Abstract
Dendritic spines, the bulbous protrusions that form the postsynaptic half of excitatory synapses, are one of the most prominent features of neurons and have been imaged and studied for over a century. In that time, changes in the number and morphology of dendritic spines have been correlated to the developmental process as well as the pathophysiology of a number of neurodegenerative diseases. Due to the sheer scale of synaptic connectivity in the brain, work to date has merely scratched the surface in the study of normal spine function and pathology. This review will highlight traditional approaches to the imaging of dendritic spines and newer approaches made possible by advances in microscopy, protein engineering, and image analysis. The review will also describe recent work that is leading researchers toward the possibility of a systematic and comprehensive study of spine anatomy throughout the brain.
Keywords: Microscopy, Image Analysis, Dendritic spines, Two-photon, Informatics, Transgenic mice
Introduction
Throughout the nervous system, dynamic changes in the number and structure of synapses are a hallmark of normal development and aging (Engert and Bonhoeffer, 1999, Maletic-Savatic et al., 1999, Bosch and Hayashi, 2011). Dendritic spines, named by Ramon y Cajal after ‘espinas’ or thorns in his native Spanish, are post-synaptic protuberances abundant in glutamate receptors (GluR) found primarily at excitatory synapses directly opposed to a presynaptic bouton. Composed of a round spine head and a thinner spine neck, dendritic spines serve as the point of contact between two neurons with an increased concentration of postsynaptic signaling components such as GluR (De Paola et al., 2006). Due to their unique size and shape, they provide essential sites for local signal integration and molecular compartmentalization, isolating rapid changes in local second messenger pathways particularly calcium (Shepherd, 1996, Yuste et al., 2000, Yuste and Bonhoeffer, 2001, Alvarez and Sabatini, 2007).
Because they are easily observable features of neuronal morphology, dendritic spines have been stained and imaged in fixed brain tissue for over a century. The availability of fixed brain samples from human patients of neurological disorders has allowed investigators to observe correlations between dendritic spine density and morphology with disease progression. Additionally, the number and health of dendritic spines is correlated to the health and functionality of synapses; in many neuronal subtypes, such as hippocampal pyramidal cells, there is a one-to-one correlation between spine number and synapse number (Nimchinsky et al., 2004, Alvarez and Sabatini, 2007). The use of animal models has allowed for the observation of dendritic spines throughout development and their response to disease treatments. More recently, the coupling of live tissue imaging, particularly with the advent of two-photon laser scanning microscopy, with electrophysiology has led to studies which examine the physiology of individual dendritic spines and correlate changes in their size and structure to changes in physiology (Kasai et al., 2010).
The gain, loss, and morphological remodeling of dendritic spines are normal processes in development, as well as in learning and memory (Engert and Bonhoeffer, 1999, Maletic-Savatic et al., 1999, Bosch and Hayashi, 2011). Spine dynamics, including spine turnover and changes in spine shape and motility, are vital for the development and function of neural circuits (Calabrese et al., 2006). Though most spines are stable over a long imaging period, a proportion of spines transiently appear and disappear and these synaptic changes, driven by novel sensory experience, underline experience-dependent remodeling of specific neuronal circuits (Knott and Holtmaat, 2008). In the accepted model of neuronal development, a postsynaptic neuron projects numerous small, thin filapodia which sample local synaptic inputs (Bhatt et al., 2009). Filipodia that receive sufficient synaptic input, mature into dendritic spines while those that do not may be pruned. More recently, it has been demonstrated that in adulthood changes in the size and shape of dendritic spines correlate with plasticity at individual synapses (Matsuzaki et al., 2004).
Distortions in the normal patterns of neuronal signaling caused by disease or substance abuse can manifest as changes in dendritic spine density and morphology, accompanied by corresponding functional changes (reviewed extensively in this issue). Dendritic spine abnormalities have been found in many pathological conditions and excitatory synapse loss, which can be observed in individual neurons as a reduction in spine density, is strongly related to cognitive impairment in neurodegenerative diseases such as Alzheimer’s or Rett’s Syndrome (Moolman et al., 2004, Bittner et al., 2010). Reduced spine density of 27% and a decrease in total dendritic length and complexity have also been observed in medium spiny neurons of the caudate nucleus and the putamen from Golgi-stained postmortem tissue of Parkinson’s patients (Stephens et al., 2005), indicating that synaptic dysfunction and degeneration is likely a common manifestation of neuronal insult and disease resulting from a range of unrelated causes. Strikingly, treatments that alleviate the cognitive symptoms of neurodegenerative disease also have been shown to reverse their respective spine pathologies (Smith et al., 2009). The specifics of the dendritic spine pathologies for a number of conditions are covered extensively in this issue.
The number, morphology, and dynamics of healthy synapses throughout the brain are excellent indicators of neuronal development and function and can provide investigators with information about specific brain regions and neuronal subtypes. Optical imaging of dendritic spines provides an attractive diagnostic tool for studies of synaptic health in clinical (post-mortem human tissues) and basic research studies (animal models of disease). A true understanding of the effects of disease states on synaptic structure on a systems level requires a comprehensive evaluation of changes in spine density and morphology throughout the entire brain. This presents a rather daunting challenge for optical image acquisition and analysis on a number of levels. First, the study of neurons throughout the entire anatomy of the brain requires sample preparation and microscopy techniques sufficient to preserve submicron imaging resolution for all neuronal subtypes in all brain regions. Second, the enormous number of neurons in the brain (on the order of 1011) and the incomprehensible level of connectivity between them (on the order of 1014 synaptic connections with ~90% of these terminating in spines) taxes the limits of our abilities to gather and analyze data (Williams and Herrup, 1988, Nimchinsky et al., 2004). As current high resolution imaging techniques allow for the collection of massive of amounts of three dimensional anatomical data from individual specimens, informatics becomes the rate limiting factor, i.e., the bottleneck in these studies has become accurate image analysis and classification of anatomical structures. Manual spine classification or computer assisted manual spine classification, long the standard approaches, are extremely labor intensive and unacceptably slow as well as subject to investigator variability and poor for 3D image analysis. Recently, a great deal of progress has been made toward the development of software tools that can accurately analyze dendritic structure and detect all classes of dendritic spines reproducibly using objective criteria (Rodriguez et al., 2006, Rodriguez et al., 2008, Fan et al., 2009, Li et al., 2010, Zhang et al., 2010, Son et al., 2011). Although to the human eye, the criteria that determine just what is a dendritic spine seem obvious, defining these criteria in an objective manner has been a challenge in algorithm development.
This review will recount traditional tissue staining and microscopy approaches that have been and remain invaluable in the post-mortem analysis of human neurological disease progression. Additionally, we will touch on more recent genetic tools and microscopy advances that have enabled researchers to study spine dynamics and correlate spine structure to neuronal physiology in animal models. Finally, we will discuss roadblocks in our study of dendritic spine pathology and future technical solutions that may soon address them.
Historical context of Golgi staining
Visualization of dendritic spines can be traced back to the drawings of Camillo Golgi, based on his observation of fixed brain tissues stained by the so-called black reaction, a staining technique invented by Golgi in his kitchen. Santiago Ramón y Cajal, the founder of Neuron theory, which states that the nervous system is composed of discrete individual cells which together form the complete neural network, acutely sensed the value of this technique and extensively used it in his work. It was Cajal who first documented dendritic spines in 1888 and demonstrated the reality of what he called collateral spines in 1895 (Garcia-Lopez et al., 2007). In addition, Cajal deliberately recorded the different morphologies of dendritic spines (sessile, mushroom, and thin), the different sizes among different brain areas and species, and distribution characteristics and even hypothesized the physiological function of this structure (Garcia-Lopez et al., 2007). All of these amazing achievements are due to the assistance of the best microscope at the time and his delicate manipulation of Golgi staining. The Nobel Prize in Physiology or Medicine 1906 was awarded jointly to Camillo Golgi and Santiago Ramón y Cajal "in recognition of their work on the structure of the nervous system."
Principle and procedures of Golgi staining to detect dendritic spines
Golgi staining is a progressive process involving formation of small, dense granules precipitated inside the nerve cells, leaving the nucleus and mitochondria unstained. In the process of impregnation, the silver or mercury chromate granules accumulate and gradually cover the surface of the nerve cell (Fairen, 2005). The basic procedures of Golgi staining include the exposure of brain tissue to dichromate and impregnation with heavy metal ions (silver or mercury). Only a few cells in the tissue can be clearly impregnated at the soma and the neurites on a transparent background of unstained structure, an important factor that makes individual spines detectable. Since the original Golgi staining was very time-consuming, two main modified protocols were developed, (i) Rapid Golgi staining. Brain tissue is fixed in an aldehyde-containing solution (Pilati et al., 2008), with relatively shorter impregnation and chromation duration. Several methodological modifications are made to improve the quality and reaction time (Raju et al., 2004, Pilati et al., 2008, Ranjan and Mallick, 2010). (ii) Golgi-Cox method. Brain tissues are immersed in solutions containing mercuric chloride and brain sections are treated with sodium carbonate or ammonia solution. Performing the impregnation reaction at 37°C can shorten the notoriously slow process (Ranjan and Mallick, 2010). Both methods are suitable for freshly prepared or lightly fixed tissues (Rosoklija et al., 2003). Melendez-Ferro et al. successfully applied the Golgi-Cox method to stain long-term frozen animal brain tissue and human brain tissue stored for up to 15 years in a freezer (Melendez-Ferro et al., 2009). Golgi-Cox provides cleaner, more consistent three-dimensional reconstructions after confocal imaging and is therefore more reliable to demonstrate the dendritic arborization of mammalian neurons, (Castano et al., 1995, Raju et al., 2004). However, for staining tissues that have been fixed in formalin for years, rapid Golgi and other modified Golgi methods such as Golgi-Kopsch are better choices than Golgi-Cox (Rosoklija et al., 2003, Melendez-Ferro et al., 2009).
The advantages of this traditional histological technique include a complete morphology of the entire neuron, a clear picture with minimal background, and that chromogen-stained dendritic spines are relatively stable (Pannese, 1999, Ranjan and Mallick, 2010). However, this method has certain inevitable disadvantages such as, random and unpredictable cell staining and partial morphological information in brain areas (Pilati et al., 2008), although the Golgi-Kopsch technique successfully improved the quality of impregnation on long-term fixed Homo sapiens tissue (Rosoklija et al., 2003). In addition, Golgi staining and widefield microscopy greatly underestimates the number of spines present on a given stretch of dendrite, presumably due to the lack of z-plane resolution and the small volume of spines relative to the parent dendrite (Chan-Palay et al., 1974, Feldman and Dowd, 1975, Harris and Stevens, 1988). Spine detection from serial analysis of electron microscopy sections shows that this underestimation can be as high as 3-fold in moderately spiny neurons such as CA1 hippocampal pyramidal cells and even higher in more spiny neurons such as cerebellar Purkinje cells (Harris and Stevens, 1988). Computational techniques have been in use for many years that attempt to compensate for the low detection rate by applying a formula based on the geometry of the parent dendrite, which give a more realistic estimate of the true spine density (Feldman and Peters, 1979, Horner and Arbuthnott, 1991). None of this discounts the value of the wealth of knowledge about synaptic degeneration gained in past and continuing Golgi experiments as the studies are most valuable for their relative, rather than absolute, spine numbers.
Current techniques for staining and imaging dendrite spines in the fixed brain tissues
Recent application of Golgi staining in fixed tissue revealed a loss of dendritic spines of pyramidal neurons in the brain from Creutzfeldt-Jakob disease patients (Landis et al., 1981), decreased density of dendritic arborization on the cerebellar and visual cortices of AD patients (Mavroudis et al., 2010, Mavroudis et al., 2011), abnormal neuroplasticity in specific brain area of gene-knockout and stress-induced social defeat animal model (Nietzer et al., 2011) and in depressed suicides (Hercher et al., 2010). Therefore, this method is still one of the most reliable histological techniques in the morphological evaluation of dendritic spines (Figure 1) in specific brain areas (Couch et al., 2010) and in the detection of the early dendritic pathology for a variety of neurodegenerative diseases (Melendez-Ferro et al., 2009, Mavroudis et al., 2011). More recent advances in tissue staining and microscopy have allowed researchers to compliment the valuable anatomical data gleaned from Golgi staining of postmortem human tissue with additional anatomical and physiological data, both in human tissue and in animal models of human disease.
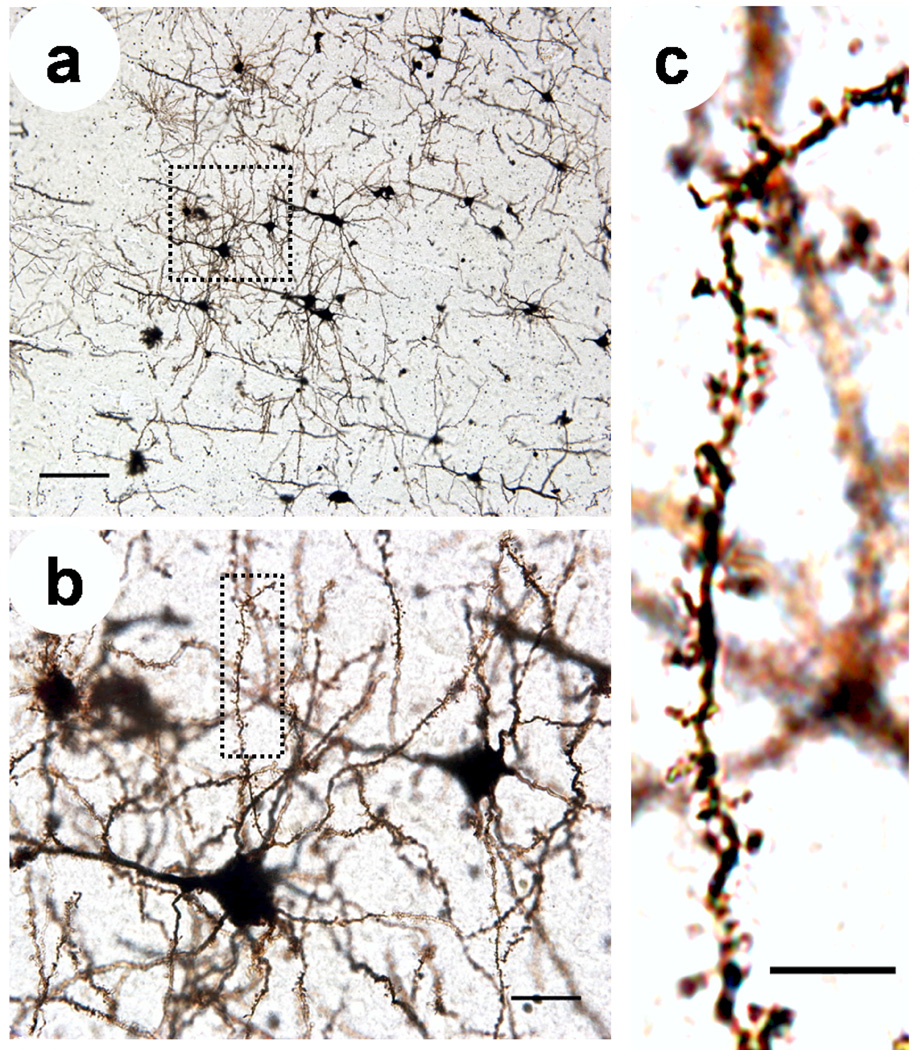
Figure 1.
Photomicrographs of a Golgi-stained mouse cortical neurons from slices, (a) Neurons in brain slices are randomly and sporadically labeled allowing visualization of individual neurons. Scale = 50 µm. (b) Enlargement of boxed area in (a) showing dendrites and spines belonging to individual neurons. Scale = 10 µm. (c) Enlargement of boxed area in (b) showing individual dendritic spines which can be counted and analyzed. Scale = 5 µm. Figure 2
Conventional brightfield microscopy is sufficient to image neurons and study the basic morphology and relative density of spines in thin slices (Perez-Costas et al., 2007) but provides insufficient resolution, particularly in the z-axis, for studies of subtle changes in spine morphology. This becomes even more relevant when studying neuronal populations that have been labeled more efficiently than is possible with the Golgi method. Widefield fluorescence provides better resolution than brightfield, but images are blurred because emitted fluorescence is detected not only from in focus photons but also from molecules excited out of the focal plane. Confocal microscopy solves this problem by using a pinhole to selectively collect emission from the focal point while excluding most light emitted out of the focal plane (Conchello and Lichtman, 2005, Oheim et al., 2006).
The emergence and widespread availability of laser scanning confocal microscopy (LCSM) has given researchers and neuroscientists a powerful tool to image changes in spine density and plasticity in brain slices and cultured hippocampal neurons with nearly diffraction-limited resolution (Moser et al., 1994, Papa et al., 1995). LSCM has been principally used to image fluorescently labeled structures and Golgi staining was not considered suitable for LCSM observations (Castano et al., 1995). More recently, a creative modification of the technique takes advantage of the reflective nature of the dense metal particles used in Golgi-impregnation. This reflection method involves selective collection of excitation light reflected from a focal point through the confocal pinhole and has been used to reconstruct high-resolution 3D structure of Golgi-Cox impregnated material (Tredici et al., 1993, Spiga et al., 2011). This additional resolution gained by this technique makes visible not only additional spines but additional parent dendrites projecting in the z-plane. Additionally, combining Golgi-Cox impregnation with immunocytochemical procedures, allows simultaneous 3D confocal visualization of both morphological and neurochemical features of neurons (Lanciego and Wouterlood, 2011, Spiga et al., 2011).
More commonly, LSCM has been paired with intracellular staining by fluorescent dyes to generate high-resolution 3-D reconstructions of neuronal anatomy. Manual injection of Lucifer yellow or other fluorescent dyes to fixed or live tissue can be used to completely label entire neurons one at time allowing 3-D confocal imaging of unambiguous neuronal structures with a detection efficiency 3x that of the traditional Golgi stain and comparable to electron microscopy studies (Buhl and Lubke, 1989, Vecellio et al., 2000, Wallace and Bear, 2004). Higher throughput cell-labeling can be obtained by application of lipophilic dyes such as DiO, Dil, and DiD (Chen et al., 2011), endocytotic dyes such as FM143 (Dhawale and Bhalla, 2008), or bioenzyme-based markers (Ryan, 2001, Couch et al., 2010, Anderson et al., 2011).
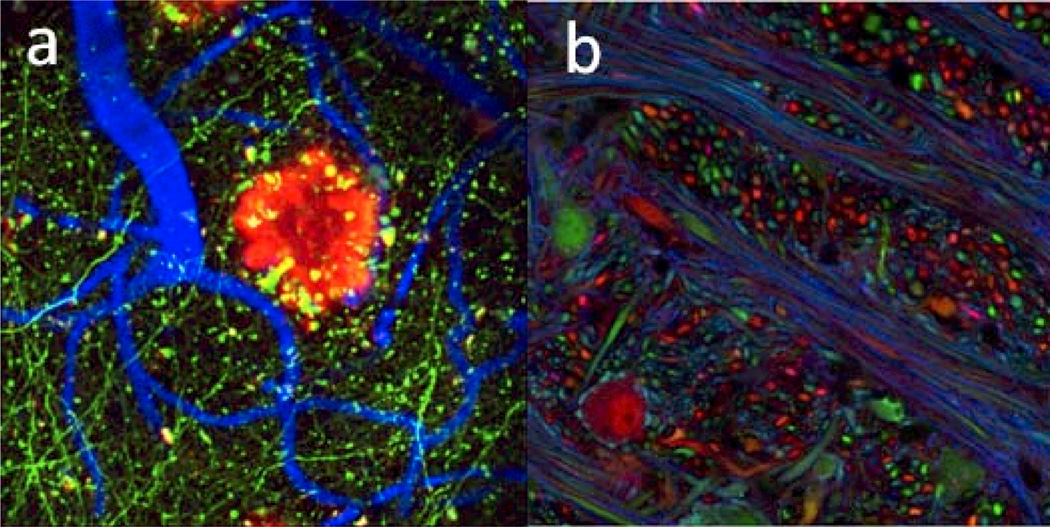
Transgenic strategies and viral expression have been used to label living neurons from animal models with fluorescent proteins in vivo, employing a variety of strategies for specific labeling of neurons of interest (Figure 2). One example is the creation of numerous lines of Thyl-YFP mice in which certain groups of neurons express YFP based on the integration site of the transgene, making possible distinction of the dendrites and spines of individual neurons from those of their unlabeled neighbors (Feng et al., 2000). Another example is labeling of individual neurons with multi-color fluorescence, based on random recombination of Cre-based transgenes for a variety of fluorescent proteins, called the Brainbow technique (Gong et al., 2003, Livet et al., 2007). Interestingly, it was only when transgenic animals that expressed fluorescent proteins sporadically were created, that this technique began to approach the usefulness of the simple Golgi stain. A number of viral vectors have been used to express fluorescent proteins in neurons for imaging including recombinant adeno-associated virus, lentivirus, and rabies virus (Chamberlin et al., 1998, Ehrengruber et al., 2001). Expression of fluorescent proteins in neurons with modified rabies virus allows investigators to image pairs of neurons known to possess direct synaptic connections through exclusively retrograde transmission of the virus (Ugolini, 2010) and when combined with anterograde tracers, which allow investigators to trace at least two layers of connectivity (Lopez et al., 2010). These techniques are valuable tools for the study of anatomical mapping, and each has specific advantages in the detection of circuit-based dendritic architecture, including spines.
Figure 2.
Transgenic labeling of neurons in vivo, (a) Cortical neurons bright express GFP allowing spine imaging and counterstaining of vasculature and Aβ plaques (data courtesy of Brad Hyman, Massachusetts General Hospital), (b) Brainbow mice display multicolor labeling allowing for tracing of processes from individual neurons over long distances (data courtesy of Jeff Lichtman, Harvard University).
The recent development of Sca/e technology to generate clear fixed biological samples in Miyawaki’s group generates a fast, simple, and inexpensive approach for optical sectioning for deep imaging and three-dimensional reconstruction of fluorescently labeled structures at subcellular resolution with minimal distortion of brain anatomy (Hama et al., 2011). Transgenic mice expressing fluorescent proteins in genetically defined neuronal populations (Feng et al., 2000, Gong et al., 2003, Livet et al., 2007) provide an ideal substrate for Sca/e. Using this tissue clearing technique, the fluorescent signal of individual dendritic spines can be discerned by multiphoton microscopy at a depth of 0.9mm under the pial surface. Furthermore, the development of a long focal length, high-NA objective (NA = 1.0, working distance = 4.0 mm) opens the possibility of spine imaging throughout the entire volume of the intact mouse brain, but work still remains to determine whether the technique causes distortions in microanatomical structures such as spines.
The morphology and protein content of dendritic spines from fixed human and animal tissues can be visualized in sections by immunofluorescence staining of synaptic proteins, for example, synaptophysin and synapsin I (presynaptic marker), PSD-95 (postsynaptic marker), or other pathological event-relevant markers (Perez-Costas et al., 2007). Such correlation of protein content and morphology is invaluable in identifying populations of spines, particularly in disease models where they may be differentially affected. Unfortunately, the differential penetration of antibodies into sections impacts immunostaining results, severely limiting its use in systematic studies of dendritic spine anatomy.
Array tomography (AT) involves immunostaining and imaging of a series of ultrathin (70–200 nm) sections using conventional fluorescence microscopy. The fluorescent images from the same region of each section are aligned and reconstructed as a 3D stack to get a precise map of antigen distribution. This powerful technique is suited for the quantitative analysis of dendritic architecture. Indeed, AT has been used to analyze glutamatergic and GABAergic synapses in mouse cortex (Micheva et al., 2010) and the glutamtergic innervations of mouse dorsal raphe nucleus (Soiza-Reilly and Commons, 2010). The advantages of AT include: (1) higher axial resolution than conventional optical imaging techniques, 70–200 nm based on slice thickness versus > 1 µm for CLSM and multiphoton, (2) multiple antigens can be detected and spatially co-localized in the same sample, (3) strippable immunofluorescence and re-immunolabeling, (4) no issue of antibody penetration, (5) reliable quantification of synapses (91% relative to EM) (Micheva et al., 2010), and (6) suitability for the studies on human clinical specimens and animal disease-model. However, this technique requires laborious sample preparation and data reconstruction, is extremely time and energy-consuming and has thus only been applied to small sample volumes.
Functional imaging of dendritic spines
A revelation of the role of dendritic spines in neuronal physiology requires functional imaging of dendritic spines in living neurons and a great deal has been learned about the functional properties of dendritic spines from work performed in neuronal culture and acute brain slices prepared from animals. Along with the development of fluorescent proteins and dyes for structural imaging has been the development of fluorescent indicators for cellular physiology. For the study of dendritic spine physiology, the most notable of these has been calcium indicator dyes. Based either on a fluorescent dye (Grynkiewicz et al., 1985, Minta et al., 1989) or protein (Nakai et al., 2001, Yu et al., 2003, Pologruto et al., 2004, Tian et al., 2009) conjugated to a calcium buffer, the indicators change their fluorescence spectra or emission intensity when bound to calcium. The coupling of confocal microscopy of fluorescent proteins and calcium indicator dyes with brain slice electrophysiology has provided a wealth of information about the role of dendritic spines in synaptic signaling and plasticity (Koester and Sakmann, 1998, Engert and Bonhoeffer, 1999, Maletic-Savatic et al., 1999, Bloodgood and Sabatini, 2005). The introduction of two-photon uncaging and imaging to brain slice electrophysiology has allowed for the direct correlation of changes in spine size with changes in synaptic strength on the single spine level (Matsuzaki et al., 2004) and revealed the dynamic nature of spine size even in the absence of synaptic activity (Yasumatsu et al., 2008).
Because fixed tissue imaging only provides a snapshot neuronal anatomy, it does not provide real spine dynamical information. An observed change in spine density cannot be attributed to a change in spine formation or pruning. Neurons imaged in culture or in brain slice preparations lack intact synaptic inputs and sensory information and therefore cannot fully address the relationship of spine plasticity to brain development and learning and memory. Directly imaging changes in individual dendritic spines in the living, intact brain addresses these points, allowing researchers to evaluate the importance of dendritic spine dynamics to the functional reorganization of neuronal circuits (Knott and Holtmaat, 2008).
The most glaring roadblock to in vivo dendritic spine imaging is that the opacity of the intact skull precludes optical imaging of neocortical neurons. Though a craniotomy window can be made to expose the cortical surface, scattering and absorption in brain tissue still restrict the use of visible excitation light to track dendritic spines to the most superficial surfaces of the cortex (Conchello and Lichtman, 2005). The strong scattering and absorption of ultraviolet (UV) and visible light used for confocal microscopy in the tissue limit optical resolution (axis resolution) and imaging depth (Denk and Svoboda, 1997). In addition, the long detection light pathway increases the loss of fluorescence. That signal loss is unacceptable for imaging of deeper brain areas (Conchello and Lichtman, 2005). Also, the use of high power visible light results in photo-bleaching and photo-damage that destroy fluorophores and shorten the duration of time lapse imaging (Oheim et al., 2006). Taken together, confocal microscopy is limited to high-resolution three-dimensional (3-D) images of spine morphology in superficial areas, tissue culture, or thin specimens (Moser et al., 1994, Papa et al., 1995).
Two-photon microscopy, based on excitation of fluorophores by the near-simultaneous absorption of two or more infrared photons, restricts the fluorescence excitation volume and can reduce photo-damage and photo-bleaching (Denk et al., 1990, Patterson and Piston, 2000, Helmchen and Denk, 2005, Oheim et al., 2006). Compared with confocal microscopy, which only collects the emitted fluorescence through the pinhole, most emission light can be collected (even scattered emission) in non-descanned mode which enhances the efficiency of fluorescence collection. Because only the fluorophore in the focal plane is excited, bleaching and photo-damage are greatly reduced (Beaurepaire et al., 2001, Oheim et al., 2006). Furthermore, the near IR excitation wavelengths used in multi-photon microscopy penetrate deeper into tissue with less light scattering and absorption, which allow for high resolution in vivo imaging as deep as 0.5 to 0.8 mm beneath the brain surface (Theer et al., 2003, Oheim et al., 2006).
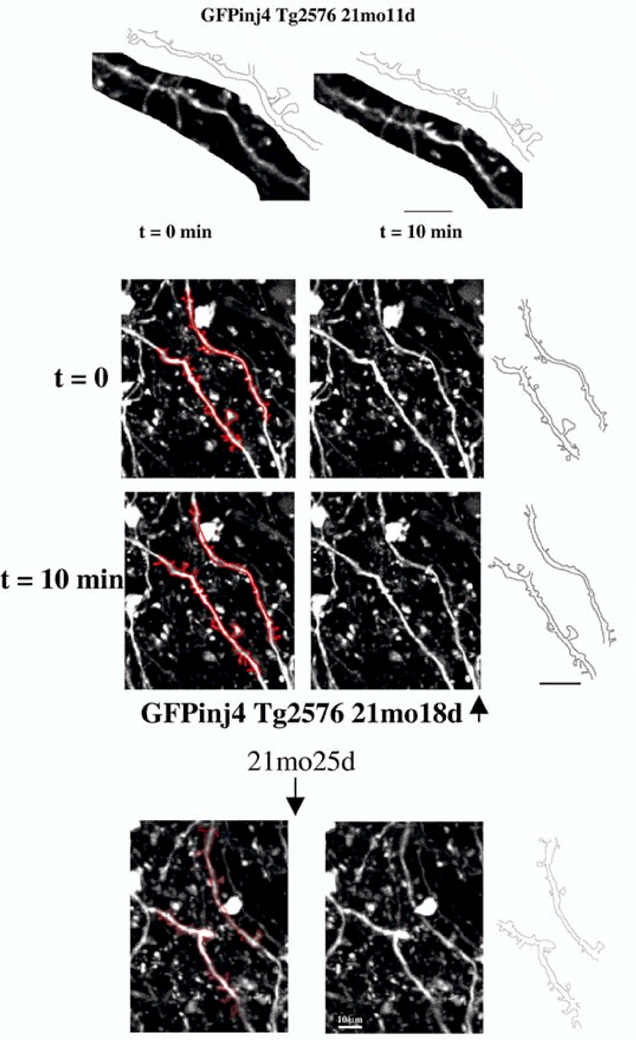
Two-photon imaging and transgenic targeting of fluorescent proteins to specific cell-types have been crucial to study the structure and dynamics of live neurons (Holtmaat and Svoboda, 2009, Wilt et al., 2009), for example, in vivo imaging studies, which can simultaneously observe dendrite growth and synaptogenesis, indicate that synapse formation can direct dendritic aborization (Niell et al., 2004). Observation of neuronal development has revealed a population of transient spines that appear and disappear over days and a population of persistent spines that increase gradually until adulthood (Holtmaat et al., 2005). Recent in vivo time-lapse imaging studies of astrocyte and dendritic spine activity indicate that astrocytic protrusive activity regulates the stabilization of individual dendritic protrusions and spine maturation (Nishida and Okabe, 2007). The advent of two-photon microscopy and the specific transgenic targeting of fluorescent proteins has given rise to a substantial number of in vivo studies imaging dendritic spine dynamics in the brain over long time periods (Figure 3) (Gray et al., 2006, Svoboda and Yasuda, 2006, Spires-Jones et al., 2007).
Figure 3.
Time lapse two-photon imaging of the changes in neurites and dendritic spines in a live mouse model of Alzheimer’s Disease during a therapeutic treatment on Aβ plaques. (Data courtesy of Brad Hyman and Tara Spires, MGH)
Still, high resolution optical imaging studies are restricted to the superficial layers of the cortex in live animals due to light scattering and absorption while many neurodegenerative disorders are correlated with changes in spine dynamics in deeper brain structures such as hippocampus. Even with the development of phytochrome-based near-infrared fluorescent proteins (Shu et al., 2009, Filonov et al., 2011), which reduce scattering and absorption (Misgeld and Kerschensteiner, 2006, Bittner et al., 2010), many brain areas are inaccessible to conventional optical microscopy in live subjects.
Micro-endoscopy microscopy
Optical microendoscopy, one promising approach to imaging deeper brain areas such as hippocampus, is accomplished by inserting an optical micro-probe directly into a specifically targeted deep structure (Jung and Schnitzer, 2003, Jung et al., 2004, Levene et al., 2004). A focal depth of more than 1 cm into the brain can be achieved by adjusting the position of the microscope objective lens relative to the microendoscope probe. Because high-resolution micro lenses provide the micron-scale resolution of a conventional water immersion objective during direct insertion into tissue (Jung et al., 2004, Levene et al., 2004, Barretto et al., 2009), imaging of spine dynamics beneath the penetration depth of conventional light microscopy is possible (Flusberg et al., 2008). Currently, sub-cellular microendoscopy has been used to track CA1 hippocampus pyramidal neuron and dendrite dynamics in adult mice over the course of weeks and indicate that the change of dendrite stability in diseased hippocampus might be a hot spot in future studies (Barretto et al., 2009, Barretto et al., 2011).
Spine identification and characterization methods
As current high resolution imaging techniques allow for the collection of massive amounts of three dimensional anatomical data or data series from individual specimens, the bottleneck in these studies has become accurate quantification and classification of anatomical structures. Manual spine segmentation or computer assisted manual spine classification, long the standard approaches, are unacceptably slow for a task of this magnitude, as well as subject to investigator variability and poor for 3D and 4D image analysis. Additionally, manual classification of dendritic spines results in an unnatural grouping of observed spines into subjectively defined groups when, in fact, dendritic spines display a continuum of shapes and sizes. It is also tedious work to correlate spines temporally in time lapse datasets. Because different pathological conditions have such varying effects on spine morphology, the criteria for these groups can change dramatically from one experiment to the next. To these ends automated spine detection and characterization algorithms have three long-term goals:
Remove subjective bias and error from spine counting.
Quantify spines based on objectively defined morphological parameters and perform automatic spine counting for different groups, rather than placing into subjective shape-based groups such as thin, stubby, and mushroom for healthy neurons.
Perform fast, automatic analysis on the large data sets potentially generated by the current and future high-resolution imaging techniques.
The general mechanism of spine analysis consists of a pipeline for dendrite/centerline extraction, spine detection, feature extraction, and spine classification (Hosokawa et al., 1995, Weaver et al., 2004, Rodriguez et al., 2008). First, because of the level of resolution necessary for accurate spine detection and characterization, most methods begin with a filtering step that removes the background noise and a deconvolution step that corrects for the point spread function (PSF) of the optical imaging system. Deconvolution can be based on an empirically determined PSF (Kawata and Ichioka, 1980) but this approach is often slow and tedious, therefore, many methods start with a faster blind deconvolution step. Next, voxels corresponding to dendritic spines are segmented or distinguished from those corresponding to cell soma, dendrites, and the background. Dendrite extraction remains the most important and challenging task of the segmentation step because spines are identified as protruberances from the parent dendrites. Centerline or skeleton extraction is most commonly employed for identifying dendrites and isolating possible spine regions. Then, spine detection is performed followed by morphological feature calculation and automatic classification.
In vivo imaging of dendritic spines allows for the study of spine dynamics rather than the simple snapshot provided by fixed tissue imaging, but automated analysis of spine dynamics adds a layer of complexity. Thanks to the recent development of image registration and segmentation techniques, time lapse dendritic spine images can be first aligned temporally by using either global or deformable registration methods and then segmented using above mentioned 3D segmentation Pre-segmentation of each time-point image could also help improve the registration accuracy, and recently, joint segmentation and registration techniques were explored to simultaneously obtain the temporal correspondences of spines and their segmentations.
The first generation of spine segmentation algorithms was effective in analysis of tissue samples, each with relative strengths and weaknesses. The method demonstrated by (Koh et al., 2002) provides automatic detection and quantification of the 3D structures of dendritic spines. Spine length, volume, density, and shape features were used for classification of static or time-lapse images of hippocampal pyramidal neurons. The method first extracted the dendritic backbone by calculating the medial axis. Then, geometric analysis was performed to detect detached and attached spines according to the shapes of each candidate spine region. Finally, multiple candidate regions could be merged into one spine according to their combined shapes. Time lapse data were first processed by using image registration. Features such as spine length and volume were used for classification. As for spine detection, 95% (sensitivity) of spines were detected automatically compared to manual results with a positive predictive value of 8%. No significant differences (p-value>0.05) were found between manual and automatic spine measures. However, the method is based on geometric constraints of spines, and led to false positives dependent on the level of the shape threshold. In later tools these problems have been addressed using adaptive thresholds and model-based methods of dendrite detection.
Of the more recent tools, NeuronStudio (Rodriguez et al., 2008) is particularly well-suited for the analysis of three-dimensional data sets derived from confocal or two-photon imaging using a Rayburst sampling algorithm (Rodriguez et al., 2006). The algorithm first filters out the dendrite that connects a series of spherical shapes along a centerline and then identifies the shapes of the objects, after removing the dendrite, based on the linear distance from an internal point to the external boundary of the object. Such spine detection is essentially a spherical model fitting for spines after dendrite extraction. NeuronStudio was comparable to manual spine counting in detecting spines and performs at a relatively fast rate, but exhibited shortcomings in identifying individual spines from clusters, which are common particularly in spiny neurons, as well as in lacking image segmentation capability to characterize spines qualitatively in terms of spine size, shape, volume, and other morphological features. Because of the limitation of the model, bigger flattened spines attached to the dendrite might be missed. Still, NeuronStudio remains an excellent tool for determination of spine number and density that is comparable to manual spine counting without the investigator bias.
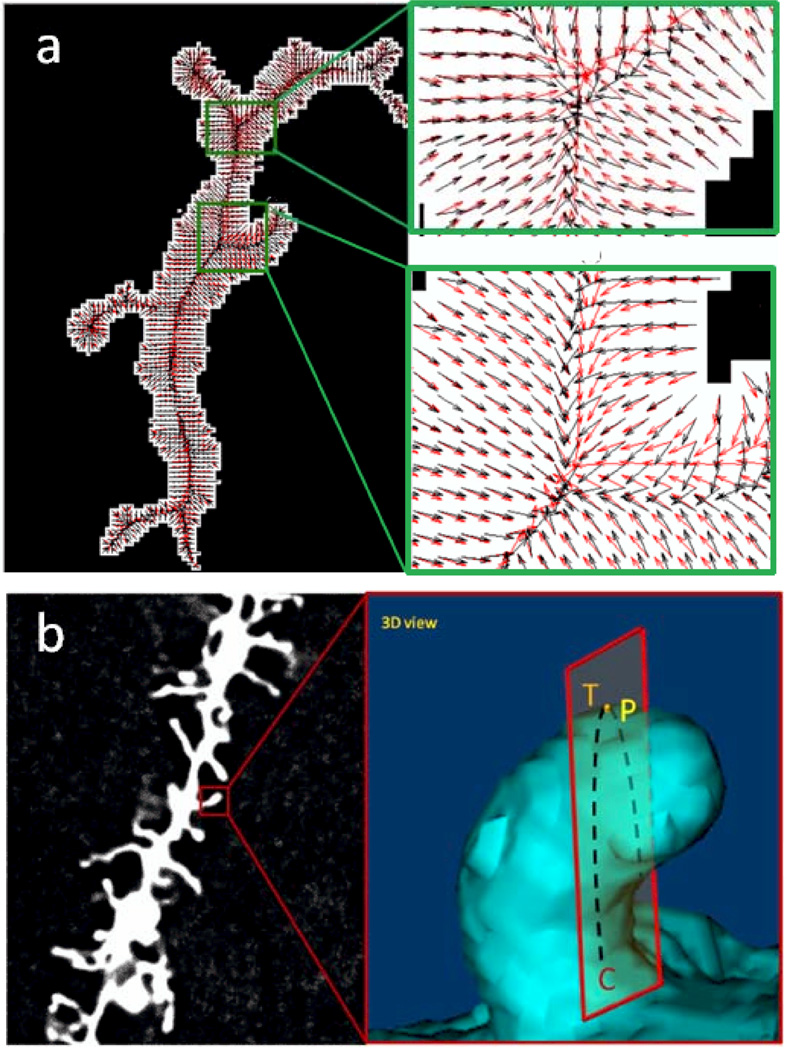
Along the spine analysis pipeline mentioned above, centerline extraction for dendrite detection and spine detection from isolated regions of the dendrite remain the critical step for spine quantification. Researchers seek automatic methods for more robust dendrite extraction and spine detection. Centerline extraction-based methods detect all the possible centerlines and treat dendritic spines as small protrusions attached to the dendrites (Janoos et al., 2009). For example, Janoos and colleagues presented a method for dendritic skeleton structure extraction using a curve-skeleton approach based on the medial geodesic function, which is defined on the reconstructed isosurfaces. The curvilinear fitting method (Zhang et al., 2007) finds the medial axis of the dendritic backbone effectively (Figure 4a).
Figure 4.
Comparison of two alternative spine detection algorithms, (a) (Zhang et al., 2010) Gradient vector fields for centerline determination using the conventional GVF method (black arrows) and proposed GVF with strong smoothing criteria (red arrows). Left, a test neuron image with gradient vector fields superimposed and right, zoomed area 1 and 2 to better visualize gradient vector fields, (b) (from He et al.) Detection of Spine Tip Area Using Minimal Cross-Sectional Curvature. Cross-sectional plane (T) of 3D dendritic spine surface when viewed from a particular angle, the cross-sectional curvature at dendritic spine surface point P is the Gaussian curvature of sectional curve C on plane T.
After determining the full diameter of the dendrite, spines are defined as structures protruding from the dendrite. Classification-based methods separate points into different groups using a classifier (Rodriguez et al., 2008), which can be used to judge whether each isolated image region is a spine and to merge neighboring regions if their combination can form a spine. Incorporating the centerline and spine detection, a publicly available software tool for spine detection and characterization called neuron image quantitator (NeuronlQ) has been developed (Cheng et al., 2007, Zhang et al., 2007). NeuronlQ is one of the rare quality tools for both determination of spine number and quantification of the spatial dimensions of detected spines via automated image segmentation. A variation of this method has been successfully employed for accurate spine detection from in vivo imaging data (Fan et al., 2009). After extracting the centerline of dendrite and isolating it, a level-set model is used to segment and detect the spines. The method has been applied to both static and time lapse images. Taking the 105 manually marked spines as the ground truth, the sensitivity was 96% with a positive predictive value of 10%. A Kolmogorov-Smirnov test also confirmed that the automated and manual spine measurements are drawn from the same distribution and thus had no significant difference. To deal with the partial volume effects or a large spacing between neighboring slices, the speed function for the well-known level set method can also be modified to allow for the segmentation of the treelike structures, such as dendrites (Rink and Tonnies, 2007).
A more recent incarnation of the spine detection methods combines both centerline detection and a geometric model of spines together using a gradient vector flow (GVF) method (Zhang et al., 2010). This method uses GVF to determine the central regions of the dendritic tree and spines. Then, eigen values of the Hessian matrix at each candidate spine central voxel are used to classify whether it belongs to different shapes such as blobs, tubes, or plates. Finally, these candidate spine central points are used as an initialization of a level-set method to segment the final spines. The major contribution is that by performing eigen analysis for the central regions of each candidate spine, false positives are eliminated by distinguishing true spines from other structures. Tested on 1123 manually segmented spines, the method yielded a sensitivity of 90% with a positive predictive value of 9.6%. This method has been shown to be accurate for 3D data analysis but, in general, such methods remain too slow for systematic analysis of large neuronal populations.
Rather than using spine segmentation directly after removing the dendrite, Janoos (Janoos et al., 2009) proposed to first use a level set method for segmentation of the entire dendrite and spine structures and then extract the surface. Then curve-skeletons were extracted using the medial geodesic function, representing the dendritic spine tree. Finally, dendrite and spine skeletons were identified in the graph space. The sensitivity and specificity of the method were 95.3% and 90.1%, respectively. The work by (Son et al., 2011) used a deformable model to segment each spine with the initial points as the tip region of each spine that can be determined from the skeleton structure. Although implemented for 2D projection images the curve-skeletons obtained from Janoos et al.’s work perfectly feed to this deformable segmentation.
In direct contrast to the above mentioned centerline extraction and spine detection pipeline is a newer method that directly uses the outside edges of the dendritic spine structures to detect spines (He et al., 2012) based on the curvature of their tips (Figure 4b). From the dendritic spine shapes, it can be seen that the commonality of spine shapes is that they have tip areas that distinguish them from dendrite and spine walls. Such a distinctive tip feature can be quantified using the minimal cross-sectional curvatures on the surfaces. For spine tip, it will be significantly larger than those on other locations. Such a new spine tip detection method provides a robust initialization for spine detection. Region-growing or gradient vector flow-based methods can then be employed to extract the entire spine regions. Experiments showed that this spine detection-based method is just as accurate (97% sensitivity) but far more computationally efficient than combining centerline extraction and gradient vector flow methods.
In addition to spine detection from 3D images, as more and more time lapse assays are available both in vitro and in vivo, longitudinally corresponding the spines becomes more and more important. As shown in a number of studies (Koh et al., 2002, Fan et al., 2009, Li et al., 2010), such automatic time lapse dendritic spine analysis generally involves non-rigid serial image registration in the spine detection procedure. The real challenge in such registration comes in identifying which spines persist over time and which spines are new, in part because the shapes of the spines themselves are so variable. Li (Li et al., 2010) developed a global non-rigid registration system that first corrects for translation and rotation of the parent dendrite over time and then uses B-Spline-based registration to correct the elastic shape deformations. Spines are then identified using a spatial similarity metric over a series of time points. The temporal spine associations are obtained through global similarity maximization. The basic assumption of the association algorithm is that the temporally corresponding spines should have similar shapes and should not move far away (after alignment) under the current longitudinal deformations. The formulation also allows one spine at a time point to correspond to no spines at a following time point or vice versa. In this way, trajectories of spine centroids are analyzed and invalid associations can be discarded. Therefore, the major contribution is post-processing after image registration in order to overcome possible errors due to image alignment.
Work over the last decade has yielded spine detection algorithms that quantify and characterize dendritic spines from 3D image stacks or time lapse images. These algorithms offer a clear objective alternative to manual counting by removing operator variability and provide reliable classification of spines based on measured size and morphology. Efforts have shown greater success extracting the dendritic centerline for isolating candidate spine regions followed by spine detection or segmentation based spine shape features or shape models. Novel image segmentation methods such as level-set and fast marching have been employed for spine segmentation. Notably, rather than starting from the dendrite backbone, spine tip detection offers an alternative approach, starting from the outside spiky points.
Recent developments toward faster computation times and reduced operator input are encouraging but current methods still require minutes rather than milliseconds to analyze anatomical data from single neurons. Additionally, to date all spine detection algorithms require a single specified region of interest ideally from the dendrites of a single neuron. Due to the wide range of pathophysiologies for dendritic spines, more sophisticated automatic software is necessary to handle multiple dendrites and spines from in vivo and in vitro image mosaics with relatively large fields of view. Although no one-size-fits-all algorithm exists to date for spine detection, ideally, automatic algorithms must recognize the dendritic arbors of a multitude of individual neurons per image set and combine as many distinct spine recognition criteria as possible in order to accurately and objectively characterize disease states.
In summary, based on the above analysis of automatic dendritic spine detection and tracking techniques and the requirements for high content study, future work should be focused on the following areas:
Continued optimization of the pipeline for dendrite and spine detection. Studies showed that each step is not independent of one other and such optimization could involve integration of the steps focusing on robustly segmenting spines using novel features, while other structures act as a constraint of non-spine regions.
Applying accurate serial image registration (rather than only two time points) and simultaneously incorporating both original image and segmented results in the registration procedure. Studies showed that such joint image segmentation and registration steps could benefit each other.
Dependent on the application, the imaging speed, and the staining technique, in time lapse images, spine shapes can change dramatically with time. It could be a challenging task to accurately register the spines with larger or more rapid morphological changes. Therefore, precisely aligning dendrite and relatively stable spines provide an invaluable opportunity to construct the temporal association of each spine and make it possible to track bigger spine changes.
Developing a sophisticated tool that can automatically process complicated images with multiple dendrites not only for in vitro but also for more complicated in vivo images is necessary for fast automatic analysis on the large data sets in high content studies.
Conclusions
Traditional studies of neuronal anatomy have provided a wealth of information about neuronal structure and particularly the changes that correspond to disease in the human brain. The workhorse of these studies has been and continues to be the Golgi stain. As recent studies have attempted to understand and correct the mechanisms of many of these diseases, researchers have started to complement these studies with newer approaches that provide systematic evaluation of brain development, aging, and pathogenesis. Three main advances have marked this transition. First, we have seen the evolution from the analog staining, eyeball detection, and manual labeling of Cajal and Golgi to digital image acquisition and automated image analysis. Second, recent advances in cell labeling and high resolution optical microscopy have allowed researchers to extend from endpoint studies performed on fixed, post-mortem tissue to real-time, in vivo, live animal dynamic studies utilizing either intravital two-photon microscopy or optical fiber microendoscopy. Third, coupling of recent transgenic animal engineering techniques with automated image acquisition enables the degree of scalability necessary to image synaptic connections in the high-throughput manner required for a complete, systematic study. The final piece needed to complete the puzzle is a robust image analysis tool capable of quantifying all of the spine information throughout these large data sets, ranging from gigabytes to terabytes. Recent work has made significant progress toward objective characterization of dendritic spines, but a great deal still remains in speeding up the analysis and expanding it to identify and characterize spines from large populations of labeled and intertwined neurons.
Highlights.
Golgi staining and manual spine identification have provided insight into neurodegeneration
Advances in microscopy and tissue labeling have allowed in vivo studies in animals
Automated spine algorithms are necessary tool for a systems level scale study
Acknowledgements
The authors would like to thank Dr. Kemi Cui of The Methodist Hospital Research Institute for his assistance in imaging and figure preparation for this review. This research is supported by NIH R01AG028928, NIH R01LM009161, Ting Tsung and Wei Fong Chao Center for Bioinformatics Research and Imaging in Neurosciences (BRAIN), and John S Dunn Research Foundation to STCWs.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alvarez VA, Sabatini BL. Anatomical and physiological plasticity of dendritic spines. Annual review of neuroscience. 2007;30:79–97. doi: 10.1146/annurev.neuro.30.051606.094222. [DOI] [PubMed] [Google Scholar]
- Anderson JC, Kennedy H, Martin KA. Pathways of attention: synaptic relationships of frontal eye field to V4, lateral intraparietal cortex, and area 46 in macaque monkey. J Neurosci. 2011;31:10872–10881. doi: 10.1523/JNEUROSCI.0622-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barretto RP, Ko TH, Jung JC, Wang TJ, Capps G, Waters AC, Ziv Y, Attardo A, Recht L, Schnitzer MJ. Time-lapse imaging of disease progression in deep brain areas using fluorescence microendoscopy. Nature medicine. 2011;17:223–228. doi: 10.1038/nm.2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barretto RP, Messerschmidt B, Schnitzer MJ. In vivo fluorescence imaging with high-resolution microlenses. Nature methods. 2009;6:511–512. doi: 10.1038/nmeth.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaurepaire E, Oheim M, Mertz J. Ultra-deep two-photon fluorescence excitation in turbid media. Opt Commun. 2001;188:25–29. [Google Scholar]
- Bhatt DH, Zhang S, Gan WB. Dendritic spine dynamics. Annual review of physiology. 2009;71:261–282. doi: 10.1146/annurev.physiol.010908.163140. [DOI] [PubMed] [Google Scholar]
- Bittner T, Fuhrmann M, Burgold S, Ochs SM, Hoffmann N, Mitteregger G, Kretzschmar H, LaFerla FM, Herms J. Multiple events lead to dendritic spine loss in triple transgenic Alzheimer's disease mice. PloS one. 2010;5:e15477. doi: 10.1371/journal.pone.0015477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloodgood BL, Sabatini BL. Neuronal activity regulates diffusion across the neck of dendritic spines. Science (New York, NY. 2005;310:866–869. doi: 10.1126/science.1114816. [DOI] [PubMed] [Google Scholar]
- Bosch M, Hayashi Y. Structural plasticity of dendritic spines. Current opinion in neurobiology. 2011 doi: 10.1016/j.conb.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhl EH, Lubke J. Intracellular lucifer yellow injection in fixed brain slices combined with retrograde tracing, light and electron microscopy. Neuroscience. 1989;28:3–16. doi: 10.1016/0306-4522(89)90227-3. [DOI] [PubMed] [Google Scholar]
- Calabrese B, Wilson MS, Halpain S. Development and regulation of dendritic spine synapses. Physiology (Bethesda, Md. 2006;21:38–47. doi: 10.1152/physiol.00042.2005. [DOI] [PubMed] [Google Scholar]
- Castano P, Gioia M, Barajon I, Rumio C, Miani A. A comparision between rapid Golgi and Golgi-Cox impregnation methods for 3-D reconstruction of neurons at the confocal scanning laser microscope. Italian journal of anatomy and embryology = Archivio italiano di anatomia ed embriologia. 1995;100(Suppl 1):613–622. [PubMed] [Google Scholar]
- Chamberlin NL, Du B, de Lacalle S, Saper CB. Recombinant adeno-associated virus vector: use for transgene expression and anterograde tract tracing in the CNS. Brain research. 1998;793:169–175. doi: 10.1016/s0006-8993(98)00169-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan-Palay V, Palay SL, Billings-Gagliardi SM. Meynert cells in the primate visual cortex. Journal of neurocytology. 1974;3:631–658. doi: 10.1007/BF01097628. [DOI] [PubMed] [Google Scholar]
- Chen X, Leischner U, Rochefort NL, Nelken I, Konnerth A. Functional mapping of single spines in cortical neurons in vivo. Nature. 2011;475:501–505. doi: 10.1038/nature10193. [DOI] [PubMed] [Google Scholar]
- Cheng J, Zhou X, Miller E, Witt RM, Zhu J, Sabatini BL, Wong ST. A novel computational approach for automatic dendrite spines detection in two-photon laser scan microscopy. Journal of neuroscience methods. 2007;165:122–134. doi: 10.1016/j.jneumeth.2007.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conchello JA, Lichtman JW. Optical sectioning microscopy. Nature methods. 2005;2:920–931. doi: 10.1038/nmeth815. [DOI] [PubMed] [Google Scholar]
- Couch BA, DeMarco GJ, Gourley SL, Koleske AJ. Increased dendrite branching in AbetaPP/PS1 mice and elongation of dendrite arbors by fasudil administration. J Alzheimers Dis. 2010;20:1003–1008. doi: 10.3233/JAD-2010-091114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Paola V, Holtmaat A, Knott G, Song S, Wilbrecht L, Caroni P, Svoboda K. Cell type-specific structural plasticity of axonal branches and boutons in the adult neocortex. Neuron. 2006;49:861–875. doi: 10.1016/j.neuron.2006.02.017. [DOI] [PubMed] [Google Scholar]
- Denk W, Strickler JH, Webb WW. Two-photon laser scanning fluorescence microscopy. Science (New York, NY. 1990;248:73–76. doi: 10.1126/science.2321027. [DOI] [PubMed] [Google Scholar]
- Denk W, Svoboda K. Photon upmanship: why multiphoton imaging is more than a gimmick. Neuron. 1997;18:351–357. doi: 10.1016/s0896-6273(00)81237-4. [DOI] [PubMed] [Google Scholar]
- Dhawale A, Bhalla US. The networks and the synapse: 100 years after Cajal. HFSP Journal. 2008;2:12–16. doi: 10.2976/1.2835214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrengruber MU, Hennou S, Bueler H, Naim HY, Deglon N, Lundstrom K. Gene transfer into neurons from hippocampal slices: comparison of recombinant Semliki Forest Virus, adenovirus, adeno-associated virus, lentivirus, and measles virus. Molecular and cellular neurosciences. 2001;17:855–871. doi: 10.1006/mcne.2001.0982. [DOI] [PubMed] [Google Scholar]
- Engert F, Bonhoeffer T. Dendritic spine changes associated with hippocampal long-term synaptic plasticity. Nature. 1999;399:66–70. doi: 10.1038/19978. [DOI] [PubMed] [Google Scholar]
- Fairen A. Pioneering a golden age of cerebral microcircuits: the births of the combined Golgi-electron microscope methods. Neuroscience. 2005;136:607–614. doi: 10.1016/j.neuroscience.2005.08.011. [DOI] [PubMed] [Google Scholar]
- Fan J, Zhou X, Dy JG, Zhang Y, Wong ST. An automated pipeline for dendrite spine detection and tracking of 3D optical microscopy neuron images of in vivo mouse models. Neuroinformatics. 2009;7:113–130. doi: 10.1007/s12021-009-9047-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman ML, Dowd C. Loss of dendritic spines in aging cerebral cortex. Anatomy and embryology. 1975;148:279–301. doi: 10.1007/BF00319848. [DOI] [PubMed] [Google Scholar]
- Feldman ML, Peters A. A technique for estimating total spine numbers on Golgi-impregnated dendrites. The Journal of comparative neurology. 1979;188:527–542. doi: 10.1002/cne.901880403. [DOI] [PubMed] [Google Scholar]
- Feng G, Mellor RH, Bernstein M, Keller-Peck C, Nguyen QT, Wallace M, Nerbonne JM, Lichtman JW, Sanes JR. Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron. 2000;28:41–51. doi: 10.1016/s0896-6273(00)00084-2. [DOI] [PubMed] [Google Scholar]
- Filonov GS, Piatkevich KD, Ting LM, Zhang J, Kim K, Verkhusha VV. Bright and stable near-infrared fluorescent protein for in vivo imaging. Nature biotechnology. 2011;29:757–761. doi: 10.1038/nbt.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flusberg BA, Nimmerjahn A, Cocker ED, Mukamel EA, Barretto RP, Ko TH, Burns LD, Jung JC, Schnitzer MJ. High-speed, miniaturized fluorescence microscopy in freely moving mice. Nature methods. 2008;5:935–938. doi: 10.1038/nmeth.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Lopez P, Garcia-Marin V, Freire M. The discovery of dendritic spines by Cajal in 1888 and its relevance in the present neuroscience. Progress in neurobiology. 2007;83:110–130. doi: 10.1016/j.pneurobio.2007.06.002. [DOI] [PubMed] [Google Scholar]
- Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, Schambra UB, Nowak NJ, Joyner A, Leblanc G, Hatten ME, Heintz N. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425:917–925. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- Gray NW, Weimer RM, Bureau I, Svoboda K. Rapid redistribution of synaptic PSD-95 in the neocortex in vivo. PLoS biology. 2006;4:e370. doi: 10.1371/journal.pbio.0040370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grynkiewicz G, Poenie M, Tsien RY. A new generation of Ca2+ indicators with greatly improved fluorescence properties. The Journal of biological chemistry. 1985;260:3440–3450. [PubMed] [Google Scholar]
- Hama H, Kurokawa H, Kawano H, Ando R, Shimogori T, Noda H, Fukami K, Sakaue-Sawano A, Miyawaki A. Scale: a chemical approach for fluorescence imaging and reconstruction of transparent mouse brain. Nature neuroscience. 2011 doi: 10.1038/nn.2928. [DOI] [PubMed] [Google Scholar]
- Harris KM, Stevens JK. Dendritic spines of rat cerebellar Purkinje cells: serial electron microscopy with reference to their biophysical characteristics. J Neurosci. 1988;8:4455–4469. doi: 10.1523/JNEUROSCI.08-12-04455.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He T, Xue Z, Kim Y, Wong ST. Three dimensional dendritic spine detection in neuropsychiatric disease model based on minimal cross-sectional curvature submitted. 2012 [Google Scholar]
- Helmchen F, Denk W. Deep tissue two-photon microscopy. Nature methods. 2005;2:932–940. doi: 10.1038/nmeth818. [DOI] [PubMed] [Google Scholar]
- Hercher C, Canetti L, Turecki G, Mechawar N. Anterior cingulate pyramidal neurons display altered dendritic branching in depressed suicides. Journal of psychiatric research. 2010;44:286–293. doi: 10.1016/j.jpsychires.2009.08.011. [DOI] [PubMed] [Google Scholar]
- Holtmaat A, Svoboda K. Experience-dependent structural synaptic plasticity in the mammalian brain. Nature reviews. 2009;10:647–658. doi: 10.1038/nrn2699. [DOI] [PubMed] [Google Scholar]
- Holtmaat AJ, Trachtenberg JT, Wilbrecht L, Shepherd GM, Zhang X, Knott GW, Svoboda K. Transient and persistent dendritic spines in the neocortex in vivo. Neuron. 2005;45:279–291. doi: 10.1016/j.neuron.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Horner CH, Arbuthnott E. Methods of estimation of spine density--are spines evenly distributed throughout the dendritic field? Journal of anatomy. 1991;177:179–184. [PMC free article] [PubMed] [Google Scholar]
- Hosokawa T, Rusakov DA, Bliss TV, Fine A. Repeated confocal imaging of individual dendritic spines in the living hippocampal slice: evidence for changes in length and orientation associated with chemically induced LTP. J Neurosci. 1995;15:5560–5573. doi: 10.1523/JNEUROSCI.15-08-05560.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janoos F, Mosaliganti K, Xu X, Machiraju R, Huang K, Wong ST. Robust 3D reconstruction and identification of dendritic spines from optical microscopy imaging. Medical image analysis. 2009;13:167–179. doi: 10.1016/j.media.2008.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung JC, Mehta AD, Aksay E, Stepnoski R, Schnitzer MJ. In vivo mammalian brain imaging using one- and two-photon fluorescence microendoscopy. Journal of neurophysiology. 2004;92:3121–3133. doi: 10.1152/jn.00234.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung JC, Schnitzer MJ. Multiphoton endoscopy. Optics letters. 2003;28:902–904. doi: 10.1364/ol.28.000902. [DOI] [PubMed] [Google Scholar]
- Kasai H, Fukuda M, Watanabe S, Hayashi-Takagi A, Noguchi J. Structural dynamics of dendritic spines in memory and cognition. Trends in neurosciences. 2010;33:121–129. doi: 10.1016/j.tins.2010.01.001. [DOI] [PubMed] [Google Scholar]
- Kawata S, Ichioka Y. Iterative image restoration for linearly degraded images. II. Reblurring procedure. J Opt Soc Am. 1980;70:768–772. [Google Scholar]
- Knott G, Holtmaat A. Dendritic spine plasticity--current understanding from in vivo studies. Brain research reviews. 2008;58:282–289. doi: 10.1016/j.brainresrev.2008.01.002. [DOI] [PubMed] [Google Scholar]
- Koester HJ, Sakmann B. Calcium dynamics in single spines during coincident pre- and postsynaptic activity depend on relative timing of back-propagating action potentials and subthreshold excitatory postsynaptic potentials. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:9596–9601. doi: 10.1073/pnas.95.16.9596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh IY, Lindquist WB, Zito K, Nimchinsky EA, Svoboda K. An image analysis algorithm for dendritic spines. Neural computation. 2002;14:1283–1310. doi: 10.1162/089976602753712945. [DOI] [PubMed] [Google Scholar]
- Lanciego JL, Wouterlood FG. A half century of experimental neuroanatomical tracing. Journal of chemical neuroanatomy. 2011;42:157–183. doi: 10.1016/j.jchemneu.2011.07.001. [DOI] [PubMed] [Google Scholar]
- Landis DM, Williams RS, Masters CL. Golgi and electronmicroscopic studies of spongiform encephalopathy. Neurology. 1981;31:538–549. doi: 10.1212/wnl.31.5.538. [DOI] [PubMed] [Google Scholar]
- Levene MJ, Dombeck DA, Kasischke KA, Molloy RP, Webb WW. In vivo multiphoton microscopy of deep brain tissue. Journal of neurophysiology. 2004;91:1908–1912. doi: 10.1152/jn.01007.2003. [DOI] [PubMed] [Google Scholar]
- Li Q, Deng Z, Zhang Y, Zhou X, Nagerl UV, Wong ST. A global spatial similarity optimization scheme to track large numbers of dendritic spines in time-lapse confocal microscopy. IEEE Trans Med Imaging. 2010;30:632–641. doi: 10.1109/TMI.2010.2090354. [DOI] [PubMed] [Google Scholar]
- Livet J, Weissman TA, Kang H, Draft RW, Lu J, Bennis RA, Sanes JR, Lichtman JW. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature. 2007;450:56–62. doi: 10.1038/nature06293. [DOI] [PubMed] [Google Scholar]
- Lopez IP, Salin P, Kachidian P, Barroso-Chinea P, Rico AJ, Gomez-Bautista V, Conte-Perales L, Coulon P, Goff LK, Lanciego JL. The added value of rabies virus as a retrograde tracer when combined with dual anterograde tract-tracing. Journal of neuroscience methods. 2010;194:21–27. doi: 10.1016/j.jneumeth.2010.01.015. [DOI] [PubMed] [Google Scholar]
- Maletic-Savatic M, Malinow R, Svoboda K. Rapid dendritic morphogenesis in CA1 hippocampal dendrites induced by synaptic activity. Science (New York, NY. 1999;283:1923–1927. doi: 10.1126/science.283.5409.1923. [DOI] [PubMed] [Google Scholar]
- Matsuzaki M, Honkura N, Ellis-Davies GC, Kasai H. Structural basis of long-term potentiation in single dendritic spines. Nature. 2004;429:761–766. doi: 10.1038/nature02617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavroudis IA, Fotiou DF, Adipepe LF, Manani MG, Njau SD, Psaroulis D, Costa VG, Baloyannis SJ. Morphological changes of the human purkinje cells and deposition of neuritic plaques and neurofibrillary tangles on the cerebellar cortex of Alzheimer's disease. American journal of Alzheimer's disease and other dementias. 2010;25:585–591. doi: 10.1177/1533317510382892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavroudis IA, Fotiou DF, Manani MG, Njaou SN, Frangou D, Costa VG, Baloyannis SJ. Dendritic pathology and spinal loss in the visual cortex in Alzheimer's disease: a Golgi study in pathology. The International journal of neuroscience. 2011;121:347–354. doi: 10.3109/00207454.2011.553753. [DOI] [PubMed] [Google Scholar]
- Melendez-Ferro M, Perez-Costas E, Roberts RC. A new use for long-term frozen brain tissue: golgi impregnation. Journal of neuroscience methods. 2009;176:72–77. doi: 10.1016/j.jneumeth.2008.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micheva KD, Busse B, Weiler NC, O'Rourke N, Smith SJ. Single-synapse analysis of a diverse synapse population: proteomic imaging methods and markers. Neuron. 2010;68:639–653. doi: 10.1016/j.neuron.2010.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minta A, Kao JP, Tsien RY. Fluorescent indicators for cytosolic calcium based on rhodamine and fluorescein chromophores. The Journal of biological chemistry. 1989;264:8171–8178. [PubMed] [Google Scholar]
- Misgeld T, Kerschensteiner M. In vivo imaging of the diseased nervous system. Nature reviews. 2006;7:449–463. doi: 10.1038/nrn1905. [DOI] [PubMed] [Google Scholar]
- Moolman DL, Vitolo OV, Vonsattel JP, Shelanski ML. Dendrite and dendritic spine alterations in Alzheimer models. Journal of neurocytology. 2004;33:377–387. doi: 10.1023/B:NEUR.0000044197.83514.64. [DOI] [PubMed] [Google Scholar]
- Moser MB, Trommald M, Andersen P. An increase in dendritic spine density on hippocampal CA1 pyramidal cells following spatial learning in adult rats suggests the formation of new synapses. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:12673–12675. doi: 10.1073/pnas.91.26.12673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai J, Ohkura M, Imoto K. A high signal-to-noise Ca(2+) probe composed of a single green fluorescent protein. Nature biotechnology. 2001;19:137–141. doi: 10.1038/84397. [DOI] [PubMed] [Google Scholar]
- Niell CM, Meyer MP, Smith SJ. In vivo imaging of synapse formation on a growing dendritic arbor. Nature neuroscience. 2004;7:254–260. doi: 10.1038/nn1191. [DOI] [PubMed] [Google Scholar]
- Nietzer SL, Bonn M, Jansen F, Heiming RS, Lewejohann L, Sachser N, Asan ES, Lesch KP, Schmitt AG. Serotonin transporter knockout and repeated social defeat stress: impact on neuronal morphology and plasticity in limbic brain areas. Behavioural brain research. 2011;220:42–54. doi: 10.1016/j.bbr.2011.01.011. [DOI] [PubMed] [Google Scholar]
- Nimchinsky EA, Yasuda R, Oertner TG, Svoboda K. The number of glutamate receptors opened by synaptic stimulation in single hippocampal spines. J Neurosci. 2004;24:2054–2064. doi: 10.1523/JNEUROSCI.5066-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida H, Okabe S. Direct astrocytic contacts regulate local maturation of dendritic spines. J Neurosci. 2007;27:331–340. doi: 10.1523/JNEUROSCI.4466-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oheim M, Michael DJ, Geisbauer M, Madsen D, Chow RH. Principles of two-photon excitation fluorescence microscopy and other nonlinear imaging approaches. Advanced drug delivery reviews. 2006;58:788–808. doi: 10.1016/j.addr.2006.07.005. [DOI] [PubMed] [Google Scholar]
- Pannese E. The Golgi Stain: invention, diffusion and impact on neurosciences. Journal of the history of the neurosciences. 1999;8:132–140. doi: 10.1076/jhin.8.2.132.1847. [DOI] [PubMed] [Google Scholar]
- Papa M, Bundman MC, Greenberger V, Segal M. Morphological analysis of dendritic spine development in primary cultures of hippocampal neurons. J Neurosci. 1995;15:1–11. doi: 10.1523/JNEUROSCI.15-01-00001.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson GH, Piston DW. Photobleaching in two-photon excitation microscopy. Biophysical journal. 2000;78:2159–2162. doi: 10.1016/S0006-3495(00)76762-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Costas E, Melendez-Ferro M, Roberts RC. Microscopy techniques and the study of synapses. In: Méndez-Vilas A, DJ, editors. Microscopy: science, technology, applications and education. 2007. pp. 164–170. [Google Scholar]
- Pilati N, Barker M, Panteleimonitis S, Donga R, Hamann M. A rapid method combining Golgi and Nissl staining to study neuronal morphology and cytoarchitecture. J Histochem Cytochem. 2008;56:539–550. doi: 10.1369/jhc.2008.950246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pologruto TA, Yasuda R, Svoboda K. Monitoring neural activity and [Ca2+] with genetically encoded Ca2+ indicators. J Neurosci. 2004;24:9572–9579. doi: 10.1523/JNEUROSCI.2854-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raju TR, Kutty BM, TN S, BS SR. The Golgi Techniques for Staining Neurons. Brain and Behanvior. 2004:108–111. [Google Scholar]
- Ranjan A, Mallick BN. A modified method for consistent and reliable Golgi-cox staining in significantly reduced time. Frontiers in neurology. 2010;1:157. doi: 10.3389/fneur.2010.00157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rink K, Tonnies K. A Level Set Bridging Force for the Segmentation of Dendritic Spines. In: Kropatsch W, et al., editors. 12th International Converence on Computer Analysis of Images and Patterns. Springer-Verlag; 2007. pp. 571–578. [Google Scholar]
- Rodriguez A, Ehlenberger DB, Dickstein DL, Hof PR, Wearne SL. Automated three-dimensional detection and shape classification of dendritic spines from fluorescence microscopy images. PloS one. 2008;3:e1997. doi: 10.1371/journal.pone.0001997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez A, Ehlenberger DB, Hof PR, Wearne SL. Rayburst sampling, an algorithm for automated three dimensional shape analysis from laser scanning microscopy images. Nature protocols. 2006;1:2152–2161. doi: 10.1038/nprot.2006.313. [DOI] [PubMed] [Google Scholar]
- Rosoklija G, Mancevski B, Ilievski B, Perera T, Lisanby SH, Coplan JD, Duma A, Serafimova T, Dwork AJ. Optimization of Golgi methods for impregnation of brain tissue from humans and monkeys. Journal of neuroscience methods. 2003;131:1–7. doi: 10.1016/j.jneumeth.2003.06.001. [DOI] [PubMed] [Google Scholar]
- Ryan TA. Presynaptic imaging techniques. Current opinion in neurobiology. 2001;11:544–549. doi: 10.1016/s0959-4388(00)00247-6. [DOI] [PubMed] [Google Scholar]
- Shepherd GM. The dendritic spine: a multifunctional integrative unit. Journal of neurophysiology. 1996;75:2197–2210. doi: 10.1152/jn.1996.75.6.2197. [DOI] [PubMed] [Google Scholar]
- Shu X, Royant A, Lin MZ, Aguilera TA, Lev-Ram V, Steinbach PA, Tsien RY. Mammalian expression of infrared fluorescent proteins engineered from a bacterial phytochrome. Science (New York, NY. 2009;324:804–807. doi: 10.1126/science.1168683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DL, Pozueta J, Gong B, Arancio O, Shelanski M. Reversal of long-term dendritic spine alterations in Alzheimer disease models. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:16877–16882. doi: 10.1073/pnas.0908706106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soiza-Reilly M, Commons KG. Quantitative analysis of glutamatergic innervation of the mouse dorsal raphe nucleus using array tomography. The Journal of comparative neurology. 2010;519:3802–3814. doi: 10.1002/cne.22734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son J, Song S, Lee S, Chang S, Kim M. Morphological change tracking of dendritic spines based on structural features. Journal of microscopy. 2011;241:261–272. doi: 10.1111/j.1365-2818.2010.03427.x. [DOI] [PubMed] [Google Scholar]
- Spiga S, Acquas E, Puddu MC, Mulas G, Lintas A, Diana M. Simultaneous Golgi-Cox and immunofluorescence using confocal microscopy. Brain structure & function. 2011;216:171–182. doi: 10.1007/s00429-011-0312-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spires-Jones TL, Meyer-Luehmann M, Osetek JD, Jones PB, Stern EA, Bacskai BJ, Hyman BT. Impaired spine stability underlies plaque-related spine loss in an Alzheimer's disease mouse model. The American journal of pathology. 2007;171:1304–1311. doi: 10.2353/ajpath.2007.070055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens B, Mueller AJ, Shering AF, Hood SH, Taggart P, Arbuthnott GW, Bell JE, Kilford L, Kingsbury AE, Daniel SE, Ingham CA. Evidence of a breakdown of corticostriatal connections in Parkinson's disease. Neuroscience. 2005;132:741–754. doi: 10.1016/j.neuroscience.2005.01.007. [DOI] [PubMed] [Google Scholar]
- Svoboda K, Yasuda R. Principles of two-photon excitation microscopy and its applications to neuroscience. Neuron. 2006;50:823–839. doi: 10.1016/j.neuron.2006.05.019. [DOI] [PubMed] [Google Scholar]
- Theer P, Hasan MT, Denk W. Two-photon imaging to a depth of 1000 microm in living brains by use of a Ti:Al2O3 regenerative amplifier. Optics letters. 2003;28:1022–1024. doi: 10.1364/ol.28.001022. [DOI] [PubMed] [Google Scholar]
- Tian L, Hires SA, Mao T, Huber D, Chiappe ME, Chalasani SH, Petreanu L, Akerboom J, McKinney SA, Schreiter ER, Bargmann CI, Jayaraman V, Svoboda K, Looger LL. Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nature methods. 2009;6:875–881. doi: 10.1038/nmeth.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tredici G, Di Francesco A, Miani A, Jr, Pizzini G. Real complete three-dimensional reconstruction of Golgi-impregnated neurons by means of a confocal laser scanning microscope. NeuroImage. 1993;1:87–93. doi: 10.1006/nimg.1993.1002. [DOI] [PubMed] [Google Scholar]
- Ugolini G. Advances in viral transneuronal tracing. Journal of neuroscience methods. 2010;194:2–20. doi: 10.1016/j.jneumeth.2009.12.001. [DOI] [PubMed] [Google Scholar]
- Vecellio M, Schwaller B, Meyer M, Hunziker W, Celio MR. Alterations in Purkinje cell spines of calbindin D-28 k and parvalbumin knock-out mice. The European journal of neuroscience. 2000;12:945–954. doi: 10.1046/j.1460-9568.2000.00986.x. [DOI] [PubMed] [Google Scholar]
- Wallace W, Bear MF. A morphological correlate of synaptic scaling in visual cortex. J Neurosci. 2004;24:6928–6938. doi: 10.1523/JNEUROSCI.1110-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver CM, Hof PR, Wearne SL, Lindquist WB. Automated algorithms for multiscale morphometry of neuronal dendrites. Neural Comput. 2004;16:1353–1383. doi: 10.1162/089976604323057425. [DOI] [PubMed] [Google Scholar]
- Williams RW, Herrup K. The control of neuron number. Annual review of neuroscience. 1988;11:423–453. doi: 10.1146/annurev.ne.11.030188.002231. [DOI] [PubMed] [Google Scholar]
- Wilt BA, Burns LD, Wei Ho ET, Ghosh KK, Mukamel EA, Schnitzer MJ. Advances in light microscopy for neuroscience. Annual review of neuroscience. 2009;32:435–506. doi: 10.1146/annurev.neuro.051508.135540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasumatsu N, Matsuzaki M, Miyazaki T, Noguchi J, Kasai H. Principles of long-term dynamics of dendritic spines. J Neurosci. 2008;28:13592–13608. doi: 10.1523/JNEUROSCI.0603-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Baird GS, Tsien RY, Davis RL. Detection of calcium transients in Drosophila mushroom body neurons with camgaroo reporters. J Neurosci. 2003;23:64–72. doi: 10.1523/JNEUROSCI.23-01-00064.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuste R, Bonhoeffer T. Morphological changes in dendritic spines associated with long-term synaptic plasticity. Annual review of neuroscience. 2001;24:1071–1089. doi: 10.1146/annurev.neuro.24.1.1071. [DOI] [PubMed] [Google Scholar]
- Yuste R, Majewska A, Holthoff K. From form to function: calcium compartmentalization in dendritic spines. Nature neuroscience. 2000;3:653–659. doi: 10.1038/76609. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Chen K, Baron M, Teylan MA, Kim Y, Song Z, Greengard P, Wong ST. A neurocomputational method for fully automated 3D dendritic spine detection and segmentation of medium-sized spiny neurons. NeuroImage. 2010;50:1472–1484. doi: 10.1016/j.neuroimage.2010.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhou X, Witt RM, Sabatini BL, Adjeroh D, Wong ST. Dendritic spine detection using curvilinear structure detector and LDA classifier. NeuroImage. 2007;36:346–360. doi: 10.1016/j.neuroimage.2007.02.044. [DOI] [PubMed] [Google Scholar]