Abstract
The prevalence and economic burden of obesity and type 2 diabetes is a driving force for the discovery of molecular targets to improve insulin sensitivity and glycemic control. Here, we review several transgenic mouse models that identify promising targets, ranging from proteins involved in the insulin signaling pathway, alterations of genes affecting energy metabolism, and transcriptional metabolic regulators. Despite the diverse endpoints in each model, a common thread that emerges is the necessity for maintenance of energy balance, suggesting pharmacotherapy must target the development of drugs that decrease energy intake, accelerate energy expenditure in a well controlled manner, or augment natural compensatory responses to positive energy balance.
Introduction
A progressive decline in the sensitivity to insulin is an early defect in the etiology of type 2 diabetes. Identifying potential therapeutic targets to improve insulin sensitivity is a major goal of academic and industry research efforts, because the prevalence and economic burden of obesity and type 2 diabetes continues to rise globally (Dixon, 2010; Farag and Gaballa, 2011). From 2008 to 2010, 728 patent applications were filed in the United States for specific antidiabetic interventions, targeting various components of the glucose metabolism and insulin signaling pathways (Carpino and Goodwin, 2010). Many of these targets were identified initially through genetic models in which a single protein had been underexpressed or overexpressed and found to generate a phenotype that is protected against diet-induced obesity and/or insulin resistance. The long-term objective is to develop and validate molecules that modulate the target protein's function with the expectation of improving glycemic control to prevent the onset and/or attenuate the progression of diabetes. However, despite enormous investment by federal agencies, private foundations, and the pharmaceutical industry, prescription drugs currently available for treating hyperglycemia are limited, have significant shortcomings in terms of efficacy, and often require combinatorial therapy to achieve accepted treatment goals (Moller, 2012). The process of new drug development has brought to light thousands of potential therapeutic molecules, >90% of which typically fail in the preclinical phases. A number of factors have contributed to the high attrition rate of pipeline molecules, not the least of which is that many seemingly promising targets have not been adequately vetted either conceptually or experimentally. The current perspective will review some of the more promising targets identified by basic research in the past several years, discuss potential common but often less appreciated mechanisms to emerge from these studies, and provide an assessment of potential therapeutic strategies for moving forward for the prevention/treatment of diabetes.
Knockout Models Involving the Insulin Signaling Pathway
Insulin Receptor Phosphorylation.
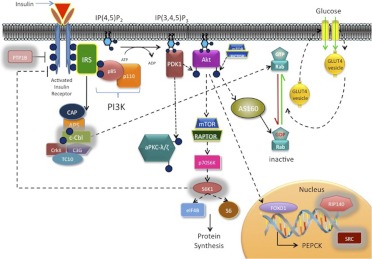
Insulin binding activates tyrosine kinase activity intrinsic to the β-subunit of the insulin receptor, eliciting tyrosine phosphorylation of the receptor itself as well as the insulin receptor substrate (IRS) docking proteins (Fig. 1). A series of phosphatases associated with the insulin receptor are responsible for “turning off” insulin-stimulated cell signaling under normal conditions (Worm et al., 1999) and display enhanced activity in numerous insulin-resistant states (McGuire et al., 1991; Hauguel-de Mouzon et al., 1993; Ahmad and Goldstein, 1995).
Fig. 1.
Downstream molecular signals of the insulin receptor pathway. Proteins outlined in gray are manipulated in knockout mouse models and discussed in detail in the current review. APS, adaptor proteins associated with pleckstrin homology and Src homology 2 domains; AS160, Akt substrate of 160 kDa; CAP, c-Cbl-associated protein; C3G, Crk Src homology 3-binding guanine nucleotide-releasing factor; mTOR, mammalian target of rapamycin; PDK1, phosphoinositide-dependent kinase 1; IP(3,4)P2, phosphatidylinositol-3,4-biphosphate; IP(3,4,5)P3, phosphatidylinositol-3,4,5-triphosphate; PKC, protein kinase C.
Protein-tyrosine phosphatase 1B (PTP1B) is a ubiquitously expressed tyrosine phosphatase that negatively regulates insulin signaling under normal conditions (Tonks, 2003). Blocking PTP1B action therefore represents a potential target for enhancing insulin action and potentially improving glycemic control in insulin-resistant states. Consistent with this hypothesis, two independently developed lines of PTP1B-deficient mice [PTP1B(−/−)] were found to have improved glucose tolerance and enhanced insulin sensitivity compared with wild-type controls, even when consuming a high-fat diet (Elchebly et al., 1999; Klaman et al., 2000). However, PTP1B(−/−) mice are also characterized by a marked increase in total daily energy expenditure and, as such, are resistant to weight gain despite a significant increase in food intake. The increase in energy expenditure seems to be mediated by a centrally driven increase in activity level, because neuronal-specific, but not muscle- or liver-specific (Delibegovic et al., 2007; Agouni et al., 2011), PTP1B(−/−) mice are more physically active than PTP1B(+/+) mice and are protected from high-fat diet-induced weight gain, adiposity, and insulin resistance (Bence et al., 2006), similar to whole-body PTP1B(−/−) mice (Klaman et al., 2000). It is noteworthy that the activity of AMP-activated protein kinase (AMPK), a ubiquitously expressed protein activated by increased energy demand, is decreased in the hypothalmic nuclei of neuronal and whole-body PTP1B(−/−) mice (Xue et al., 2009), raising the exciting possibility that a disruption in fuel sensing in neuronal cells induced by PTP1B inhibition may alter physical activity behavior. In this same study (Xue et al., 2009), AMPK activity and the expression of oxidative metabolism genes were increased in skeletal muscle. Curiously, these responses were attributed to altered neuronal signaling leading to an increase in AMP in muscle and were proposed to at least partially account for the increase in energy expenditure (Xue et al., 2009), seemingly overlooking the more plausible explanation that the adaptive responses in muscle were simply driven by the increased activity level in these animals (i.e., similar to exercise training). Regardless, PTB1B inhibitors are currently being developed as potential pharmacological therapy for type 2 diabetes (Johnson et al., 2002; Thareja et al., 2012). However, the improvements in glucose homeostasis induced by whole-body or neuronal PTP1B deficiency in mice seem to be most likely mediated by PTP1B actions in the brain controlling physical activity rather than the consequence of a direct effect in muscle.
Insulin Signaling to Protein Synthesis.
Insulin signaling also leads to the activation of protein synthesis, which is mediated in part by S6 kinase 1 (S6K1), an effector of the mammalian target of rapamycin (Um et al., 2006). S6K1 has also been implicated as a negative regulator of insulin signaling by leading either directly or indirectly to inhibitory serine phosphorylation of IRS1 during nutrient overload (Um et al., 2004; Tremblay et al., 2005). Targeting inhibition of negative regulators of insulin signaling, such as S6K1, could lead to enhanced insulin sensitivity and thus is a potentially attractive strategy for the treatment of insulin resistance. Indeed, S6K1(−/−) mice were found to be protected from high-fat diet-induced obesity and insulin resistance despite similar or even slightly greater rates of food intake. Insulin signaling was maintained in muscle, liver, and adipose tissue in S6K1(−/−) mice, which was attributed to enhanced β-oxidation caused by increased mitochondrial content in muscle and associated elevated metabolic rate (Um et al., 2004).
Casitas b-lineage lymphoma (c-Cbl) is a multiadaptor protein with several protein-protein interaction domains, including a tyrosine kinase binding domain, a Src homology 2 domain, and a RING finger domain containing intrinsic E3 ubiquitin ligase activity that is known to target tyrosine kinase growth factor receptors for degradation (Schmidt and Dikic, 2005; Thien and Langdon, 2005). c-Cbl has also been identified as a key protein in the alternate pathway of insulin signaling to glucose transporter type 4 (GLUT4) translocation [i.e., non-phosphoinositide 3-kinase (PI3K)-Akt] in 3T3-L1 adipocytes (Liu et al., 2003; Saltiel and Pessin, 2003), seemingly counter to its role as a negative regulator of growth factor signaling (Fig. 1). c-Cbl(−/−) mice have decreased adiposity despite hyperphagia and improved whole-body insulin action resulting from increased muscle glucose uptake (Molero et al., 2004). When fed a high-fat diet, c-Cbl(−/−) mice are protected from diet-induced weight gain and insulin resistance (Molero et al., 2006b). Although these findings are consistent with the notion of increased insulin receptor stability caused by loss of a putative negative regulator, c-Cbl(−/−) mice are also characterized by a profound increase in energy expenditure that is paralleled by increased mitochondrial density and respiratory capacity (Molero et al., 2004). It is noteworthy that transgenic c-Cbl mice with mutated RING fingers, but not mice with mutated PI3K binding domains, still display increased energy expenditure and are protected from high-fat diet-induced obesity and insulin resistance (Molero et al., 2006a). Increased muscle AMPK activity and whole-body flux through β-oxidation were also evident and interpreted as driving the increase in energy expenditure, implying activation of a thermogenic mechanism (Molero et al., 2006a). However, these mice also display a marked increase in activity level (Molero et al., 2004, 2006b).
Alterations of Genes Affecting Energy Metabolism
The phosphoenolpyruvate carboxykinase (PEPCK) gene is expressed primarily in liver and kidney where it catalyzes the first highly regulated step in gluconeogenesis, converting oxalacetate to phosphoenolpyruvate. Offspring of mice with the PEPCK gene ablated [PEPCK(−/−)] survive for only 2 days after birth (She et al., 2000). Perinatal studies revealed PEPCK(−/−) mice were severely hypoglycemic with depleted hepatic glycogen stores (Hakimi et al., 2005), which is congruent with results in a hepatic-specific PEPCK(−/−) line (She et al., 2000, 2003), demonstrating PEPCK expression in liver is essential for glucose homeostasis.
Fasting hyperglycemia occurs with insulin resistance, and the decline of insulin sensitivity in the liver leads to unregulated transcription of PEPCK and elevated hepatic glucose production (Hanson and Reshef, 1997). So, it is not surprising that transgenic mice that overexpress PEPCK are hyperglycemic with impaired glucose tolerance (Valera et al., 1994). PEPCK involvement in regulating gluconeogenesis in the liver is well established. However, although not widely discussed, PEPCK activity is present at low levels in other tissues that are not involved in the production of glucose (e.g., adipose tissue, skeletal muscle). In these tissues, PEPCK is thought to play a role in glyceroneogeneisis by synthesizing glycerol 3-phosphate, a precursor required for re-esterficiation of free fatty acids, ultimately leading to fat storage (Ballard et al., 1967). Franckhauser et al. (2002) developed a transgenic mouse model that overexpressed PEPCK in adipose tissue, hypothesizing that the increase in glyceroneogenesis and fat storage would lower circulating free fatty acid. These mice did, in fact, weigh more and showed increased fat pad mass compared with their wild-type littermate counterparts, despite similar food intake. Even with the obese phenotype, glucose tolerance was normal and insulin sensitivity was slightly increased compared with wild-type controls. The adipose-specific overexpression of PEPCK essentially created an obese model in the absence of insulin resistance. However, when challenged with a high-fat diet the transgenic mice showed the same glucose intolerance and insulin resistance as the high-fat fed controls (Franckhauser et al., 2006). Although mice that overexpressed PEPCK in adipose tissue were not protected from diet-induced insulin resistance, studies involving the overexpression of PEPCK in skeletal muscle revealed an entirely different phenotype.
The muscle-specific overexpressor of cytosolic PEPCK (PEPCK-Cmus) created a transgenic mouse line of endurance athletes. These transgenic mice display a remarkable endurance capacity, running 30 times longer than their wild-type counterparts during a treadmill test to exhaustion (Hakimi et al., 2007). The PEPCK-Cmus mice are also characterized by extremely high rates of spontaneous activity; they consume 60% more food but weigh nearly 50% less with 10% the body fat of wild-type littermates. They have five times higher triglyceride content in their skeletal muscle and yet are extremely insulin sensitive (Hakimi et al., 2007), reminiscent of the “athletes' paradox” that is evident in endurance athletes (Goodpaster et al., 2001). The overexpressing PEPCK-Cmus mice rely primarily on fatty acid for fuel and have much higher levels of mitochondria in their skeletal muscle. The mechanism by which overexpression of PEPCK in muscle produces such a remarkable phenotype is unknown. It is noteworthy that rats bred for high-endurance running capacity are also consistently more active and have elevated PEPCK expression in muscle (Waters et al., 2008; Novak et al., 2010). It is tempting to speculate elevated PEPCK-Cmus in muscle activates some type of cataplerotic futile cycle that accelerates energy turnover, which, in turn, is communicated to the brain to stimulate locomotor activity. This would explain the increase in mitochondrial biogenesis and increased endurance capacity. It has also been suggested that because the brains of highly active rats are more sensitive and/or responsive to neuropeptides that stimulate activity, it is possible that skeletal muscle may produce a factor that modulates the brain circuitry controlling activity (Novak et al., 2010).
Modifications of Transcriptional Metabolic Regulators
Obesity Susceptibility Genes.
Single-nucleotide polymorphism (SNP) variants in the fat mass and obesity-associated (FTO) gene are correlated with obesity and increased body mass index (Scuteri et al., 2007). FTO is ubiquitiously expressed, but highly concentrated in the arcuate nucleus of the hypothalamus, the center for appetite regulation (Wardle et al., 2008). The FTO gene encodes for a AlkB-like 2-oxoglutarate-dependent nucleic acid demethylase (Gerken et al., 2007), but the direct role FTO plays in metabolism is still unclear.
Patients with high-risk FTO alleles have one of the SNP variants on intron 1 of the FTO gene, which correlates with increased body weight and increased daily energy intake or appetite (Speakman et al., 2008; Wardle et al., 2008). Over the past several years FTO has became a high-interest “obesity gene target,” dubbed the most common obesity susceptibility gene in the white population (Wardle et al., 2008). In normal mice, FTO mRNA levels in the arcuate nucleus are decreased after a 48-h fast and elevated after feeding (Gerken et al., 2007), so it was unclear whether FTO was positively or negatively regulated by energy intake. Transgenic mice that overexpress FTO by one (FTO-3) or two (FTO-4) additional copies have increased body weight, food intake, and fat mass compared with their wild-type littermates (Church et al., 2010). When placed on a high-fat diet, FTO overexpressors have a greater percentage increase in food intake, with a direct relationship between number of additional copies of FTO and increased body weight and fat mass.
It is noteworthy that two loss-of-function models also suggest that FTO plays an important role in regulating food intake and energy expenditure. Deleting the FTO gene protects against the development of obesity. Mice deficient in FTO [FTO(−/−)], or mice harboring a dominant missense mutation that partially alters function (i.e., analogous to humans carrying at risk SNPs in the FTO gene), have reduced body weight and fat mass with similar or even higher rates of food intake (Church et al., 2009; Fischer et al., 2009). These mice are also resistant to high-fat diet-induced obesity and insulin resistance, apparently as a consequence of a marked increase in energy expenditure (elevated oxygen consumption rate) in the face of similar or even reduced spontaneous locomotor activity. The increase in energy expenditure could not be attributed to any difference in thyroid hormone or expression of uncoupling protein (UCP) 1 in brown adipose tissue, but was associated with increased sympathetic tone (Fischer et al., 2009). Collectively, findings from these various mouse models suggest that elevated FTO activity is associated with increased body weight that is mediated primarily by increased food intake, whereas loss or partial loss of FTO activity protects against diet-induced obesity by increasing energy expenditure, potentially by inducing some type of futile energy cycle. It is worth reiterating that the SNPs identified within the FTO gene most strongly associated with the increased risk of obesity are located within the first intron of the gene (Speakman et al., 2008; Wardle et al., 2008), the functional impact of which is unknown. It is noteworthy that in two other studies in which dietary fat intake and physical activity were factored in failed to find any association between the high-risk FTO alleles and obesity or body mass index in humans (Sonestedt et al., 2009; Razquin et al., 2010), suggesting lifestyle factors, or increases in energy demand, can modify the susceptibility to obesity despite unfavorable genetic predisposition.
Transcriptional Coregulators.
The balance of transcriptional coregulators (enhancers and repressors) plays a large role in regulating the expression of genes involved in metabolic pathways, ultimately controlling energy homeostasis. In general, transcriptional coregulators serve to enhance or repress the remodeling of chromatin and/or the assembly of the transcriptional machinery in the promoter region of target genes. The peroxisome proliferator-activated receptor γ coactivator 1α (PGC-1α) is perhaps the best-known coactivator because of its well defined role in promoting adaptive thermogenesis in brown adipose tissue and metabolic programs that promote oxidative metabolism in other tissues, including muscle and liver (Puigserver and Spiegelman, 2003). In contrast to PGC-1α, receptor interacting protein 140 (RIP140) is a nuclear receptor corepressor that represses transcription of metabolic genes through histone deacetylase-dependent and -independent mechanisms (Christian et al., 2006). RIP140 is constitutively expressed at high levels in metabolic tissues including adipose tissue, muscle, and liver (Leonardsson et al., 2004), suggesting that repression of RIP140 could promote increased expression of a favorable metabolic gene program in these tissues. Indeed, RIP140 null mice are characterized by up-regulation of numerous genes involved in oxidative metabolism in both white adipose tissue and skeletal muscle, have reduced body fat and total body weight on chow or high-fat diets despite no difference in food intake, and are protected from both aging and high-fat diet-induced insulin resistance (Leonardsson et al., 2004; Powelka et al., 2006; Seth et al., 2007). As predicted from the apparent change in energy homeostasis, RIP140-null mice display a consistently elevated rate of oxygen consumption and energy expenditure in both the light and dark cycle relative to wild-type littermates (Seth et al., 2007). The mechanism by which the absence of RIP140 increases energy expenditure is unclear. It is noteworthy, however, that several genes whose expression is normally restricted to brown adipose tissue, including UCP1, dramatically increase in expression in white adipose tissue (Leonardsson et al., 2004), raising the possibility that activation of thermogenesis in white adipose tissue could account for at least a portion of the increase in energy expenditure. The absence of RIP140 also elicits a marked increase in oxidative enzyme gene expression in the normally glycolytic fast-twitch muscles, increasing overall mitochondrial respiratory capacity (Seth et al., 2007). Although it is tempting to speculate that the up-regulation of genes involved in oxidative metabolism may account for the elevated energy expenditure in RIP140-null mice (Rosell et al., 2011), it is important to recognize that energy expenditure is a demand-driven process and an increase in oxidative capacity will increase, not decrease, energetic efficiency (Muoio and Neufer, 2012). The extent to which such changes in the metabolic profile in muscle may influence spontaneous locomotor activity and thus contribute to the increase in energy expenditure was, unfortunately, not determined. Nevertheless, suppression of RIP140 has been suggested as a potential therapeutic target for the treatment in metabolic disorders associated with nutritional overload (Rosell et al., 2011).
Transcriptional coregulators interact directly with transcription factors to either enhance or repress transcription. Many of the coregulators facilitate the remodeling of chromatin structure to allow specific transcription factors and the basal transcription machinery to gain access to the promoter region of target genes. Steroid receptor coactivators (SRCs), also known as p160 proteins, interact with nuclear hormone receptors upon ligand binding (Aranda and Pascual, 2001). Three members of the SRC family, SRC-1 (also known as NcoA-1), SRC-2 (also known as TIF2, GRIP1, and NCOA2), and SRC-3 (also known as p/CIP, AIB1, ACTR, RAC3, and TRAM-1), have been implicated in the control of energy balance, primarily by regulating the interaction with transcription factors, such as peroxisome proliferator-activated receptor γ and other coregulators, such as PGC-1α, that control the expression of genes involved in energy metabolism (Picard et al., 2002). It is noteworthy that SRC-2(−/−) and SRC-3(−/−) mice are protected from diet-induced obesity, whereas SRC-1(−/−) mice are more susceptible to weight gain (Picard et al., 2002; Coste et al., 2008). Whole-body SRC-2(−/−) and SRC-3(−/−) mice, as well as skeletal muscle-specific SRC-2 knockout mice, are characterized by increased energy expenditure and higher body temperatures, responses that seem to be mediated by increased UCP expression and activity in mitochondria of both brown adipose tissue and skeletal muscle (Picard et al., 2002; Coste et al., 2008; Duteil et al., 2010). Normally, SRC-2 and SRC-3 help to maintain optimal efficiency between mitochondrial respiration and ATP synthesis by antagonizing the actions of SRC-1 and/or PGC-1α, both of which promote UCP expression and thermogenesis. Thus, the absence of SRC-2 or SRC-3 leaves SRC-1 unopposed, generating an increase in energy expenditure and activation of mitochondrial biogenesis. In fact, knocking out SRC-2 specifically in muscle is sufficient to protect against high-fat diet-induced obesity and insulin resistance, suggesting that decreasing SRC-2 and/or increasing SRC-1 in skeletal muscle may represent viable strategies for counteracting nutritional overload metabolic disorders (Duteil et al., 2010).
Conclusions and Perspectives
The transgenic models highlighted in this review are just a few examples of genetic manipulations that have been found to protect against diet-induced insulin resistance and/or obesity. The general premise has been that developing pharmaceutical approaches that target those insulin signaling, metabolic, and/or transcriptional regulatory proteins found to be protective represents a viable strategy for improving insulin-stimulated glucose uptake and glycemic control in insulin-resistant or diabetic patients. It is noteworthy, however, that despite the diversity of proteins and cellular processes targeted by genetic manipulation, an increase in energy expenditure, either via thermogenesis, futile energy cycling, or increased activity level, has emerged as a common mechanism to explain the protection against diet-induced metabolic dysfunction in these studies. In fact, increased energy expenditure was identified more than 10 years ago as a common underlying protective mechanism in various genetic models developed up to that point (Chen and Farese, 2001; Reitman, 2002). Thus collectively, these findings are entirely consistent with what seems intuitively obvious, that if energy surplus is the underlying cause of metabolic disease, then restoration of energy balance is ultimately going to be required to reverse these diseases.
This is consistent with the principles of cellular bioenergetics, that energy expenditure in all cells is determined by the rate of energy demand, not energy supply. Available evidence to date suggest that when cells consistently experience nutritional overload the elevated fuel supply creates an increase in reducing pressure that is sensed by the mitochondria and counterbalanced by an increase in mitochondrial H2O2 emission, which, via redox signaling networks, leads to a decrease in cellular insulin sensitivity (Fisher-Wellman and Neufer, 2012). Thus for insulin-sensitive tissues, the development of insulin resistance is a natural defense mechanism against nutritional overload. This implies targeting a single insulin signaling protein, transcription factor/coregulator, or metabolic reaction to increase insulin-stimulated glucose uptake, in the absence of an increase in metabolic demand, will ultimately place a greater metabolic burden on those tissues. Compensatory strategies to either attenuate oxidant production or supplement existing redox buffering systems represent potential therapeutic approaches to improving insulin sensitivity. Metformin, for example, the most widely prescribed drug for the treatment of type 2 diabetes, is thought to improve insulin sensitivity by partially activating the energy-sensing kinase AMPK and/or by partially attenuating mitochondrial H2O2 emission (Zhou et al., 2001; Batandier et al., 2006; Kane et al., 2010). Regardless of the target, however, compensatory approaches by definition do not block the underlying problem and therefore are unlikely to reverse metabolic disease in the long run unless metabolic balance at the cellular and whole-body level is also restored. Therefore, strategies to restore metabolic balance by targeting energy supply (i.e., appetite control, nutrient absorption) and/or energy expenditure (i.e., uncouplers, futile cycles, activity behavior) systems probably represent the most viable pharmacological approaches for those individuals that are unable to sustain lifestyle modifications.
We are supported by the National Institutes of Health National Institute of Diabetes and Digestive and Kidney Diseases [Grants R01 DK073488, R01 DK074825].
Article, publication date, and citation information can be found at http://jpet.aspetjournals.org.
- IRS
- insulin receptor substrate
- PTP1B
- protein-tyrosine phosphatase 1B
- AMPK
- AMP-activated protein kinase
- S6K1
- S6 kinase 1
- c-Cbl
- Casitas b-lineage lymphoma
- PEPCK
- phosphoenolpyruvate carboxykinase
- PEPCK-Cmus
- muscle-specific overexpressor of cytosolic PEPCK
- FTO
- fat mass and obesity associated
- GLUT4
- glucose transporter type 4
- SNP
- single-nucleotide polymorphism
- RIP140
- receptor interacting protein 140
- SRC
- steroid receptor coactivator
- PI3K
- phosphoinositide 3-kinase
- PGC-1α
- peroxisome proliferator-activated receptor γ coactivator 1α
- UCP
- uncoupling protein.
Authorship Contributions
Wrote or contributed to the writing of the manuscript: Gilliam and Neufer.
References
- Agouni A, Mody N, Owen C, Czopek A, Zimmer D, Bentires-Alj M, Bence KK, Delibegović M. (2011) Liver-specific deletion of protein tyrosine phosphatase (PTP) 1B improves obesity- and pharmacologically induced endoplasmic reticulum stress. Biochem J 438:369–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad F, Goldstein BJ. (1995) Alterations in specific protein-tyrosine phosphatases accompany insulin resistance of streptozotocin diabetes. Am J Physiol Endocrinol Metab 268:E932–E940 [DOI] [PubMed] [Google Scholar]
- Aranda A, Pascual A. (2001) Nuclear hormone receptors and gene expression. Physiol Rev 81:1269–1304 [DOI] [PubMed] [Google Scholar]
- Ballard FJ, Hanson RW, Leveille GA. (1967) Phosphoenolpyruvate carboxykinase and the synthesis of glyceride-glycerol from pyruvate in adipose tissue. J Biol Chem 242:2746–2750 [PubMed] [Google Scholar]
- Batandier C, Guigas B, Detaille D, El-Mir MY, Fontaine E, Rigoulet M, Leverve XM. (2006) The ROS production induced by a reverse-electron flux at respiratory-chain complex 1 is hampered by metformin. J Bioenerg Biomembr 38:33–42 [DOI] [PubMed] [Google Scholar]
- Bence KK, Delibegovic M, Xue B, Gorgun CZ, Hotamisligil GS, Neel BG, Kahn BB. (2006) Neuronal PTP1B regulates body weight, adiposity and leptin action. Nat Med 12:917–924 [DOI] [PubMed] [Google Scholar]
- Carpino PA, Goodwin B. (2010) Diabetes area participation analysis: a review of companies and targets described in the 2008–2010 patent literature. Expert Opin Ther Pat 20:1627–1651 [DOI] [PubMed] [Google Scholar]
- Chen HC, Farese RV., Jr (2001) Turning WAT into BAT gets rid of fat. Nat Med 7:1102–1103 [DOI] [PubMed] [Google Scholar]
- Christian M, White R, Parker MG. (2006) Metabolic regulation by the nuclear receptor corepressor RIP140. Trends Endocrinol Metab 17:243–250 [DOI] [PubMed] [Google Scholar]
- Church C, Lee S, Bagg EA, McTaggart JS, Deacon R, Gerken T, Lee A, Moir L, Mecinović J, Quwailid MM, et al. (2009) A mouse model for the metabolic effects of the human fat mass and obesity associated FTO gene. PLoS Genet 5:e1000599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church C, Moir L, McMurray F, Girard C, Banks GT, Teboul L, Wells S, Brüning JC, Nolan PM, Ashcroft FM, et al. (2010) Overexpression of Fto leads to increased food intake and results in obesity. Nat Genet 42:1086–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coste A, Louet JF, Lagouge M, Lerin C, Antal MC, Meziane H, Schoonjans K, Puigserver P, O'Malley BW, Auwerx J. (2008) The genetic ablation of SRC-3 protects against obesity and improves insulin sensitivity by reducing the acetylation of PGC-1α. Proc Natl Acad Sci U S A 105:17187–17192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delibegovic M, Bence KK, Mody N, Hong EG, Ko HJ, Kim JK, Kahn BB, Neel BG. (2007) Improved glucose homeostasis in mice with muscle-specific deletion of protein-tyrosine phosphatase 1B. Mol Cell Biol 27:7727–7734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JB. (2010) The effect of obesity on health outcomes. Mol Cell Endocrinol 316:104–108 [DOI] [PubMed] [Google Scholar]
- Duteil D, Chambon C, Ali F, Malivindi R, Zoll J, Kato S, Geny B, Chambon P, Metzger D. (2010) The transcriptional coregulators TIF2 and SRC-1 regulate energy homeostasis by modulating mitochondrial respiration in skeletal muscles. Cell Metabolism 12:496–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elchebly M, Payette P, Michaliszyn E, Cromlish W, Collins S, Loy AL, Normandin D, Cheng A, Himms-Hagen J, Chan CC, et al. (1999) Increased insulin sensitivity and obesity resistance in mice lacking the protein tyrosine phosphatase-1B gene. Science 283:1544–1548 [DOI] [PubMed] [Google Scholar]
- Farag YM, Gaballa MR. (2011) Diabesity: an overview of a rising epidemic. Nephrol Dial Transplant 26:28–35 [DOI] [PubMed] [Google Scholar]
- Fischer J, Koch L, Emmerling C, Vierkotten J, Peters T, Brüning JC, Rüther U. (2009) Inactivation of the Fto gene protects from obesity. Nature 458:894–898 [DOI] [PubMed] [Google Scholar]
- Fisher-Wellman KH, Neufer PD. (2012) Linking mitochondrial bioenergetics to insulin resistance via redox biology. Trends Endocrinol Metab 23:142–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franckhauser S, Muñoz S, Elias I, Ferre T, Bosch F. (2006) Adipose overexpression of phosphoenolpyruvate carboxykinase leads to high susceptibility to diet-induced insulin resistance and obesity. Diabetes 55:273–280 [DOI] [PubMed] [Google Scholar]
- Franckhauser S, Muñoz S, Pujol A, Casellas A, Riu E, Otaegui P, Su B, Bosch F. (2002) Increased fatty acid re-esterification by PEPCK overexpression in adipose tissue leads to obesity without insulin resistance. Diabetes 51:624–630 [DOI] [PubMed] [Google Scholar]
- Gerken T, Girard CA, Tung YC, Webby CJ, Saudek V, Hewitson KS, Yeo GS, McDonough MA, Cunliffe S, McNeill LA, et al. (2007) The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science 318:1469–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodpaster BH, He J, Watkins S, Kelley DE. (2001) Skeletal muscle lipid content and insulin resistance: evidence for a paradox in endurance-trained athletes. J Clin Endocrinol Metab 86:5755–5761 [DOI] [PubMed] [Google Scholar]
- Hakimi P, Johnson MT, Yang J, Lepage DF, Conlon RA, Kalhan SC, Reshef L, Tilghman SM, Hanson RW. (2005) Phosphoenolpyruvate carboxykinase and the critical role of cataplerosis in the control of hepatic metabolism. Nutr Metab (Lond) 2:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakimi P, Yang J, Casadesus G, Massillon D, Tolentino-Silva F, Nye CK, Cabrera ME, Hagen DR, Utter CB, Baghdy Y, et al. (2007) Overexpression of the cytosolic form of phosphoenolpyruvate carboxykinase (GTP) in skeletal muscle repatterns energy metabolism in the mouse. J Biol Chem 282:32844–32855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson RW, Reshef L. (1997) Regulation of phosphoenolpyruvate carboxykinase (GTP) gene expression. Annu Rev Biochem 66:581–611 [DOI] [PubMed] [Google Scholar]
- Hauguel-de Mouzon S, Peraldi P, Alengrin F, Van Obberghen E. (1993) Alteration of phosphotyrosine phosphatase activity in tissues from diabetic and pregnant rats. Endocrinology 132:67–74 [DOI] [PubMed] [Google Scholar]
- Johnson TO, Ermolieff J, Jirousek MR. (2002) Protein tyrosine phosphatase 1B inhibitors for diabetes. Nat Rev Drug Discov 1:696–709 [DOI] [PubMed] [Google Scholar]
- Kane DA, Anderson EJ, Price JW, 3rd, Woodlief TL, Lin CT, Bikman BT, Cortright RN, Neufer PD. (2010) Metformin selectively attenuates mitochondrial H2O2 emission without affecting respiratory capacity in skeletal muscle of obese rats. Free Radic Biol Med 49:1082–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaman LD, Boss O, Peroni OD, Kim JK, Martino JL, Zabolotny JM, Moghal N, Lubkin M, Kim YB, Sharpe AH, et al. (2000) Increased energy expenditure, decreased adiposity, and tissue-specific insulin sensitivity in protein-tyrosine phosphatase 1B-deficient mice. Mol Cell Biol 20:5479–5489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonardsson G, Steel JH, Christian M, Pocock V, Milligan S, Bell J, So PW, Medina-Gomez G, Vidal-Puig A, White R, et al. (2004) Nuclear receptor corepressor RIP140 regulates fat accumulation. Proc Natl Acad Sci U S A 101:8437–8442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, DeYoung SM, Hwang JB, O'Leary EE, Saltiel AR. (2003) The roles of Cbl-b and c-Cbl in insulin-stimulated glucose transport. J Biol Chem 278:36754–36762 [DOI] [PubMed] [Google Scholar]
- McGuire MC, Fields RM, Nyomba BL, Raz I, Bogardus C, Tonks NK, Sommercorn J. (1991) Abnormal regulation of protein tyrosine phosphatase activities in skeletal muscle of insulin-resistant humans. Diabetes 40:939–942 [DOI] [PubMed] [Google Scholar]
- Molero JC, Jensen TE, Withers PC, Couzens M, Herzog H, Thien CB, Langdon WY, Walder K, Murphy MA, Bowtell DD, et al. (2004) c-Cbl-deficient mice have reduced adiposity, higher energy expenditure, and improved peripheral insulin action. J Clin Invest 114:1326–1333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molero JC, Turner N, Thien CB, Langdon WY, James DE, Cooney GJ. (2006a) Genetic ablation of the c-Cbl ubiquitin ligase domain results in increased energy expenditure and improved insulin action. Diabetes 55:3411–3417 [DOI] [PubMed] [Google Scholar]
- Molero JC, Waring SG, Cooper A, Turner N, Laybutt R, Cooney GJ, James DE. (2006b) Casitas b-lineage lymphoma-deficient mice are protected against high-fat diet-induced obesity and insulin resistance. Diabetes 55:708–715 [DOI] [PubMed] [Google Scholar]
- Moller DE. (2012) Metabolic disease drug discovery- “hitting the target” is easier said than done. Cell Metab 15:19–24 [DOI] [PubMed] [Google Scholar]
- Muoio DM, Neufer PD. (2012) Lipid-induced mitochondrial stress and insulin action in muscle. Cell Metab 15:595–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak CM, Escande C, Burghardt PR, Zhang M, Barbosa MT, Chini EN, Britton SL, Koch LG, Akil H, Levine JA. (2010) Spontaneous activity, economy of activity, and resistance to diet-induced obesity in rats bred for high intrinsic aerobic capacity. Horm Behav 58:355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard F, Géhin M, Annicotte J, Rocchi S, Champy MF, O'Malley BW, Chambon P, Auwerx J. (2002) SRC-1 and TIF2 control energy balance between white and brown adipose tissues. Cell 111:931–941 [DOI] [PubMed] [Google Scholar]
- Powelka AM, Seth A, Virbasius JV, Kiskinis E, Nicoloro SM, Guilherme A, Tang X, Straubhaar J, Cherniack AD, Parker MG, et al. (2006) Suppression of oxidative metabolism and mitochondrial biogenesis by the transcriptional corepressor RIP140 in mouse adipocytes. J Clin Invest 116:125–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puigserver P, Spiegelman BM. (2003) Peroxisome proliferator-activated receptor-γ coactivator 1α (PGC-1α): transcriptional coactivator and metabolic regulator. Endocr Rev 24:78–90 [DOI] [PubMed] [Google Scholar]
- Razquin C, Martinez JA, Martinez-Gonzalez MA, Bes-Rastrollo M, Fernandez-Crehuet J, Marti A. (2010) A 3-year intervention with a Mediterranean diet modified the association between the rs9939609 gene variant in FTO and body weight changes. Int J Obes (Lond) 34:266–272 [DOI] [PubMed] [Google Scholar]
- Reitman ML. (2002) Metabolic lessons from genetically lean mice. Annu Rev Nutr 22:459–482 [DOI] [PubMed] [Google Scholar]
- Rosell M, Jones MC, Parker MG. (2011) Role of nuclear receptor corepressor RIP140 in metabolic syndrome. Biochim Biophys Acta 1812:919–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saltiel AR, Pessin JE. (2003) Insulin signaling in microdomains of the plasma membrane. Traffic 4:711–716 [DOI] [PubMed] [Google Scholar]
- Schmidt MH, Dikic I. (2005) The Cbl interactome and its functions. Nat Rev Mol Cell Biol 6:907–918 [DOI] [PubMed] [Google Scholar]
- Scuteri A, Sanna S, Chen WM, Uda M, Albai G, Strait J, Najjar S, Nagaraja R, Orrú M, Usala G, et al. (2007) Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genet 3:e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth A, Steel JH, Nichol D, Pocock V, Kumaran MK, Fritah A, Mobberley M, Ryder TA, Rowlerson A, Scott J, et al. (2007) The transcriptional corepressor RIP140 regulates oxidative metabolism in skeletal muscle. Cell Metab 6:236–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- She P, Burgess SC, Shiota M, Flakoll P, Donahue EP, Malloy CR, Sherry AD, Magnuson MA. (2003) Mechanisms by which liver-specific PEPCK knockout mice preserve euglycemia during starvation. Diabetes 52:1649–1654 [DOI] [PubMed] [Google Scholar]
- She P, Shiota M, Shelton KD, Chalkley R, Postic C, Magnuson MA. (2000) Phosphoenolpyruvate carboxykinase is necessary for the integration of hepatic energy metabolism. Mol Cell Biol 20:6508–6517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonestedt E, Roos C, Gullberg B, Ericson U, Wirfält E, Orho-Melander M. (2009) Fat and carbohydrate intake modify the association between genetic variation in the FTO genotype and obesity. Am J Clin Nutr 90:1418–1425 [DOI] [PubMed] [Google Scholar]
- Speakman JR, Rance KA, Johnstone AM. (2008) Polymorphisms of the FTO gene are associated with variation in energy intake, but not energy expenditure. Obesity (Silver Spring) 16:1961–1965 [DOI] [PubMed] [Google Scholar]
- Thareja S, Aggarwal S, Bhardwaj TR, Kumar M. (2012) Protein tyrosine phosphatase 1B inhibitors: a molecular level legitimate approach for the management of diabetes mellitus. Med Res Rev 32:459–517 [DOI] [PubMed] [Google Scholar]
- Thien CB, Langdon WY. (2005) c-Cbl and Cbl-b ubiquitin ligases: substrate diversity and the negative regulation of signalling responses. Biochem J 391:153–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonks NK. (2003) PTP1B: from the sidelines to the front lines! FEBS Lett 546:140–148 [DOI] [PubMed] [Google Scholar]
- Tremblay F, Krebs M, Dombrowski L, Brehm A, Bernroider E, Roth E, Nowotny P, Waldhäusl W, Marette A, Roden M. (2005) Overactivation of S6 kinase 1 as a cause of human insulin resistance during increased amino acid availability. Diabetes 54:2674–2684 [DOI] [PubMed] [Google Scholar]
- Um SH, D'Alessio D, Thomas G. (2006) Nutrient overload, insulin resistance, and ribosomal protein S6 kinase 1, S6K1. Cell Metab 3:393–402 [DOI] [PubMed] [Google Scholar]
- Um SH, Frigerio F, Watanabe M, Picard F, Joaquin M, Sticker M, Fumagalli S, Allegrini PR, Kozma SC, Auwerx J, et al. (2004) Absence of S6K1 protects against age- and diet-induced obesity while enhancing insulin sensitivity. Nature 431:200–205 [DOI] [PubMed] [Google Scholar]
- Valera A, Pujol A, Pelegrin M, Bosch F. (1994) Transgenic mice overexpressing phosphoenolpyruvate carboxykinase develop non-insulin-dependent diabetes mellitus. Proc Natl Acad Sci U S A 91:9151–9154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardle J, Carnell S, Haworth CM, Farooqi IS, O'Rahilly S, Plomin R. (2008) Obesity associated genetic variation in FTO is associated with diminished satiety. J Clin Endocrinol Metab 93:3640–3643 [DOI] [PubMed] [Google Scholar]
- Waters RP, Renner KJ, Pringle RB, Summers CH, Britton SL, Koch LG, Swallow JG. (2008) Selection for aerobic capacity affects corticosterone, monoamines and wheel-running activity. Physiol Behav 93:1044–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worm D, Vinten J, Beck-Nielsen H. (1999) The significance of phosphotyrosine phosphatase (PTPase) 1B in insulin signalling. Diabetologia 42:1146–1149 [DOI] [PubMed] [Google Scholar]
- Xue B, Pulinilkunnil T, Murano I, Bence KK, He H, Minokoshi Y, Asakura K, Lee A, Haj F, Furukawa N, et al. (2009) Neuronal protein tyrosine phosphatase 1B deficiency results in inhibition of hypothalamic AMPK and isoform-specific activation of AMPK in peripheral tissues. Mol Cell Biol 29:4563–4573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou G, Myers R, Li Y, Chen Y, Shen X, Fenyk-Melody J, Wu M, Ventre J, Doebber T, Fujii N, et al. (2001) Role of AMP-activated protein kinase in mechanism of metformin action. J Clin Invest 108:1167–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]