Abstract
Homozygosity for the naturally occurring Δ32 deletion in the HIV co-receptor CCR5 confers resistance to HIV-1 infection. We generated an HIV-resistant genotype de novo using engineered zinc-finger nucleases (ZFNs) to disrupt endogenous CCR5. Transient expression of CCR5 ZFNs permanently and specifically disrupted ~50% of CCR5 alleles in a pool of primary human CD4+ T cells. Genetic disruption of CCR5 provided robust, stable and heritable protection against HIV-1 infection in vitro and in vivo in a NOG model of HIV infection. HIV-1-infected mice engrafted with ZFN-modified CD4+ T cells had lower viral loads and higher CD4+ T-cell counts than mice engrafted with wild-type CD4+ T cells, consistent with the potential to reconstitute immune function in individuals with HIV/AIDS by maintenance of an HIV-resistant CD4+ T-cell population. Thus adoptive transfer of ex vivo expanded CCR5 ZFN–modified autologous CD4+ T cells in HIV patients is an attractive approach for the treatment of HIV-1 infection.
CCR5, a seven-transmembrane chemokine receptor, is the major co-receptor for HIV-1 entry1,2. Since the discovery that the homozygous Δ32 deletion in CCR5 confers resistance to HIV-13-5, CCR5 has been intensely studied and validated as a target for HIV therapy6,7. Recently, small-molecule approaches that block the CCR5-HIV interaction have shown promise in clinical trials8. However, the small-molecule approach has resulted in the development of resistance by selection for escape mutants, which continue to use CCR5 for viral entry9. These results, taken together with experience from individuals heterozygous for the Δ32 allele, point to the importance of a genetic knockout of CCR5 for phenotypic penetrance and long-term resistance to infection rather than its knock-down by approaches based on small molecules, intrabodies, antisense or RNA interference (RNAi)10-15. Therefore, we sought to permanently disrupt the endogenous CCR5 and thus make a phenocopy of the Δ32CCR5 null genotype in primary human CD4+ T cells by the application of engineered ZFNs.
Previously, we have shown that reconstituting CD4+ helper T-cell activity through adoptive transfer of costimulated CD4+ T cells may augment natural immunity to HIV-1 infection13. Here we show that engineered ZFNs targeting human CCR5 efficiently generate a double-strand break at a predetermined site in the CCR5 coding region upstream of the natural CCR5Δ32 mutation. The CCR5 ZFNs promote efficient and permanent disruption of CCR5 in primary human CD4+ T lymphocytes and confer robust protection against HIV-1 infection both in vitro and in an in vivo mouse model of HIV-1 infection. Combining the two approaches may provide further benefit to patients with HIV-1 in future clinical trials.
RESULTS
Design of ZFNs targeted against CCR5 (CCR5 ZFN)
We designed and optimized a large series of ZFNs targeted to human CCR5 using a previously described approach16. For both target sites two zinc-finger protein (ZFP) DNA-binding domains, each containing four zinc-finger motifs (recognizing a total of 24 base pairs), were assembled from an archive of ZFP DNA-binding modules17,18. These ZFPs were coupled to the DNA cleavage domain of the type IIS restriction enzyme, FokI, to produce novel ZFNs in which the location of DNA cleavage is determined by the DNA-binding specificity of the engineered ZFP domains, as previously shown16,17,19. Targeting a double-strand break to a specific site in the genome with ZFNs has been used to disrupt permanently the genomic sequence surrounding the ZFN target site in a variety of eukaryotic organisms20,21 via imperfect repair by nonhomologous end joining (NHEJ)22,23. To exploit this property of double-strand break repair, we elected to focus our ZFN designs upon the DNA sequence encoding the first transmembrane domain (TM1 spans residues Arg31 to Asn57) of the CCR5 co-receptor. We reasoned that this location, upstream of the Δ32 mutation, would display substantial structural sensitivity in the context of the CCR5 protein. Thus mutations introduced during repair via NHEJ would be predicted to result in truncated or nonfunctional gene products that would fail to be expressed on the cell surface, in a manner analogous to the naturally occurring Δ32 mutant allele3,4. The lead ZFN pair binds the sequence flanking the codon for Leu55 (within TM1) of human CCR5 (Fig. 1a), and is referred to throughout as ZFN-215. A variant of these ZFNs (ZFN-224) was generated that incorporates engineered FokI domains that function as obligate heterodimers and thereby improve ZFN specificity. The complete sequence of the ZFN pair is shown in FASTA format (Supplementary Fig. 1 online).
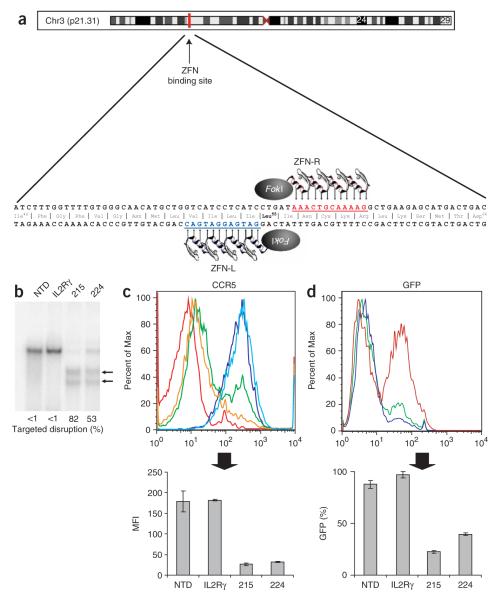
Figure 1.
ZFN-mediated disruption of CCR5 and protection from HIV-1 infection in GHOST-CCR5 cells. (a) Schematic of the CCR5 coding region showing the genomic DNA sequences targeted by CCR5 ZFNs 215/224. (b) Level of targeted gene disruption in GHOST-CCR5 cells transduced with an Ad5/35 vector encoding ZFNs targeting either CCR5 or IL-2Rγ as assessed by the Surveyor assay (Supplementary Fig. 2). Lower migrating products (arrows) are a direct measure of ZFN-mediated gene disruption. NTD, nontransduced cells. (c) Decreased CCR5 surface expression measured by flow cytometry (light blue, IL2Rγ ZFN; green CCR5 ZFN-224; orange, CCR5 ZFN-215; dark blue, nontransduced cells; red, unstained cells). (d) Protection from HIV-1BAL measured by flow cytometry 48 h after HIV-1 challenge of CCR5 ZFN-215 and CCR5 ZFN-224-modified cells compared to IL2Rγ ZFN and control GHOST-CCR5 cells (red, IL2Rγ ZFN; green, CCR5 ZFN-224; blue, CCR5 ZFN-215). GFP fluorescence indicates HIV-1 entry and is plotted as average percent infected relative to positive control. MFI, mean fluorescence intensity. Histograms of CCR5 (c) and GFP expression (d) show one replicate for each condition; bar graphs below represent averages (±s.d.) of triplicates. Expression of CCR5 and HIV-1 infection frequency of CCR5 ZFN-treated cells is less than IL2Rγ ZFN or nontransduced cells (P < 0.001).
Entry inhibition of CCR5-tropic HIV-1 by ZFN-targeted disruption
To determine whether transient expression of the CCR5 ZFNs would alter CCR5 expression levels and HIV-1 entry, we transduced GHOST-CCR5 cells, a reporter cell line for HIV-1 infection containing multiple (approximately four) copies of an autologous CCR5 expression cassette and an inducible green fluorescent protein (GFP) marker gene under the control of the HIV-2 long terminal repeat (LTR)24, with CCR5 ZFNs and subjected them to CCR5-tropic HIV-1 challenge. To achieve transient yet high-efficiency ZFN delivery, we transduced GHOST-CCR5 cells with an adenovirus (Ad5/35) vector25 encoding the lead CCR5 ZFNs. First, we confirmed and quantified the generation of ZFN-induced mutations at the target site using an assay based upon the mismatch-sensitive Surveyor nuclease (Supplementary Fig. 2 online). DNA analysis using this assay revealed high-efficiency (50–80%) target gene mutation in the population of GHOST-CCR5 cells transduced with CCR5 ZFNs (Fig. 1b). This result was CCR5 ZFN dependent as neither nontransduced control cells, nor cells transduced with an Ad5/35 vector encoding interleukin (IL)-2Rγ–specific ZFNs16 showed detectable CCR5 modifications (Fig. 1b).
These transduced cell populations were maintained in culture and 1 week later were infected with HIV-1BAL, a prototype CCR5-tropic HIV-1 isolate. Immediately before HIV-1 infection, CCR5 surface expression was analyzed and found to be reduced by more than tenfold in the pool of CCR5 ZFN–transduced cells compared with control IL-2Rγ-ZFN–treated cells (Fig. 1c). Consistent with this reduction in CCR5 expression, HIV-1BAL challenge demonstrated a substantial decrease in HIV-1 infection in CCR5 ZFN-treated samples, as measured by loss of GFP induction 48 h after infection (Fig. 1d). Genetic modification at the intended target site within CCR5 was confirmed by sequencing of genomic DNA from ZFN-treated GHOST-CCR5 cells (data not shown). CCR5 reconstitution experiments were carried out to confirm the mechanism of resistance to HIV-1 infection in the experiments described above. Single cell–derived clones from the ZFN-treated GHOST-CCR5 population were isolated and shown to be completely resistant to HIV-1 infection using CCR5-tropic HIV-1BAL in contrast to cloned GHOST-CCR5 cells that retained unmodified CCR5 genes (Supplementary Fig. 3a online). CCR5 reconstitution experiments demonstrated that resistance to HIV-1 infection was mediated exclusively by a defect in viral entry through ZFN-mediated CCR5 disruption (Supplementary Fig. 3b). Taken together these results demonstrate that the CCR5 ZFNs can efficiently cleave their DNA target site in CCR5 and confirm that a high proportion of ZFN-induced mutations prevent CCR5 cell-surface expression, resulting in resistance to CCR5-tropic HIV-1 infection.
Survival advantage of ZFN-modified CD4+ T cells in vitro
Next, we evaluated whether ZFN-mediated disruption of CCR5 would confer the long-term resistance to HIV-1 expected from a permanent genetic change. PM1 cells, a CD4+ T-cell line with similar levels of CCR5 expression to primary CD4+ T cells (data not shown), were electroporated with sub-optimal amounts of CCR5 ZFN expression plasmids. Analysis of the DNA from this population of treated cells demonstrated a baseline endogenous CCR5 disruption level of 2.4% of the alleles (Fig. 2a). On day 7, this ZFN-treated cell population was infected with HIV-1BAL or mock infected. The cells were expanded in continuous culture for 70 d, and the proportion of ZFN-modified alleles measured by DNA analysis on the indicated days (Fig. 2a). By day 52 of infection, the HIV-1–infected PM1 culture had undergone a B30-fold enrichment for ZFN-modified CCR5 alleles (Fig. 2a). The level of enrichment observed by day 52 approaches the upper limit of detection of the Surveyor assay and may underestimate the true frequency of disruption (Supplementary Fig. 2b). In contrast, the mock-infected population showed stable persistence of the ZFN-disrupted CCR5 alleles at ~2.3% frequency, indicating no adverse consequences in growth rates for cells carrying a ZFN–modified allele in the absence of selective pressure (Fig. 2a). PM1 cells electroporated with control ZFN expression plasmids were susceptible to HIV-1 infection and showed no evidence of CCR5 disruption (data not shown). These data demonstrate that HIV-1 infection provides a potent selective advantage for CCR5 ZFN-modified cells, an advantage that is maintained long-term in culture.
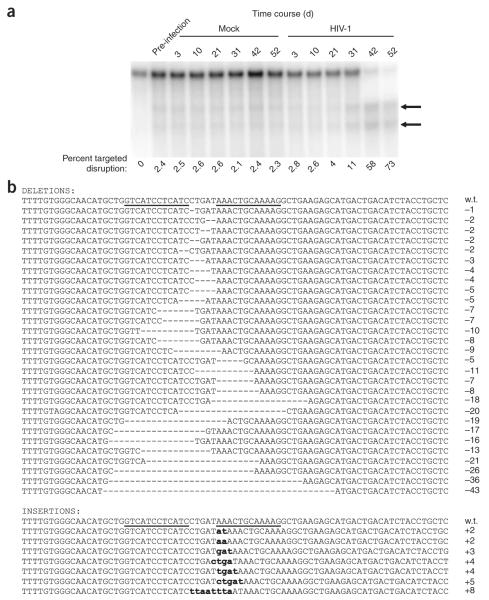
Figure 2.
In vitro selection of CCR5-disrupted cells following HIV-1 challenge of the CD4+ T-cell line, PM1. (a) The level of ZFN-disrupted CCR5 alleles was determined at the indicated times post-HIV-1 challenge with R5-tropic HIV-1BAL or after mock HIV-1 infection. Disrupted CCR5 alleles remained at stable levels in mock-infected cultures but were enriched in the presence of HIV-1. Similar results were obtained in two experiments, each extending for over 2 months. (b) The genomic CCR5 ZFN target site in ZFN-treated PM1 cells at day 52 post-HIV-1 challenge was amplified, cloned and sequenced to confirm ZFN cleavage as the molecular basis of HIV-1 resistance. Sequence alignment revealed distinct ZFN-induced insertions and deletions within the target region of CCR5.
To determine the molecular identity of the ZFN-mediated mutations in CCR5 that were selected by HIV-1 infection, we amplified by PCR the CCR5 ZFN target region from genomic DNA harvested at day 52 after HIV-1 infection from ZFN-224–transfected PM1 cells. Sequencing of this product revealed numerous molecularly distinct short deletions and insertions in 78% of sequence reads (63 out of 81 sequences), indicating that we did not select for any specific rare event (Fig. 2b). All of the mutations found mapped to the core of the ZFN recognition site, suggesting that the permanent modifications to CCR5 were the product of ZFN-cleavage and subsequent repair via NHEJ. Although a broad range of different deletion and insertion mutations was observed, a specific 5-bp insertion (a duplication of the sequence between the ZFN binding sites) represented >30% of all modified sequences (data not shown). This particular modification resulted in the introduction of two stop codons immediately after Isoleucine56, generating a truncation of the wild-type protein at this residue.
Superinfection experiments confirmed that CCR5 ZFN–modified PM1 cells remained susceptible to CXCR4-tropic HIV-1, and maintained a selective advantage when reinfected with CCR5-tropic virus (Supplementary Fig. 4a online). Viral evolution toward CXCR4 co-receptor usage was not detected in supernatants collected at early and late time points from CCR5 ZFN–treated and HIV-1–infected cultures (Supplementary Fig. 4b). Analysis of viral envelope V3 sequences isolated from the cultures supports this finding (Supplementary Fig. 4c,d), as viruses isolated from both GFP- and CCR5 ZFN–treated cells matched most closely to the CCR5-tropic consensus envelope sequence. Together, these results demonstrate that transient expression of CCR5 ZFNs can establish stable and selective resistance to CCR5-tropic HIV-1, consistent with expectations based on individuals carrying the naturally occurring CCR5Δ32 mutation3-5,26.
Selective advantage of CCR5 ZFN–modified primary CD4+ T cells during HIV-1 infection
To determine the efficacy of CCR5 ZFNs in primary human cells, we transduced CD4+ T cells from healthy wild-type CCR5 donors with an Ad5/35 vector encoding CCR5 ZFNs to provide transient yet high-efficiency ZFN delivery. Multiplicities of infection (MOI)-dependent levels of ZFN-mediated CCR5 disruption (reaching 40–60% of the CCR5 alleles) were observed in multiple experiments using cells isolated from different donors (Fig. 3a and data not shown). The population-doubling rate of the modified primary CD4+ T cells was indistinguishable from nontransduced cells, with the proportion of CCR5-modified alleles remaining stable for at least 1 month during in vitro culture (Fig. 3b and data not shown).
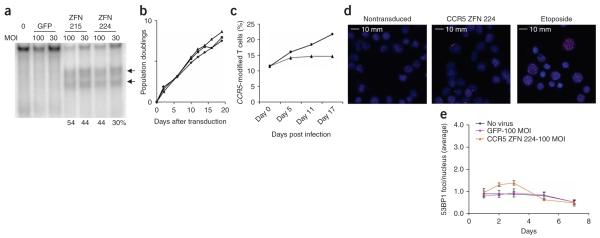
Figure 3.
Enrichment of CCR5 ZFN-modified primary CD4+ T cells during in vitro HIV-1 challenge. (a) Primary CD4+ T cells from CCR5 wild-type anonymous healthy donor were transduced with Ad5/35 vector expressing CCR5 ZFN-215, ZFN-224 or GFP at MOI of 30 and 100; percentage of total alleles are indicated below each lane. (b) Population doubling rate for CCR5 ZFN- and GFP control-transduced CD4+ T cells (triangle, nontransduced; square, CCR5 ZFN-224; diamond, GFP transduced). (c) Enrichment of ZFN-215-transduced CD4+ T cells over time following in vitro challenge with CCR5-tropic HIV-1US1 compared to mock (square, HIV-1 infected; triangle, mock infected). An ~10% starting level of ZFN-disrupted CCR5 alleles was obtained by mixing Ad5/35-transduced CD4+ T cells from a 1 in 3 with unmodified CD4+ T cells. (d) Intranuclear 53BP1 immunostaining and epifluorescence microscopy 2 d after CD4+ T cells were transduced with Ad5/35 vectors expressing CCR5 ZFN pair 224, nontransduced (negative control) or 1 μM etoposide (positive control). Representative images are shown in the panels. (e) The mean (± s.d.) numbers of foci over time is shown following Ad5/35 vector transduction with CCR5 ZFN-224, GFP and nontransduced CD4+ T cells. Significant elevation in the number of foci was observed for CCR5 ZFN-224 treated cells on days 2 (P = 0.03) to 3 post treatment (P = 0.004, unpaired t-test, n = 4), whereas GFP-transduced cells were statistically indistinguishable from nontransduced control lymphocytes.
Infection of a bulk CCR5 ZFN–transduced CD4+ T-cell population with the CCR5-tropic HIV-1US1 resulted in a twofold enrichment of gene-edited cells with ZFN-disrupted CCR5 alleles over 17 d of culture, whereas mock-infected control populations maintained a stable level of ZFN-disrupted CCR5 alleles (Fig. 3c). CD4+ T cells transduced with an Ad5/35 GFP control vector showed no detectable disruption of CCR5 (data not shown).
After demonstrating a high frequency of mutagenesis at the CCR5 locus, we determined what percentage of CD4+ T cells have both alleles modified. CD4+ T cells were infected with the Ad5/35 CCR5 ZFN vector and cloned by flow sorting. Fifty-two T-cell clones were obtained, and 12/52 (23%) had CCR5 modification as judged by the Surveyor nuclease assay, confirming the results found within the bulk population (28–30%) before cloning. The 12 clones with disrupted CCR5 loci were then genotyped by directly sequencing the CCR5 alleles. Four out of the 12 (33%) mutant clones were homozygous for CCR5 disruption. This experiment was done under nonselecting conditions; we expect the frequency of homozygous disruption to be higher in the presence of HIV-1 challenge.
Specificity of CCR5 ZFNs in primary CD4+ T cells
The efficacy and tolerability of CCR5 ZFN–driven genome editing in primary human cells supports the potential clinical application of autologous ZFN-modified CD4+ T cells in patients with HIV. However, in developing such an approach it is critical to verify the specificity of ZFN action. Cleavage at a given target site requires binding of two ZFNs in a specific spatial orientation relative to each other, that is, on opposite strands and separated by 5 or 6 bp to facilitate the dimerization of the FokI domains necessary for DNA cleavage27,28 (Fig. 1a). This serves to restrict the induction of a double-strand break to those positions in the genome where binding sites for two ZFNs are found in the required juxtaposition. To quantify the number of double-strand breaks generated after ZFN expression, we conducted intranuclear staining for genome-wide double-strand breaks via immunodetection of p53 binding protein 1 (53BP1) foci as an unbiased measure of ZFN action throughout the nucleus. 53BP1 is recruited to the sites of double-strand breaks early in the repair response and is required for NHEJ29. The genomic integrity of CD4+ T cells was assessed at several time points after transduction with Ad5/35 expressing the indicated CCR5 ZFN by enumeration of the number of 53BP1 foci per nucleus (Fig. 3d). We observed a transient 1.4–1.6-fold increase on days 2 and 3 of culture in the mean number of intranuclear 53BP1 foci when comparing ZFN-224 transduced to nontransduced or GFP-transduced CD4+ T cells (Fig. 3e). In contrast, etoposide-treated positive control cells had a 4.2-fold increase in 53BP1 staining over control cells that persisted for at least a week (data not shown). No significant difference in the mean perimeter of 53BP1 foci was observed among all conditions (data not shown).
To confirm the specificity of ZFN-224 action, we experimentally determined the consensus ZFN binding site by SELEX (Supplementary Fig. 5a online). The experimentally derived consensus matches the unique intended target sequence in CCR5. To expand this analysis, we extended the consensus to allow up to two mismatches per ZFN binding site and identified the top 15 putative alternate cleavage sites throughout the genome with the highest similarity (Supplementary Fig. 5b). Surveyor nuclease assays revealed no detectable ZFN activity (1% limit of detection) at any of these sites with the exception of CCR2, the closest relative of CCR5 in the human genome. We observed 4.1% modification of CCR2 alleles in the population under conditions that revealed 35.6% ZFN-modified CCR5 alleles (Supplementary Fig. 5c). Note that CCR5 and CCR2 are found juxtaposed to one another on the same chromosome, a fact that may have rendered CCR2 more susceptible to cleavage30,31. The close proximity of these two genes eliminates the possibility of visualizing two independent 53BP1 foci by intranuclear immunodetection (Fig. 3d). Loss of CCR2 in CD4+ T cells is predicted to be well tolerated as CCR2−/− mice display phenotypes that are not disabling32. Mutant alleles of CCR2 have been correlated with delayed progression to AIDS in HIV-infected individuals, although no influence on the incidence of HIV-1 infection was observed33. Thus, parallel mutation of a small proportion of CCR2 in CD4+ T cells ex vivo is unlikely to be deleterious and may increase protection of modified CD4+ T cells to HIV infection. The combination of ZFP consensus-binding-site-directed analysis of the most similar off-target sites in the genome together with the unbiased intranuclear staining for genome-wide double-strand break generation suggests that ZFN-224 is a specific engineered nuclease with measurable activity at only CCR5 and, to an approximately tenfold lesser extent, the CCR5 homolog CCR2.
To increase the probability of detecting rare off-target events at potential genome-wide alternative targets, we performed ultra-deep pyrosequencing34 on bulk CD4+ T cells after ZFN-224 treatment. This approach permits targeted deep sequencing of heterogeneous DNA material. PCR probes for the top 15 sites identified by SELEX (shown in Supplementary Fig. 5b) were designed, and a multiplex PCR assay combined with subsequent 454 pyrosequencing was carried out. Approximately 40,000 sequences were recovered from each site. Under conditions where CCR5 was modified at 36% efficiency, there were 1,995 probable NHEJ events of 37,028 sequences at the CCR2 locus that were read, so that measured disruption frequency by 454 pyrosequencing was 5.39%. Of the other 13 sites, the only additional off-target site found was in an intron of ABLIM2 on chromosome 4, which had a frequency of two mutations in 38,023 sequences (Supplementary Fig. 5d). Low-frequency intronic mutations in ABLIM2 would be expected to have minimal effect on ABLIM2 expression or function. Thus, except for CCR2 (5.39%), and rare (~1/20,000) events at ABLIM2, all the remaining sites showed no evidence of NHEJ, given a threshold level of detection of ~1 in 10,000 sequences or better, dependent on the number of reads in a given sample. Taken together, the 454 pyrosequencing data, the Surveyor nuclease data, the 53BP1 immunostaining and the preservation of biologic and replicative properties of the cells after transduction support the conclusion that the CCR5 ZFNs are specific in CD4+ T cells.
Reduced viremia and selection of CCR5 ZFN-modified primary CD4+ T cells during HIV-1 infection in vivo
To explore the feasibility, safety and therapeutic potential of this approach, we used a NOG mouse35 model of HIV-1 infection to test adoptive transfer and protection from HIV-1 infection of the ZFN-modified CD4+ T cells in vivo. Primary CD4+ T cells were transduced with Ad5/35 vectors expressing the CCR5 ZFNs or GFP, and expanded in culture using anti-CD3/anti-CD28–coated magnetic beads in the presence of IL-2. NOG mice were randomly assigned to two treatment groups, which received CCR5 ZFN–transduced ex-vivo expanded primary human CD4+ T cells and either noninfected or HIV-1–infected phytohemagglutinin A (PHA)-blasted peripheral blood mononuclear cells (PBMC) (Fig. 4a). Peripheral blood sampling was performed on the indicated days after adoptive transfer (Fig. 4a) and analyzed for engraftment by flow cytometry for human CD45, CD4 and CD8 expression. All groups showed equal engraftment, although in the HIV-infected groups, but not in mock-infected controls, we noted a reduced CD4+ to CD8+ T-cell ratio in vivo relative to that infused, consistent with HIV-induced CD4+ T-cell depletion (Supplementary Fig. 6a online).
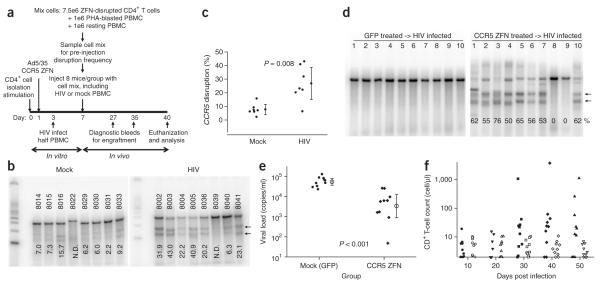
Figure 4.
Reduction in viremia and selection for CCR5 ZFN-modified CD4+ T cells in the presence of HIV-1 challenge in vivo. (a) Experimental outline for adoptive transfer of modified CD4+ T cells and in vivo HIV-1 challenge in NOG mice. CCR5 ZFN-transduced CD4+ T-cell population pre-mix (day 3); CCR5 disruption level, 33%. Injected mixed samples baseline CCR5 disruption level 15% in control (mock infected) and 14% in HIV-infected group (day 7). (b) Level of ZFN-disrupted CCR5 alleles in CD4+ T cells isolated on day 40 from spleens of control or HIV-infected mice. Percent disruption indicated at base of each lane. One mouse from each group (HIV-infected mouse no. 8039 and control mouse no. 8022) excluded for later analysis due to inadequate CD4+ T-cell DNA recovery and purification. N.D., not determined. (c) Plot of in vivo disruption frequencies in spleens on day 40. Results for each group (n = 7) averaged and analyzed using an unpaired t-test with mean ± 95% confidence intervals indicated. (d–f) In an independent experiment, mice were engrafted with CCR5 ZFN-transduced CD4+ T cells (51% disruption) or GFP-transduced cells (mock) and followed for 50 d post HIV-1 infection. Enrichment for CCR5-disrupted CD4+ T cells in peripheral blood on day 50 post infection (Surveyor assay); lower migrating products (arrows) are a direct measure of ZFN-mediated gene disruption (d). Plasma viremia in mice day 10 post infection. HIV-1 viral RNA (copies/ml) is plotted for the individual mice; the mean ± 95% confidence interval is shown (e). The CCR5 ZFN-treated mice had a significantly lower viral load (P < 0.001; Mann Whitney test). Engraftment of CD4+ T cells in peripheral blood from days 10 to 50 post infection. The CD4+ T-cell counts (Trucount assay) for the mice engrafted with CCR5 ZFN-(solid symbols) and GFP-modified cells (open symbols) are plotted (f). Mice engrafted with CCR5 ZFN-treated cells had higher CD4+ T-cell counts on days 30–50 post infection (P = 0.04).
After a month of HIV-1 infection in vivo, mice were killed and genomic DNA from human CD4+ T lymphocytes purified from the spleen was used for analysis of ZFN-mediated CCR5 disruption with the Surveyor nuclease assay (Fig. 4b). Of the CD4+ T-cell DNA preparations that passed quality control (Supplementary Fig. 6b), we found an approximately threefold enrichment for ZFN-disrupted CCR5 alleles in the HIV-infected group (27.5% average CCR5 disruption), compared with animals receiving the identical starting population of ZFN-treated CD4+ T cells in the absence of HIV infection (mock group 8.5%, P = 0.008) (Fig. 4c). An independent experiment was carried out to further determine whether CCR5-modified cells have a protective effect on CD4+ T-cell depletion and on viremia (Fig. 4d-f). Mice were engrafted and infected as in Figure 4a, and followed for 50 d after infection. By day 50 after infection, 8 of 10 HIV-infected mice had >50% CCR5-disrupted CD4+ T cells in peripheral blood (Fig. 4d). The HIV-infected mice had increased numbers of CD4+ T cells in peripheral blood on days 30 to 50 after infection; however, the early engraftment was not different (Fig. 4f). In addition, mice given CCR5 ZFN–treated cells had substantially lower plasma viremia (mean viral load 8,300 copies/ml) than mice populated with the mock CD4+ T cells (mean viral load 60,100); this demonstrates highly significant protection (P < 0.001, n = 10 mice per group; Fig. 4e). Thus, the modified cells confer resistance to HIV-1 infection in vivo as measured by preferential expansion, viral load and CD4+ T-cell counts. Furthermore, these results demonstrate normal engraftment and growth of these same ZFN-transduced cells even in the absence of this selective pressure.
The above studies indicate that the function of CCR5 is abrogated by ZFN treatment as expected. One issue regarding the possible immunogenicity of autologous CCR5-modified CD4+ T cells is whether there is expression of any type of CCR5 fragment after modification. From the DNA sequence data shown in Figure 2b, we determined the predicted protein sequence up to the first stop codon for the insertions and deletions identified in PM-1 cells (Supplementary Fig. 7 online). To further address this concern, we used western blot analysis of CD4+ T-cell lysates following modification by adenoviral infection with the CCR5 ZFNs (Supplementary Fig. 8 online). Nontransduced CD4+ T cells and naturally occurring CCR5Δ32 null mutation CD4+ T cells were used as negative and positive controls, respectively. Using antibodies that bind to the N and C termini of the unmutagenized CCR5 product, we observed a dose-dependent decrease in wild-type CCR5 expression. Notably, no products appeared that were not present in wild-type CD4+ T cells. Together, these data indicate that the transient delivery of engineered ZFNs succeeded in mimicking the selective advantage of the naturally occurring CCR5Δ32 null mutation. Furthermore, the in vivo data demonstrates genome editing to introduce a disease-resistance genotype at therapeutic levels of efficiency.
DISCUSSION
To our knowledge, genome editing that is sufficiently robust to support therapy in an animal model has not been shown previously. The ZFN-guided genomic editing was highly specific and well tolerated, as revealed by examination of the stability, growth and engraftment characteristics of the genome-modified sub-population even in the absence of selection. The fidelity of ZFN action was further supported by direct staining for intranuclear double-strand break–induced 53BP1 foci, testing for cleavage at the most similar putative off-target genomic sites and deep pyrosequencing. Moreover, in the presence of a selective pressure in the form of active HIV-1 infection, ZFN modification conferred a significant survival advantage during CCR5-tropic, but not CXCR4-tropic, HIV-1 challenge assays in vitro to levels comparable to those obtained with naturally occurring homozygous CCR5Δ32 cells. We also observed a threefold enrichment of the ZFN-modified primary human CD4+ T cells and protection from viremia in a NOG mouse model of active HIV-1 infection. As predicted for a genetically determined trait, the ZFN-modified cells demonstrated stable and heritable resistance in progeny cells to HIV-1 infection both in vitro and in vivo. These results demonstrate that ZFN-mediated genome editing can be used to reproduce a CCR5 null genotype in primary human cells.
The selection of CCR5 as the focus of this work stemmed from the earlier discovery of healthy individuals naturally homozygous for the CCR5Δ32 allele and thus possessing a CCR5 null genotype with consequent resistance to CCR5-tropic HIV infection3-5. This finding established CCR5 as a promising target for HIV treatment. Strategies based on small molecules, intrabodies, antisense or RNAi involve partial subtraction or blockade of CCR5 at an mRNA or protein level. For example, gene therapy approaches using RNAi and/or ribozymes targeting CCR5 have been used to prevent HIV-1 infection in vitro11,14 and have now progressed to nonhuman primate models of HIV infection36. The advantage of the present approach is that one can genetically replicate the CCR5 null cell and avoid the use of integrating viral vectors, which have been associated with insertional mutagenesis. It should be noted, however, that the random nature of the ZFN-induced repair process results in a broad range of insertions and deletions. It remains formally possible that some of these mutated alleles encode novel CCR5 epitopes that could be recognized as foreign and eliminated by the host immune system. We note, however, that a specific 5-bp insertion (a duplication of the sequence between the ZFN binding sites) represented >30% of all modified sequences and results in the introduction of two stop codons immediately after Isoleucine56, thus generating a simple truncation of the wild-type protein at this residue.
One limitation of the present work is the availability and validity of animal models to predict the impact of novel biotherapeutics in patients. We have used an acute HIV infection in the NOG mouse, which has the advantage of directly testing the modified human CD4+ T cells and the Ad5/35 CCR5 ZFN virus that will be used clinically. A limitation of this model is that it is an assay of the resistance to infection, in that it demonstrates that the ZFN-modified CD4+ T cells have been made HIV resistant in vivo, but does not extend to modeling the chronic phase of infection or to issues pertaining to the remaining T-cell repertoire in immunodepleted patients. A complementary approach would be to test ZFN-modified CD4+ T cells in the SHIV and SIV nonhuman primate models. However, the sequence of CCR5 within the ZFN binding site in macaques is not conserved with humans and, thus, this experiment would require the design and assembly of a distinct ZFN binding set for testing in SIV infection.
ZFN-modified cells are permanently CCR5 negative, preferentially survive HIV-1 infection and give rise to daughter cells resistant to HIV-1 infection. In practice, such an approach could theoretically complement the use of small-molecule CCR5 inhibitors, which may lead to the emergence of escape variants that retain tropism for CCR537. A number of gene transfer studies in HIV infection have been conducted, demonstrating safety and some evidence of antiviral efficacy38. Autologous CD4+ T cells transduced with a lentiviral vector expressing an anti-HIV antisense molecule have already shown promise in clinical trials of HIV patients failing antiretroviral therapy, despite the limitations of post-entry blockade and the need for random integration of the viral vector into the genome39. ZFN-mediated permanent genetic modification eliminates viral entry without the requirement for the integration of any foreign DNA into the genome, as all of the results obtained here used only transient delivery and expression of the CCR5 ZFNs. Such a selective advantage in vivo could augment the enhancement of CD4+ T-cell counts and possibly anti-HIV immune effects already observed in phase 1/2 clinical trials of adoptive transfer of ex vivo-expanded, costimulated CD4+ T cells in HIV patients.
In summary, the present results support the clinical development of adoptive immunotherapy in the setting of HIV-1 infection to reconstitute or preserve the memory cell pool of HIV-infected patients with ZFN-modified ex vivo expanded, polyclonal CD4+ T cells that are intrinsically resistant to HIV infection. In our recent preclinical studies we have successfully adapted this process to large scale, yielding 1 × 1010 ZFN-modified CD4+ T cells (data not shown), a number sufficient in principle to support clinical trials. The existence of memory T cells with stem cell-like qualities and the capability for extensive self-renewal40,41 further supports the rationale for this approach to replenish the memory T-cell pool. Finally, although there would be additional safety considerations in extending this work to stem cells, recent work indicates that it is possible to apply ZFN-based approaches to stem cells42, so that it is conceivable that the framework presented here could be applied to a number of monogenic congenital and acquired diseases.
METHODS
CCR5 ZFN construct assembly
We designed a series of ZFNs targeted to human CCR5 using a previously described approach16. ZFPs were optimized against the coding sequence of CCR5 and were assembled from an archive of in-vitro-selected modules18,43, assembled as described44, and after α-helix optimization, yielded the following ZFP moieties (target gene; ZFP name; target sequence; recognition α-helices): CCR5; ZFN-R; AAACTGCAAAAG; RSDNLSV, QKINLQV, RSDVLSE, QRNHRTT and CCR5; ZFN-L;GATGAG GATGAC; DRSNLSR, ISSNLNS, RSDNLAR, TSGNLTR. Assembled ZFPs were cloned in-frame as NH2-terminal fusions to the catalytic domain of FokI45-47, and cloned into pVax1 (Invitrogen). The ZFN-224 pair was generated by incorporating engineered FokI domains that function as obligate heterodimers shown previously to improve ZFN specificity48. The ZFNs were cloned into the pAdEasy-1/F35 vector using a 2A sequence and a cytomegalovirus internal promoter, and the Ad5/35 virus was generated as described49.
Surveyor nuclease assay
Genomic DNA was extracted from modified and control cells using the MasturePureTM DNA purification kit (Epicentre Biotechnologies). After radioactive PCR amplification of the CCR5 ZFN binding site, the Surveyor nuclease (Surveyor mutation detection kit; Transgenomic) was used according to the manufacturer. Products were resolved by PAGE and bands quantified by phosphorimager. Ratio of cleaved to uncleaved products was calculated (Supplementary Fig. 2) as a measure of frequency of gene disruption. The assay is sensitive enough to detect single-nucleotide changes induced by NHEJ and has a detection limit of ~1%.
Cell culture
PM150, CXCR4- and CCR5-GHOST24 cells were obtained from the National Institutes of Health (NIH) AIDS Research and Reference Reagent Program; CCR5 GHOST cells were originally produced by KewalRamani and Littman51. Anonymous healthy donors donated lymphocytes at the University of Pennsylvania Apheresis Unit after informed consent under an Institutional Review Board-approved protocol, and cells were processed at the Center for AIDS Research Immunology Core of the University of Pennsylvania. CD4+ T cells were purified from the PBMC using the Miltenyi column bead purification system. CD4+ T cells were maintained at a density of 0.8-1e6 cells/ml in X-Vivo medium with 10% FCS, 1% penicillin-streptomycin and 0.9% N-acetylcysteine and IL-2 at 300 IU/ml after bead stimulation.
Ex vivo targeted gene disruption
PM1 cells were grown according to the suppliers’ instructions and transfected by Nucleofection (Solution V, Program T16, Amaxa Biosystems) according to the manufacturer’s protocol. Cell lines or Miltenyi column-purified CD4+ T cells from healthy donors were activated on day 0 and transduced 24 h later in 12-well plates by the addition of Ad5/35 vector at the specified MOI. GFP control vector routinely resulted in a transduction efficiency of ≥50% at an MOI of 30.
In vitro HIV-1 infection challenges
CCR5 tropic strains, US-1 (gift from J. Mascola) and Bal-1 (gift from S. Gartner), of HIV-1 were used for in vitro challenge infections. CXCR4 tropic HIV-1BK132 was from J. Mascola and used as an X4 control where appropriate. Infections were initiated with MOI from 0.01 (BAL-1) to 0.1 (US-1). Viruses were obtained from the NIH AIDS Research and Reference Reagent Program and propagated in CD8-depleted PBMC to generate working stocks. CCR5 detection was done by flow cytometry using anti-CCR5 monoclonal antibodies 2D7 and 3A9 (Becton Dickinson).
In vivo HIV-1 infection challenges
Primary CD4+ T cells were transduced with the Ad5/35 vectors and expanded in culture using anti-CD3/anti-CD28–coated magnetic beads in the presence of IL-2. NOG mice (7–9 weeks old) were randomly assigned to two treatment groups (n = 8 to 10 mice per group) with an equal mix of males and females in each group. These mice were maintained in a defined flora animal facility at the University of Pennsylvania with approval of our institutional animal care and use committee. Both groups received an intraperitoneal injection of 100 μl of PBS containing 7.5 million CCR5 ZFN exvivo expanded primary human CD4+ T cells and 1 million of resting, autologous PBMCs to promote engraftment in combination. In the experiment shown in Figure 4a–c, the mock-treated animals received 1 million noninfected PHA-blasted autologous PBMCs whereas the infected group of animals received 1 million CCR5-tropic HIV-1US1 infected PHA-blasted PBMCs. In the experiment shown in Figure 4d–f, the mice were injected either with CCR5 ZFN–treated CD4 cells or with GFP-transduced CD4 cells, and all mice were also injected with HIV-1US1–infected PHA-blasted PBMCs.
To assess engraftment, peripheral blood sampling was performed at 10-d intervals after adoptive transfer and analyzed for engraftment by flow cytometry for human CD45, CD4 and CD8. After 4.5 to 7 weeks, mice were killed and CD4+ T lymphocytes from peripheral blood and spleen were purified using the Miltenyi MACS separation kit. Only samples with >75% purity were used for the final analysis. HIV-1 RNA viral loads were determined in mouse plasma at the Contra Costa Public Health Lab using the COBAS Ampliprep/COBAS Taqman HIV-1 test (Roche Diagnostics).
To determine CCR5 disruption frequency, a modified Surveyor nuclease assay was performed by performing a nested PCR approach to fully remove contaminating mouse genomic DNA. The DNA from purified splenic CD4+ T cells was amplified first using 50 pmols of outside primers (R5-det-out-F1: CTGCCTCATAAGGTTGCCCTAAG; C5_HDR_R: CCAGCAATAGATGATC CAACTCAAATTCC) for 25 cycles (95 1C 30 s, 58 1C 30 s and 68 1C 3 min), the resulting material was gel purified, and the Surveyor nuclease assay was performed on the purified product as per the manufacturers’ recommendations.
Microscopy
Intranuclear stain for 53BP1 was performed by collecting CD4+ T cells at the indicated times post-transduction. Slides were prepared by attaching the cells using a cytospin (Thermo Scientific), and fixing the cells with methanol. The cells were then permeabilized by treatment with 0.5% Triton X-100 buffer (0.5% Triton X-100, 1% BSA, 0.02% NaN3, PBS) at 25 1C for 5 min. Cells were then incubated with anti-53BP1 rabbit polyclonal antibodies (Bethyl Laboratories) in the presence of 5% goat serum to block nonspecific staining followed by incubation with Alexa Fluor 594-conjugated secondary antibodies (Invitrogen-Molecular Probes). Slides were mounted in the presence of DAPI (Invitrogen-Molecular Probes) to counterstain cell nuclei and examined under an immunofluorescence microscope (Nikon Eclipse 80i). Images were acquired with a CCD camera, and the data was analyzed using the SimplePCI software (Compix). Analysis of discrete regions of 53BP1 fluorescence was performed by adjusting exposure time and thresholds to minimize autofluorescence. Individual regions identified were then enumerated and measured. Only green fluorescent regions that colocalized with DAPI fluorescence were included in the final analyses.
Pyrosequencing
Pyrosequencing-based technology34 was used to measure low frequency DNA mutational events (454 Sequencing). This technology permits rapid (4 h) sequencing of a large number (250,000) of distinct DNA molecules permitting the ultra-deep sequencing of the bulk CD4+ T-cell population modified by ZFN treatment. The 454 pyrosequencing technology operates to provide sequence information from no fewer than 400 K DNA molecules with an average read length of 100 bp in a single run. To multiplex this assay and monitor ZFN at any of the predicted top 15 off-target sites, we made use of the fact that the 454 process uses an initial PCR step—the target locus is amplified before sequencing. Therefore, the designed PCR primer pairs for each locus serve as distinct sequence tags to uniquely identify PCR products originating from each potential off-target site. For comparison, we sequenced the region of the CCR5 locus targeted by CCR5 ZFN-224. PCR products from these different samples were pooled and analyzed simultaneously in the context of a single 454 sequencing run. This strategy for multiplexing the results from a single sequencing run has been validated in several cellular proof-of-concept studies, and the sensitivity of the data indicates that this method detects a mutation frequency of >1:10,000 cells.
Statistical analysis
Data from at least three sets of samples were used for statistical analysis. Mean ± s.d. are shown. Statistical significance was calculated by Student’s t-test, or Mann-Whitney U test if indicated. P-values <0.05 were considered significant.
Supplementary Material
ACKNOWLEDGMENTS
Research supported in part by National Institutes of Health, a grant from ATP NIST and the Abramson Family Cancer Research Institute. Elena Perez was supported by K08AI062468 for this work. The authors are grateful for constructive comments from Frederick Bushman, for help by Anthony Secreto and other lab members, for support from the Center for AIDS Research Cores, for advice from Bruce Levine and Gwen Binder, for bioinformatics support from Beilin Zhang, for analysis of the V3 loop data by Toby Dylan Hocking, and for experimental assistance from Gwenn-aël H. Danet-Desnoyers and the Xenograft Core Facility at the University of Pennsylvania School of Medicine, Erica Moehle, Jeremy Rock, Lei Zhang, Shuyuan Yao, Nhu Tran, Matthew Mendel, Deng Xia and Sarah Hinkley and members of the Sangamo production group, Melody Hung-Fan and the Contra Costa Public Health Lab for HIV RNA analyses, for pAdEasy-1/F35 vector provided by Xiaolong Fong, and at Bioqual Inc., Mark Lewis and Jake Yalley-Ogunro. CXCR4 tropic HIV-1BK132 and CCR5 tropic strains, US-1 were from John Mascola (Vaccine Research Center, NIH, Bethesda, Maryland), and Bal-1 was from Suzanne Gartner (Johns Hopkins, Baltimore). The following reagent was obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: (GHOST (3) Hi-5 and GHOST (3) CXCR4) from Vineet N. KewalRamani and Dan R. Littman.
Footnotes
Requests for reagents. mholmes@sangamo.com.
Note: Supplementary information is available on the Nature Biotechnology website.
AUTHOR CONTRIBUTIONS
E.E.P., Y.J., J.W., O.L., C.C., K.A.K., J.S.O., J.C.M., V.V.B., D.Y.G., I.R., A.J.W., Y.-L.L., N.W., G.L., F.D.U. and E.J.R. designed and performed experiments; R.G.C., D.A. and P.D.G. assisted with experimental design; J.L.R., M.C.H., P.D.G. and C.H.J. are co-senior authors; E.E.P., M.C.H., P.D.G. and C.H.J. wrote the manuscript.
COMPETING INTERESTS STATEMENT
The authors declare competing financial interests: details accompany the full-text HTML version of the paper at http://www.nature.com/naturebiotechnology/.
References
- 1.Deng HK, et al. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 2.Alkhatib G, et al. Cc Ckrs: A Rantes, Mip-1 Alpha, Mip-1 Beta Receptor As A Fusion Cofactor for Macrophage-Tropic HIV-1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 3.Liu R, et al. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell. 1996;86:367–377. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- 4.Samson M, et al. Resistance to HIV-1 infection in Caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature. 1996;382:722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 5.Huang YX, et al. The role of a mutant CCR5 allele in HIV-1 transmission and disease progression. Nat. Med. 1996;2:1240–1243. doi: 10.1038/nm1196-1240. [DOI] [PubMed] [Google Scholar]
- 6.Lederman MM, et al. Prevention of vaginal SHIV transmission in rhesus macaques through inhibition of CCR5. Science. 2004;306:485–487. doi: 10.1126/science.1099288. [DOI] [PubMed] [Google Scholar]
- 7.Mosier DE, et al. Highly potent RANTES analogues either prevent CCR5-using human immunodeficiency virus type 1 infection in vivo or rapidly select for CXCR4-using variants. J. Virol. 1999;73:3544–3550. doi: 10.1128/jvi.73.5.3544-3550.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fatkenheuer G, et al. Efficacy of short-term monotherapy with maraviroc, a new CCR5 antagonist, in patients infected with HIV-1. Nat. Med. 2005;11:1170–1172. doi: 10.1038/nm1319. [DOI] [PubMed] [Google Scholar]
- 9.Kuhmann SE, et al. Genetic and phenotypic analyses of human immunodeficiency virus type 1 escape from a small-molecule CCR5 inhibitor. J. Virol. 2004;78:2790–2807. doi: 10.1128/JVI.78.6.2790-2807.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abad JL, et al. Novel interfering bifunctional molecules against the CCR5 coreceptor are efficient inhibitors of HIV-1 infection. Mol. Ther. 2003;8:475–484. doi: 10.1016/s1525-0016(03)00202-8. [DOI] [PubMed] [Google Scholar]
- 11.Bai JR, et al. Characterization of anti-CCR5 ribozyme-transduced CD34(+) hematopoietic progenitor cells in vitro and in a SCID-hu mouse model in vivo. Mol. Ther. 2000;1:244–254. doi: 10.1006/mthe.2000.0038. [DOI] [PubMed] [Google Scholar]
- 12.Barassi C, et al. Induction of murine mucosal CCR5-reactive antibodies as an anti-human immunodeficiency virus strategy. J. Virol. 2005;79:6848–6858. doi: 10.1128/JVI.79.11.6848-6858.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levine BL, et al. Adoptive transfer of costimulated CD4(+) T cells induces expansion of peripheral T cells and decreased CCR5 expression in HIV infection. Nat. Med. 2002;8:47–53. doi: 10.1038/nm0102-47. [DOI] [PubMed] [Google Scholar]
- 14.Qin XF, An DS, Chen ISY, Baltimore D. Inhibiting HIV-1 infection in human T cells by lentiviral-mediated delivery of small interfering RNA against CCR5. Proc. Natl. Acad. Sci. USA. 2003;100:183–188. doi: 10.1073/pnas.232688199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Steinberger P, Andris-Widhopf J, Buhler B, Torbett BE, Barbas CF. Functional deletion of the CCR5 receptor by intracellular immunization produces cells that are refractory to CCR5-dependent HIV-1 infection and cell fusion. Proc. Natl. Acad. Sci. USA. 2000;97:805–810. doi: 10.1073/pnas.97.2.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Urnov FD, et al. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- 17.Moore M, Choo Y, Klug A. Design of polyzinc finger peptides with structured linkers. Proc. Natl. Acad. Sci. USA. 2001;98:1432–1436. doi: 10.1073/pnas.98.4.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jamieson AC, Miller JC, Pabo CO. Drug discovery with engineered zinc-finger proteins. Nat. Rev. Drug Discov. 2003;2:361–368. doi: 10.1038/nrd1087. [DOI] [PubMed] [Google Scholar]
- 19.Smith J, et al. Requirements for double-strand cleavage by chimeric restriction enzymes with zinc finger DNA-recognition domains. Nucleic Acids Res. 2000;28:3361–3369. doi: 10.1093/nar/28.17.3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bibikova M, Golic M, Golic KG, Carroll D. Targeted chromosomal cleavage and mutagenesis in Drosophila using zinc-finger nucleases. Genetics. 2002;161:1169–1175. doi: 10.1093/genetics/161.3.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lloyd A, Plaisier CL, Carroll D, Drews GN. Targeted mutagenesis using zinc-finger nucleases in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2005;102:2232–2237. doi: 10.1073/pnas.0409339102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jasin M. Genetic manipulation of genomes with rare-cutting endonucleases. Trends Genet. 1996;12:224–228. doi: 10.1016/0168-9525(96)10019-6. [DOI] [PubMed] [Google Scholar]
- 23.Valerie K, Povirk LF. Regulation and mechanisms of mammalian double-strand break repair. Oncogene. 2003;22:5792–5812. doi: 10.1038/sj.onc.1206679. [DOI] [PubMed] [Google Scholar]
- 24.Morner A, et al. Primary human immunodeficiency virus type 2 (HIV-2) isolates, like HIV-1 isolates, frequently use CCR5 but show promiscuity in coreceptor usage. J. Virol. 1999;73:2343–2349. doi: 10.1128/jvi.73.3.2343-2349.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schroers R, et al. Gene transfer into human T lymphocytes and natural killer cells by Ad5/F35 chimeric adenoviral vectors. Exp. Hematol. 2004;32:536–546. doi: 10.1016/j.exphem.2004.03.010. [DOI] [PubMed] [Google Scholar]
- 26.Hung CS, Vander Heyden N, Ratner L. Analysis of the critical domain in the V3 loop of human immunodeficiency virus type 1 gp120 involved in CCR5 utilization. J. Virol. 1999;73:8216–8226. doi: 10.1128/jvi.73.10.8216-8226.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bibikova M, et al. Stimulation of homologous recombination through targeted cleavage by chimeric nucleases. Mol. Cell. Biol. 2001;21:289–297. doi: 10.1128/MCB.21.1.289-297.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bitinaite J, Wah DA, Aggarwal AK, Schildkraut I. FokI dimerization is required for DNA cleavage. Proc. Natl. Acad. Sci. USA. 1998;95:10570–10575. doi: 10.1073/pnas.95.18.10570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schultz LB, Chehab NH, Malikzay A, Halazonetis TD. p53 Binding protein 1 (53BP1) is an early participant in the cellular response to DNA double-strand breaks. J. Cell Biol. 2000;151:1381–1390. doi: 10.1083/jcb.151.7.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thiriet C, Hayes JJ. Chromatin in need of a fix: Phosphorylation of H2AX connects chromatin to DNA repair. Mol. Cell. 2005;18:617–622. doi: 10.1016/j.molcel.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 31.Tsukuda T, Fleming AB, Nickoloff JA, Osley MA. Chromatin remodelling at a DNA double-strand break site in Saccharomyces cerevisiae. Nature. 2005;438:379–383. doi: 10.1038/nature04148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peters W, Dupuis M, Charo IF. A mechanism for the impaired IFN-gamma production in C-C chemokine receptor 2 (CCR2) knockout mice: Role of CCR2 in linking the innate and adaptive immune responses. J. Immunol. 2000;165:7072–7077. doi: 10.4049/jimmunol.165.12.7072. [DOI] [PubMed] [Google Scholar]
- 33.Smith MW, et al. CCR2 chemokine receptor and AIDS progression. Nat. Med. 1997;3:1052–1053. doi: 10.1038/nm1097-1052c. [DOI] [PubMed] [Google Scholar]
- 34.Margulies M, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Watanabe S, et al. Hematopoietic stem cell-engrafted NOD/SCID/IL2Rgamma null mice develop human lymphoid systems and induce long-lasting HIV-1 infection with specific humoral immune responses. Blood. 2007;109:212–218. doi: 10.1182/blood-2006-04-017681. [DOI] [PubMed] [Google Scholar]
- 36.An DS, et al. Stable reduction of CCR5 by RNAi through hematopoietic stem cell transplant in non-human primates. Proc. Natl. Acad. Sci. USA. 2007;104:13110–13115. doi: 10.1073/pnas.0705474104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trkola A, et al. HIV-1 escape from a small molecule, CCR5-specific entry inhibitor does not involve CXCR4 use. Proc. Natl. Acad. Sci. USA. 2002;99:395–400. doi: 10.1073/pnas.012519099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rossi JJ, June CH, Kohn DB. Genetic therapies against HIV. Nat. Biotechnol. 2007;25:1444–1454. doi: 10.1038/nbt1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Levine BL, et al. Gene transfer in humans using a conditionally replicating lentiviral vector. Proc. Natl. Acad. Sci. USA. 2006;103:17372–17377. doi: 10.1073/pnas.0608138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sallusto F, Lenig D, Forster R, Lipp M, Lanzavecchia A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature. 1999;401:708–712. doi: 10.1038/44385. [DOI] [PubMed] [Google Scholar]
- 41.Zhang Y, Joe G, Hexner E, Zhu J, Emerson SG. Host-reactive CD8(+) memory stem cells in graft-versus-host disease. Nat. Med. 2005;11:1299–1305. doi: 10.1038/nm1326. [DOI] [PubMed] [Google Scholar]
- 42.Lombardo A, et al. Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nat. Biotechnol. 2007;25:1298–1306. doi: 10.1038/nbt1353. [DOI] [PubMed] [Google Scholar]
- 43.Isalan M, Klug A, Choo Y. A rapid, generally applicable method to engineer zinc fingers illustrated by targeting the HIV-1 promoter. Nat. Biotechnol. 2001;19:656–660. doi: 10.1038/90264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Isalan M, Choo Y. Rapid, high-throughput engineering of sequence-specific zinc finger DNA-binding proteins. Methods Enzymol. 2001;340:593–609. doi: 10.1016/s0076-6879(01)40444-7. [DOI] [PubMed] [Google Scholar]
- 45.Bibikova M, Beumer K, Trautman JK, Carroll D. Enhancing gene targeting with designed zinc finger nucleases. Science. 2003;300:764. doi: 10.1126/science.1079512. [DOI] [PubMed] [Google Scholar]
- 46.Porteus MH, Baltimore D. Chimeric nucleases stimulate gene targeting in human cells. Science. 2003;300:763. doi: 10.1126/science.1078395. [DOI] [PubMed] [Google Scholar]
- 47.Smith J, Berg JM, Chandrasegaran S. A detailed study of the substrate specificity of a chimeric restriction enzyme. Nucleic Acids Res. 1999;27:674–681. doi: 10.1093/nar/27.2.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miller JC, et al. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat. Biotechnol. 2007;25:778–785. doi: 10.1038/nbt1319. [DOI] [PubMed] [Google Scholar]
- 49.Nilsson M, et al. Development of an adenoviral vector system with adenovirus serotype 35 tropism; efficient transient gene transfer into primary malignant hematopoietic cells. J. Gene Med. 2004;6:631–641. doi: 10.1002/jgm.543. [DOI] [PubMed] [Google Scholar]
- 50.Lusso P, et al. Growth of macrophage-tropic and primary human-immunodeficiency virus type 1 (HIV-1) isolates in a unique CD4+ T-cell clone (PM1): failure to down-regulate CD4 and to interfere with cell-line-tropic HIV-1. J. Virol. 1995;69:3712–3720. doi: 10.1128/jvi.69.6.3712-3720.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Morner A, et al. Primary human immunodeficiency virus type 2 (HIV-2) isolates, like HIV-1 isolates, frequently use CCR5 but show promiscuity in coreceptor usage. J. Virol. 1999;73:2343–2349. doi: 10.1128/jvi.73.3.2343-2349.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.