Summary
Bacteria of the Bordetella genus cause respiratory tract infections. Both broad host range (e.g. Bordetella bronchiseptica) and human-adapted (e.g. Bordetella pertussis) strains produce a surface-exposed and secreted protein called filamentous haemagglutinin (FHA) that functions in adherence and immunomodulation. Previous studies using B. pertussis and cultured mammalian cells identified several FHA domains with potential roles in host cell interactions, including an Arg-Gly-Asp (RGD) triplet that was reported to bind integrins on epithelial cells and monocytes to activate host signalling pathways. We show here that, in contrast to our previous report, the fhaB genes of B. pertussis and B. bronchiseptica are functionally interchangeable, at least with regard to the various in vitro and in vivo assays investigated. This result is significant because it indicates that information obtained studying FHA using B. bronchiseptica and natural-host animal models should apply to B. pertussis FHA as well. We also show that the C-terminus of mature FHA, which we name the MCD, mediates adherence to epithelial and macrophage-like cells and is required for colonization of the rat respiratory tract and modulation of the inflammatory response in mouse lungs. We could not, however, detect a role for the RGD in any of these processes.
Introduction
Pertussis, or whooping cough, is an acute respiratory disease that is increasing in incidence despite widespread vaccine coverage (Deville et al., 1995; Campos-Outcalt, 2005; Wood and McIntyre, 2008). The causative agents, Bordetella pertussis and Bordetella parapertussishu, are Gram-negative bacteria that infect only humans. Phylogenetic analyses indicate that these bacteria diverged independently and relatively recently from Bordetella bronchiseptica or a B. bronchiseptica-like ancestor and that all three ‘species’ are so closely related that they should be considered subspecies or strains of the same species (Arico et al., 1987; Bemis, 1992; van der Zee et al., 1997; Parkhill et al., 2003; Cummings et al., 2004; Diavatopoulos et al., 2005). Despite this remarkable similarity, B. bronchiseptica differs significantly from B. pertussis and B. parapertussishu by displaying a broad host range that includes animals commonly studied in the laboratory such as rabbits, rats, guinea pigs and mice (Bemis, 1992).
Filamentous haemagglutinin (FHA) was one of the first B. pertussis virulence factors to be discovered (Arai and Sato, 1976; Sato et al., 1981). It is a large, rod-shaped, highly immunogenic protein that is both surface-associated and secreted and is a primary component of acellular pertussis vaccines (Arai and Sato, 1976; Sato et al., 1981; Jacob-Dubuisson et al., 1999; 2000; Sato and Sato, 1999). A prototypical member of the Two Partner Secretion (TPS) pathway family, FHA is first synthesized as an ~370 kDa preproprotein called FhaB that loses its 71-amino-acid (aa) signal sequence during Sec-dependent secretion across the cytoplasmic membrane (Jacob-Dubuisson et al., 1996; Chevalier et al., 2004). The N-terminal ~250 aa ‘TPS domain’ targets the proprotein in the periplasm to FhaC, an outer membrane, β-barrel, channel-forming protein that is required for translocation of FhaB to the cell surface (Clantin et al., 2004; 2007; Hodak et al., 2006; Meli et al., 2006). At some point during translocation, FhaB is cleaved in an SphB1-dependent manner to form the mature ~240 kDa FHA protein (Coutte et al., 2001; Mazar and Cotter, 2006). SphB1-dependent cleavage occurs in at least two locations, one somewhere between aa 2362 and 2372 (based on the aa numbering of FhaB predicted from the fhaB gene of B. pertussis Tohama 1) and the other approximately 100 aa N-terminal to that site (Coutte et al., 2001; Mazar and Cotter, 2006). The importance of cleavage at one site versus the other is unknown. The ~130 kDa C-terminal prodomain that is released upon cleavage cannot be detected in whole cell lysates (WCLs) or concentrated supernatants, presumably due to its rapid degradation (Renauld-Mongenie et al., 1996; Mazar and Cotter, 2006). X-ray crystallography, high-resolution electron microscopy, and modelling studies indicate that mature FHA is shaped like a horseshoe nail, with the N-terminal ~2000 aa forming a β-helix that makes up the ‘shaft’ and the C-terminal ~500 aa forming a globular domain (which we call the MCD, for mature C-terminal domain) at one end (Kajava et al., 2001; Clantin et al., 2004). We showed recently that mature cell-associated FHA is oriented such that its C-terminus (the MCD), and not its N-terminus, is located distally from the cell surface and is accessible to antibodies (Mazar and Cotter, 2006).
In vitro studies using B. pertussis or FHA purified from B. pertussis have identified three putative functional domains (see Fig. 3 for a schematic showing their relative locations). A heparin binding domain (HBD) located near the N-terminus of FHA has been reported to mediate attachment to sulphated polysaccharides (Hannah et al., 1994). A carbohydrate recognition domain (CRD) located near the centre of the protein has been reported to mediate adherence to respiratory epithelial cells and macrophages (Prasad et al., 1993) and an arg-gly asp (RGD) triplet, located just N-terminal to the CRD, has been reported to interact with the leucocyte response integrin/ integrin-associated protein (LRI/IAP) complex on monocytes/macrophages, resulting in upregulation of complement receptor 3 (CR3) binding activity (Ishibashi et al., 1994), and with very late antigen 5 (VLA-5) on epithelial cells to stimulate the upregulation of intercellular adhesion molecule 1 (ICAM-1) (Ishibashi and Nishikawa, 2002; 2003). FHA has also been reported to inhibit antigen-dependent CD4+ T cell proliferation and to induce apoptosis when incubated with monocytes/macrophages (Boschwitz et al., 1997a; Abramson et al., 2001) and to induce immunosuppressive effects on murine macrophages and dendritic cells by downregulating production of IL-12 in an IL-10-dependent manner (McGuirk and Mills, 2000; McGuirk et al., 2002). Based on these and other studies, FHA has been proposed to function as an adhesin and an immunomodulator.
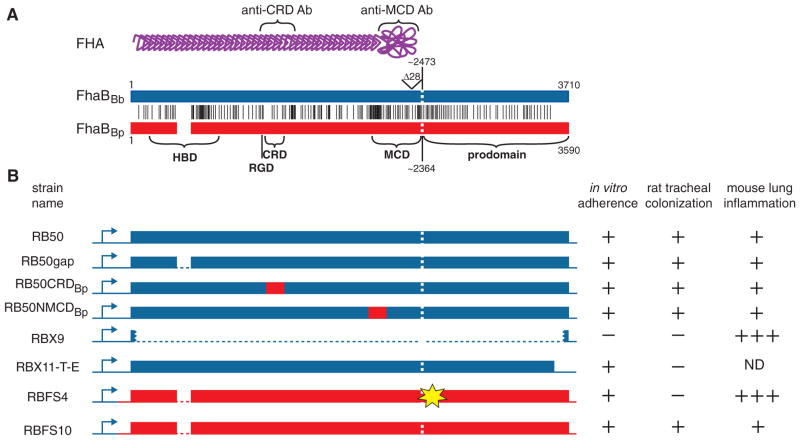
Fig. 3. A. Comparison of the FhaB proteins predicted for B. bronchiseptica RB50 (FhaBBb, blue) and B. pertussis Tohama 1 (FhaBBp, red). Vertical black lines represent the positions of amino acid differences. FhaBBb contains six additional 19 aa repeats near the N-terminus (white space in FhaBBp). The vertical white dashed line represents the site of SphB1-dependent maturation. The various domains are designated across the bottom. A schematic of mature FHA is shown at the top in purple. The regions used for the production of the anti-CRD and anti-MCD antibodies are indicated.
B. Schematic of the various ‘chimeric’ strains constructed and their phenotypes in adherence to L2 cells, tracheal colonization in Wistar rats, and lung inflammation in the lungs of BALB/c mice. Dark blue represents B. bronchiseptica RB50 DNA, red represents B. pertussis Tohama 1 DNA. The yellow ‘star’ in RBFS4 represents the location of the insertion mutation. ND, not determined. The tabular part of part B summarizes many animal experiments. Those involving strains RB50, RBX9, RBX11-T-E, RBFS4 and RBFS10 have been performed many times. Rat and mouse experiments using RB50gap, RB50CRDBp and RB50NMCDBp were performed once.
Studies aimed at determining roles for FHA in vivo using B. pertussis and mouse models have yielded conflicting data, with most failing to reveal any difference between wild-type and FHA-deficient bacteria (Weiss and Goodwin, 1989; Goodwin and Weiss, 1990; Kimura et al., 1990; Roberts et al., 1993; Khelef et al., 1994; Alonso et al., 2001; McGuirk et al., 2002). Lack of a clear phenotype for fhaB mutants in these studies may be due to the fact that mice are not natural-hosts for B. pertussis.
The FhaB protein produced by B. bronchiseptica strain RB50 is predicted to be 90% identical and 93% similar to FhaB of B. pertussis strain Tohama 1 (Parkhill et al., 2003) and in vitro studies have shown FHA to be both necessary and sufficient for mediating adherence of B. bronchiseptica to a variety of epithelial and macrophage cell lines (Cotter et al., 1998; Mattoo et al., 2000; Inatsuka et al., 2005; Julio and Cotter, 2005). By contrast with studies using B. pertussis, animal experiments revealed dramatic differences between wild-type and FHA-deficient B. bronchiseptica. For example, while wild-type B. bronchiseptica persistently colonizes both the nasal cavity and trachea of rats and mice inoculated with a relatively small number of bacteria delivered in a small volume to the nares, ΔfhaB mutants are only able to colonize the nasal cavity, and often with decreased efficiency (Cotter et al., 1998; Mattoo et al., 2000; Julio and Cotter, 2005). Mice inoculated with a large number of FHA-deficient B. bronchiseptica in a large volume that deposits bacteria into the lungs produce a robust inflammatory response that is often fatal while those inoculated with the same number of wild-type bacteria remain healthy (Inatsuka et al., 2005). These data suggest that FHA contributes to colonization of the lower respiratory tract by allowing B. bronchiseptica to suppress the inflammatory response. We reported previously that the fhaB gene of B. pertussis (fhaBBp) could not substitute for the fhaB gene of B. bronchiseptica (fhaBBb) during infection (Inatsuka et al., 2005). The goals of the current study were to determine the molecular basis for the lack of fhaBBp and fhaBBb interchangeability in vivo and to investigate the roles of the RGD triplet and the C-terminus of the mature FHA protein (the MCD) in pathogenesis.
Results
Construction and in vitro characterization of a B. pertussis Tohama 1 derivative expressing fhaB from B. bronchiseptica RB50
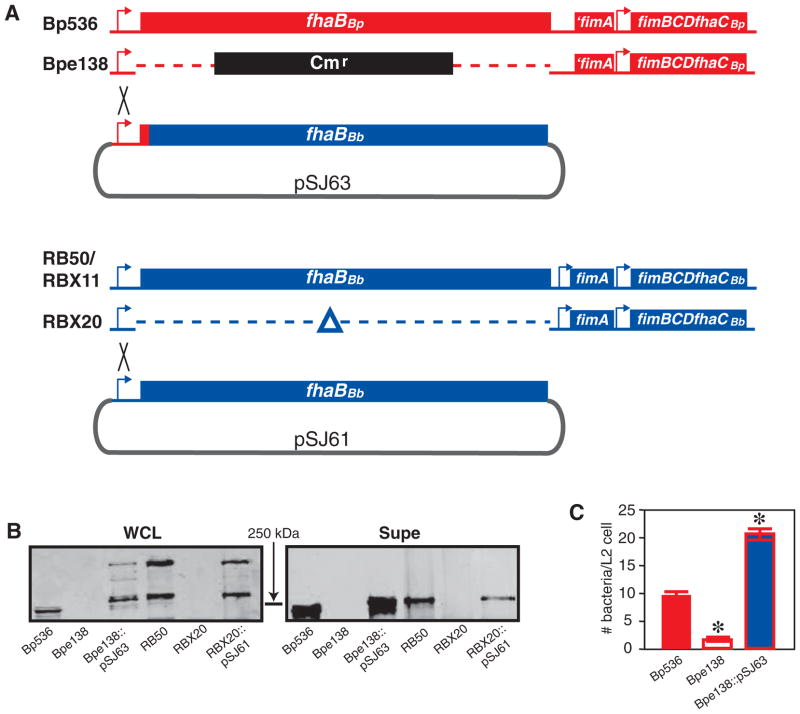
We reported previously that fhaBBp could substitute for fhaBBb in vitro but not in vivo (Inatsuka et al., 2005). To determine if fhaBBb could substitute for fhaBBp, we constructed a B. pertussis strain expressing fhaBBb. Bp536 is a streptomycin-resistant derivative of Tohama 1, the B. pertussis strain for which the genome sequence was determined (Parkhill et al., 2003). We used allelic exchange to construct a derivative of Bp536, called Bpe138, in which the entire fhaBBp coding sequence was replaced with a gene encoding chloramphenicol resistance (Fig. 1A). Plasmid pSJ63 contains all but the first 70 codons of fhaBBb from RB50 fused to the fhaBBp promoter region plus the first 70 codons of fhaBBp. The first 70 aa encoded by fhaBBp and fhaBBb differ at only two positions and because the FhaB signal sequence is 71 aa long, neither of those aa are present in the FhaB proprotein or mature FHA. Integration of pSJ63 into the chromosome of Bpe138 via recombination within the promoter region (confirmed by PCR) resulted in a B. pertussis strain (Bpe138::pSJ63) that expressed fhaBBb (with the first 70 codons from fhaBBp) from the native B. pertussis fhaB promoter. Expression of genes 3′ to fhaB (fimBCD and fhaC) is expected to be the same in this strain as in Bp536 and Bpe138. [Note that fimA in B. pertussis (‘fimA) lacks transcription and translation initiation sequences and the fimBCDfhaC operon is transcribed from the fimB promoter (Boschwitz et al., 1997b).]
Fig. 1. Expression of fhaBBb in B. pertussis.
A. Schematic of strains used. The B. pertussis fhaB locus is shown in red and the B. bronchiseptica fhaB locus is shown in blue. The regions of integration of pSJ63 and pSJ61 into the chromosomes of Bpe138 and RBX20 to form Bpe138::pSJ63 and RBX20::pSJ61, respectively, are indicated by the black crosses.
B. Western blot showing FhaB (~370 kDa) and FHA (~250 kDa) in whole cell lysates (WCLs) and concentrated supernatants (Supe) of the various B. pertussis and B. bronchiseptica strains as indicated below each lane. Blots were probed with the anti-CRD antibody. The position of the 250 kDa molecular mass marker is shown.
C. Adherence of wild-type and mutant B. pertussis strains to L2 cells (moi = 200). Asterisks indicate a statistically significant (P < 0.05).
Western blot analysis showed that Bpe138 did not produce FHA and that Bpe138::pSJ63 produced and secreted FHABb, which is slightly larger than FHABp (Fig. 1B). [The fact that FHA can be detected in strain Bpe138::pSJ63 provides functional evidence that genes downstream of fhaB are expressed because neither FhaB nor FHA can be detected in fhaC mutants (Willems et al., 1994; Jacob-Dubuisson et al., 1997; Julio and Cotter, 2005).] As shown previously, maturation of the ~370 kDa FhaB proprotein to mature ~250 kDa FHA appears to be more efficient in B. pertussis Tohama 1 and its derivatives than in B. bronchiseptica RB50 and its derivatives because the ~370 kDa FhaB proprotein is visible in WCLs of RB50 but not Tohama 1 derivatives such as Bp536 (Fig. 1B) and BPSM (Mazar and Cotter, 2006). Also as shown previously, Tohama 1 derivatives release more FHA into culture supernatants than RB50 and its derivatives (Fig. 1B and Mazar and Cotter, 2006). Both strain background and the specific aa sequences of the FhaB proteins apparently contribute to both phenotypes because the amount of the ~370 kDa FhaB proprotein visible in WCLs of Bpe138::pSJ63 and the amount of FHA visible in culture supernatants of Bpe138::pSJ63 are intermediate between those of Bp536 and RB50 (note that the supernatant sample used for Bp536 was diluted twofold compared with the others) (Fig. 1B).
We and others have shown that FHA contributes to adherence of B. pertussis to epithelial and macrophage-like cell lines in vitro (Relman et al., 1990; Inatsuka et al., 2005). Consistent with these previous data, Bpe138 was less able to adhere to rat lung epithelial (L2) cells than Bp536 (Fig. 1C). Expression of fhaBBb in Bpe138 restored its ability to adhere to L2 cells (Fig. 1C) and J774A.1 macrophage-like cells (data not shown). The FHABb protein produced in Bpe138::pSJ63 is therefore functional with regard to mediating adherence to cultured cells.
Substitution of fhaB in B. pertussis Tohama 1 with fhaB from B. bronchiseptica RB50 does not alter the ability of B. pertussis Tohama 1 to cause respiratory infection in rats or mice
Inoculation of Wistar rats with as few as 20 colony-forming units (cfu) of B. bronchiseptica results in colonization of the nasal cavities and tracheas with high numbers of bacteria by day 10 post inoculation (Akerley et al., 1995). B. bronchiseptica fhaB mutants are unable to colonize the rat trachea and show decreased ability to colonize the nasal cavity (Cotter et al., 1998; Mattoo et al., 2000; Inatsuka et al., 2005; Julio and Cotter, 2005). Wild-type B. pertussis, by contrast, is unable to colonize either the nasal cavity or trachea of rats, even when inoculated at a dose of 106 cfu (our unpublished observation). To determine if FHABb could increase the ability of B. pertussis to establish respiratory infection in rats, we inoculated Wistar rats intranasally with 1 × 106 cfu of Bp536, Bpe138 and Bpe138::pSJ63. No Bordetella were recovered from the nasal cavities or tracheas of any B. pertussis-inoculated animal at 14 days post inoculation (data not shown). As a control to demonstrate that the fhaBBb gene contained on plasmid pSJ63 encodes an FHA protein that is functional in vivo, we also inoculated rats with B. bronchiseptica strains RBX11, RBX20 and RBX20::pSJ61. RBX11 is an RB50 derivative containing a large in-frame deletion mutation in fhaS. fhaS is an fhaB homologue that plays no discernible role in the infection of rats or mice by B. bronchiseptica (Julio and Cotter, 2005), but which, due to its high degree of nucleotide identity with fhaB, complicates genetic manipulation of the fhaB gene. RBX20 is an RB50 derivative containing large in-frame deletion mutations in fhaB and fhaS. pSJ61 is the parent plasmid of pSJ63. It contains the entire fhaB gene and promoter region from RB50 (Fig. 1A). At day 14 post inoculation with 1000 cfu, RBX11 and RBX20::pSJ61 were recovered from the nasal cavities and tracheas at high numbers while RBX20 was not recovered from any trachea and was recovered at low numbers from the nasal cavities of inoculated animals (data not shown). Together, these results indicate that the fhaB gene on pSJ61 (and hence pSJ63) encodes an FHA protein that is functional in vivo, but that production of this protein is not sufficient to allow B. pertussis to establish respiratory infection in rats.
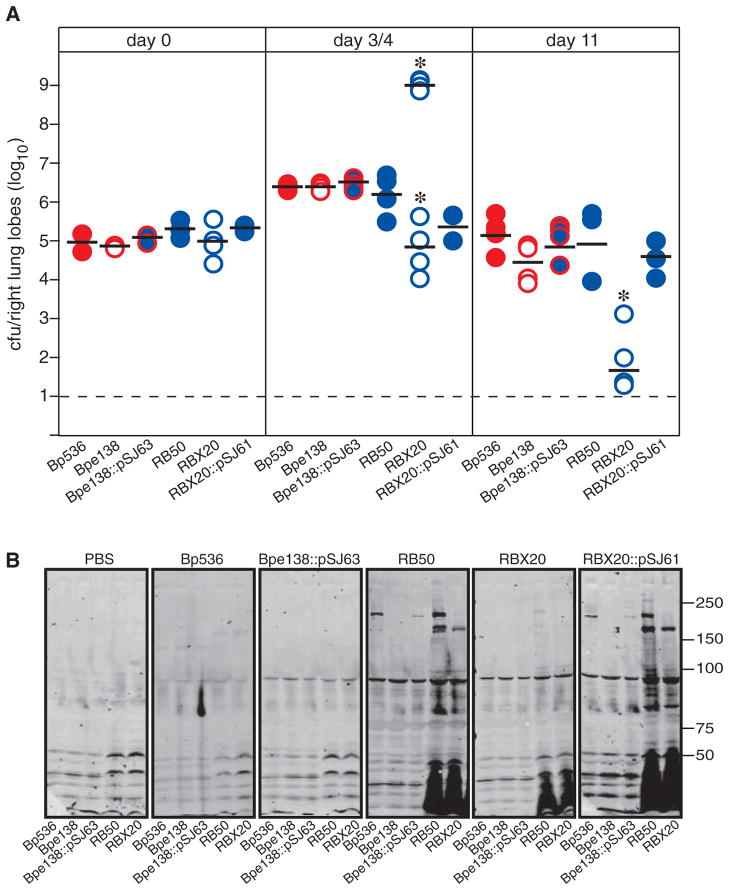
Although it does not reflect a natural course of infection, inoculation of mice intranasally with a large number of bacteria (5 × 105 cfu) delivered in a large volume (50 μl) has proven to be a useful model for investigating the ability of Bordetella to induce an inflammatory response and to resist clearance by inflammatory cells (Harvill et al., 1999a,b; Inatsuka et al., 2005). Consistent with our previously published results (Inatsuka et al., 2005), ~1 × 106 cfu of RB50 were recovered from the lungs at day 4 post inoculation and ~1 × 105 cfu of RB50 were recovered at day 11 post inoculation using this protocol (Fig. 2A). Also consistent with our previous results, inoculation with the ΔfhaB strain, RBX20, resulted in a bimodal response; half of the animals became moribund by day 4 and very high numbers (~1 × 109) of cfu were recovered from their lungs and half of the animals remained healthy and the number of cfu recovered from their lungs was slightly lower than the number recovered from the lungs of RB50-inoculated animals (Fig. 2A). For RBX20 therefore the LD50 is ~5 × 105 cfu when inoculated using this protocol. In animals that remained healthy, the number of cfu recovered from the lungs at day 11 post inoculation was very low. The lungs of animals that appeared healthy at day 4 post inoculation showed only mild inflammation as assessed by microscopic examination of haematoxylin and eosin (H&E)-stained tissue sections, while the lungs of moribund animals showed massive infiltration of inflammatory cells that included primarily neutrophils and lymphocytes and areas of infarction and fluid accumulation were also observed (data not shown and Inatsuka et al., 2005). We have interpreted these data to indicate that FHA functions to modulate the robustness of the inflammatory response; without FHA, a (hyper)inflammatory response is induced that either clears the bacteria quickly, or causes local tissue damage that promotes increased bacterial growth, more inflammation, more damage and ultimately death of the mouse (Inatsuka et al., 2005). RBX20::pSJ61 was recovered from the lungs at numbers similar to RB50 at all time points (Fig. 2A) demonstrating that the fhaBBb gene contained on pSJ61 (and pSJ63) encodes a protein that is capable of allowing B. bronchiseptica to modulate the inflammatory response in mice. When inoculated using this protocol, the number of cfu of B. pertussis recovered from the lungs of mice also increases at days 3–7 post inoculation, then decreases over time, but differences between wild-type and fhaB mutant strains have not been consistently observed and the bimodal response that we observed for the ΔfhaB strain of B. bronchiseptica has not been reported for fhaB mutant strains of B. pertussis (Kimura et al., 1990; Khelef et al., 1994; Harvill et al., 1999a; 2000; Alonso et al., 2001). To determine if fhaBBb alters the interaction between B. pertussis and the murine respiratory tract, we inoculated mice with our various B. pertussis strains. Consistent with previous reports (Kimura et al., 1990; Khelef et al., 1994; Harvill et al., 1999a; 2000; Alonso et al., 2001), ~3 × 106 and ~2 × 105 cfu of Bp536 were recovered from the lungs at days 4 and 11 post inoculation respectively (Fig. 2A). Also consistent with previous reports (Kimura et al., 1990; Khelef et al., 1994; Alonso et al., 2001), the number of cfu of the ΔfhaB B. pertussis strain (Bpe138) recovered from the lungs was not significantly different from the number of cfu of its wild-type parental strain (Bp536) recovered at any time point. Moreover, examination of H&E-stained lung sections revealed only mild inflammation in all B. pertussis-inoculated animals (data not shown). In contrast to the case with B. bronchiseptica therefore, there was no evidence that a more robust inflammatory response was induced in animals inoculated with ΔfhaB B. pertussis strains compared with wild-type B. pertussis. The number of cfu recovered from the lungs of Bpe138::pSJ61-inoculated animals at all time points was not different from those recovered from Bp536- and Bpe138-inoculated animals (Fig. 2A).
Fig. 2. Expression of fhaBBb in B. pertussis does not alter its ability to infect mice.
A. The number of cfu recovered from the lungs of BALB/c mice at days 0, 3 or 4, and 11 post inoculation with 50 μl PBS containing 5 × 105 cfu of the indicated strains is shown. Each circle represents the number of cfu recovered from a single animal. The horizontal black bar is the geometric mean for each group. For RBX20 at day 3/4, the means of numbers from moribund animals (~109) and healthy animals (~105) were calculated separately as shown. The dashed line represents the lower limit of detection. Asterisks indicate a statistically significant difference compared with RB50 (P < 0.05). This experiment has been performed at least twice with similar results. The data from one experiment are shown.
B. Western blots of whole cell lysates of the strains indicated across the bottom probed with serum from mice inoculated with PBS or the strains indicated across the top. The sera were collected 21 days post inoculation. Molecular mass markers are shown on the right.
As an additional assessment of the ability of the various strains to establish infection, we compared the serum antibody responses of inoculated animals by Western blot analysis. As shown previously (Harvill et al., 1999a), sera from RB50-inoculated mice contain antibodies that recognize Bordetella-specific antigens, such as lipopolysaccharide, adenylate cyclase and FHA (Fig. 2B). Also consistent with our previous results (Inatsuka et al., 2005), the antibody response generated in animals inoculated with FHA-deficient B. bronchiseptica is weak. The Western blot results obtained using sera from Bp536- and Bpe138::pSJ63-inoculated animals did not differ from those obtained using sera from phosphate-buffered saline (PBS)-inoculated animals (Fig. 2B). These results indicate, consistent with previous results (Harvill et al., 1999a), that inoculation of mice with B. pertussis does not result in an antibody response that can be detected by Western blot, and that although expression of FHABb contributes to the ability of B. bronchiseptica to induce a strong antibody response, it does not alter the antibody response induced by B. pertussis.
Taken together, these results indicate that expression of fhaBBb in B. pertussis does not alter its ability to establish respiratory infection in rats, to induce an inflammatory response in mice, to resist clearance by inflammatory cells in mice, or to induce adaptive immunity in mice. FHABb therefore does not improve the ability of B. pertussis to infect rats and mice. Whether FHABb can actually substitute for FHABp in vivo, however, cannot be concluded from these experiments because these in vivo assays, which use an animal not normally infected by B. pertussis, could not distinguish ΔfhaB B. pertussis from wild-type B. pertussis.
FHABp can substitute for FHABb with regard to rat and mouse infection
We reported previously that FHABp could not substitute for FHABb in B. bronchiseptica in vivo (Inatsuka et al., 2005). The strain used in that study, RBFS4, was believed to contain and express an intact wild-type fhaB allele from B. pertussis: it produced FHABp of the expected mature size that was localized on the surface of the bacteria and was also secreted in a manner similar to wild-type B. bronchiseptica. Like RB50, RBFS4 was able to adhere to epithelial cells and macrophages (albeit with somewhat reduced ability), but like RBX9 (the B. bronchiseptica ΔfhaB derivative), RBFS4 was unable to colonize the tracheas of rats and it elicited a hyperinflammatory response in mouse lungs. Because a majority of the aa differences between FHABb and FHABp are within the HBD, the CRD and the N-terminal half of the MCD, we constructed B. bronchiseptica strains producing chimeric FHA proteins (Fig. 3) to determine if differences in one of these regions could account for the different phenotypes displayed by RB50 and RBFS4. All of these strains were indistinguishable from RB50 in their ability to adhere to cultured cells and to infect rats and mice (data summarized in Fig. 3). At the same time that we were conducting these experiments, we found that deletions in the region of fhaB encoding the C-terminal prodomain (which is removed from the mature FHA protein at some point in the secretion process and which cannot be detected as a separate protein in WCLs or culture supernatants) resulted in strains that produced and secreted mature FHA but were unable to colonize the tracheas of rats (Mazar and Cotter, 2006). One of these strains (RBX11-T-E) was able to adhere to epithelial and macrophage-like cell lines, but with slightly reduced ability compared with RB50 (Fig. 3 and Mazar and Cotter, 2006). This strain therefore displayed a phenotypic profile identical to that of RBFS4 (although the inflammatory response to RBX11-T-E was not investigated in the previous study). In light of this information, and the fact that expression of fhaBBb in B. pertussis did not alter its ability to infect rats or mice, we resequenced the DNA region in RBFS4 encoding the prodomain. This new sequence information revealed three thymidines instead of two at nucleotide position 7126–7127 (relative to the adenosine in the translational start codon of fhaBBb), which is located about 10 codons 3′ to the region encoding the putative primary SphB1-dependent maturation site. The additional thymidine is predicted to shift the reading frame such that it is followed by 42-aa-encoding codons and then a stop codon. Using allelic exchange, we ‘repaired’ the frame-shift mutation in RBFS4 to create RBFS10. RBFS10 was indistinguishable from RB50 in its ability to adhere to epithelial and macrophage-like cells, to colonize the tracheas of rats, and to colonize the lungs of mice with the induction of only a mild inflammatory response (Fig. 3). These results show that, in contrast to our previously published report, the fhaB gene from B. pertussis can substitute for that of B. bronchiseptica with regard to these in vitro and in vivo phenotypes. These data also show that individual FHA domains are also interchangeable, not just the entire molecules.
The FHA RGD triplet does not appear to play a role in the ability of B. bronchiseptica RB50 to cause respiratory infection in rats and mice
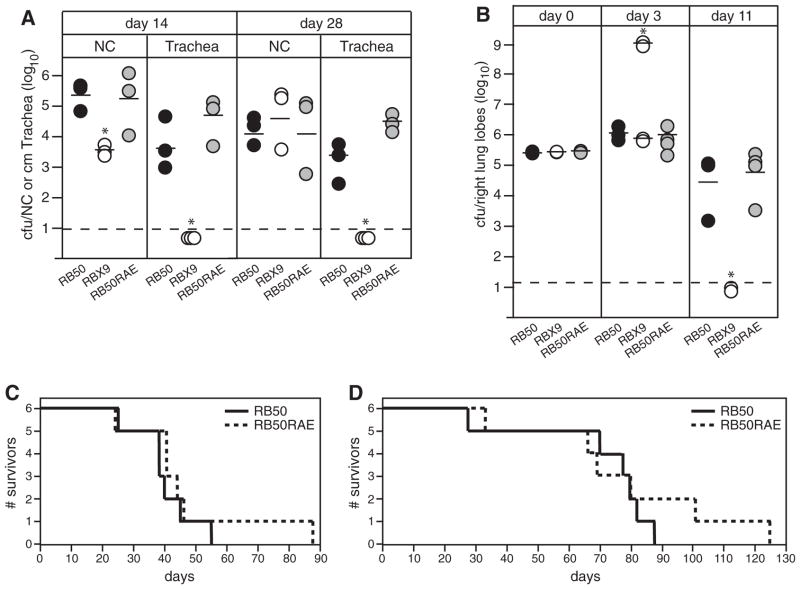
The results described above indicate that FHABb and FHABp are functionally interchangeable, at least with regard to the in vitro and in vivo phenotypes that we have studied. Information gleaned about FHA function from studies using B. bronchiseptica may therefore apply to FHA function in B. pertussis as well. Several reports using B. pertussis, FHA purified from B. pertussis, and various cell culture models have suggested a role for the RGD triplet located near the middle of the mature FHA protein in pathogenesis (Relman et al., 1990; Ishibashi et al., 1994; 2001; 2002; Ishibashi and Nishikawa, 2002; 2003). To investigate the role of the RGD triplet in vivo, we constructed B. bronchiseptica strains producing FHA proteins in which the RGD was replaced with RAD, RGE or RAE. These substitutions were chosen because although they are relatively conservative changes, each individually has been shown to abrogate cell-attachment activity of RGD containing proteins (Pierschbacher and Ruoslahti, 1984). All of the strains produced and secreted FHA proteins that were indistinguishable in size and amount from RB50 and all of the strains adhered to L2 cells and J774A.1 cells in a manner indistinguishable from RB50 (data not shown). We hypothesized that the RAE substitution, which differed the most from the wild-type sequence, would be the most likely to alter FHA function and therefore we used only the strain expressing this FHA mutant (which we called RB50RAE) in all subsequent experiments. RB50RAE was indistinguishable from RB50 in its ability to colonize the nasal cavities and tracheas of rats (Fig. 4A) and it was recovered at the same numbers as RB50 from the lungs of mice at days 3 and 11 post inoculation (Fig. 4B). Lung tissues from mice infected with RB50 and RB50RAE that were sectioned and stained with H&E were indistinguishable (data not shown). RB50RAE was also similar to RB50 in its ability to cause a lethal infection in immunodeficient SCID/Bg mice following both low-dose and high-dose inoculation (Fig. 4C and D). These results indicate that the RGD motif does not contribute to the ability of B. bronchiseptica to establish respiratory infections in rats, to induce or suppress an inflammatory response in mice, or to resist inflammation-mediated clearance in mice, at least not in an way that can be distinguished using this repertoire of animal models.
Fig. 4. Assessment of the FHA RGD triplet in B. bronchiseptica pathogenesis.
A. Wistar rats were inoculated intranasally with 10 μl PBS containing 1000 cfu of B. bronchiseptica (strains indicated across the bottom) and the number of cfu recovered from the nasal cavity (NC) and 1 cm trachea determined at days 14 and 28 post inoculation. Each circle represents the number of cfu recovered from a single animal. The horizontal line shows the geometric mean for each group. The dashed line represents the lower limit of detection. y-axis is log scale. Asterisks indicate a statistically significant difference compared with RB50 (P < 0.05).
B. BALB/c mice were inoculated intranasally with 50 μl PBS containing 5 × 105 cfu of B. bronchiseptica (strains indicated across the bottom) and the number of cfu in the right lung lobes was determined 60 min (day 0), 3 and 11 days post inoculation. Each circle represents the number of cfu recovered from a single animal. The horizontal line shows the geometric mean for each group. The dashed line represents the lower limit of detection. For RBX9, the means at day 3 for moribund (~109) and healthy (~106) animals were calculated separately as indicated. y-axis is log scale. Asterisks indicate a statistically significant difference compared with RB50 (P < 0.05).
C and D. SCID/Bg mice were inoculated intranasally with 50 μl PBS containing 5 × 105 (for C) or 10 μl containing 1000 (for D) cfu of RB50 (solid line) or RB50RAE (dashed line) and the number of survivors plotted over time. The mean time to death was not significantly different between RB50 and RB50RAE for either experiment, as determined by the Log Rank (Mantel-Cox) test. RB50 and RBX9 have been compared in these models many times with similar results. Each type of animal experiment that included the RB50RAE strain was performed once and not repeated in the interest of minimizing the number of animals used because no difference between RB50RAE and RB50 was detected in any of the experiments.
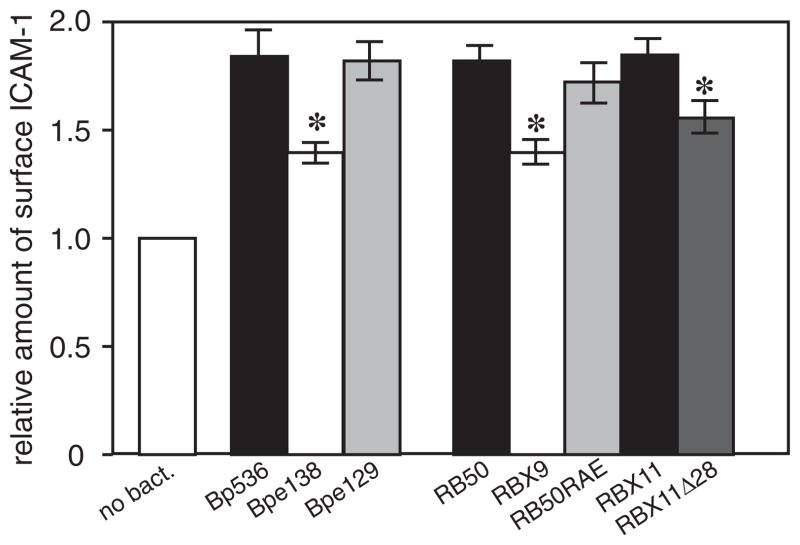
The FHA RGD triplet does not appear to contribute to Bordetella-induced ICAM-1 surface expression in respiratory epithelial cells
Lack of a difference between RB50 and RB50RAE in rats and mice was somewhat unexpected given previous publications (Relman et al., 1990; Ishibashi et al., 1994; 2001; 2002; Ishibashi and Nishikawa, 2002; 2003). Among the activities attributed to the FHA RGD is the upregulation of ICAM-1 on the surface of epithelial cells (Ishibashi and Nishikawa, 2002). We therefore determined if B. bronchiseptica could induce surface expression of ICAM-1 in BEAS-2B cells, and, if so, if FHA, and specifically the RGD triplet of FHA, was required for this ability. We incubated BEAS-2B cells with RB50, RBX9, RB50RAE and also the B. pertussis strains used in previous studies, at a multiplicity of infection (moi) of 100 and measured surface ICAM-1 by flow cytometry. Both B. bronchiseptica and B. pertussis caused a modest increase in surface expression of ICAM-1 in an FHA-dependent manner, consistent with previous studies with B. pertussis (Fig. 5) (Ishibashi and Nishikawa, 2002). By contrast with those studies, however, the RGD motif was not required for this activity because the amount of ICAM-1 on the surface of BEAS-2B cells following incubation with the B. pertussis RAD mutant and the B. bronchiseptica RAE mutant was the same as following incubation with either wild-type strain (Fig. 5). These data indicate that the FHA RGD motif does not contribute to the increased surface expression of ICAM-1 that occurs in epithelial cells in response to exposure to either B. bronchiseptica or B. pertussis.
Fig. 5.
ICAM-1 induction. BEAS-2B cells were incubated with the indicated strains of B. bronchiseptica or B. pertussis at a moi of 100 and the amount of surface ICAM-1 was determined by flow cytometry. (Bp536 = wild-type B. pertussis, Bpe138 = B. pertussis ΔfhaB, Bpe129 = B. pertussis RAD mutant, RB50 = wild-type B. bronchiseptica, RBX9 = RB50ΔfhaB, RB50RAE = RB50 RAE mutant, RBX11 = RB50ΔfhaS, RBX11Δ28 = RB50 containing the FHA ‘Δ28’ mutation.) Fluorescence of cells incubated with PBS alone was set as a value of one and relative levels of fluorescence in samples incubated with bacteria are shown. Asterisks indicate a statistically significant difference compared with the wild-type parental strain (P < 0.05).
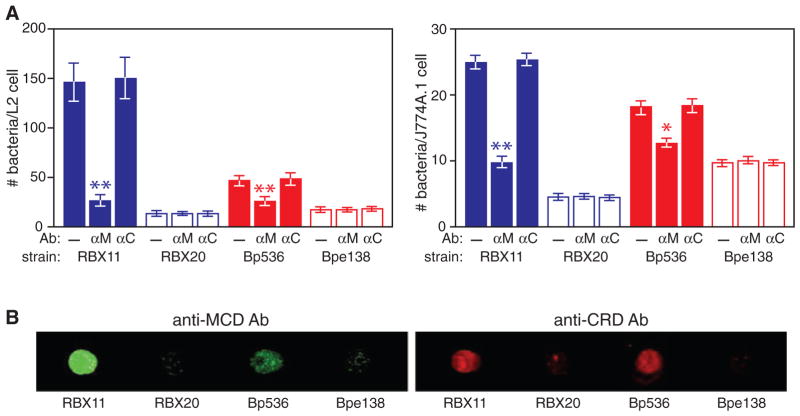
Antibodies against the MCD, but not those against the CRD, block FHA-mediated adherence of B. pertussis and B. bronchiseptica to epithelial and macrophage-like cell lines
We showed recently that the C-terminus of mature FHA (the MCD) is exposed distally on the cell surface (Mazar and Cotter, 2006). The CRD and the RGD, however, have been suggested to mediate interactions with host cells (Relman et al., 1990; Prasad et al., 1993; Ishibashi et al., 1994; 2002; Ishibashi and Nishikawa, 2002; 2003). To investigate the importance of these regions in adherence, we incubated bacteria with anti-CRD or anti-MCD antibodies, or buffer alone, washed away unbound antibody, and then determined the ability of these bacteria to adhere to L2 cells and J774A.1 macrophage-like cells. Pre-incubation with anti-MCD antibodies resulted in adherence levels of B. pertussis and B. bronchiseptica that were dramatically lower than adherence in the absence of incubation with antibody (Fig. 6A). By contrast, pre-incubation of bacteria with anti-CRD antibodies had no effect on the level of adherence. (Note that the polypeptide used to generate these antibodies included the RGD triplet.) Aliquots of the same bacteria that were used in the adherence assay were also incubated with fluorophore-conjugated secondary antibodies, washed, spotted onto membranes and examined for fluorescence. Both the anti-CRD and anti-MCD antibodies were detected on FHA+ but not FHA− B. bronchiseptica and B. pertussis (Fig. 6B). These results provide evidence that the MCD mediates adherence to epithelial and macrophage-like cells and that at least portions of the CRD are not required for this activity.
Fig. 6. Antibodies to the MCD, but not the CRD, block adherence to L2 and J774A.1 cells.
A. B. pertussis and B. bronchiseptica strains (indicated) were incubated with anti-MCD (αM) or anti-CRD (αC) antibodies then tested for adherence to L2 or J774A.1 cells. The number of bacteria per cell is shown. Asterisks indicate statistically significant differences in adherence compared with wild-type bacteria that were not incubated with antibody. *P < 0.05 and **P < 0.01.
B. Aliquots of the bacteria used in the adherence assays were incubated with goat anti-chicken antibody conjugated to IRdye 680 (for anti-CRD) or goat anti-rabbit conjugated to IRdye 800 (for anti-MCD), washed, then spotted into nitrocellulose and fluorescence was visualized using an Odyssey imager.
The C-terminal domain of mature FHA is required for FHA function in vivo
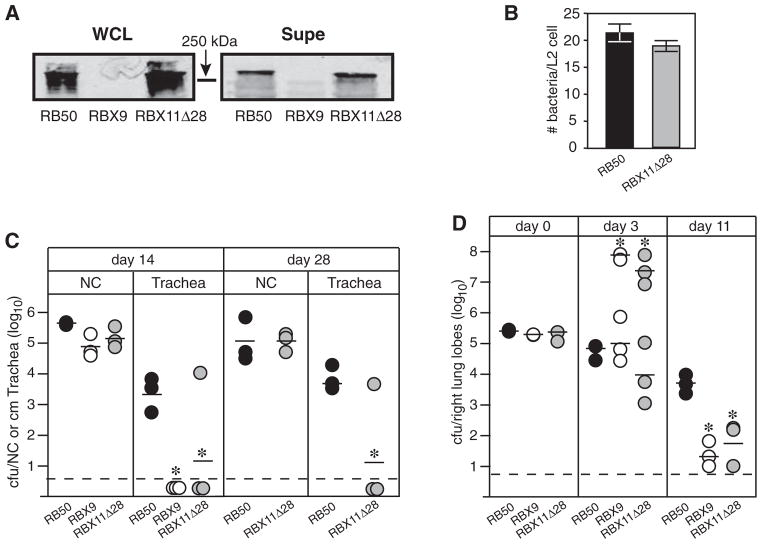
To investigate the importance of the FHA MCD in vivo, we constructed a B. bronchiseptica strain containing a deletion mutation in fhaB that removed 28 codons corresponding to aa 2429–2456 of the preproprotein (centred approximately 30 codons 5′ to the region encoding the primary FHA maturation site). Western blot analysis showed that FHA was produced in this strain, called RBX11Δ28, in amounts similar to that in wild-type B. bronchiseptica (Fig. 7A). RBX11Δ28 was able to adhere to L2 cells (Fig. 7B) and macrophage-like cells (data not shown) in vitro, but was defective in its ability to colonize the tracheas of rats (Fig. 7C). This strain also displayed a ‘bimodal phenotype’ in mice (identical to that displayed by the ΔfhaB strain) in which half of the mice became moribund and contained very high numbers of bacteria in their lungs at day 3 post inoculation and half of the animals remained healthy and contained similar or slightly lower numbers of bacteria in their lungs than animals infected with wild-type B. bronchiseptica at day 3 post inoculation (Fig. 7D). Consistent with previous observations, lung sections of moribund animals showed massive infiltration of inflammatory cells while lung sections from animals that appeared healthy showed only mild inflammation (data not shown). These results indicate that an intact MCD is required for FHA function in vivo. RBX11Δ28 was also similar to RBX9 (the ΔfhaB strain) in its ability to induce surface expression of ICAM-1 in BEAS-2B cells in vitro, providing additional evidence that it is the MCD that is important in FHA-dependent host cell interactions (Fig. 5).
Fig. 7. The MCD is required for FHA function in vivo.
A. Western blot showing FHA (~250 kDa) in whole cell lysates (WCLs) and concentrated supernatants (Supe) of the indicated B. bronchiseptica strains. Blots were probed with the anti-CRD antibody.
B. Adherence of RB50 and RBX11Δ28 to L2 cells is shown. Moi = 200. The difference between the values was not statistically significant.
C. Wistar rats were inoculated intranasally with 10 μl PBS containing 1000 cfu of B. bronchiseptica (strains indicated across the bottom) and the number of cfu recovered from the nasal cavity and 1 cm trachea determined at days 14 and 28 post inoculation. Each circle represents the number of cfu recovered from a single animal. The horizontal line shows the geometric mean for each group. The dashed line represents the lower limit of detection. y-axis is log scale. *P < 0.05.
D. BALB/c mice were inoculated intranasally with 50 μl PBS containing 5 × 105 cfu of B. bronchiseptica (strains indicated across the bottom) and the number of cfu in the right lung lobes was determined 60 min (day 0), 3 and 11 days post inoculation. Each circle represents the number of cfu recovered from a single animal. The horizontal line shows the geometric mean for each group. The dashed line represents the lower limit of detection. For RBX9 and RBX11Δ28, the means at day 3 for moribund (~108) and healthy (~104–5) animals were calculated separately as indicated. y-axis is log scale. *P < 0.05. The rat and mouse experiments using RBX11Δ28 were performed at least twice with similar results and the data from one of each type of experiment are shown.
Discussion
We reported previously that FHABp could not substitute for FHABb in vivo based on characterization of RBFS4, a B. bronchiseptica strain in which the native fhaBBb gene was replaced with fhaBBp (Inatsuka et al., 2005). Replacement of fhaBBp with fhaBBb in B. pertussis, however, did not alter the ability of B. pertussis to infect rats or mice and ‘swapping’ experiments, including those shown in Fig. 2, failed to identify a domain within mature FHABp responsible for the inability of B. bronchiseptica strain RBFS4 to colonize the tracheas of rats or to modulate the immune response in the lungs of mice. These results, together with the discovery that the FhaB prodomain is not required for FHA production, maturation or secretion but is required for rat tracheal colonization (Mazar and Cotter, 2006), led us to re-examine the prodomain-encoding region of fhaBBp in RBFS4. Repeated sequencing efforts through a particularly difficult section of fhaBBp DNA in RBFS4 ultimately revealed an additional thymidine approximately 10 codons 3′ to the primary SphB1-dependent maturation site. ‘Repair’ of the mutation resulted in a strain, RBFS10, that is indistinguishable from RB50 in its ability to adhere to epithelial cells and macrophages, to colonize the tracheas of rats, and to modulate the inflammatory response in the lungs of mice, supporting the conclusion that FHABp can in fact substitute for FHABb in B. bronchiseptica in vivo. We regret our failure to find this mutation initially and our reporting of erroneous conclusions based on the characterization of RBFS4. Re-interpretation of the data obtained with RBFS4 in light of this new information, however, provides some insight into FHA secretion, maturation and function.
The RB50 derivatives that revealed a role for the pro-domain in FHA function (characterized in Mazar and Cotter, 2006) produced FhaBBb proproteins that were 2588 and 3371 aa in length, i.e. they contained at least the N-terminal ~97 aa of the prodomain. The truncated FhaBBp proprotein produced in RBFS4 contains only the N-terminal 7–15 aa of the prodomain, followed by 42 aa that differ from those of the native protein, yet RBFS4, like the mutants characterized previously, produced an FHA protein that was translocated to the cell surface, processed in an SphB1-dependent manner, and released into the extracellular milieu in a manner indistinguishable from RB50, and was similarly unable to colonize the tracheas of rats (Inatsuka et al., 2005; Mazar and Cotter, 2006). Characterization of RBFS4 therefore shows that even the very N-terminal region of the prodomain is not required for FHA secretion and processing but is required for FHA-mediated rat tracheal colonization. Moreover, characterization of RBFS4 also showed that the FhaB prodomain is required for B. bronchiseptica to modulate the inflammatory response in the lungs of mice, which we had not investigated in our previous study. These data, along with those demonstrating the importance of the MCD in vivo (discussed below), support the hypothesis that the role of the prodomain is to control secretion-dependent folding of the MCD and that proper folding of the MCD is critical for FHA function. Consistent with this hypothesis, we have recently obtained evidence from native gel electrophoresis and cysteine availability experiments that conformation of the MCD differs in a prodomain-dependent manner (C.R. Noel, J.A. Sexton, J. Mazar and P.A. Cotter, in preparation).
The fact that the ‘repaired’ strain, RBFS10, was indistinguishable from RB50 in vitro (using both human and non-human cell lines) and in vivo indicates that FHABp and FHABb are functionally interchangeable, at least with regard to the phenotypes that we have investigated. The fact that the B. bronchiseptica strains producing chimeric FHA proteins were also indistinguishable from RB50 in these assays indicates that specific domains within FHA are also functionally interchangeable, arguing against the possibility that specific domains (at least those investigated in this study) of FHABb or FHABp require other domains from the same protein (FHABb or FHABp) to function properly. We cannot conclude that FHABb can substitute for FHABp in B. pertussis during the natural course of infection because of the limited host range of this species. However, our results suggest that information obtained studying FHABb in B. bronchiseptica using natural-host animal models may provide insight into the function of FHABp in B. pertussis during human infection.
Of the FHA domains identified in vitro, the RGD triplet has received the most attention. Reported activities include binding to VLA-5 on epithelial cells to cause increased surface expression of ICAM-1 (Ishibashi et al., 1994; Ishibashi and Nishikawa, 2002; 2003) and binding to LRI/IAP complexes on monocytes to cause enhancement of CR3 binding activity (Ishibashi et al., 1994; 2002). Because wild-type and ΔfhaB mutant B. pertussis strains are indistinguishable in the mouse model, the role of specific FHA domains cannot be investigated in this context. We reasoned, however, that the sensitive animal models available for use with B. bronchiseptica would allow us to identify the role(s) of the FHA RGD during infection, and that the results obtained would reflect the function of both FHABb and FHABp. We were surprised to find that a strain producing an FHA protein containing an RAE triplet instead of RGD was indistinguishable from wild-type B. bronchiseptica in its ability to colonize the respiratory tracts of rats, to modulate the inflammatory response in the lungs of mice, and to cause a lethal systemic infection in immunodeficient mice. Evaluation of our B. bronchiseptica strains, and re-evaluation of the B. pertussis strains used in previous studies, confirmed that FHA is required for a modest but reproducible induction of ICAM-1 surface expression in epithelial cells, but a role for the RGD motif was not apparent from these experiments. Together, our data indicate that if the RGD triplet contributes to FHA function, its contribution is not evident in these models and assays. The RGD motif of aggregation substance of Enterococcus faecalis was similarly shown not to be responsible for the interaction of this pathogen with host cells, despite previous suggestive reports to the contrary (Waters et al., 2003). The degree to which RGD motifs in proteins of bacterial pathogens or symbionts contribute to bacterial–host interactions remains unknown.
Despite the fact that the C-terminal ~500 aa of mature FHA (the MCD) has been shown to be immunodominant in inoculated or immunized mice and rabbits (Delisse-Gathoye et al., 1990; Wilson et al., 1998) and, importantly, in humans recovering from pertussis (Leininger et al., 1997; Piatti, 1999), a role for the MCD in FHA function has not been investigated, or even proposed, previously. Failure to recognize the MCD as a potentially important domain likely stemmed from the assumed ‘N-terminus out’ topology of cell-associated FHA. Our recent discovery that the MCD is located distally from the cell surface and is accessible by antibodies prompted us to consider its contribution to FHA function (Mazar and Cotter, 2006). Our current study shows that a strain producing an FHA protein lacking 28 aa near the C-terminus of the MCD is unable to colonize the tracheas of rats and to modulate the inflammatory response in mice; it displays a phenotype in these animals that is identical to ΔfhaB strains. This result demonstrates that aa within the MCD are required for FHA function in vivo. Whether these aa are required because they interact directly with a host cell receptor(s) or because they are required for the proper conformation of the FHA protein cannot be distinguished from our data. However, it seems unlikely that a deletion of 28 aa near the C-terminus of the MCD, which is proposed to have a globular structure, would affect the conformation of the β-helical shaft portion of the long, rod-shaped FHA molecule. We are therefore focusing our current efforts on the MCD and are conducting experiments to investigate the potential importance of specific aa within the MCD more thoroughly.
The ability to adhere to primary cells or immortalized cell lines is frequently regarded as an indication of bacterial virulence potential. FHA contributes to the ability of B. pertussis to adhere to epithelial cells and macrophages and is both necessary and sufficient for B. bronchiseptica to adhere to these cell types in vitro (Relman et al., 1990; Ishibashi et al., 1994; Cotter et al., 1998; Mattoo et al., 2000; Inatsuka et al., 2005). B. bronchiseptica mutants that do not adhere to cells in vitro are defective for tracheal colonization and overcoming inflammatory clearance in vivo (Cotter et al., 1998; Inatsuka et al., 2005). These observations suggest that the in vitro adherence assay may be a good predictor of FHA function in vivo and could potentially be used to investigate the molecular interactions in which FHA participates. Consistent with this logic, our antibody blocking experiments implicate a role for the MCD, and not the CRD, in FHA function. However, some B. bronchiseptica FHA mutants, such as RBX11Δ28 and RBX11-T-E, can adhere to mammalian cells in vitro but display phenotypes in rats and mice that are identical to those of ΔfhaB strains. Thus, while lack of adherence in vitro may predict lack of FHA function in vivo, the reverse is not true; adherence in vitro does not necessarily reflect in vivo functionality. There are several possible explanations for this apparent discrepancy. It is possible that the receptors present on cells used in vitro represent only a subset of those that are present on relevant cells types in vivo and that the FHA proteins produced by RBX11Δ28 and RBX11-T-E are capable of binding to the former but not the latter. It is also possible that it is a matter of affinity; that the FHA proteins produced by RBX11Δ28 and RBX11-T-E bind receptors with an affinity that is sufficient to mediate adherence in vitro, but below that required for interactions that lead to tracheal colonization and immunomodulation in vivo. Additional scenarios can be envisaged. Regardless of the mechanistic bases, these results highlight the fact that the complexity of the in vivo environment cannot be accurately modelled in vitro at present, illustrate the challenges associated with identifying biologically relevant host cell receptors for FHA, and underscore the importance of interpreting data obtained solely from in vitro experiments with caution.
Our results indicate that data obtained from in vivo experiments deserve cautious interpretation as well. Murine lung inflammation models have proven to be useful for studying the ability of a variety of pathogens, even those causing enteric infections, to influence and/or overcome innate immunity (Philpott et al., 2000; Fullner et al., 2002), and they have provided insight into the function of some B. pertussis virulence factors (Andreasen and Carbonetti, 2008). However, the fact that ΔfhaB mutants are indistinguishable from wild-type B. pertussis in the murine lung inflammation model and the fact that B. pertussis fails to induce a significant anti-Bordetella antibody response in mice indicate that the usefulness of mouse models for studying Bordetella pathogenesis using B. pertussis is limited.
The ultimate goals of our studies on FHA are to understand how this large complex molecule contributes to the ability of bordetellae to cause respiratory disease specifically and how inflammation is controlled generally. While identifying the MCD as an important functional domain represents a significant step towards achieving those goals, it has practical implications as well. It would likely be easier and more cost-effective to produce the MCD rather than the entire mature FHA protein for inclusion in acellular pertussis vaccines. Efforts to determine if the MCD alone can be produced in large quantities, be immunogenic, and be efficacious as a vaccine are apparently already underway (Lee et al., 2002; Knight et al., 2006). The ability to focus on the MCD will also facilitate studies aimed at identifying host cell receptors to which FHA binds and determining the consequences of those binding events. If, as the current data suggest, FHA interacts with multiple receptors, the optimal vaccine component may be one that binds a limited set, or possibly none, of those receptors – or binds them in a way that promotes the development of a robust adaptive immune response while minimizing the inflammatory response at the immunization site. Increased incidence of pertussis in recent years attests to the need for continued vaccine development and therefore a more complete understanding of how FHA, and its various domains, function during infection.
Experimental procedures
Bacterial strains, plasmids and growth media
All strains and relevant plasmids used in this study are listed in Table S1. Bordetella strains were grown in Stainer–Scholte broth (Stainer and Scholte, 1970) supplemented with 100 μg μl−1 (2,6-O-dimethyl)-β-cyclodextrin or on Bordet–Gengou agar (Becton Dickinson Microbiology systems) supplemented with defibrinated sheep blood at a concentration of 7.5% (to grow B. bronchiseptica strains) or 15% (to grow B. pertussis strains). Escherichia coli DH5α was used for all cloning experiments and was grown in Luria–Bertani (LB) agar or broth. E. coli SM10λpir was used for conjugations and was grown on LB agar. Where appropriate, antibiotics were used at the following concentrations: streptomycin, 20 μg ml−1; gentamicin, 20 μg ml−1; ampicillin, 100 μg ml−1.
To create a B. pertussis strain expressing FHA from B. bronchiseptica (Bpe138::pSJ63), the entire fhaB gene and promoter from B. bronchiseptica was cloned into a pBR322 derivative that can be used as a suicide vector in Bordetella, to create pSJ61. To create a plasmid in which the B. bronchiseptica fhaB gene was driven by the B. pertussis fhaB promoter, plasmid pSJ61 was digested with HindIII and Bsu36I, removing a 1.1 kb DNA fragment corresponding to the fhaB promoter and first 230 nucleotides of the fhaB gene. This fragment was replaced with the corresponding chromosomal DNA fragment from B. pertussis Tohama 1, to create pSJ63. This plasmid therefore contains the B. pertussis fhaB promoter and the first 230 nucleotides of B. pertussis fhaB fused in frame to the B. bronchiseptica fhaB gene, which results in a change of only 2 aa within the signal sequence of the B. bronchiseptica FhaB protein. pSJ63 was introduced into Bpe138 (a B. pertussis strain containing a deletion of the fhaB gene) and was determined via PCR analysis to have integrated into the chromosome at the fhaB promoter (data not shown).
The construction of strains exhibiting unmarked chromosomal deletions, rearrangements or mutations in the fhaB gene was done using an allelic exchange method that employs the Bacillus subtilis sacB gene for counterselection (Akerley et al., 1995). RB50gap contains an in-frame deletion mutation of nucleotides 1449–1788 in fhaB. It was constructed by allelic exchange using a plasmid containing a 1 kb fragment from fhaBBp that corresponds to a nearly identical sequence in fhaBBb but omits the DNA corresponding to nucleotides 1449–1788 in fhaBBb (because this segment is naturally missing in fhaBBp). RB50CRDBp contains a substitution of nucleotides 3764–4178 of fhaBBb with nucleotides 3422–3836 from fhaBBp. It was constructed using two consecutive allelic exchanges. In the first, a plasmid was constructed that contained a segment of fhaBBb in which, near the centre of this fragment, nucleotides 3764–4178 were deleted. This plasmid was used to create a strain containing a deletion of nucleotides 3764–4178 in fhaBBb. A second allelic exchange plasmid was constructed containing a fragment of fhaBBp corresponding to nucleotides 3422–3836 (these nucleotides represent the homologous sequence from fhaBBp that had been deleted in fhaBBb in the first allelic exchange event). Allelic exchange with this plasmid replaced the deleted nucleotides with the homologous sequence from fhaBBp. In a similar manner, RB50NMCDBp was constructed. It contains a substitution of nucleotides 5305–6366 of fhaBBb with nucleotides 4961–6036 from fhaBBp. In the first allelic exchange, nucleotides 5305–6366 were deleted and in the second, nucleotides 4961–6036 from fhaBBp replaced the deleted nucleotides. To create RBFS10, a plasmid was constructed that contains a 1 kb DNA fragment of wild-type sequence from fhaBBp centred around the site that contains the thymidine insertion mutation in RBFS4. Using allelic exchange with this plasmid, the thymidine insertion mutation was replaced by wild-type sequence, forming RBFS10. RB50RAE was constructed by allelic exchange such that the codons corresponding to aa 1213 and 1214 were changed to encode alanine and glutamic acid respectively. To create RBX11Δ28, a plasmid was constructed containing a 1 kb fragment of DNA in which nucleotides 7287–7368 of fhaBBb, located in the centre of the fragment, were deleted in frame. Using allelic exchange with this plasmid, nucleotides 7287–7368 were deleted in fhaBBb, forming RBX11Δ28. Strain RBX11-T-E was constructed as described previously (Mazar and Cotter, 2006). All strains were confirmed to be constructed as intended by PCR and DNA sequence analysis.
Immunoblotting
Immunoblots were performed as described previously (Julio and Cotter, 2005). To evaluate expression of FHA, proteins were prepared from approximately 8.0 × 108 cfu for WCLs and 1.5 × 1010 cfu for supernatant fractions as described (Martinez de Tejada et al., 1998; Julio and Cotter, 2005) and separated using 3–8% linear gradient sodium-dodecyl sulphate polyacrylamide gels, transferred to nitrocellulose (Schleicher and Schuell Bioscience), and probed with a chicken polyclonal antibody that was generated against a polypeptide corresponding to the CRD of B. bronchiseptica FHA (Julio and Cotter, 2005). Goat anti-chicken secondary antibody conjugated to IRdye 800 (Molecular Probes) was used to detect antigen–antibody complexes. For immunoblots evaluating the humoral response in rats and mice, whole cell proteins were run on 4–12% linear gradient SDS polyacrylamide gels, transferred to nitrocellulose, and probed with sera that had been collected 21 days post inoculation from PBS-exposed or Bordetella-infected rats or mice. The rat and mouse sera were diluted 1:2500. Goat anti-rat or goat anti-mouse secondary antibodies conjugated to IRdye 700 (Molecular Probes) were used to detect antigen–antibody complexes and were used at a dilution of 1:5000. Blots were visualized using an Odyssey infrared imaging system (LiCor Biosciences).
Adherence assays
Adherence of Bordetella to immortalized rat lung epithelial (L2) cells was performed as described (Cotter et al., 1998). Bacteria were added to L2 monolayers at a moi of 200, and adherence was quantified by averaging the total number of bacterial cells and eukaryotic nuclei in two separate microscopic fields from two independent experiments. Adherence of Bordetella to J774A.1 macrophage-like cells was performed as described (Cotter et al., 1998; Inatsuka et al., 2005). For experiments using anti-CRD or anti-MCD antibodies, antibodies (diluted 1:100) were incubated with stationary phase bacteria previous to performing the adherence assay. In all adherence assays, the bacteria were observed to adhere as single cells attaching to epithelial cells or macrophages, although sometimes bacteria would be adjacent to each other. Agglutination of bacteria due to the anti-CRD or anti-MCD antibodies was not observed.
Immunostaining of whole cells
Bacterial cells from stationary phase cultures were incubated with anti-CRD (Julio and Cotter, 2005) or anti-MCD antibodies. The anti-MCD antibodies were generated in rabbits using a polypeptide corresponding to the entire MCD of FHABb, which was produced in and purified from E. coli. Bacterial cell-antibody complexes were spotted onto a nitro-cellulose membrane (Schleicher and Schuell BioScience). The membranes were probed with either goat anti-chicken secondary antibody conjugated to IRdye 680 (for experiments using anti-CRD antibody) or goat anti-rabbit antibody conjugated to IRdye 800 (for experiments using anti-MCD antibody).
Measurement of ICAM-1 levels
Human bronchial epithelial (BEAS-2B) cells were incubated with Bordetella for 2 h at a moi of 100. Following several washes using PBS, the adherent bacteria were killed by gentamicin treatment for an additional 22 h. BEAS-2B cells were resuspended in PBS/3% FBS and incubated with mouse anti-human ICAM-1 conjugated to phycoerythrin (BD Pharmigen) for 30 min at 4°C. The cells were washed twice in PBS/3% PBS, and resuspended in PBS/0.5% formaldehyde and incubated overnight at 4°C. Before analysis, cells were resuspended in PBS, filtered through a 35 μm nylon mesh, and analysed on a FACS-Aria flow cytometer (Becton Dickinson).
Animal experiments
To evaluate bacterial colonization of the respiratory tract, 3- to 4-week old female Wistar rats (Charles River Laboratories) were inoculated with 1000 cfu of B. bronchiseptica or 106 cfu of B. pertussis in a volume of 10 μl to the external nares. Animals were sacrificed 14 or 28 days post infection, and the number of cfu in the nasal septa and trachea was enumerated as described (Mattoo et al., 2000). To evaluate the ability of bacteria to resist inflammation-mediated clearance, 3- to 4-week-old BALB/c mice (Charles River Laboratories) were inoculated intranasally with 5 × 105 cfu of B. bronchiseptica or B. pertussis in a 50 μl volume. The lungs were harvested at either 1 h, 3 days or 11 days post inoculation, and the number of cfu was enumerated from the right lung lobes as described (Mattoo et al., 2000). The left lung lobes were inflated with formalin, embedded in paraffin, sectioned, stained with H&E, and examined by microscopy. SCID-Beige mice (3–4 weeks old; Charles River Laboratories) were inoculated with either 5 × 105 or 1000 cfu of B. bronchiseptica delivered in 50 μl or 10 μl respectively, and monitored for signs of morbidity. Mouse lung inflammation experiments have been performed multiple times for Bp536, RB50 and RBX20 and at least twice for the other strains shown in Fig. 2 with similar results. The data from one experiment are shown. We performed various animal experiments with RB50RAE once (always including RB50 and RBX9, which we have tested many times in each model) and data from a subset of those experiments are shown in Fig. 4. In every case, the results obtained with RB50RAE were indistinguishable from those obtained for RB50. In the interest of minimizing the number of animals used, we did not repeat each type of experiment with the RB50RAE mutant because we felt that, collectively, this strain had been compared with wild-type B. bronchiseptica multiple times and in no case was a difference apparent. RBX11Δ28 has been compared with RB50 and RBX9 in the rat and mouse models at least twice with similar results. The data from one experiment each are shown in Fig. 7. In the interest of minimizing the number of animals used, experiments with RBX9 were not carried out to day 28.
Statistical analyses
The unpaired Student’s t-test was used for all statistical analyses except comparison of the survival data (Fig. 4C and D), for which the Log Rank (Mantel-Cox) test was used.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institutes of Health (AI43876) and the University of California Superfund Basic Research and Education Program to P.A.C. and Phillip Morris USA, Inc. and Phillip Morris International (to S.M.J.).
Footnotes
Additional supporting information may be found in the online version of this article.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Abramson T, Kedem H, Relman DA. Proinflamatory and proaoptotic activities associated with Bordetella pertussis filamentous hemagglutinin. Infect Immun. 2001;69:2650–2658. doi: 10.1128/IAI.69.4.2650-2658.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akerley BJ, Cotter PA, Miller JF. Ectopic expression of the flagellar regulon alters development of the Bordetella–host interaction. Cell. 1995;80:611–620. doi: 10.1016/0092-8674(95)90515-4. [DOI] [PubMed] [Google Scholar]
- Alonso S, Pethe K, Mielcarek N, Raze D, Locht C. Role of adp-ribosyltransferase activity of pertussis toxin in toxin-adhesin redundancy with filamentous hemagglutinin during Bordetella pertussis infection. Infect Immun. 2001;69:6038–6043. doi: 10.1128/IAI.69.10.6038-6043.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreasen C, Carbonetti NH. Pertussis toxin inhibits early chemokine production to delay neutrophil recruitment in response to Bordetella pertussis respiratory tract infection in mice. Infect Immun. 2008;76:5139–5148. doi: 10.1128/IAI.00895-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai H, Sato Y. Separation and characterization of two distinct hemagglutinins contained in purified leukocytosis-promoting factor from Bordetella pertussis. Biochim Biophys Acta. 1976;444:765–782. doi: 10.1016/0304-4165(76)90323-8. [DOI] [PubMed] [Google Scholar]
- Arico B, Gross R, Smida J, Rappuoli R. Evolutionary relationships in the genus Bordetella. Mol Microbiol. 1987;1:301–308. doi: 10.1111/j.1365-2958.1987.tb01936.x. [DOI] [PubMed] [Google Scholar]
- Bemis DA. Bordetella and mycoplasma respiratory infections in dogs and cats. Vet Clin North Am. 1992;22:1173–1186. doi: 10.1016/s0195-5616(92)50308-4. [DOI] [PubMed] [Google Scholar]
- Boschwitz JS, Batanghari JW, Kedem H, Relman DA. Bordetella pertussis infection of human monocytes inhibits antigen-dependent cd4 t cell proliferation. J Infect Dis. 1997a;176:678–686. doi: 10.1086/514090. [DOI] [PubMed] [Google Scholar]
- Boschwitz JS, van der Heide HGJ, Mooi FR, Relman DA. Bordetella bronchiseptica expresses the fimbrial structural subunit gene fima. J Bacteriol. 1997b;179:7882–7885. doi: 10.1128/jb.179.24.7882-7885.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos-Outcalt D. Pertussis: a disease re-emerges. J Fam Pract. 2005;54:699–703. [PubMed] [Google Scholar]
- Chevalier N, Moser M, Koch HG, Schimz KL, Willery E, Locht C, et al. Membrane targeting of a bacterial virulence factor harbouring an extended signal peptide. J Mol Microbiol Biotechnol. 2004;8:7–18. doi: 10.1159/000082076. [DOI] [PubMed] [Google Scholar]
- Clantin B, Hodak H, Willery E, Locht C, Jacob-Dubuisson F, Villeret V. The crystal structure of filamentous hemagglutinin secretion domain and its implications for the two-partner secretion pathway. Proc Natl Acad Sci USA. 2004;101:6194–6199. doi: 10.1073/pnas.0400291101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clantin B, Delattre AS, Rucktooa P, Saint N, Meli AC, Locht C, et al. Structure of the membrane protein fhac: a member of the omp85-tpsb transporter superfamily. Science. 2007;317:957–961. doi: 10.1126/science.1143860. [DOI] [PubMed] [Google Scholar]
- Cotter PA, Yuk MH, Mattoo S, Akerley BJ, Boschwitz J, Relman DA, et al. The filamentous hemagglutinin (fha) of Bordetella bronchiseptica is required for efficient establishment of tracheal colonization. Infect Immun. 1998;66:5921–5929. doi: 10.1128/iai.66.12.5921-5929.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutte L, Antoine R, Drobecq H, Locht C, Jacob-Dubuisson F. Subtilisin-like autotransporter serves as maturation protease in a bacterial secretion pathway. EMBO J. 2001;20:5040–5048. doi: 10.1093/emboj/20.18.5040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings CA, Brinig MM, Lepp PW, van de Pas S, Relman DA. Bordetella species are distinguished by patterns of substantial gene loss and host adaptation. J Bacteriol. 2004;186:1484–1492. doi: 10.1128/JB.186.5.1484-1492.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delisse-Gathoye AM, Locht C, Jacob F, Raaschou-Nielsen M, Heron I, Ruelle JL, et al. Cloning, partial sequence, expression, and antigenic analysis of the filamentous hemagglutinin gene of Bordetella pertussis. Infect Immun. 1990;58:2895–2905. doi: 10.1128/iai.58.9.2895-2905.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deville JG, Cherry JD, Christenson PD, Pineda E, Leach CT, Kuhls TL, et al. Frequency of unrecognized Bordetella pertussis infections in adults. Clin Infect Dis. 1995;21:639–642. doi: 10.1093/clinids/21.3.639. [DOI] [PubMed] [Google Scholar]
- Diavatopoulos DA, Cummings CA, Schouls LM, Brinig MM, Relman DA, Mooi FR. Bordetella pertussis, the causative agent of whooping cough, evolved from a distinct, human-associated lineage of B. bronchiseptica. PLoS Pathog. 2005;1:e45. doi: 10.1371/journal.ppat.0010045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullner KJ, Boucher JC, Hanes MA, Haines GK, 3rd, Meehan BM, Walchle C, et al. The contribution of accessory toxins of Vibrio cholerae o1 el tor to the proinflammatory response in a murine pulmonary cholera model. J Exp Med. 2002;195:1455–1462. doi: 10.1084/jem.20020318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin MS, Weiss AA. Adenylate cyclase toxin is critical for colonization and pertussis toxin is critical for lethal infection by Bordetella pertussis in infant mice. Infect Immun. 1990;58:3445–3447. doi: 10.1128/iai.58.10.3445-3447.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannah JH, Menozzi FD, Renauld G, Locht C, Brennan MJ. Sulfated glycoconjugate receptors for the Bordetella pertussis adhesin filamentous hemagglutinin (fha) and mapping of the heparin-binding domain on fha. Infect Immun. 1994;62:5010–5019. doi: 10.1128/iai.62.11.5010-5019.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvill ET, Cotter PA, Miller JF. Pregenomic comparative analysis between Bordetella bronchiseptica rb50 and Bordetella pertussis tohama i in murine models of respiratory tract infection. Infect Immun. 1999a;67:6109–6118. doi: 10.1128/iai.67.11.6109-6118.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvill ET, Cotter PA, Miller JF. Probing the function of a bacterial virulence factor by manipulating host immunity. Infect Immun. 1999b;67:1493–1500. doi: 10.1128/iai.67.3.1493-1500.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvill ET, Preston A, Cotter PA, Allen AG, Maskell DJ, Miller JF. Multiple roles for Bordetella lipopolysaccharide molecules during respiratory tract infection. Infect Immun. 2000;68:6720–6728. doi: 10.1128/iai.68.12.6720-6728.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodak H, Clantin B, Willery E, Villeret V, Locht C, Jacob-Dubuisson F. Secretion signal of the filamentous haemagglutinin, a model two-partner secretion substrate. Mol Microbiol. 2006;61:368–382. doi: 10.1111/j.1365-2958.2006.05242.x. [DOI] [PubMed] [Google Scholar]
- Inatsuka CS, Julio SM, Cotter PA. Bordetella filamentous hemagglutinin plays a critical role in immunomodulation, suggesting a mechanism for host specificity. Proc Natl Acad Sci USA. 2005;102:18578–18583. doi: 10.1073/pnas.0507910102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishibashi Y, Nishikawa A. Bordetella pertussis infection of human respiratory epithelial cells up-regulates intercellular adhesion molecule-1 expression: Role of filamentous hemagglutinin and pertussis toxin. Microb Pathog. 2002;33:115. doi: 10.1006/mpat.2002.0517. [DOI] [PubMed] [Google Scholar]
- Ishibashi Y, Nishikawa A. Role of nuclear factor-kappa b in the regulation of intercellular adhesion molecule 1 after infection of human bronchial epithelial cells by Bordetella pertussis. Microb Pathog. 2003;35:169–177. doi: 10.1016/s0882-4010(03)00113-x. [DOI] [PubMed] [Google Scholar]
- Ishibashi Y, Claus S, Relman DA. Bordetella pertussis filamentous hemagglutinin interacts with a leukocyte signal transduction complex and stimulates bacterial adherence to monocyte cr3 (cd11b/cd18) J Exp Med. 1994;180:1225–1233. doi: 10.1084/jem.180.4.1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishibashi Y, Relman DA, Nishikawa A. Invasion of human respiratory epithelial cells by Bordetella pertussis: possible role for a filamentous hemagglutinin arg-gly-asp sequence and alpha5beta1 integrin. Microb Pathog. 2001;30:279–288. doi: 10.1006/mpat.2001.0432. [DOI] [PubMed] [Google Scholar]
- Ishibashi Y, Yoshimura K, Nishikawa A, Claus S, Laudanna C, Relman DA. Role of phosphatidylinositol 3-kinase in the binding of Bordetella pertussis to human monocytes. Cell Microbiol. 2002;4:825–833. doi: 10.1046/j.1462-5822.2002.00235.x. [DOI] [PubMed] [Google Scholar]
- Jacob-Dubuisson F, Buisine C, Mielcarek N, Clement E, Menozzi FD, Locht C. Amino-terminal maturation of the Bordetella pertussis filamentous haemagglutinin. Mol Microbiol. 1996;19:65–78. doi: 10.1046/j.1365-2958.1996.349883.x. [DOI] [PubMed] [Google Scholar]
- Jacob-Dubuisson F, Buisine C, Willery E, Renauld-Mongenie G, Locht C. Lack of functional complementation between Bordetella pertussis filamentous hemagglutinin and proteus mirabilis hpma hemolysin secretion machineries. J Bacteriol. 1997;179:775–783. doi: 10.1128/jb.179.3.775-783.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob-Dubuisson F, El-Hamel C, Saint N, Guedin S, Willery E, Molle G, et al. Channel formation by fhac, the outer membrane protein involved in the secretion of the Bordetella pertussis filamentous hemagglutinin. J Biol Chem. 1999;274:37731–37735. doi: 10.1074/jbc.274.53.37731. [DOI] [PubMed] [Google Scholar]
- Jacob-Dubuisson F, Kehoe B, Willery E, Reveneau N, Locht C, Relman DA. Molecular characterization of Bordetella bronchiseptica filamentous haemagglutinin and its secretion machinery. Microbiology. 2000;146:1211–1221. doi: 10.1099/00221287-146-5-1211. [DOI] [PubMed] [Google Scholar]
- Julio SM, Cotter PA. Characterization of the filamentous hemagglutinin-like protein fhas in Bordetella bronchiseptica. Infect Immun. 2005;73:4960–4971. doi: 10.1128/IAI.73.8.4960-4971.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajava AV, Cheng N, Cleaver R, Kessel M, Simon MN, Willery E, et al. Beta-helix model for the filamentous haemagglutinin adhesin of Bordetella pertussis and related bacterial secretory proteins. Molmicrobiol. 2001;42:279–292. doi: 10.1046/j.1365-2958.2001.02598.x. [DOI] [PubMed] [Google Scholar]
- Khelef N, Bachelet CM, Vargaftig BB, Guiso N. Characterization of murine lung inflammation after infection with parental Bordetella pertussis and mutants deficient in adhesins or toxins. [Published erratum appears in Infect Immun 1994 dec; 62 (12): 5707] Infect Immun. 1994;62:2893–2900. doi: 10.1128/iai.62.7.2893-2900.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura A, Mountzouros KT, Relman DA, Falkow S, Cowell JL. Bordetella pertussis filamentous hemagglutinin: evaluation as a protective antigen and colonization factor in a mouse respiratory infection model. Infect Immun. 1990;58:7–16. doi: 10.1128/iai.58.1.7-16.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight JB, Huang YY, Halperin SA, Anderson R, Morris A, Macmillan A, et al. Immunogenicity and protective efficacy of a recombinant filamentous haemagglutinin from Bordetella pertussis. Clin Exp Immunol. 2006;144:543–551. doi: 10.1111/j.1365-2249.2006.03097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SF, Halperin SA, Knight JB, Tait A. Purification and immunogenicity of a recombinant Bordetella pertussis s1s3fha fusion protein expressed by streptococcus gordonii. Appl Environ Microbiol. 2002;68:4253–4258. doi: 10.1128/AEM.68.9.4253-4258.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leininger E, Bowen S, Renauld-Mongenie G, Rouse JH, Menozzi FD, Locht C, et al. Immunodominant domains present on the Bordetella pertussis vaccine component filamentous hemagglutinin. J Infect Dis. 1997;175:1423–1431. doi: 10.1086/516475. [DOI] [PubMed] [Google Scholar]
- McGuirk P, Mills KH. Direct anti-inflammatory effect of a bacterial virulence factor: Il-10-dependent suppression of il–12 production by filamentous hemagglutinin from Bordetella pertussis. Eur J Immunol. 2000;30:415–422. doi: 10.1002/1521-4141(200002)30:2<415::AID-IMMU415>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- McGuirk P, McCann C, Mills KH. Pathogen-specific t regulatory 1 cells induced in the respiratory tract by a bacterial molecule that stimulates interleukin 10 production by dendritic cells: a novel strategy for evasion of protective t helper type 1 responses by Bordetella pertussis. J Exp Med. 2002;195:221–231. doi: 10.1084/jem.20011288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez de Tejada G, Cotter PA, Heininger U, Camilli A, Akerley BJ, Mekalanos JJ, et al. Neither the bvg- phase nor the vrg6 locus of Bordetella pertussis are required for respiratory infection in mice. Infect Immun. 1998;66:2762–2768. doi: 10.1128/iai.66.6.2762-2768.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattoo S, Miller JF, Cotter PA. Role of Bordetella bronchiseptica fimbria in tracheal colonization and development of a humoral immune response. Infect Immun. 2000;68:2024–2033. doi: 10.1128/iai.68.4.2024-2033.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazar J, Cotter PA. Topology and maturation of filamentous haemagglutinin suggest a new model for two-partner secretion. Mol Microbiol. 2006;62:641–654. doi: 10.1111/j.1365-2958.2006.05392.x. [DOI] [PubMed] [Google Scholar]
- Meli AC, Hodak H, Clantin B, Locht C, Molle G, Jacob-Dubuisson F, et al. Channel properties of tpsb transporter fhac point to two functional domains with a c-terminal protein-conducting pore. J Biol Chem. 2006;281:158–166. doi: 10.1074/jbc.M508524200. [DOI] [PubMed] [Google Scholar]
- Parkhill J, Sebaihia M, Preston A, Murphy LD, Thomson N, Harris DE, et al. Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. Nat Genet. 2003;35:32–40. doi: 10.1038/ng1227. [DOI] [PubMed] [Google Scholar]
- Philpott DJ, Edgeworth JD, Sansonetti PJ. The pathogenesis of Shigella flexneri infection: lessons from in vitro and in vivo studies. Philos Trans R Soc Lond B Biol Sci. 2000;355:575–586. doi: 10.1098/rstb.2000.0599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatti G. Identification of immunodominant epitopes in the filamentous hemagglutinin of Bordetella pertussis. FEMS Immunol Med Microbiol. 1999;23:235–241. doi: 10.1111/j.1574-695X.1999.tb01244.x. [DOI] [PubMed] [Google Scholar]
- Pierschbacher MD, Ruoslahti E. Variants of the cell recognition site of fibronectin that retain attachment-promoting activity. Proc Natl Acad Sci USA. 1984;81:5985–5988. doi: 10.1073/pnas.81.19.5985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad SM, Yin Y, Rodzinski E, Tuomanen EI, Masure HR. Identification of a carbohydrate recognition domain in filamentous hemagglutinin from Bordetella pertussis. Infect Immun. 1993;61:2780–2785. doi: 10.1128/iai.61.7.2780-2785.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relman D, Tuomanen E, Falkow S, Golenbock DT, Saukkonen K, Wright SD. Recognition of a bacterial adhesion by an integrin: Macrophage cr3 (alpha m beta 2, cd11b/cd18) binds filamentous hemagglutinin of Bordetella pertussis. Cell. 1990;61:1375–1382. doi: 10.1016/0092-8674(90)90701-f. [DOI] [PubMed] [Google Scholar]
- Renauld-Mongenie G, Cornette J, Mielcarek N, Menozzi FD, Locht C. Distinct roles of the n-terminal and c-terminal precursor domains in the biogenesis of the Bordetella pertussis filamentous hemagglutinin. J Bacteriol. 1996;178:1053–1060. doi: 10.1128/jb.178.4.1053-1060.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts M, Cropley I, Chatfield S, Dougan G. Protection of mice against respiratory Bordetella pertussis infection by intranasal immunization with, p.69 and fha. Vaccine. 1993;11:866–872. doi: 10.1016/0264-410x(93)90363-3. [DOI] [PubMed] [Google Scholar]
- Sato Y, Sato H. Development of acellular pertussis vaccines. Biologicals. 1999;27:61–69. doi: 10.1006/biol.1999.0181. [DOI] [PubMed] [Google Scholar]
- Sato Y, Izumiya K, Sato H, Cowell JL, Manclarck CR. Role of antibody to leukocytosis-promoting factor hemagglutinin and to filamentous hemagglutinin in immunity to pertussis. Infect Immun. 1981;31:1223–1231. doi: 10.1128/iai.31.3.1223-1231.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stainer DW, Scholte MJ. A simple chemically defined medium for the production of phase i Bordetella pertussis. J Gen Microbiol. 1970;63:211–220. doi: 10.1099/00221287-63-2-211. [DOI] [PubMed] [Google Scholar]
- Waters CM, Wells CL, Dunny GM. The aggregation domain of aggregation substance, not the rgd motifs, is critical for efficient internalization by ht-29 enterocytes. Infect Immun. 2003;71:5682–5689. doi: 10.1128/IAI.71.10.5682-5689.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss AA, Goodwin MS. Lethal infection by Bordetella pertussis mutants in the infant mouse model. Infect Immun. 1989;57:3757–3764. doi: 10.1128/iai.57.12.3757-3764.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems RJ, Geuijen C, van der Heide HG, Renauld G, Bertin P, van den Akker WM, et al. Mutational analysis of the Bordetella pertussis fim/fha gene cluster: identification of a gene with sequence similarities to haemolysin accessory genes involved in export of fha. Mol Microbiol. 1994;11:337–347. doi: 10.1111/j.1365-2958.1994.tb00314.x. [DOI] [PubMed] [Google Scholar]
- Wilson DR, Siebers A, Finlay BB. Antigenic analysis of Bordetella pertussis filamentous hemagglutinin with phage display libraries and rabbit anti-filamentous hemagglutinin polyclonal antibodies. Infect Immun. 1998;66:4884–4894. doi: 10.1128/iai.66.10.4884-4894.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood N, McIntyre P. Pertussis: review of epidemiology, diagnosis, management and prevention. Paediatr Respir Rev. 2008;9:201–211. doi: 10.1016/j.prrv.2008.05.010. [DOI] [PubMed] [Google Scholar]
- van der Zee A, Mooi F, Van Embden J, Musser J. Molecular evolution and host adaptation of Bordetella spp. Phylogenetic analysis using multilocus enzyme electrophoresis and typing with three insertion sequences. J Bacteriol. 1997;179:6609–6617. doi: 10.1128/jb.179.21.6609-6617.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.