SUMMARY
The oncogenic transcription factor TAL1/SCL is aberrantly expressed in over 40% of cases of human T-cell acute lymphoblastic leukemia (T-ALL), emphasizing its importance in the molecular pathogenesis of T-ALL. Here we identify the core transcriptional regulatory circuit controlled by TAL1 and its regulatory partners HEB, E2A, LMO1/2, GATA3 and RUNX1. We show that TAL1 forms a positive interconnected auto-regulatory loop with GATA3 and RUNX1, and that the TAL1 complex directly activates the MYB oncogene, forming a positive feed-forward regulatory loop that reinforces and stabilizes the TAL1-regulated oncogenic program. One of the critical downstream targets in this circuitry is the TRIB2 gene, which is oppositely regulated by TAL1 and E2A/HEB and is essential for the survival of T-ALL cells.
INTRODUCTION
In human T-cell acute lymphoblastic leukemia (T-ALL), the normal molecular events contributing to thymocyte development are interrupted by genetic lesions that induce arrested differentiation, dysregulated proliferation and aberrant survival, leading to clonal expansion of the fully transformed leukemic cells (Armstrong and Look, 2005; Look, 1997). TAL1/SCL, one of the most prevalent oncogenic transcription factors in T-ALL, is overexpressed in 40–60% of T-ALL cases, owing to chromosomal translocations, an activating interstitial deletion (SIL-TAL1 deletion), or undefined trans-acting mechanisms (Brown et al., 1990; Ferrando and Look, 2000; Ferrando et al., 2002). Results from gene expression analysis have demonstrated that TAL1 overexpression is associated with a differentiation block at the CD4+CD8+ double-positive (DP) stage of thymocytes in both human tumors and murine models (Ferrando et al., 2002; Larson et al., 1996; Tremblay et al., 2010).
TAL1 is a class II basic helix-loop-helix (bHLH) transcription factor that forms an obligate heterodimer with the class I bHLH E-proteins, which include TCF3/E2A and TCF12/HEB (Hsu et al., 1991; Hsu et al., 1994). In hematopoietic cells, TAL1 regulates the transcription of its target genes by binding to E-box motifs and nucleating a large complex that includes the E-proteins, GATA family members, and several non-DNA-binding LMO proteins (Lecuyer et al., 2002; Wadman et al., 1997; Xu et al., 2003). TAL1 expression is normally silenced during early thymocyte development (Herblot et al., 2000); thus, its expression and the regulatory complex it recruits to its direct target genes are clearly aberrant in DP thymocytes. By contrast, E-proteins act as homo- or heterodimers to regulate gene expression (Kee, 2009) and are required for thymocyte development in a stage-specific manner (Bernard et al., 1998; Herblot et al., 2000). A number of genes have been implicated as direct targets of TAL1 and its regulatory partners in human T-ALL (Bernard et al., 1998; Herblot et al., 2000; Ono et al., 1998; Palii et al., 2011b; Palomero et al., 2006). Studies in murine models have shown that E2a and Heb are highly dosage dependent, in that haplo-insufficiency for either gene accelerates the onset of TAL1-induced T-ALL (O’Neil et al., 2004). Hence, although E2A and HEB are critical for the formation of the TAL1 transcriptional complex, a change in the relative dosage of each member can affect the onset and severity of leukemia, through mechanisms that remain to be elucidated.
Recently, several groups have identified high level expression of genes encoding multiple transcription factors in the TAL1-overexpressing T-ALL subgroup, including RUNX1, MYB, NKX3-1 and ETS family members, such as ERG and ETS1 (Clappier et al., 2007; Kusy et al., 2010; Lahortiga et al., 2007; O’Neil et al., 2007; Thoms et al., 2011). However, it has been difficult to integrate them into a unified network of altered gene regulation that promotes thymocyte transformation. The current study was designed to elucidate the role of TAL1 in this aberrant transcriptional circuitry.
RESULTS
TAL1 binding is highly overlapping in multiple T-ALL cell lines and primary T-ALL cells
We generated high-resolution maps of the genome-wide occupancy of TAL1 by chromatin immunoprecipitation coupled to massively parallel DNA sequencing (ChIP-seq; Table S1). Multiple TAL1-expressing T-ALL samples were analyzed, which included two cell lines (Jurkat and CCRF-CEM; Figures S1A and 1B) and two “primagraft” samples (“Prima 2” and “Prima 5”) derived from primary T-ALL cells expanded in immunocompromised mice without any exposure to in vitro culture. Aberrant expression of TAL1 in both primagrafts and the CCRF-CEM cell line is due to a ~90 kb SIL-TAL deletion. The activity of the TAL1 antibody used was validated by ChIP followed by Western blot analysis with a different specific antibody (Figure S1C). Of note, TAL1 was enriched in chromatin precipitated with anti-HEB and anti-E2A antibodies (Figure S1C) that do not cross react (Figure S1D), consistent with its ability to heterodimerize with each of these E-proteins.
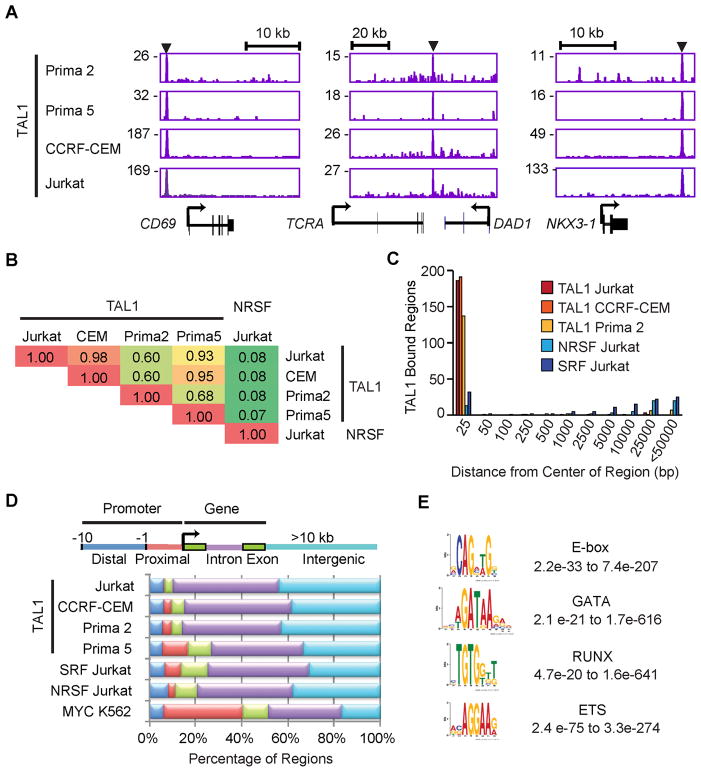
We first examined the results for known TAL1 target genes, including CD69, TCRA enhancer and NKX3-1 (Bernard et al., 1998; Kusy et al., 2010; Palii et al., 2011b), and detected TAL1 binding at sites in the regulatory regions of each gene (Figure 1A). Our results agree with previously reported TAL1 binding sites except for NKX3-1 (Kusy et al., 2010), where we find that TAL1 occupied a region consistent with a candidate distal enhancer in each of the four T-ALL samples, but not the previously identified promoter region (Figure 1A, right). We then investigated the relative overlap of high-confidence TAL1-bound regions across all four T-ALL samples. Pairwise comparisons of the top 200 TAL1-bound regions showed a high degree of agreement, compared with the results for NRSF-bound regions in Jurkat cells as a negative control (Figure 1B). Nearest-neighbor analysis confirmed this result (Figure 1C). When we compared the relative distribution of TAL1-bound regions with the locations of protein-coding genes, the majority of the bound regions were within the gene body and intergenic regions of known protein-coding genes, consistent with the location of enhancer elements, as opposed to sites in the proximal or distal promoter (Figure 1D). Hence, the TAL1-binding sites identified in multiple T-ALL cell samples overlapped substantially at known and candidate regulatory elements.
Figure 1. TAL1 occupancy is highly consistent across T-ALL cell lines and primagraft samples.
(A) Gene tracks represent binding of TAL1 in T-ALL primagrafts (Prima 2 and Prima 5) and cell lines (CCRF-CEM and Jurkat) at the CD69, TCRA and NKX3-1 loci. The x-axis indicates the linear sequence of genomic DNA, and the y-axis the total number of mapped reads. The black horizontal bar above each gene example indicates the genomic scale in kilobases (kb). Black boxes in the gene map represent exons, and arrows indicate the location and direction of the transcriptional start site. Arrowheads denote regions bound by TAL1. (B) Pairwise comparison of the TAL1-bound regions found in T-ALL primagrafts and cell lines. Fractions of the top 200 TAL1-bound regions in each cell type (rows) that were occupied by TAL1 in the other cell types (columns) are shown as a matrix. Regions occupied by the unrelated transcription factor NRSF in Jurkat served as a negative control. (C) Distances from the center of the TAL1-bound region (top 200) in Prima 5 to the center of the nearest bound region of the indicated transcription factors were determined and grouped into bins (x-axis); the heights of bars represent the sum of the bound regions in each bin (y-axis). (D) Each region bound by TAL1 was mapped to the closest Refseq gene: distal (blue) and proximal (red) promoters, exon (green), intron (violet) and intergenic regions (light blue) more than 10 kb from the gene. (E) The representative position weight matrix for each motif enriched in TAL1 by ChIP-seq and the range of E-values found across different T-ALL cell types are shown. See also Figure S1 and Table S1.
We next sought to identify DNA motifs that were statistically overrepresented within 200 bp of the peak of TAL1 binding in each T-ALL sample. Four transcription factor binding motifs were enriched in TAL1-bound regions, including E-box (5′-CAG[CG]TG-3′), GATA (5′-AGATAA-3′), RUNX (5′-TGTGGTC-3′), and motifs recognized by the ETS family of transcription factors (5′-GGAA-3′)(Figure 1E). This complement of motifs is highly similar to the TAL1 motifs identified by ChIP-seq in normal murine hematopoietic progenitors and red cells (Kassouf et al., 2010; Wilson et al., 2010) and in human hematopoietic cells (Novershtern et al., 2011; Palii et al., 2011b; Tijssen et al., 2011). We expect that TAL1 is likely co-regulating its target genes in T-ALL in a complex analogous to that identified in normal hematopoietic cells.
TAL1 complex controls genes involved in T-cell homeostasis
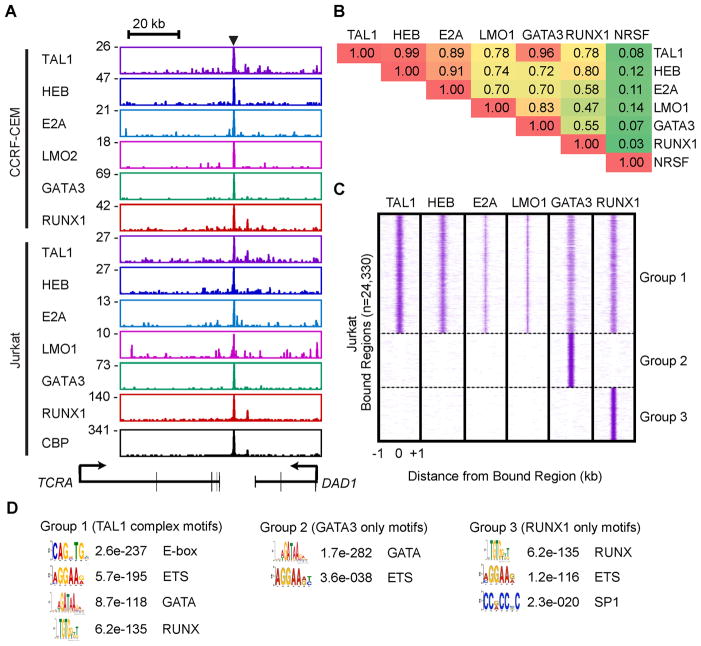
To identify the regulatory network controlled by the TAL1 transcriptional complex in human T-ALL cells, we performed ChIP-seq analysis for TAL1 and its regulatory partners HEB, E2A, GATA3, RUNX1 and LMO1/2 in Jurkat and CCRF-CEM cells, which express high levels of LMO1 and LMO2, respectively (Figures S1A–C). The genomic sites occupied in T-ALL samples showed remarkable concordance for TAL1, its regulatory partners and the transcriptional co-activator CBP, as illustrated for a known TAL1 complex target, the TCRA enhancer (Bernard et al., 1998; Hollenhorst et al., 2007)(Figure 2A). The E2A-bound regions we identified included a number of E2A target genes reported in murine thymocytes, including PTCRA, NOTCH3, RAG1, RAG2 and GFI1 (Miyazaki et al., 2011).
Figure 2. TAL1 co-occupies genomic sites with HEB, E2A, LMO1/2, GATA3 and RUNX1 in T-ALL cells.
(A) Gene tracks represent binding of TAL1, HEB, E2A, LMO1 or LMO2, GATA3 and RUNX1 at the TCRA locus in human T-ALL cell lines. See Figure 1A legend for details. (B) Pairwise comparison of transcription factor-bound regions in Jurkat cells. Fraction of the top 200 bound regions for each transcription factor (rows) co-occupied by the all bound regions of the other transcription factors (columns) are shown as a matrix. (C) Distributions of TAL1, HEB, E2A, LMO1, GATA3 and RUNX1 in regions bound by these transcription factors in Jurkat cells: Group 1 (bound by TAL1, HEB, E2A, LMO1, GATA3 or RUNX1), Group 2 (bound by GATA3 alone) or Group 3 (bound by RUNX1 alone). For each region (y-axis), the sequence density centered at 0 indicates overlapping bound regions. (D) DREME-based motif analysis of the regions shown in Figure 2C. The results are presented as position-weight matrices with the E-value significance and the name of the motif family within searched sequences. See also Figure S2 and Tables S2–4.
Examination of the overlap among regions enriched for TAL1, HEB, E2A, LMO1/2, GATA3 or RUNX1 revealed that TAL1 binds to the majority of HEB- and E2A-enriched regions, which frequently overlap with the LMO1/2-, GATA3- and RUNX1-enriched regions (Figures 2B and S2A). We next investigated the relative binding overlap of these factors within high-confidence enriched regions (Table S2), identifying three different classes of regulatory elements in both Jurkat (Figure 2C) and CCRF-CEM cells (Figure S2B). One showed concordant enrichment for multiple TAL1 complex members (Group 1), a second was predominantly occupied by GATA3 alone (Group 2), while a third was mainly occupied by RUNX1 alone (Group 3).
Several groups have reported that the TAL1 complex frequently contains ETS family members (e.g., ETS1) in multiple hematopoietic cell types (Palii et al., 2011b; Soler et al., 2010; Wilson et al., 2010). By comparing the ChIP-seq binding profiles for TAL1 complex members determined in our study with those described by three other groups (Hollenhorst et al., 2009; Palii et al., 2011b; Valouev et al., 2008), we determined that ETS1 occupancy correlates with the binding of other members of the TAL1 complex, as compared with two non-ETS transcription factors, SRF and NRSF (Figure S2C). ERG, an ETS family member, is also highly expressed and regulated by the TAL1 complex in human T-ALL cells (Thoms et al., 2011).
Analysis by the de novo motif discovery method, based on regions we have segregated into three groups, identified E-box, GATA and RUNX motifs as well as known motifs for the ETS family of transcription factors in Group 1, GATA and ETS motifs in Group 2, and RUNX, ETS and SP1 motifs in Group 3 (Figure 2D). The distribution of consensus motifs for E-box, GATA, RUNX and ETS motifs was centered under the peak of ChIP-seq enrichment within these three groups (Figure S2D). Moreover, the majority of enriched regions in Group 1 contained at least one E-box or one GATA motif within 200 bp of the center (n=8,245/12,748 in Jurkat and n=9,005/14,745 in CCRF-CEM). Many of the motifs enriched in E2A-bound regions had been identified in murine lymphocytes (Lin et al., 2010), where they also were frequently associated with ETS and RUNX binding sites.
To determine the dominant functions of the genes occupied by the TAL1 complex, we compared target genes (Table S3) to identify overrepresented functional groups (Figures S2E–G and Table S4). TAL1 complex targets were enriched in molecular pathways that regulate cell development, growth and death, as well as those known to contribute to cancer, hematological and immune diseases (Figures S2E and S2F). Importantly, enriched genes include those regulating T-cell development, differentiation, morphology, cell number and activation (Figure S2G). These observations indicate that in transformed thymocytes, the TAL1 complex sits at the apex of a network that drives aberrant proliferation, differentiation and survival.
TAL1, GATA3 and RUNX1 form a positive interconnected auto-regulatory loop
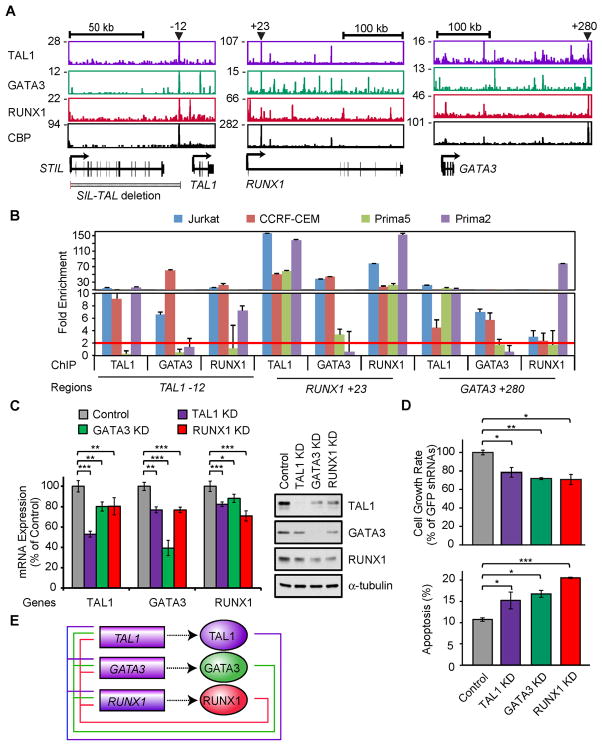
Importantly, we found that members of the TAL1 complex frequently occupy known and candidate regulatory regions of their own and each other’s genes (Figure 3A). For example, members of the TAL1 complex co-occupy enhancers that have been identified by others in normal hematopoietic cells, including the GATA3 3′ T-cell-specific enhancer (Hosoya-Ohmura et al., 2011), the RUNX1 +23 enhancer (Nottingham et al., 2007)(Figure 3A) and the LMO2 −24 enhancer (Landry et al., 2009). A candidate enhancer 12 kb upstream of the TAL1 gene, which is within the region between SIL and TAL1 that is frequently deleted in TAL1-positive T-ALL cases and is distinct from the regulatory elements previously described for normal hematopoietic cells (Gottgens et al., 2010), is co-occupied by TAL1 complex members and CBP in Jurkat cells (Figure 3A, left). Using ChIP-PCR, we found that these known and candidate regulatory elements for the TAL1, RUNX1 and GATA3 loci are also frequently co-occupied by TAL1 complex members in primary T-ALL cases (Figure 3B).
Figure 3. TAL1, GATA3 and RUNX1 form a positive interconnected auto-regulatory loop.
(A) Gene tracks represent binding of TAL1, GATA3, RUNX1 and CBP at the TAL1 (right), RUNX1 (middle) and GATA3 (left) loci in Jurkat cells. See Figure 1A legend for details. (B) Co-occupancy by TAL1, GATA3 and RUNX1 at the TAL1, RUNX1 and GATA3 gene loci in multiple T-ALL cells. Enrichment of regions indicated in panel A (TAL1 enhancer, RUNX1 +23 and GATA3 +280) in four T-ALL cell samples (Jurkat, CCRF-CEM, Prima 2 and Prima 5) was analyzed by ChIP-PCR. The NANOG promoter was used as a negative control. The error bars represent the standard deviation (SD) of the fold enrichment. The red line represents the two-fold enrichment detection for the negative control. (C) mRNA (left) and protein (right) levels of TAL1, GATA3 and RUNX1 after knockdown (KD) of each of these factors in Jurkat cells. The data are means ± SD of duplicate experiments; *p<0.05, **p<0.01 and ***p<0.001 by two sample, two-tailed t-test. (D) Growth inhibition and apoptosis induction after knockdown of TAL1, GATA3 and RUNX1 in Jurkat cells. Cell viability was measured after 3 and 7 days of lentivirus infection. The growth rate (day7/day3) is reported as means ± SD percentage of that for control shRNAs (GFP and Luc) in triplicate experiments. Apoptosis was analyzed at day 4 after lentiviral infection by flow cytometric analysis of cells stained with AnnexinV-FITC. The values are means ± SD of duplicate experiments. Asterisks denote p-values as described for panel C. (E) Positive interconnected auto-regulatory loop formed by TAL1, GATA3 and RUNX1. Genes are presented as rectangles and proteins as ovals. See also Figure S3 and Table S5.
To further dissect the function of the TAL1 complex, we used two independent short-hairpin RNAs (shRNAs) to knockdown the level of TAL1 expression in T-ALL cell lines (Figure S3A). In this context, there was significant downregulation of a known target gene, ALDH1A2 (Ono et al., 1998), which was rescued by re-expression of the TAL1 cDNA (Figure S3B). Several cell surface markers related to T-cell activation (CD69 and CD84) or cell survival in vivo (CD47) were also downregulated upon TAL1 knockdown (Figure S3C and Table S5). Analysis of gene expression changes after TAL1 knockdown in Jurkat cells showed that GATA3 and RUNX1 were significantly downregulated (Figure 3C). Similarly, knockdown of GATA3 or RUNX1 (Figure S3A) resulted in the coordinate downregulation of expression of each of these factors (Figure 3C). We observed the same relationships in CCRF-CEM cells (Figure S3D), except that knockdown of GATA3 or RUNX1 did not affect the levels of TAL1 expression, which was expected because in this cell line TAL1 expression is controlled by the SIL promoter. These alterations in expression after gene knockdown demonstrate the presence of a positive interconnected auto-regulatory loop involving TAL1, GATA3 and RUNX1, and support the critical role of TAL1 overexpression in initiating the formation of the loop.
The knockdown of TAL1 in Jurkat cells reduced the growth rate of these cells (Figures 3D and S3E), consistent with our previous study using TAL1 shRNA #2 (Palomero et al., 2006) and published results of Palii and coworkers (Palii et al., 2011b). TAL1 shRNA #1 induced similar growth inhibition of multiple T-ALL cell lines expressing TAL1 (Figure S3F and Table S5). We also detected increased fractions of apoptotic cells after transduction with shRNA #1, but not with shRNA #2 (Figure S3G), indicating that higher levels of TAL1 knockdown are needed to detect cell death. Importantly, knockdown of GATA3 and RUNX1 also inhibited cell growth and induced apoptosis (Figure 3D), indicating that each of these three components of the auto-regulatory loop is required for cell growth and survival. Taken together, our results indicate that components of the TAL1 complex positively regulate each other in T-ALL and act to promote the growth and survival of T-ALL cells (Figure 3E), underscoring the importance of this positive interconnected auto-regulatory loop in maintaining the malignant state.
Expression levels of high-confidence TAL1 targets classify T-ALL subtypes
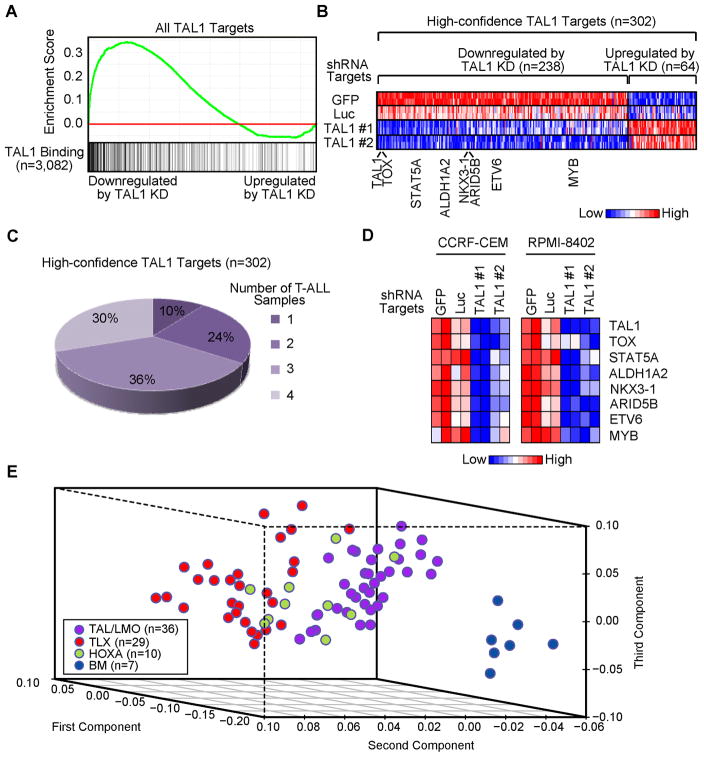
We next performed microarray gene expression analysis after knockdown of each transcription factor gene in Jurkat cells (Table S6). Gene Set Enrichment Analysis (GSEA) revealed that genes enriched for TAL1 binding by ChIP-seq analysis were more likely to be downregulated upon TAL1 knockdown than genes lacking TAL1 occupancy (Figure 4A). This difference was statistically significant (p<2.2E-16 by the Kolmogorov-Smirnov test), indicating that TAL1 acts predominantly as a positive regulator of the expression of its direct target genes in T-ALL.
Figure 4. Expression of high-confidence TAL1 target genes classify T-ALL subtypes.
(A) Gene set enrichment analysis (GSEA) to determine the correlation of DNA binding with gene expression change upon knockdown (KD) of TAL1. GSEA plot indicates the degree to which TAL1 targets are overrepresented at the extreme left (downregulated by knockdown) or right (upregulated by knockdown) of the entire ranked list. Solid bars represent bound genes. (B) Heatmap images representing the relative expression levels of high-confidence TAL1 targets in Jurkat cells with or without knockdown of TAL1. Two independent shRNAs targeting TAL1 as well as two control shRNAs (GFP and Luc) were transduced in Jurkat cells. Each row corresponds to a gene and is normalized across the row. (C) TAL1 binding to high-confidence target genes (n=302) in four T-ALL cell samples (Jurkat, CCRF-CEM, Prima 2 and Prima 5). The percentages of high-confidence TAL1 targets bound in Jurkat only (n=1) or multiple T-ALL samples (n=2–4) are indicated. (D) Gene expression changes of TAL1 targets upon TAL1 knockdown in CCRF-CEM and RPMI-8402 cells. Relative expression values (TAL1, TOX, STAT5A, ALDH1A2, NKX3-1, ARID5B, ETV6 and MYB) compared to GAPDH were calculated, normalized for each row and shown as a heatmap. For all genes, the changes were significant at p<0.05 by two sample, two-tailed t-test. (E) Principal component analysis of 75 primary T-ALL samples and seven bone marrow (BM) samples based on the expression profile of high-confidence TAL1 target genes. Primary T-ALL samples were classified into three groups (TAL/LMO, TLX or HOXA) based on the genetic alterations reported in the original article (Homminga et al., 2011). The analysis was performed with a set of 238 genes that are bound by TAL1 and significantly downregulated after TAL1 knockdown (see Figure 4B, left). See also Figure S4, Tables S6 and S7.
Further analyses were based on high-confidence TAL1 target genes (n=302) that showed TAL1 binding and significant alterations in expression (p<0.05 with an absolute log2 fold-change ≥0.24) upon TAL1 knockdown (Figure 4B and Table S7). Since the TAL1 gene was reduced by 1.89-fold, we were expecting modest but consistent expression changes in other target genes. The distribution of expression changes of genes selected was statistically different when compared to all genes (p<2.2E-16; also see Figures S4A–C for details). The percentage of genes within the high-confidence targets occupied by TAL1 across the four T-ALL cells (Jurkat, CCRF-CEM, Prima 2 and 5) showed that 90% of genes occupied in Jurkat cells were occupied by TAL1 in at least one of the other T-ALL samples (Figure 4C). To address whether the target genes that are directly regulated by TAL1 in Jurkat cells are similarly regulated in TAL1-positive T-ALL cells, we analyzed gene expression profiles in other TAL1-positive T-ALL cell lines and primary T-ALL samples. We observed consistent downregulation of selected TAL1 target genes upon TAL1 knockdown in two additional T-ALL cell lines (CCRF-CEM and RPMI-8402; Figure 4D). Principal component analysis of gene expression levels in 75 primary T-ALL samples with well-defined genetic alterations as well as 7 normal bone marrow (BM) samples (Homminga et al., 2011), based on expression levels of the positively regulated high-confidence target genes (n=238; Figure 4B, left), clearly distinguished the TAL/LMO-positive T-ALL subgroup from other subgroups (TLX- or HOXA-positive) and BM samples (Figure 4E), confirming the importance of these TAL1 target genes in TAL1-positive T-ALL cases. High levels of expression of the TAL1 targets were observed in primary TAL1-positive T-ALL samples (Figure S4D).
TAL1 positively regulates target genes in concert with GATA3 and RUNX1
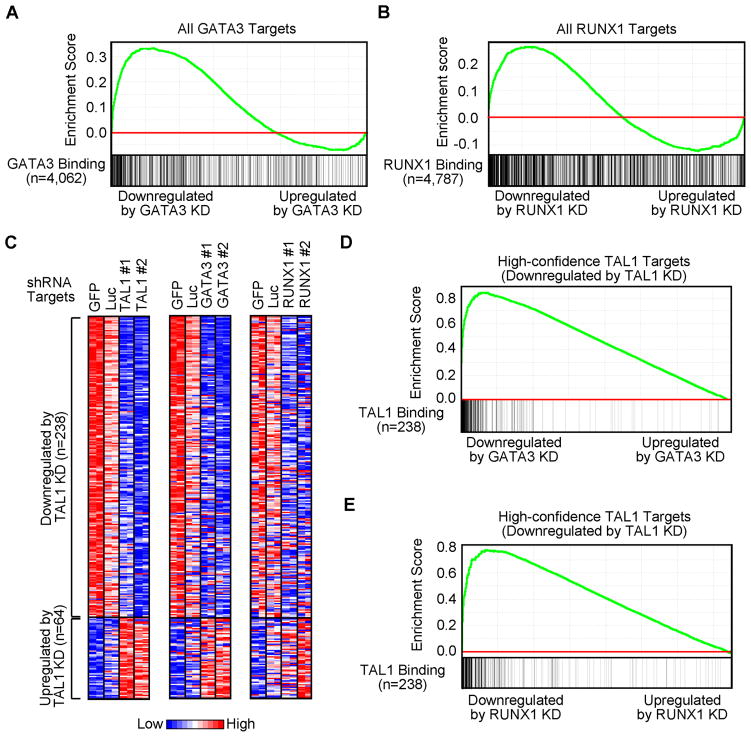
An important question in this study was whether GATA3 and RUNX1 coordinately regulate gene expression with TAL1 in T-ALL cells. When genes enriched for GATA3 binding were analyzed by GSEA, most of them were downregulated after GATA3 knockdown (Figure 5A), implicating GATA3 as a positive regulator of their expressions in T-ALL. By contrast, direct targets of RUNX1 were equally likely to be up- or downregulated when RUNX1 was depleted (Figure 5B).
Figure 5. TAL1 positively regulates target genes with GATA3 and RUNX1 in Jurkat cells.
(A, B) GSEA to determine the correlation of DNA binding with gene expression changes upon knockdown (KD) of GATA3 or RUNX1, respectively. See Figure 4A legend for details. (C) Heatmap images representing the relative expression levels of high-confidence TAL1 targets in Jurkat cells with or without knockdown of TAL1 (left), GATA3 (middle) or RUNX1 (right). See Figure 4B legend for details. (D, E) GSEA of expression changes of high-confidence TAL1 targets upon knockdown of GATA3 or RUNX1, respectively. TAL1 target genes (n=238) that were significantly downregulated by TAL1 knockdown (Figure 4B, left) were used as a gene set.
Interrogation of the effects of GATA3 or RUNX1 depletion on the expression of the 238 high-confidence TAL1 target genes revealed that the genes downregulated upon loss of TAL1 expression were generally also downregulated by the loss of GATA3 or RUNX1 expression (Figures 5C–E), indicating that TAL1 acts in concert with GATA3 and RUNX1 to positively regulate the majority of its direct target genes in T-ALL.
MYB oncogene coordinately regulates TAL1 target genes
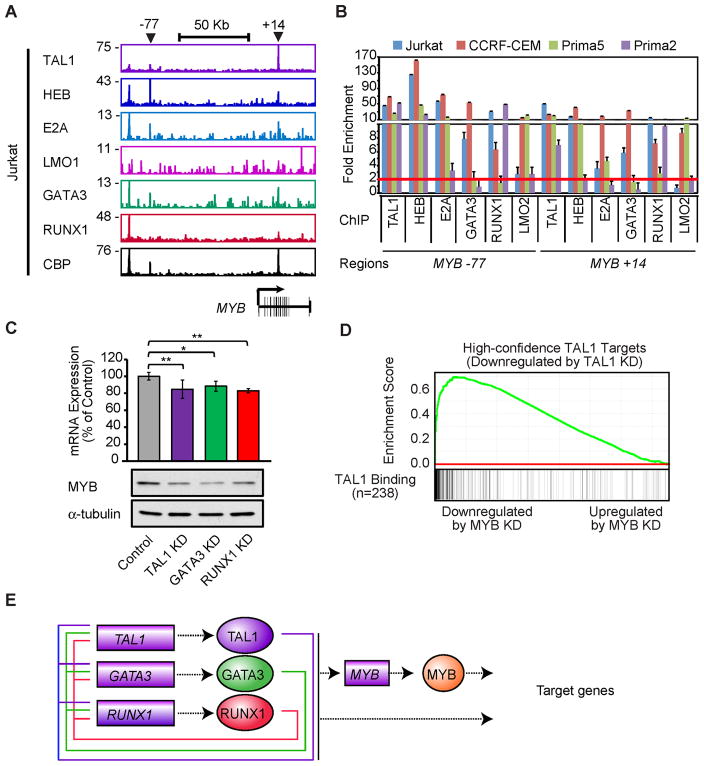
MYB overexpression, mediated in part through gene duplication, is commonly found in TALL (Clappier et al., 2007; Lahortiga et al., 2007; O’Neil et al., 2007)(Figure S5A). Interrogation of ChIP-seq data across the MYB locus indicated that MYB was a common target of the TAL1 complex in TAL1-positive T-ALL cell lines (Figure 6A), indicating that it is directly upregulated by the TAL1 complex due to the aberrant expression of TAL1. The TAL1-occupied regions included the known locus control region located 77 kb upstream of Myb in mice (Ramsay and Gonda, 2008) and a candidate regulatory element within intron 8 (+14) co-occupied by the TAL1 complex in T-ALL cells (Figure 6A). Co-occupancy of the TAL1 complex was observed at this candidate regulatory element in T-ALL primagrafts and cell lines by ChIP-PCR (Figure 6B). In all cases, MYB gene expression was sensitive to the reduction of multiple TAL1 complex members in Jurkat and CCRF-CEM cells (Figures 6C and S5B).
Figure 6. TAL1 positively regulates the MYB oncogene, which coordinately regulates TAL1 target genes.
(A) Gene tracks represent binding of TAL1, HEB, E2A, LMO1, GATA3, RUNX1 and CBP at the MYB gene locus in Jurkat cells. See Figure 1A legend for details. (B) Co-occupancy by the TAL1 complex at the MYB gene in multiple TALL cell samples by ChIP-PCR. See Figure 3B legend for details. (C) mRNA (top) and protein (bottom) levels of MYB after knockdown (KD) of TAL1, GATA3 and RUNX1 in Jurkat cells. The data are means ± SD of duplicate experiments; *p<0.05 and **p<0.01 by two sample, two-tailed t-test. (D) GSEA of expression changes of high-confidence TAL1 targets upon MYB knockdown. TAL1 target genes (n=238) that were significantly downregulated by TAL1 knockdown were used as a gene set. See Figure 4A legend for details. (E) Positive feed-forward loop formed by the TAL1 complex and MYB that controls the gene expression program of T-ALL cells. MYB is bound and activated by the TAL1 complex and, in turn, regulates the same set of genes. See also Figure S5.
Since MYB is a known transcriptional regulator of normal and malignant hematopoiesis (Ramsay and Gonda, 2008), a reduction of its expression could indicate the transcriptional relationship between the TAL1 complex and this gene in T-ALL. We therefore performed knockdown analysis of MYB (Figure S5C), followed by microarray gene expression analysis. The result showed that many TAL1 targets were also significantly downregulated by MYB knockdown (Figures 6D and S5D), indicating that MYB is not only induced by TAL1, but also acts to coordinately upregulate overlapping sets of target genes controlled by TAL1. Thus, the MYB oncogene appears to reinforce the activities of the TAL1-regulated oncogenic network through a feed-forward circuit that maintains the gene expression program in T-ALL cells (Figure 6E).
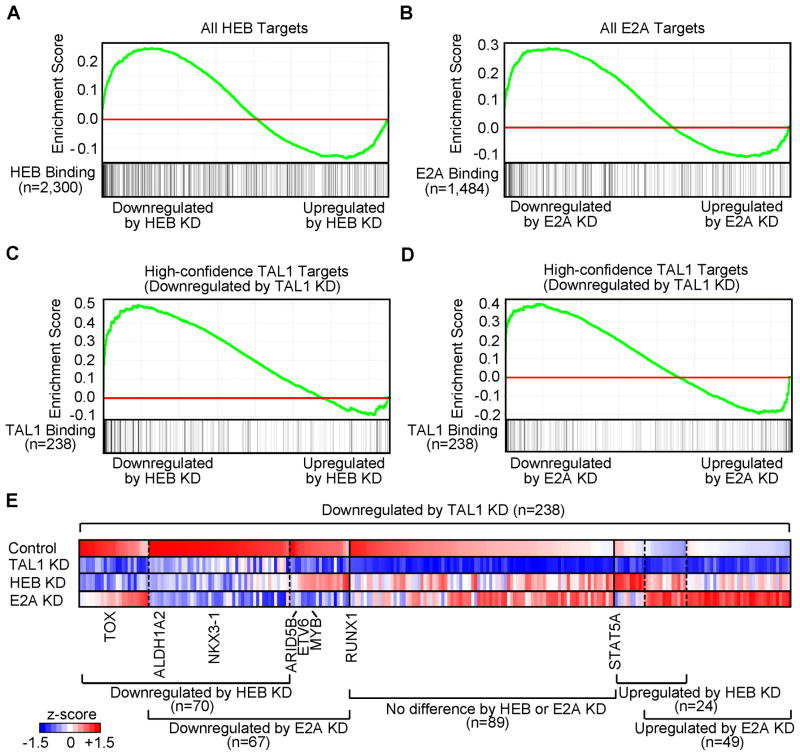
HEB and E2A oppose positive regulation by TAL1 at critical target genes
Several groups have postulated that the ectopic expression of TAL1 in normal thymocytes may antagonize the physiologic activities of E2A or HEB in thymocyte development by recruiting them into the TAL1 complex, where they could deregulate key target genes (Bain et al., 1997; Herblot et al., 2000; O’Neil et al., 2004). Hence, we next interrogated the effects of HEB and E2A knockdown (Figure S6) on the expression of their direct target genes. GSEA revealed that genes enriched for HEB or E2A binding by ChIP-seq analysis could be either down- or upregulated after knockdown of each gene (Figures 7A and 7B). As expected, 40% of the TAL1 target genes that were downregulated after TAL1 knockdown were also downregulated by the knockdown of either E2A or HEB (Figures 7C–7E, left). By contrast, 25% of the TAL1 target genes that were downregulated by TAL1 knockdown were upregulated by the loss of either E2A or HEB expression (Figures 7C–7E, right), indicating that they can act to repress gene expression in the absence of TAL1. Only 50 of the 238 genes that were downregulated after TAL1 knockdown were significantly upregulated after knockdown of HEB and/or E2A and an even smaller group of seven TAL1 target genes showed the inverse relationship (Table S8).
Figure 7. TAL1 positively regulates the expression of a specific subset of genes that are negatively regulated by HEB and E2A.
(A, B) GSEA to determine the correlation of DNA binding with gene expression changes upon knockdown (KD) of HEB or E2A, respectively. See Figure 4A legend for details. (C, D) GSEA of expression changes of high-confidence TAL1 targets upon knockdown of HEB or E2A, respectively. TAL1 target genes (n=238) that were significantly downregulated by TAL1 knockdown (Figure 4B, left) were used as a gene set. (E) Heatmap image representing expression levels of the responsive TAL1 targets in Jurkat cells upon knockdown of TAL1, HEB or E2A. Relative gene expression levels normalized for each gene are illustrated. See also Figure S6 and Table S8.
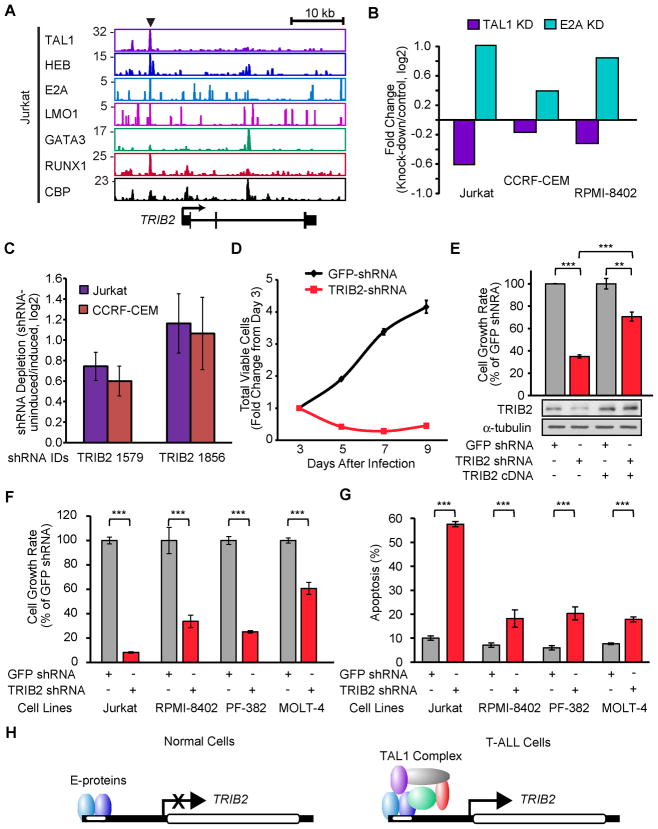
TRIB2 is a critical target of TAL1 and is required for the survival of T-ALL cells
We also conducted an inducible RNA interference screen (Ngo et al., 2006) in two TAL1-positive T-ALL cell lines (Jurkat and CCRF-CEM) and identified four genes that were required for T-ALL growth among the high-confidence targets: STAT5A, TNFSF4, PI3KC2B and TRIB2 (Table S9). An especially attractive candidate target of the TAL1 complex is TRIB2 (Figure 8A), whose expression was downregulated by TAL1 knockdown and upregulated by E2A knockdown in multiple TAL1-positive T-ALL cell lines (Figure 8B). After 3 weeks of culture, Jurkat or CCRF-CEM cells expressing two different shRNAs targeting TRIB2 were depleted (Figure 8C). This result was validated by knockdown of TRIB2 using another shRNA (Figure 8D), which was partially rescued by the ectopic expression of the TRIB2 cDNA (Figure 8E). TRIB2 knockdown inhibited the growth of additional TAL1-positive T-ALL cell lines (Figure 8F) and induced apoptosis in each cell line (Figure 8G), but did not affect the cell cycle phase distribution (Figure S7), indicating that this factor is required for the survival of T-ALL cells. A reasonable hypothesis is that TRIB2 is transcriptionally repressed by E-proteins and the aberrant expression of TAL1 by translocation or intrachromosomal rearrangement upregulates TRIB2 expression by heterodimerizing with E2A and HEB, thus actively opposing their repressive effects on the TRIB2 locus (Figure 8H).
Figure 8. TRIB2 gene is required for the survival of T-ALL cells.
(A) Gene tracks represent binding of TAL1, HEB, E2A, LMO1, GATA3, RUNX1 and CBP at the TRIB2 gene locus in Jurkat cells. See Figure 1A legend for details. (B) Comparison of mRNA expression of TRIB2 gene in three TAL1-positive T-ALL cell lines transduced with shRNAs targeting TAL1, E2A or control shRNAs and analyzed by qRT-PCR. Mean fold-changes (knockdown/controls, log2) are shown. (C) shRNA screen with 12,500 inducible shRNAs that target 1,050 genes, performed on two TAL1-positive T-ALL cell lines (Jurkat and CCRF-CEM). Depletion of TRIB2 shRNAs from the cell population was calculated as uninduced/induced, log2, and shown as the mean ± standard error of the mean of four independent experiments. (D) Growth inhibition by TRIB2 knockdown in Jurkat cells. Cell viability was measured after 3, 5, 7 and 9 days of lentivirus infection with control (GFP) and TRIB2 shRNA. The growth rate (fold-change) compared to day 3 is indicated. Values are means ± SD of triplicate experiments. (E) cDNA containing the wild-type TRIB2 coding region was transduced by retroviral infection of Jurkat cells, followed by lentivirus-mediated transduction of infected cells with control GFP or TRIB2 shRNA. The growth rate (day7/day3) after lentivirus infection was assessed for TRIB2 shRNA relative to GFP shRNA and is shown as the means ± SD of triplicate experiments; ** p<0.01 and *** p<0.001 by two sample, two-tailed t-test. (F) Growth rate (day 7/day 3) was assessed for each TRIB2 shRNA relative to control GFP shRNA in each cell line (Jurkat, RPMI-8402, PF-382 or MOLT-4) and is reported as the means ± SD of triplicate experiments. (G) Apoptosis was measured in four T–ALL cell lines after 4 days of lentiviral infection by flow cytometric analysis of cells stained with AnnexinV-FITC. The values are means ± SD of triplicate experiments. (H) Model of differential regulation of E-protein (HEB and E2A) targets in normal versus malignant T-cells. TRIB2 that is repressed by HEB and E2A in normal cells (left) is upregulated by the TAL1 complex in T-ALL (right). See also Figure S7 and Table S9.
DISCUSSION
We have identified a set of transcriptional regulators that collaborate with TAL1 to generate a “core” regulatory circuit that contributes to the initiation and maintenance of human T-ALL. Critically, we found that TAL1, GATA3 and RUNX1 co-occupy elements of their own and each other’s genes, forming a positive interconnected auto-regulatory loop. This network structure provides the means by which aberrant expression of a single oncogenic transcription factor can have sustained effects on an entire program of gene expression that contributes to malignant transformation. Indeed, such regulatory structures have been shown to reinforce and increase the stability of gene expression programs (Alon, 2007), which in turn act to establish and maintain critical cell states, such as pluripotency in embryonic stem cells (Young, 2011).
Expression of GATA3 and RUNX1 is required in normal T-lineage commitment to CD4 or CD8 single-positive cells, respectively (Collins et al., 2009; Ho et al., 2009). TAL1-sustained upregulation of GATA3 and RUNX1 may therefore contribute to the differentiation block at the DP stage. Thus, overexpression of RUNX1 appears to contribute to thymocyte transformation in TAL1-positive T-ALL, in marked contrast to its role as a tumor suppressor whose loss of function promotes the onset of acute myeloid leukemia (AML) and myelodysplastic syndromes. Two groups have also recently reported inactivating RUNX1 mutations in the early thymocyte precursor (ETP) subgroup of T-ALL (Della Gatta et al., 2012; Grossmann et al., 2011). ETP leukemia is distinct from TAL1-positive T-ALL, which generally shows a block at the DP stage of thymocyte development, and involves different aberrant molecular pathways leading to transformation (Coustan-Smith et al., 2009; Gutierrez et al., 2010; Zhang et al., 2012). Thus, despite the presence of inactivating RUNX1 mutations in ETP T-ALL, our data show that in T-ALLs blocked in a later DP stage of thymocyte development, RUNX1 serves as a key member of an interconnected auto-regulatory loop involved in reinforcing and stabilizing the malignant cell state.
We also identified an oncogenic transcription factor gene, MYB, as a direct target of TAL1 that is expressed at high levels in TAL1-positive T-ALL cases. It integrates with the TAL1-controlled transcriptional network through a positive feed-forward loop that likely acts to stabilize the TAL1 oncogenic program, similar to mechanisms first identified in model organisms (Alon, 2007). The Myb gene was reported as a Tal1 target in murine hematopoietic progenitor cells (Wilson et al., 2009), which may reflect an important regulatory module shared by leukemic and normal stem cells. MYB has recently been implicated in the control of aberrant self-renewal programs in AML (Zuber et al., 2011), reinforcing its potential importance as the target of a feed-forward regulatory motif mediated by the TAL1 complex in T-ALL. This transcriptional regulatory circuit is also implicated in normal hematopoiesis (Novershtern et al., 2011) and presumably contributes to the establishment and stability of the transformed state in TAL1-overexpressing thymocytes.
It will be important to confirm and extend our results with shRNA knockdown as we have done with ChIP-seq analysis in primary T-ALL cells. Several groups have reported success with shRNA gene knockdown in primary T-ALL cells (Gerby et al., 2009; Kusy et al., 2010; Palii et al., 2011a; Palii et al., 2011b). These procedures involve both improved lentivirus production using the new-generation plasmids (Palii et al., 2011a; Palii et al., 2011b), or the new pseudotyping vector, resulting in IL-7-displaying lentiviral vectors that promote efficient gene transfer into primary T-cells (Gerby et al., 2009; Kusy et al., 2010; Verhoeyen et al., 2003). We are now attempting to optimize these procedures for shRNA transduction into primary T-ALL cells explanted from our primagraft models. When established, this approach should be helpful in tracing the regulatory circuits in de novo TALL leukemias.
We also present a comprehensive analysis of the gene set that is differentially regulated by TAL1 and its partners, HEB and E2A. Many of the direct targets of these three interacting proteins are coordinately upregulated as a consequence of binding to key regulatory regions. Only a small subset of these direct targets are differentially regulated physiologically when E2A and HEB are co-expressed compared to the cell state attained when TAL1 is aberrantly overexpressed. This aspect of our study is crucial to understanding T-ALL pathogenesis, because it directly addresses in vivo data indicating that haploinsufficiency for either E2a or Heb markedly accelerates the onset of T-ALL in Tal1-transgenic mice (O’Neil et al., 2004). Among the relatively small set of genes directly targeted by TAL1 in T-ALL cells, only those activated by TAL1 but repressed by E2A and HEB would produce this phenotype. Some of these genes are likely inconsequential in terms of a contribution to the malignant phenotype, such as those normally expressed only in activated mature T-cells (e.g., CD28 and GMZA) as well as genes that are not associated at all with the T-cell lineage (e.g., KRT1 and KRT2).
Mammalian genes of the Tribbles family (TRIB1, 2 and 3) encode proteins that contain pseudo-kinase domains that are unable to directly phosphorylate target proteins, but rather appear to act as adaptors that negatively regulate key cellular signaling pathways (Yokoyama and Nakamura, 2011). Overexpression of TRIB2 by retroviral transduction in murine hematopoietic stem cells identified it as an oncogene that contributes to AML (Keeshan et al., 2006). Our data indicate that TRIB2 is required for the survival of T-ALL cells. Until our analysis, there had been no evidence implicating TRIB2 in T-ALL pathogenesis, although this role was not entirely unexpected given the expression of TRIB2 in a specific subset of AML cases that shared characteristics with T-cells and the status of this gene as a target of NOTCH1 (Wouters et al., 2007). Since TRIB2 is a direct target for upregulation by both the NOTCH1 and TAL1 transcription factors, our data suggest that a progressive increase in its expression levels contribute to the collaboration between aberrant TAL1 expression and mutationally activated NOTCH1 in the pathogenesis of T-ALL.
EXPERIMENTAL PROCEDURES
T-ALL Cell Samples
Human T-ALL cell lines were maintained in RPMI-1640 medium (Invitrogen, Carlsbad, CA) supplemented with 10% FBS (Sigma-Aldrich, St Louis, MO), L-glutamine and penicillin/streptomycin (Invitrogen). Diagnostic T-ALL samples were obtained with informed consent and institutional review board (IRB) approval from children treated on Dana-Farber Cancer Institute study 05-01 and were used on an existing IRB approval by the University of California, San Diego (UCSD) Human Research Protections Program, entitled “Protocol 070678: Permission to Collect Blood and/or Bone Specimens and/or Tumor Samples and/or Saliva from Patients with Hematology Problems for Research (Adult)”. Human CD34+ T-ALL cells were transplanted into Rag2−/−γc−/− mice to propagate the cells as “primagrafts”. This study was carried out in strict accordance with the recommendations of the Institutional Animal Care and Use Committee at the UCSD. The protocol was approved by the Committee under Animal Use Protocol Number S06015. Human leukemia cells were isolated from these primagrafts and used in TAL1 ChIP-seq analysis.
Chromatin Immunoprecipitation (ChIP)
ChIP was performed according to previously described methods (Lee et al., 2006). The antibodies and detailed ChIP conditions can be found in Supplemental Experimental Procedures. For ChIP-seq analysis, Solexa/Illumina sequencing and analysis were done according to the protocol described in (Marson et al., 2008).
shRNA Knockdown Analysis
shRNA sequences were cloned into the lentiviral vector pLKO.1-puro. Each construct was cotransfected into 293T cells with delta 8.9 and VSV-G using FuGENE 6 reagent (Roche, Indianapolis, IN). Supernatants containing the lentivirus were collected, filtered and added to T-ALL cell lines in the presence of polybrene. The level of knockdown was verified by qRT-PCR for RNA or by Western blot.
RNA Extraction, cDNA and Expression Analysis
We extracted mRNA by Trizol (Invitrogen) followed by column purification using the RNeasy Mini Kit (QIAGEN, Valencia, CA). Purified RNA was reverse-transcribed using QuantiTect (Qiagen). Quantitative real-time qPCR was performed on the AB7300 Detection System (Applied Biosystems, Foster City, CA) using gene-specific primers and Power SYBR Green PCR Master Mix (Applied Biosystems).
Microarray Expression Analysis
Total RNA samples from two biological replicates performed in Jurkat cells were used to assess gene expression change for two target shRNAs per transcription factor versus two control shRNA performed in duplicate on Affymetrix HG U133 2.0 plus microarrays. The detailed analysis can be found in Supplemental Experimental Procedures. GSEA (Broad Institute, Cambridge, MA) was performed for direct targets identified by ChIP-seq by comparing control samples with knockdown samples. The genes, which are direct TAL1 targets identified by ChIP-seq based on a significant gene expression change upon shRNA knockdown (absolute log2-fold-change ≥0.24; p-value <0.05), were defined as the high-confidence TAL1 targets and used as a gene set.
Supplementary Material
SIGNIFICANCE.
Studies in embryonic stem cells have revealed “core” transcriptional regulatory circuits that control pluripotency and self-renewal, raising the possibility that similar mechanisms might contribute to malignant transformation. Here we identify and dissect a core transcriptional regulatory circuit controlled by the aberrant expression of TAL1 in T-ALL cells. Our findings demonstrate that TAL1 and its binding partners GATA3 and RUNX1 form a positive auto-regulatory loop, reinforced by a feed-forward mechanism involving the MYB oncoprotein, which likely contributes to the initiation and maintenance of the malignant state in thymocytes. Moreover, we implicate TRIB2, which is oppositely regulated by TAL1 and E2A/HEB and required for T-ALL cell survival.
HIGHLIGHTS.
TAL1, GATA3 and RUNX1 form a positive interconnected auto-regulatory loop
TAL1 core regulatory circuit controls genes involved in T-cell homeostasis
The TAL1 complex recruits MYB in a positive feed-forward regulatory loop
TRIB2 is oppositely regulated by TAL1 and E-proteins and is crucial for cell survival
Acknowledgments
We thank members of the Look and Young laboratories for discussions and critical review, and Jennifer O’Neil for initial experimentation and TAL1 plasmids. We are grateful to Garrett Frampton and Dave Orlando for the development of ChIP-seq analysis tools and continued support with the analysis. We are also grateful to the Whitehead Genome Technology Core (V. Dhanapal, J.-A. Kwon, J. Love, S. Gupta and T. Volkert) for assistance with ChIP-Seq and expression array hybridization, and to Bingbing Yuan from BaRC for developing the phenotype ontology analysis. We acknowledge the RNAi Consortium for providing lentivirus shRNA constructs and thank John R. Gilbert for editing and critical review of the manuscript. This research was supported by grants from the National Cancer Institute (5P01CA109901, 5P01CA68484 and 1K99CA157951) and by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. T.S. is supported by grants from the William Lawrence and Blanche Hughes Foundation, the Children’s Leukemia Research Association and the Japan Society for the Promotion of Science. W.M. and C. J. are supported by the CIRM leukemia team grant.
Footnotes
ACCESSION NUMBERS
Expression data and ChIP-seq can be found at http://www.ncbi.nlm.nih.gov/geo/ under superseries accession numbers GSE29181.
Supplemental Information includes Supplemental Experimental Procedures, 7 figures, and 9 tables and can be found with this article online.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alon U. Network motifs: theory and experimental approaches. Nat Rev Genet. 2007;8:450–461. doi: 10.1038/nrg2102. [DOI] [PubMed] [Google Scholar]
- Armstrong SA, Look AT. Molecular genetics of acute lymphoblastic leukemia. J Clin Oncol. 2005;23:6306–6315. doi: 10.1200/JCO.2005.05.047. [DOI] [PubMed] [Google Scholar]
- Bain G, Engel I, Robanus Maandag EC, te Riele HP, Voland JR, Sharp LL, Chun J, Huey B, Pinkel D, Murre C. E2A deficiency leads to abnormalities in alphabeta T-cell development and to rapid development of T-cell lymphomas. Mol Cell Biol. 1997;17:4782–4791. doi: 10.1128/mcb.17.8.4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard M, Delabesse E, Smit L, Millien C, Kirsch IR, Strominger JL, Macintyre EA. Helix-loop-helix (E2–5, HEB, TAL1 and Id1) protein interaction with the TCRalphadelta enhancers. Int Immunol. 1998;10:1539–1549. doi: 10.1093/intimm/10.10.1539. [DOI] [PubMed] [Google Scholar]
- Brown L, Cheng JT, Chen Q, Siciliano MJ, Crist W, Buchanan G, Baer R. Site-specific recombination of the tal-1 gene is a common occurrence in human T cell leukemia. EMBO J. 1990;9:3343–3351. doi: 10.1002/j.1460-2075.1990.tb07535.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clappier E, Cuccuini W, Kalota A, Crinquette A, Cayuela JM, Dik WA, Langerak AW, Montpellier B, Nadel B, Walrafen P, et al. The C-MYB locus is involved in chromosomal translocation and genomic duplications in human T-cell acute leukemia (T-ALL), the translocation defining a new T-ALL subtype in very young children. Blood. 2007;110:1251–1261. doi: 10.1182/blood-2006-12-064683. [DOI] [PubMed] [Google Scholar]
- Collins A, Littman DR, Taniuchi I. RUNX proteins in transcription factor networks that regulate T-cell lineage choice. Nat Rev Immunol. 2009;9:106–115. doi: 10.1038/nri2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coustan-Smith E, Mullighan CG, Onciu M, Behm FG, Raimondi SC, Pei D, Cheng C, Su X, Rubnitz JE, Basso G, et al. Early T-cell precursor leukaemia: a subtype of very high-risk acute lymphoblastic leukaemia. Lancet Oncol. 2009;10:147–156. doi: 10.1016/S1470-2045(08)70314-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Della Gatta G, Palomero T, Perez-Garcia A, Ambesi-Impiombato A, Bansal M, Carpenter ZW, De Keersmaecker K, Sole X, Xu L, Paietta E, et al. Reverse engineering of TLX oncogenic transcriptional networks identifies RUNX1 as tumor suppressor in T-ALL. Nat Med. 2012 doi: 10.1038/nm.2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrando AA, Look AT. Clinical implications of recurring chromosomal and associated molecular abnormalities in acute lymphoblastic leukemia. Semin Hematol. 2000;37:381–395. doi: 10.1016/s0037-1963(00)90018-0. [DOI] [PubMed] [Google Scholar]
- Ferrando AA, Neuberg DS, Staunton J, Loh ML, Huard C, Raimondi SC, Behm FG, Pui CH, Downing JR, Gilliland DG, et al. Gene expression signatures define novel oncogenic pathways in T cell acute lymphoblastic leukemia. Cancer Cell. 2002;1:75–87. doi: 10.1016/s1535-6108(02)00018-1. [DOI] [PubMed] [Google Scholar]
- Gerby B, Armstrong F, de la Grange PB, Medyouf H, Calvo J, Verhoeyen E, Cosset FL, Bernstein I, Amselem S, Boissel N, et al. Optimized gene transfer into human primary leukemic T cell with NOD-SCID/leukemia-initiating cell activity. Leukemia. 2009;24:646–649. doi: 10.1038/leu.2009.235. [DOI] [PubMed] [Google Scholar]
- Gottgens B, Ferreira R, Sanchez MJ, Ishibashi S, Li J, Spensberger D, Lefevre P, Ottersbach K, Chapman M, Kinston S, et al. cis-Regulatory remodeling of the SCL locus during vertebrate evolution. Molecular and cellular biology. 2010;30:5741–5751. doi: 10.1128/MCB.00870-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossmann V, Kern W, Harbich S, Alpermann T, Jeromin S, Schnittger S, Haferlach C, Haferlach T, Kohlmann A. Prognostic relevance of RUNX1 mutations in T-cell acute lymphoblastic leukemia. Haematologica. 2011;96:1874–1877. doi: 10.3324/haematol.2011.043919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez A, Dahlberg SE, Neuberg DS, Zhang J, Grebliunaite R, Sanda T, Protopopov A, Tosello V, Kutok J, Larson RS, et al. Absence of biallelic TCRgamma deletion predicts early treatment failure in pediatric T-cell acute lymphoblastic leukemia. J Clin Oncol. 2010;28:3816–3823. doi: 10.1200/JCO.2010.28.3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herblot S, Steff AM, Hugo P, Aplan PD, Hoang T. SCL and LMO1 alter thymocyte differentiation: inhibition of E2A-HEB function and pre-T alpha chain expression. Nat Immunol. 2000;1:138–144. doi: 10.1038/77819. [DOI] [PubMed] [Google Scholar]
- Ho IC, Tai TS, Pai SY. GATA3 and the T-cell lineage: essential functions before and after T-helper-2-cell differentiation. Nat Rev Immunol. 2009;9:125–135. doi: 10.1038/nri2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenhorst PC, Chandler KJ, Poulsen RL, Johnson WE, Speck NA, Graves BJ. DNA specificity determinants associate with distinct transcription factor functions. PLoS Genet. 2009;5:e1000778. doi: 10.1371/journal.pgen.1000778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenhorst PC, Shah AA, Hopkins C, Graves BJ. Genome-wide analyses reveal properties of redundant and specific promoter occupancy within the ETS gene family. Genes & development. 2007;21:1882–1894. doi: 10.1101/gad.1561707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homminga I, Pieters R, Langerak AW, de Rooi JJ, Stubbs A, Verstegen M, Vuerhard M, Buijs-Gladdines J, Kooi C, Klous P, et al. Integrated transcript and genome analyses reveal NKX2–1 and MEF2C as potential oncogenes in T cell acute lymphoblastic leukemia. Cancer Cell. 2011;19:484–497. doi: 10.1016/j.ccr.2011.02.008. [DOI] [PubMed] [Google Scholar]
- Hosoya-Ohmura S, Lin YH, Herrmann M, Kuroha T, Rao A, Moriguchi T, Lim KC, Hosoya T, Engel JD. An NK and T cell enhancer lies 280 kilobase pairs 3′ to the gata3 structural gene. Molecular and cellular biology. 2011;31:1894–1904. doi: 10.1128/MCB.05065-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu HL, Cheng JT, Chen Q, Baer R. Enhancer-binding activity of the tal-1 oncoprotein in association with the E47/E12 helix-loop-helix proteins. Mol Cell Biol. 1991;11:3037–3042. doi: 10.1128/mcb.11.6.3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu HL, Wadman I, Baer R. Formation of in vivo complexes between the TAL1 and E2A polypeptides of leukemic T cells. Proc Natl Acad Sci U S A. 1994;91:3181–3185. doi: 10.1073/pnas.91.8.3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassouf MT, Hughes JR, Taylor S, McGowan SJ, Soneji S, Green AL, Vyas P, Porcher C. Genome-wide identification of TAL1’s functional targets: Insights into its mechanisms of action in primary erythroid cells. Genome Res. 2010 doi: 10.1101/gr.104935.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee BL. E and ID proteins branch out. Nat Rev Immunol. 2009;9:175–184. doi: 10.1038/nri2507. [DOI] [PubMed] [Google Scholar]
- Keeshan K, He Y, Wouters BJ, Shestova O, Xu L, Sai H, Rodriguez CG, Maillard I, Tobias JW, Valk P, et al. Tribbles homolog 2 inactivates C/EBPalpha and causes acute myelogenous leukemia. Cancer Cell. 2006;10:401–411. doi: 10.1016/j.ccr.2006.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusy S, Gerby B, Goardon N, Gault N, Ferri F, Gerard D, Armstrong F, Ballerini P, Cayuela JM, Baruchel A, et al. NKX3.1 is a direct TAL1 target gene that mediates proliferation of TAL1-expressing human T cell acute lymphoblastic leukemia. The Journal of experimental medicine. 2010;207:2141–2156. doi: 10.1084/jem.20100745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahortiga I, De Keersmaecker K, Van Vlierberghe P, Graux C, Cauwelier B, Lambert F, Mentens N, Beverloo HB, Pieters R, Speleman F, et al. Duplication of the MYB oncogene in T cell acute lymphoblastic leukemia. Nat Genet. 2007;39:593–595. doi: 10.1038/ng2025. [DOI] [PubMed] [Google Scholar]
- Landry JR, Bonadies N, Kinston S, Knezevic K, Wilson NK, Oram SH, Janes M, Piltz S, Hammett M, Carter J, et al. Expression of the leukemia oncogene Lmo2 is controlled by an array of tissue-specific elements dispersed over 100 kb and bound by Tal1/Lmo2, Ets, and Gata factors. Blood. 2009;113:5783–5792. doi: 10.1182/blood-2008-11-187757. [DOI] [PubMed] [Google Scholar]
- Larson RC, Lavenir I, Larson TA, Baer R, Warren AJ, Wadman I, Nottage K, Rabbitts TH. Protein dimerization between Lmo2 (Rbtn2) and Tal1 alters thymocyte development and potentiates T cell tumorigenesis in transgenic mice. The EMBO journal. 1996;15:1021–1027. [PMC free article] [PubMed] [Google Scholar]
- Lecuyer E, Herblot S, Saint-Denis M, Martin R, Begley CG, Porcher C, Orkin SH, Hoang T. The SCL complex regulates c-kit expression in hematopoietic cells through functional interaction with Sp1. Blood. 2002;100:2430–2440. doi: 10.1182/blood-2002-02-0568. [DOI] [PubMed] [Google Scholar]
- Lee TI, Johnstone SE, Young RA. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat Protoc. 2006;1:729–748. doi: 10.1038/nprot.2006.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YC, Jhunjhunwala S, Benner C, Heinz S, Welinder E, Mansson R, Sigvardsson M, Hagman J, Espinoza CA, Dutkowski J, et al. A global network of transcription factors, involving E2A, EBF1 and Foxo1, that orchestrates B cell fate. Nature immunology. 2010;11:635–643. doi: 10.1038/ni.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Look AT. Oncogenic transcription factors in the human acute leukemias. Science. 1997;278:1059–1064. doi: 10.1126/science.278.5340.1059. [DOI] [PubMed] [Google Scholar]
- Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, Newman J, et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki M, Rivera RR, Miyazaki K, Lin YC, Agata Y, Murre C. The opposing roles of the transcription factor E2A and its antagonist Id3 that orchestrate and enforce the naive fate of T cells. Nat Immunol. 2011;12:992–1001. doi: 10.1038/ni.2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo VN, Davis RE, Lamy L, Yu X, Zhao H, Lenz G, Lam LT, Dave S, Yang L, Powell J, Staudt LM. A loss-of-function RNA interference screen for molecular targets in cancer. Nature. 2006;441:106–110. doi: 10.1038/nature04687. [DOI] [PubMed] [Google Scholar]
- Nottingham WT, Jarratt A, Burgess M, Speck CL, Cheng JF, Prabhakar S, Rubin EM, Li PS, Sloane-Stanley J, Kong ASJ, de Bruijn MF. Runx1-mediated hematopoietic stem-cell emergence is controlled by a Gata/Ets/SCL-regulated enhancer. Blood. 2007;110:4188–4197. doi: 10.1182/blood-2007-07-100883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novershtern N, Subramanian A, Lawton LN, Mak RH, Haining WN, McConkey ME, Habib N, Yosef N, Chang CY, Shay T, et al. Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell. 2011;144:296–309. doi: 10.1016/j.cell.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neil J, Shank J, Cusson N, Murre C, Kelliher M. TAL1/SCL induces leukemia by inhibiting the transcriptional activity of E47/HEB. Cancer Cell. 2004;5:587–596. doi: 10.1016/j.ccr.2004.05.023. [DOI] [PubMed] [Google Scholar]
- O’Neil J, Tchinda J, Gutierrez A, Moreau L, Maser RS, Wong KK, Li W, McKenna K, Liu XS, Feng B, et al. Alu elements mediate MYB gene tandem duplication in human T-ALL. J Exp Med. 2007;204:3059–3066. doi: 10.1084/jem.20071637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono Y, Fukuhara N, Yoshie O. TAL1 and LIM-only proteins synergistically induce retinaldehyde dehydrogenase 2 expression in T-cell acute lymphoblastic leukemia by acting as cofactors for GATA3. Mol Cell Biol. 1998;18:6939–6950. doi: 10.1128/mcb.18.12.6939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palii CG, Pasha R, Brand M. Lentiviral-mediated knockdown during ex vivo erythropoiesis of human hematopoietic stem cells. J Vis Exp. 2011a doi: 10.3791/2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palii CG, Perez-Iratxeta C, Yao Z, Cao Y, Dai F, Davison J, Atkins H, Allan D, Dilworth FJ, Gentleman R, et al. Differential genomic targeting of the transcription factor TAL1 in alternate haematopoietic lineages. The EMBO journal. 2011b;30:494–509. doi: 10.1038/emboj.2010.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palomero T, Odom DT, O’Neil J, Ferrando AA, Margolin A, Neuberg DS, Winter SS, Larson RS, Li W, Liu XS, et al. Transcriptional regulatory networks downstream of TAL1/SCL in T-cell acute lymphoblastic leukemia. Blood. 2006;108:986–992. doi: 10.1182/blood-2005-08-3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay RG, Gonda TJ. MYB function in normal and cancer cells. Nat Rev Cancer. 2008;8:523–534. doi: 10.1038/nrc2439. [DOI] [PubMed] [Google Scholar]
- Soler E, Andrieu-Soler C, de Boer E, Bryne JC, Thongjuea S, Stadhouders R, Palstra RJ, Stevens M, Kockx C, van Ijcken W, et al. The genome-wide dynamics of the binding of Ldb1 complexes during erythroid differentiation. Genes Dev. 2010;24:277–289. doi: 10.1101/gad.551810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoms JA, Birger Y, Foster S, Knezevic K, Kirschenbaum Y, Chandrakanthan V, Jonquieres G, Spensberger D, Wong JW, Oram SH, et al. ERG promotes T-acute lymphoblastic leukemia and is transcriptionally regulated in leukemic cells by a stem cell enhancer. Blood. 2011;117:7079–7089. doi: 10.1182/blood-2010-12-317990. [DOI] [PubMed] [Google Scholar]
- Tijssen MR, Cvejic A, Joshi A, Hannah RL, Ferreira R, Forrai A, Bellissimo DC, Oram SH, Smethurst PA, Wilson NK, et al. Genome-wide analysis of simultaneous GATA1/2, RUNX1, FLI1, and SCL binding in megakaryocytes identifies hematopoietic regulators. Developmental cell. 2011;20:597–609. doi: 10.1016/j.devcel.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay M, Tremblay CS, Herblot S, Aplan PD, Hebert J, Perreault C, Hoang T. Modeling T-cell acute lymphoblastic leukemia induced by the SCL and LMO1 oncogenes. Genes Dev. 2010;24:1093–1105. doi: 10.1101/gad.1897910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valouev A, Johnson DS, Sundquist A, Medina C, Anton E, Batzoglou S, Myers RM, Sidow A. Genome-wide analysis of transcription factor binding sites based on ChIP-Seq data. Nat Methods. 2008;5:829–834. doi: 10.1038/nmeth.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhoeyen E, Dardalhon V, Ducrey-Rundquist O, Trono D, Taylor N, Cosset FL. IL-7 surface-engineered lentiviral vectors promote survival and efficient gene transfer in resting primary T lymphocytes. Blood. 2003;101:2167–2174. doi: 10.1182/blood-2002-07-2224. [DOI] [PubMed] [Google Scholar]
- Wadman IA, Osada H, Grutz GG, Agulnick AD, Westphal H, Forster A, Rabbitts TH. The LIM-only protein Lmo2 is a bridging molecule assembling an erythroid, DNA-binding complex which includes the TAL1, E47, GATA-1 and Ldb1/NLI proteins. EMBO J. 1997;16:3145–3157. doi: 10.1093/emboj/16.11.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson NK, Foster SD, Wang X, Knezevic K, Schutte J, Kaimakis P, Chilarska PM, Kinston S, Ouwehand WH, Dzierzak E, et al. Combinatorial transcriptional control in blood stem/progenitor cells: genome-wide analysis of ten major transcriptional regulators. Cell Stem Cell. 2010;7:532–544. doi: 10.1016/j.stem.2010.07.016. [DOI] [PubMed] [Google Scholar]
- Wilson NK, Miranda-Saavedra D, Kinston S, Bonadies N, Foster SD, Calero-Nieto F, Dawson MA, Donaldson IJ, Dumon S, Frampton J, et al. The transcriptional program controlled by the stem cell leukemia gene Scl/Tal1 during early embryonic hematopoietic development. Blood. 2009;113:5456–5465. doi: 10.1182/blood-2009-01-200048. [DOI] [PubMed] [Google Scholar]
- Wouters BJ, Jorda MA, Keeshan K, Louwers I, Erpelinck-Verschueren CA, Tielemans D, Langerak AW, He Y, Yashiro-Ohtani Y, Zhang P, et al. Distinct gene expression profiles of acute myeloid/T-lymphoid leukemia with silenced CEBPA and mutations in NOTCH1. Blood. 2007;110:3706–3714. doi: 10.1182/blood-2007-02-073486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Huang S, Chang LS, Agulnick AD, Brandt SJ. Identification of a TAL1 target gene reveals a positive role for the LIM domain-binding protein Ldb1 in erythroid gene expression and differentiation. Mol Cell Biol. 2003;23:7585–7599. doi: 10.1128/MCB.23.21.7585-7599.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama T, Nakamura T. Tribbles in disease: Signaling pathways important for cellular function and neoplastic transformation. Cancer Sci. 2011 doi: 10.1111/j.1349-7006.2011.01914.x. [DOI] [PubMed] [Google Scholar]
- Young RA. Control of the embryonic stem cell state. Cell. 2011;144:940–954. doi: 10.1016/j.cell.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Ding L, Holmfeldt L, Wu G, Heatley SL, Payne-Turner D, Easton J, Chen X, Wang J, Rusch M, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481:157–163. doi: 10.1038/nature10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber J, Rappaport AR, Luo W, Wang E, Chen C, Vaseva AV, Shi J, Weissmueller S, Fellmann C, Taylor MJ, et al. An integrated approach to dissecting oncogene addiction implicates a Myb-coordinated self-renewal program as essential for leukemia maintenance. Genes & development. 2011;25:1628–1640. doi: 10.1101/gad.17269211. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.