Malignant pleural mesothelioma (MPM) is a highly aggressive tumor.1 Biomarker information for MPM is lacking and could prove useful for understanding MPM metastasis and proliferation, and providing potential targets for therapy. The Dishevelled (Dvl) family of proteins are membrane-proximal signaling intermediates in the Wnt pathway.2 Moreover, we previously found that the Wnt pathway is activated in mesothelioma through Dvl3 overexpression.3 Excitatory amino acid transporters (EAATs) are known to function as glutamate carriers 4, 5. Among the EAATs, EAAT1 is a ubiquitous subtype. Glutamine synthetase (GS) is another important enzyme that plays a role in the metabolic pathway of glutamate.6 In this study, we evaluated the possible association between expression levels of Dvl3, EAAT1 and GS in MPM to determine possible biomarkers and therapeutic targets.
Clinical data was obtained from 39 patients with primary MPM between 1997 and 2009. The diagnosis of malignant mesothelioma was confirmed by review of the pathology reports in all cases. We also used human mesothelioma cell lines H28, 211H, H2052, MS-1, H290, H513 and human normal mesothelial cell line LP9 in this research.
Thirty-eight mesothelioma samples (seven that included adjacent normal pleural tissues to serve as controls) were obtained from the patients who were mentioned above. Samples for one patient were not available for EAAT1 staining, three were not available for GS staining and six were not available for Dvl3 staining. All human tissue samples were obtained and analyzed in accordance with procedures approved by the institutional review board of the University of California, San Francisco (IRB H8714-22942-01).
The tissue microarray sections contained normal pleura tissue samples from another three patients. The samples in the sections were done immunohistochemistry staining with primary anti-EAAT1, anti-GS or anti-Dvl3 polyclonal antibodies, and biotin-labeled secondary antibody. Mesothelioma cell lines and normal mesothelial cell line were immunostained according to the same procedure.7
Total RNA from cell lines H28, 211H, H2052, MS-1, H290, H513 and LP9 were obtained. RT-PCR was carried out using the One-Step RT-PCR kit (Invitrogen, Carlsbad, CA). The primers of human EAAT1, GS and GAPDH were shown in Table 1. Western blot analysis was done with the primary EAAT1 antibody (1:1000). GAPDH was used as a control.
Table 1.
Sequences of oligonucleiotides used in the study
| Primer | Sequence | Product (bp) |
|---|---|---|
| EAAT1 | F: 5′-GTTGTCTTCTCCATGTGCTTCG-3′ | 326 |
| R: 5′-GCTTGCAGCAACCCTCCA-3′ | ||
| GS | F: 5′-GACCTTGTGAAGGAATCAGCATGG-3′ | 598 |
| R: 5′-CCCTTTGAGTTACAATCGGGACAA-3′ | ||
| GAPDH | F: 5′-GAAGGTCGGAGTCAACGGATTT-3′ | 212 |
| R: 5′-ATGGGTGGAATCATATTGGAAC-3′ |
Multiple correlation analyses, Kaplan-Meyer curve, statistical analysis (log rank) and Cox regression analysis were performed using SPSS 13.0 (SPSS Inc, Chicago, USA).
Disease stages, histological types, surgical procedures and survival timess of these patients were showed in Table 2. Disease stage correlated significantly with survival time (p<0.01), but surgical procedure did not.
Table 2.
Characteristics of primary human mesothelioma samples
| Sample number | IHC of anti-EAAT1 | IHC of anti-GS | IHC of anti-DVL3 | Pathologic type | Stage | Surgical procedure | Overall survival (months) |
|---|---|---|---|---|---|---|---|
| N233 | − | + | ND | ND | ND | ND | ND |
| N237 | + | + | ++ | ND | ND | ND | ND |
| T237 | ++ | + | +++ | Mesothelioma-Epithelioid | III | PLE | 26.67 |
| T241 | ++ | ++ | ND | Mesothelioma-Epithelioid | III | PLE | 13.70 |
| N242 | + | + | − | ND | ND | ND | ND |
| T242 | ++ | + | ND | Mesothelioma-Epithelioid | ND | PLE | 7.23 |
| N244 | ++ | + | − | ND | ND | ND | ND |
| T246 | +++ | ++ | ++ | Mesothelioma-Epithelioid | III | PLE | 3.07 |
| T264 | + | ++ | + | Mesothelioma-Epithelioid | IB | PLE | 109.90 |
| T300 | ++ | ND | ND | Mesothelioma-Epithelioid | II | PLE | 34.37 |
| T324 | ++ | + | ND | Mesothelioma-Epithelioid | II | PLE | 27.43 |
| N331 | − | − | +++ | ND | ND | ND | ND |
| T331 | + | − | + | Mesothelioma-Epithelioid | II | PLE | 28.97 |
| T370 | ++ | + | +++ | Mesothelioma-Epithelioid | III | PLE | 11.77 |
| T374 | +++ | ND | ++ | Mesothelioma-Epithelioid | ND | PLE | 9.43 |
| T383 | ND | ND | ND | Mesothelioma-Epithelioid | II | PLE | 9.60 |
| T392 | ++ | + | +++ | Mesothelioma-Epithelioid | IV | PLE | 10.07 |
| T394 | ++ | ++ | + | Mesothelioma-Epithelioid | IB | BIO | 45.60 |
| T396 | +++ | ++ | +++ | Mesothelioma-Epithelioid | III | PLE | 11.47 |
| T401 | − | ND | + | Mesothelioma-Epithelioid | II | CWR | 45.93 |
| T410 | + | ++ | +++ | Mesothelioma-Epithelioid | IV | PLE | 28.33 |
| T416 | +++ | ++ | ++ | Mesothelioma-sarcomatoid | IV | PLE | 3.47 |
| T439 | + | + | ++ | Mesothelioma-Epithelioid | ND | PLE | 1.53 |
| T453 | +++ | ++ | ++ | Mesothelioma-Epithelioid | II | PLE | 14.47 |
| N472 | − | + | ND | ND | ND | ND | ND |
| T472 | + | + | ++ | Mesothelioma-biphasic | III | PLE | 5.33 |
| T473 | ++ | + | +++ | Mesothelioma-Epithelioid | IV | PLE | 0.57 |
| T484 | +++ | +++ | +++ | Mesothelioma-Epithelioid | III | PLE | 7.87 |
| T501 | ++ | + | +++ | Mesothelioma-Epithelioid | III | PLE | 9.37 |
| T514 | ++ | ++ | ++ | Mesothelioma-Epithelioid | II | PLE | 24.97 |
| T713 | +++ | +++ | +++ | Mesothelioma-Epithelioid | ND | PLE | 51.47 |
| T737 | ++ | + | + | Mesothelioma-Epithelioid | III | EPP | 32.23 |
| N745 | ++ | ++ | + | ND | ND | ND | ND |
| T773 | + | + | ND | Mesothelioma-Epithelioid | III | PLE | 42.10 |
| N775 | − | + | − | ND | ND | ND | ND |
| T775 | ++ | ++ | ++ | Mesothelioma-Epithelioid | III | EPP | 32.33 |
| N777 | + | + | − | ND | ND | ND | ND |
| T777 | +++ | ++ | ++ | Mesothelioma-sarcomatoid | III | EPP | 4.37 |
| T795 | + | + | ++ | Mesothelioma-biphasic | III | PLE | 13.13 |
| T809 | +++ | ++ | +++ | Mesothelioma-Epithelioid | III | EPP | 4.93 |
| T813 | + | ++ | ND | Mesothelioma-Epithelioid | III | EPP | 13.47 |
| T829 | ++ | ++ | +++ | Mesothelioma-Epithelioid | III | EPP | 8.17 |
| T839 | + | − | + | Mesothelioma-Epithelioid | II | PLE | 13.60 |
| T858 | + | ++ | +++ | Mesothelioma-Epithelioid | ND | PLE | 2.87 |
| T862 | + | + | + | Mesothelioma-Epithelioid | IB | EPP | 47.83 |
| N869 | − | − | ND | ND | ND | ND | ND |
| T869 | ++ | +++ | +++ | Mesothelioma-Epithelioid | II | PLE | 10.73 |
| T905 | + | − | ++ | Mesothelioma-Epithelioid | ND | ND | 5.03 |
| T936 | ND | ++ | − | Mesothelioma-Epithelioid | II | EPP | 42.00 |
N, normal tissue; T, tumor tissue; IHC, immunohistochemistry; PLE, pleurectomy; BIO, biopsy; CWR, chest-wall resection; EPP, extrapleural pneumonectomy; −, no stain; +, weak stain; ++, moderate stain; +++, strong stain; ND, not done. Information on pathologic type, stage, surgical procedure and survival was not available for every sample.
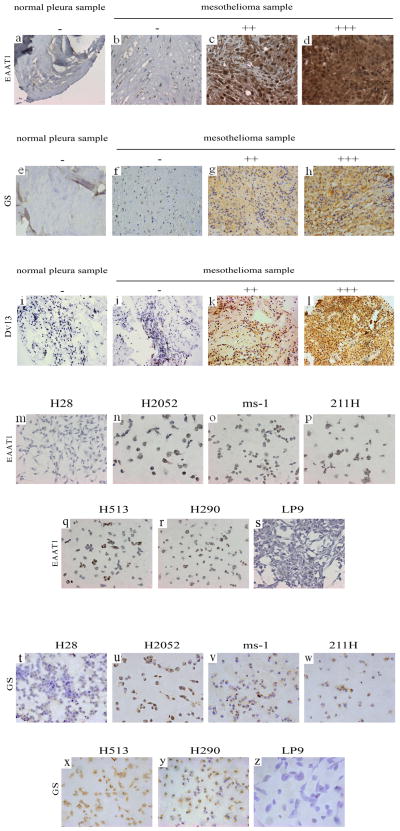
The immunohistochemistry staining results were showed in Figure 1, EAAT1 staining was positive for 97.3% (36/37) of the mesothelioma samples and 50% (5/10) of the normal pleura samples. GS staining was positive for 91.4% (32/35) of mesothelioma samples and 80% (8/10) of normal pleura samples. Dvl3 staining was positive for 96.9% (31/32) of the mesothelioma samples and 43% (3/7) of the normal pleura samples. Staining for anti-EAAT1 and anti-GS antibodies was positive in H2052, H513, MS-1, 211H and H290, and negative in H28 and LP9.
FIG. 1.
Immunohistochemistry of EAAT1, GS and Dvl3 staining of mesothelioma tissue samples, normal pleura samples mesothelioma cell lines and normal mesothelial cell line. a~d: EAAT1 staining results in mesothelioma samples and normal pleura samples. a: normal pleura sample, negative. b~d: mesothelioma samples with negative (b), moderate (c) and strong (d) staining. e~h: GS staining results in mesothelioma samples and normal pleura samples. e: normal pleura sample, negative. f~h: mesothelioma samples, with negative (f), moderate (g) and strong (h) staining. i~l: Dvl3 staining results in mesothelioma samples and normal pleura samples. i: normal pleura sample, negative. j~l: mesothelioma samples, with negative (j), moderate (k) and strong (l) staining. m~s: EAAT1 staining results. m~r: mesothelioma cell lines. m: H28, negative. n: H2052, positive. o: MS-1, positive. p: 211H, positive. q: H513, positive. r: H290, positive. s: normal mesothelial cell line LP9, negative. t~z: GS staining results. t~y: mesothelioma cell lines. t: H28, negative. u: H2052, positive. v: MS-1, positive. w: 211H, positive. x: H513, positive. y: H290, positive. z: normal mesothelial cell line LP9, negative.
The degree of EAAT1 staining associated significantly with survival time (p = 0.028) (Fig. 2a). The degree of GS staining correlated significantly with that of EAAT1 staining (p = 0.002) and Dvl3 staining (p = 0.036), but the association with survival time was not of statistical significance (p= 0.353) (Fig. 2b). The degree of Dvl3 staining correlated with degree of EAAT1 staining (p = 0.035) and associated with survival time (p = 0.001) (Fig. 2c). The result of Cox regression analysis showed the degree of Dvl3 staining was significantly correlated with survival time (p = 0.007).
FIG. 2.
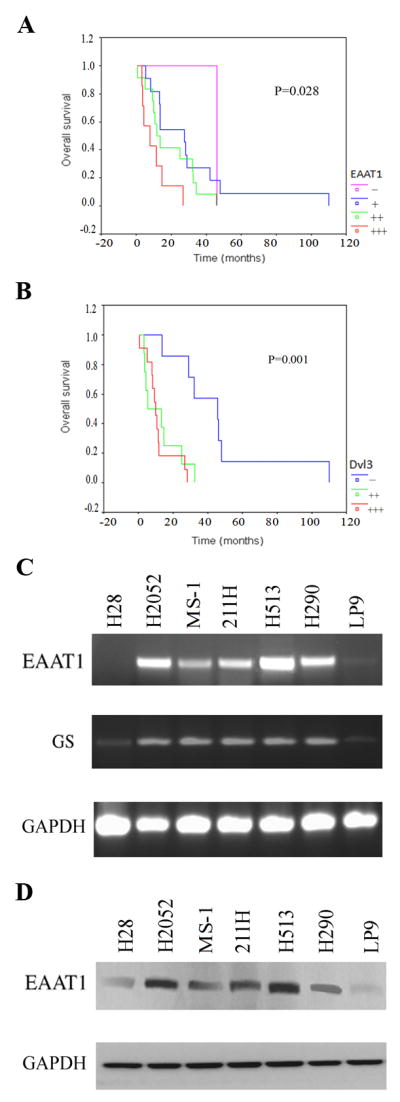
Survival curves and expression of EAAT1 and GS. a~c: Survival curves of MPM patients correlated with EAAT1, GS and Dvl3. Horizontal axis shows time (months); vertical axis shows survival rate. a: survival curves correlated with EAAT1. In Log Rank, the statistic is 9.13, the df is 3 and the significance (p) is 0.028. b: survival curves correlated with GS. In Log Rank, the statistic is 3.26, the df is 3 and the significance (p) is 0.353. c: survival curves correlated with Dvl3. In Log Rank, the statistic is 13.51, the df is 2 and the significance (p) is 0.001.d and e: Expression of EAAT1 and GS in mesothelioma and normal mesothelial cell lines. d: RT-PCR result for EAAT1, GS and GAPDH (as control) in 6 mesothelioma cell lines (H28, H2052, MS-1, 211H, H513 and H290) and 1 normal mesothelial cell line LP9. e: Western blot results of EAAT1 and GAPDH (as control) in 6 mesothelioma cell lines (H28, H2052, MS-1, 211H, H513 and H290) and 1 normal mesothelial cell line, LP9.
The RT-PCR results indicated that EAAT1 was over-expressed inH2052, MS-1, 211H, H513 and H290, and was weakly expressed in LP9 (Fig. 2d). GS was over-expressed in H2052, MS-1, 211H, H513 and H290 and was weakly expressed in H28 and LP9 (Fig. 2d). GAPDH was used as an internal control. Western blot analysis showed that EAAT1 protein expression was high in H2052, MS-1, 211H, H513 and H290, but low in H28 and LP9. GAPDH was detected in all of the 7 cell lines as expected (Fig. 2e).
According to the results in this study, our findings indicate that MPM cells express more EAAT1 than normal cells. Moreover, the survival time of MPM patients was inversely correlated with the degree of EAAT1 staining—the stronger the staining, the shorter the survival time.
GS was expressed in the same five mesothelioma cell lines as EAAT1, as shown by staining, RT-PCR and western blot. These results suggest that GS is downstream of EAAT1 in mesothelioma cells and that EAAT1 and GS are important in MPM. The subcellular co-localization of GS and the EAAT1 allows immediate conversion of transported glutamate into glutamine. Moreover, the function of the glutamate-glutamine cycle in MPM cell growth, proliferation, metastases and survival needs further investigation.
Cadoret and colleagues found a strong correlation between β-catenin and GS.8 This finding suggests a link between activation of the β-catenin signaling and the control of glutamine metabolism. β-catenin is known as downstream molecular of Dvl3. Therefore, Dvl3 also correlates with EAAT1 by the linking of GS. Our result suggested that EAAT1, GS and Dvl3 counterbalance the protective and trophic properties in MPM.
In summary, our study shows that further studies are needed to evaluate EAAT1, Dvl3 and GS in mesothelioma. They may also serve as potential therapeutic targets of MPM in future.
Acknowledgments
We are grateful for support from the Kazan, McClain, Abrams, Fernandez, Lyons, Greenwood, Harley & Oberman Foundation, Inc.; the Estate of Robert Griffiths; the Jeffrey and Karen Peterson Family Foundation; Paul and Michelle Zygielbaum; the Estate of Norman Mancini; and the Barbara Isackson Lung Cancer Research Fund. We thank Lorretta Chan, Rick Baehnev and Mulan Jin for their important guidance in staining. We also thank Pamela Derish and David Hsieh in the UCSF Department of Surgery for editorial assistance with the manuscript.
Funding:
The present work was supported by NIH grant R01 CA140654-01A1 (L. Y.).
Footnotes
Conflict of Interest Statement:
The authors have declared that no competing interests exist.
Statement of Author Contributions Conceived and designed the experiments: TL SH DJ LY. Performed the experiments: TL YCW LY BY. Analyzed the data: JM SH XL JLT AA. Contributed reagents/materials/analysis tools: JM XL JN ZX. Wrote the paper: TL SH DJ YL. Reviewed and revised the manuscript: JLT XL SH LY AA. Gave important directions to the study and revised the manuscript: DJ.
References
- 1.Yang H, Testa JR, Carbone M. Mesothelioma epidemiology, carcinogenesis, and pathogenesis. Curr Treat Options Oncol. 2008;9:147–157. doi: 10.1007/s11864-008-0067-z. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sokol SY. Analysis of Dishevelled signalling pathways during Xenopus development. Curr Biol. 1996;6:1456–1467. doi: 10.1016/s0960-9822(96)00750-6. [DOI] [PubMed] [Google Scholar]
- 3.Uematsu K, Kanazawa S, You L, et al. Wnt Pathway Activation in Mesothelioma: Evidence of Dishevelled Overexpression and Transcriptional Activity of β-Catenin. Cancer Res. 2003;63:4547–4551. [PubMed] [Google Scholar]
- 4.Amara SG, Fontana AC. Excitatory amino acid transporters: keeping up with glutamate. Neurochem Int. 2002;41:313–318. doi: 10.1016/s0197-0186(02)00018-9. [DOI] [PubMed] [Google Scholar]
- 5.Pissimissis N, Papageorgiou E, Lembessis P, et al. The Glutamatergic System Expression in Human PC-3 and LNCaP Prostate Cancer Cells. Anticancer Res. 2009;29:371–377. [PubMed] [Google Scholar]
- 6.Martinez-Hernandez A, Bell KP, Norenberg MD. Glutamine synthetase: glial localization in brain. Science. 1977;195:1356–1358. doi: 10.1126/science.14400. [DOI] [PubMed] [Google Scholar]
- 7.Li T, Li H, Wang Y, et al. The expression of CXCR4, CXCL12 and CXCR7 in malignant pleural mesothelioma. J Pathol. 2011;223:519–530. doi: 10.1002/path.2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cadoret A, Ovejero C, Terris B, et al. New targets of β-catenin signaling in the liver are involved in the glutamine metabolism. Oncogene. 2002;21:8293–8301. doi: 10.1038/sj.onc.1206118. [DOI] [PubMed] [Google Scholar]