Summary
VP4, the major structural protein of the haloarchaeal pleomorphic virus, HRPV-1, is glycosylated. To define the glycan structure attached to this protein, oligosaccharides released by β-elimination were analysed by mass spectrometry and nuclear magnetic resonance spectroscopy. Such analyses showed that the major VP4-derived glycan is a pentasaccharide comprising glucose, glucuronic acid, mannose, sulphated glucuronic acid and a terminal 5-N-formyllegionaminic acid residue. This is the first observation of legionaminic acid, a sialic acid-like sugar, in an archaeal-derived glycan structure. The importance of this residue for viral infection was demonstrated upon incubation with N-acetylneuraminic acid, a similar monosaccharide. Such treatment reduced progeny virus production by half 4 h post infection. LC-ESI/MS analysis confirmed the presence of pentasaccharide precursors on two different VP4-derived peptides bearing the N-glycosylation signal, NTT. The same sites modified by the native host, Halorubrum sp. strain PV6, were also recognized by the Haloferax volcanii N-glycosylation apparatus, as determined by LC-ESI/MS of heterologously expressed VP4. Here, however, the N-linked pentasaccharide was the same as shown to decorate the S-layer glycoprotein in this species. Hence, N-glycosylation of the haloarchaeal viral protein, VP4, is host-specific. These results thus present additional examples of archaeal N-glycosylation diversity and show the ability of Archaea to modify heterologously expressed proteins.
Introduction
N-glycosylation is a post-translational modification experienced by proteins in all three domains of life. While the steps involved in the eukaryal and bacterial versions of this universal protein-processing event are relatively well defined, far less is known of the N-glycosylation pathway in Archaea (Helenius and Aebi, 2004; Eichler and Adams, 2005; Szymanski and Wren, 2005; Weerapana and Imperiali, 2006; Abu-Qarn et al., 2008; Calo et al., 2010a; Dell et al., 2010; Larkin and Imperiali, 2011).
Of late, studies of several archaeal species have begun to provide insight into archaeal N-glycosylation. While such efforts reveal the use of pathways similar to their eukaryal or bacterial counterparts, other aspects of the process are unique to Archaea (Yurist-Doutsch et al., 2008; Calo et al., 2010a). As in Eukarya, the N-linked glycan is assembled on a phosphodolichol carrier in Archaea, rather than the undecaprenol pyrophosphate carrier used in bacterial N-glycosylation (Behrens and Leloir, 1970; Hartmann and Konig, 1989; Kuntz et al., 1997; Burda and Aebi, 1999; Linton et al., 2005; Jones et al., 2009; Guan et al., 2010). Moreover, in the halophile Haloferax (Hfx.) volcanii, multiple dolichol phosphate carriers participate in N-glycosylation as in Eukarya (Burda and Aebi, 1999; Guan et al., 2010). On the other hand, in contrast to eukaryal dolichol phosphate, which is only saturated at the a-position isoprene position (Swiezewska and Danikiewicz, 2005; Jones et al., 2009), archaeal dolichol phosphate is saturated at both the α- and the ω-position isoprenes (Kuntz et al., 1997; Guan et al., 2010), and in some cases, at more internal positions (Guan et al., 2011). At the same time, the archaeal N-glycosylation process shares traits with its bacterial counterpart. For instance, as in Bacteria, the archaeal oligosaccharide transferase comprises a single subunit, AglB (Chaban et al., 2006; Abu-Qarn et al., 2007; Igura et al., 2008), rather than the multimeric complex found in Eukarya (Kelleher and Gilmore, 2006). Yet, structural and mechanistic considerations assign many archaeal oligosaccharide transferases to a distinct subclass of the enzyme (Maita et al., 2010). Moreover, AglB from the haloarchaeon, Halobacterium (Hbt.) salinarum, apparently recognizes two distinct glycans attached to dolichol phosphate carriers that differ in both the degree of phosphorylation and in terms of the linking sugar (Lechner and Wieland, 1989). Finally, it has been shown that in Hfx. volcanii, the protein-bound glycan can be further processed, as also occurs in eukaryal (but not bacterial) N-glycosylation (Calo et al., 2011a). These and other observations, together with the unparalleled diversity seen in archaeal N-linked glycans (Calo et al., 2010a; Schwarz and Aebi, 2011), point to the archaeal N-glycosylation pathway as being highly versatile and indeed, malleable, as suggested by recent glyco-engineering efforts in Hfx. volcanii (Calo et al., 2010b; 2011b).
In this study, we defined the composition of the major glycan species N-linked to the Halorubrum (Hrr.) sp. pleomorphic virus 1 (HRPV-1) VP4 protein, one of the two major structural proteins of this haloarchaeal virus (Pietilä et al., 2009). VP4, a 53 kDa spike protein protruding from and C-terminally anchored to the viral membrane, is suggested to be responsible for host recognition by the virus (Pietilä et al., 2009; 2010). We determined the structure of the major glycan species decorating VP4 obtained directly from the purified virion produced by its original host, Hrr. sp. PV6. In doing so, we not only present the first report of legionaminic acid in an archaeal-derived glycoprotein but show the importance of this sugar for HRPV-1 infectivity. Finally, we compared the structure of the glycan decorating VP4 expressed in Hrr. sp. PV6 with the structure of the glycan attached to a recombinant version of the protein expressed in Hfx. volcanii. The results reveal not only the ability of haloarchaea to N-glycosylate non-native proteins but also that the composition of N-linked glycans added to such targets is host-specific.
Results
Isolation and mass spectrometry (MS) analysis of HRPV-1 VP4 glycans
Earlier efforts relying on glycostaining approaches, combined with chemical deglycosylation, revealed that the HRPV-1 VP4 protein, a major structural component of the virus, is glycosylated (Pietilä et al., 2010). Now, efforts were directed at describing the composition of the glycan that decorates VP4.
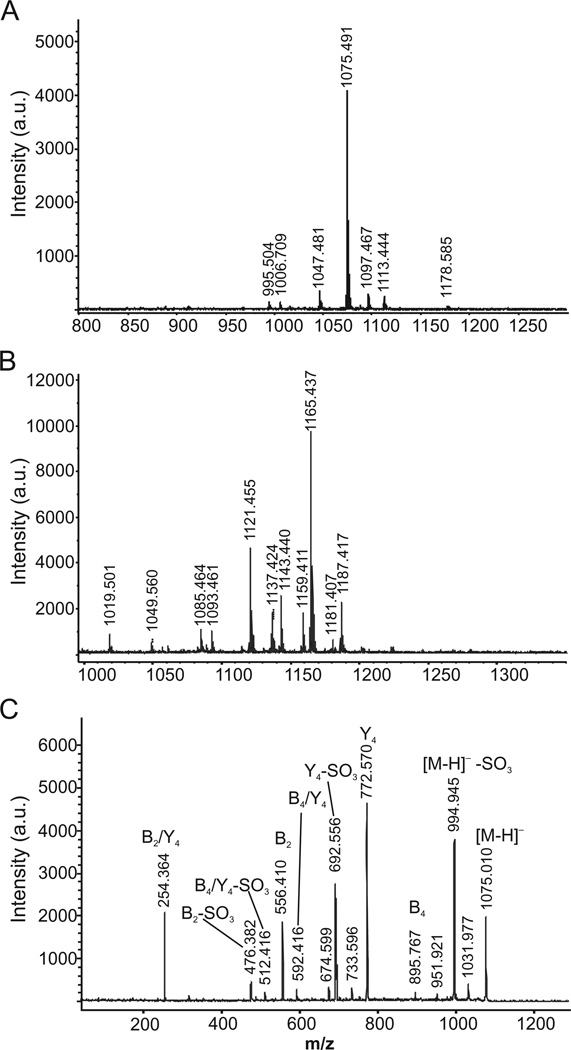
Initially, the VP4 protein was released and separated from virions by a method described in Experimental procedures. Isolated VP4 protein was subjected to non-reductive β-elimination so as to detach the oligosaccharide chains from the protein backbone. The liberated glycan pool was desalted using a graphitized carbon cartridge, which also allows for separation of neutral and acidic components. No glycans were recovered in the neutral fraction, as revealed by matrix-assisted laser desorption ionization-time of flight MS (MALDI-TOF MS) analysis (not shown). The acidic fraction, however, revealed one major glycan component in the negative ion mode at m/z 1075.24 ([M-H]−; Fig. 1A). In the positive ion mode, a complex pattern of signals was observed (Fig. 1B). These signals were assignable to cationized counterparts of the major species observed in the negative ion mode, namely m/z 1121 [M-H+2Na]+, m/z 1143 [M-2H+3Na]+, m/z 1165 [M-3H+4Na]+ and m/z 1187 [M-4H+5Na]+. In addition, potassium adducts (+16 Da) were observed for many of the signals. These data imply that the major glycan observed carries four acidic groups (typically carboxyl, sulphate and/or phosphate groups).
Fig. 1.
Mass spectra of VP4 protein-derived glycans.
A. Glycans detected in negative ion mode.
B. Glycans detected in positive ion mode.
C. MS/MS profile of the major species (m/z 1075) detected in the negative ion mode. The fragments are denoted according to the nomenclature of Domon and Costello (1988).
The major glycan species (m/z 1075.24) was subjected to tandem MS (MS/MS) in the negative ion mode to gain insight into the structural features of the VP4-derived oligosaccharide (Fig. 1C). The major fragments observed were 80 Da and 302 Da lighter than this major glycan species, implying the presence of a sulphate/phosphate group and a more complex monosaccharide unit respectively. By comparing these data to the MS/MS profile of the NaBH4-reduced counterpart, i.e. following treatment that causes the reducing end fragments to shift +2 Da (not shown), it was concluded that the m/z 254.4, 476.4 and 556.4 fragments were all derived from the non-reducing terminus of the glycan. Losses of the 80 Da and 302 Da from the m/z 556.4 fragment were also observed, implying that the terminal structure carries a sulphated/phosphated hexuronic acid unit linked to a 302 Da component. As the major loss from the mother m/z 1075 ion was 302 Da, it was concluded that the 302 Da component is the glycan terminal unit. The MS/MS data also point to the reducing end residue being a hexose, as shown by the 180 Da reduction of the mother ion to m/z 895.8. By calculating the masses of the identified components described above, the remaining unidentified units may correspond to a hexose and a hexuronic acid. Hence, MS showed the VP4-derived glycan to correspond to a pentasaccharide comprising (a 302 Da subunit)-(a sulphated/phosphated hexuronic acid)-(hexuronic acid + hexose)-hexose.
Nuclear magnetic resonance (NMR) analysis reveals the presence of 5-N-formyl-legionaminic acid in the VP4-derived glycan
A set of two-dimensional (2D) homo- and heteronuclear spectroscopic approaches [double quantum filtered correlation spectroscopy (DQF-COSY), total correlation spectroscopy (TOCSY), nuclear Overhauser effect spectroscopy (NOESY), heteronuclear single quantum correlation spectroscopy (HSQC) and heteronuclear multiple bond correlation spectroscopy (HMBC)] were employed for the assignment of 1H and 13C resonances of the VP4-derived glycan. Coupling constants were measured from one-dimensional (1D) 1H, DQF-COSY and TOCSY spectra. Resonance assignments are listed in Table 1. As some heterogeneity was present in the sample, only structural characterization of the major glycoform is considered here.
Table 1.
1H and 13C chemical shifts for the VP4-derived glycan.
| 1H chemical shifts | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Residue | H1 | H2 | H3 | H4 | H5 | H6 | H7 | H8 | H9 | CH3CON | NFo |
| A | – | – | 1.758ax 2.799eq |
3.641 | 3.709 | 3.897 | 3.826 | 4.040 | 1.169 | 2.115 | 8.111 |
| B | 4.710 | 4.132 | 3.764 | 4.187 | 3.930 | – | – | – | – | – | – |
| C | 5.491 | 4.164 | 3.825 | 3.823 | 3.661 | 3.79a | – | – | – | – | – |
| D | 4.548 | 3.427 | 3.696 | 3.825 | 3.931 | – | – | – | – | – | – |
| E | 4.986 | 3.439 | 3.711 | 3.649 | 3.666 | 3.827/3.934 | – | – | – | – | – |
| 13C chemical shifts | |||||||||||
| Residue | C1 | C2 | C3 | C4 | C5 | C6 | C7 | C8 | C9 | CH3CON/CH3CON | CFo |
| A | n.d. | 101.4 | 42.0 | 70.1 | 52.5 | 73.7 | 55.7 | 68.4 | 20.4 | 24.2/175.6 | 166.0 |
| B | 101.0 | 81.5 | 75.4 | 75.1 | 77.5 | 175.8 | – | – | – | – | – |
| C | 99.7 | 79.7 | 71.4 | 68.1 | 75.0 | 62.6 | – | – | – | – | – |
| D | 104.1 | 75.0 | 77.8 | 78.3 | 77.1 | 176.0 | – | – | – | – | – |
| E | 80.9 | 73.3 | 76.7 | 79.8 | 78.2 | 61.6 | – | – | – | – | – |
Overlapping signals centred on 3.79 ppm.
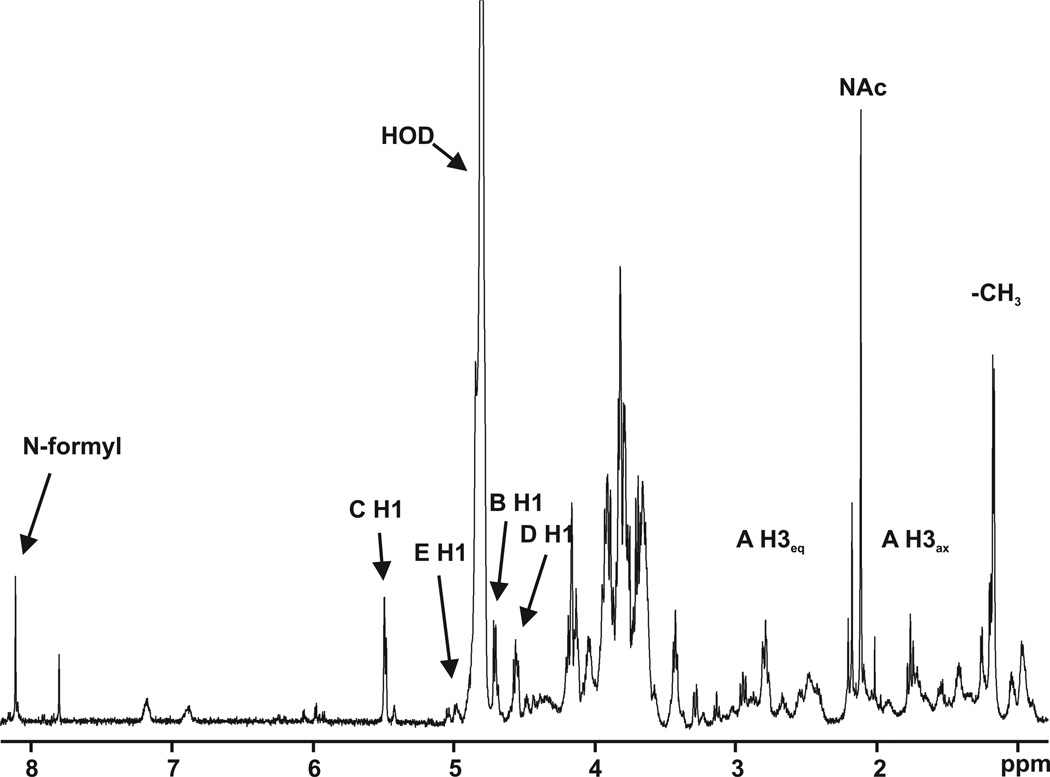
Inspection of the 1D 1H spectrum (Fig. 2) shows a low field singlet at 8.1 ppm. Four signals in the anomeric region at 5.49, 4.99, 4.70 and 4.55 ppm are also detected. Based on the three-bond homonuclear (JH,H)coupling constants, we propose that the first signal corresponds to the α anomeric configuration (monosaccharide C), while the other three signals reflect the β configuration, with a coupling constant of approximately 7.5 Hz (monosaccharides E, B and D respectively). The two signals at 2.796 and 1.757 ppm suggest the presence of a sialic acid-like monosaccharide. A singlet with an intensity corresponding to three protons at 2.045 ppm is typical for saccharides containing an N-acetyl group. In addition, a doublet in the methyl region of the spectrum is visible at 1.167 ppm.
Fig. 2.
The proton spectrum of the HRPV-1 VP4-derived glycan. The data were obtained using an 800 MHz spectrometer.
The protons resonating at 2.796 and 1.757 ppm are methylene protons, as expected for a sialic acid-like sugar and are respectively assigned as the equatorial and axial H3 protons of monosaccharide A. Monosaccharide A is, however, not a sialic acid. Carbon C7 of monosaccharideA resonates at 55.71 ppm, a value typical of aliphatic ring carbons bound to nitrogen. There is a correlation in the HMBC spectrum between proton H7 and a carbonyl carbon, which, in turn, correlates with the N-acetyl protons at 2.045 ppm, indicating that monosaccharide A bears an N-acetyl group on carbon C7. In addition, carbon C9 is a methyl carbon (20.4 ppm), suggesting that monosaccharide A could be either pseudaminic acid or legionaminic acid (5,7-diacetamido-3,5,7,9-tetradeoxy-D-glycero-D-galacto-nonulosonic acid). The 1D projection of the TOCSYspectrum emerging from H3ax is shown in Fig. 3A. The H5 signal appears as a triplet with J4,5 and J5,6 coupling constants of approximately 10 Hz, demonstrating that H4, H5 and H6 are axial protons. Monosaccharide A has the same absolute configuration as does legionaminic acid because H5 in pseudaminic acid is equatorial and its J4,5 = 4 Hz and J5,6 = 2 Hz (Knirel et al., 1987). The possibility that monosaccharide A is 4-epilegionaminic acid can be ruled out, as this sugar contains an equatorial H5. Likewise, the J7,8 coupling constant of 8-epilegionaminic acid is 6.4 Hz (Knirel et al., 2009), making it unlikely that this species is monosaccharide A. Instead, monosaccharide A likely corresponds to legionaminic acid, with a J7,8 coupling constant of 9.5 Hz (Tsvetkov et al., 2001). C5 of monosaccharide A is also bound to nitrogen, according to its chemical shift of 52.5 ppm. Correlation exists in the HMBCspectrum between C5 and the low field proton at 8.1 ppm, a value typical of N-formyl protons. In addition, H5 correlates with the formyl carbonyl carbon at 166.0 ppm. This demonstrates that C5 of the legionaminic acid (monosaccharide A) carries an N-formyl group. The chemical shift difference between H3ax and H3eq in sialic acid-like sugars can be used to determine the anomeric configuration (Tsvetkov et al., 2001). The large observed difference of 1.04 ppm corresponds to an axial carboxylic acid group at C2, indicating that the 5-formyl-legionaminic acid at position A of the VP4-derived glycan is found in the a-conformation (5FmLegα).
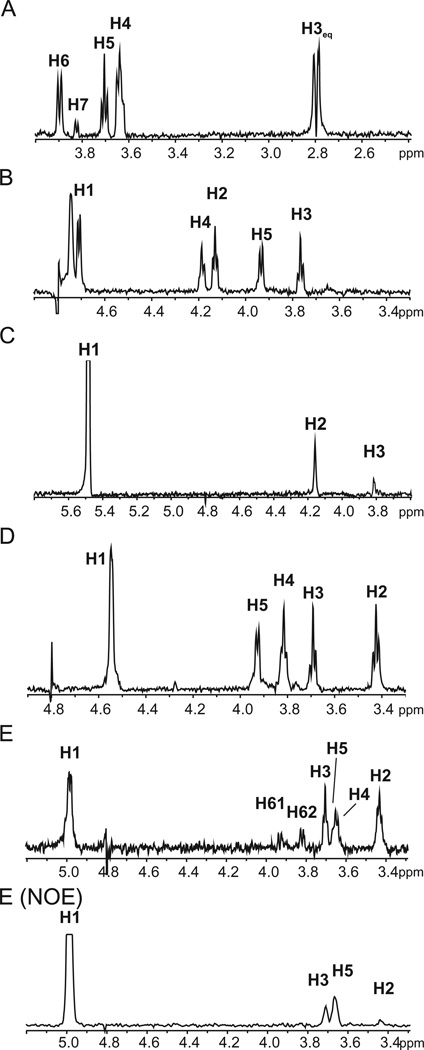
Fig. 3.
1D projections from the TOCSY spectrum for spin systems of monosaccharides A to E (A–E) and 1D projection from the NOESY spectrum emerging from monosaccharide E H1 [E (NOE)]. See text for details.
As seen from the 1D projections of the TOCSY spectra (Fig. 3B and D), the spin systems of monosaccharides B and D reveal species that each contain five protons, while the coupling constants indicate that these monosaccharides are found in the glucose configuration. In addition, correlation in the HMBC spectrum between H5 and a carbon with a chemical shift typical of a carboxylic acid carbon is seen for both monosaccharides. It is thus concluded that these two monosaccharides are β-glucuronic acids. Interestingly, the carbon C2 and proton H2 of the glucuronic acid at position B resonate at a particularly low field, while the HMBC spectrum shows that C2 is not involved in a glycosidic linkage. As sulphation moves chemical shifts towards the low field (Kogelberg and Rutherford, 1994), together with the MS analysis indicating that the glycan is sulphated, it can be concluded that carbon C2 on the glucuronic acid at position B of the VP4-derived glycan is sulphated.
Monosaccharide C was designated as α-mannopyranoside. In mannose, the vicinal coupling constants, J1,2 and J2,3, are small. Consistently, only the signals of H2 and H3 were detected in the 1D projection of TOCSY emerging from H1 of C1 (Fig. 3C), even with a mixing time of 140 ms. The anomericity of a mannose residue can be determined from the 1JC,H value for C1 (Kärcher et al., 1993). With a 1JC,H = 172 Hz obtained from a HSQC spectrum without proton decoupling, monosaccharide C apparently assumes the α-configuration.
Glycan release by non-reductive β-elimination yields a glycan with a glycosylamine at the reducing end, which is consistent with the 80.9 ppm 13C chemical shift of monosaccharide E C1, showing that it is bound to nitrogen. This shift is considerably lower than the typical value for anomeric carbons bound to oxygen of around 100 ppm. Signals up to H6s are visible in the 1D projection of TOCSY of the spin system of monosaccharide E, suggesting that this sugar is found in the β-gluco-configuration (Fig. 3E). In addition, strong nuclear Overhauser effects (NOEs) were observed between EH1, EH3 and EH5, confirming that monosaccharide E is β-glucosylamine (Fig. 1E – NOE).
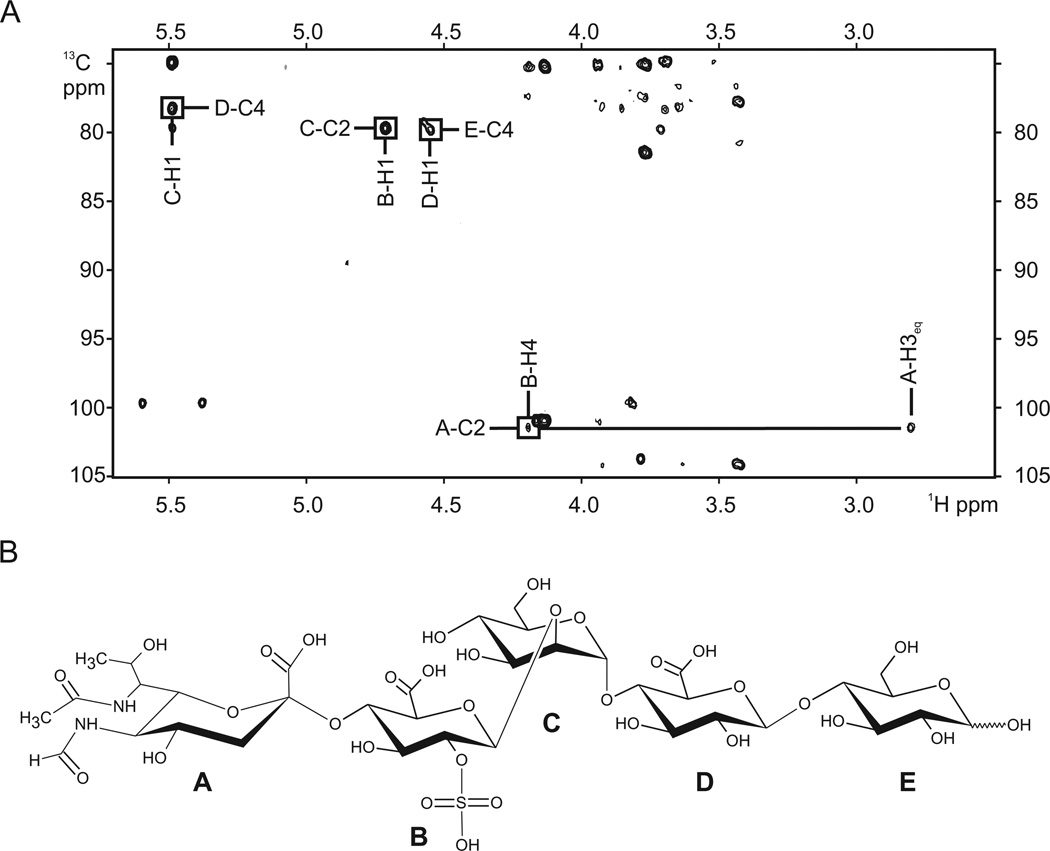
The positions of the glycosidic linkages were determined from the HMBC spectrum (Fig. 4A). HMBC-based correlations between the monosaccharide A quaternary carbon C2 (not detectable in the HSQC spectrum) and monosaccharide B H4, monosaccharide B H1 and monosaccharide C C2, monosaccharide C H1 and monosaccharide D C4, monosaccharide D H1 and monosaccharide E C4 are observed, indicating that the structure of glycan is 5FmLegaα-(2-4)-GlcA(2SO3)β-(1-2)-Manα-(1-4)-GlcAβ-(1-4)–Glc1Nβ.
Fig. 4.
Characterization of the HRPV-1 VP4-derived glycan using MALDI-TOF/MS and NMR.
A. 13C, 1H HMBC spectrum of HRPV-1 VP4-derived glycan.
B. The structure of the major glycan species of HRPV-1 VP4, as determined by MALDI-TOF/MS and NMR. The identity of each monosaccharide (A–E) is provided in the text.
VP4 isolated from the HRPV-1 virus is N-glycosylated
To determine whether VP4 released from the virus produced in Hrr. sp. strain PV6 is N-glycosylated, trypsin-generated fragments of the protein were examined by liquid chromatography-electrospray ionizationMS(LC-ESI MS). Such analysis revealed a peak of m/z 945.44 (Fig. 5A), corresponding to the [M+2H]2+ ion of the VP4-derived peptide, 171TNSPDYSLAYSNTTEK187 (calculated mass, m/z 945.43), containing the putative N-glycosylation site, Asn-182 (Pietilä et al., 2009). Peaks of m/z 1026.47, 1148.47, 1195.51 and 1323.50 were also detected, consistent with calculated masses of the Asn-182-containing peptide modified by a hexose (m/z 1026.43), a hexose and a hexuronic acid (m/z 1114.43), a hexose, a hexuronic acid and a hexose (m/z 1195.43) and a hexose, a hexuronic acid, a hexose and a sulphated hexuronic acid (m/z 1323.43) (Fig. 5B–E respectively). Similarly, a m/z 1381.15 peak, corresponding to the [M+2H]2+ ion of the Asn-427-containing VP4 tryptic peptide (425TANTTELLEVQNQLIELR442; calculated mass, m/z 1043.06) modified by the same tetrasaccharide lacking the sulphate group on the final hexuronic acid (calculated mass, m/z 1381.06), was also detected, as were peaks corresponding to the precursor tri-, di- and monosaccharide-modified peptide m/z 1293.13 (calculated mass, m/z 1293.06), a m/z 1212.11 (calculated mass, m/z 1212.06) and a m/z 1124.09 (calculated mass, m/z 1124.06) respectively (not shown).
Fig. 5.
VP4 Asn-182 is glycosylated. LC-ESI/MS analysis of the Asn-182-containing tryptic peptide derived from VP4 from HRPV1-infected Hrr. sp. strain PV6 cells was performed. Shown are doubly charged [M+2H]2+ ion peaks corresponding to (A) the 171TNSPDYSLAYSNTTEK187 peptide (m/z 945.44), and the same peptide successively modified by (B) a hexose (m/z 1026.47), (C) a hexuronic acid (m/z 1148.47), (D) a hexose (m/z 1195.51) and (E) a sulphated/phosphorylated hexuronic acid (m/z 1323.50). In each panel, the inset shows the N-glycosylation status of the peptide, where ‘N’ corresponds to Asn-182. Full circles correspond to hexose residues, while the full squares correspond to hexuronic acid residues.
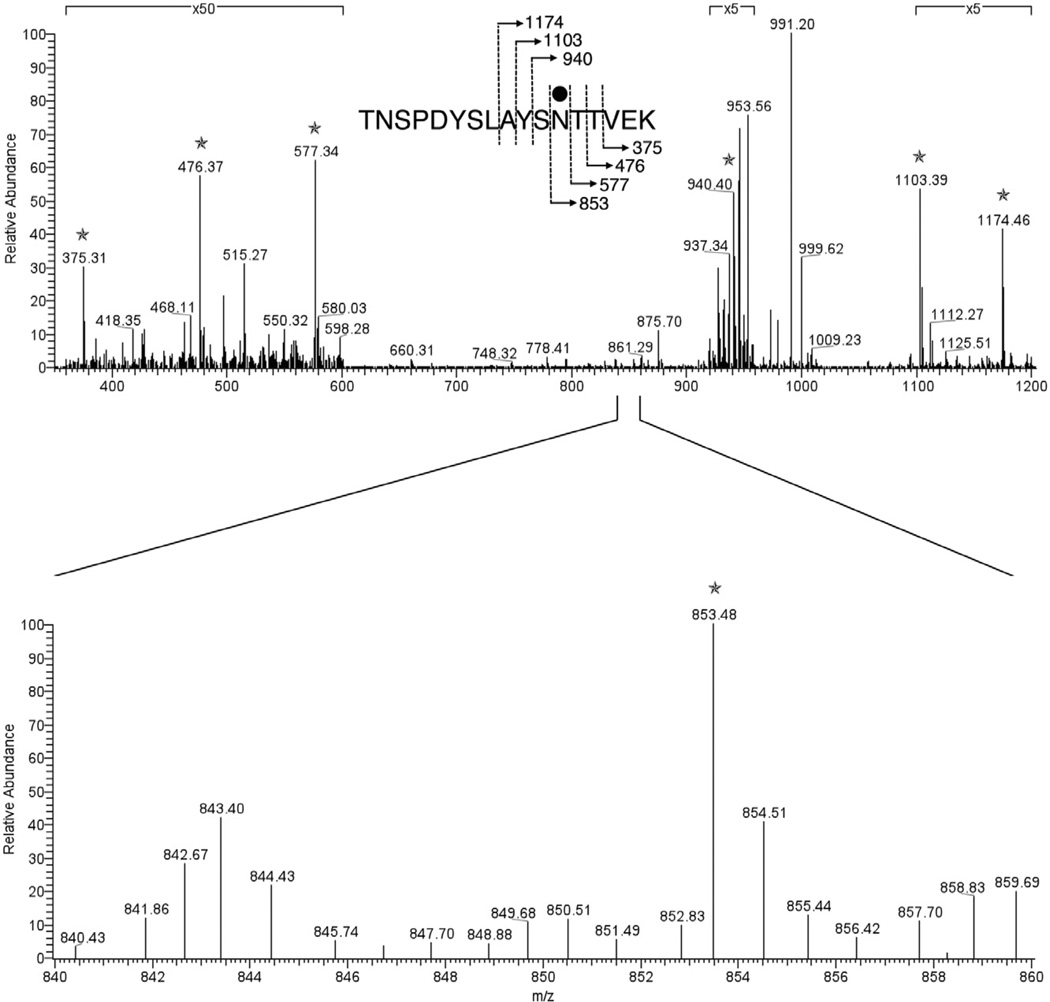
Glycosylation of Asn-182 was verified by MS/MS analysis of the [M+2H] 2+ base peak of the hexose-modified VP4-derived peptide, 171TNSPDYSLAYSNTTEK187, observed at m/z 1026.97. As shown in Fig. 6, the product ion spectrum contains a series of y-ion fragments that allowed unambiguous determination of the hexose modification at the Asn-182 residue.
Fig. 6.
MS/MS verification of Asn-182 glycosylation. To localize the glycosylation of the hexose-modified VP4-derived peptide, 171TNSPDYSLAYSNTTEK187, the MS/MS spectrum of the [M+2H]2+ base peak of this peptide, observed at m/z 1026.97, was acquired through mass-dependent acquisition by the LTQ Orbitrap XL mass spectrometer. The C-terminus-containing y-ion fragments, indicated by stars, allowed the localization of the hexose modification at the Asn-182 residue. The inset in the top panel shows the fragmentation scheme, with the bound hexose being represented by the full circle. Those regions of the spectrum magnified ×5 or ×50 are indicated. The lower panel shows an expansion of the region between m/z 840 and 860.
N-glycosylation of VP4 expressed in Hfx. volcanii
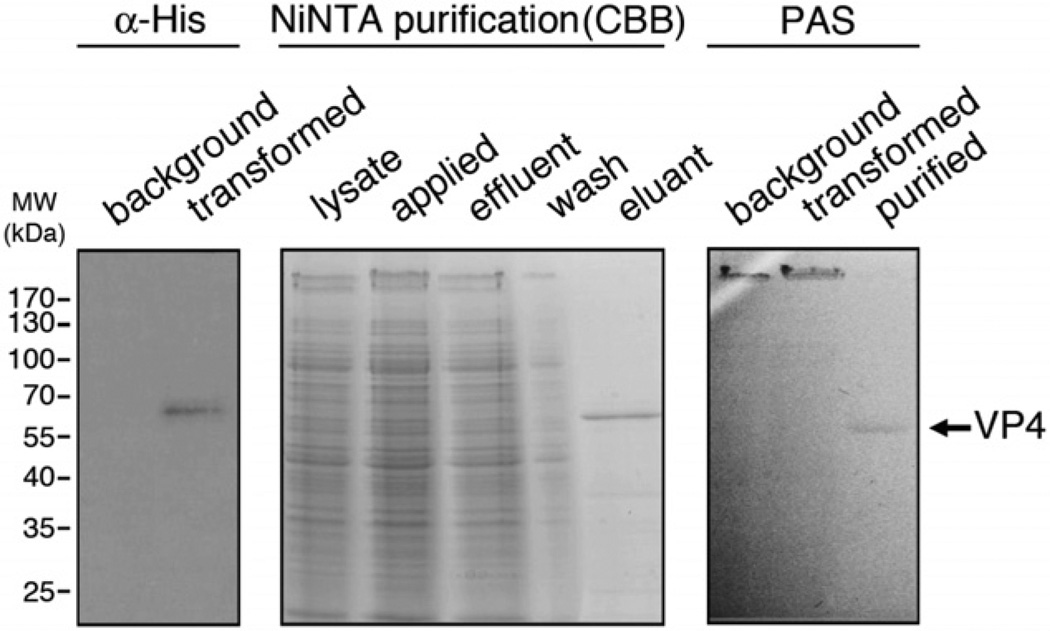
In recent years, substantial progress has been made in delineating the N-glycosylation pathway of the haloarchaeon, Hfx. volcanii (cf. Calo et al., 2010a). To determine whether a non-native protein, shown to be N-glycosylated in the native host, also undergoes this post-translational modification when expressed in Hfx. volcanii, a polyhistidine-tagged version of VP4 was introduced into Hfx. volcanii cells. Successful expression of VP4 in the foreign host was verified by immunoblot using anti-His antibodies (Fig. 7, left panel). The His-tagged protein was subsequently purified from the transformed Hfx. volcanii cells on NiNTA resin (Fig. 7, middle panel). Finally, the glycosylation of VP4 expressed in Hfx. volcanii was verified by glycostaining using periodic acid/Schiff’s reagent (PAS) (Fig. 7, right panel).
Fig. 7.
Purification of VP4 expressed in Hfx. volcanii. (Left) Immunoblotting with anti-histidine antibodies (α-His) identifies a polyhistidine-tagged version of VP4 in transformed Hfx. volcanii cells. (Middle) Polyhistidine-tagged VP4 was purified on NiNTA resin. Coomassie Brilliant Blue (CBB)-stained samples from the various chromatographic steps are shown. (Right) PAS staining of background and transformed cells, as well as NiNTA-treated VP4, is shown. In the lanes containing samples of the background and transformed cells, the S-layer glycoprotein (> 170 kDa) is stained.
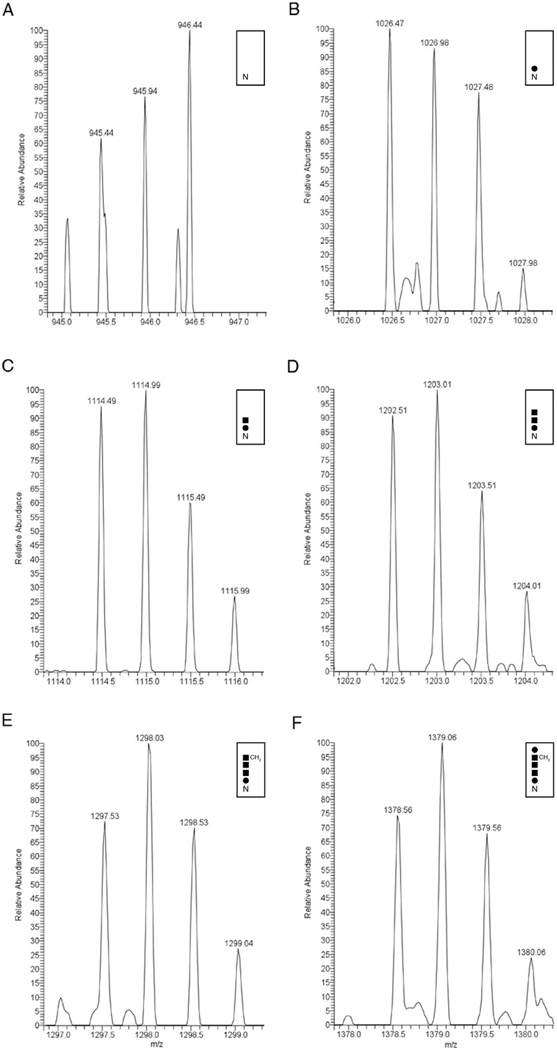
VP4 purified from transformed Hfx. volcanii cells was next subjected to LC-ESI MS analysis. A peak corresponding to the [M+2H]2+ ion of the VP4-derived peptide, 171TNSPDYSLAYSNTTEK187 (m/z 954.44; calculated mass, m/z 945.43), containing the putative N-glycosylation site, Asn-182 (Pietilä et al., 2009), shown to undergo modification in Hrr. sp. strain PV6 (see VP4 isolated from the HRPV-1 virus is N-glycosylated above), was observed (Fig. 8A). In addition, m/z 1026.47, 1114.49, 1202.51, 1297.53 and 1378.56 peaks were also detected, corresponding to the masses of the Asn-182-containing peptide modified by a hexose (calculated mass, m/z 1026.43), a hexose and a hexuronic acid (calculated mass, m/z 1114.43), a hexose and two hexuronic acids (calculated mass, m/z 1202.43), a hexose, two hexuronic acids and a methyl ester of hexuronic acid (calculated mass, m/z 1297.43) and a hexose, two hexuronic acids, a methyl ester of hexuronic acid and a hexose (calculated mass, m/z 1378.43) respectively (Fig. 8B–F respectively). At the same time, a m/z 1476.18 peak, corresponding to the [M+2H]2+ ion of the Asn-427-containing VP4-derived tryptic peptide that is N-glycosylated in Hrr. sp. strain PV6 (425TANTTELLEVQNQLIELR442; m/z 1043.07; calculated mass, m/z 1043.06), now modified by the same Hfx. volcanii pentasaccharide (calculated mass, m/z 1476.06) as N-linked to Asn-182, was also detected (not shown). Likewise, peaks corresponding to the precursor tetra-, tri-, diand monosaccharide-modified peptide at m/z 1395.15 (calculated mass, m/z 1395.06), at m/z 1300.13 (calculated mass, m/z 1300.06), at m/z 1212.11 (calculated mass, m/z 1212.06) and at m/z 1124.09 (calculated mass, m/z 1124.06), respectively, were also observed (not shown).
Fig. 8.
VP4 expressed in Hfx. volcanii is N-glycosylated by the same pentasaccharide N-linked to the S-layer glycoprotein. LC-ESI/MS analysis of the Asn-182-containing tryptic peptide derived from VP4 expressed in Hfx. volcanii cells was performed. Shown are doubly charged [M+2H]2+ ion peaks corresponding to (A) the 171TNSPDYSLAYSNTTEK187 peptide (m/z 945.44), and the same peptide successively modified by (B) a hexose (m/z 1026.47), (C) a hexuronic acid (m/z 1114.49), (D) a second hexuronic acid (m/z 1202.51), (E) a methyl ester of hexuronic acid (m/z 1297.53) and (F) a hexose (m/z 1378.56). In each panel, the inset shows the N-glycosylation status of the peptide, where ‘N’ corresponds to Asn-182. Full circles correspond to hexose residues, while the full squares correspond to hexuronic acid residues.
As such, VP4 expressed in Hfx. volcanii is modified by the same pentasaccharide as is N-linked to the S-layer glycoprotein, a native reporter of N-glycosylation in this species (Sumper et al., 1990; Abu-Qarn et al., 2007; Magidovich et al., 2010).
Inhibition of HRPV-1 infection by N-acetylneuraminic acid
Protruding from the HRPV-1 membrane, the VP4 spike protein is proposed to participate in viral host recognition (Pietilä et al., 2009). Given that molecular recognition events are often mediated by sugar-based interactions, it is possible that 5FmLeg, the terminal monosaccharide of the major VP4-derived glycan, plays a role in the initial recognition of the host cell by HRPV-1. This hypothesis was tested in an infection inhibition assay using N-acetylneuraminic acid (NeuAc), a monosaccharide closely resembling 5FmLeg.
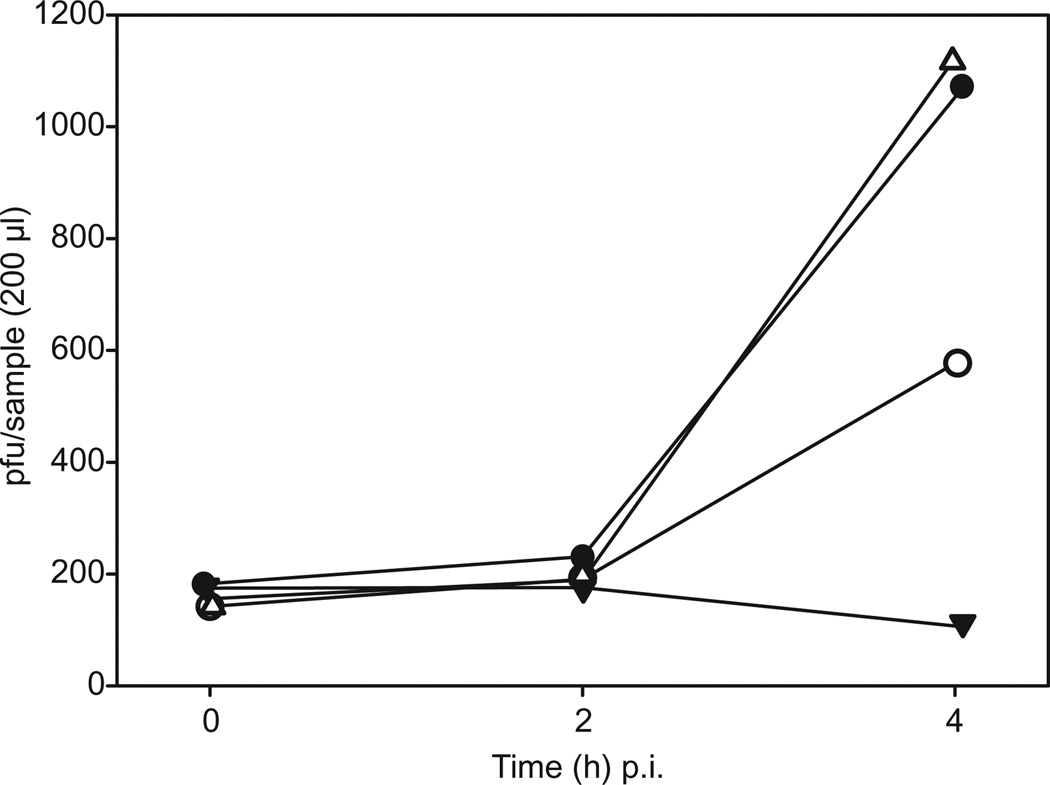
The adsorption rate of HRPV-1 is too low to be detected reliably (Pietilä et al., 2009). Inhibition of adsorption, however, can be indirectly shown by the inhibition of virus production by newly infected cells approximately 2–4 h post infection (p.i.). As shown in Fig. 9, the amount of infectious HRPV-1 particles in the control sample increased more than fivefold 4 h p.i., whereas in the sample containing NeuAc, the amount of new progeny viruses was approximately one half of that in the control sample. When the same infection inhibition assay was performed using glucuronic acid at the same concentration as NeuAc, progeny virus production was not prevented. The amount of infectious viruses incubated under the same conditions but in the absence of host cells remained relatively stable, only decreasing slightly towards the end of the experiment.
Fig. 9.
Inhibition of HRPV-1 infection by NeuAc. Hrr. sp. PV6 cells (1 ml) in logarithmic growth phase were infected with approximately 1000 infectious virus particles. Samples of 200 µl were taken 0, 2 and 4 h p.i. The amounts of infectious viral particles are shown for the control sample (Hrr. sp. PV6 and HRPV-1; filled circles), the sample containing N-acetylneuraminic acid (Hrr. sp. PV6, 2.5 mg ml−1 NeuAc and HRPV-1; open circles), the sample containing glucuronic acid (Hrr. sp. PV6, 2.5 mg ml−1 GlcA and HRPV-1; open triangles) and the virus control (2.5 mg ml−1 NeuAc and HRPV-1; filled triangles).
Discussion
Glycosylation is the most abundant post-translational modification that proteins undergo (Larkin and Imperiali, 2011). Despite a relatively limited degree of investigation, archaeal protein N-glycosylation has been shown to occur more frequently than in Bacteria, to rely on biosynthetic steps similar to what has been seen in Eukarya, and to present more structural diversity than seen in cells belonging to either of these domains (Calo et al., 2010a). Still, detailed data on the archaeal N-glycosylation process and the generated glycan structures have been obtained from only a few model organisms, including the halophiles Hfx. volcanii and Hbt. salinarum.
In the present study, we have added to this body of knowledge by characterizing the main glycan structure of the viral protein VP4 of the virus HRPV-1 that infects Hrr. sp. PV6. HRPV-1 is a pleomorphic virus, which in analogy to some enveloped animal viruses, is thought to assemble into the virion on specific assembly sites on the host cytoplasmic membrane and subsequently leave the host cell via budding (Pietilä et al., 2009). As part of this process, VP4 is translated as a signal sequence-bearing precursor. Following signal sequence processing by the host secretion machinery, translocated VP4, anchored to membrane via a C-terminal transmembrane domain, is exposed to the surface at viral assembly sites. It is presumably at this point that VP4 is glycosylated. Accordingly, VP4 is hypothesized to participate in the recognition of the host and in the initial stages of infection (Pietilä et al., 2009; 2010; Roine and Oksanen, 2011).
Glycans were directly released from HRPV-1 VP4 using both reductive and non-reductive β-elimination. MALDITOF MS/MS as well as NMR analyses were able to show that the major glycan decorating the viral VP4 protein is the pentasaccharide, 5FmLegα(2-4)-GlcA(2SO3)β-(1-2)-Manα-(1-4)-GlcAβ-(1-4)–Glc. Moreover, LC-ESI MS analysis of two VP4-derived tryptic peptides revealed species corresponding to the peptide bearing a N-glycan structure up to the tetrasaccharide, and in some cases lacking the sulphate group on the glucuronic acid residue at position four. Several scenarios can explain why only a truncated peptide-bound glycan was detected by LC-ESI MS. Tryptic peptides containing the complete glycan may be difficult to detect because of the highly negatively charged nature of several component sugars. Alternatively, 5-N-formyl-legionaminic acid and the sulphate group on glucuronic acid may have been lost during generation, work-up and analysis of the tryptic peptides. It is also conceivable that the presence of these moieties interfered with effective tryptic digestion of the protein. When, however, VP4 was heterologously expressed in Hfx. volcanii and the same tryptic peptides as addressed above were considered, they were shown to be decorated by a pentasaccharide previously shown to modify select Asn residues of the Hfx. volcanii S-layer glycoprotein (Abu-Qarn et al., 2007; Magidovich et al., 2010). The distinct modification of VP4 in the different species shows that in Archaea, the composition of N-linked glycans is host-specific. Moreover, the finding that VP4 expressed in Hfx. volcanii undergoes N-glycosylation is important for efforts aimed at exploiting this species as a platform for glyco-engineering efforts. Previous studies have shown that components of the native N-glycosylation pathway can be replaced by homologues from other haloarchaea, leading to the appearance of novel N-glycans (Calo et al., 2010b; 2011b). The ability of Hfx. volcanii to N-glycosylate heterologous proteins offers additional support for such endeavours.
The detection of 5FmLeg in VP4 represents the first report of this monosaccharide in a glycan modifying an archaeal-derived protein. Legionaminic acid is a sialic acid-like sugar. Sialic acids are a group of nine-carbon sugars widely found in animal cells, where they serve a variety of important functions, including presumed roles in cell differentiation, cell adhesion and disease, and are utilized by many bacterial and eukaryotic pathogens for host cell recognition (Angata and Varki, 2002). Initially identified as a component of Legionella pneumophila serogroup 1 lipopolysaccharide (Knirel et al., 1994), the causative agent of Legionnaires’ disease (Cianciotto, 2001), legionaminic acid and its derivatives were subsequently also found to be O-linked to Campylobacter coli flagellin (McNally et al., 2007). To our knowledge, legionaminic acid had not been reported as a component of a N-linked glycan prior to the present report. The detection of the sialic acid-like sugar, 5FmLeg, as the terminal saccharide unit of the N-glycan decorating VP4 is in agreement with earlier observations showing that sialic acids are typically located at the terminal positions of glycoconjugates (Lehmann et al., 2006).
The abundance of negative charges in the VP4-bound glycan synthesized in the natural host, provided by the highly acidic terminal 5-N-formyl-legionaminic acid subunit, along with hexuronic acid and a sulphated version of the same sugar, likely facilitates the survival of Hrr. sp. PV6 in the hypersaline environment in which it exists. An excess of negative surface charge is thought to stabilize proteins in such surroundings, possibly via the formation of an energetically favourable protein–water–salt hydration network, as proposed to explain the highly acidic nature of the N-glycans decorating the Hbt. salinarum S-layer glycoprotein (Mengele and Sumper, 1992). In addition, the glycosylation of haloarchaeal proteins may influence the relative abundance of negatively charged amino acids in the polypeptide. In HRPV-1 VP4, the relative amount of aspartic and glutamic acid residues is lower than in VP4 of Haloarcula hispanica virus 1 (HHPV-1; Roine et al., 2010), a non-glycosylated homologue. Future research will address the interplay between the acidic amino acid content and acidic glycan modification of halophilic proteins.
As noted, the detection of 5FmLeg in VP4 represents the first report of this saccharide in a glycan structure of an archaeal-derived protein and confirms that sialic acid-like protein modification occurs in cells belonging to all three domains of life. This suggests that the genes involved in the glycan modifications, in general, and in legionaminic acid synthesis, in particular, may have been horizontally transferred between the organisms belonging to the three domains of life. It has also been proposed that, at least between the eukaryotes and prokaryotes, sialic acid synthesis and modification genes arose as a result of convergent evolution rather than horizontal gene transfer (Varki et al., 2009). Presently, testing the validity of these hypotheses is limited not only by the scarce characterization of these genes in different organisms but also by the lack of functional analysis approaches. Indeed, no genes involved in Hrr. sp. PV6-mediated N-glycosylation of VP4 have been described. Earlier studies, however, identified aglB, encoding the archaeal oligosaccharide transferase, in Halorubrum lacusprofundi, in addition to two predicted glycosyltransferases found in close proximity in the genome (Magidovich and Eichler, 2009).
Protein-bound glycans serve numerous roles in a variety of biological processes that require specific recognition, including receptor binding by N-glycosylated viral proteins (Vigerust and Shepherd, 2007). As such, glycosylation of HRPV-1 VP4 by a highly acidic pentasaccharide would not only serve to stabilize the protein in the high-salinity environment in which the host lives but could also serve a major role in the receptor binding and infectivity of this virus. Our finding that virus infection could be partially inhibited by NeuAc, a monosaccharide closely related to 5FmLeg, the terminal monosaccharide of the N-linked pentasaccharide N-linked to VP4, supports this hypothesis. Moreover, based on these results, identification of the host cell receptor in closer detail and description of the recognition process in an environment characterized by high salinity is possible.
Experimental procedures
Strains and growth conditions
Halorubrum sp. PV6 was grown at 37°C in modified growth medium (MGM), prepared as described (Nuttall and Dyall-Smith, 1993). A 30% stock of medium prepared with 240 g of NaCl, 30 g of MgCl2·6H2O, 35 g of MgSO4·7H2O, 7 g of KCl, 5 ml of 1 M CaCl2·2H2O and 80 ml of 1 M Tris-HCl, pH 7.2, per litre of water was diluted to 23% (broth), 20% (solid) and 18% (top-layer agar) and 5 g of peptone (Oxoid) and 1 g of Bacto yeast extract (Difco) were added per litre. Solid and top-layer agar MGM contained 14 g and 4 g of Bacto agar (Difco) per litre respectively. Hfx. volcanii was grown in medium containing 3.4 M NaCl, 0.15 M MgSO4·7H2O, 1 mM MnCl2, 4 mM KCl, 3 mM CaCl2, 0.3% (w/v) yeast extract, 0.5% (w/v) tryptone, 50 Mm Tris-HCl, pH 7.2, at 42°C (Mevarech and Werczberger, 1985). The HRPV-1 stock solution was prepared as described inPietilä et al. (2009). Escherichia coli BL21 (Promega) was used for sub-cloning.
Purification of HRPV-1 VP4 protein
The HRPV-1 VP4 protein was released from ‘1×’ purified viral material at low ionic strength at 60°C, as previously described (Pietilä et al., 2010). Briefly, purified viral material in HRPV-1 buffer (20 mM Tris-HCl, pH 7.5, 1.5 M NaCl, 100 mM MgCl2, 2 mM CaCl2) was suspended at a concentration of 95 µg protein ml−1 in 20 mM Tris-HCl, pH 7.5 and the final NaCl concentration was adjusted to 50 mM. The viral preparation was incubated at 60°C for 1 h. Dissociation products were separated by rate zonal centrifugation (Sorvall AH629, 81 400 g, 7 h 10 min, 15°C) in a linear 5–20% sucrose gradient (in 20 mM Tris-HCl, pH 7.5, 50 mM NaCl). Soluble VP4 was collected from the top of the gradient, concentrated using Amicon Ultra 15 concentrators (MWCO 10 000) and washed with 20 mM Tris-HCl, pH 7.5, 50 mM NaCl.
Reductive β-elimination
Purified and concentrated VP4 protein preparation was dried under vacuum and resuspended in 1 M NaBH4 in 0.1 M NaOH and incubated approximately 42 h at 37°C. The reaction was terminated by addition of 2 M acetic acid until the pH dropped between 4 and 5. The released glycans were purified from protein remnants by solid-phase extraction on C-18 silica and then isolated by graphitized carbon chromatography, essentially as described (Packer et al., 1998). In brief, the neutralized β-elimination reaction was passed through a 150 mg column of graphitized carbon (Alltech), washed with water and eluted with 25% aqueous acetonitrile (neutral fraction) and with 25% acetonitrile in aqueous 0.1% trifluoroacetic acid (acidic fraction). The eluates were dried prior to MS and NMR analysis.
Non-reductive β-elimination
Purified and concentrated VP4 protein was dried under vacuum and dissolved in saturated ammonium carbonate in 25% ammonia (Huang et al., 2001). The reaction was allowed to proceed for 18 h at 60°C, and then dried under reduced pressure with several additions of water to eliminate salts. The released glycans were purified by C-18 silica and graphitized carbon chromatography, essentially as described above.
MALDI-TOF MS of the VP4-derived glycan
MALDI-TOF mass spectra of the VP4-derived glycan were collected using a Bruker Ultraflex TOF/TOF mass spectrometer, as previously described (Heiskanen et al., 2009). MS/MS spectra of the major species were obtained in the negative ion mode.
NMR spectroscopy of the VP4-derived glycan
Prior to NMR analyses, VP4-derived glycan samples were once dried from 99.9% deuterium oxide (Aldrich), and then dissolved in 240 µl of 99.996% deuterium oxide (Cambridge Isotope Laboratories) and transferred to a Shigemi NMR tube. All NMR experiments were carried out at 23°C on a Varian Unity INOVA 800 MHz spectrometer equipped with a 5 mm 15N/13C/1H z-gradient triple-resonance cold probe. In recording 1D proton spectrum pre-saturation was used for water suppression. For the DQF-COSY, TOCSY and NOESY experiments, matrices of 4096 × 512 points were collected. In TOCSY, spin-lock times of 120 and 140 ms and in NOESY, a mixing time of 500 ms was used. For HSQC and HMBC, spectra matrices of 2048 × 512 points were recorded. The average one- and three-bond 1H–13C couplings were estimated to be 140 Hz and 8 Hz, and 1H–13C transfer delays for HSQC and HMBC were set to 3.57 and 62.5 ms respectively. The 1H and 13C chemical shifts were referenced to internal acetone as 2.225 ppm and 31.55 ppm respectively. The VNMRJ 2.1 software package was used for recording and processing of the spectra. Spectrum analysis was carried out using Sparky 3.110 (Goddard and Kneller).
Purification of polyhistidine-tagged VP4 from transformed Hfx. volcanii
To introduce VP4 bearing a C-terminal polyhistidine tag into Hfx. volcanii, PCR amplification of the HRPV-1 vp4 sequence (Pietilä et al., 2009) was achieved using a construct containing the desired sequence in plasmid pHRPV1-VP4.2 (E. Roine, unpublished) as template together with primers designed to introduce an NdeI site at the 5′-end and a XhoI site at the 3′-end of the fragment (forward: ccccatATGTCTGTGAATCGCTCGTC; reverse: gggctcgagGTGGTGGTGGTGGTGGTGGC; introduced restriction sites in lower case letters). The fragment was ligated into the pGEM-T Easy vector (Promega) and sequenced.
The fragment was then excised by digestion with NdeI and XhoI and introduced into the pET24b+ vector (Novagen), previously cleaved with NdeI and XhoI. Following ligation, the fragment-containing plasmids were treated with NdeI and BlpI so as to release the vp4 sequence encoding a polyhistidine tag at the 3′-end. These fragments were introduced into plasmid pJAM202 (Reuter et al., 2004), pre-cleaved with NdeI and BlpI, to generate plasmid pJAM202-VP4. The Hfx. volcanii WR536 parent strain was then transformed with plasmid pJAM202-VP4, as described previously (Cline et al., 1989), and selected in Hfx. volcanii medium supplemented with 1 µg ml−1 novobiocin.
To purify VP4, transformed Hfx. volcanii cells (100 ml) were centrifuged for 10 min at 9000 g and resuspended in 8 ml of 20 mM imidazole, 2 M NaCl, 50 mM Tris-HCl, pH 7.2. The cells were disrupted by sonication (35% output, three times for 30 s, 2 s on and 1 s off; Misonix XL2020 ultrasonicator, Farmington, NY, USA), and 1 ml of 1% Triton X-100 was added. The lysate was applied to Ni–NTA resin (Qiagen), previously equilibrated with 20 mM imidazole, 2 M NaCl, 50 mM Tris-HCl, pH 7.2. Following a 1 h shaking incubation at 4°C, unbound proteins were removed by washing with the equilibration buffer. Specifically bound protein was eluted upon addition of 500 mM imidazole, 2 M NaCl, 50 mM Tris-HCl, pH 7.2. The identity of the eluted proteins was confirmed by immunoblotting. For immunoblotting, proteins separated by 10% SDS-PAGE were transferred to nitrocellulose (0.45 µm; Whatman, Dassel, Germany). The membrane was then probed with anti-polyhistidine horseradish peroxidase-conjugated monoclonal antibodies (1:500; Sigma) in PBS containing 0.5% Tween-20 and 5% low-fat milk powder. Antibody binding was detected using an enhanced chemiluminescence kit (Amersham, Buckingham, UK). Glycostaining of VP4 by PAS was performed as previously described (Dubray and Bezard, 1982).
LC-ESI/MS analysis of VP4-derived tryptic fragments
For LC-ESI/MS analysis of VP4 released from the HRPV-1 virus or produced heterologously in Hfx. volcanii, HRPV-1 VP4 or the protein contents of Hfx. volcanii cells were separated on 7.5% polyacrylamide gels and stained with Coomassie R-250 (Fluka). For in-gel digestion of VP4, the protein band was excised, destained in 400 µl of 50% (v/v) acetonitrile (Sigma) in 40 mM NH4HCO3, pH 8.4, dehydrated with 100% acetonitrile, and dried using a SpeedVac drying apparatus. The glycoprotein was reduced with 10 mM dithiothreitol (Sigma) in 40 mM NH4HCO3 at 56°C for 60 min and then alkylated for 45 min at room temperature with 55 mM iodoacetamide in 40 mM NH4HCO3. The gel pieces were washed with 40 mM NH4HCO3 for 15 min, dehydrated with 100% acetonitrile, and SpeedVac dried. The gel slices were rehydrated with 12.5 ng µl−1 of MS-grade Trypsin Gold (Promega) in 40 mM NH4HCO3. The protease-generated peptides were extracted with 0.1% (v/v) formic acid in 20 mM NH4HCO3, followed by sonication for 20 min at room temperature, dehydration with 50% (v/v) acetonitrile, and additional sonication. After three rounds of extraction, the gel pieces were dehydrated with 100% acetonitrile, dried completely with a SpeedVac, resuspended in 5% (v/v) acetonitrile containing 1% formic acid (v/v) and infused into the mass spectrometer using static nanospray Econotips (New Objective, Woburn, MA, USA). The protein digests were separated online by nano-flow reverse-phase liquid chromatography by loading onto a 150 mm by 75 µm (internal diameter) by 365 µm (external diameter) Jupifer pre-packed fused silica 5 µm C18 300Å reverse-phase column (Thermo Fisher Scientific, Bremen, Germany). The sample was eluted into the LTQ Orbitrap XL mass spectrometer (Thermo Fisher Scientific) using a 60 min linear gradient of 0.1% formic acid (v/v) in acetonitrile/0.1% formic acid (1:19, by volume) to 0.1% formic acid in acetonitrile/0.1% formic acid (4:1, by volume) at a flow rate of 300 nl min−1.
Infection inhibition test
Halorubrum sp. PV6 cells in mid-exponential growth phase (approximately 4 × 108 cfu ml−1) were harvested (Sorvall SA-600, 10 000 g, 20 min, 15°C) and resuspended in the same volume of fresh growth medium. To all samples, sodium hydroxide was added to 10 mM to compensate for the possible drop in pH because of the presence of acidic monosaccharides. For cell samples, 1 ml of host cell suspension was infected with 100 µl of a diluted HRPV-1 stock solution that contained approximately 1000 infectious particles (multiplicity of infection 0.0000025). NeuAc or glucuronic acid was freshly dissolved in MGM and added to the appropriate samples at a concentration of 2.5 mg ml−1 prior to the addition of the virus. For the virus control, an equal amount of viruses was added to MGM broth that also contained 2.5 mg ml−1 NeuAc (8 mM). All samples were incubated at 37°C with shaking and 200 µl of aliquots was taken at 0, 2 and 4 h p.i. Cells were harvested (16 000 g, 7 min) and the amount of viruses in the supernatant was determined by plaque assay (Pietilä et al., 2009). The cell pellet was resuspended in 200 µl of fresh growth medium and the amount of viruses was determined by plaque assay. The results shown in Fig. 9 represent one experiment of three experiments all giving similar results.
Acknowledgements
Sari Korhonen is acknowledged for excellent technical assistance. This work was supported by Helsinki University three year grant 2010–2012 (E. R.) and Academy Professor research funding (Academy of Finland grants 255342 and 256518 to D. H. B.). We thank University of Helsinki for the support to EU ESFRI Instruct Associate Centre for Virus Production and Purification used in this study. The MS facility in the Department of Biochemistry of the Duke University Medical Center and Z. G. are supported by the LIPID MAPS Large Scale Collaborative Grant number GM-069338 from NIH. P. P. was supported by Academy of Finland (131144). J. E. is supported by the Israel Science Foundation (grant 8/11) and the US Army Research Office (W911NF-11-1-520).
Footnotes
The authors have no conflict of interest.
References
- Abu-Qarn M, Yurist-Doutsch S, Giordano A, Trauner A, Morris HR, Hitchen P, et al. Haloferax volcanii AglB and AglD are involved in N-glycosylation of the S-layer glycoprotein and proper assembly of the surface layer. J Mol Biol. 2007;14:1224–1236. doi: 10.1016/j.jmb.2007.10.042. [DOI] [PubMed] [Google Scholar]
- Abu-Qarn M, Eichler J, Sharon N. Not just for Eukarya anymore: N-glycosylation in Bacteria and Archaea. Curr Opin Struct Biol. 2008;18:544–550. doi: 10.1016/j.sbi.2008.06.010. [DOI] [PubMed] [Google Scholar]
- Angata T, Varki A. Chemical diversity in the sialic acids and related alpha-keto acids: an evolutionary perspective. Chem Rev. 2002;102:439–469. doi: 10.1021/cr000407m. [DOI] [PubMed] [Google Scholar]
- Behrens NH, Leloir LF. Dolichol monophosphate glucose: an intermediate in glucose transfer in liver. Proc Natl Acad Sci USA. 1970;66:153–159. doi: 10.1073/pnas.66.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burda P, Aebi M. The dolichol pathway of N-linked glycosylation. Biochim Biophys Acta. 1999;1426:239–257. doi: 10.1016/s0304-4165(98)00127-5. [DOI] [PubMed] [Google Scholar]
- Calo D, Kaminski L, Eichler J. Protein glycosylation in Archaea: sweet and extreme. Glycobiology. 2010a;20:1065–1079. doi: 10.1093/glycob/cwq055. [DOI] [PubMed] [Google Scholar]
- Calo D, Eilam Y, Lichtenstein RG, Eichler J. Towards glyco-engineering in Archaea: replacing Haloferax volcanii AglD with homologous glycosyltransferases from other halophilic archaea. Appl Environ Microbiol. 2010b;76:5684–5692. doi: 10.1128/AEM.00681-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calo D, Guan Z, Eichler J. Glyco-engineering in Archaea: differential N-glycosylation of the S-layer glycoprotein in a transformed Haloferax volcanii strain. Microb Biotechnol. 2011a;4:461–470. doi: 10.1111/j.1751-7915.2011.00250.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calo D, Guan Z, Naparstek S, Eichler J. Different routes to the same ending: comparing the N-glycosylation processes of Haloferax volcanii and Haloarcula marismortui two halophilic archaea from the Dead Sea. Mol Microbiol. 2011b;81:1166–1177. doi: 10.1111/j.1365-2958.2011.07781.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaban B, Voisin S, Kelly J, Logan SM, Jarrell KF. Identification of genes involved in the biosynthesis and attachment of Methanococcus voltae N-linked glycans: insight into N-linked glycosylation pathways in Archaea. Mol Microbiol. 2006;61:259–268. doi: 10.1111/j.1365-2958.2006.05226.x. [DOI] [PubMed] [Google Scholar]
- Cianciotto NP. Pathogenicity of Legionella pneumophila . Int J Med Microbiol. 2001;291:331–343. doi: 10.1078/1438-4221-00139. [DOI] [PubMed] [Google Scholar]
- Cline SW, Lam WL, Charlebois RL, Schalkwyk LC, Doolittle WF. Transformation methods for halophilic archaebacteria. Can J Microbiol. 1989;35:148–152. doi: 10.1139/m89-022. [DOI] [PubMed] [Google Scholar]
- Dell A, Galadari A, Sastre F, Hitchen P. Similarities and differences in the glycosylation mechanisms in prokaryotes and eukaryotes. Int J Microbiol. 2010;2010:148178. doi: 10.1155/2010/148178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domon B, Costello CE. A systematic nomenclature for carbohydrate fragmentations in FAB-MS/MS spectra of glycoconjugates. Glycoconj J. 1988;5:397–409. [Google Scholar]
- Dubray G, Bezard G. A highly sensitive periodic acid-silver stain for 1,2-diol groups of glycoproteins and polysaccharides in polyacrylamide gels. Anal Biochem. 1982;119:325–329. doi: 10.1016/0003-2697(82)90593-0. [DOI] [PubMed] [Google Scholar]
- Eichler J, Adams MWW. Posttranslational protein modification in Archaea. Microbiol Mol Biol Rev. 2005;69:393–425. doi: 10.1128/MMBR.69.3.393-425.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard TD, Kneller DG. Sparky 3. San Francisco, CA: University of California [WWW document]; URL http://www.cgl.ucsf.edu/home/sparky/. [Google Scholar]
- Guan Z, Naparstek S, Kaminski L, Konrad Z, Eichler J. Distinct glycan-charged phosphodolichol carriers are required for the assembly of the pentasaccharide N-linked to the Haloferax volcanii S-layer glycoprotein. Mol Microbiol. 2010;78:1294–1303. doi: 10.1111/j.1365-2958.2010.07405.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Z, Meyer BM, Albers SV, Eichler J. The thermoacidophilic archaeon Sulfolobus acidocaldarius contains a novel, highly reduced dolichyl phosphate. Biochim Biophys Acta – Mol Cell Biol Lipids. 2011;1811:607–616. doi: 10.1016/j.bbalip.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann E, Konig H. Uridine and dolichyl diphosphate activated oligosaccharides are intermediates in the biosynthesis of the S-layer glycoprotein of Methanothermus fervidus . Arch Microbiol. 1989;151:274–281. [Google Scholar]
- Heiskanen A, Hirvonen T, Salo H, Impola U, Olonen A, Laitinen A, et al. Glycomics of bone marrowderived mesenchymal stem cells can be used to evaluate their cellular differentiation stage. Glycoconj J. 2009;26:367–384. doi: 10.1007/s10719-008-9217-6. [DOI] [PubMed] [Google Scholar]
- Helenius A, Aebi M. Roles of N-linked glycans in the endoplasmic reticulum. Annu Rev Biochem. 2004;73:1019–1049. doi: 10.1146/annurev.biochem.73.011303.073752. [DOI] [PubMed] [Google Scholar]
- Huang Y, Mechref Y, Novotny MV. Microscale nonreductive release of O-linked glycans for subsequent analysis through MALDI mass spectrometry and capillary electrophoresis. Anal Chem. 2001;73:6063–6069. doi: 10.1021/ac015534c. [DOI] [PubMed] [Google Scholar]
- Igura M, Maita N, Kamishikiryo J, Yamada M, Obita T, Maenaka K, Kohda D. Structure-guided identification of a new catalytic motif of oligosaccharyltransferase. EMBO J. 2008;27:234–243. doi: 10.1038/sj.emboj.7601940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MB, Rosenberg JN, Betenbaugh MJ, Krag SS. Structure and synthesis of polyisoprenoids used in N-glycosylation across the three domains of life. Biochim Biophys Acta. 2009;1790:485–494. doi: 10.1016/j.bbagen.2009.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kärcher U, Schröder H, Haslinger E, Allmaier G, Schreiner R, Wieland F, et al. Primary structure of the heterosaccharide of the surface glycoprotein of Methanothermus fervidus . J Mol Biol. 1993;268:26821–26826. [PubMed] [Google Scholar]
- Kelleher DJ, Gilmore R. An evolving view of the eukaryotic oligosaccharyltransferase. Glycobiology. 2006;16:47R–62R. doi: 10.1093/glycob/cwj066. [DOI] [PubMed] [Google Scholar]
- Knirel YA, Kocharova NA, Shashkov AS, Dmitriev BA, Kochetkov NK, Stanislavsky ES, Mashilova GM. Somatic antigens of Pseudomonas aeruginosa The structure of O-specific polysaccharide chains of the lipopolysaccharides from P. aeruginosa O5 (Lanyi) and immunotype 6 (Fisher) Eur J Biochem. 1987;163:639–652. doi: 10.1111/j.1432-1033.1987.tb10913.x. [DOI] [PubMed] [Google Scholar]
- Knirel YA, Rietschel ET, Marre E, Zähringer U. The structure of the O-specific chain of Legionella pneumophila serogroup 1 lipopolysaccharide. Eur J Biochem. 1994;221:239–245. doi: 10.1111/j.1432-1033.1994.tb18734.x. [DOI] [PubMed] [Google Scholar]
- Knirel YA, Senchenkova SN, Shashkov AS, Shevelev SD, Perepelov AV, Bin L, et al. First isolation and identification of a Di-N-Acyl derivate of 5,7,-Diamino- 3,5,7,9-tetradeoxy-L-glycero-D-galacto-non-2-ulosonic (8-Epilegionaminic) acid. Adv Sci Lett. 2009;2:384–387. [Google Scholar]
- Kogelberg H, Rutherford TJ. Studies on the three-dimensional behaviour of the selectin ligands Lewis(a) and sulphated Lewis(a) using NMR spectroscopy and molecular dynamics simulations. Glycobiology. 1994;4:49–57. doi: 10.1093/glycob/4.1.49. [DOI] [PubMed] [Google Scholar]
- Kuntz C, Sonnenbichler J, Sonnenbichler I, Sumper M, Zeitler R. Isolation and characterization of dolichol-linked oligosaccharides from Haloferax volcanii . Glycobiology. 1997;7:897–904. doi: 10.1093/glycob/7.7.897. [DOI] [PubMed] [Google Scholar]
- Larkin A, Imperiali B. The expanding horizons of asparagine-linked glycosylation. Biochemistry. 2011;50:4411–4426. doi: 10.1021/bi200346n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner J, Wieland F. Structure and biosynthesis of prokaryotic glycoproteins. Annu Rev Biochem. 1989;58:173–194. doi: 10.1146/annurev.bi.58.070189.001133. [DOI] [PubMed] [Google Scholar]
- Lehmann F, Tiralongo E, Tiralongo J. Sialic acid-specific lectins: occurrence, specificity and function. Cell Mol Life Sci. 2006;63:1331–1354. doi: 10.1007/s00018-005-5589-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linton D, Dorrell N, Hitchen PG, Amber S, Karlyshev AV, Morris HR, et al. Functional analysis of the Campylobacter jejuni N-linked protein glycosylation pathway. Mol Microbiol. 2005;55:1695–1703. doi: 10.1111/j.1365-2958.2005.04519.x. [DOI] [PubMed] [Google Scholar]
- McNally DJ, Aubry AJ, Hui JPM, Khieu NH, Whitfield D, Ewing CP, et al. Targeted metabolomics analysis of Campylobacter coli VC167 reveals legionaminic acid derivatives as novel flagellar glycans. J Biol Chem. 2007;282:14463–14475. doi: 10.1074/jbc.M611027200. [DOI] [PubMed] [Google Scholar]
- Magidovich H, Eichler J. Glycosyltransferases and oligosaccharyltransferases in Archaea: putative components of the N-glycosylation pathway in the third domain of life. FEMS Microbiol Lett. 2009;300:122–130. doi: 10.1111/j.1574-6968.2009.01775.x. [DOI] [PubMed] [Google Scholar]
- Magidovich H, Yurist-Doutsch S, Konrad Z, Ventura VV, Hitchen PG, Dell A, Eichler J. AglP is a S-adenosyl-L-methionine-dependent methyltransferase that participates in the N-glycosylation pathway of Haloferax volcanii . Mol Microbiol. 2010;76:190–199. doi: 10.1111/j.1365-2958.2010.07090.x. [DOI] [PubMed] [Google Scholar]
- Maita N, Nyirenda J, Igura M, Kamishikiryo J, Kohda D. Comparative structural biology of eubacterial and archaeal oligosaccharyltransferases. J Biol Chem. 2010;285:4941–4950. doi: 10.1074/jbc.M109.081752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengele R, Sumper M. Drastic differences in glycosylation of related S-layer glycoproteins from moderate and extreme halophiles. J Biol Chem. 1992;267:8182–8185. [PubMed] [Google Scholar]
- Mevarech M, Werczberger R. Genetic transfer in Halobacterium volcanii . J Bacteriol. 1985;162:461–462. doi: 10.1128/jb.162.1.461-462.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuttall SD, Dyall-Smith ML. HF1 and HF2: novel bacteriophages of halophilic archaea. Virology. 1993;197:678–684. doi: 10.1006/viro.1993.1643. [DOI] [PubMed] [Google Scholar]
- Packer N, Lawson MA, Jardine DR, Redmond JW. A general approach to desalting oligosaccharides released from glycoproteins. Glycoconj J. 1998;15:737–747. doi: 10.1023/a:1006983125913. [DOI] [PubMed] [Google Scholar]
- Pietilä MK, Roine E, Paulin L, Kalkkinen N, Bamford DH. An ssDNA virus infecting archaea: a new lineage of viruses with a membrane envelope. Mol Microbiol. 2009;72:307–319. doi: 10.1111/j.1365-2958.2009.06642.x. [DOI] [PubMed] [Google Scholar]
- Pietilä MK, Laurinavičius S, Sund J, Roine E, Bamford DH. The single-stranded DNA genome of novel archaeal virus halorubrum pleomorphic virus 1 is enclosed in the envelope decorated with glycoprotein spikes. J Virol. 2010;84:788–798. doi: 10.1128/JVI.01347-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter CJ, Kaczowka SJ, Maupin-Furlow JA. Differential regulation of PanA and PanB proteasome-activating nucleotidase and 20S proteasome proteins of the haloarchaeon Haloferax volcanii . J Bacteriol. 2004;186:7763–7772. doi: 10.1128/JB.186.22.7763-7772.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roine E, Oksanen HM. Viruses from the hypersaline environment. In: Ventosa A, Oren A, Ma Y, editors. Halophiles and Hypersaline Environments: Current Research and Future Trends. Heidelberg: Springer; 2011. pp. 153–172. [Google Scholar]
- Roine E, Kukkaro P, Paulin L, Laurinavičius S, Domanska A, Somerharju P, Bamford DH. New, closely related haloarchaeal viral elements with different nucleic acid types. J Virol. 2010;84:3682–3689. doi: 10.1128/JVI.01879-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz F, Aebi M. Mechanisms and principles of N-linked protein glycosylation. Curr Opin Struct Biol. 2011;21:576–582. doi: 10.1016/j.sbi.2011.08.005. [DOI] [PubMed] [Google Scholar]
- Sumper M, Berg E, Mengele R, Strobel I. Primary structure and glycosylation of the S-layer protein of Haloferax volcanii . J Bacteriol. 1990;172:7111–7118. doi: 10.1128/jb.172.12.7111-7118.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiezewska E, Danikiewicz W. Polyisoprenoids: structure, biosynthesis and function. Prog Lipid Res. 2005;44:235–258. doi: 10.1016/j.plipres.2005.05.002. [DOI] [PubMed] [Google Scholar]
- Szymanski CM, Wren BW. Protein glycosylation in bacterial mucosal pathogens. Nat Rev Microbiol. 2005;3:225–237. doi: 10.1038/nrmicro1100. [DOI] [PubMed] [Google Scholar]
- Tsvetkov YE, Shashkov AS, Knirel YA, Zähringer U. Synthesis and NMR spectroscopy of nine stereoisomeric 5,7,diacetamido-3,5,7,9-tetradeoxynon-2- ulosonic acids. Carbohydr Res. 2001;335:221–243. doi: 10.1016/s0008-6215(01)00235-x. [DOI] [PubMed] [Google Scholar]
- Varki A, Freeze HH, Gagneux P. Evolution of glycan diversity. In: Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, et al., editors. Essentials of Glycobiology. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2009. pp. 281–292. [PubMed] [Google Scholar]
- Vigerust DJ, Shepherd VL. Virus glycosylation: role in virulence and immune interactions. Trends Microbiol. 2007;15:211–218. doi: 10.1016/j.tim.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weerapana E, Imperiali B. Asparagine-linked protein glycosylation: from eukaryotic to prokaryotic systems. Glycobiology. 2006;16:91R–101R. doi: 10.1093/glycob/cwj099. [DOI] [PubMed] [Google Scholar]
- Yurist-Doutsch S, Chaban B, VanDyke D, Jarrell KF, Eichler J. Sweet to the extreme: protein glycosylation in Archaea. Mol Microbiol. 2008;68:1079–1084. doi: 10.1111/j.1365-2958.2008.06224.x. [DOI] [PubMed] [Google Scholar]