blot against α-tubulin and by Ponceau Red staining. The following primary antibodies were from Abcam: CAPN1-NH2 (ab28257), CAPN2-NH2 (ab39165), CAPN1-COOH (ab39170), calnexin (ab22595), glyceraldehyde-3-phosphate dehydrogenase (ab8245), AIF (ab1998), LAMP2 (ab13524) and ATP6V1B2 (ab73404). The following primary antibodies from Santa Cruz were used: TGN38 (trans-Golgi network protein 2) (sc-33784), cytochrome c (sc-7159) and α-tubulin (sc-5286). Finally, CAPN2-COOH (3372) was from BioVision (San Francisco, CA, USA), LAMP2A (PA1-655) antibody was from Pierce Antibodies (Rockford, IL, USA), cleaved caspase-3 (Asp 175) was from Cell Signaling (9661) and an antibody against nucleoporin p62 (MAb414) was from Covance (Princeton, NJ, USA).
CAPN activity assay
CAPN activity was measured using Suc-LLVY-AMC as the fluorogenic substrate provided in the ‘calpain activity assay kit' (QIA-120, Calbiochem, Billerica, MA, USA). Proteins (30 μg) obtained from total, lysosomal or mitochondrial extracts were solubilized in cell lysis buffer. CAPN activity was determined by subtracting the activity obtained using a CAPN-inhibition buffer containing BAPTA (a calcium chelator that completely blocks CAPN activity) from the activity detected with the activation buffer containing calcium and a reducing agent (TCEP) according to the manufacturer's instructions.
Subcellular fractionation: isolation of lysosomes and mitochondria
For lysosomal isolation a ‘lysosome enrichment kit' (#89839, Thermo Scientific, Waltham, MA, USA) was used according to the manufacturer's instructions. Briefly, inguinal mammary glands were minced and homogenized in a Dounce glass homogenizer and centrifuged at 500 × g for 10 min to remove cell debris. The supernatant was subjected to a discontinuous iodixanol gradient (17–30%) and centrifuged at 145 000 × g in an Optima-XL Ultracentifuge (Beckman Coulter, Brea, CA, USA). The lysosomal fraction was removed from the top of the gradients, and then washed with PBS and stored at −80 °C until use.
Mammary gland mitochondria were obtained by differential centrifugation, as described.39 Briefly, minced mammary glands were homogenized and centrifuged at 1000 × g for 10 min. Supernatants were then centrifuged at 10 000 × g for 10 min to purify the mitochondria. Mitochondrial pellets were then solubilized in a buffer solution containing 0.3 M sucrose, 1 mM EGTA, 5 mM MOPS, 14.7 μM BSA and 5 mM KH2PO4, pH 7.4, and stored at −80 °C.
In vitro CAPN assay in mitochondria and lysosomes
Analysis of mitochondrial permeabilization was performed using mitochondrial extracts, isolated as mentioned above. These were incubated for 15 min at 37 °C with different units of recombinant CAPN1 in the presence of calcium (0.5 mM). After the incubation, samples were centrifuged at 10 000 × g for 15 min at 4 °C. Supernatants were carefully removed and analyzed by western blot.
To analyze lysosomal LAMP2a cleavage, different units of recombinant CAPN1 and 0.5 mM calcium were incubated with enriched lysosomal extracts for 15 min at 37 °C in a solubilization buffer (HEPES-KOH 20 mM, sucrose 250 mM, KCl 10 mM, MgCl2 1.5 mM, EDTA 1 mM, EGTA 1 mM, DTT 8 mM and PMSF 1 mM, at pH 7.5). After incubation, lysosomes were pelleted at 17 000 × g for 15 min at 4 °C and resuspended in CHAPS buffer prior to western blot analysis. The same procedure was followed for CAPN2 supplemented with calcium (5 mM). Finally, to assess whether Lamp2a was cleaved by CAPNs or by cathepsins, different CAPN and cathepsin inhibitors were used. Lysosome extracts were incubated as described above, but supplemented with different concentrations of Ca-074Me (5, 20 μM), calpeptin (15, 50 μM), CAPN inhibitor VI (50, 100 μM) or ALLN (50, 100 μM).
Two-dimensional gel electrophoresis
A DIGE approach was used to identify endogenous CAPN substrates in enriched lysosomal fractions from lactating mice. In all, 10 U of recombinant CAPN2 was incubated with half of the preparation for 1 h at 37 °C in the presence of 5 mM calcium. Control samples were incubated in the same conditions but without CAPN2. After incubation, CAPN-treated samples and control counterparts were labeled alternatively with Cy-3 or Cy-5 and recombined. A pool of both samples was labeled with Cy-2 for normalization. The recombined sample was subjected to first-dimension isoelectric focusing on immobilized pH gradient strips (pH 3–7, BioRad, Hercules, CA, USA) in a Protean IEF Cell. The IPG strips were then loaded at the top of a 12.5% SDS–PAGE. For Sypro Ruby staining, the gels were stained following the manufacturer's instructions. Relative quantification of gel features was achieved using DeCyder software (v7.0, Amersham Biosciences, GE Healthcare). Images from separate gels were matched with the BVA software using the Cy-2 labeled image on each gel for normalization. Quantitative differences were only accepted when at least a 1.5-fold change was confirmed in three independent experiments. The statistical significance of the differences was calculated using Student's t-test and accepted when the value was P<0.05. MS/MS analysis was performed as previously described40 by the Proteomics and Bioinformatics Unit, Centre for Applied Medical Research (CIMA) at the University of Navarra (Pamplona, Spain).
Calpeptin administration in vivo
CAPN inhibitor studies were carried out in 10-day lactating mice, in which pups were removed prior to the first administration. Calpeptin (#03-34-0051 from Merck Chemicals, Nottingham, UK) was dissolved in DMSO and further diluted in physiological saline (10 mg/ml). A quantity of 40 mg/kg was administered by intraperitoneal injection every 12 h during 3 days. A second group of control mice received vehicle alone. After 3 days, inguinal mammary glands were removed, fixed in p-formaldehyde or freeze-clamped in liquid nitrogen until further use.
Depletion of CAPN1 in mouse mammary gland
Lactating mice at the peak of lactation were forced weaned and treated with CAPN1 siRNAs from Sigma-Aldrich (Mission esiRNA, EMU057001) during 3 days. siRNA (5 μg per 24 h during 3 days) was injected locally to inguinal mammary glands mixed with L-PEI (in vivo-jetPEI, PolyPlus-Transfection, Illkirch, France), following the manufacturer's instructions. Successful transfection of CAPN1 siRNAs was confirmed by western blotting analysis of mammary tissue homogenates.
Determination of oligonucleosomal DNA fragments
The presence of soluble histone–DNA complexes was measured by the ‘Cell Death Detection Assay' (#11774425001, Roche Diagnostics, Mannheim, Germany). Cell death enzyme-linked immunosorbent assays were performed according to the manufacturer's instructions. Specific enrichment of mono- and oligonucleosomes released into the cytoplasm (enrichment factor) was calculated as the ratio between the absorbance values of the samples obtained from 72-h-weaned mice (calpeptin-treated and untreated) and 10-day-lactation controls.
Measurement of caspase-3 activity
Caspase-3 activity was measured in mammary tissue lysates using the ‘Caspase-3/CPP32 Colorimetric Assay Kit' (#K106, BioVision, Mountain View, CA, USA) as previously described.5
Statistics
Data are presented as means±S.E.M. and were analyzed by a one-way ANOVA. Significant differences were determined by Tukey–Kramer test. Differences were considered significant at P<0.05, and different superscript letters indicate significant differences in the figures. The letter ‘a' always represents the lowest value within the group. Independent experiments were conducted with a minimum of three replicates per condition to allow for statistical comparison. In Figure 7c, Student's t-test with Bonferroni correction was used for comparison between the calpeptin-treated and control groups.
Acknowledgments
This study was supported by grants from Ministerio de Ciencia e Innovación (BFU2010-18253 to JRV and PS09-02360 to ERG-T), Consellería de Educación (GVPROMETEO 2010-075) and Consellería de Sanidad (AP-085/11 to RZ). TA is the recipient of a pre-doctoral fellowship from Ministerio de Educación and IF-V is funded by Consellería de Educación (GVPROMETEO 2010-075). We thank Estefanía Fernández for technical advice on proteomic studies, Eva Serna for microarray experiments, Sonia Priego for acquiring confocal microscopy images, and Jaime Ferrer and Elisa Alonso-Yuste for tissue histology.
Glossary
- LAMP
lysosomal-associated membrane protein
- NF-κB
nuclear factor kappaB
- ChIP
chromatin immunoprecipitation
- CAPN
calpain
- RNApol II
RNA polymerase II
- PMCA2
plasma membrane calcium ATPase
- COX IV
cytochrome c oxidase IV
- VATB2
vacuolar-type proton ATPase subunit B
- v-ATPase
vacuolar H+-ATPase
- TGF-β1
transforming growth factor beta 1
- TGN38
trans-Golgi network protein 2
- RT-PCR
reverse transcriptase-PCR
- RT-qPCR
real-time quantitative PCR
- siRNA
small interfering RNA
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by H-U Simon
Supplementary Material
References
- Clarkson RWE, Wayland MT, Lee J, Freeman T, Watson CJ. Gene expression profiling of mammary gland development reveals putative roles for death receptors and immune mediators in post-lactational regression. Breast Cancer Res. 2004;6:92–109. doi: 10.1186/bcr754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein T, Morris JS, Davies CR, Weber-Hall SJ, Duffy MA, Heath VJ, et al. Involution of the mouse mammary gland is associated with an immune cascade and an acute-phase response, involving LBP, CD14 and STAT3. Breast Cancer Res. 2004;6:75–91. doi: 10.1186/bcr753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaragozá R, Miralles VJ, Rus AD, García C, Carmena R, García-Trevijano ER, et al. Weaning induces NOS-2 expression through NF-kappa modulation in the lactating mammary gland: importance of GSH. Biochem J. 2005;391:581–588. doi: 10.1042/BJ20050507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarkson RW, Heeley JL, Chapman R, Aillet F, Hay RT, Wyllie A, et al. NF-κB inhibits apoptosis in murine mammary epithelia. J Biol Chem. 2000;275:12737–12742. doi: 10.1074/jbc.275.17.12737. [DOI] [PubMed] [Google Scholar]
- Zaragozá R, Bosch A, García C, Sandoval J, Serna E, Torres L, et al. Nitric oxide triggers mammary gland involution after weaning: remodelling is delayed but not impaired in mice lacking inducible nitric oxide synthase. Biochem J. 2010;428:451–462. doi: 10.1042/BJ20091091. [DOI] [PubMed] [Google Scholar]
- Connelly L, Barham W, Pigg R, Saint-Jean L, Sherrill T, Cheng DS, et al. Activation of nuclear factor kappa B in mammary epithelium promotes milk loss during mammary development and infection. J Cell Physiol. 2010;222:73–81. doi: 10.1002/jcp.21922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres L, Serna E, Bosch A, Zaragozá R, García C, Miralles VJ, et al. NF-κB as node for signal amplification during weaning. Cell Physiol Biochem. 2011;28:833–846. doi: 10.1159/000335797. [DOI] [PubMed] [Google Scholar]
- Goll DE, Thompson VF, Li H, Wei W, Cong J. The calpain system. Physiol Rev. 2003;83:731–801. doi: 10.1152/physrev.00029.2002. [DOI] [PubMed] [Google Scholar]
- Suzuki K, Hata S, Kawabata Y, Sorimachi H. Structure, activation, and biology of calpain. Diabetes. 2004;53:12–18. doi: 10.2337/diabetes.53.2007.s12. [DOI] [PubMed] [Google Scholar]
- Franco SJ, Huttenlocher A. Regulating cell migration: calpains make the cut. J Cell Sci. 2005;118:3829–3838. doi: 10.1242/jcs.02562. [DOI] [PubMed] [Google Scholar]
- Tompa P, Buzder-Lantos P, Tantos A, Farkas A, Szilágyi A, Bánóczi Z, et al. On the sequential determinants of calpain cleavage. J Biol Chem. 2004;279:20775–20785. doi: 10.1074/jbc.M313873200. [DOI] [PubMed] [Google Scholar]
- Gil-Parrado S, Fernández-Montalván A, Assfalg-Machleidt I, Popp O, Bestvater F, Holloschi A. Ionomycin-activates calpain triggers apoptosis. J Biol Chem. 2002;277:27217–27226. doi: 10.1074/jbc.M202945200. [DOI] [PubMed] [Google Scholar]
- Orrenius S, Zhivotovsky B, Nicotera P. Regulation of cell death: the calcium-apoptosis link. Nat Rev Mol Cell Biol. 2003;4:552–565. doi: 10.1038/nrm1150. [DOI] [PubMed] [Google Scholar]
- Garcia M, Bondada V, Geddes JW. Mitochondrial localization of mu-calpain. Biochem Biophys Res Commun. 2005;338:1241–1247. doi: 10.1016/j.bbrc.2005.10.081. [DOI] [PubMed] [Google Scholar]
- Cao G, Xing J, Liou AKF, Yin X-M, Clark RSB, Graham SH, et al. Critical role of calpain I in mitochondrial release of apoptosis-inducing factor in ischemic neuronal injury. J Neurosci. 2007;27:9278–9293. doi: 10.1523/JNEUROSCI.2826-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kar P, Chakraborti T, Samanta K, Chakraborti S. mu-calpain mediated cleavage of the Na+/Ca2+ exchanger in isolated mitochondria under A23187 induced Ca2+ stimulation. Arch Biochem Biophys. 2009;482:66–76. doi: 10.1016/j.abb.2008.11.024. [DOI] [PubMed] [Google Scholar]
- Norberg E, Orrenius S, Zhivotovsky B. Mitochondrial regulation of cell death: processing of apoptosis-inducing factor (AIF) Biochem Biophys Res Commun. 2010;396:95–100. doi: 10.1016/j.bbrc.2010.02.163. [DOI] [PubMed] [Google Scholar]
- Sahara S, Yamashima T. Calpain-mediated Hsp70.1 cleavage in hippocampal CA1 neuronal death. Biochem Biophys Res Commun. 2010;393:806–811. doi: 10.1016/j.bbrc.2010.02.087. [DOI] [PubMed] [Google Scholar]
- Tofighi R, Johansson C, Goldoni M, Ibrahim WN, Gogvadze V, Mutti A, et al. Hippocampal neurons exposed to the environmental contaminants methylmercury and polychlorinated biphenyls undergo cell death via parallel activation of calpains and lysosomal proteases. Neurotox Res. 2011;19:183–194. doi: 10.1007/s12640-010-9159-1. [DOI] [PubMed] [Google Scholar]
- Bevers MB, Neumar RW. Mechanistic role of calpains in postischemic neurodegeneration. J Cereb Blood Flow Metab. 2008;28:655–673. doi: 10.1038/sj.jcbfm.9600595. [DOI] [PubMed] [Google Scholar]
- Reinhardt TA, Lippolis JD. Mammary gland involution is associated with rapid down regulation of major mammary Ca2+-ATPases. Biochem Biophys Res Commun. 2009;378:99–102. doi: 10.1016/j.bbrc.2008.11.004. [DOI] [PubMed] [Google Scholar]
- Kreuzaler PA, Staniszewska AD, Li W, Omidvar N, Kedjouar B, Turkson J, et al. Stat3 controls lysosomal-mediated cell death in vivo. Nat Cell Biol. 2011;13:303–309. doi: 10.1038/ncb2171. [DOI] [PubMed] [Google Scholar]
- VanHouten J, Sullivan C, Bazinet C, Ryoo T, Camp R, Rimm DL, et al. PMCA2 regulates apoptosis during mammary gland involution and predicts outcome in breast cancer. Proc Natl Acad Sci USA. 2010;107:11405–11410. doi: 10.1073/pnas.0911186107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baki A, Tompa P, Alexa A, Molnar O, Friedrich P. Autolysis parallels activation of mu-calpain. Biochem J. 1996;318:897–901. doi: 10.1042/bj3180897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosenko E, Poghosyan A, Kaminsky Y. Subcellular compartimentalization of proteolytic enzymes in brain regions and the effects of chronic β-amyloid treatment. Brain Res. 2011;1369:184–193. doi: 10.1016/j.brainres.2010.10.078. [DOI] [PubMed] [Google Scholar]
- Fehrenbacher N, Bastholm L, Kirkegaard-Sørensen T, Rafn B, Bøttzauw T, Nielsen C, et al. Sensitization to the lysosomal cell death pathway by oncogene-induced down-regulation of lysosome-associated membrane proteins 1 and 2. Cancer Res. 2008;68:6623–6633. doi: 10.1158/0008-5472.CAN-08-0463. [DOI] [PubMed] [Google Scholar]
- DuVerle DA, Ono Y, Sorimachi H, Mamitsuka H. Calpain cleavage prediction using multiple kernel learning. PLoS One. 2011;6:e19035. doi: 10.1371/journal.pone.0019035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyenbach KW, Wieczorek H. The V-type H+ ATPase: molecular structure and function, physiological roles and regulation. J Exp Biol. 2006;209:577–589. doi: 10.1242/jeb.02014. [DOI] [PubMed] [Google Scholar]
- Nozaki K, Das A, Ray SK, Banik NL. Calpeptin attenuated apoptosis and intracellular inflammatory changes in muscle cells. J Neurosci Res. 2011;89:536–543. doi: 10.1002/jnr.22585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien J, Hansen K, Barkan D, Green J, Schedin P. Non-steroidal anti-inflammatory drugs target the pro-tumorigenic extracellular matrix of the postpartum mammary gland. Int J Dev Biol. 2011;55:745–755. doi: 10.1387/ijdb.113379jo. [DOI] [PubMed] [Google Scholar]
- Booth BW, Jhappan C, Merlino G, Smith GH. TGFbeta1 and TGFalpha contrarily affect alveolar survival and tumorigenesis in mouse mammary epithelium. Int J Cancer. 2007;120:493–499. doi: 10.1002/ijc.22310. [DOI] [PubMed] [Google Scholar]
- Zhang F, Wang Q, Ye L, Feng Y, Zhang X. Hepatitis B virus X protein upregulates expression of calpain small subunit 1 via nuclear factor-kappaB/p65 in hepatoma cells. J Med Virol. 2010;82:920–928. doi: 10.1002/jmv.21753. [DOI] [PubMed] [Google Scholar]
- Liu L, Xing D, Chen WR. Micro-calpain regulates caspase-dependent and apoptosis inducing factor-mediated caspase-independent apoptotic pathways in cisplatin-induced apoptosis. Int J Cancer. 2009;125:2757–2766. doi: 10.1002/ijc.24626. [DOI] [PubMed] [Google Scholar]
- Kar P, Samanta K, Shaikh S, Chowdhury A, Chakraborti T, Chakraborti S. Mitochondrial calpain system: an overview. Arch Biochem Biophys. 2010;495:1–7. doi: 10.1016/j.abb.2009.12.020. [DOI] [PubMed] [Google Scholar]
- Cuervo AM, Dice JF. Regulation of lamp2a levels in the lysosomal membrane. Traffic. 2000;1:570–583. doi: 10.1034/j.1600-0854.2000.010707.x. [DOI] [PubMed] [Google Scholar]
- McHenry P, Wang W-LW, Devitt E, Kluesner N, Davisson VJ, McKee E, et al. Iejimalides A and B inhibit lysosomal vacuolar H+-ATPase (V-ATPase) activity and induce S-phase arrest and apoptosis in MCF-7 cells. J Cell Biochem. 2010;109:634–642. doi: 10.1002/jcb.22438. [DOI] [PubMed] [Google Scholar]
- Nakashima S, Hiraku Y, Tada-Oikawa S, Hishita T, Gabazza EC, Tamaki S, et al. Vacuolar H+-ATPase inhibitor induces apoptosis via lysosomal dysfunction in the human gastric cancer cell line MKN-1. J Biochem. 2003;134:359–364. doi: 10.1093/jb/mvg153. [DOI] [PubMed] [Google Scholar]
- Zaragozá R, Gimeno A, Miralles VJ, García-Trevijano ER, Carmena R, García C, et al. Retinoids induce MMP-9 expression through RARa during mammary gland remodeling. Am J Physiol (Endocrinol Metab) 2007;292:1140–1148. doi: 10.1152/ajpendo.00463.2006. [DOI] [PubMed] [Google Scholar]
- Borrás C, Gambini J, López-Grueso R, Pallardó FV, Viña J. Direct antioxidant and protective effect of estradiol on isolated mitochondria. Biochim Biophys Acta. 2010;1802:205–211. doi: 10.1016/j.bbadis.2009.09.007. [DOI] [PubMed] [Google Scholar]
- Muñoz J, Fernández-Irigoyen J, Santamaría E, Parbel A, Obeso J, Corrales FJ. Mass spectrometric characterization of mitochondrial complex I NDUFA10 variants. Proteomics. 2008;8:1898–1908. doi: 10.1002/pmic.200701085. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.