SUMMARY
To achieve cell entry, many nonenveloped viruses must transform from a dormant to a primed state. In contrast to the membrane fusion mechanism of enveloped viruses (e.g., influenza virus), this membrane penetration mechanism is poorly understood. Here, using single-particle cryo-electron microscopy, we report a 3.3 Å structure of the primed, infectious subvirion particle of aquareovirus. The density map reveals side-chain densities of all types of amino acids (except glycine), enabling construction of a full-atom model of the viral particle. Our structure and biochemical results show that priming involves autocleavage of the membrane penetration protein and suggest that Lys84 and Glu76 may facilitate this autocleavage in a nucleophilic attack. We observe a myristoyl group, covalently linked to the N terminus of the penetration protein and embedded in a hydrophobic pocket. These results suggest a well-orchestrated process of nonenveloped virus entry involving autocleavage of the penetration protein prior to exposure of its membrane-insertion finger.
INTRODUCTION
Without a lipid bilayer, nonenveloped viruses invade cells by penetrating the cell membrane. They do so through a poorly understood process that is accomplished by viral membrane-penetration proteins (Banerjee and Johnson, 2008). In contrast, the mechanism of membrane fusion used by enveloped viruses like the human immunodeficiency virus (HIV) and the influenza virus is well understood. In those cases, the viral lipid bilayer fuses with the cell membrane (White et al., 2008). Turreted double-stranded RNA (dsRNA) viruses of the Reoviridae family have been extensively studied and serve as ideal model systems for studying the mechanism of cell entry by the nonenveloped viruses (Chandran and Nibert, 2003). The functional states of the viral membrane penetration protein are precisely controlled during cell entry. To survive in a harsh environment, the viral membrane “penetration protein” is kept in a dormant, stable state by binding to a “protection protein” in the virion (Chandran and Nibert, 2003). However, to penetrate the cell membrane during infection, this viral membrane penetration protein must be converted to a metastable, primed state by proteolytic removal of its protection protein, resulting in formation of an infectious subvirion particle (ISVP) (Chandran and Nibert, 2003). For that reason, mutations that prevent such conversion of the penetration protein result in loss of infectivity (Agosto et al., 2007; Middleton et al., 2007).
Structural information of both the dormant and primed states is needed in order to gain insight into the mechanism of this dormant-to-primed conversion of the membrane penetration proteins of the turreted dsRNA reoviruses. To date, only the atomic structure in a stable dormant state has been determined (i.e., the crystal structure of the σ33μ13 heterohexamer of orthoreovirus; σ3 and μ1 are the protection and the membrane penetration protein, respectively) (Liemann et al., 2002). In the absence of an atomic structure at the metastable, primed state of the membrane penetration protein, the molecular details of this dormant-to-primed conversion are unknown.
Due to the metastable property of the ISVP, cryo-electron microscopy (cryo-EM) is particularly suitable for its structural determination because no crystal is required and because freshly prepared samples can be preserved in their native state by flash freezing. Recent advances in single-particle cryo-EM have pushed the limit to near atomic resolution (Jiang et al., 2008; Yu et al., 2008; Zhang et al., 2008). However, at near atomic resolution, only protein backbone models can be built, significantly limiting the value of such models for functional interpretation. Through improving data quality (Figures 1A and 1B) by including, for example, the use of parallel illumination and better microscope alignment, we have pushed the resolution limit of single-particle cryo-EM to ~3.3 Å, allowing ab initio model building of full-atom structures and functional interpretation of the resulting models (Figures 1C-1E). In so doing, we obtained a 3.3 Å resolution structure of the primed penetration protein.
Figure 1. Cryo-EM and 3D Reconstruction of Infectious Subvirion Particle of Aquareovirus.
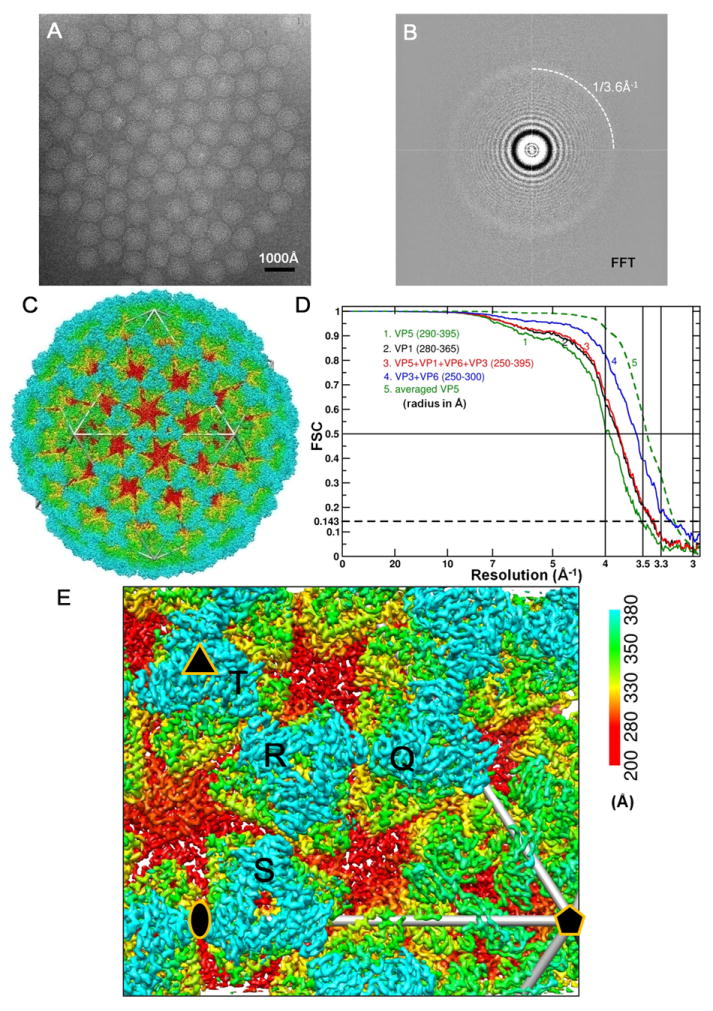
(A) Representative cryo-EM image of the ISVP at 1.2 μm underdefocus. There is no carbon support film, and viral particles are suspended in vitreous ice.
(B) Fourier-transformed spectrum of the image in (A), showing contrast transfer function rings visible beyond 1/4 Å-1 and a strong signal at ~1/3.6 Å-1 (dashed arc), which is likely due to H2O (Dowell and Rinfret, 1960).
(C) Density map of the aquareovirus ISVP at 3.3 Å resolution, colored according to radius. An enlarged view of an asymmetric unit is shown in Figure 1E. See also Movie S1.
(D) Fourier shell cross-correlation coefficients (FSC), indicating the effective resolution to be 3.3 Å for VP3, VP6 and the averaged VP5, and 3.5 Å for VP1 according to the reference-based criterion as defined by Rosenthal and Henderson (Cref=0.5 or FSC=0.143) (Rosenthal and Henderson, 2003). Different curves represent different protein shells: the whole protein shell (red), the shell containing VP1 (black), the shell of VP3 and VP6 (blue) and the VP5 layer (green: the solid line for the original VP5 layer and the dashed line for the averaged VP5).
(E) Enlarged view of the cryo-EM density of the primed ISVP, showing an asymmetric unit in Figure1C. Four pseudo equivalent VP5 trimers are labeled (Q, R, S and T), and positions of 2- (ellipse), 3- (triangle) and 5-fold (pentagon) axes are indicated. See also Figure S1 and Movie S1.
Our atomic structure of the primed aquareovirus ISVP reveals that conversion of VP5 is accompanied by an autocleavage at the Asn42-Pro43 bond, probably due to a nucleophilic attack facilitated by nearby Lys84 and Glu76 residues. The atomic structure also reveals the density of an N-terminal covalently-linked myristoyl group, the membrane insertion “finger”, which is sheltered in a hydrophobic pocket. Release of this myristoyl group from the pocket will be required in a later step during membrane penetration.
RESULTS AND DISCUSSION
Cryo-EM Structure Determination and Model Building
To gain insight into mechanism of the dormant-to-primed state conversion, we obtained a 3.3 Å resolution structure of the metastable, primed aquareovirus ISVP by single-particle cryo-EM. The primed virus was prepared by removing the outermost VP7 protection protein of the aquareovirus virion (see the Experimental Procedures), leaving just four different structural proteins in the ISVP. We collected a total of 650 films in a FEI Titan Krios microscope, and obtained a three-dimensional (3D) reconstruction by use of an integrative approach of multiple software packages (see the Experimental Procedures). The effective resolution of the 3D reconstruction is estimated to be ~3.3 Å, based on both the resolving power/feature-based criterion (Figures 1E and 2; Figures S1, S2 and S3 and Movies S1, S2 and S3 available online) and the reference-based Fourier shell correlation criterion (Cref=0.5 or FSC=0.143) (Figure 1D) (Rosenthal and Henderson, 2003; Zhou, 2008). For example, our map shows continuous densities with clear bumps every ~3.8 Å, corresponding to the distance between Cα atoms (Figures 2A-2D, Figures S1, S2, and S3, and Movies S2 and S3) and distinctive side-chain densities of most residues, including those like alanine with small side chains (Figures 2B and 2D, Figures S1, S2 and S3, and Movie S2). Notably, bumpy densities corresponding to some carbonyl oxygen atoms can also be identified (Figures 2D and S2C), and different types of aromatic side chains can be distinguished from their densities alone (Figure 2D).
Figure 2. Atomic Model of the Aquareovirus ISVP.
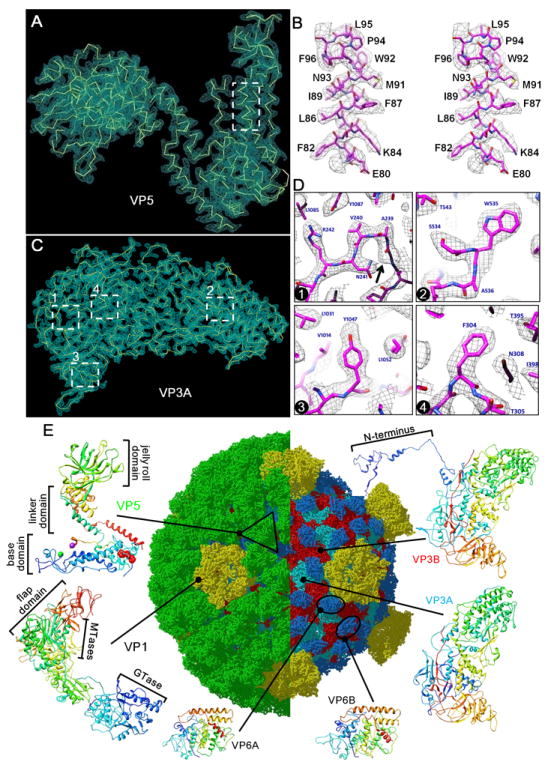
(A) Cryo-EM density (cyan mesh) of a VP5 monomer superimposed on its atomic model (yellow; only backbone shown).
(B) Stereo view of the box from (A). Cryo-EM densities (gray mesh) are superimposed on its atomic model (magenta).
(C) Cryo-EM density (cyan mesh) of VP3A superimposed on its atomic model (yellow; only backbone shown).
(D) Cryo-EM densities (gray mesh) of the four boxed regions in (C) showing representative side chains. Cryo-EM densities (gray mesh) are superimposed on its atomic model (magenta). Box 1 also shows density for a carboxyl oxygen atom (arrow). The different aromatic amino acids, Trp, Tyr and Phe, in boxes 2-4 are readily distinguished.
(E) Complete atomic model of the ISVP. In the right half, removal of the VP5 coat reveals core proteins. Ribbon models of the atomic structures of the six conformers from four structural proteins are shown in the periphery: two VP3 conformers (Figure S1), two VP6 conformers (Figure S2 and Movie S2) one VP1 (Figure S3 and Movie S2), and one VP5. These atomic models are color-coded according to amino acid sequence from blue (N terminus) through green and yellow to red (C-terminus). The black triangle demarcates a VP5 trimer. See also Figure S2 and Movie S2.
Careful comparison indicates that the resolving power of our map is comparable to that of crystal structures at similar resolution (e.g., the ABC transporter at 3.2 Å [Locher et al., 2002], mitochondrial creatine kinase at 3.2 Å [Fritz-Wolf et al., 1996], human hepatitis B virus core at 3.3 Å [Wynne et al., 1999] and the bacterial ribosome at 3.3 Å [Schluenzen et al., 2000]). Solely based on our cryo-EM density map, we built an ab initio atomic model for the ISVP by using model-building procedures commonly used in X-ray crystallography (see details in the Experimental Procedures). The quality of our models is confirmed both by the agreement of the model with the density map (Figures 2A-D and Figures S1, S2 and S3) and by other criteria such as the Ramachandran plot and the psudocrystallographic R factor/Rfree (Table 1).
Table 1.
Statistics for VP5 Model and Pseudocrystallographic Refinement
| Parameter names | Parameter Values |
|---|---|
| Cell dimension a, b, c (Å) | 158.4, 165.0, 176.0 |
| Cell angle α, β, γ (°) | 90.0, 90.0, 90.0 |
| Resolution (Å) | 20–3.3 |
| Rworka | 28.40% |
| Rfreeb | 29.20% |
| Number of protein atoms (residues 2–646) | 4797 |
| Number of nonprotein atoms | 15 |
| Average B factor | 50.6 |
|
| |
| Rms Deviations | Deviation Values |
|
| |
| Bond lengths (Å) | 0.01 |
| Bond angles (°) | 1.6 |
| Dihedral angles (°) | 23 |
| Improper angles (°) | 1.26 |
|
| |
| Ramachandran Plot (From PROCHECK) | Percentage |
|
| |
| Most favored + additional allowed regions | 93.9% |
| Generously allowed regions | 4.3% |
| Disallowed regions | 1.8% |
Besides the penetration protein VP5, we also obtained full atomic models for all three core proteins of the ISVP: VP1, VP3 and VP6. Our final model of the ISVP contains six conformers of four proteins with a total of 4990 residues: two VP3 (VP3A: residues 188-1211 of 1214; VP3B: residues 15-1214 of 1214), two VP6 (VP6A and VP6B, both residues 2-412 of 412), one VP1 (residues 1-1299 of 1299) and one VP5 (residues 2-646 of 648, Table 1) (Figure 2E). (For the inner shell protein VP3A, the N-terminal 187 residuals are disordered, probably as a result of their interaction with the genome or with other proteins inside the “core” shell.)
Overall Structure of the ISVP
Each ISVP has an RNA-containing “core” (Cheng et al., 2008), enclosed by a “coat” (Figure 2E). Unlike the spiky appearance of the aquareovirus virion, which is due to the presence of the outermost VP7 protein (Cheng et al., 2008), the aquareovirus ISVP (VP7 removed) has a smooth coat surface formed by a network of VP5 trimers (Figures 1C and 2E and Movie S1). The final atomic model of the ISVP contains six conformers of four proteins: two VP3, two VP6, one VP1 on the core and one VP5 on the coat (Figure 2E). The coat is made up of 200 VP5 trimers organized into an incomplete T=13 lattice. The exposed VP5 trimers in the ISVP are well situated to interact with cell membrane during membrane penetration (Figure 1C). The core is a T=1 particle with 12 VP1 pentameric turrets decorating a shell of 120 VP3 monomers, which are clamped together by 120 VP6 monomers (Figure 2E) (Cheng et al., 2008). With 900 protein molecules, a total molecular weight of ~72MDa and a diameter of ~800Å, our ISVP structure by single-particle cryo-EM represents the largest atomic structure ever determined by any structural biology techniques.
Below, we first describe the finding of the autocleavage of the membrane penetration protein and then present the observation of the myristoyl group, both in the context of the dormant-to-primed conversion of the penetration protein. At the end, we briefly summarize the structures of the core proteins, which are highly conserved with their homologs in other members of the Reoviridae family.
Primed Membrane Penetration Protein
For our primed, membrane penetration protein VP5, the ten quasiequivalent subunits in an asymmetric unit are almost identical, so we averaged their densities to further improve map quality (Figures 2A and 2B). The atomic model of primed VP5, based on this averaged VP5 density map, has a “Z” shape (Figures 2A and 2E). It can be divided into three domains: a jelly-roll domain at the tip of the Z, a base domain at the bottom, and a linker domain joining the two (Figure 2E). The jelly-roll domain (residues 287-484, Figure 2E), at the outer surface, mainly consists of a β hairpin on the top, a five-stranded β sheet in the middle, and a four-stranded β sheet at the bottom. The base domain (residues 2-242, Figure 2E) at the bottom consists of four α helices and several long loops. The linker domain (residues 243-286 and 485-648, Figure 2E) is composed of six α helices and several short loops.
The VP5 protein of aquareovirus shares a high degree of sequence similarity (38.4%) to the μ1 protein of orthoreovirus and their structures in their virions (dormant state) were found to be very similar (Cheng et al., 2008). In contrast, we find a conformational difference between the primed and dormant states of the penetration protein by comparing our primed VP5 protein of aquareovirus to the dormant μ1 protein in complex with the protection protein σ3 of orthoreovirus (Figure 3) (Liemann et al., 2002). In vivo, the conformational change is probably induced by removal of the aquareovirus protection protein VP7 during the residence of the virus in the endosome. In our study, priming was accomplished by proteolytic removal of VP7 (see the Experimental Procedures). We find different conformations between the dormant and primed proteins for some of the loops in the helix-rich region and similar conformations for the remainder of that region (Figure 3). We also find different conformations between the two proteins in the β hairpin and the middle five-stranded β sheet in the tip region of the jelly-roll domain but similar conformations in the bottom four-stranded β sheet of that region (Figure 3). Indeed, in orthoreovirus, the correct initial folding of the membrane penetration protein μ1 requires protection protein σ3 (Shing and Coombs, 1996; Tillotson and Shatkin, 1992). Thus, it is not surprising that removal of the protection protein would permit a conformational change of the penetration protein as part of the priming process for cell entry.
Figure 3. Structural Comparison of Primed and Dormant Penetration Proteins.
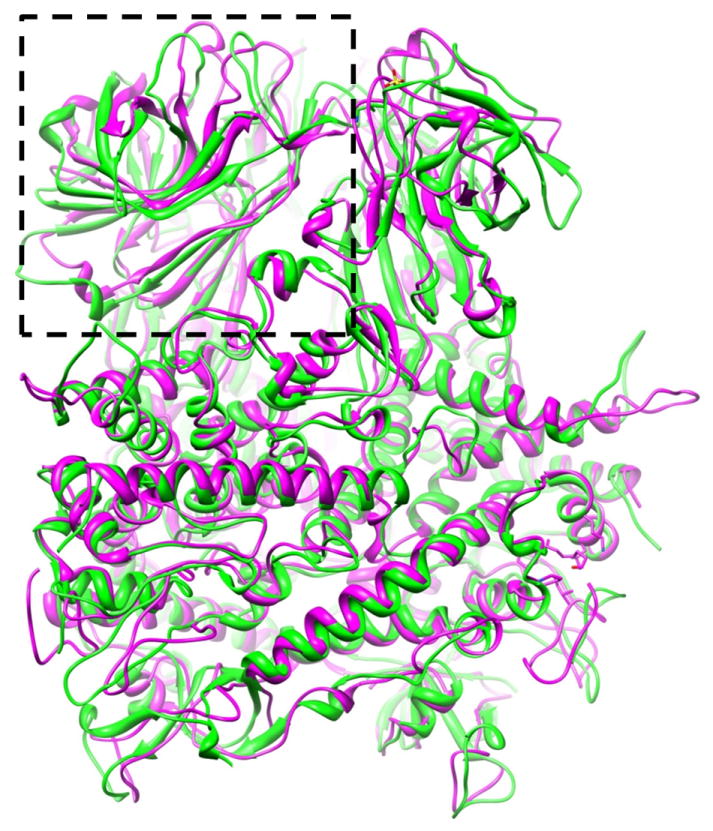
Atomic model of a primed VP5 trimer (magenta ribbon) is superimposed on the atomic model of a dormant μ1 trimer of orthoreovirus (green ribbon), showing different conformations for some of the loops in the helix-rich region but similar conformations for the remainder of the helix-rich region. The jelly-roll domain, indicated by a dashed box.
Autocleavage in the Primed Membrane Penetration Protein
We find a single gap in the backbone density of the base domain of VP5 in every one of the ten VP5 molecules in an asymmetric unit of the ISVP. Atomic modeling unambiguously locates the formerly bonded amino acids Asn42 and Pro43 to the two sides of this gap (Figures 4A and 4B). Asn42 is located at the C-terminus of a short helix that is buried in the interior, and Pro43 is located at the N-terminus of a loop that is more exposed to the bottom surface of VP5 (Figures 4C). The distance between the Cα atom of Asn42 and the Cα atom of Pro43 is 11.7 Å, significantly larger than the standard Cα-Cα bonded distance of 3.8Å (Figure 4B). From our structural results we conclude that the Asn42-Pro43 bond in the primed VP5 of the ISVP is cleaved.
Figure 4. Cleavage Sites of the Membrane Penetration Protein VP5.
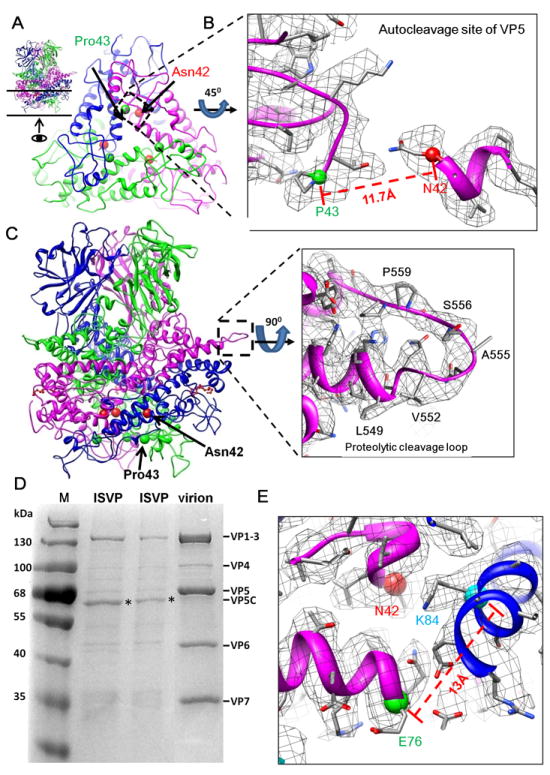
(A) VP5 trimer reviewed from side (inset; see also Movie S4) and from inside of ISVP (indicated by an arrow in the inset). Each monomer of the trimer contains an autocleavage site at Asn42-Pro43. The Cα atoms of Asn42 (red ball) and Pro43 (green ball) are indicated. Only a 30 Å thick slab (indicated by the two horizontal lines in the inset) at the trimer base of the trimer is shown for clarity.
(B) A enlarged view of the boxed region of one VP5 monomer in (A), except the cryo-EM densities are also shown (gray mesh). Autocleavage of the Asn42-Pro43 bond is indicated by an 11.7Å gap between their Cα atoms.
(C) Side view of the ribbon model of a VP5 trimer and the position of the intact δ-ϕ proteolytic cleavage loop (residues: 550-561) (dashed box). Cryo-EM density (gray mesh) of the boxed region of one VP5 monomer is also shown, demonstrating an uncleaved loop from residues 551 to 560.
(D) Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) of aquareovirus virion (dormant) and ISVP (primed) from different bands of the same CsCl gradient, showing intact VP5 in virions and autocleaved VP5 (i.e., VP5C, bands indicated by asterisks) in ISVPs. Lane (M), molecular weight markers.
(E) Lys84 (cyan ball) near Asn42 (red ball) at the autocleavage site, and an adjacent Glu76 (green ball). See also Movie S3.
For the penetration protein μ1 in the orthoreovirus virion, both the cryo-EM structure at 7 Å (Zhang et al., 2005a) and biochemistry (Nibert et al., 2005) show that it is not cleaved in the dormant state. The orthoreovirus literature shows that, on the one hand, the penetration protein is cleaved in its crystal structure in complex with a protection protein – unable to penetrate membranes and presumed dormant – with the cleavage site also between Asn42 and Pro43 residues (Liemann et al., 2002). On the other hand, it has been stated that this cleavage was artificially induced during crystallization (Liemann et al., 2002; Nibert et al., 2005). For this reason, the timing of the autocleavage was not identified. Nonetheless, this cleavage was proposed to take place in a later stage, after priming (Agosto et al., 2008; Tsai, 2007).
However, for the aquareovirus penetration protein VP5, by comparing the results of sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) of our ISVP and virion samples, we confirm that VP5 is cleaved in the ISVP and show that it is intact in the whole virion (Figure 4D). (We know that the cleavage in the ISVP is not caused by some artifact in preparation because we obtained the ISVP and virion samples from two different bands of a single CsCl gradient separation after partial proteolytic attack [by α-chymotrypsin] on whole virions in the same preparation [see the Experimental Procedures].) The SDS-PAGE results also suggest that the cleavage is caused by removal of protection protein VP7 since, aside from cleavage of VP5, the gel shows that loss of VP7 is the only difference between the ISVP and the dormant virion samples (Figure 4D).
We further conclude that this cleavage in VP5 of the primed ISVP is an autocleavage because the Asn42-Pro43 bond in the ISVP is located in the interior of the VP5 layer, inaccessible to any external proteases (Figure 4C). This autocleavage is a critical step in cell entry of turreted reoviruses (Nibert et al., 1991a) like aquareovirus, but it has been proposed to occur during a later conversion from the primed ISVP to a further disassembled particle (called ISVP*) (Agosto et al., 2008; Tsai, 2007). Here, given that the penetration protein is intact in the dormant virions of both orthoreovirus (Nibert et al., 2005) and aquareovirus (Figure 4D), our structure (Figure 4B) and biochemistry (Figure 4D) reveal that autocleavage of the penetration protein occurs during the conversion from the dormant to the primed state, not at later stages.
However, a second Asn-Pro bond (N587-P588) in the orthoreovirus μ1 protein is not cleaved, even in the presence of protease (Liemann et al., 2002; Nibert and Fields, 1992; Tillotson and Shatkin, 1992), suggesting that the pairing of Asn-Pro is not by itself sufficient for autocleavage and that neighboring residues are required. Indeed, careful examination of our atomic structure for VP5 supports a mechanism for the Asn42-Pro43 autocleavage through nucleophilic attack, as proposed for other viruses such as orthoreovirus (Liemann et al., 2002), Flock House virus (FHV) (Reddy et al., 2004) and Nudaurelia capensis ω virus (NωV) (Taylor and Johnson, 2005). In FHV and NωV, an Asp or Glu plays a key role by activating Asn at the cleavage site for nucleophilic attack.
In our structure, we observed Glu76 and Lys84 near the Asn42 (Figure 4E). On the same subunit of Asn42-Pro43 and ~11 Å away from Asn42, Glu76 might facilitate the autocleavage by activating Asn42, similar to what was found in FHV (Reddy et al., 2003) and NωV (Taylor and Johnson, 2005). In addition, on an adjacent VP5 subunit, only ~4 Å away from Asn42, Lys84 could facilitate the autocleavage by stabilizing the negatively charged transition intermediate, similar to what occurs in several enzymatic reactions (Bartlett et al., 2002; Greenleaf et al., 2008). Notably, Lys84 – as well as Asn42 and Pro43 – are highly conserved among aqua-, mammalian and avian reoviruses (Noad et al., 2006).
A related observation has been made for dormant orthoreovirus, in which it was noted that Lys113 on an adjacent subunit and Glu105 on the same subunit as Asn42-Pro43 are likely participants in the autocleavage reaction (Liemann et al., 2002). In that dormant structure, Lys113 and Glu105 form a salt bridge (Liemann et al., 2002). For these two residues, we note that aquareovirus VP5 has a counterpart Lys84 on an adjacent subunit and a Glu76 on the same subunit. In our primed structure, these two amino acids are far apart (~13 Å) (Figure 4E). We suggest that in the dormant state these two residues should be close and connected by a salt bridge, making both Glu76 and Lys84 unavailable to promote the autocleavage. During priming, this salt bridge would break due to the conformational change of the penetration protein induced by removing the protection protein VP7 (Figure 3), thus allowing Glu76 and Lys84 to facilitate the autocleavage reaction (Figure 4E).
Direct Observation of a Myristoyl Group in the Primed Membrane Penetration Protein
Biochemical data has shown that there is a myristoyl group linked to the N-terminal amino acid of reovirus μ1 protein (Nibert et al., 1991b). In the cryo-EM density map of our primed VP5 protein, a rod-shaped density extends from the N-terminal end, specifically from Gly2 into a pocket in the base domain of all ten VP5 molecules of each asymmetric unit of the ISVP (Figure 5A and 5B and Movies S3 and S4). The rod-like density, without side-chain features, can be modeled as a myristoyl group. The continuity of the myristoyl group density with Gly2 indicates that it is covalently linked to the N-terminus of the VP5 protein (Figures 5B and 5C and Movie S3). (Met1 is known to be removed by amino-peptidase prior to adding of the myristoyl group by a N-myristoyl transferase [Towler et al., 1988].)
Figure 5. Myristoyl Group of the Primed VP5 in the ISVP.
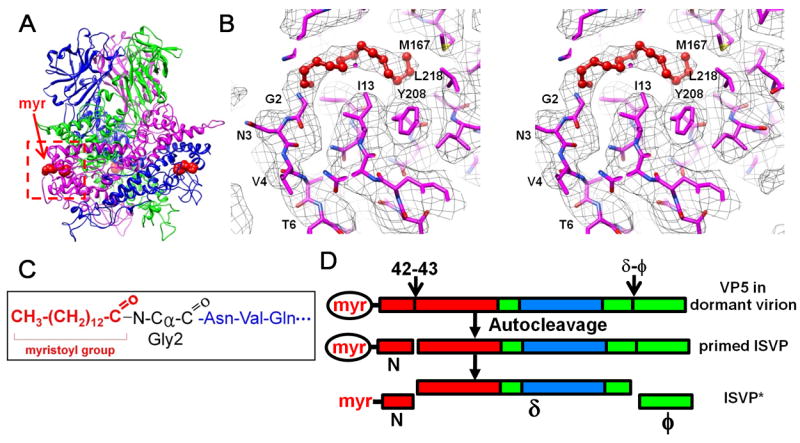
(A) Positions of three myristoyl groups (red balls) near the base of the VP5 trimer. See also Movie S4.
(B) Cryo-EM density (gray mesh) of the base region in Figure 5A (red box), superimposed with its atomic model (magenta), showing the myristoyl group (red balls and sticks) inside a hydrophobic pocket. See also Movie S4.
(C) Amide linkage of the myristoyl group to Gly2.
(D) Changes of the penetration protein VP5 from a stable, dormant state to a metastable, primed ISVP state and subsequently to the ISVP* state. Red, base domain (2-242); green, linker domain (243-286 & 485-648) and cyan, jelly-roll domain (287-484). The myristoyl group (myr) in the hydrophobic pocket is enclosed by an ellipse; the released myristoyl group has no ellipse.
The myristoyl group was not observed in the crystal structure of the orthoreovirus μ13σ33 heterohexamer, since the myristoyl group plus the N-terminal nine residuals were disordered in that crystal structure. However, in the crystal structure, a detergent molecule – β-octyl glucoside (added for crystallization purpose) – was observed in a hydrophobic pocket. Based on this observation, the authors proposed that this pocket is likely the site where the anticipated myristoyl group resides (Liemann et al., 2002).
Here, we directly observe the myristoyl group in this hydrophobic pocket in our cryo-EM structure. The depth of the pocket is ~15Å, formed mainly by hydrophobic residues (Ile13, Leu163, Met 167, Trp183, Tyr208, Ala211 and Leu218) that are well positioned to interact with the buried alkyl chain of the myristoyl group (Figure 5B). The entrance of the pocket is formed by Thr14, Lys191, Leu201 and Pro204. Outside the pocket, the string of residues Gly2-Val8 appears to be relatively flexible, as suggested by the relatively low densities of their backbone and side chains as compared to the rest of the protein. Such flexibility might be required for the myristoyl group to move out of the hydrophobic pocket to serve its role as a membrane insertion “finger” by interacting with cell membrane. These results directly reveal the presence of a covalently linked myristoyl group of the penetration protein inside the hydrophobic pocket (Figure 5B). Indeed, it is known that the myristoyl group is required for the correct folding of the penetration protein and the autocleavage (Tillotson and Shatkin, 1992).
Proteolytic Cleavage Site
Modulating the activity of the myristoyl insertion finger, the ϕ-δ proteolytic cleavage site, as distinct from the autocleavage site, is present in the linker domain of the aquareovirus and orthoreovirus penetration proteins, loop 551-560 for the former (Figure 4C) (Fang et al., 2008) and loop 579-588 for the latter (Chandran and Nibert, 1998; Fang et al., 2008). This proteolytic cleavage increases pore-forming activity by the myristoylated N terminus of the penetration protein during membrane penetration and releases the ϕ fragment (Figure 5D) that may be involved in recruiting virus cores to newly penetrated membrane pores (Ivanovic et al., 2008). In addition, the ϕ fragment released into the cytoplasm can induce apoptosis, which is required for the maximum growth of progeny viruses (Clarke et al., 2005; Danthi et al., 2008). This proteolytic cleavage was proposed to occur before the autocleavage (Agosto et al., 2008; Ivanovic et al., 2008). In contrast, our cryo-EM structure shows that this loop is intact in the primed penetration protein VP5 of the ISVP, thus indicating that the ϕ-δ proteolytic cleavage occurs at a later step after the autocleavage.
Atomic Models of Core Proteins and Conservation among Homologs in the Reoviridae
The structures of individual domains of the mRNA capping protein VP1 (Figure 2E and Figure S3) resemble those of the λ2 protein of the orthoreovirus core (Reinisch et al., 2000), but the relative orientation of the flap domains with respect to the turret channel axis in the primed aquareovirus ISVP is different from that in the orthoreovirus virion and the aquareovirus virion (Figure 2E and Figure S3). In addition, the aquareovirus VP1 turret adopts a more closed conformation (26 Å diameter) in the primed ISVP (Figure S3C) than the open conformation (33 Å diameter) in the virion (Figure S3D) (Cheng et al., 2008).
In contrast to VP1 and VP5, the VP3 and VP6 proteins in the primed ISVP show no obvious structural differences from their structures in the stable dormant state in the virion (Cheng et al., 2010) and from their homologs in the core of orthoreovirus (Reinisch et al., 2000). Thus, inner shell protein VP3 has a plate-shaped, helix-rich structure that resembles the inner shell proteins of other dsRNA viruses (Grimes et al., 1998; Reinisch et al., 2000) (Figures 2C and 2E and Figure S1). In addition, the cementing protein VP6 is oval shaped with eight major helices (Figure 2E and Figure S2). However, this protein binds on the outer surface of the VP3 inner shell at two instead of three positions as in orthoreoviruses (Nason et al., 2000; Zhang et al., 2005b).
Implications into the Processes of Cell Entry of Non-enveloped Virus
Our results demonstrate that after the conversion from the dormant to the primed state the myristoyl groups remain embedded in the hydrophobic pockets of the penetration protein (Figures 5A and 5B). As a membrane insertion “finger”, the buried myristoyl group must be released from the pocket during perforation of the membrane (Nibert et al., 1991b; Odegard et al., 2004). Our result indicates that the release of the myristoyl group from the pocket occurs during a later conversion of ISVP to ISVP* (Figure 5D). This step was previously proposed to be coupled to the autocleavage of the penetration protein (Agosto et al., 2008; Liemann et al., 2002; Tsai, 2007). Instead, our results reveal that the processes of autocleavage (during dormant to primed ISVP) and release of the myristoyl group (during primed ISVP to ISVP*) are not coupled (Figure 5D).
Structural Differences among VP5 Homologs Suggest Adaptations for Infection at Different Temperatures
The aquatic environment in which aquareoviruses establish infection differs significantly in temperature from that encountered by mammalian orthoreovirus. Comparison of our atomic structure for aquareovirus with that for orthoreovirus reveals a possible structural basis for their adaptation to these different environments. Consistent with sequence alignment results (Noad et al., 2006; Zhang et al., 2005a), our atomic structure of aquareovirus VP5 does not have two major structural segments found in the orthoreovirus penetration protein, a helix (residues 72-96) and a hub structure (residues 675-708). These segments in orthoreovirus interact with neighboring penetration trimers to further stabilize the lattice of the penetration proteins (Zhang et al., 2005a). Because the μ1 lattice is disrupted in the orthoreovirus ISVP* particle (Zhang et al., 2006), this stabilization should apply to the ISVP, thus providing an extra energy barrier for the ISVP-to-ISVP* conversion and requiring a higher conversion temperature. The absence of these stabilization segments in aquareovirus may be related to its adaptation to infect cold-blooded, aquatic animals at a lower temperature. Indeed, unlike orthoreovirus, which only infects cells at relatively high temperature (e.g. ≥32°C) (Chandran et al., 2002), aquareovirus can establish active infection at 4°C (Figure 6) and 2.5°C (Rivas et al., 1998).
Figure 6. Infection and Replication of Aquareovirus at 4°C.
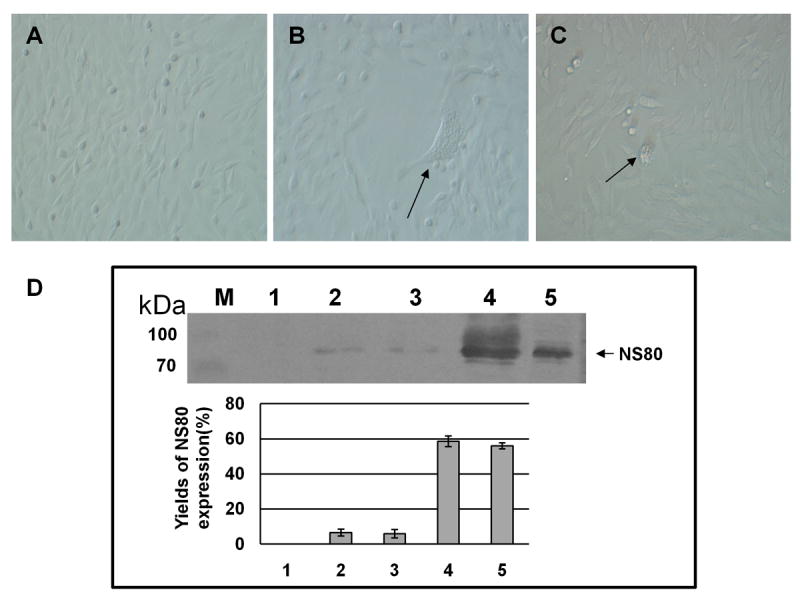
(A) Mock infected CIK.
(B) and (C) aquareovirus infected CIK cell at 4°C and 3 days post infection. Arrows indicate the cytopathic effects (CPE) in infected cells.
(D) Immunoblot analysis of viral NS80 expression. Lane (M), Molecular weight markers; Lane 1, Mock infected CIK cell lysate showing the NS80 protein expression level of 0% as negative control (measured by relative optical density: 0%, no absorbance; 100%, complete absorbance); Lanes 2 and 3, aquareovirus infected CIK cell lysate at 4°C showing the NS80 protein expression levels of 6.6% and 5.8% at 5 days post infection; Lane 4, aquareovirus infected CIK cell lysate at 28°C showing the NS80 protein expression level of 58.7% at 2 days post infection; Lane 5, E. coli expressed NS80 at level of 56% as positive control. Experiments were performed in triplets, and the mean optical density of the triplet immunoblots were shown (error bars represent standard deviation).
In summary, we report a 3.3 Å resolution structure determined by single-particle cryo-EM and the building of an atomic model of a metastable complex from its cryo-EM density map alone. The structure of the aquareovirus particle reveals an autocleavage of the membrane penetration protein during viral conversion from a dormant to a primed state. The density map directly reveals the membrane insertion “finger”, a myristoyl group, sheltered in a hydrophobic pocket during the dormant-to-primed conversion. Our results demonstrate that single-particle cryo-EM is now capable of determining the atomic structure of a large complex in its native environment. Therefore, this approach should open the door to atomic structural studies of metastable or even transient functional intermediates without crystallization, such as the nuclear pore complex, spliceosome and virus-receptor complexes, and will have broad applications in general.
EXPERIMENTAL PROCEDURES
Virus Propagation and Purification
Grass carp aquareovirus (GCRV) was propagated in CIK cell cultures as described previously (Fang et al., 2005). The virus particles were recovered from the infected cell-culture supernatants by low-speed centrifugation (3~6000 rpm, SW28 rotor) for 20 min to remove cell debris, and ultracentrifugation (26,000 rpm, SW28 rotor) for 2 hr to pellet virus. To prepare ISVP, the pelleted virions of GCRV were suspended very carefully in virion buffer (150 mM NaCl, 10 mM MgCl2, 10 mM Tris, pH 7.5) at a concentration of ~2.5mg/ml and then digested with 200 mg/ml of α-chymotrypsin (Bovine Pancreas, Sigma) at 28 °C for 15 min, when the digestion was stopped by cooling the digestion mixtures to 0°C. Further CsCl gradient (20%-50%) purification of intact virion, ISVP and core were performed as described previously (Cheng et al., 2008; Fang et al., 2008; Furlong et al., 1988). ISVP particles collected from gradients were dissolved in10 mM phosphate-buffered saline and checked by negative-stain transmission electron microscopy to confirm the presence of highly purified ISVP.
Confirmation of Intact and Cleaved VP5 by SDS-PAGE and Infectivity Assay at Low Temperature by Western Blotting
To verify that VP5 is uncleaved in the virion and cleaved in the ISVP and to test if any in vitro procedure could result in autocleavage of the VP5 protein during the ISVP preparation, we performed SDS-PAGE of virion and ISVP samples from different bands of the same CsCl gradient after a limited protease digestion (see the above Experimental Procedures) according to methods described previously (Laemmli, 1970; Nibert et al., 2005).
To estimate the level of viral infection at 4°C and 28°C, CIK cells were infected and incubated at these temperatures for 3 and 2 days, respectively. Cells were collected and viral protein levels were either detected by staining with Coomassie brilliant blue R-250 (Sigma) or by Western blotting. In the latter case, the sample was transferred to a polyvinylidene fluoride (PVDF) transfer membrane by a semidry transfer cell following the manufacturer’s instruction (Bio-Rad), and proteins were visualized by Western blotting (Figure 6). The aquareovirus NS80 specific rabbit antisera raised against His-tag fusion protein (C. Fan & Q.F., unpublished data) were employed at a 1:1,000 dilution as primary antibodies. The alkaline phosphatase (Sigma) coupled goat anti-rabbit IgG was used as second antibody at a 1:2000 dilution. Antibody binding was detected with NBT/BCIP AP substrate solution (Promega) according to the manufacturer’s instructions.
Cryo-EM Imaging, Image Processing and Resolution Assessment
An aliquot of 2.5 μl purified ISVP sample was applied to a 400 mesh Quantifoil grid (1.2 μm hole size) and blotted inside an FEI vitrobot using 7 s blotting time with 100% humidity. Cryo-EM images were collected at liquid nitrogen temperature in an FEI Titan Krios cryo-electron microscope operated at 300kV using parallel illumination. Before data collection, the microscope was carefully aligned to reveal visible contrast transfer function (CTF) rings beyond 1/3Å-1 in the Fourier transform of carbon film images recorded under the same low-dose condition. Beam tilt was minimized by the coma-free alignment. Images were recorded on Kodak SO-163 films at a magnification of 59,000x and dosage of ~25 e-/Å2. Micrographs were digitized with Nikon Coolscan 9000ED scanner at a step size of 6.35 μm/pixel, giving a pixel size of 1.075 Å/pixel on specimen. The final pixel size of the density map was calibrated to be 1.1 Å/pixel using Tobacco Mosaic Virus (TMV) as a standard.
From a total of 650 micrographs, we selected 247 for further processing, all with visible CTF rings beyond 1/5.5-Å-1 in their Fourier transforms. A total of 20,473 particles were boxed from these 247 micrographs. The underdefocus values of these micrographs were determined as 0.4 μm to 2.7 μm using CTFFIND (Mindell and Grigorieff, 2003). Initial structure was obtained using IMIRS (Liang et al., 2002), and further alignment and reconstruction were accomplished with Frealign (Grigorieff, 2007) and G3D, a GPU-based reconstruction program (X. Zhang, X.Z. and Z.H.Z., unpublished data). After each cycle, the effective resolution was estimated and used as a resolution cutoff in the next alignment cycle until no further improvement can be obtained. The final map was reconstructed using the top 18,646 of the 20,473 particles based on their phase residuals. The quality of the map of the membrane penetration protein VP5 was further improved by averaging the ten VP5 monomers (one conformer) in each asymmetric unit (Zhang et al., 2008). A reverse B factor was estimated using a trial-and-error method by examining the noise level, backbone density continuity and side-chain densities, and we chose a final B factor of 240 Å2 (conventional definition) for the reconstruction reported here.
The effective resolution of the cryo-EM density map of aquareovirus ISVP was estimated both by the reference-based Fourier shell correlation (Cref=0.5 or FSC=0.143) (Figures 1D) according to the definition of Rosenthal and Henderson (Rosenthal and Henderson, 2003), and by examining structural features that were expected to show up at this resolution (Figures 1E, 2 and Figures S1, S2, and S3). Based on these facts, the effective resolution was estimated as 3.3 Å for the inner shell proteins VP6 and VP3 (Figures 1D and Figures S1 and S2), 3.3 Å for the membrane penetration protein VP5 after an additional 10-fold averaging (Figures 1D, 2A and 2B), and 3.5-Å for the turret protein VP1 (Figure 1D and Figure S3). This radial difference of resolution may be due to the different influence of alignment error at different radii and the different effect of Ewald sphere at different radii. These estimates were consistent with the resolution assessment based on a structural feature-based approach (Zhou, 2008), which compares the resolving power and structural features of our map with X-ray structures determined to similar resolutions (See the Results). The final density map, the source of all of the density maps of individual proteins in this report, was low-pass filtered to 1/3.2 Å-1 with a B factor of 240 Å2 imposed.
Atomic Model Building and Refinement
At 3.3 Å resolution, the cryo-EM density is good enough for ab initio full-atomic model building. Taking advantage of clear bumps for Cα atoms, we first built Cα models using the Baton_build utility in crystallographic program O (Jones et al., 1991) and Coot (Emsley and Cowtan, 2004). Amino-acid registration was accomplished solely based on the clear densities for “landmark” stretches of residues. We then built full-atom models with the help of a computational tool, REMO (Li and Zhang, 2009).
These coarse full-atom models were then refined in a pseudocrystallographic manner using CNS (Brunger et al., 1998). This procedure is used only to improve atomic models, not to modify the 3.3 Å cryo-EM density map. In this method, densities for individual proteins were segmented and put in artificial crystal lattices, and then structure factors were calculated using the utility program em_map_to_hkl.inp in CNS (Brunger et al., 1998). The amplitudes of these structure factors were used as pseudoexperimental diffraction data for model refinement by crystallographic conjugate gradient minimization and simulated annealing refinement in CNS. Model/map fit was further improved by iterative cycles of manual model rebuilding using O/COOT and pseudocrystallographic refinement using CNS. Eventually, we obtained full-atom protein models which could match the cryo-EM density well and also have good geometry. The quality of our models was confirmed by visual check of our models and the density map, and also by the psudocrystallographic R factor/Rfree (Table 1). Protein model geometry was checked by PROCHECK (Laskowski et al., 1993). It is worthy to note that the phases of these structure factors were not used in the CNS refinement, and therefore the future refinement of cryo-EM atomic models should incorporate this phase information as constraints since the phase information of cryo-EM map is more reliable than amplitudes due to deteriorating effects of modulation functions such as CTF on the amplitudes.
Cryo-EM density map was visualized and segmented in Chimera (Pettersen et al., 2004). Figures were prepared using xmgrace (http://plasma-gate.weizmann.ac.il/Grace), Chimera (Pettersen et al., 2004), Molscript (Kraulis, 1991), Pymol (DeLano, 2002), O (Jones et al., 1991) and Signature (Chen and Grigorieff, 2007). Movies were prepared using Chimera (Pettersen et al., 2004).
Supplementary Material
Acknowledgments
We thank Stan Schein for editing our manuscript, Xiaokang Zhang for assistance in programming, Xuekui Yu for advice in sample purification, Hongrong Liu and Lingpeng Cheng for assistance in film development, and Connie Huang for film digitization. This project is supported in part by grants from the National Institutes of Health (NIH; GM071940 and AI069015 to Z.H.Z.) and Abraxis BioScience. We acknowledge the use of the cryo-EM facility at the Electron Imaging Center for NanoMachines supported by NIH (1S10RR23057 to Z.H.Z.). This project is also supported by the National Basic Research Program of China (973 Program, #2009CB118701 to Q.F.), the National Natural Scientific Foundation of China (#30671615 and #30871940 to Q.F.) and the Chinese Academy of Sciences (KSCX2-YW-N-021 to Q.F.).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes three figures and can be found with this article online at doi:10.1016/j.cell.2010.03.041.
ACCESSION NUMBERS
The cryo-EM densities of the aquareovirus ISVP, the averaged membrane penetration protein VP5, and the atomic models were deposited with accession codes EMD-5160 and PDB-3IYL.
References
- Agosto MA, Middleton JK, Freimont EC, Yin J, Nibert ML. Thermolabilizing pseudoreversions in reovirus outer-capsid protein micro 1 rescue the entry defect conferred by a thermostabilizing mutation. J Virol. 2007;81:7400–7409. doi: 10.1128/JVI.02720-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agosto MA, Myers KS, Ivanovic T, Nibert ML. A positive-feedback mechanism promotes reovirus particle conversion to the intermediate associated with membrane penetration. Proc Natl Acad Sci U S A. 2008;105:10571–10576. doi: 10.1073/pnas.0802039105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee M, Johnson JE. Activation, exposure and penetration of virally encoded, membrane-active polypeptides during non-enveloped virus entry. Curr Protein Pept Sci. 2008;9:16–27. doi: 10.2174/138920308783565732. [DOI] [PubMed] [Google Scholar]
- Bartlett GJ, Porter CT, Borkakoti N, Thornton JM. Analysis of catalytic residues in enzyme active sites. J Mol Biol. 2002;324:105–121. doi: 10.1016/s0022-2836(02)01036-7. [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Chandran K, Farsetta DL, Nibert ML. Strategy for nonenveloped virus entry: a hydrophobic conformer of the reovirus membrane penetration protein micro 1 mediates membrane disruption. J Virol. 2002;76:9920–9933. doi: 10.1128/JVI.76.19.9920-9933.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandran K, Nibert ML. Protease cleavage of reovirus capsid protein mu1/mu1C is blocked by alkyl sulfate detergents, yielding a new type of infectious subvirion particle. J Virol. 1998;72:467–475. doi: 10.1128/jvi.72.1.467-475.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandran K, Nibert ML. Animal cell invasion by a large nonenveloped virus: reovirus delivers the goods. Trends Microbiol. 2003;11:374–382. doi: 10.1016/s0966-842x(03)00178-1. [DOI] [PubMed] [Google Scholar]
- Chen JZ, Grigorieff N. SIGNATURE: a single-particle selection system for molecular electron microscopy. J Struct Biol. 2007;157:168–173. doi: 10.1016/j.jsb.2006.06.001. [DOI] [PubMed] [Google Scholar]
- Cheng L, Fang Q, Shah S, Atanasov IC, Zhou ZH. Subnanometer-resolution structures of the grass carp reovirus core and virion. J Mol Biol. 2008;382:213–222. doi: 10.1016/j.jmb.2008.06.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Zhu J, Hui WH, Zhang X, Honig B, Fang Q, Zhou ZH. Backbone model of an aquareovirus virion by cryo-electron microscopy and bioinformatics. J Mol Biol. 2010;397:852–863. doi: 10.1016/j.jmb.2009.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke P, Richardson-Burns SM, DeBiasi RL, Tyler KL. Mechanisms of Apoptosis During Reovirus Infection. Current Topic In Microbiology and Immunology. 2005;289:1–24. doi: 10.1007/3-540-27320-4_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danthi P, Kobayashi T, Holm GH, Hansberger MW, Abel TW, Dermody TS. Reovirus apoptosis and virulence are regulated by host cell membrane penetration efficiency. J Virol. 2008;82:161–172. doi: 10.1128/JVI.01739-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL. The PyMOL User’s Manual. DeLano Scientific; Palo Alto, CA, USA: 2002. [Google Scholar]
- Dowell LG, Rinfret AP. Low-temperature Forms of Ice Studied by X-ray diffraction. Nature. 1960;188:1144–1148. [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Fang Q, Seng EK, Ding QQ, Zhang LL. Characterization of infectious particles of grass carp reovirus by treatment with proteases. Arch Virol. 2008;153:675–682. doi: 10.1007/s00705-008-0048-3. [DOI] [PubMed] [Google Scholar]
- Fang Q, Shah S, Liang Y, Zhou ZH. 3D reconstruction and capsid protein characterization of grass carp reovirus. Sci China C Life Sci. 2005;48:593–600. doi: 10.1360/062004-105. [DOI] [PubMed] [Google Scholar]
- Fritz-Wolf K, Schnyder T, Wallimann T, Kabsch W. Structure of mitochondrial creatine kinase. Nature. 1996;381:341–345. doi: 10.1038/381341a0. [DOI] [PubMed] [Google Scholar]
- Furlong D, Nilbert M, Fields B. Sigma 1 protein of mammalian reoviruses extends from the surfaces of viral particles. Journal of Virology. 1988;62:246–256. doi: 10.1128/jvi.62.1.246-256.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenleaf WB, Shen J, Gai D, Chen XS. Systematic study of the functions for the residues around the nucleotide pocket in simian virus 40 AAA+ hexameric helicase. J Virol. 2008;82:6017–6023. doi: 10.1128/JVI.00387-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigorieff N. FREALIGN: high-resolution refinement of single particle structures. J Struct Biol. 2007;157:117–125. doi: 10.1016/j.jsb.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Grimes JM, Burroughs JN, Gouet P, Diprose JM, Malby R, Zientara S, Mertens PP, Stuart DI. The atomic structure of the bluetongue virus core. Nature. 1998;395:470–478. doi: 10.1038/26694. [DOI] [PubMed] [Google Scholar]
- Ivanovic T, Agosto MA, Zhang L, Chandran K, Harrison SC, Nibert ML. Peptides released from reovirus outer capsid form membrane pores that recruit virus particles. Embo J. 2008;27:1289–1298. doi: 10.1038/emboj.2008.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Baker ML, Jakana J, Weigele PR, King J, Chiu W. Backbone structure of the infectious epsilon15 virus capsid revealed by electron cryomicroscopy. Nature. 2008;451:1130–1134. doi: 10.1038/nature06665. [DOI] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Kraulis PJ. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- Li Y, Zhang Y. REMO: A new protocol to refine full atomic protein models from C-alpha traces by optimizing hydrogen-bonding networks. Proteins. 2009;76:665–676. doi: 10.1002/prot.22380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Ke EY, Zhou ZH. IMIRS: a high-resolution 3D reconstruction package integrated with a relational image database. J Struct Biol. 2002;137:292–304. doi: 10.1016/s1047-8477(02)00014-x. [DOI] [PubMed] [Google Scholar]
- Liemann S, Chandran K, Baker TS, Nibert ML, Harrison SC. Structure of the reovirus membrane-penetration protein, Mu1, in a complex with is protector protein, Sigma3. Cell. 2002;108:283–295. doi: 10.1016/s0092-8674(02)00612-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locher KP, Lee AT, Rees DC. The E. coli BtuCD structure: a framework for ABC transporter architecture and mechanism. Science. 2002;296:1091–1098. doi: 10.1126/science.1071142. [DOI] [PubMed] [Google Scholar]
- Middleton JK, Agosto MA, Severson TF, Yin J, Nibert ML. Thermostabilizing mutations in reovirus outer-capsid protein mu1 selected by heat inactivation of infectious subvirion particles. Virology. 2007;361:412–425. doi: 10.1016/j.virol.2006.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- Nason EL, Samal SK, Prasad BV. Trypsin-induced structural transformation in aquareovirus. J Virol. 2000;74:6546–6555. doi: 10.1128/jvi.74.14.6546-6555.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nibert ML, Fields BN. A carboxy-terminal fragment of protein mu 1/mu 1C is present in infectious subvirion particles of mammalian reoviruses and is proposed to have a role in penetration. J Virol. 1992;66:6408–6418. doi: 10.1128/jvi.66.11.6408-6418.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nibert ML, Furlong DB, Fields BN. Mechanisms of viral pathogenesis. Distinct forms of reoviruses and their roles during replication in cells and host. J Clin Invest. 1991a;88:727–734. doi: 10.1172/JCI115369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nibert ML, Odegard AL, Agosto MA, Chandran K, Schiff LA. Putative autocleavage of reovirus mu1 protein in concert with outer-capsid disassembly and activation for membrane permeabilization. J Mol Biol. 2005;345:461–474. doi: 10.1016/j.jmb.2004.10.026. [DOI] [PubMed] [Google Scholar]
- Nibert ML, Schiff LA, Fields BN. Mammalian reoviruses contain a myristoylated structural protein. J Virol. 1991b;65:1960–1967. doi: 10.1128/jvi.65.4.1960-1967.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noad L, Shou J, Coombs KM, Duncan R. Sequences of avian reovirus M1, M2 and M3 genes and predicted structure/function of the encoded mu proteins. Virus Res. 2006;116:45–57. doi: 10.1016/j.virusres.2005.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odegard AL, Chandran K, Zhang X, Parker JS, Baker TS, Nibert ML. Putative autocleavage of outer capsid protein micro1, allowing release of myristoylated peptide micro1N during particle uncoating, is critical for cell entry by reovirus. J Virol. 2004;78:8732–8745. doi: 10.1128/JVI.78.16.8732-8745.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Reddy V, Schneemann A, Johnson JE. Nodavirus Endopeptidase. (2) 2004;1 Handbook of Proteolytic Enzymes. [Google Scholar]
- Reinisch KM, Nibert ML, Harrison SC. Structure of the reovirus core at 3.6 Å resolution. Nature. 2000;404:960–967. doi: 10.1038/35010041. [DOI] [PubMed] [Google Scholar]
- Rivas C, Noya M, Cepeda C, Bandin I, Barja JL, Dopazo CP. Replication and morphogenesis of the turbot aquareovirus (TRV) in cell culture. Aquaculture. 1998;160:47–62. [Google Scholar]
- Rosenthal PB, Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol. 2003;333:721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- Schluenzen F, Tocilj A, Zarivach R, Harms J, Gluehmann M, Janell D, Bashan A, Bartels H, Agmon I, Franceschi F, Yonath A. Structure of functionally activated small ribosomal subunit at 3.3 angstroms resolution. Cell. 2000;102:615–623. doi: 10.1016/s0092-8674(00)00084-2. [DOI] [PubMed] [Google Scholar]
- Shing M, Coombs KM. Assembly of the reovirus outer capsid requires mu 1/sigma 3 interactions which are prevented by misfolded sigma 3 protein in temperature-sensitive mutant tsG453. Virus Res. 1996;46:19–29. doi: 10.1016/s0168-1702(96)01372-x. [DOI] [PubMed] [Google Scholar]
- Taylor DJ, Johnson JE. Folding and particle assembly are disrupted by single-point mutations near the autocatalytic cleavage site of Nudaurelia capensis omega virus capsid protein. Protein Sci. 2005;14:401–408. doi: 10.1110/ps.041054605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillotson L, Shatkin AJ. Reovirus polypeptide sigma 3 and N-terminal myristoylation of polypeptide mu 1 are required for site-specific cleavage to mu 1C in transfected cells. J Virol. 1992;66:2180–2186. doi: 10.1128/jvi.66.4.2180-2186.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towler DA, Gordon JI, Adams SP, Glaser L. The biology and enzymology of eukaryotic protein acylation. Annu Rev Biochem. 1988;57:69–99. doi: 10.1146/annurev.bi.57.070188.000441. [DOI] [PubMed] [Google Scholar]
- Tsai B. Penetration of nonenveloped viruses into the cytoplasm. Annu Rev Cell Dev Biol. 2007;23:23–43. doi: 10.1146/annurev.cellbio.23.090506.123454. [DOI] [PubMed] [Google Scholar]
- White JM, Delos SE, Brecher M, Schornberg K. Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit Rev Biochem Mol Biol. 2008;43:189–219. doi: 10.1080/10409230802058320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynne SA, Crowther RA, Leslie AG. The crystal structure of the human hepatitis B virus capsid. Mol Cell. 1999;3:771–780. doi: 10.1016/s1097-2765(01)80009-5. [DOI] [PubMed] [Google Scholar]
- Yu X, Jin L, Zhou ZH. 3.88 A structure of cytoplasmic polyhedrosis virus by cryo-electron microscopy. Nature. 2008;453:415–419. doi: 10.1038/nature06893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Chandran K, Nibert ML, Harrison SC. Reovirus mu1 structural rearrangements that mediate membrane penetration. J Virol. 2006;80:12367–12376. doi: 10.1128/JVI.01343-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Ji Y, Zhang L, Harrison SC, Marinescu DC, Nibert ML, Baker TS. Features of reovirus outer capsid protein mu1 revealed by electron cryomicroscopy and image reconstruction of the virion at 7.0 Angstrom resolution. Structure. 2005a;13:1545–1557. doi: 10.1016/j.str.2005.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Settembre E, Xu C, Dormitzer PR, Bellamy R, Harrison SC, Grigorieff N. Near-atomic resolution using electron cryomicroscopy and single-particle reconstruction. Proc Natl Acad Sci U S A. 2008;105:1867–1872. doi: 10.1073/pnas.0711623105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Tang J, Walker SB, O’Hara D, Nibert ML, Duncan R, Baker TS. Structure of avian orthoreovirus virion by electron cryomicroscopy and image reconstruction. Virology. 2005b;343:25–35. doi: 10.1016/j.virol.2005.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou ZH. Towards atomic resolution structural determination by single-particle cryo-electron microscopy. Curr Opin Struct Biol. 2008;18:218–228. doi: 10.1016/j.sbi.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


