Abstract
Diabetic kidney disease (DKD) is a microvascular complication of type 1 and 2 diabetes with a devastating impact on individuals with the disease, their families and society as a whole. DKD is the single most frequent cause of incident chronic kidney disease (CKD) cases and accounts for over 40% of the population with end stage renal disease (ESRD). Contributing factors for the high prevalence are the increase in obesity and subsequent diabetes combined with an improved long–term survival with diabetes. Environment and genetic variations contribute to DKD susceptibility and progressive loss of kidney function. How the molecular mechanisms of genetic and environmental exposures interact during DKD initiation and progression are the focus of ongoing research efforts. The development of standardized, unbiased high throughput profiling technologies of human DKD samples opens new avenues in capturing the multiple layers of DKD pathobiology. These techniques routinely interrogate analytes on a genome–wide scale generating comprehensive DKD associated fingerprints. Linking the molecular fingerprints to deep clinical phenotypes may ultimately elucidate the intricate molecular interplay in a disease stage and subtype specific manner. This insight will form the basis for accurate prognosis and facilitate targeted therapeutic interventions. In this review, we present ongoing efforts from large scale data integration translating “–omics” research efforts into improved and individualized health care in DKD.
Keywords: diabetic nephropathy, systems biology, high throughput molecular profiling, transcriptional regulatory networks, proteomics, metabolomics
Introduction
Diabetic kidney disease (DKD) is a common microvascular complication of diabetes mellitus (DM) that is associated with progressive loss of kidney function, systemic endocrine and cardiovascular complications, and premature death. In addition to the personal impact, DKD imposes a great socio–economic burden and constitutes a significant public health challenge. As the epidemic of DM increases worldwide, developing countries will account for ~70% of diabetic patients placing substantial demands on already scarce resources [1,2]. DKD affects roughly 20–40% of individuals suffering from both type 1 (T1DM) and type 2 DM (T2DM) over a 10-year disease period. DKD is therefore not only the single leading cause of incident chronic kidney disease (CKD) but also responsible for over 40% of patients initiating renal replacement therapy due to end stage renal disease (ESRD) in the United States, 18–40% across Europe according to European Registry Data, and 35% and 51% in Australia and New Zealand respectively [3–8]. The increase in DKD induced ESRD is widely attributed to the worldwide growing obesity and T2DM epidemic, accounting for almost 85–90% of DKD cases overall. Furthermore, progressive DKD is an independent risk factor that amplifies morbidity and mortality in patients with diabetes [9–11].
Advances in our understanding of renal physiology and DKD pathogenesis have led to the identification of Renin–Angiotensin–Aldosterone System (RAAS) inhibition, blood glucose and blood pressure control as the pillars of DKD management. These interventions can slow, but usually do not stop progression to ESRD, leaving a large unmet need for novel therapeutic strategies in DKD for more than two decades. Even for established interventions, robust prognostic biomarkers to detect patients at risk for DKD, predicting DKD trajectory and response to therapeutic intervention, are widely lacking. Current classification and treatment strategies primarily rely on a descriptive clinical phenotyping most likely lumping patients with different disease mechanism and stages together. Ongoing research efforts have uncovered potential genetic, behavioral and environmental contributions to disease manifestation in a focused hypothesis driven manner. The development of molecular medicine with genome–wide profiling capabilities of human biosamples offers an opportunity to capture a holistic picture of DKD initiation and progression. Critical questions with this strategy are: How can we effectively exploit these new technologies to facilitate robust and reproducible molecular phenotyping and disease dissection? What proportion of disease depends upon genetic variation? What are the interrelationships between the different puzzle pieces captured by the–omics profiles? How can we integrate this knowledge to define biomarkers of mechanism based disease prediction? How can we distinguish the drivers of pathogenesis from associated, but not causal “passengers” of the disease process? Integrating some of the answers into our current disease understanding and classification will enable us to develop a detailed molecular taxonomy, which identifies specific patient subgroups according to prognostic relevance and necessary therapeutic intervention.
Background
The multifactorial DKD pathogenesis is a consequence of an intricate interplay between genetic factors, environmental exposure and lifestyle. On a molecular level, disease pathogenesis is driven by the hallmark of DM – hyperglycemia, which is caused by incremental lack of insulin in T1DM and target tissue insulin resistance in T2DM [12]. Current understanding suggests that hyperglycemia–driven generation of reactive oxygen species (ROS) and formation of long lasting glycated protein products defined as advanced glycation end products (AGEs) lead to substantial tissue damage. The downstream effects include activation of the intracellular stress response machinery, increased activity of protein kinase C (PKC) pathway, activation of inflammatory signaling mechanisms converging on augmented transcriptional activity of NF–κB, JAK/STAT signaling and a plethora of other effectors [13–15]. Upon initial kidney tissue injury and release of chemoattractants like macrophage chemoattractant MCP–1, infiltrating monocytes/macrophages are a primary source of tumor necrosis factor α (TNF α) release, although resident kidney cells such as endothelial and tubular epithelial cells possess the capability to synthesize this cytokine [16]. TNF–α driven receptor activation changes expression patterns of growth factors, adhesion molecules and other inflammatory molecules. In this hyperglycemic environment TNF–α mediated processes are unleashed from their regulatory constraints, perpetuating an increase of proinflammatory cytokines sustaining further the process of kidney damage. TNF–α has also been implicated in the unfavorable glomerular hemodynamics that characterize DKD e.g. increasing expression of vasoconstrictor endothelin–1, and augmenting glomerular permeability as well as further glomerular and interstitial recruitment of inflammatory cells in DKD [17–22]. DKD associated vascular stress triggers local release of highly vasoactive hormones and peptides influencing activity of the RAAS and endothelin signaling [13]. Activation of the autonomic nervous system [23] can contribute to kidney injury through neuroimmune interaction, modulating interstitial macrophage recruitment and local TNF-α expression during the progressive phase of DKD and CKD in general [24]. Concurrent to the inflammatory processes, there is activation of potent profibrotic mechanisms, which eventually lead to replacement of functional kidney tissue with extracellular matrix, aggravating the progressive phase of DKD. One dominant protagonist is TGF–β (transforming growth factor β) and its downstream mediator CTGF (connective tissue growth factor) [25–27]. This persistent activation of pathogenic mechanisms is not matched by an efficient recruitment of protective pathways (e.g. nitric oxide pathway) leading over time to the specific and well characterized morphological changes and functional deterioration of DKD.
The need for a molecular–based disease definition
The need for early predictors
Diabetic kidney disease develops only in a proportion of patients after decades with DM [8]. Eventually, a subset of those will further progress to ESRD. The current clinical definition of DKD solely relies upon urinary excretion of albumin (usually determined by the urinary albumin–to–creatinine ratio, ACR), using empirically defined thresholds, where ACR < 30 mg/g is termed normalbuminuria (NAlb), 30 ≤ ACR < 300 mg/g defines microalbuminuria (μAlb) and ACR values ≥ 300 mg/g as macroalbuminuria (MAlb) and changes in glomerular filtration rate (GFR) in the presence of T1DM or T2DM. ACR and in particular GFR, are subject to substantial measurement variability, show a heterogeneous pattern and do not necessarily correlate with physiopathology and predict renal function decline [28–31]. Since albuminuria is an early feature of most non–diabetic kidney diseases and can increase with diseases of non–renal origin, it is not very helpful in establishing early diagnosis or in predicting prospective events [32,33]. More than 10 years ago, Caramori and colleagues already emphasized that in context of T1DM GFR changes as well as detection of albuminuria are not biomarkers of early disease stage but rather a sign of significant tissue damage [34]. As such, ACR’s and GFR’s capability to detect and closely monitor early pathogenic inflammatory and metabolic processes involved in DKD has significant limitations [34–36]. ACR is the predominant clinical “biomarker” for establishing DKD diagnosis and estimating risk of progression based on observational and interventional studies [37]. However, recent analysis from subjects with T1DM enrolled in the Joslin Study of the Natural History of Microalbuminuria suggests that progression of DKD occurs to a substantial proportion even in the absence of μAlb, consequently challenging ACR as a mandatory feature of DKD [35,38–40]. This finding has been corroborated in population–based studies of DKD such as the National Health and Nutrition Examination Study (NHANES) and the Developing Education on Microalbuminuria for Awareness of Renal and Cardiovascular Risk in Diabetes study (DEMAND) [8,41]. Although, the risk of DKD progression is highest among those T1DM individuals with longer diabetes duration, higher HbA1c and increasing albuminuria, early renal function decline strongly supports the concept that renal damage does not begin with or after development of albuminuria stage, but can occur much earlier. These observations have been corroborated in Native Americans with T2DM–DKD [42]. Additionally, Karalliedde et al. pointed out the complex and multifaceted pathophysiology of albuminuria [43]. The phenotype of diabetic albuminuria is further confounded by the substantial ACR variation occasionally creating phenotypic ambiguity [44]. These data clearly show the limits of our current understanding of early changes in DKD pathogenesis and underline the need for research to define the fundamental disease mechanisms and to develop potentially nephroprotective strategies. Furthermore, ethnic differences not only influence the prevalence of DM but also have a substantial impact on the prevalence of diabetes–associated complications and further disease progression. For instance, DKD incidence is lowest in patients of Caucasian descent, followed by Asians, Hispanics, Native Americans and African Americans [45]. On top of racial disparities, DKD susceptibility and rate of progression is clustered in certain pedigrees as results from The Family Investigation of Nephropathy and Diabetes (FIND) study [46,47] and the Genetics of Kidneys in Diabetes (GoKinD) Study showed over the last years [48–54].
Rationale for a systems level definition of DKD
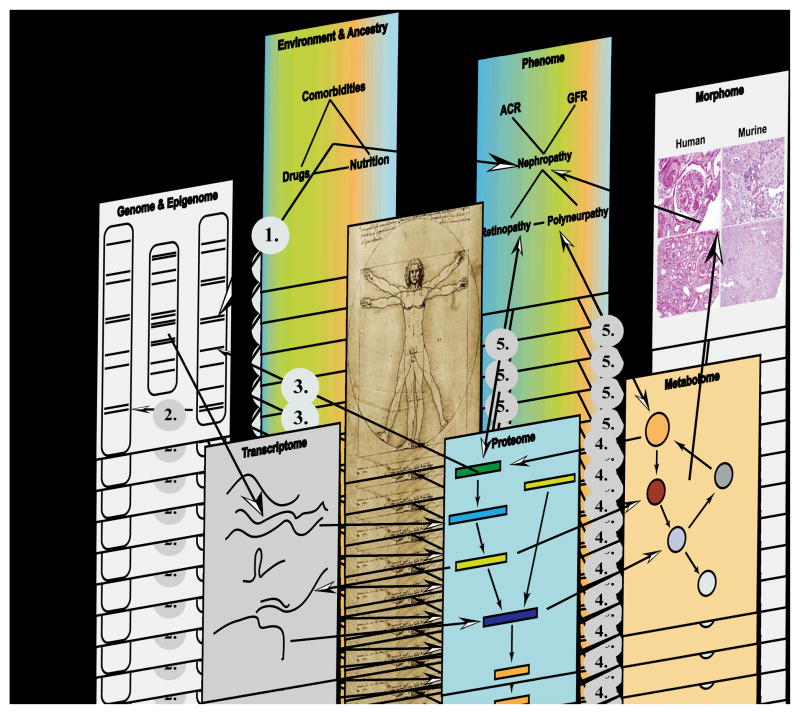
DM and its end organ damage, including DKD, are diseases affecting multiple interrelated end organ systems, requiring a research strategy matching the systemic nature of the disease. Over the last decade both data generation and data analysis approaches have matured to a level that we can start to explore DM and DKD in patients on a systems level, generating a network view of relevant disease processes [55,56]. The term “Systems Biology” in the context of this review is used to describe the integration of large–scale data sets into regulatory networks, using the concept of “Plurality of cause and effect” as introduced by Sauer [57]. Molecular medicine has matured to a level that we are now in a position to generate information from patients covering the entire spectrum from genetic variations such as single nucleotide polymorphisms (SNPs) or copy number variations (CNVs) (Genomics), along epigenetic modifications (methylation and histone modifications; Epigenomics), coding and non-coding RNAs (mRNA, miRNA, lincRNA, tRNA, and rRNA; Transcriptomics), down to the level of proteins (Proteomics) and metabolites (Metabolomics), which are further refined by detailed morphologic characterization (Histo–/Morphogenomics) and in depth ethno–demographics as well as clinical phenotyping (Phenomics) along the “genome–phenome continuum” [58–60] (see Figure 1). This approach has already facilitated a more comprehensive understanding of disease pathogenesis in oncology and developmental biology [61]. This is the necessary step towards individualized medicine – “the right drug, at the right time, at the right dose, for the right person” [62] – in nephrology. However, in order to achieve this goal of a tailored and risk adapted therapy we have to implement a systematic approach to define the impact of the disease along the genotype–phenotype continuum [63]. This approach can be broken down in four equally challenging steps:
Figure 1. From Genome–to–Phenome – An Integrative Systems Biology Perspective on Diabetic Kidney Disease.
Each biological system (e.g. diabetic kidney) can be regarded as being composed of discrete components. Each component can be independently inquired through unbiased high throughput “–omics” profiling or imaging technologies (ultrasound, nuclear magnetic resonance imaging, positron emission tomography), but individual level data is embedded in an interaction network that connects and integrates the various components in a functional context. Potential interactions between two or more components are indicated through arrows e.g. gene–by–environment interaction (1), gene–by–gene interaction (2), protein–DNA interaction (3), metabolite–protein interaction (4) e.g. allosteric regulation of enzymes by small metabolites and reciprocal Phenome–“omes” interaction (5). Technologies applied to generate –omics level data are: Genome/Epigenome (SNPs, CNV, promoters): e.g. genome sequencing, high–density SNP chips, methylation assays (MeDIP), histone modifications; Transcriptome (mRNA, miRNA, lncRNA, piRNA, tRNA, rRNA): microarray(s) and RNA sequencing; Proteome (Phosphoproteome, Glycoproteome, Acetylome): yeast two–hybrid, coimmunoprecipitation, 2D differential gel electrophoresis; Metabolome (sugars, lipids, amino acids, small molecules): nuclear magnetic resonance spectroscopy and mass spectrometry; Morphome (tissue damage/integrity): histology, immunohistochemistry; Phenome (ACR, GFR, retinal microaneurysm): clinical chemistry, clearance measurement, imaging studies; Environment and Ancestry (Drugs, Nutrition, Comorbidities): nutritional assessment, environmental exposure and socio–economic assessment.
Establish DKD cohorts with deep phenotypes and matching comprehensive biorepositories for genome wide analysis.
Map genomic landscape in representative DKD tissue samples (e.g. kidney biopsy, blood and urine).
Integrate generated large–scale data sets to define key disease drivers active across multiple levels of the genotype–phenotype continuum.
Test hypotheses derived from III. in model systems and in human interventional trials.
Several DKD cohorts, established over the last two decades, allow capturing of regulatory networks on a genome wide scale, but further investment into patient cohorts as key resources for systems biology are urgently needed. Several strategies are currently discussed to overcome this obstacle ranging from protocol biopsies cohorts to postmortem acquisition of tissue in well phenotyped patients. A recent workshop focused on the discussion of this very key issue pertaining to DKD research [64]. A detailed discussion of this topic warrants an independent review.
Mapping the genomic landscape in DKD
Genetics, Epigenetics and the concept of “metabolic memory” in DKD
Fueled by the discovery of monogenic defects causing rare hereditary forms of diabetes and kidney diseases, strategies were developed to detect the genetic underpinning of DKD. First reports suggesting a genetic contribution to disease risk were based on T1DM sib–pair analysis where DKD subjects aggregated [65,66]. Soon thereafter, reports followed suggesting a heritable component of DKD in T2DM as well [67,68]. Hallmark studies in DKD revealed numerous genetic variations, with some of them replicated across different ethnicities, in T1DM – and T2DM–DKD and some specific for the analyzed cohort and phenotype ([46,69] and Metaanalysis in [70]). The studies published to date identified genetic loci associated with DKD as starting point for functional analysis into disease pathogenesis. However, the overall explained heritability is of low effect size, shared with many other complex traits [71,72]. The very modest genetic effect size confirms the polyetiological nature of the disease. Finding significantly associated genetic variations at a genome wide level for DKD is only the first step. Validation by subsequent resequencing and fine mapping studies to determine the actual causal variant that is in linkage disequilibrium with the tagging SNP, replication in an independent cohort and generation of a testable hypothesis of the mechanistic impact of the polymorphism are critical next steps [73–75]. In oncology, Pomerantz et al. recently highlighted such a successful strategy and the importance of experimental follow up [76]. In DKD, the currently available information on genetic risk loci are not of sufficient effect size and robustly replicated as to be developed for clinical decision support, despite the fact that several loci confer a statistically significant increase in disease risk [70,77,78].
Long–term outcome data from DCCT/EDIC and the UKPDS study showing a lasting impact of early intervention on the incidence of diabetic end organ complications [79,80], led to the resurrection of the metabolic memory concept, first described by Engerman and Kern in the dog retina of alloxan–induced diabetes [81]. In addition to long–lasting protein modification like generation of AGEs, epigenetic modifications of the genome are prime candidate memory mechanisms, due to their partial heritability and high plasticity in response to internal and environmental challenges such as drugs, toxins, hyperglycemia and inflammation [82–87]. The area of epigenomics in context of diabetic complications is currently under intense investigation [88–91]. Integrating the epigenetic regulation of the genome with the emerging information of genetic risk loci of DKD might allow defining the individual genetic risk at a clinically relevant level [92]. Rapidly dropping sequencing costs will further facilitate comprehensive sequence based analysis in populations at risk of DKD [93].
Transcriptomics and the next generation profiling of diabetic kidney disease
Transcriptomic analysis, the comprehensive mRNA expression analysis, was the first genome wide profiling approach available for broad application in human tissues [94]. Initially, microarrays using predefined oligonucleotides or cDNAs were employed. These array technologies are currently starting to be replaced by unbiased RNA Sequencing (RNA–Seq.) technology. Expression profiles can be simultaneously interrogated in kidney tissue, blood and urine, offering the chance to compare across organ boundaries and evaluate their potential for non–invasive surrogate marker. With the adoption of a standardized technique of specimen handling and RNA isolation developed by the European Renal cDNA Bank, a robust protocol was established taking advantage of diagnostic renal biopsies for molecular profiling studies in an international multicenter setting [95,96] and the approach has been adopted by multiple networks around the globe. Initial studies focused on targeted analysis of candidate transcripts identified by the analysis of monogenetic renal diseases [97] or model systems [98], followed by studies comparing in vitro responses to proteinuria to the in vivo setting [99]. Baelde and colleagues compared the glomerular transcriptome from two cadaveric healthy donors with two DKD cases and where able to partially recapitulate findings from prior studies investigating gene expression changes in cell culture, animal models and human biopsy sections [100]. A first comprehensive transcriptional network analysis combined with promoter modeling approaches identified a specific NF–κB model as a key driver in progressive DKD compared to early DKD and other proteinuric diseases [101]. Additionally, the mechanism involving hypoxic gene regulation, vascular plasticity as well as JAK/STAT signaling pathway show temporal and compartment specific gene expression patterns in human DKD [15,102]. Furthermore, transcriptomic profiling not only validated prior documented significance of complement pathway and inflammatory signaling pathways but also described new pathways involved and/or affected by DKD, which haven’t been suspected yet or underestimated e.g. caveolar–mediated endocytosis and lipid metabolism [103]. Current limitations of kidney tissue specific transcriptomics relate primarily to the limited available sample size and the cross sectional design with limited inference of causality. Most studies using clinical biopsies represent established DKD stages with impaired eGFR and high degree of proteinuria and describe the inflammatory and fibrotic response seen with progression to ESRD [101,103]. A significant concern is the selection bias for patients with suspected DKD undergoing biopsy, as kidney biopsy in most centers is not part of the routine DKD diagnostic evaluation, except in cases of sudden unexplained rise in serum creatinine, drop in eGFR or increased proteinuria [104], requiring careful evaluation of clinical and histological features to select cases of DKD without secondary disease manifestation.
Several groups have implemented renal research or protocol biopsies in early stages of T1DM– and T2DM–DKD [105,106]. A low incidence in kidney biopsy related complications in carefully selected patients and improved diagnostic value for disease entities potentially amenable to treatment have been reported with modern biopsy technologies [107,108]. The unique renal macro– and microarchitecture with multiple cell lineages arranged in a defined order and spacing along the nephron (e.g. podocytes, mesangial cells) contribute to substantial heterogeneity. This can be addressed to some extent by manual microdissection of the nephron compartments [109,110] or “in silico” microdissection using cell–lineage specific transcripts [97]. Methods using the concept of cell lineage specific transcripts on a genome scale to deconvolve the transcriptome of complex tissues are currently being developed [111].
As with all genome wide profiling approaches, validating diagnostic candidate markers of molecular pathways in an independent replication cohort is a critical step to address the inherent danger of overfitting large–scale data on small sample sizes (see [112] and a detailed discussion by the Institute of Medicine [113]). Generating the necessary cohorts and subsequent data analysis will require further integration of existing efforts and expansion of international collaborative networks. As most groups use comparable tissue procurement, processing and analysis platforms for transcriptional profiling, data integration and comparative analysis are an achievable goal.
Proteomics and Metabolomics in DKD
Advances in high throughput profiling technologies facilitated an unprecedented explosion in generating large–scale molecular data that is not any longer constrained to the genome and transcriptome level, but also leads to significant improvements on the level of proteomics and metabolomics. The proteomic studies start to highlight the substantial role of posttranscriptional regulatory mechanisms and simultaneously reveal how gene expression affects actual protein abundance and function [114–116]. The various layers of posttranscriptional and posttranslational regulation add an additional level of complexity in proteomic studies. Major advances in analytical chemistry, including improved chromatographic and mass spectrometry (MS) platforms supported by sophisticated computational algorithms are finally starting to bring proteomics towards a level of protein coverage for widespread use in systems biology [117,118].
Proteomics
Since ACR does not accurately predict clinical endpoints in DKD, several investigators have focused on urinary and plasma proteome patterns from healthy volunteers, T1DM and T2DM patients with and without various degrees of albuminuria and different ranges of GFR [119–125]. Three recent examples highlight the potential of an untargeted proteomics approach. Otu and colleagues using a nested case–control design and surface–enhanced laser desorption/ionization time–of–flight mass spectrometry (SELDI–TOF/MS) of urine from type 2 DM Pima Indians were able to distinguish initially NAlb subjects developing DKD within a 10 year follow up period based on a 12–peak urine peptide signature detected in urine collected at time the individuals were still NAlb [126]. Rossing and co–workers compared gender and age matched healthy controls, and T1DM patients from the Steno Diabetes Center with varying degrees of albuminuria (NAlb, μAlb and MAlb) [124]. They evaluated the potential of Capillary electrophoresis (CE) MS to define urinary polypeptides signatures an identified increased albumin fragments, decreased uromodulin and collagen type I fragments. Overgaard et al. utilized isobaric mass tags (iTRAQ) coupled with liquid chromatography (LC) MS and SELDI–TOF/MS to identify plasma protein/peptides in a cross–sectional analysis of 123 T1DM cases previously classified as NAlb, μAlb and MAlb. This strategy yielded many promising candidates including transthyretin, apolipoprotein A1, apolipoprotein C1 and cystatin c. Interestingly, unidentified plasma protein peaks showed promising features in improving predictability of DKD [127,128]. Other studies applied CE–MS and confirmed urinary peptides such as collagen fragments as robust predictors of CKD and DKD respectively [129–133]. One of the limitations of these studies is that majority of the identified fragments arise from abundant plasma proteins. As the glomerular filtration barrier fails with DKD, non–selective, albumin predominant pattern ensues which may skew detected polypeptides to abundant proteins [134]. Methods effectively addressing the detection of low abundance proteins have recently been developed, in order to provide a higher degree of disease specificity and unravel low–abundant proteins which are likely to be of mechanistic significance [135]. Immunodepletion and glycoprotein enrichment techniques allow effective depletion of albumin, immunoglobulins and other highly abundant urinary and plasma proteins while providing a more in depth analysis of a sub fraction of the inquired proteome. As glycosylated proteins are critical for cellular interactions and signaling cascades, disease states are likely to cause early and specific alterations in urinary glycoprotein excretion. Indeed, glycoproteins are now important markers of autoimmunity and malignancy [136,137]. More recently, the plasma glycoproteome has been used to predict nephropathy in diabetic subjects [138]. Despite this promising role as a non–invasive and specific biomarker of disease, little is known about the urinary glycoproteome in DKD. Consequently, we hypothesized that the urinary glycoproteome would be altered in patients with CKD compared to healthy controls, and that specific glycoprotein alterations might be useful in predicting CKD progression. We performed an exploratory analysis of the urine glycoproteins in patients with CKD that included DKD subjects [139]. We utilized a hydrazide enrichment technique combined with tandem MS identification of glycoproteins. In urine, this strategy identified several differentially expressed proteins compared to healthy controls, including proteins with endopeptidase inhibitor activity, protein–binding functions, and acute–phase/immune–stress response activity that have been previously linked to kidney disease and therefore supporting the existing hypotheses that inflammation plays a key role in renal disease pathogenesis. Additional renal specific sources of proteins include analysis of excreted urinary exosomes (30–90 nm vesicles), which consist of an evolutionary conserved tissue– and cell–type specific set of proteins. Due to its tissue specificity, exosomes analysis might facilitate a more accurate representation of renal pathology [140,141]. Despite these advances, several challenges remain in proteomic analysis. These include: a) impaired detection of low–abundant proteins and skewing to high–abundant proteins especially in blood and urine (while strategies involving depletion of high–abundant proteins and enrichment of low–abundant proteins have improved detection, coverage still lacks depth); b) obstacles dependent on employed techniques and software data analysis issues [142] and c) difficulty in predicting the functional context of the protein. For example protein abundance may vary but functionality may not change as it may depend on posttranslational modifications such phosphorylation, acetylation and glycosylation. These issues have been recently addressed in a position statement including recommendations for quality and reporting metrics, data deposition and quantification methods [143–145], similar to the MIAME reporting guidelines for transcriptomic analysis [146]. Collectively, these observations suggest that the human proteome and in particular the glycoproteome may serve as a discovery source for novel disease mechanism–based biomarkers not only in DKD but also in renal diseases in general but challenges remain in translating these findings to functional context.
Metabolomics
Metabolomics is the rapidly evolving field that attempts to systematically capture, identify and quantify endogenous and exogenous metabolites within a biological sample. The small molecules represent the result of complex biological processes in a given tissue or organ and the final functional read–out of a biological system. Therefore, metabolites form attractive candidates as biomarkers, as they are closer to the final phenotype. The chemical diversity of the metabolites makes analysis a formidable challenge. The rapid development of a variety of MS and nuclear magnetic resonance (NMR) based platforms have enabled detection, separation, characterization and quantification of such chemically diverse low–molecular weight structures such as lipids, amino acids, nucleic acids and organic acids (reviewed in [118]). Metabolite profiling strategies have been implemented successfully to identify metabolic markers of disease in cancer, cardiovascular disease (CVD) and diabetes [147–155]. We recently utilized an untargeted metabolomic strategy to explore underlying unifying mechanisms of disease progression and treatment response in order to identify novel metabolite biomarkers of progressive murine DKD [156]. The urine from control DBA/2J and streptozotocin–induced diabetic mice as well as mice treated for 10 weeks with rosiglitazone (which reverses DKD phenotype) was analyzed with LC electrospray ionization (ESI) TOF/MS [156]. In total 56 features showed up– or down–regulation by > 2–fold in the diabetic animals. Of the 56 molecular features, 32 were identified by database searching. Rosiglitazone reversed nine of these 32 compounds back to baseline and may therefore serve as potential biomarkers for response to treatment and reversal of rodent DKD phenotype. For compound identification, we leveraged the publicly available METLIN Metabolite Database [157] and the Human Metabolome Database [158]. The pathophysiology of the metabolic changes that include ubiquinone and indoxyl sulfate is currently under intense investigation. Interestingly, a study from Barreto et al. reported that serum indoxyl sulfate correlates inversely with renal function and might have a direct relationship with aortic calcification and pulse wave velocity in patients with CKD [159]. In survival analyses, the highest indoxyl sulfate tertile was a powerful predictor of overall and cardiovascular mortality. The predictive power of indoxyl sulfate for death was retained after adjustment for age, gender, diabetes, albumin, hemoglobin, phosphate and aortic calcification. Other clinical studies highlight the potential application of metabolomics in DKD research. The study from Zhang and colleagues utilized Ultra performance liquid chromatography (UPLC) coupled with TOF/MS to distinguish the serum metabolic profiles of 25 healthy volunteers, 33 T2DM and 8 DKD patients [160]. Significant changes in the serum level of leucine, dihydrosphingosine and phytosphingosine were noted, indicating perturbations of amino acid metabolism and phospholipid metabolism. Another study investigated 52 T1DM patients recruited in the FinnDiane Cohort who were prospectively followed over 5–6 years, during which one half developed proteinuria (progressors) and the other half did not (non–progressor) [161]. Metabolite profiles of baseline 24–h urine samples were analyzed by gas chromatography–mass spectrometry (GC-MS) and liquid chromatography–mass spectrometry (LC-MS). Multivariate logistic regression modeling of the GC MS data resulted in a classifier profile that had an overall accuracy of 75% and a precision of 73% for a binary classification model of progressor vs. non–progressor. The discriminating metabolites included acyl–carnitines, acyl–glycines, and metabolites related to tryptophan metabolism. Other studies of DKD yielded metabolic profiles indicating alterations in carbohydrate, lipid, nucleic acid, and amino acid metabolism [121,162]. The biomarker potential for these metabolic markers for predicting clinical phenotypes need to be validated in large–scale clinical trials.
Systems Genetics: linking genetic variance to clinical outcomes via intermediary phenotypes
Genome wide association studies (GWAS) of complex traits have been very effective in defining statistically significant risk loci by mapping genome wide genetic variance in large cohorts onto clinical traits. However, this approach is agnostic concerning the molecular mechanism and intermediate regulatory cascades responsible for the manifestation of the clinical phenotype. To close the molecular knowledge gap, systems genetics employs as a key tool association studies linking genotypic information with intermediary molecular traits of interest, e.g. transcript level or metabolite level (expression/metabolic Quantitative Trait Locus, eQTL/mQTL) [163–165]. Subsequent integration of molecular QTLs with association studies aids to identify the molecular impact of genetic variance associated with a disease phenotype.
For instance, the metabolome, as a close “proxy” of the final phenotype in metabolic diseases like DM, can be associated with the genetic variance seen in a population to establish links between genetic variance and metabolite levels in a defined biosample during a disease process [166–169]. A key study by Ferrara et al. linked tissue metabolite levels to genetic variation. The authors performed transcriptome analysis and whole genome sequencing of liver samples from a diabetes–resistant C57BL/6 leptinob/ob and diabetes–susceptible BTBR leptinob/ob mouse strain and further refined their analysis by integrating quantitative metabolite levels [168]. Various groups of liver metabolites significantly associated with distinct chromosomal regions suggesting that they were under the control of genetic variations in those regions. Suhre et al. reported on the genetic association of urinary metabolites detected by NMR spectroscopy in urine of 862 male participants from the epidemiological Study of Health in Pomerania (SHIP) [170]. An independent validation in additional 2,031 samples (1,039 independent SHIP and 992 samples from the Cooperative Health Research in the Augsburg Region (KORA) study) revealed consistent genome wide significant loci tagging SLC7A9 and NAT2 which have been already associated with CKD and drug–induced liver toxicity respectively [171–173]. Kettunen et al. utilized NMR spectroscopy based detection of serum metabolites of over 8,000 genotyped Finish individuals and were able to ascertain a high degree of heritability for metabolic phenotypes, ranging from 40% up to 60% [174]. The quantitative traits of various body fluids are currently being mapped for intra– and inter–individual comparison on a population scale [175,176]. While these represent early stage discoveries, they already help to bring genetic variance closer to disease phenotypes.
Building regulatory networks in health and disease
Networks are increasingly used to display complex dependencies in biological systems. A network is a graph consisting of nodes and edges. Nodes represent specific entities or objects for instance a gene, a protein or trait of interest and the edges represent a defined relationship for connected nodes (e.g. protein–protein interaction). These relationships can be directed, where node A interacts and activates node B, or without a specific directionality merely describing an interaction. As each level of information has its own merits and weaknesses, the integration of multiple –omics layers might circumvent or minimize those drawbacks and deliver a more accurate picture of the ongoing processes [177–181]. One such way is to generate and visualize integrated, functional networks by combining genomic information, transcriptomic and proteomic data obtained from the same individual tissue. This approach aids in understanding how DNA sequence variation (e.g. from GWAS) and transcriptional events lead to corresponding changes in pathway functionality. A key aspect of network Employing such a strategy enables the characterization of genetically determined transcriptional regulatory networks in health and their perturbation in disease [101,182]. Moreover, by combining results from time series experiments one can expand and track dynamic changes within those networks over time and detect driving core components and even distinguish between early or late events in the disease process [183,184]. Leveraging this uniform integrative strategy allows combining the strengths and compensates for the limitations on each of the multiple lines of evidence. The resulting hierarchical and comprehensive “disease map”, defines key molecular functionalities, highlighting points of conversion that might be more amenable for further exploration and therapeutic intervention [185]. Through a combination of causal and probabilistic methods across mouse and humans, Schadt and co–workers were able to pin point key functionalities with high confidence and provide a framework for targeted downstream experimental evaluation. Due to the unbiased nature of these strategies, integrative approaches are capable of uncovering new disease specific relationships and can shed additional light into cellular and organismal pathophysiology [184,186,187].
A crucial step in advancing integrative network biology analysis into the clinical arena will rely on a robust and comprehensive visualization of these multilayered interactions [188,189]. Cytoscape, the versatile open source bioinformatics platform is the most common and powerful tool used to visualize and analyze networks [190]. Even though network representation has proven useful in the past years, networks are still compressing multidimensional data sets into two or maximal three dimensions [191] and represent therefore a highly simplified version of the true underlying dynamic processes. This “static” network concept has to be expanded to capture dynamic interactions and to adapt to environmental and genetic constraints over time in order to gain full insight into cellular dynamics [184]. Defining human health and disease on a genome wide scale has already yielded very exciting results across the genome–phenome continuum. The concept of integrated personal –omics profiling (iPOP) is increasingly gaining attraction [192], but challenges remain: a) discovery of rare variants and variants with smaller effect sizes, b) understanding the link between genetic variations and perturbation of pathophysiology, and finally c) translating –omics results efficiently into improved patient care.
Biomarker: the surrogates for diagnosis, risk assessment and prognosis
In order to facilitate an accurate diagnosis, predict risk of progression and response to drug/dietary intervention clinical medicine relies on specific, disease–associated biomarkers. Biomarkers are objective, quantitatively or qualitatively measured characteristics indicating normal or pathogenic biological processes or response to therapeutic intervention [193–195]. Sensitivity, specificity and positive or negative predictive value of those markers influence our therapeutic decision–making and patient counseling. However, the available biomarkers for DKD, such as ACR and GFR, harbor significant limitations and can be misleading in the research and clinical setting (see above). The optimal biomarker should reflect consistency, strength of relationship between “marker” and outcome in question, i.e. for DKD progression to ESRD. Biomarkers strength are of higher impact if an adequate characterization of disease dynamics can be established a priori e.g. the marker precedes the desired outcome, outperforms the already established disease predictors and responds favorably to therapeutic interventions that are known to have salutary effects on disease progression. The degree of confidence can be augmented further if the biomarker does not only show a pure “guilt by association” characteristic but is also an integral part of the pathophysiologic process in question [43,196]. Biomarkers can be classified according to their purpose: diagnostic, disease staging and prognostic, as well as source based – genomic, transcriptomic, proteomic and metabolomic marker(s). The field of biomarkers in DKD and kidney disease in general is rapidly evolving and has been extensively reviewed in [119,197,198].
Challenges and perspectives
What we can learn from Oncology
Over the past years, we witnessed a substantial progress in uncovering new pathophysiologic mechanisms of neoplastic disease and their explicit characterization on a molecular and network level. The advent of profiling technology enabled the transition from a pure experimental screening method to implementation of tumor specific fingerprinting in prospective clinical trials guiding therapeutic decision [199]. However, recent studies showed that vigorous analytical assessment of generated data as well as documentation of robust reproducibility in independent cohorts is of paramount importance, as ambiguous results can impact the decision making process leading to misassignment of targeted therapeutics and severely harm patients [200,201]. One such instance that served as a wakeup call for the entire –omics research community is the experience from a recent case involving scientific misconduct in translating –omics research results to clinical patient stratification without rigorous validation [202,203]. After retraction of a series of articles in Journal of Clinical Oncology, Nature Medicine and New England Journal of Medicine [204–206], and at request of the National Cancer Institute (NCI) the recently released report from the Institute of Medicine (IOM) pertaining to the topic of “translational OMICS” establishes guidelines to assure reproducible omics–based test technology grounded on sound scientific conduct [113]. Therefore, robust validation strategies need to be employed, including oversight by federal regulatory agencies such as the Food and Drug Administration (FDA), when the intended use of developed tests will extend into clinical trials [207].
Thus far, the limited studies using a data mining strategy on genome wide DKD data sets have not only confirmed established pathophysiological mechanism, but also described novel mechanisms associated with disease pathogenesis. One major aspect contributing to the limited capability of robust mechanistic discoveries and uncovering potential confounding factors is the availability of high quality samples for –omics analysis in conjunction with in–depth clinical phenotyping data in sufficiently sized cohorts. However, increasing the sample size is and can only be a first step in the process of dissecting the underlying molecular mechanisms of DKD. Curtis et al. demonstrated the power of integrative –omics data analysis in combination with longitudinal follow up of over 2,000 clinically annotated breast cancer specimens [208]. Based on experimental, observational and interventional data DKD can be considered as a spectrum of “diseases” with extreme phenotypes at each end e.g. fast progressors vs. non–progressors regardless of intervention (“resistant to the effect of poor metabolic control” [209]). The Joslin 50–year Medalist Study has established a longstanding cohort of T1DM patients with no apparent end organ damage after decades of DM [210]. The hope is that these cohorts will allow to capture the genetic and environmental sources of phenotypic heterogeneity in DKD.
An additional source of mechanistic heterogeneity seen in DKD relates to disease process activation in a stage specific manner. Pathogenic disease processes such as ROS generation, inflammation and profibrotic pathway activation may contribute differently at various disease stages and might not be equally affected by therapeutic interventions. Loss of differentiation and inflammation might play a more significant role during initiation of complications, but might be superseded in late stages of progression by profibrotic mechanisms. To distinguish these events sufficient large cohorts with clinical and structural information on disease stage are essential. However, even modest samples size can allow to define a compartment and stage–specific regulation, as has been demonstrated for the JAK/STAT pathway with activation during early DKD in the glomerular compartment, followed by the tubulointerstitial compartment during the progressive phase of the disease [15].
Tackling critical steps in a collaborative effort in order to assemble necessary cohorts for multilevel outcome analysis is a crucial challenge in DKD. These strategies are already implemented in other glomerular diseases i.e. as realized for Nephrotic Syndrome with the Nephrotic Syndrome Study Network (NEPTUNE, [211]) where a multidisciplinary team of scientists work together with patients and their families towards a comprehensive picture of glomerular failure. To ensure effective use of large–scale data sets by the renal research community requires innovative concepts of data processing and sharing between different knowledge domains (for a formal analysis of this collaborative interdisciplinary process see [212]). We need easily accessible platforms that allow data storage in reasonable accessible repositories, facilitate data exchange between involved researchers, and allow dissemination to the research community. First tools serving these functionalities in defined knowledge domains are emerging (i.e. Nephromine for renal transcriptional data sets [213]) and more comprehensive solutions concerning this key challenge of large–scale data research are currently pursued [214].
Concluding remarks
DKD develops as a consequence of systemic disease, diabetes, driving the intrarenal disease progression via a multitude of interrelated molecular pathways. DKD and other diabetic complications, particularly CVD, interact via multiple feedback loops leading to the demise of the entire organism. Molecular medicine strategies described in this review allow for the first time to capture the complexity of these individual regulatory events in a genome wide manner. The emerging tools for network–based analysis of biological systems will provide a starting point to understand the regulatory cascades of DKD on a tissue and organismal level. Matching the complexity of DKD disease process with a comprehensive research approach should allow defining the key drivers across disease stages and organs. This will define the critical entry points for molecular based diagnostics, prognostics and targeted therapy.
Acknowledgments
This work was supported by grants from the Juvenile Diabetes Research Foundation to M.K. and S.P., the German Research Foundation to C.V.K. (KO 4266/1-1), the National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Disease (P30-DK-081943 to F.C.B. and M.K, DK083912 to M.K., DK089503, DK094292 to S.P., DK082841 to M.K. and S.P.). The authors appreciate the assistance of Dr. Jeff B. Hodgin (Department of Pathology, University of Michigan) for providing the histological images. We further thank Chandra Hall for carefully proofreading the manuscript and the editor and referees for helpful comments and suggestions on an earlier version of the manuscript. The authors apologize to those colleagues whose work we were unable to cite in this review due to space limitation.
Abbreviations
- GWAS
genome wide association study
- SNP
single nucleotide polymorphism
- CNV
copy number variation
- NF–κB
nuclear factor κ–light–chain–enhancer of activated B cells
- JAK/STAT
Janus kinase/Signal transducer and activator of transcription
- MCP–1/CCL2
macrophage chemoattractant protein 1
- FDA
U.S. Food and Drug Administration
- DCCT/EDIC
The Diabetes Control and Complications Trial/Epidemiology of Diabetes Interventions and Complications Study
- UKPDS
The United Kingdom Prospective Diabetes Study
- ERCB
European Renal cDNA Bank–Kröner–Fresenius Biospy Bank
- MIAME
Minimum Information About a Microarray Experiment
Footnotes
Conflict of interest: The authors declare no competing financial interest.
References
- 1.Wild S, Roglic G, Green A, Sicree R, King H. Global Prevalence of Diabetes. Diabetes Care. 2004;27 (5):1047–1053. doi: 10.2337/diacare.27.5.1047. [DOI] [PubMed] [Google Scholar]
- 2.Shaw JE, Sicree RA, Zimmet PZ. Global estimates of the prevalence of diabetes for 2010 and 2030. Diabetes Res Clin Pract. 2010;87 (1):4–14. doi: 10.1016/j.diabres.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 3.Coresh J, Selvin E, Stevens LA, Manzi J, Kusek JW, Eggers P, Van Lente F, Levey AS. Prevalence of Chronic Kidney Disease in the United States. JAMA. 2007;298 (17):2038–2047. doi: 10.1001/jama.298.17.2038. [DOI] [PubMed] [Google Scholar]
- 4.Foley R, Collins A. The growing economic burden of diabetic kidney disease. Curr Diab Rep. 2009;9 (6):460–465. doi: 10.1007/s11892-009-0075-9. [DOI] [PubMed] [Google Scholar]
- 5.The Australia and New Zealand Dialysis and Transplant Registry. [Accessed: 3/30/2012];The 34th Annual ANZDATA Report 2011 - Data to 2010. 2011 http://wwwanzdataorgau/v1/report_2011html.
- 6.European Renal Association - European Dialysis and Transplant (ERA-EDTA) Registry. [Accessed: 3/30/2012];ERA-EDTA Registry Annual Report 2009. 2011 http://wwwera-edta-regorg/files/annualreports/pdf/AnnRep2009_newpdf.
- 7.United States Renal Data System. USRDS 2011 Annual Data Report: Atlas of Chronic Kidney Disease and End-Stage Renal Disease in the United States. Vol. 2011. National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases; Bethesda, MD: 2011. http://www.usrds.org. [Google Scholar]
- 8.de Boer IH, Rue TC, Hall YN, Heagerty PJ, Weiss NS, Himmelfarb J. Temporal Trends in the Prevalence of Diabetic Kidney Disease in the United States. JAMA. 2011;305 (24):2532–2539. doi: 10.1001/jama.2011.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haffner SM, Lehto S, Rönnemaa T, Pyörälä K, Laakso M. Mortality from Coronary Heart Disease in Subjects with Type 2 Diabetes and in Nondiabetic Subjects with and without Prior Myocardial Infarction. New Engl J Med. 1998;339 (4):229–234. doi: 10.1056/NEJM199807233390404. [DOI] [PubMed] [Google Scholar]
- 10.Valmadrid CT, Klein R, Moss SE, Klein BEK. The Risk of Cardiovascular Disease Mortality Associated With Microalbuminuria and Gross Proteinuria in Persons With Older-Onset Diabetes Mellitus. Arch Intern Med. 2000;160 (8):1093–1100. doi: 10.1001/archinte.160.8.1093. [DOI] [PubMed] [Google Scholar]
- 11.Sarnak MJ, Levey AS, Schoolwerth AC, Coresh J, Culleton B, Hamm LL, McCullough PA, Kasiske BL, Kelepouris E, Klag MJ, Parfrey P, Pfeffer M, Raij L, Spinosa DJ, Wilson PW. Kidney Disease as a Risk Factor for Development of Cardiovascular Disease. Hypertension. 2003;42 (5):1050–1065. doi: 10.1161/01.HYP.0000102971.85504.7c. [DOI] [PubMed] [Google Scholar]
- 12.Brownlee M. Biochemistry and molecular cell biology of diabetic complications. Nature. 2001;414 (6865):813–820. doi: 10.1038/414813a. [DOI] [PubMed] [Google Scholar]
- 13.Forbes JM, Fukami K, Cooper ME. Diabetic Nephropathy: Where Hemodynamics Meets Metabolism. Exp Clin Endocrinol Diabetes. 2007;115 (2):69–84. doi: 10.1055/s-2007-949721. [DOI] [PubMed] [Google Scholar]
- 14.Figueroa-Romero C, Sadidi M, Feldman E. Mechanisms of disease: The oxidative stress theory of diabetic neuropathy. Rev Endocr Metab Disord. 2008;9 (4):301–314. doi: 10.1007/s11154-008-9104-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berthier CC, Zhang H, Schin M, Henger A, Nelson RG, Yee B, Boucherot A, Neusser MA, Cohen CD, Carter-Su C, Argetsinger LS, Rastaldi MP, Brosius FC, Kretzler M. Enhanced Expression of Janus Kinase and Signal Transducer and Activator of Transcription Pathway Members in Human Diabetic Nephropathy. Diabetes. 2009;58 (2):469–477. doi: 10.2337/db08-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Navarro JF, Mora-Fernández C. The role of TNF-α in diabetic nephropathy: Pathogenic and therapeutic implications. Cytokine Growth Factor Rev. 2006;17 (6):441–450. doi: 10.1016/j.cytogfr.2006.09.011. [DOI] [PubMed] [Google Scholar]
- 17.Williams ME. Diabetic Nephropathy: The Proteinuria Hypothesis. Am J Nephrol. 2005;25 (2):77–94. doi: 10.1159/000084286. [DOI] [PubMed] [Google Scholar]
- 18.Schena FP, Gesualdo L. Pathogenetic Mechanisms of Diabetic Nephropathy. J Am Soc Nephrol. 2005;16 (3_suppl_1):S30–33. doi: 10.1681/asn.2004110970. [DOI] [PubMed] [Google Scholar]
- 19.Galkina E, Ley K. Leukocyte Recruitment and Vascular Injury in Diabetic Nephropathy. J Am Soc Nephrol. 2006;17 (2):368–377. doi: 10.1681/ASN.2005080859. [DOI] [PubMed] [Google Scholar]
- 20.Nawroth PP, Isermann B. Mechanisms of Diabetic Nephropathy – Old Buddies and Newcomers Part 2. Exp Clin Endocrinol Diabetes. 2010;118 (10):667–672. doi: 10.1055/s-0030-1253440. [DOI] [PubMed] [Google Scholar]
- 21.Nawroth PP, Isermann B. Mechanisms of Diabetic Nephropathy – Old Buddies and Newcomers Part 1. Exp Clin Endocrinol Diabetes. 2010;118 (9):571–576. doi: 10.1055/s-0030-1255051. [DOI] [PubMed] [Google Scholar]
- 22.Kanwar YS, Sun L, Xie P, Liu F-y, Chen S. A Glimpse of Various Pathogenetic Mechanisms of Diabetic Nephropathy. Annu Rev Pathol. 2011;6 (1):395–423. doi: 10.1146/annurev.pathol.4.110807.092150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.DiBona GF, Kopp UC. Neural control of renal function. Physiol Rev. 1997;77 (1):75–197. doi: 10.1152/physrev.1997.77.1.75. [DOI] [PubMed] [Google Scholar]
- 24.Augustyniak RA, Tuncel M, Zhang W, Toto RD, Victor RG. Sympathetic overactivity as a cause of hypertension in chronic renal failure. J Hypertens. 2002;20 (1):3–9. doi: 10.1097/00004872-200201000-00002. [DOI] [PubMed] [Google Scholar]
- 25.Grotendorst GR. Connective tissue growth factor: a mediator of TGF-β action on fibroblasts. Cytokine Growth Factor Rev. 1997;8 (3):171–179. doi: 10.1016/s1359-6101(97)00010-5. [DOI] [PubMed] [Google Scholar]
- 26.Böttinger EP, Bitzer M. TGF-β Signaling in Renal Disease. J Am Soc Nephrol. 2002;13 (10):2600–2610. doi: 10.1097/01.asn.0000033611.79556.ae. [DOI] [PubMed] [Google Scholar]
- 27.Liu Y. Renal fibrosis: New insights into the pathogenesis and therapeutics. Kidney Int. 2006;69 (2):213–217. doi: 10.1038/sj.ki.5000054. [DOI] [PubMed] [Google Scholar]
- 28.Froissart M, Rossert J, Jacquot C, Paillard M, Houillier P. Predictive Performance of the Modification of Diet in Renal Disease and Cockcroft-Gault Equations for Estimating Renal Function. J Am Soc Nephrol. 2005;16 (3):763–773. doi: 10.1681/ASN.2004070549. [DOI] [PubMed] [Google Scholar]
- 29.Stevens LA, Coresh J, Greene T, Levey AS. Assessing Kidney Function — Measured and Estimated Glomerular Filtration Rate. New Engl J Med. 2006;354 (23):2473–2483. doi: 10.1056/NEJMra054415. [DOI] [PubMed] [Google Scholar]
- 30.Stevens LA, Levey AS. Measured GFR as a Confirmatory Test for Estimated GFR. J Am Soc Nephrol. 2009;20 (11):2305–2313. doi: 10.1681/ASN.2009020171. [DOI] [PubMed] [Google Scholar]
- 31.Stevens LA, Schmid CH, Greene T, Zhang Y, Beck GJ, Froissart M, Hamm LL, Lewis JB, Mauer M, Navis GJ, Steffes MW, Eggers PW, Coresh J, Levey AS. Comparative Performance of the CKD Epidemiology Collaboration (CKD-EPI) and the Modification of Diet in Renal Disease (MDRD) Study Equations for Estimating GFR Levels Above 60 mL/min/1.73 m2. Am J Kidney Dis. 2010;56 (3):486–495. doi: 10.1053/j.ajkd.2010.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Levey AS, Cattran D, Friedman A, Miller WG, Sedor J, Tuttle K, Kasiske B, Hostetter T. Proteinuria as a Surrogate Outcome in CKD: Report of a Scientific Workshop Sponsored by the National Kidney Foundation and the US Food and Drug Administration. Am J Kidney Dis. 2009;54 (2):205–226. doi: 10.1053/j.ajkd.2009.04.029. [DOI] [PubMed] [Google Scholar]
- 33.Julian BA, Suzuki H, Suzuki Y, Tomino Y, Spasovski G, Novak J. Sources of urinary proteins and their analysis by urinary proteomics for the detection of biomarkers of disease. Proteomics Clin Appl. 2009;3 (9):1029–1043. doi: 10.1002/prca.200800243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Caramori ML, Fioretto P, Mauer M. The need for early predictors of diabetic nephropathy risk: is albumin excretion rate sufficient? Diabetes. 2000;49 (9):1399–1408. doi: 10.2337/diabetes.49.9.1399. [DOI] [PubMed] [Google Scholar]
- 35.Perkins BA, Ficociello LH, Silva KH, Finkelstein DM, Warram JH, Krolewski AS. Regression of Microalbuminuria in Type 1 Diabetes. New Engl J Med. 2003;348 (23):2285–2293. doi: 10.1056/NEJMoa021835. [DOI] [PubMed] [Google Scholar]
- 36.Fioretto P, Mauer M. Histopathology of Diabetic Nephropathy. Semin Nephrol. 2007;27 (2):195–207. doi: 10.1016/j.semnephrol.2007.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.American Diabetes Association. Standards of Medical Care in Diabetes - 2011. Diabetes Care. 2011;34 (Supplement 1):S11–S61. doi: 10.2337/dc11-S011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perkins BA, Ficociello LH, Ostrander BE, Silva KH, Weinberg J, Warram JH, Krolewski AS. Microalbuminuria and the Risk for Early Progressive Renal Function Decline in Type 1 Diabetes. J Am Soc Nephrol. 2007;18 (4):1353–1361. doi: 10.1681/ASN.2006080872. [DOI] [PubMed] [Google Scholar]
- 39.Perkins BA, Ficociello LH, Roshan B, Warram JH, Krolewski AS. In patients with type 1 diabetes and new-onset microalbuminuria the development of advanced chronic kidney disease may not require progression to proteinuria. Kidney Int. 2010;77 (1):57–64. doi: 10.1038/ki.2009.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.MacIsaac R, Jerums G. Diabetic kidney disease with and without albuminuria. Curr Opin Nephrol Hypertens. 2011;20 (3):246–257. doi: 10.1097/MNH.0b013e3283456546. [DOI] [PubMed] [Google Scholar]
- 41.Dwyer JP, Parving HH, Hunsicker LG, Ravid M, Remuzzi G, Lewis JB. Renal Dysfunction in the Presence of Normoalbuminuria in Type 2 Diabetes: Results from the DEMAND Study. Cardiorenal Medicine. 2012;2 (1):1–10. doi: 10.1159/000333249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pavkov ME, Knowler WC, Lemley KV, Mason CC, Myers BD, Nelson RG. Early Renal Function Decline in Type 2 Diabetes. Clin J Am Soc Nephrol. 2012;7 (1):78–84. doi: 10.2215/CJN.07610711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Karalliedde J, Viberti G. Proteinuria in Diabetes: Bystander or Pathway to Cardiorenal Disease? J Am Soc Nephrol. 2010;21 (12):2020–2027. doi: 10.1681/ASN.2010030250. [DOI] [PubMed] [Google Scholar]
- 44.Tryggvason K, Pettersson E. Causes and consequences of proteinuria: the kidney filtration barrier and progressive renal failure. J Intern Med. 2003;254 (3):216–224. doi: 10.1046/j.1365-2796.2003.01207.x. [DOI] [PubMed] [Google Scholar]
- 45.Iyengar SK, Schelling JR, Sedor JR. Approaches to understanding susceptibility to nephropathy: From genetics to genomics. Kidney Int. 2002;61 (Suppl 1):S61–S67. doi: 10.1046/j.1523-1755.2002.0610s1061.x. [DOI] [PubMed] [Google Scholar]
- 46.Iyengar SK, Abboud HE, Goddard KAB, Saad MF, Adler SG, Arar NH, Bowden DW, Duggirala R, Elston RC, Hanson RL, Ipp E, Kao WHL, Kimmel PL, Klag MJ, Knowler WC, Meoni LA, Nelson RG, Nicholas SB, Pahl MV, Parekh RS, Quade SRE, Rich SS, Rotter JI, Scavini M, Schelling JR, Sedor JR, Sehgal AR, Shah VO, Smith MW, Taylor KD, Winkler CA, Zager PG, Freedman BI. Genome-Wide Scans for Diabetic Nephropathy and Albuminuria in Multiethnic Populations: The Family Investigation of Nephropathy and Diabetes (FIND) Diabetes. 2007;56(6):1577–1585. doi: 10.2337/db06-1154. [DOI] [PubMed] [Google Scholar]
- 47.Igo JRP, Iyengar SK, Nicholas SB, Goddard KAB, Langefeld CD, Hanson RL, Duggirala R, Divers J, Abboud H, Adler SG, Arar NH, Horvath A, Elston RC, Bowden DW, Guo X, Ipp E, Kao WHL, Kimmel PL, Knowler WC, Meoni LA, Molineros J, Nelson RG, Pahl MV, Parekh RS, Rasooly RS, Schelling JR, Shah VO, Smith MW, Winkler CA, Zager PG, Sedor JR, Freedman BI The Family Investigation of Nephropathy and Diabetes Research Group . Genomewide Linkage Scan for Diabetic Renal Failure and Albuminuria: The FIND Study. Am J Nephrol. 2011;33 (5):381–389. doi: 10.1159/000326763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gohda T, Tanimoto M, Watanabe-Yamada K, Matsumoto M, Kaneko S, Hagiwara S, Shiina K, Shike T, Funabiki K, Tomino Y. Genetic susceptibility to type 2 diabetic nephropathy in human and animal models. Nephrology. 2005;10 (Supplement s2):S22–S25. doi: 10.1111/j.1440-1797.2005.00452.x. [DOI] [PubMed] [Google Scholar]
- 49.Tanaka N, Babazono T. Assessing genetic susceptibility to diabetic nephropathy. Nephrology. 2005;10 (Supplement s2):S17–S21. doi: 10.1111/j.1440-1797.2005.00451.x. [DOI] [PubMed] [Google Scholar]
- 50.Mueller PW, Rogus JJ, Cleary PA, Zhao Y, Smiles AM, Steffes MW, Bucksa J, Gibson TB, Cordovado SK, Krolewski AS, Nierras CR, Warram JH. Genetics of Kidneys in Diabetes (GoKinD) Study: A Genetics Collection Available for Identifying Genetic Susceptibility Factors for Diabetic Nephropathy in Type 1 Diabetes. J Am Soc Nephrol. 2006;17 (7):1782–1790. doi: 10.1681/ASN.2005080822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Freedman BI, Bostrom M, Daeihagh P, Bowden DW. Genetic Factors in Diabetic Nephropathy. Clin J Am Soc Nephrol. 2007;2 (6):1306–1316. doi: 10.2215/CJN.02560607. [DOI] [PubMed] [Google Scholar]
- 52.Conway BR, Maxwell AP. Genetics of Diabetic Nephropathy: Are There Clues to the Understanding of Common Kidney Diseases? Nephron Clin Pract. 2009;112 (4):c213–c221. doi: 10.1159/000224787. [DOI] [PubMed] [Google Scholar]
- 53.Pezzolesi MG, Poznik GD, Mychaleckyj JC, Paterson AD, Barati MT, Klein JB, Ng DPK, Placha G, Canani LH, Bochenski J, Waggott D, Merchant ML, Krolewski B, Mirea L, Wanic K, Katavetin P, Kure M, Wolkow P, Dunn JS, Smiles A, Walker WH, Boright AP, Bull SB, Doria A, Rogus JJ, Rich SS, Warram JH, Krolewski AS the DCCT/EDIC Research Group. Genome-Wide Association Scan for Diabetic Nephropathy Susceptibility Genes in Type 1 Diabetes. Diabetes. 2009;58 (6):1403–1410. doi: 10.2337/db08-1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pezzolesi MG, Skupien J, Mychaleckyj JC, Warram JH, Krolewski AS. Insights to the Genetics of Diabetic Nephropathy Through a Genome-Wide Association Study of the GoKinD Collection. Semin Nephrol. 2010;30 (2):126–140. doi: 10.1016/j.semnephrol.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schadt EE, Sachs A, Friend S. Embracing Complexity, Inching Closer to Reality. Sci STKE. 2005;2005(295):pe40. doi: 10.1126/stke.2952005pe40. [DOI] [PubMed] [Google Scholar]
- 56.Dobrin R, Zhu J, Molony C, Argman C, Parrish M, Carlson S, Allan M, Pomp D, Schadt E. Multi-tissue coexpression networks reveal unexpected subnetworks associated with disease. Genome Biol. 2009;10 (5):R55. doi: 10.1186/gb-2009-10-5-r55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sauer U, Heinemann M, Zamboni N. Getting Closer to the Whole Picture. Science. 2007;316 (5824):550–551. doi: 10.1126/science.1142502. [DOI] [PubMed] [Google Scholar]
- 58.Chuang HY, Hofree M, Ideker T. A Decade of Systems Biology. Annu Rev Cell Dev Biol. 2010;26 (1):721–744. doi: 10.1146/annurev-cellbio-100109-104122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Houle D, Govindaraju DR, Omholt S. Phenomics: the next challenge. Nat Rev Genet. 2010;11 (12):855–866. doi: 10.1038/nrg2897. [DOI] [PubMed] [Google Scholar]
- 60.Haring R, Wallaschofski H. Diving Through the “-Omics”: The Case for Deep Phenotyping and Systems Epidemiology. OMICS: J Integrative Biol. 2012:16. doi: 10.1089/omi.2011.0108. (Epub ahead of print: February 9, 2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R, Lee C, Montie JE, Shah RB, Pienta KJ, Rubin MA, Chinnaiyan AM. Recurrent Fusion of TMPRSS2 and ETS Transcription Factor Genes in Prostate Cancer. Science. 2005;310 (5748):644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 62.Hamburg MA, Collins FS. The Path to Personalized Medicine. New Engl J Med. 2010;363 (4):301–304. doi: 10.1056/NEJMp1006304. [DOI] [PubMed] [Google Scholar]
- 63.Allison M. Is personalized medicine finally arriving? Nat Biotechnol. 2008;26 (5):509–517. doi: 10.1038/nbt0508-509. [DOI] [PubMed] [Google Scholar]
- 64.http://www2.niddk.nih.gov/News/Calendar/HumanTissues2011.htm.
- 65.Seaquist ER, Goetz FC, Rich S, Barbosa J. Familial clustering of diabetic kidney disease: Evidence for genetic susceptibility to diabetic nephropathy. New Engl J Med. 1989;320 (18):1161–1165. doi: 10.1056/NEJM198905043201801. [DOI] [PubMed] [Google Scholar]
- 66.Borch-Johnsen K, Norgaard K, Hommel E, Mathiesen ER, Jensen JS, Deckert T, Parving HH. Is diabetic nephropathy an inherited complication? Kidney Int. 1992;41 (4):719–722. doi: 10.1038/ki.1992.112. [DOI] [PubMed] [Google Scholar]
- 67.Imperatore G, Knowler WC, Pettitt DJ, Kobes S, Bennett PH, Hanson RL. Segregation analysis of diabetic nephropathy in Pima Indians. Diabetes. 2000;49 (6):1049–1056. doi: 10.2337/diabetes.49.6.1049. [DOI] [PubMed] [Google Scholar]
- 68.Knowler WC, Coresh J, Elston RC, Freedman BI, Iyengar SK, Kimmel PL, Olson JM, Plaetke R, Sedor JR, Seldin MF. The Family Investigation of Nephropathy and Diabetes (FIND): Design and methods. J Diabetes Complications. 2005;19 (1):1–9. doi: 10.1016/j.jdiacomp.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 69.Pezzolesi MG, Poznik GD, Skupien J, Smiles AM, Mychaleckyj JC, Rich SS, Warram JH, Krolewski AS. An intergenic region on chromosome 13q33.3 is associated with the susceptibility to kidney disease in type 1 and 2 diabetes. Kidney Int. 2011;80 (1):105–111. doi: 10.1038/ki.2011.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mooyaart A, Valk E, van Es L, Bruijn J, de Heer E, Freedman B, Dekkers O, Baelde H. Genetic associations in diabetic nephropathy: a meta-analysis. Diabetologia. 2011;54 (3):544–553. doi: 10.1007/s00125-010-1996-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, Cho JH, Guttmacher AE, Kong A, Kruglyak L, Mardis E, Rotimi CN, Slatkin M, Valle D, Whittemore AS, Boehnke M, Clark AG, Eichler EE, Gibson G, Haines JL, Mackay TFC, McCarroll SA, Visscher PM. Finding the missing heritability of complex diseases. Nature. 2009;461 (7265):747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zuk O, Hechter E, Sunyaev SR, Lander ES. The mystery of missing heritability: Genetic interactions create phantom heritability. Proc Natl Acad Sci U S A. 2012;109 (4):1193–1198. doi: 10.1073/pnas.1119675109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, Ioannidis JPA, Hirschhorn JN. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet. 2008;9 (5):356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 74.Genovese G, Friedman DJ, Ross MD, Lecordier L, Uzureau P, Freedman BI, Bowden DW, Langefeld CD, Oleksyk TK, Uscinski Knob AL, Bernhardy AJ, Hicks PJ, Nelson GW, Vanhollebeke B, Winkler CA, Kopp JB, Pays E, Pollak MR. Association of Trypanolytic ApoL1 Variants with Kidney Disease in African Americans. Science. 2010;329 (5993):841–845. doi: 10.1126/science.1193032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lusis AJ. Life After GWAS. Atertio Thromb Vasc Biol. 2012;32 (2):169–169. doi: 10.1161/ATVBAHA.111.243543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pomerantz MM, Ahmadiyeh N, Jia L, Herman P, Verzi MP, Doddapaneni H, Beckwith CA, Chan JA, Hills A, Davis M, Yao K, Kehoe SM, Lenz HJ, Haiman CA, Yan C, Henderson BE, Frenkel B, Barretina J, Bass A, Tabernero J, Baselga J, Regan MM, Manak JR, Shivdasani R, Coetzee GA, Freedman ML. The 8q24 cancer risk variant rs6983267 shows long-range interaction with MYC in colorectal cancer. Nat Genet. 2009;41 (8):882–884. doi: 10.1038/ng.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pearson ER. Pharmacogenetics in diabetes. Curr Diab Rep. 2009;9 (2):172–181. doi: 10.1007/s11892-009-0028-3. [DOI] [PubMed] [Google Scholar]
- 78.McCarthy MI. Genomics, Type 2 Diabetes, and Obesity. New Engl J Med. 2010;363 (24):2339–2350. doi: 10.1056/NEJMra0906948. [DOI] [PubMed] [Google Scholar]
- 79.The Diabetes Control and Complications Trial Research Group. The Effect of Intensive Treatment of Diabetes on the Development and Progression of Long-Term Complications in Insulin-Dependent Diabetes Mellitus. New Engl J Med. 1993;329 (14):977–986. doi: 10.1056/NEJM199309303291401. [DOI] [PubMed] [Google Scholar]
- 80.UK Prospective Diabetes Study (UKPDS) Group. Intensive blood-glucose control with sulphonylureas or insulin compared with conventional treatment and risk of complications in patients with type 2 diabetes (UKPDS 33) The Lancet. 1998;352 (9131):837–853. [PubMed] [Google Scholar]
- 81.Engerman RL, Kern TS. Progression of incipient diabetic retinopathy during good glycemic control. Diabetes. 1987;36 (7):808–812. doi: 10.2337/diab.36.7.808. [DOI] [PubMed] [Google Scholar]
- 82.Ihnat MA, Thorpe JE, Ceriello A. Hypothesis: the metabolic memory’, the new challenge of diabetes. Diabet Med. 2007;24 (6):582–586. doi: 10.1111/j.1464-5491.2007.02138.x. [DOI] [PubMed] [Google Scholar]
- 83.El-Osta A, Brasacchio D, Yao D, Pocai A, Jones PL, Roeder RG, Cooper ME, Brownlee M. Transient high glucose causes persistent epigenetic changes and altered gene expression during subsequent normoglycemia. J Exp Med. 2008;205 (10):2409–2417. doi: 10.1084/jem.20081188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Feinberg AP. Epigenetics at the Epicenter of Modern Medicine. JAMA. 2008;299 (11):1345–1350. doi: 10.1001/jama.299.11.1345. [DOI] [PubMed] [Google Scholar]
- 85.Ceriello A, Ihnat MA, Thorpe JE. The “Metabolic Memory”: Is More Than Just Tight Glucose Control Necessary to Prevent Diabetic Complications? J Clin Endocrinol Metab. 2009;94 (2):410–415. doi: 10.1210/jc.2008-1824. [DOI] [PubMed] [Google Scholar]
- 86.Dwivedi RS, Herman JG, McCaffrey TA, Raj DSC. Beyond genetics: epigenetic code in chronic kidney disease. Kidney Int. 2011;79 (1):23–32. doi: 10.1038/ki.2010.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Reddy MA, Natarajan R. Epigenetics in Diabetic Kidney Disease. J Am Soc Nephrol. 2011;22 (12):2182–2185. doi: 10.1681/ASN.2011060629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Villeneuve LM, Natarajan R. Epigenetics of diabetic complications. Expert Rev Endocrinol Metab. 2010;5 (1):137–148. doi: 10.1586/eem.09.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pirola L, Balcerczyk A, Okabe J, El-Osta A. Epigenetic phenomena linked to diabetic complications. Nat Rev Endocrinol. 2010;6 (12):665–675. doi: 10.1038/nrendo.2010.188. [DOI] [PubMed] [Google Scholar]
- 90.Mohtat D, Susztak K. Fine Tuning Gene Expression: The Epigenome. Semin Nephrol. 2010;30 (5):468–476. doi: 10.1016/j.semnephrol.2010.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Reddy MA, Natarajan R. Epigenetic mechanisms in diabetic vascular complications. Cardiovasc Res. 2011;90 (3):421–429. doi: 10.1093/cvr/cvr024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Thomas MC, Groop PH, Tryggvason K. Towards understanding the inherited susceptibility for nephropathy in diabetes. Curr Opin Nephrol Hypertens. 2012;21 (2):195–202. doi: 10.1097/MNH.0b013e328350313e. [DOI] [PubMed] [Google Scholar]
- 93.Wetterstrand K. [Accessed: 3/20/2012];DNA Sequencing Costs: Data from the NHGRI Large-Scale Genome Sequencing Program. available at. http://wwwgenomegov/sequencingcosts.
- 94.Schena M, Shalon D, Davis RW, Brown PO. Quantitative Monitoring of Gene Expression Patterns with a Complementary DNA Microarray. Science. 1995;270 (5235):467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 95.Kretzler M, Cohen CD, Doran P, Henger A, Madden S, Gröne EF, Nelson PJ, Schlöndorff D, Gröne HJ. Repuncturing the Renal Biopsy: Strategies for Molecular Diagnosis in Nephrology. J Am Soc Nephrol. 2002;13 (7):1961–1972. doi: 10.1097/01.asn.0000020390.29418.70. [DOI] [PubMed] [Google Scholar]
- 96.Cohen CD, Frach K, Schlondorff D, Kretzler M. Quantitative gene expression analysis in renal biopsies: A novel protocol for a high-throughput multicenter application. Kidney Int. 2002;61 (1):133–140. doi: 10.1046/j.1523-1755.2002.00113.x. [DOI] [PubMed] [Google Scholar]
- 97.Schmid H, Henger A, Cohen CD, Frach K, Gröne HJ, Schlöndorff D, Kretzler M. Gene Expression Profiles of Podocyte-Associated Molecules as Diagnostic Markers in Acquired Proteinuric Diseases. J Am Soc Nephrol. 2003;14 (11):2958–2966. doi: 10.1097/01.asn.0000090745.85482.06. [DOI] [PubMed] [Google Scholar]
- 98.Kieran NE, Doran PP, Connolly SB, Greenan MC, Higgins DF, Leonard M, Godson C, Taylor CT, Henger A, Kretzler M, Burne MJ, Rabb H, Brady HR. Modification of the transcriptomic response to renal ischemia/reperfusion injury by lipoxin analog. Kidney Int. 2003;64 (2):480–492. doi: 10.1046/j.1523-1755.2003.00106.x. [DOI] [PubMed] [Google Scholar]
- 99.Henger A, Kretzler M, Doran P, Bonrouhi M, Schmid H, Kiss E, Cohen CD, Madden S, Porubsky S, Grone EF, Schlondorff D, Nelson PJ, Grone HJ. Gene expression fingerprints in human tubulointerstitial inflammation and fibrosis as prognostic markers of disease progression. Kidney Int. 2004;65 (3):904–917. doi: 10.1111/j.1523-1755.2004.00499.x. [DOI] [PubMed] [Google Scholar]
- 100.Baelde HJ, Eikmans M, Doran PP, Lappin DWP, de Heer E, Bruijn JA. Gene expression profiling in glomeruli from human kidneys with diabetic nephropathy. Am J Kidney Dis. 2004;43 (4):636–650. doi: 10.1053/j.ajkd.2003.12.028. [DOI] [PubMed] [Google Scholar]
- 101.Schmid H, Boucherot A, Yasuda Y, Henger A, Brunner B, Eichinger F, Nitsche A, Kiss E, Bleich M, Gröne HJ, Nelson PJ, Schlöndorff D, Cohen CD, Kretzler M. Modular Activation of Nuclear Factor-κB Transcriptional Programs in Human Diabetic Nephropathy. Diabetes. 2006;55 (11):2993–3003. doi: 10.2337/db06-0477. [DOI] [PubMed] [Google Scholar]
- 102.Lindenmeyer MT, Kretzler M, Boucherot A, Berra S, Yasuda Y, Henger A, Eichinger F, Gaiser S, Schmid H, Rastaldi MP, Schrier RW, Schlondorff D, Cohen CD. Interstitial Vascular Rarefaction and Reduced VEGF-A Expression in Human Diabetic Nephropathy. J Am Soc Nephrol. 2007;18 (6):1765–1776. doi: 10.1681/ASN.2006121304. [DOI] [PubMed] [Google Scholar]
- 103.Woroniecka KI, Park ASD, Mohtat D, Thomas DB, Pullman JM, Susztak K. Transcriptome Analysis of Human Diabetic Kidney Disease. Diabetes. 2011;60 (9):2354–2369. doi: 10.2337/db10-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lin YL, Peng SJ, Ferng SH, Tzen CY, Yang CS. Clinical indicators which necessitate renal biopsy in type 2 diabetes mellitus patients with renal disease. Int J Clin Pract. 2009;63 (8):1167–1176. doi: 10.1111/j.1742-1241.2008.01753.x. [DOI] [PubMed] [Google Scholar]
- 105.Mauer SM, Steffes MW, Ellis EN, Sutherland DE, Brown DM, Goetz FC. Structural-functional relationships in diabetic nephropathy. J Clin Invest. 1984;74 (4):1143–1155. doi: 10.1172/JCI111523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Pagtalunan ME, Miller PL, Jumping-Eagle S, Nelson RG, Myers BD, Rennke HG, Coplon NS, Sun L, Meyer TW. Podocyte loss and progressive glomerular injury in type II diabetes. J Clin Invest. 1997;99 (2):342–348. doi: 10.1172/JCI119163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Moutzouris DA, Herlitz L, Appel GB, Markowitz GS, Freudenthal B, Radhakrishnan J, D’Agati VD. Renal Biopsy in the Very Elderly. Clin J Am Soc Nephrol. 2009;4 (6):1073–1082. doi: 10.2215/CJN.00990209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zhang PP, Ge YC, Li SJ, Xie HL, Li LS, Liu ZH. Renal biopsy in type 2 diabetes: Timing of complications and evaluating of safety in Chinese patients. Nephrology. 2011;16 (1):100–105. doi: 10.1111/j.1440-1797.2010.01369.x. [DOI] [PubMed] [Google Scholar]
- 109.Cohen CD, Grone HJ, Grone EF, Nelson PJ, Schlondorff D, Kretzler M. Laser microdissection and gene expression analysis on formaldehyde-fixed archival tissue. Kidney Int. 2002;61 (1):125–132. doi: 10.1046/j.1523-1755.2002.00112.x. [DOI] [PubMed] [Google Scholar]
- 110.Hodgin JB, Borczuk AC, Nasr SH, Markowitz GS, Nair V, Martini S, Eichinger F, Vining C, Berthier CC, Kretzler M, D’Agati VD. A Molecular Profile of Focal Segmental Glomerulosclerosis from Formalin-Fixed, Paraffin-Embedded Tissue. Am J Pathol. 2010;177 (4):1674–1686. doi: 10.2353/ajpath.2010.090746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Shen-Orr SS, Tibshirani R, Khatri P, Bodian DL, Staedtler F, Perry NM, Hastie T, Sarwal MM, Davis MM, Butte AJ. Cell type-specific gene expression differences in complex tissues. Nat Methods. 2010;7 (4):287–289. doi: 10.1038/nmeth.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kaiser J. Biomarker Tests Need Closer Scrutiny, IOM Concludes. Science. 2012;335 (6076):1554–1554. doi: 10.1126/science.335.6076.1554. [DOI] [PubMed] [Google Scholar]
- 113.Consensus Report Institute of Medicine. [Accessed: 3/23/2012];Evolution of Translational OMICS: Lessons Learned and the Path Forward. 2012 http://wwwiomedu/Reports/2012/Evolution-of-Translational-Omics.aspx. [PubMed]
- 114.Gygi SP, Rochon Y, Franza BR, Aebersold R. Correlation between Protein and mRNA Abundance in Yeast. Mol Cell Biol. 1999;19 (3):1720–1730. doi: 10.1128/mcb.19.3.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Cravatt BF, Simon GM, Yates JR., III The biological impact of mass-spectrometry-based proteomics. Nature. 2007;450 (7172):991–1000. doi: 10.1038/nature06525. [DOI] [PubMed] [Google Scholar]
- 116.Vogel C, Marcotte EM. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet. 2012;13 (4):227–232. doi: 10.1038/nrg3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sébédio JL, Pujos-Guillot E, Ferrara M. Metabolomics in evaluation of glucose disorders. Curr Opin Clin Nutr Metab Care. 2009;12 (4):412–418. doi: 10.1097/MCO.0b013e32832c97c3. [DOI] [PubMed] [Google Scholar]
- 118.Wang JH, Byun J, Pennathur S. Analytical Approaches to Metabolomics and Applications to Systems Biology. Semin Nephrol. 2010;30 (5):500–511. doi: 10.1016/j.semnephrol.2010.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ben Ameur R, Molina L, Bolvin C, Kifagi C, Jarraya F, Ayadi H, Molina F, Granier C. Proteomic approaches for discovering biomarkers of diabetic nephropathy. Nephrol Dial Transplant. 2010;25 (9):2866–2875. doi: 10.1093/ndt/gfq258. [DOI] [PubMed] [Google Scholar]
- 120.Thongboonkerd V. Study of diabetic nephropathy in the proteomic era. Contrib Nephrol. 2011;170:172–183. doi: 10.1159/000325657. [DOI] [PubMed] [Google Scholar]
- 121.Mäkinen VP, Tynkkynen T, Soininen P, Peltola T, Kangas AJ, Forsblom C, Thorn LM, Kaski K, Laatikainen R, Ala-Korpela M, Groop PH. Metabolic Diversity of Progressive Kidney Disease in 325 Patients with Type 1 Diabetes (the FinnDiane Study) J Proteome Res. 2012;11 (3):1782–1790. doi: 10.1021/pr201036j. [DOI] [PubMed] [Google Scholar]
- 122.Thongboonkerd V, Malasit P. Renal and urinary proteomics: Current applications and challenges. Proteomics. 2005;5 (4):1033–1042. doi: 10.1002/pmic.200401012. [DOI] [PubMed] [Google Scholar]
- 123.Decramer S, de Peredo AG, Breuil B, Mischak H, Monsarrat B, Bascands JL, Schanstra JP. Urine in Clinical Proteomics. Mol Cell Proteomics. 2008;7 (10):1850–1862. doi: 10.1074/mcp.R800001-MCP200. [DOI] [PubMed] [Google Scholar]
- 124.Rossing K, Mischak H, Dakna M, Zurbig P, Novak J, Julian BA, Good DM, Coon JJ, Tarnow L, Rossing P on behalf of the PREDICTIONS Network. Urinary Proteomics in Diabetes and CKD. J Am Soc Nephrol. 2008;19 (7):1283–1290. doi: 10.1681/ASN.2007091025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Merchant ML, Perkins BA, Boratyn GM, Ficociello LH, Wilkey DW, Barati MT, Bertram CC, Page GP, Rovin BH, Warram JH, Krolewski AS, Klein JB. Urinary Peptidome May Predict Renal Function Decline in Type 1 Diabetes and Microalbuminuria. J Am Soc Nephrol. 2009;20 (9):2065–2074. doi: 10.1681/ASN.2008121233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Otu HH, Can H, Spentzos D, Nelson RG, Hanson RL, Looker HC, Knowler WC, Monroy M, Libermann TA, Karumanchi SA, Thadhani R. Prediction of Diabetic Nephropathy Using Urine Proteomic Profiling 10 Years Prior to Development of Nephropathy. Diabetes Care. 2007;30 (3):638–643. doi: 10.2337/dc06-1656. [DOI] [PubMed] [Google Scholar]
- 127.Overgaard A, Hansen H, Lajer M, Pedersen L, Tarnow L, Rossing P, McGuire J, Pociot F. Plasma proteome analysis of patients with type 1 diabetes with diabetic nephropathy. Proteome Sci. 2010;8 (1):4. doi: 10.1186/1477-5956-8-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Overgaard AJ, Thingholm TE, Larsen MR, Tarnow L, Rossing P, McGuire JN, Pociot F. Quantitative iTRAQ-Based Proteomic Identification of Candidate Biomarkers for Diabetic Nephropathy in Plasma of Type 1 Diabetic Patients. Clin Proteomics. 2010;6 (4):105–114. doi: 10.1007/s12014-010-9053-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Coon JJ, Zürbig P, Dakna M, Dominiczak AF, Decramer S, Fliser D, Frommberger M, Golovko I, Good DM, Herget-Rosenthal S, Jankowski J, Julian BA, Kellmann M, Kolch W, Massy Z, Novak J, Rossing K, Schanstra JP, Schiffer E, Theodorescu D, Vanholder R, Weissinger EM, Mischak H, Schmitt-Kopplin P. CE-MS analysis of the human urinary proteome for biomarker discovery and disease diagnostics. Proteomics Clin Appl. 2008;2 (7–8):964–973. doi: 10.1002/prca.200800024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Jantos-Siwy J, Schiffer E, Brand K, Schumann G, Rossing K, Delles C, Mischak H, Metzger J. Quantitative Urinary Proteome Analysis for Biomarker Evaluation in Chronic Kidney Disease. J Proteome Res. 2009;8 (1):268–281. doi: 10.1021/pr800401m. [DOI] [PubMed] [Google Scholar]
- 131.Maahs DM, Siwy J, Argilés À, Cerna M, Delles C, Dominiczak AF, Gayrard N, Iphöfer A, Jänsch L, Jerums G, Medek K, Mischak H, Navis GJ, Roob JM, Rossing K, Rossing P, Rychlík I, Schiffer E, Schmieder RE, Wascher TC, Winklhofer-Roob BM, Zimmerli LU, Zürbig P, Snell-Bergeon JK. Urinary Collagen Fragments Are Significantly Altered in Diabetes: A Link to Pathophysiology. PLoS ONE. 2010;5 (9):e13051. doi: 10.1371/journal.pone.0013051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Alkhalaf A, Zürbig P, Bakker SJL, Bilo HJG, Cerna M, Fischer C, Fuchs S, Janssen B, Medek K, Mischak H, Roob JM, Rossing K, Rossing P, Rychlík I, Sourij H, Tiran B, Winklhofer-Roob BM, Navis GJ for the PREDICTIONS Group. Multicentric Validation of Proteomic Biomarkers in Urine Specific for Diabetic Nephropathy. PLoS ONE. 2010;5 (10):e13421. doi: 10.1371/journal.pone.0013421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Good DM, Zurbig P, Argiles A, Bauer HW, Behrens G, Coon JJ, Dakna M, Decramer S, Delles C, Dominiczak AF, Ehrich JHH, Eitner F, Fliser D, Frommberger M, Ganser A, Girolami MA, Golovko I, Gwinner W, Haubitz M, Herget-Rosenthal S, Jankowski J, Jahn H, Jerums G, Julian BA, Kellmann M, Kliem V, Kolch W, Krolewski AS, Luppi M, Massy Z, Melter M, Neususs C, Novak J, Peter K, Rossing K, Rupprecht H, Schanstra JP, Schiffer E, Stolzenburg JU, Tarnow L, Theodorescu D, Thongboonkerd V, Vanholder R, Weissinger EM, Mischak H, Schmitt-Kopplin P. Naturally occurring human urinary peptides for use in diagnosis of chronic kidney disease. Mol Cell Proteomics. 2010;9 (11):2424–2437. doi: 10.1074/mcp.M110.001917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Jain S, Rajput A, Kumar Y, Uppuluri N, Arvind AS, Tatu U. Proteomic analysis of urinary protein markers for accurate prediction of diabetic kidney disorder. J Assoc Physicians India. 2005 Jun;53 :513–520. [PubMed] [Google Scholar]
- 135.Fisher WG, Lucas JE, Mehdi UF, Qunibi DW, Garner HR, Rosenblatt KP, Toto RD. A method for isolation and identification of urinary biomarkers in patients with diabetic nephropathy. Proteomics Clin Appl. 2011;5 (11–12):603–612. doi: 10.1002/prca.201000156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lowe JB. Glycosylation, Immunity, and Autoimmunity. Cell. 2001;104 (6):809–812. doi: 10.1016/s0092-8674(01)00277-x. [DOI] [PubMed] [Google Scholar]
- 137.Yang N, Feng S, Shedden K, Xie X, Liu Y, Rosser CJ, Lubman DM, Goodison S. Urinary Glycoprotein Biomarker Discovery for Bladder Cancer Detection Using LC/MS-MS and Label-Free Quantification. Clin Cancer Res. 2011;17 (10):3349–3359. doi: 10.1158/1078-0432.CCR-10-3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ahn JM, Kim BG, Yu MH, Lee IK, Cho JY. Identification of diabetic nephropathy-selective proteins in human plasma by multi-lectin affinity chromatography and LC-MS/MS. Proteomics Clin Appl. 2010;4 (6–7):644–653. doi: 10.1002/prca.200900196. [DOI] [PubMed] [Google Scholar]
- 139.Vivekanandan-Giri A, Slocum JL, Buller CL, Basrur V, Ju W, Pop-Busui R, Lubman DM, Kretzler M, Pennathur S. Urine Glycoprotein Profile Reveals Novel Markers for Chronic Kidney Disease. Int J Proteomics. 2011;2011 (Article ID 214715):18. doi: 10.1155/2011/214715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Pisitkun T, Shen RF, Knepper MA. Identification and proteomic profiling of exosomes in human urine. Proc Natl Acad Sci U S A. 2004;101 (36):13368–13373. doi: 10.1073/pnas.0403453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Gonzales PA, Pisitkun T, Hoffert JD, Tchapyjnikov D, Star RA, Kleta R, Wang NS, Knepper MA. Large-Scale Proteomics and Phosphoproteomics of Urinary Exosomes. J Am Soc Nephrol. 2009;20 (2):363–379. doi: 10.1681/ASN.2008040406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Konvalinka A, Scholey JW, Diamandis EP. Searching for New Biomarkers of Renal Diseases through Proteomics. Clin Chem. 2012;58 (2):353–365. doi: 10.1373/clinchem.2011.165969. [DOI] [PubMed] [Google Scholar]
- 143.Mischak H, Apweiler R, Banks RE, Conaway M, Coon J, Dominiczak A, Ehrich JHH, Fliser D, Girolami M, Hermjakob H, Hochstrasser D, Jankowski J, Julian BA, Kolch W, Massy ZA, Neusuess C, Novak J, Peter K, Rossing K, Schanstra J, Semmes OJ, Theodorescu D, Thongboonkerd V, Weissinger EM, Van Eyk JE, Yamamoto T. Clinical proteomics: A need to define the field and to begin to set adequate standards. Proteomics Clin Appl. 2007;1 (2):148–156. doi: 10.1002/prca.200600771. [DOI] [PubMed] [Google Scholar]
- 144.Good DM, Thongboonkerd V, Novak J, Bascands JL, Schanstra JP, Coon JJ, Dominiczak A, Mischak H. Body Fluid Proteomics for Biomarker Discovery: Lessons from the Past Hold the Key to Success in the Future. J Proteome Res. 2007;6 (12):4549–4555. doi: 10.1021/pr070529w. [DOI] [PubMed] [Google Scholar]
- 145.Kinsinger CR, Apffel J, Baker M, Bian X, Borchers CH, Bradshaw R, Brusniak MY, Chan DW, Deutsch EW, Domon B, Gorman J, Grimm R, Hancock W, Hermjakob H, Horn D, Hunter C, Kolar P, Kraus HJ, Langen H, Linding R, Moritz RL, Omenn GS, Orlando R, Pandey A, Ping P, Rahbar A, Rivers R, Seymour SL, Simpson RJ, Slotta D, Smith RD, Stein SE, Tabb DL, Tagle D, Yates JR, Rodriguez H. Recommendations for mass spectrometry data quality metrics for open access data (corollary to the Amsterdam principles) Proteomics Clin Appl. 2011;5 (11–12):580–589. doi: 10.1002/prca.201100097. [DOI] [PubMed] [Google Scholar]
- 146.Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, Causton HC, Gaasterland T, Glenisson P, Holstege FCP, Kim IF, Markowitz V, Matese JC, Parkinson H, Robinson A, Sarkans U, Schulze-Kremer S, Stewart J, Taylor R, Vilo J, Vingron M. Minimum information about a microarray experiment (MIAME)—toward standards for microarray data. Nat Genet. 2001;29 (4):365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- 147.Sreekumar A, Poisson LM, Rajendiran TM, Khan AP, Cao Q, Yu J, Laxman B, Mehra R, Lonigro RJ, Li Y, Nyati MK, Ahsan A, Kalyana-Sundaram S, Han B, Cao X, Byun J, Omenn GS, Ghosh D, Pennathur S, Alexander DC, Berger A, Shuster JR, Wei JT, Varambally S, Beecher C, Chinnaiyan AM. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature. 2009;457 (7231):910–914. doi: 10.1038/nature07762. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 148.Kim K, Aronov P, Zakharkin SO, Anderson D, Perroud B, Thompson IM, Weiss RH. Urine Metabolomics Analysis for Kidney Cancer Detection and Biomarker Discovery. Mol Cell Proteomics. 2009;8 (3):558–570. doi: 10.1074/mcp.M800165-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Newgard CB, An J, Bain JR, Muehlbauer MJ, Stevens RD, Lien LF, Haqq AM, Shah SH, Arlotto M, Slentz CA, Rochon J, Gallup D, Ilkayeva O, Wenner BR, Yancy WS, Jr, Eisenson H, Musante G, Surwit RS, Millington DS, Butler MD, Svetkey LP. A Branched-Chain Amino Acid-Related Metabolic Signature that Differentiates Obese and Lean Humans and Contributes to Insulin Resistance. Cell Metabolism. 2009;9 (4):311–326. doi: 10.1016/j.cmet.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Suhre K, Meisinger C, Döring A, Altmaier E, Belcredi P, Gieger C, Chang D, Milburn MV, Gall WE, Weinberger KM, Mewes HW, Hrabé de Angelis M, Wichmann HE, Kronenberg F, Adamski J, Illig T. Metabolic Footprint of Diabetes: A Multiplatform Metabolomics Study in an Epidemiological Setting. PLoS ONE. 2010;5 (11):e13953. doi: 10.1371/journal.pone.0013953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Wang TJ, Larson MG, Vasan RS, Cheng S, Rhee EP, McCabe E, Lewis GD, Fox CS, Jacques PF, Fernandez C, O’Donnell CJ, Carr SA, Mootha VK, Florez JC, Souza A, Melander O, Clish CB, Gerszten RE. Metabolite profiles and the risk of developing diabetes. Nat Med. 2011;17 (4):448–453. doi: 10.1038/nm.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Rhee EP, Cheng S, Larson MG, Walford GA, Lewis GD, McCabe E, Yang E, Farrell L, Fox CS, O’Donnell CJ, Carr SA, Vasan RS, Florez JC, Clish CB, Wang TJ, Gerszten RE. Lipid profiling identifies a triacylglycerol signature of insulin resistance and improves diabetes prediction in humans. J Clin Invest. 2011;121 (4):1402–1411. doi: 10.1172/JCI44442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Ganti S, Weiss RH. Urine metabolomics for kidney cancer detection and biomarker discovery. Urol Oncol. 2011;29 (5):551–557. doi: 10.1016/j.urolonc.2011.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Rhee EP, Gerszten RE. Metabolomics and Cardiovascular Biomarker Discovery. Clin Chem. 2012;58 (1):139–147. doi: 10.1373/clinchem.2011.169573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Weiss RH, Kim K. Metabolomics in the study of kidney diseases. Nat Rev Nephrol. 2012;8 (1):22–33. doi: 10.1038/nrneph.2011.152. [DOI] [PubMed] [Google Scholar]
- 156.Zhang H, Saha J, Byun J, Schin M, Lorenz M, Kennedy RT, Kretzler M, Feldman EL, Pennathur S, Brosius FC. Rosiglitazone reduces renal and plasma markers of oxidative injury and reverses urinary metabolite abnormalities in the amelioration of diabetic nephropathy. Am J Physiol Renal Physiol. 2008;295 (4):F1071–F1081. doi: 10.1152/ajprenal.90208.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Smith CA, Maille GO, Want EJ, Qin C, Trauger SA, Brandon TR, Custodio DE, Abagyan R, Siuzdak G. METLIN: A Metabolite Mass Spectral Database. Ther Drug Monit. 2005;27 (6):747–751. doi: 10.1097/01.ftd.0000179845.53213.39. [DOI] [PubMed] [Google Scholar]
- 158.Wishart DS, Tzur D, Knox C, Eisner R, Guo AC, Young N, Cheng D, Jewell K, Arndt D, Sawhney S, Fung C, Nikolai L, Lewis M, Coutouly MA, Forsythe I, Tang P, Shrivastava S, Jeroncic K, Stothard P, Amegbey G, Block D, Hau DD, Wagner J, Miniaci J, Clements M, Gebremedhin M, Guo N, Zhang Y, Duggan GE, MacInnis GD, Weljie AM, Dowlatabadi R, Bamforth F, Clive D, Greiner R, Li L, Marrie T, Sykes BD, Vogel HJ, Querengesser L. HMDB: the Human Metabolome Database. NAR. 2007;35 (suppl 1):D521–D526. doi: 10.1093/nar/gkl923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Barreto FC, Barreto DV, Liabeuf S, Meert N, Glorieux G, Temmar M, Choukroun G, Vanholder R, Massy ZA on behalf of the European Uremic Toxin Work G. Serum Indoxyl Sulfate Is Associated with Vascular Disease and Mortality in Chronic Kidney Disease Patients. Clin J Am Soc Nephrol. 2009;4 (10):1551–1558. doi: 10.2215/CJN.03980609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Zhang J, Yan L, Chen W, Lin L, Song X, Yan X, Hang W, Huang B. Metabonomics research of diabetic nephropathy and type 2 diabetes mellitus based on UPLC–oaTOF-MS system. Anal Chim Acta. 2009;650 (1):16–22. doi: 10.1016/j.aca.2009.02.027. [DOI] [PubMed] [Google Scholar]
- 161.van der Kloet F, Tempels F, Ismail N, van der Heijden R, Kasper P, Rojas-Cherto M, van Doorn R, Spijksma G, Koek M, van der Greef J, Mäkinen V, Forsblom C, Holthöfer H, Groop P, Reijmers T, Hankemeier T. Discovery of early-stage biomarkers for diabetic kidney disease using ms-based metabolomics (FinnDiane study) Metabolomics. 2012;8 (1):109–119. doi: 10.1007/s11306-011-0291-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Xia JF, Liang QL, Liang XP, Wang YM, Hu P, Li P, Luo GA. Ultraviolet and tandem mass spectrometry for simultaneous quantification of 21 pivotal metabolites in plasma from patients with diabetic nephropathy. J Chromatogr B Analyt Technol Biomed Life Sci. 2009;877 (20–21):1930–1936. doi: 10.1016/j.jchromb.2009.05.047. [DOI] [PubMed] [Google Scholar]
- 163.Sieberts S, Schadt EE. Moving toward a system genetics view of disease. Mamm Genome. 2007;18 (6):389–401. doi: 10.1007/s00335-007-9040-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Emilsson V, Thorleifsson G, Zhang B, Leonardson AS, Zink F, Zhu J, Carlson S, Helgason A, Walters GB, Gunnarsdottir S, Mouy M, Steinthorsdottir V, Eiriksdottir GH, Bjornsdottir G, Reynisdottir I, Gudbjartsson D, Helgadottir A, Jonasdottir A, Jonasdottir A, Styrkarsdottir U, Gretarsdottir S, Magnusson KP, Stefansson H, Fossdal R, Kristjansson K, Gislason HG, Stefansson T, Leifsson BG, Thorsteinsdottir U, Lamb JR, Gulcher JR, Reitman ML, Kong A, Schadt EE, Stefansson K. Genetics of gene expression and its effect on disease. Nature. 2008;452 (7186):423–428. doi: 10.1038/nature06758. [DOI] [PubMed] [Google Scholar]
- 165.Ioannidis JPA, Thomas G, Daly MJ. Validating, augmenting and refining genome-wide association signals. Nat Rev Genet. 2009;10 (5):318–329. doi: 10.1038/nrg2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Keurentjes JJB, Fu J, de Vos CHR, Lommen A, Hall RD, Bino RJ, van der Plas LHW, Jansen RC, Vreugdenhil D, Koornneef M. The genetics of plant metabolism. Nat Genet. 2006;38 (7):842–849. doi: 10.1038/ng1815. [DOI] [PubMed] [Google Scholar]
- 167.Gieger C, Geistlinger L, Altmaier E, Hrabé de Angelis M, Kronenberg F, Meitinger T, Mewes HW, Wichmann HE, Weinberger KM, Adamski J, Illig T, Suhre K. Genetics Meets Metabolomics: A Genome-Wide Association Study of Metabolite Profiles in Human Serum. PLoS Genet. 2008;4 (11):e1000282. doi: 10.1371/journal.pgen.1000282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Ferrara CT, Wang P, Neto EC, Stevens RD, Bain JR, Wenner BR, Ilkayeva OR, Keller MP, Blasiole DA, Kendziorski C, Yandell BS, Newgard CB, Attie AD. Genetic networks of liver metabolism revealed by integration of metabolic and transcriptional profiling. PLoS Genet. 2008;4 (3):e1000034. doi: 10.1371/journal.pgen.1000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Shah SH, Hauser ER, Bain JR, Muehlbauer MJ, Haynes C, Stevens RD, Wenner BR, Dowdy ZE, Granger CB, Ginsburg GS, Newgard CB, Kraus WE. High heritability of metabolomic profiles in families burdened with premature cardiovascular disease. Mol Syst Biol. 2009;5:Article number: 258. doi: 10.1038/msb.2009.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Suhre K, Wallaschofski H, Raffler J, Friedrich N, Haring R, Michael K, Wasner C, Krebs A, Kronenberg F, Chang D, Meisinger C, Wichmann HE, Hoffmann W, Volzke H, Volker U, Teumer A, Biffar R, Kocher T, Felix SB, Illig T, Kroemer HK, Gieger C, Romisch-Margl W, Nauck M. A genome-wide association study of metabolic traits in human urine. Nat Genet. 2011;43 (6):565–569. doi: 10.1038/ng.837. [DOI] [PubMed] [Google Scholar]
- 171.Daly AK. Drug-induced liver injury: past, present and future. Pharmacogenomics. 2010;11 (5):607–611. doi: 10.2217/pgs.10.24. [DOI] [PubMed] [Google Scholar]
- 172.Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, Pirruccello JP, Ripatti S, Chasman DI, Willer CJ, Johansen CT, Fouchier SW, Isaacs A, Peloso GM, Barbalic M, Ricketts SL, Bis JC, Aulchenko YS, Thorleifsson G, Feitosa MF, Chambers J, Orho-Melander M, Melander O, Johnson T, Li X, Guo X, Li M, Shin Cho Y, Jin Go M, Jin Kim Y, Lee JY, Park T, Kim K, Sim X, Twee-Hee Ong R, Croteau-Chonka DC, Lange LA, Smith JD, Song K, Hua Zhao J, Yuan X, Luan Ja, Lamina C, Ziegler A, Zhang W, Zee RYL, Wright AF, Witteman JCM, Wilson JF, Willemsen G, Wichmann HE, Whitfield JB, Waterworth DM, Wareham NJ, Waeber G, Vollenweider P, Voight BF, Vitart V, Uitterlinden AG, Uda M, Tuomilehto J, Thompson JR, Tanaka T, Surakka I, Stringham HM, Spector TD, Soranzo N, Smit JH, Sinisalo J, Silander K, Sijbrands EJG, Scuteri A, Scott J, Schlessinger D, Sanna S, Salomaa V, Saharinen J, Sabatti C, Ruokonen A, Rudan I, Rose LM, Roberts R, Rieder M, Psaty BM, Pramstaller PP, Pichler I, Perola M, Penninx BWJH, Pedersen NL, Pattaro C, Parker AN, Pare G, Oostra BA, O’Donnell CJ, Nieminen MS, Nickerson DA, Montgomery GW, Meitinger T, McPherson R, McCarthy MI, McArdle W, Masson D, Martin NG, Marroni F, Mangino M, Magnusson PKE, Lucas G, Luben R, Loos RJF, Lokki ML, Lettre G, Langenberg C, Launer LJ, Lakatta EG, Laaksonen R, Kyvik KO, Kronenberg F, Konig IR, Khaw KT, Kaprio J, Kaplan LM, Johansson A, Jarvelin MR, Cecile JW, Janssens A, Ingelsson E, Igl W, Kees Hovingh G, Hottenga JJ, Hofman A, Hicks AA, Hengstenberg C, Heid IM, Hayward C, Havulinna AS, Hastie ND, Harris TB, Haritunians T, Hall AS, Gyllensten U, Guiducci C, Groop LC, Gonzalez E, Gieger C, Freimer NB, Ferrucci L, Erdmann J, Elliott P, Ejebe KG, Doring A, Dominiczak AF, Demissie S, Deloukas P, de Geus EJC, de Faire U, Crawford G, Collins FS, Chen Y-dI, Caulfield MJ, Campbell H, Burtt NP, Bonnycastle LL, Boomsma DI, Boekholdt SM, Bergman RN, Barroso I, Bandinelli S, Ballantyne CM, Assimes TL, Quertermous T, Altshuler D, Seielstad M, Wong TY, Tai ES, Feranil AB, Kuzawa CW, Adair LS, Taylor HA, Jr, Borecki IB, Gabriel SB, Wilson JG, Holm H, Thorsteinsdottir U, Gudnason V, Krauss RM, Mohlke KL, Ordovas JM, Munroe PB, Kooner JS, Tall AR, Hegele RA, Kastelein JJP, Schadt EE, Rotter JI, Boerwinkle E, Strachan DP, Mooser V, Stefansson K, Reilly MP, Samani NJ, Schunkert H, Cupples LA, Sandhu MS, Ridker PM, Rader DJ, van Duijn CM, Peltonen L, Abecasis GR, Boehnke M, Kathiresan S. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466 (7307):707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Köttgen A, Pattaro C, Boger CA, Fuchsberger C, Olden M, Glazer NL, Parsa A, Gao X, Yang Q, Smith AV, O’Connell JR, Li M, Schmidt H, Tanaka T, Isaacs A, Ketkar S, Hwang SJ, Johnson AD, Dehghan A, Teumer A, Pare G, Atkinson EJ, Zeller T, Lohman K, Cornelis MC, Probst-Hensch NM, Kronenberg F, Tonjes A, Hayward C, Aspelund T, Eiriksdottir G, Launer LJ, Harris TB, Rampersaud E, Mitchell BD, Arking DE, Boerwinkle E, Struchalin M, Cavalieri M, Singleton A, Giallauria F, Metter J, de Boer IH, Haritunians T, Lumley T, Siscovick D, Psaty BM, Zillikens MC, Oostra BA, Feitosa M, Province M, de Andrade M, Turner ST, Schillert A, Ziegler A, Wild PS, Schnabel RB, Wilde S, Munzel TF, Leak TS, Illig T, Klopp N, Meisinger C, Wichmann HE, Koenig W, Zgaga L, Zemunik T, Kolcic I, Minelli C, Hu FB, Johansson A, Igl W, Zaboli G, Wild SH, Wright AF, Campbell H, Ellinghaus D, Schreiber S, Aulchenko YS, Felix JF, Rivadeneira F, Uitterlinden AG, Hofman A, Imboden M, Nitsch D, Brandstatter A, Kollerits B, Kedenko L, Magi R, Stumvoll M, Kovacs P, Boban M, Campbell S, Endlich K, Volzke H, Kroemer HK, Nauck M, Volker U, Polasek O, Vitart V, Badola S, Parker AN, Ridker PM, Kardia SLR, Blankenberg S, Liu Y, Curhan GC, Franke A, Rochat T, Paulweber B, Prokopenko I, Wang W, Gudnason V, Shuldiner AR, Coresh J, Schmidt R, Ferrucci L, Shlipak MG, van Duijn CM, Borecki I, Kramer BK, Rudan I, Gyllensten U, Wilson JF, Witteman JC, Pramstaller PP, Rettig R, Hastie N, Chasman DI, Kao WH, Heid IM, Fox CS. New loci associated with kidney function and chronic kidney disease. Nat Genet. 2010;42 (5):376–384. doi: 10.1038/ng.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Kettunen J, Tukiainen T, Sarin AP, Ortega-Alonso A, Tikkanen E, Lyytikainen LP, Kangas AJ, Soininen P, Wurtz P, Silander K, Dick DM, Rose RJ, Savolainen MJ, Viikari J, Kahonen M, Lehtimaki T, Pietilainen KH, Inouye M, McCarthy MI, Jula A, Eriksson J, Raitakari OT, Salomaa V, Kaprio J, Jarvelin MR, Peltonen L, Perola M, Freimer NB, Ala-Korpela M, Palotie A, Ripatti S. Genome-wide association study identifies multiple loci influencing human serum metabolite levels. Nat Genet. 2012;44 (3):269–276. doi: 10.1038/ng.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Suhre K, Shin SY, Petersen AK, Mohney RP, Meredith D, Wagele B, Altmaier E, Gram Cardio, Deloukas P, Erdmann J, Grundberg E, Hammond CJ, de Angelis MH, Kastenmuller G, Kottgen A, Kronenberg F, Mangino M, Meisinger C, Meitinger T, Mewes H-W, Milburn MV, Prehn C, Raffler J, Ried JS, Romisch-Margl W, Samani NJ, Small KS, Erich Wichmann H, Zhai G, Illig T, Spector TD, Adamski J, Soranzo N, Gieger C. Human metabolic individuality in biomedical and pharmaceutical research. Nature. 2011;477 (7362):54–60. doi: 10.1038/nature10354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Nicholson G, Rantalainen M, Li JV, Maher AD, Malmodin D, Ahmadi KR, Faber JH, Barrett A, Min JL, Rayner NW, Toft H, Krestyaninova M, Viksna J, Neogi SG, Dumas ME, Sarkans U, Donnelly P, Illig T, Adamski J, Suhre K, Allen M, Zondervan KT, Spector TD, Nicholson JK, Lindon JC, Baunsgaard D, Holmes E, McCarthy MI, Holmes CC, The Mol PC. A Genome-Wide Metabolic QTL Analysis in Europeans Implicates Two Loci Shaped by Recent Positive Selection. PLoS Genet. 2011;7 (9):e1002270. doi: 10.1371/journal.pgen.1002270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Barabasi AL, Oltvai ZN. Network biology: understanding the cell’s functional organization. Nat Rev Genet. 2004;5 (2):101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 178.Barabasi AL, Gulbahce N, Loscalzo J. Network medicine: a network-based approach to human disease. Nat Rev Genet. 2011;12 (1):56–68. doi: 10.1038/nrg2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Emmert-Streib F, Glazko GV. Network biology: a direct approach to study biological function. Wiley Interdiscip Rev Syst Biol Med. 2011;3 (4):379–391. doi: 10.1002/wsbm.134. [DOI] [PubMed] [Google Scholar]
- 180.Keller BJ, Martini S, Sedor JR, Kretzler M. A systems view of genetics in chronic kidney disease. Kidney Int. 2012;81 (1):14–21. doi: 10.1038/ki.2011.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.He JC, Chuang PY, Ma’Ayan A, Iyengar R. Systems biology of kidney diseases. Kidney Int. 2012;81 (1):22–39. doi: 10.1038/ki.2011.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Schadt E, Zhang B, Zhu J. Advances in systems biology are enhancing our understanding of disease and moving us closer to novel disease treatments. Gen. 2009;136 (2):259–269. doi: 10.1007/s10709-009-9359-x. [DOI] [PubMed] [Google Scholar]
- 183.Ostrowski J, Wyrwicz LS. Integrating genomics, proteomics and bioinformatics in translational studies of molecular medicine. Expert Rev Mol Diagn. 2009;9 (6):623–630. doi: 10.1586/erm.09.41. [DOI] [PubMed] [Google Scholar]
- 184.Ideker T, Krogan NJ. Differential network biology. Mol Syst Biol. 2012;8:Article number: 565. doi: 10.1038/msb.2011.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Schadt EE, Molony C, Chudin E, Hao K, Yang X, Lum PY, Kasarskis A, Zhang B, Wang S, Suver C, Zhu J, Millstein J, Sieberts S, Lamb J, GuhaThakurta D, Derry J, Storey JD, Avila-Campillo I, Kruger MJ, Johnson JM, Rohl CA, van Nas A, Mehrabian M, Drake TA, Lusis AJ, Smith RC, Guengerich FP, Strom SC, Schuetz E, Rushmore TH, Ulrich R. Mapping the Genetic Architecture of Gene Expression in Human Liver. PLoS Biol. 2008;6 (5):e107. doi: 10.1371/journal.pbio.0060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Mirel B, Eichinger F, Keller BJ, Kretzler M. A cognitive task analysis of a visual analytic workflow: Exploring molecular interaction networks in systems biology. J Biomed Discov Collab. 2011;6:1–33. doi: 10.5210/disco.v6i0.3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Bergholdt R, Brorsson C, Palleja A, Berchtold LA, Fløyel T, Bang-Berthelsen CH, Frederiksen KS, Jensen LJ, Størling J, Pociot F. Identification of Novel Type 1 Diabetes Candidate Genes by Integrating Genome-Wide Association Data, Protein-Protein Interactions, and Human Pancreatic Islet Gene Expression. Diabetes. 2012;61 (4):954–962. doi: 10.2337/db11-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Friend SH, Ideker T. POINT: Are we prepared for the future doctor visit? Nat Biotech. 2011;29 (3):215–218. doi: 10.1038/nbt.1794. [DOI] [PubMed] [Google Scholar]
- 189.Kohane IS, Margulies DM. COUNTERPOINT: Do not opine before it’s time. Nat Biotech. 2011;29 (3):218–219. doi: 10.1038/nbt.1797. [DOI] [PubMed] [Google Scholar]
- 190.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003;13 (11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Bhavnani S, Ganesan A, Hall T, Maslowski E, Eichinger F, Martini S, Saxman P, Bellala G, Kretzler M. Discovering hidden relationships between renal diseases and regulated genes through 3D network visualizations. BMC Res Notes. 2010;3 (1):296. doi: 10.1186/1756-0500-3-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Chen R, Mias George I, Li-Pook-Than J, Jiang L, Lam Hugo YK, Chen R, Miriami E, Karczewski Konrad J, Hariharan M, Dewey Frederick E, Cheng Y, Clark Michael J, Im H, Habegger L, Balasubramanian S, O’Huallachain M, Dudley Joel T, Hillenmeyer S, Haraksingh R, Sharon D, Euskirchen G, Lacroute P, Bettinger K, Boyle Alan P, Kasowski M, Grubert F, Seki S, Garcia M, Whirl-Carrillo M, Gallardo M, Blasco Maria A, Greenberg Peter L, Snyder P, Klein Teri E, Altman Russ B, Butte AJ, Ashley Euan A, Gerstein M, Nadeau Kari C, Tang H, Snyder M. Personal Omics Profiling Reveals Dynamic Molecular and Medical Phenotypes. Cell. 2012;148 (6):1293–1307. doi: 10.1016/j.cell.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Atkinson AJ, Colburn WA, DeGruttola VG, DeMets DL, Downing GJ, Hoth DF, Oates JA, Peck CC, Schooley RT, Spilker BA, Woodcock J, Zeger SL Biomarkers Definitions Working Group. Biomarkers and surrogate endpoints: Preferred definitions and conceptual framework. Clin Pharmacol Ther. 2001;69 (3):89–95. doi: 10.1067/mcp.2001.113989. [DOI] [PubMed] [Google Scholar]
- 194.Mischak H, Allmaier G, Apweiler R, Attwood T, Baumann M, Benigni A, Bennett SE, Bischoff R, Bongcam-Rudloff E, Capasso G, Coon JJ, D’Haese P, Dominiczak AF, Dakna M, Dihazi H, Ehrich JH, Fernandez-Llama P, Fliser D, Frokiaer J, Garin J, Girolami M, Hancock WS, Haubitz M, Hochstrasser D, Holman RR, Ioannidis JPA, Jankowski J, Julian BA, Klein JB, Kolch W, Luider T, Massy Z, Mattes WB, Molina F, Monsarrat B, Novak J, Peter K, Rossing P, Sánchez-Carbayo M, Schanstra JP, Semmes OJ, Spasovski G, Theodorescu D, Thongboonkerd V, Vanholder R, Veenstra TD, Weissinger E, Yamamoto T, Vlahou A. Recommendations for Biomarker Identification and Qualification in Clinical Proteomics. Sci Transl Med. 2010;2(46):46ps42. doi: 10.1126/scitranslmed.3001249. [DOI] [PubMed] [Google Scholar]
- 195.Matheis K, Laurie D, Andriamandroso C, Arber N, Badimon L, Benain X, Bendjama K, Clavier I, Colman P, Firat H, Goepfert J, Hall S, Joos T, Kraus S, Kretschmer A, Merz M, Padro T, Planatscher H, Rossi A, Schneiderhan-Marra N, Schuppe-Koistinen I, Thomann P, Vidal JM, Molac B. A generic operational strategy to qualify translational safety biomarkers. Drug Discov Today. 2011;16 (13–14):600–608. doi: 10.1016/j.drudis.2011.04.011. [DOI] [PubMed] [Google Scholar]
- 196.Hill AB. The Environment and Disease: Association or Causation? Proc R Soc Med. 1965;58 (5):295–300. [PMC free article] [PubMed] [Google Scholar]
- 197.Slocum JL, Heung M, Pennathur S. Marking renal injury: can we move beyond serum creatinine? Transl Res. 2012;159 (4):277–289. doi: 10.1016/j.trsl.2012.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Ju W, Smith S, Kretzler M. Genomic biomarkers for chronic kidney disease. Transl Res. 2012;159 (4):290–302. doi: 10.1016/j.trsl.2012.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Sotiriou C, Piccart MJ. Taking gene-expression profiling to the clinic: when will molecular signatures become relevant to patient care? Nat Rev Cancer. 2007;7 (7):545–553. doi: 10.1038/nrc2173. [DOI] [PubMed] [Google Scholar]
- 200.Dabbs DJ, Klein ME, Mohsin SK, Tubbs RR, Shuai Y, Bhargava R. High False-Negative Rate of HER2 Quantitative Reverse Transcription Polymerase Chain Reaction of the Oncotype DX Test: An Independent Quality Assurance Study. J Clin Oncol. 2011;29 (32):4279–4285. doi: 10.1200/JCO.2011.34.7963. [DOI] [PubMed] [Google Scholar]
- 201.Ignatiadis M, Sotiriou C. Breast cancer: Should we assess HER2 status by Oncotype DX®? Nat Rev Clin Oncol. 2012;9 (1):12–14. doi: 10.1038/nrclinonc.2011.188. [DOI] [PubMed] [Google Scholar]
- 202.Coombes KR, Wang J, Baggerly KA. Microarrays: retracing steps. Nat Med. 2007;13 (11):1276–1277. doi: 10.1038/nm1107-1276b. [DOI] [PubMed] [Google Scholar]
- 203.Baggerly KA, Coombes KR. Deriving chemosensitivity from cell lines: Forensic bioinformatics and reproducible research in high-throughput biology. Ann Appl Stat. 2009;3 (4):1309–1334. [Google Scholar]
- 204.RETRACTION for Hsu DS, Balakumaran BS, Acharya CR, Vlahovic V, Walters KS, Garman K, Anders C, Riedel RF, Lancaster J, Harpole D, Dressman HK, Nevins JR, Febbo PG, Potti A. Retraction. Pharmacogenomic Strategies Provide a Rational Approach to the Treatment of Cisplatin-Resistant Patients With Advanced Cancer. J Clin Oncol. 2010;28 (35):5229–5229. doi: 10.1200/JCO.2007.11.0593.
- 205.Potti A, Mukherjee S, Petersen R, Dressman HK, Bild A, Koontz J, Kratzke R, Watson MA, Kelley M, Ginsburg GS, West M, Harpole DH, Nevins JR. Retraction: A Genomic Strategy to Refine Prognosis in Early-Stage Non–Small-Cell Lung Cancer. N Engl J Med 2006;355:570–80. New Engl J Med. 2011;364 (12):1176–1176. doi: 10.1056/NEJMc1101915. [DOI] [PubMed] [Google Scholar]
- 206.Potti A, Dressman HK, Bild A, Riedel RF, Chan G, Sayer R, Cragun J, Cottrill H, Kelley MJ, Petersen R, Harpole D, Marks J, Berchuck A, Ginsburg GS, Febbo P, Lancaster J, Nevins JR. Retraction: Genomic signatures to guide the use of chemotherapeutics. Nat Med. 2011;17 (1):135–135. doi: 10.1038/nm0111-135. [DOI] [PubMed] [Google Scholar]
- 207.Hayden EC. Lapses in oversight compromise omics results. US board calls for tighter control of test-based data. Nature. 2012 Mar 23;2012 [Google Scholar]
- 208.Curtis C, Shah SP, Chin S-F, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, Graf S, Ha G, Haffari G, Bashashati A, Russell R, McKinney S, Langerod A, Green A, Provenzano E, Wishart G, Pinder S, Watson P, Markowetz F, Murphy L, Ellis I, Purushotham A, Borresen-Dale A-L, Brenton JD, Tavare S, Caldas C, Aparicio S. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012 doi: 10.1038/nature10983. [Published online: 18 April 2012] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Monnier VM, Vishwanath V, Frank KE, Elmets CA, Dauchot P, Kohn RR. Relation between Complications of Type I Diabetes Mellitus and Collagen-Linked Fluorescence. New Engl J Med. 1986;314 (7):403–408. doi: 10.1056/NEJM198602133140702. [DOI] [PubMed] [Google Scholar]
- 210.Sun JK, Keenan HA, Cavallerano JD, Asztalos BF, Schaefer EJ, Sell DR, Strauch CM, Monnier VM, Doria A, Aiello LP, King GL. Protection From Retinopathy and Other Complications in Patients With Type 1 Diabetes of Extreme Duration. The Joslin 50-Year Medalist Study. Diabetes Care. 2011;34 (4):968–974. doi: 10.2337/dc10-1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.https://rarediseasesnetwork.epi.usf.edu/NEPTUNE/ (NCT01209000 at http://clinicaltrials.gov/ct2/home).
- 212.Mirel B, Eichinger F, Nair V, Kretzler M. Integrating automated workflows, human intelligence and collaboration. Summit on Translat Bioinforma. 2009;2009:79–83. [PMC free article] [PubMed] [Google Scholar]
- 213.http://www.nephromine.org (freely accessible for academic users).
- 214.Derry JMJ, Mangravite LM, Suver C, Furia MD, Henderson D, Schildwachter X, Bot B, Izant J, Sieberts SK, Kellen MR, Friend SH. Developing predictive molecular maps of human disease through community-based modeling. Nat Genet. 2012;44 (2):127–130. doi: 10.1038/ng.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]