Abstract
Toward the expansion of the genetic alphabet of DNA, several artificial third base pairs (unnatural base pairs) have been created. Synthetic DNAs containing the unnatural base pairs can be amplified faithfully by PCR, along with the natural A–T and G–C pairs, and transcribed into RNA. The unnatural base pair systems now have high potential to open the door to next generation biotechnology. The creation of unnatural base pairs is a consequence of repeating “proof of concept” experiments. In the process, initially designed base pairs were modified to address their weak points. Some of them were artificially evolved to ones with higher efficiency and selectivity in polymerase reactions, while others were eliminated from the analysis. Here, we describe the process of unnatural base pair development, as well as the tests of their applications.
Keywords: unnatural base pair, genetic alphabet, replication, transcription, translation, PCR
Introduction
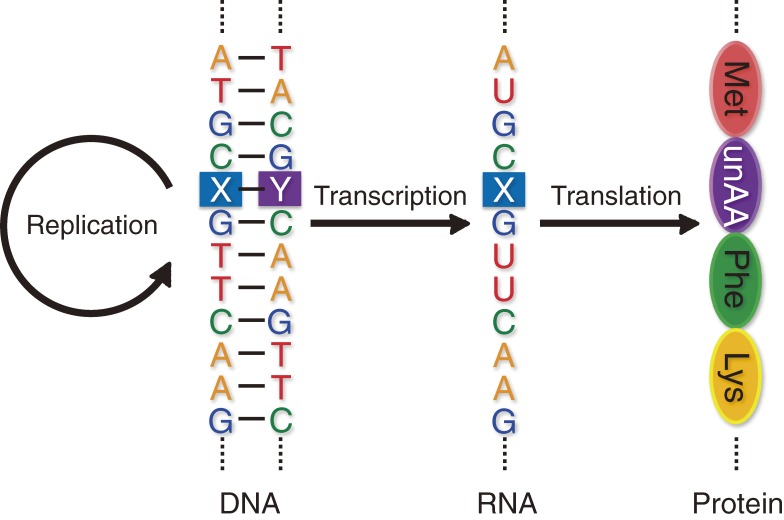
Through molecular biology studies, from the discovery of the double helix structure of DNA in 19531) to the deciphering of the genetic code in the 1960s,2–4) researchers revealed that the complementarity of the A–T(U) and G–C base pairs is the fundamental rule of the genetic information flow of terrestrial life (Fig. 1). With surprising foresight, during that period, Alexander Rich proposed the possibility of an artificial, third base pair between isoguanine (isoG, 6-amino-2-ketopurine) and isocytosine (isoC, 2-amino-4-ketopyrimidine) in his article in 1962 (Fig. 2a).5) The artificially designed isoG–isoC pair has three hydrogen bonds with specific geometry of the proton donor and acceptor combinations, which are different from those in the A–T and G–C pairs. If the third base pair (unnatural base pair), along with the natural base pairs, selectively functions in the central dogma comprising replication, transcription, and translation, then the genetic alphabet and code could be expanded, providing a new method for the site-specific incorporation of novel components into nucleic acids and proteins with increased functionality (Fig. 3). In addition, unnatural base pair studies could also provide new aspects for biological mechanisms related to the genetic information flow and for elucidating the origin of the A–T(U) and G–C pairs in the prebiotic era on Earth.6–11) Furthermore, the creation of unnatural base pairs is a challenging theme for synthetic biology researchers seeking the possibility of artificially rebuilding the present life system to a further evolved one.
Figure 1.
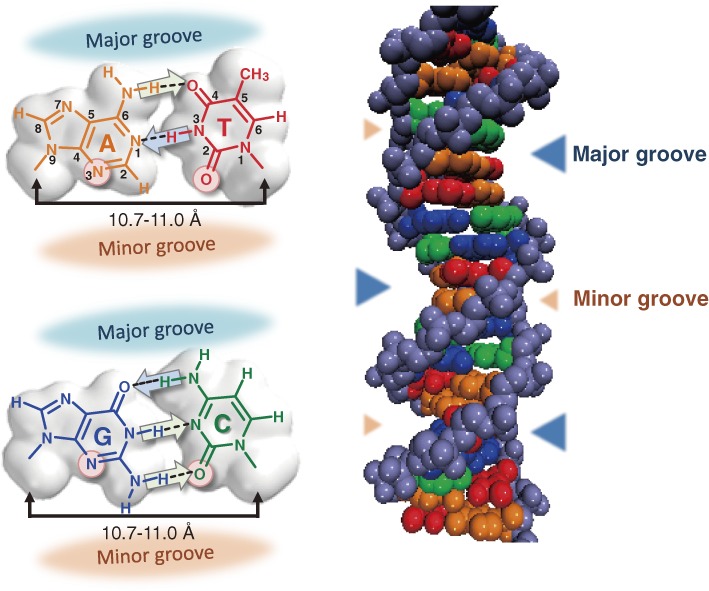
Structures of the natural A–T and G–C pairs and the DNA duplex. The common properties of the natural base pairs, including the proton acceptor residues on the minor groove side (circles) for recognition by polymerases, the hydrogen-bonding patterns (arrows, from a proton donor to a proton acceptor residue), and the distances between the glycosidic bond positions of each base pair are indicated (left). The major and minor grooves in the DNA duplex are indicated by triangles (right).
Figure 2.
Chemical structures of Benner’s base pairs. (a) The unnatural isoG–isoC pair. (b) The unnatural X–κ pair. (c) The non-cognate isoG (enol)–T pair. (d) The non-cognate isoG (enol)–TS pair. (e) The cognate A–TS pair.
Figure 3.
Unnatural base pair systems toward the expansion of the genetic alphabet and code. The unnatural base pair (X–Y), which works together with the natural A–T and G–C base pairs in the central dogma, allows the site-specific incorporation of extra unnatural nucleotides (X and Y) into DNA and RNA, and unnatural amino acids (unAA) into proteins.
In 1989, Benner’s group reported the chemical synthesis of the nucleoside derivatives of isoG and isoC, and tested the isoG–isoC pair in in vitro replication and transcription.12) They also designed other unnatural base pairs with different hydrogen bonding geometries, such as X–κ (Fig. 2b).13) In 1992, they demonstrated an in vitro translation system using the isoG–isoC pair for the site-specific incorporation of a non-standard amino acid, 3-iodotyrosine, into a peptide, by creating a new isoCAG codon.14) Through these experiments, the possibility and potential of unnatural base pairs were examined, and recently, the isoG–isoC pair was utilized in a multiplex real-time PCR system to detect a target nucleic acid sequence.15) However, the pairing selectivities of these unnatural base pairs were not sufficient in polymerase reactions, and thus their applications have been severely limited.16–19)
In the mid-1990s, Kool’s group synthesized a non-hydrogen-bonded natural base pair analog between 4-methylbenzimidazole (Z) and 2,4-difluorotoluene (F), as a steric isostere of the A–T pair (Figs. 4a and 4b), to investigate the contribution of hydrogen bonds to the base pair selectivity in replication.20,21) The steric shapes of the Z and F bases mimic those of the A and T bases, respectively. The hydrogen bonding residues and atoms in A and T were replaced with non-hydrogen bonding ones in Z and F. For example, the 3-imino group and the 2- and 4-keto groups in T were replaced with a C-H group and fluorine atoms, respectively, in F. In the base pairing, the proton acceptor ability of a fluorine atom is approximately ten times lower than that of the keto group.22,23) Replication studies using the Z–F pair revealed that the Z and F bases were equally replaceable with the A and T bases, respectively. These results suggested the importance of the shape complementarity of the pairing bases, rather than the hydrogen bonding interactions. This suggestion initiated some controversy, since it contradicted the traditional fundamental role of the base pair selectivity.23–26) However, another key point of Kool’s discovery was that non-hydrogen-bonded hydrophobic base analogs could be potential candidates for creating unnatural third base pairs.
Figure 4.
Chemical structures of Kool’s base pairs. (a) The hydrophobic unnatural Z–F pair. (b) The natural A–T pair. (c) The hydrophobic unnatural Q–F pair.
In a genetic expansion system, highly exclusive selectivity of the unnatural base pair is required for it to function as a third base pair in polymerase reactions, such as PCR amplification and transcription. The error rates for natural base pairing by proofreading-deficient DNA polymerases (without 3′-exonuclease activity) range from 10−3 to 10−6 error/bp. This means that the incorporation selectivity of the natural base substrates opposite their complementary base in templates ranges from 99.9 to 99.9999% in replication. Since replicated DNA fragments also act as templates for further replication, mutations of bases in replicated DNA fragments are accumulated by repeated amplification, such as increased numbers of PCR cycles. If the selectivity of an unnatural base pair is 99.9% per replication, then 98% of the unnatural base pair could be retained at a specific position in its amplified DNA after 20 cycles of PCR (0.99920 = ∼0.98 in a theoretical exponential amplification). However, when using an unnatural base pair with 99.0% replication selectivity, the retention of the unnatural base pair in its amplified DNA after 20-cycle PCR would decrease to 82% (0.9920 = ∼0.82).
Since the late 1990s, the creation of unnatural base pairs for new biological systems has competitively and rapidly advanced.6,11,18,27–29) At present, three types of unnatural base pairs have been developed by Benner’s,30,31) Romesberg’s32,33) and our groups,34–37) and DNA fragments containing these unnatural base pairs can be efficiently and faithfully amplified by PCR with high selectivity (>99%). However, in the early stages, when the difficulties with the isoG–isoC pair were revealed, most researchers were skeptical about the possibility of artificially introducing an unnatural base pair into the present sophisticated biology systems that have evolved over several billion years. Here, we will describe the developmental process of unnatural base pairs and their applications, by focusing on our research as well as the progress made by other groups.
I. Base pairing complementarity in polymerase reactions
In general, the creation of unnatural base pairs is initiated by designing the molecular structures or modifying the A–T and G–C pairs, based on a certain idea or concept underlying the mechanism of the selective complementarity of the natural base pairings in polymerase reactions. The natural base pairs comprise two pairs between purine (A, G) and pyrimidine (T, C) bases, in which the distances between the glycoside bond positions of the pairing bases are constant (10.7–11.0 Å) (Fig. 1). The constant distance in double-stranded DNA is essential to allow the access of polymerases. Each pair is formed by hydrogen bonds, and the hydrogen bonding patterns of the proton donor and acceptor combinations are different from each other (Fig. 1). Until Kool’s experiments, the hydrogen-bonding patterns seemed to be the only means by which the bases recognize their cognate pairing bases (A–T and G–C). Now, the structural complementarity of the shapes between pairing bases is also considered as an important factor for the base pair selectivity in replication. From our experiments using several base pair analogs with different numbers of hydrogen bonds, we consider both the shape complementarity and the strength of the hydrogen-bonding interaction as affecting the base pair formation in replication.38)
Other factors, such as polarity, electrical repulsion, and hydrophobicity between pairing bases, and minor groove interactions with polymerases, should also be considered when creating unnatural base pairs. X-ray crystallography of the ternary complexes among DNA templates, triphosphate substrates, and polymerases revealed that the 3-nitrogen atoms of A and G and the 2-keto groups of C and T on the minor groove side (Fig. 1) interact with the side chains of amino acids in polymerases directly or via a water molecule.39,40) Replication experiments using base analogs proved the importance of these minor groove interactions. The nucleoside triphosphates of pyrimidine analogs (2-aminopyridin-5-yl and 3-methyl-2-pyridon-5-yl), which lack the 2-keto group of C and T, strongly inhibited PCR amplification by Taq DNA polymerase or the Klenow fragment of Escherichia coli DNA polymerase I (KF).41) Kool’s group developed a modified Z base (9-methyl-1-H-imidazo[(4,5)-b]pyridine, Q), in which the carbon of the Z base, corresponding to position 3 of purines, was replaced with nitrogen (Fig. 4c), and found the increased efficiency of the Q–F pairing in replication using KF, as compared to that of the Z–F pairing.42) Both the oxygen of the pyrimidine 2-keto groups and the purine 3-nitrogen are located in similar relative positions from the nucleic acid backbone, and thus polymerases can equally recognize all four bases with the same interaction mechanism (Fig. 1). However, the position and the orientation of the lone-pair electrons of the oxygen in the pyrimidines are slightly different from those of the nitrogen in the purines. Therefore, slight fluctuations of these proton donor atom positions are acceptable. As for interactions with polymerases, the higher hydrophobicity of the substrate and template bases is favorable. X-ray crystallographic analyses of DNA and RNA polymerases revealed that these bases are stacked with aromatic amino acid residues in the polymerases.40,43,44) For the creation of unnatural base pairs, these factors should also be considered, besides the hydrogen-bonding pattern and shape-complementarity of the base pairings.
II. Developmental process of unnatural base pairs
The creation of unnatural base pairs has been achieved by repeating “proof of concept” experiments. First, an unnatural base pair is designed by considering the known factors mentioned above. Second, nucleoside 5′-triphosphates (as substrates in replication and transcription)45,46) and amidite derivatives (for automated DNA synthesis) of the designed unnatural bases are chemically synthesized. Third, replication and transcription using the unnatural base substrates and DNA templates are examined in a test tube. Then, the designed base pairs are modified or new pairs are redesigned, by addressing the problems revealed by the experiments. This process is repeated over and over, until satisfactory base pairs are created for practical use in replication, transcription, and/or translation systems.
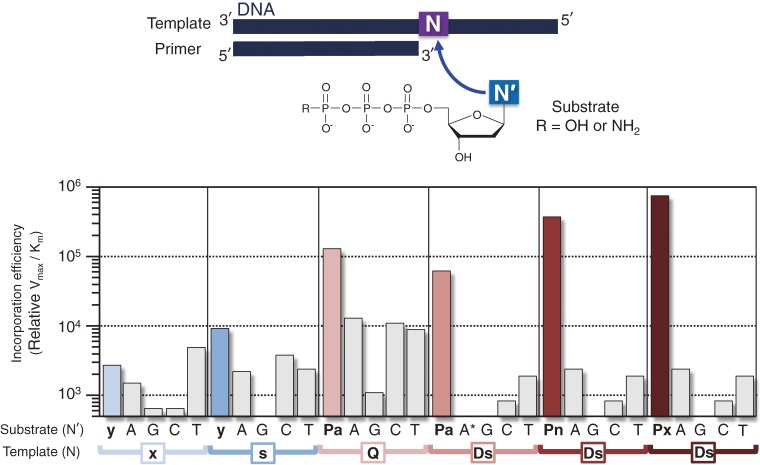
The enzymatic incorporation efficiency and selectivity of designed unnatural base pairs are assessed in in vitro polymerase reactions. For replication studies, the steady-state kinetic parameters (Vmax/KM or kcat/KM) of cognate and non-cognate base pairings are determined by a conventional single-nucleotide insertion experiment,47–49) which is useful to assess the enzymatic incorporation efficiency of a substrate, opposite a template base, into a primer strand (see Fig. 15). The single-nucleotide insertion experiments are performed by using combinations of each substrate with each template base in a primer/template complex, and KF without 3′-exonuclease activity. Usually, a gel-based analysis using radioactive labeled-primers is employed to determine the parameters of all possible combinations of cognate and non-cognate natural and unnatural base pairs. However, the gel-based analysis is tremendously laborious and time-consuming. Therefore, we improved the method by using an automated DNA sequencer with dye-labeled primers.50) This method has greatly accelerated our repetitive ‘proof of concept’ experiments.
Figure 15.
Single-nucleotide incorporation efficiencies and selectivities of our unnatural base pairs by KF. A* corresponds to the γ-amidotriphosphate of A.
Primer extension and PCR amplification in the presence of all substrates of natural and unnatural bases are more practical assessments of unnatural base pairings in replication. In vitro transcription using T7 RNA polymerase is also often used to assess the site-specific incorporation of unnatural bases into RNA. In these experiments, new methods should be developed for determining the incorporation positions and selectivities of unnatural base pairs in the replicated DNA and transcribed RNA fragments. In the following sections, we describe several methods associated with the process of unnatural base pair development.
III. Hydrogen-bonded, unnatural base pairs
1. Early stage of unnatural base pair studies.
Benner’s group pioneered the examination of the abilities of hydrogen-bonded unnatural base pairs, such as isoG–isoC and X–κ (xanthine–2,6-diaminopyrimidine) (Figs. 2a and 2b).12,13) Although their efforts revealed the possibility of a genetic expansion system utilizing unnatural base pairs, they also highlighted several weaknesses of their base pairs for practical use. First, in contrast to the natural bases, the isoC, X and κ bases have proton-donor 2-amino (in isoC and κ) or 3-imino (in X) groups on the minor groove side, which are unfavorable for polymerase recognition, thus decreasing their incorporation efficiencies.16,17) Second, the isoC nucleoside is chemically unstable under alkaline and acidic conditions, which makes it difficult to prepare isoC-containing oligonucleotides by chemical synthesis.18,51) Third, hydrogen-bonded unnatural base pairs sometimes suffer from the instability of the hydrogen-bonding geometry, due to tautomerism. Especially, isoG undergoes keto-enol tautomerization, depending on the solvent polarity, and the enol-form of isoG mispairs with T (Fig. 2c), greatly decreasing the isoG–isoC pairing selectivity.16,19,52,53) Due to these shortcomings, unnatural base pair studies slowed down for a period in the mid-1990s.
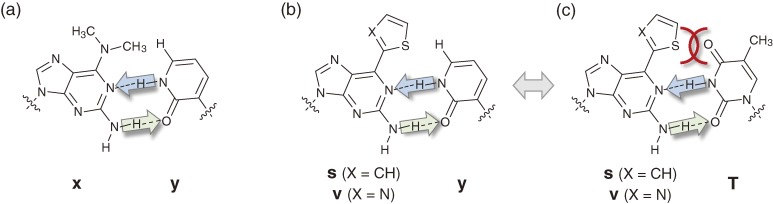
We initiated our unnatural base pair studies in 1997, by addressing the base pair problems identified by Benner’s group. By combining Benner’s hydrogen-bonding geometry concept with a steric hindrance idea, we first designed and synthesized a series of unnatural base pairs between 2-amino-6-modifiedpurines and pyridine-2-one (y), such as 2-amino-6-(N,N-dimethylamino)purine (x) and y,54,55) 2-amino-6-(2-thienyl)purine (s) and y,56,57) and 2-amino-6-(2-thiazoyl)purine (v) and y (Figs. 5a and 5b).58) In these unnatural base pairs, the bulky substituents at the 6-position of the x, s and v bases exclude the mis-pairing with the natural pyrimidines, by sterically clashing with the 4-keto group of T(U) and the 4-amino group of C (Fig. 5c), but in the cognate pairing with y, the bulky 6-substituents can accommodate the 6-hydrogen atom of y, corresponding to position 4 of the pyrimidines. The x–y, s–y and v–y pairs function well in transcription, and T7 RNA polymerase site-specifically incorporates the substrate of y (yTP) into RNA opposite x, s or v in chemically-synthesized DNA templates.
Figure 5.
Hydrogen-bonded, unnatural base pairs developed by our group. (a) The unnatural x–y pair. (b) The unnatural s–y or v–y pair. (c) The non-cognate pairing of s or v with T.
Among these unnatural base pairs, the order of the efficiency and selectivity of the unnatural base pairing in T7 transcription is v–y > s–y > x–y. In comparison with the freely rotated 6-dimethylamino group of x, the planarity and the electronegative atoms of the 6-heterocycles, thiazolyl in v and thienyl in s, efficiently exclude the pairing with the natural bases, especially T or U, by the electron negative repulsion with the 4-keto group of T or U, as well as the steric clash. In addition, the DNA duplexes containing the s–y or v–y pair exhibit higher thermal stability than those with the x–y pair. This is because the 6-heterocycles stack with their neighboring natural bases in the duplex DNA. However, the nucleoside derivatives of v are chemically less stable under basic conditions, relative to those of s. Thus, among them, the s–y pair is the most useful unnatural base pair for the transcription system. Interestingly, we found that the s and v bases are fluorescent.59)
2. Transcription system using the s–y pair.
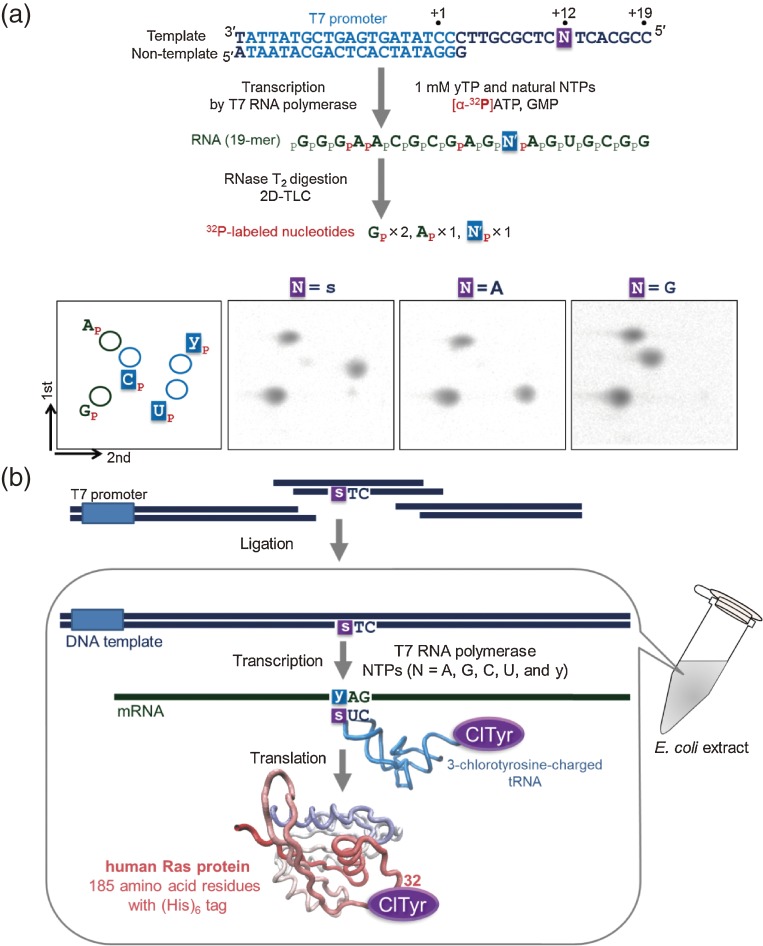
The site-specific incorporation of yTP into RNA was confirmed by a 2D-TLC nucleotide-composition analysis of transcripts (Fig. 6a).57) For the analysis, we used partially double-stranded 37-mer DNA templates, from which 19-mer RNA fragments were transcribed. T7 transcription was performed in the presence of [α-32P]ATP, besides the natural and unnatural substrates. Since the following base of the unnatural base was T in the templates (position +13 of the template in Fig. 6a), the 5′-32P-labeled A was incorporated into the opposite position after the y incorporation in the transcripts. Thus, the 3′-phosphate of y was radioactively labeled, and by RNase T2 digestion of the transcripts, the 3′-labeled nucleotides of y and some other labeled natural bases were obtained. These labeled nucleotides were analyzed and quantified by 2D-TLC. The analysis revealed that more than 90% of yTP was site-specifically incorporated into RNA, opposite s in templates, by T7 transcription in the presence of 1 mM each substrate (see TLC for N = s in Fig. 6a). In addition, no significant misincorporations of yTP opposite the natural bases were observed (see TLC for N =A and G in Fig. 6a).
Figure 6.
Transcription using the unnatural s–y pair. (a) Scheme of the transcription experiments and 2D-TLC for nucleotide composition analyses of transcripts. (b) The coupled transcription–translation system involving the s–y pair. The DNA template containing a CTs sequence and the 3-chlorotyrosine-charged tRNA were prepared separately and added to the transcription-coupled translation system. The yAG codon–CUs anticodon interaction was used for the site-specific incorporation of 3-chlorotyrosine at position 32 of the human Ras protein.
This specific transcription involving the s–y pair was combined with an in vitro translation system, and in 2002, we succeeded in the site-specific incorporation of a non-standard amino acid, 3-chlorotyrosine, into the human Ras protein (185 amino acid residues with a (His)6 tag) by a coupled T7 transcription and E. coli translation system (Fig. 6b).57) For the transcription of the 747-mer y-containing RNA, we prepared DNA templates containing a CTs sequence, as a new yAG codon for a non-standard amino acid, by the chemical synthesis of a short DNA fragment containing s, followed by enzymatic ligation with 5′- and 3′-ras gene fragments. Separately, a 3-chlorotyrosine-charged tRNA containing a CUs anticodon, was prepared by a combination of chemical synthesis and enzymatic ligation, followed by aminoacylation of 3-chlorotyrosine using an aminoacyl-tRNA synthetase (Fig. 7). We chose the sequence of Saccharomyces cerevisiae tRNATyr for the CUs-anticodon tRNA (tRNACUs), since it is a very poor substrate for any E. coli aminoacyl-tRNA synthetase. In contrast, a mutated S. cerevisiae tyrosyl-tRNA synthetase60) efficiently aminoacylated tRNACUs with 3-chlorotyrosine. The in vitro coupled transcription and translation system was tested, using 3-chlorotyrosyl-tRNACUs and the DNA template containing the CTs sequence with yTP. A mass spectrometric analysis of the obtained protein confirmed the site-specific incorporation of 3-chlorotyrosine into position 32 of the Ras protein. This was the first achievement of the direct synthesis of a protein containing a non-standard amino acid at a specific position by the transcription and translation system, using its long DNA template.
Figure 7.
Preparation of 3-chlorotyrosyl tRNACUs. The tRNA was prepared by ligation of the 5′-half fragment derived from the native S. cerevisiae tyrosine tRNA and the chemically synthesized 3′-half fragment containing the CUs anticodon. The aminoacylation with 3-chlorotyrosine was performed by using a mutated S. cerevisiae tyrosyl-tRNA synthetase.
In addition, the specific transcription involving the s–y pair was also applied to generate RNA molecules with extra functional components at desired positions. In general, the site-specific modification of RNA molecules is achieved by chemical RNA synthesis, post-transcriptional modification, and enzymatic incorporation of nucleotide analogs as substrates. Although chemical synthesis is commonly used, it is only practical for preparing small RNA molecules. Post-transcriptional modification61,62) and enzymatic incorporation63–66) are restricted to only the 5′- or 3′-termini of RNA molecules, and the incorporation efficiency and selectivity are not always high. In contrast, the transcription mediated by the s–y and v–y pairs allows the selective site-specific incorporation of a wide variety of modified y bases, which are linked with functional groups of interest via a linker, into desired positions of RNA transcripts. A series of modified yTPs, such as iodo-, amino-, biotin- and fluorophore-linked y bases, were chemically synthesized (Fig. 8a).67–71) These modified yTPs can be site-specifically incorporated into RNA, opposite s or v in templates, by T7 transcription.
Figure 8.
Site-specific modification of RNA molecules using the unnatural s–y and v–y pairs. (a) Scheme for the site-specific modification of RNA molecules at the 3′ terminal region. The DNA templates containing s or v at specific positions can be prepared by PCR, using a 5′ primer including the T7 promoter and a 3′ primer containing s or v. Functional substrates of modified y bases are shown in the yellow box. (b) Site-specific fluorescent labeling of an anti-theophylline aptamer. FAM-y was introduced into position 6, in place of U6, in the aptamer.
The iodo-y base is a photosensitive component, capable of cross-linking with proximal reactive residues by irradiation at 312 nm.68) RNA molecules containing iodo-y at specific positions can be used for studying RNA-RNA and RNA-protein interactions and for fixing the interactions by photo-crosslinking, as reported in RNA photoaptamers.72,73) We demonstrated the photo-crosslinking ability using the iodo-y base, by introducing it into an RNA aptamer and examining the interaction of the aptamer and its target protein.67,74) We also demonstrated the 3′-terminal labeling of RNA molecules by T7 transcription using modified yTPs. DNA templates containing s or v at the terminal position can be easily prepared by PCR using s- or v-containing 3′-primers (Fig. 8a). By T7 transcription using fluorescein- or biotin-linked y substrates (FAM-yTP, FAM-hx-yTP or Biotin-yTP in Fig. 8a), we performed both the fluorescent labeling and immobilization of an RNA aptamer.69,70)
By rational design based on structural information, the site-specific modified-y incorporation could be a powerful tool to endow RNA molecules with new functions. To show this, we developed a fluorescence sensor system using an anti-theophylline RNA aptamer.69) We introduced FAM-y into a specific position of the anti-theophylline aptamer, on the basis of the tertiary structure of the aptamer–theophylline complex (Fig. 8b). Upon binding to theophylline, the aptamer forms a specific tertiary structure with a unique base triplet, U6-U23-A28, adjacent to the theophylline binding site. In the complex structure, the 4-keto group of U6 is not involved in triplet formation, and thus, U6 can be replaced with FAM-y. Then, we introduced FAM-y into the U6 position of the aptamer by T7 transcription. Interestingly, the FAM fluorescence intensity of the aptamer increased in the presence of theophylline. This is because the FAM moiety of the y base is stacked with other bases inside the non-structured aptamer in the absence of theophylline, but upon theophylline binding, it moves outside by base triplet formation, resulting in the increased fluorescence. This detection system was theophylline specific, and the fluorescence did not change in the presence of caffeine, which has a similar structure to theophylline. By using this system, 50 pmol of theophylline (500 nM in 0.1 ml) can be detected.
3. Further improvement of the s–y pair.
The s–y pair transcription system is useful for the incorporation of y into RNA opposite s in DNA templates. However, the opposite case, namely, the s incorporation into RNA opposite y, is less effective. Although the 6-thienyl group of s efficiently prevents the mispairing with the natural pyrimidine bases, y has no such functional group to exclude mispairing with the natural purine bases. In addition, y possesses good shape complementarily with A as well as s (Figs. 9a and 9b). Thus, the misincorporation of ATP opposite y is the main problem, and the s–y pair is only used as a unidirectional third base pair for transcription.
Figure 9.
Structures of the hydrogen-bonded, unnatural base pairs designed by our group. (a) The cognate s–y pair. (b) The non-cognate A–y pair. (c) The cognate s–z pair. (d) The non-cognate A–z pair. Space filling models of the base alone (with a methyl group in place of the ribose) are shown.
To address this problem, we redesigned the pairing partner of s, instead of y, and developed imidazolin-2-one (z) (Fig. 9c).75) The five-membered ring of z reduces the shape complementarity of the pairing with A (Fig. 9d), relative to that of y. Usually, hydrophilic bases are hydrated with surrounding waters, and a pairing partner with less shape complementarity cannot exclude the waters and form the base pair.76) In addition, the small ring of z is accommodated well in the s–z pair. We synthesized the nucleoside derivatives of z, and examined the efficiency and selectivity of the s–z pairing in replication and transcription, by comparison to those of the s–y pairing. As expected, the steady-state kinetics of single-nucleotide insertion experiments using KF revealed that the selectivity of the non-cognate A–z pair (Vmax/KM = 1.6 × 104 for the dATP incorporation opposite z) was greatly reduced, as compared to that of the non-cognate A–y pair (Vmax/KM = 1.5 × 105 for the dATP incorporation opposite y). In T7 transcription, z is superior to y as a template base for the site-specific s incorporation into RNA.75)
However, the z incorporation opposite s into DNA and RNA was less effective, relative to the y incorporation opposite s. This might be due to the lower hydrophobicity of the z base, which would reduce the interaction with polymerases. From the single-nucleotide insertion experiments using KF, the KM value for the z incorporation opposite s (KM = 600 µM) was much larger than that for the s incorporation opposite z (KM = 95 µM), indicating that the low stacking ability of the z substrate, because of the low hydrophobicity, reduced the affinity between the DNA polymerase and the z substrate. Like DNA polymerase, T7 RNA polymerase also prefers hydrophobic bases as substrates. Similarly, the thermal stability of duplex DNA fragments containing the s–z pair was lower, as compared to those with the s–y pair. Thus, for a new codon and anticodon in translation, the s–y pair is better than the s–z pair. For all of these reasons, a two-unnatural base pair system of the s–y and s–z pairs could be used as a coupled transcription and translation system for the site-specific incorporation of non-standard amino acids into proteins. The s–y pair can be used for preparing mRNA containing y, and the s–z pair is useful for preparing tRNA containing z by T7 transcription.75)
Throughout our studies so far, the hydrogen-bonded, unnatural base pairs function well as a third base pair in transcription and translation. However, in replication, the selectivity of our unnatural base pairs is still insufficient. Some of the hydrogen-bonding residues and atoms of the s, y and z bases attract the natural hydrophilic bases in replication. In addition, small, hydrophilic molecules, such as the z base, are not favorable for interactions with polymerases. Therefore, we shifted the focus of our studies to the creation of hydrophobic, unnatural base pairs.
IV. Hydrophobic, unnatural base pairs
1. Struggling with hydrophobic base pairs.
From Kool’s study in the mid-1990s, non-hydrogen bonded, hydrophobic base analogues became candidates for a third base pair. In addition, hydrophobic base analogs are well recognized by polymerases, because an aromatic amino acid residue in the polymerases stacks with an incoming substrate base. Our first attempt to design a hydrophobic base pair was the self-pair between 4-methylpyridine-2-one (4MP) bases (Fig. 10a).77) Kool’s study of the Z–F pair also revealed slight mispairing between the F bases in replication using KF,21) and thus we designed the 4MP base from the F base. Unlike F, the 4MP base has a 4-methyl group to prevent mispairing with A by clashing between the 4-methyl group of 4MP and the 6-amino group of A, and it has the 2-keto group for the minor groove interaction with polymerases. We synthesized the amidite and triphosphate of 4MP, and tested the 4MP–4MP self-pairing in replication using KF. However, contrary to our expectations, the 4MP substrate was not incorporated opposite 4MP at all, although we found that 4MP has unique dual specificity for pairing with A or G.77)
Figure 10.
Hydrophobic, unnatural base pairs. (a) The hypothetical self-pair between 4-methylpyridin-2-one (4MP) bases. (b) The self-pair between 6-propynylcarbostyril (PICS) bases. (c) The Q–F pair. The clash of the hydrogen atoms in the center of the Q–F pairing surface is indicated in red. (d) The Q–Pa pair.
Meanwhile, in 1999, Romesberg’s group reported their successful hydrophobic self-pair between the 7-propynylisocarbostyril (PICS) bases in single-nucleotide insertion experiments (Fig. 10b).78) The PICS substrate was efficiently incorporated into DNA, opposite PICS in templates, by KF. Furthermore, DNA duplexes containing the PICS–PICS pair exhibited high thermal stability, relative to those containing non-cognate pairs between PICS and each of the natural bases. However, after the PICS incorporation opposite PICS, the following extension was prevented in replication. The PICS–PICS pair is too large for accommodation in the 10.7–11.0 Å distance of the DNA duplex, and thus, the large and hydrophobic PICS molecules stack on each other in the pairing, and this unusual stacked structure cannot be recognized by polymerases during primer extension.27)
In 2003, while considering Kool’s Z–F and Q–F pairs, which are hydrophobic isosteres of the natural A–T pair, we found that the shape complementarity of these base pair is still imperfect, and the hydrogen atoms in the center on the pairing surface clash with each other (Fig. 10c). We noticed that this clash could be eliminated by employing a five-membered ring base analog, in place of the six-membered F base. Therefore, we designed pyrrole-2-carbaldehyde (Pa) (Fig. 10d), as a pairing partner of Kool’s Q base.79) Like the effective cognate s–z and less effective non-cognate A–z pairs, the hydrophobic, five-membered Pa base was expected to reduce the shape complementarity with A in the A–Pa pair, relative to that of the A–F pair. In addition, we added an aldehyde group to Pa, to increase the minor groove interaction with polymerases.
We synthesized the substrate and amidite of Pa, and examined the efficiency and selectivity of the Q–Pa pairing in replication by KF. As expected, the single-nucleotide insertion experiments using KF revealed the improved selectivity of the cognate Q–Pa pair (see Fig. 15), relative to those of the Q–F pair.79) We also determined the tertiary structure of a DNA duplex containing the Q–Pa pair by NMR, and confirmed that the geometry of the Q–Pa pair in the duplex closely resembles those of the natural base pairs,79) rather than that of the Z–F pair. From the study, we realized that the fine-tuning of the shape complementarity of unnatural base pairs is important to create a third base pair.
Although we made a favorable start in our analysis of hydrophobic base pairs, the Q–Pa pair still did not work complementarily as a third base pair in replication. Since the Q base is an isostere of A, Q also pairs with T. In addition, we found that dQTP was self-complementarily incorporated into DNA opposite Q in templates, and like the PICS–PICS pair,78) after the Q–Q pair formation, the following extension was paused.
In 2005, Benner’s group reported a new genetic expansion system using the isoG–isoC, G–C and A–2-thiothymine (TS) pairs, by addressing the tautomerism problem of isoG.80) To prevent the pairing of the enol form of isoG with T, they replaced T with TS, in which the large 2-sulfur atom clashes with the 2-hydroxy group of the enol form of isoG (Figs. 2d and 2e). This new set of three base pairs functions in PCR: the selectivity of the isoG–isoC pair reached 98% per PCR cycle, while the isoG–isoC selectivity without the help of TS was about 93% per PCR cycle. Although the 98% selectivity of the unnatural base pairing is not sufficient for practical use in replication (the retention rate of the isoG–isoC pair in 20-cycle PCR amplified DNA fragments becomes around 67% (0.9820 = ∼0.67)), this is a unique expansion system for PCR amplification. At this point, we had no further breakthrough ideas other than the Q–Pa pair. As a desperate measure, we collected all of the efficiency and selectivity data of our unnatural base pairs, and examined all possible pairing combinations comprehensively, using the unnatural bases that we had developed thus far.
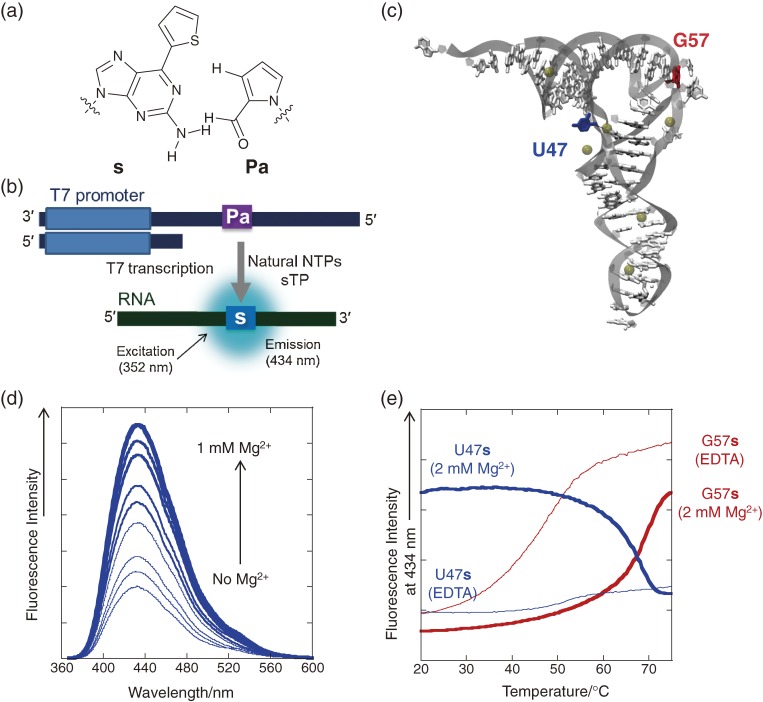
The comprehensive experiments fortunately revealed that the base pair between s and Pa functioned well in the site-specific incorporation of sTP into RNA opposite Pa, with higher efficiency than that of the s–z pair, in T7 transcription (Figs. 11a and 11b).81) Despite the lower shape complementarity of the s–Pa pair, relative to the s–z pair, the incorporation efficiency of sTP opposite Pa was higher than that opposite z in templates. This might be due to the higher hydrophobicity of Pa than that of z. Although the s–Pa pair cannot be used in replication, this finding led to a new idea for creating an unnatural base pair. Furthermore, since the s base is fluorescent, the efficient, site-specific incorporation of the s base into RNA provides a new tool for the local structural analysis of functional RNA molecules. The s base is an analog of the well-known fluorescent 2-aminopurine base, and the ribonucleoside of s exhibits fluorescence emission centered at 434 nm by excitation at 352 nm in phosphate buffer (pH 7).59) The fluorescent intensity of the s base in RNA molecules varies, depending on its structural environment. The s fluorescence is reduced by quenching when the s base stacks with neighboring bases in a tertiary RNA structure. Thus, the site-specific s labeling can be used as a probe for analyzing the local structural features of RNA molecules and their intra- or intermolecular interactions. We demonstrated the site-specific s labeling of an RNA hairpin and tRNA molecules by T7 transcription mediated by the s–Pa pair (Figs. 11c–11e). By introducing the s base at several positions within the tRNA, the magnesium ion dependency and the thermal stability of the L-shape structure formation were examined by the changes in the fluorescence of the s base at each position.81,82)
Figure 11.
Site-specific fluorescent labeling of RNA molecules using the unnatural s–Pa pair. (a) The structure of the s–Pa pair. (b) Scheme for the site-specific incorporation of the fluorescent s base (excitation: 352 nm; emission: 434 nm) into RNA opposite Pa in the DNA template by T7 transcription. (c) The tertiary structure of yeast tRNAPhe. The positions substituted with s, U47 and G57, are indicated in blue and red, respectively. The yellow sphere represents Mg2+. (d) The fluorescent spectra of tRNA containing s at position 47 at different MgCl2 concentrations with excitation at 352 nm. (e) The fluorescent intensity profiles of tRNA containing s at position 47 (blue) or 57 (red), in the presence of 0.1 mM EDTA (thin line) and 2 mM MgCl2 (bold line), at different temperatures.
2. Hydrophobic, unnatural base pairs functioning in PCR.
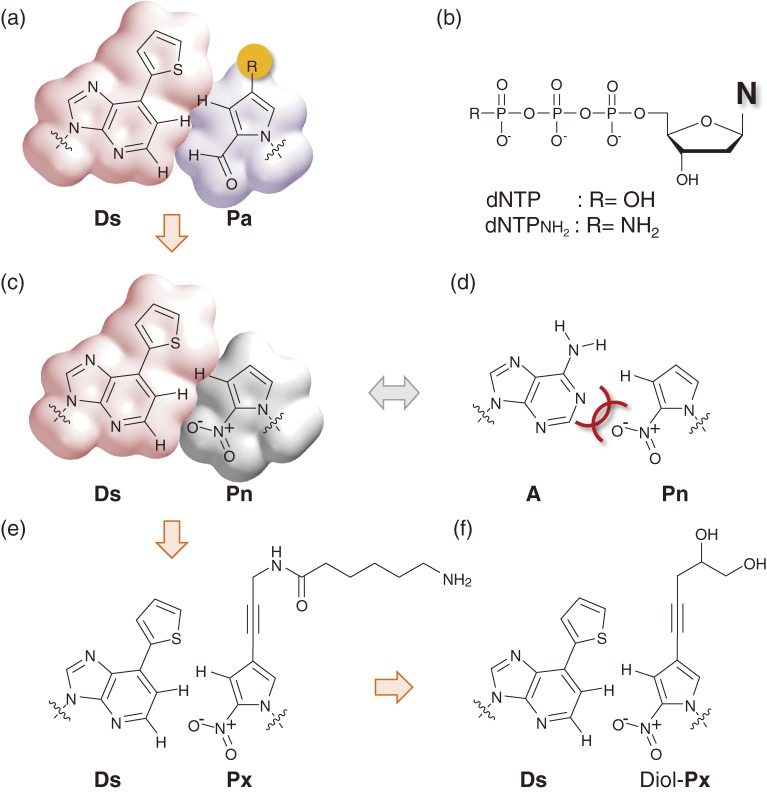
More importantly, the s–Pa pair gave us a new idea for creating further unnatural base pairs. The problem with using it in replication was that the hydrogen-bonding residues and atoms in the s base attract one of the natural bases. Therefore, we replaced the 1-nitrogen of s with C-H and removed the 2-amino group, and designed the hydrophobic base, 7-(2-thienyl)imidazo[4,5-b]pyridine (Ds), as a new pairing partner of Pa (Fig. 12a).34) This Ds–Pa pair was also derived by improving the Q–Pa pair. To prevent the Q–T mispair, the methyl group of Q was replaced with a thienyl group, like that of the s base. We then chemically synthesized the substrates and amidite of Ds. However, at that point, we missed the disturbing possibility of the Ds–Ds self-pairing in replication, like the PICS–PICS and Q–Q pairings. Unfortunately, it was true that the efficiency of the Ds incorporation opposite Ds was as high as that opposite Pa in replication using KF, and after the Ds incorporation opposite Ds, the extension was paused. This was a difficult problem specific to hydrophobic, unnatural base pairs. To address the same problem with the PICS–PICS pair, Romesberg’s group engineered DNA polymerases by an evolutional technique, to develop mutated polymerases capable of continuous extension after the PICS–PICS pairing.83,84)
Figure 12.
Hydrophobic, unnatural base pairs designed by our group. (a) The unnatural Ds–Pa pair. (b) The usual triphosphate (R=OH) and γ-amidotriphosphate (R=NH2) used as substrates in the unnatural Ds–Pa pair system for PCR. (c) The unnatural Ds–Pn pair. (d) The non-cognate A–Pn pair. (e) The unnatural Ds–Px pair. (f) The unnatural Ds–Diol-Px pair.
Serendipitously, we solved this Ds–Ds self-pairing problem in a different way from Romesberg’s approach. We accidentally synthesized a modified Ds substrate, the γ-amidotriphosphate of Ds (dDsTPNH2, Fig. 12b), instead of the usual Ds triphosphate substrate by modifying the synthetic route, as described below. We tested this unique substrate out of curiosity,34) and thereby found that DNA polymerases sensitively recognize dDsTPNH2 as a pairing partner of Pa, but not of Ds, in templates. In single-nucleotide insertion experiments using KF, the incorporation efficiency of dDsTPNH2 opposite Ds (Vmax/KM = 9.9 × 103) was lower than those of both dDsTP opposite Ds (Vmax/KM = 2.0 × 105) and dDsTPNH2 opposite Pa (Vmax/KM = 6.7 × 104). Furthermore, we found that this γ-amidotriphosphate effect has broad utility. Thus, we also employed the γ-amidotriphosphate of A (dATPNH2), because the slight misincorporation of dATP opposite Pa is another problem. In 2006, we achieved an efficient genetic expansion PCR system involving the Ds–Pa pair with a combination of the usual (dPaTP, dGTP, dCTP and dTTP) and modified (dDsTPNH2 and dATPNH2) substrates.34)
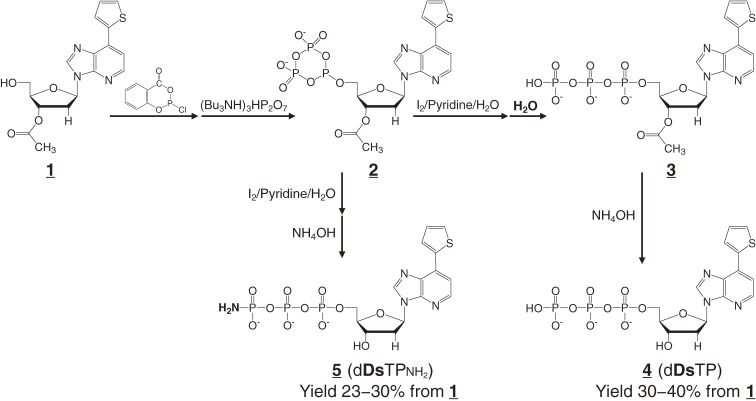
The γ-amidotriphosphate derivatives were synthesized by a modification of a Eckstein’s standard method (Fig. 13). In the conventional method,45) 2′-deoxyribonucleoside 5′-triphosphates (compound 4, dDsTP in Fig. 13) are prepared from 3′-protected deoxyribonucleosides (compound 1) via 5′-dioxocyclotriphosphite derivatives (compound 2). Compound 2 is oxidized with iodine, and the resulting cyclic phosphates are usually hydrolyzed with water to yield their triphosphates (compound 3). The 3′-protected acyl group is then removed by hydrolysis with concentrated ammonia water. One day, one of our researchers omitted the water hydrolysis of the cyclic phosphates, and directly treated them with concentrated ammonia water, to simplify the triphosphate synthesis. However, this process primarily generated the γ-amidotriphosphate derivatives (compound 5, dDsTPNH2).
Figure 13.
Chemical syntheses of the usual 2′-deoxyribonucleoside triphosphate of Ds (dDsTP, 4) and its γ-amidotriphosphate (dDsTPNH2, 5).
We still do not know the mechanism underlying the improved fidelity of the unnatural base pairing using the γ-amidotriphosphates. The use of γ-modified triphosphates, γ-P-aminonaphthalene-5-sulfonate triphosphates, also reportedly improves the fidelity of the natural base pairing in reverse transcription.85) In our study, the γ-amidotriphosphates also exhibited reduced incorporation efficiency even in the cognate base pairings, although the efficiency is still much higher than those of the non-cognate base pairings. Thus, DNA polymerases might sensitively recognize the γ-amidotriphosphate moiety of the substrates, and the geometrical fluctuation caused by non-cognate pairings, such as Ds–Ds and A–Pa, significantly reduces the interaction between the amino acid residues in the polymerase and the γ-amidotriphosphate moiety, resulting in the loss of the function of the amidodiphosphate part as a leaving group, like the pyrophosphate moiety of triphosphate substrates.
In the unique PCR system using the γ-amidotriphosphates of Ds and A, the Ds–Pa pair exhibited high selectivity (>99% per replication), and about 97% of the Ds–Pa pair was retained in the amplified DNA fragments after 20 cycles of PCR, using the exonuclease-proficient Vent DNA polymerase.34) The incorporation site and retention rate of the Ds–Pa pair in the amplified DNA fragments after PCR amplification were determined by two dideoxy-dye terminator cycle-sequencing methods in the presence and absence of dPa′TP (Pa′ = 4-propynylpyrrole-2-carbaldehyde, which is a modified Pa base, and the incorporation efficiency of dPa′TP opposite Ds is higher than that of dPaTP.) (Fig. 14).34) In the presence of dPa′TP (Method 1), the unnatural base position can be identified as a gap in the sequencing peak patterns, because we do not add the dideoxy-dye terminator corresponding to Pa′. In contrast, in the absence of dPa′TP (Method 2), there are no substrates for the incorporation opposite Ds in the sequencing reaction, and the polymerase pauses at the unnatural base position. Thus, the sequencing peaks disappear after the unnatural base position. Method 1 is useful for the confirmation of the unnatural base position in the DNA fragments, and Method 2 is used to determine the retention rate of the unnatural base pair in the amplified DNA fragments. If the misincorporation of the natural base substrates opposite unnatural bases occurs at the original unnatural base position, then the sequencing peak heights after the unnatural base position in Method 2 increase, in correlation with the misincorporation rates. Using Method 2, we developed a quantification method to determine the misincorporation rate of the natural bases at the Ds–Pa pair position, and assessed the selectivity (>99% per replication) of the Ds–Pa pairing in PCR using Vent DNA polymerase (exo+).
Figure 14.
Sequencing of DNA fragments containing Ds using the Ds–Pa′ pair. In Method 1, using Ds-containing DNA fragments supplemented with dPa′TP, the unnatural base position of DNA fragments is identified as a gap on the sequencing peak pattern. In Method 2, using Ds-containing DNA fragments in the absence of dPa′TP, all sequencing peaks after the unnatural base positions disappear, due to the termination of sequencing at the unnatural base position.
The hydrophobic Ds–Pa pair also functions complementarily in T7 transcription. Both DsTP and PaTP are site-specifically incorporated into RNA, opposite Pa and Ds, respectively, in templates, with more than 94% selectivities by T7 RNA polymerase.34,86,87) Furthermore, we chemically synthesized modified-Pa substrates, in which functional groups, such as biotin and fluorophores, can be attached to the Pa base via a propynyl linker, and these substrates are also site-specifically incorporated into RNA by T7 transcription. We compared the selectivity of the Ds–Pa pairing and the misincorporation of the unnatural base substrates opposite the natural bases in transcription with those of the natural A–U pairing, by using biotin-linked Pa and U substrates (Biotin-PaTP and Biotin-UTP). Although the selectivity of the Biotin-PaTP incorporation opposite Ds (90%) was lower than that of the Biotin-UTP incorporation opposite A (100%), the misincorporation rate of Biotin-PaTP opposite any natural bases (0.06%) was lower than that of Biotin-UTP opposite G, C, or U (0.14% per base).34) We recently chemically synthesized and tested various modified PaTPs for their site-specific incorporation in large RNA molecules by T7 transcription.86)
3. Successful unnatural base pair, Ds–Px.
The Ds–Pa pair is the first third base pair that selectively functions in PCR along with the natural base pairs. However, the application of the Ds–Pa pair system is still limited, because of the use of the γ-amidotriphosphates of Ds and A. The use of the modified substrates decreases the PCR amplification efficiency and restricts the range of in vivo applications. Therefore, we further improved the unnatural base pair system to bypass the need for the γ-amidotriphosphates, while maintaining the high selectivity (more than 99%) of the unnatural base pairing, in PCR. We first replaced the aldehyde group of Pa with a nitro group and synthesized 2-nitropyrrole (Pn) (Fig. 12c), to avoid the use of the γ-amidotriphosphate of A by preventing the A–Pa mispairing.35) The nitro group of Pn reduces the misincorporation of A opposite Pa, by the electrostatic repulsion between the oxygen of the nitro group and the 1-nitrogen of A (Fig. 12d). Next, we added a propynyl group to position 4 of the Pn base, and developed 4-[3-(6-aminohexanamido)-1-propynyl]-2-nitropyrrole (Px) as a pairing partner of Ds (Fig. 12e). Increasing the hydrophobicity and the stacking ability of the unnatural base strengthens the affinity for polymerases, allowing higher incorporation efficiency of dPxTP opposite Ds than those of dPaTP opposite Ds and dPnTP opposite Ds (Fig. 15), as well as dDsTP opposite Ds, in the single-nucleotide insertion experiments using KF. Thus, in 2009, we reported the efficient PCR amplification system with the expanded genetic alphabet, by introducing the Ds–Px pair.36)
In the absence of the γ-amidotriphosphates, DNA fragments containing the Ds–Px pair were amplified 107-fold by 30 cycles of PCR. In the amplified DNA fragments, more than 99% of the unnatural base pair was retained, and thus, the selectivity of the Ds–Px pair exceeded 99.7% per replication. In this PCR amplification, we employed DeepVent DNA polymerase with 3′-exonuclease activity. Detailed experiments revealed that the exonuclease activity of the polymerase is essential to eliminate the misincorporated unnatural bases in the strands.37) Recently, we performed 100-cycle PCR of DNA fragments containing the Ds–Px pair, to determine how much of the unnatural base survived in a huge amount of amplified DNA fragments. DNA amplification is saturated during 20–30 cycles of PCR in general, and thus, to maintain the exponential amplification throughout 100 cycles of PCR, the process of a 10-cycle PCR and the dilution of the PCR solution was repeated ten times. After 100 cycles of PCR using DeepVent DNA polymerase (exo+), the DNA fragment containing the Ds–Px pair was amplified 5 × 1027-fold, and more than 97% of the unnatural base pair was retained in the amplified DNA fragments. The selectivity of the Ds–Px pair reached 99.97% per replication in the PCR system.37)
To increase the versatility of the new biotechnology, we enhanced the sophistication of the PCR system involving the Ds–Px pair. Through our studies, we found that the efficiency and selectivity of the Ds–Px pair in PCR depend on the natural base sequence contexts around the unnatural bases.37) For example, DNA templates containing 3′-purine-Ds-purine-5′ sequences are less effective and selective, as compared to those containing 3′-pyrimidine-Ds-pyrimidine-5′ sequences. However, by fine-tuning the PCR conditions, we diminished the sequence dependency. Furthermore, we found that a certain modification of the side chain in the Px base reduced the misincorporation rate of the Px substrate opposite the natural bases in templates, without decreasing the incorporation efficiency and selectivity of the Ds and modified Px base pair. The fidelity of unnatural base pairing in the polymerase reaction is determined by both the selectivity of the unnatural base substrates opposite the unnatural pairing partner bases and the misincorporation rate of the unnatural base substrates opposite the natural bases. From extensive experiments with the Ds–Px pair, we finally developed an unnatural base pair between Ds and the dihydroxyethyl derivatives of Px (Diol-Px) (Fig. 12f), for which the selectivity is 99.77–99.92% per replication, depending on the sequence contexts, and the misincorporation rate of the unnatural base substrates is 0.005% per base per replication, in PCR amplification using DeepVent DNA polymerase (exo+).37) The intrinsic mispairing error rate among the natural bases of DeepVent DNA polymerase (exo+) is around 2 × 10−5 error per base pair, which corresponds to ∼0.002% per base per replication for the misincorporation rate of the natural bases (from New England BioLabs data). Therefore, the misincorporation rate of the Ds–Diol-Px pair is very close to that of the natural base pairs.
Summary and perspectives
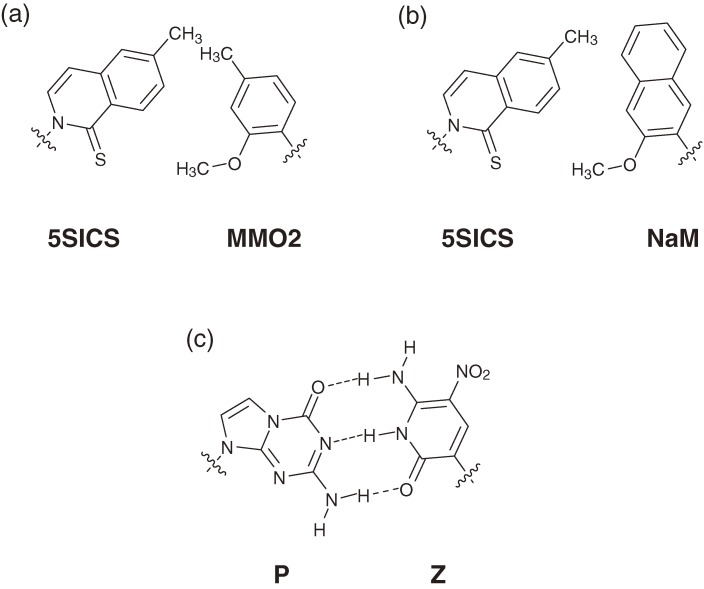
During the past fifteen years, we created a series of unnatural base pairs that can be used as a third base pair in replication, transcription, and/or translation. By repeating the ‘proof of concept’ experiments, the efficiency and selectivity of the unnatural base pairs in replication were gradually improved (Fig. 15). Finally, we developed the Ds–Px pair, which exhibits extremely high efficiency and selectivity in PCR. The selectivity of the Ds–modified-Px pairing is as high as 99.8–99.9% per replication, and the misincorporation rate of unnatural bases opposite the natural bases is 0.005% per base per replication. Recently, Benner’s and Romesberg’s groups have also developed unnatural base pairs for practical use in PCR amplification as a third base pair. In 2009, Romesberg’s group reported two types of hydrophobic base pairs (5SICS–MMO2 and 5SICS–NaM pairs) (Figs. 16a and 16b), by improving the shape complementarily of their initial PICS–PICS pair.32,33) The best selectivity of the 5SICS–NaM pairs reached 99.8% per replication in PCR. Benner’s group addressed the shortcomings of their initial isoG–isoC pair, and developed a hydrogen-bonded base pair between 2-aminoimidazo[1,2-a]-1,3,5-triazin-4(8H)-one (P) and 6-amino-5-nitro-2(1H)-pyridone (Z) (Fig. 16c).30,31) The selectivity and misincorporation rate of the P–Z pair in PCR are 99.8% per replication and 0.2% per base per replication, respectively. Representative of unnatural base pairs that function as a third base pair in replication, transcription, and/or translation are summarized in Table 1. These unnatural base pairs exhibit unique specificity and can be practically used for the site-specific incorporations of extra components into nucleic acids and/or proteins. Thus, over the past twenty years, researchers have made progress toward the possible artificial rebuilding of the central dogma by introducing a third base pair, at least in a test tube.
Figure 16.
Unnatural base pairs that function in PCR with more than 99% selectivity.
Table 1.
Unnatural base pairs that can be used as a third base pair in in vitro biology systems
| Unnatural base pair | Structure | Design concept | Functions | Remarks | ||
|---|---|---|---|---|---|---|
| Replication | Transcription | Translation | ||||
| isoG–isoC | 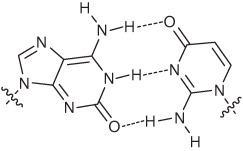 |
Hydrogen- bonding- pattern | Less selective PCR Selectivity: 93% per PCR cycle | Modified isoG incorporation | Peptide synthesis | Real-time PCR (Plexor) Weak points: Chemical instability of isoC Tautomerism of isoG |
| s–y | 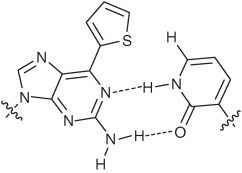 |
Hydrogen- bonding- pattern Steric hindrance | Less selective | Modified y incorporation | Protein synthesis coupled with transcription | Aptamer functionalization Weak points: Less selective for s-incorporation in replication and transcription |
| s–Pa | 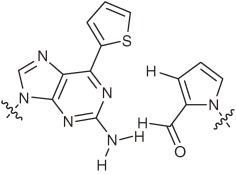 |
Non- hydrogen- bonded | Less selective | s incorporation | Not reported | Fluorescent s labeling of functional RNA molecules for local structural analysis Weak points: Only useful for s-labeling of RNA |
| Ds–Pa |  |
Hydrophobic Shape-fitting | PCR with γ-amido- triphosphates of Ds and A Selectivity: >99% per replication | Ds and modified Pa incorporation | Not reported | First unnatural base pair for PCR Weak points: Need γ-amidotriphosphates for PCR |
| Ds–Px |  |
Hydrophobic Shape-fitting | PCR Selectivity: 99.8–99.9% per replication Misincorporation rate: <0.01% per base per replication | Not reported | Not reported | Exhibits highest fidelity in PCR Weak points: Slight chemical instability of Px under basic conditions |
| 5SICS–NaM | 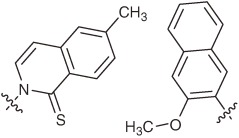 |
Hydrophobic | PCR Selectivity: 98.0–99.8% per replication | NaM and modified 5SICS incorporation | Not reported | High thermal stability |
| P–Z | 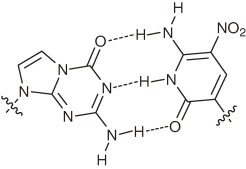 |
Hydrogen- bonding- pattern | PCR Selectivity: ∼99.8% per PCR cycle Misincorporation rate: ∼0.2% per base per PCR cycle | Not reported | Not reported | Consecutive multiple incorporation in PCR Weak points: Slight pH dependency of the pairing selectivity |
| Dss–Px |  |
Hydrophobic Shape-fitting | PCR Selectivity: >99.5% per replication | Dss incorporation (opposite Pa) | Not reported | Fluorescent (Dss) and quencher (Px) pair Weak points: Slight chemical instability of Px under basic conditions |
By using these unnatural base pair systems, a wide range of new applications for manipulating nucleic acids is anticipated. As shown here, any functional groups of interest can be attached to unnatural bases, allowing researchers to endow nucleic acids with desired new functionalities. In addition, some unnatural bases have unique properties themselves (e.g., the fluorescent s and hydrophobic Ds bases). Recently, we developed a strongly fluorescent unnatural base, 7-(2,2′-bithien-5-yl)-imidazo[4,5-b]pyridine (Dss) (Fig. 17a),88) by attaching an extra thienyl group to the Ds base. Similar to Ds, Dss retained the same shape complementarity toward the pyrrole base analogs of Pa, Pn, and Px, and these Dss–Pa, Dss–Pn, and Dss–Px pairs also functioned as a third base pair in replication and transcription. Furthermore, during the development, we found that the 2-nitropyrrole of Pn and Px functions as a fluorescence quencher (Figs. 17a and 17b).89) Therefore, the Dss–Pn and Dss–Px pairs are a third base pair between fluorophore and quencher base analogs. We applied these unique unnatural base pairs to function as a molecular beacon (Fig. 17c) and in real-time quantitative PCR to detect a target nucleic acid sequence.89,90) Hydrophobic unnatural bases are also useful, because the natural nucleic acids are relatively hydrophilic. We recently developed a method for generating DNA aptamers containing the hydrophobic Ds bases, which tightly bind to target proteins (unpublished data).
Figure 17.
The unnatural base pair between fluorophore and quencher analogs. (a) The unnatural Dss (fluorophore)–Pn/Px (quencher) pair. (b) Fluorescence quenching of Dss by Pn in double-stranded 12-mer DNA fragments. The fluorescence of the DNA fragments was detected upon irradiation at 365 nm. (c) Scheme for target DNA detection using molecular beacons containing the Dss–Pn pair. The Dss fluorescence of the molecular beacon was observed in the presence of target DNA upon irradiation at 365 nm.
Researchers now have several types of unnatural base pairs with the potential for use in in vitro systems (Table 1). The next goal is to apply unnatural base pairs to in vivo systems, by which the present genetic recombination techniques would be changed to a new genetic expansion technology. This new technology could provide safer containment technology than the present recombination technology. Since unnatural base pairs cannot be synthesized in a metabolic pathway, their nucleoside materials must be supplied as a nutrient from the outside, to maintain the artificial genes containing the unnatural base pairs in the cell. Thus, a cell lacking the nutrient cannot live. In the artificial cells, a specific gene and its expression can be traced by labeling with unnatural bases, and new proteins containing non-standard amino acids can be efficiently produced. In the in vivo system, mRNA and tRNA species containing unnatural bases could be transcribed from DNA templates and translated to proteins containing non-standard amino acids. By the translation system, we could generate unique proteins with increased functionality. To realize this, the unnatural base pair systems require further refinement. Codon and anticodon interactions are composed of only three base pairings, and thus the unnatural base pairings require thermal stabilities as high as those of the natural base pairs, for the development of an efficient translation system. In addition, we will need to learn more about the abilities and dynamics of unnatural base pairs in the cell, which in turn will provide the new synthetic biology tool of unnatural base pair systems, to elucidate molecular mechanisms in living organisms. We look forward to further advancements in this area of synthetic biology, toward next generation biotechnologies.
Acknowledgements
We thank all of our colleagues who contributed to our research described herein. We are especially grateful to Dr. S. Yokoyama, who always provided opportunities to proceed with the unnatural base studies. This work was supported by Grants-in-Aid for Scientific Research [KAKENHI 19201046 and 15350097 to I.H., 20710176 and 18710197 to M.K.] from the Ministry of Education, Culture, Sports, Science and Technology, the Targeted Proteins Research Program and the RIKEN Structural Genomics/Proteomics Initiative, the National Project on Protein Structural and Functional Analyses, Ministry of Education, Culture, Sports, Science and Technology of Japan.
Biographies
Profile
Ichiro Hirao was born in Shizuoka, Japan, in 1956. He graduated from Numazu National College of Technology in 1976, and received his B.S. (1978) degree from the Faculty of Engineering, Shizuoka University, and his M.S. (1980) and Ph.D. (1983) degrees in the chemical synthesis of 2′-5′ oligonucleotides and their structures from the Faculty of Science, Tokyo Institute of Technology. In 1984, he joined Dr. Kin-ichiro Miura’s laboratory in the Faculty of Engineering, The University of Tokyo, as a research associate, where he discovered extraordinarily thermostable DNA mini-hairpin structures. In 1992, he became an associate professor at Tokyo University of Pharmacy and Life Sciences. To expand his research areas from organic chemistry and structural biology to molecular biology and evolutional engineering, in 1995, he moved to Dr. Andrew D. Ellington’s laboratory in the Department of Chemistry, Indiana University. In 1997, to start the unnatural base pair studies, he returned to Japan and joined Dr. Shigeyuki Yokoyama’s project, ERATO, Japan Science and Technology Agency, as a group leader. In 2002, he continued his work as both a Professor at the Research Center for Advanced Science and Technology, The University of Tokyo and as a senior visiting scientist at the RIKEN Genomic Sciences Center. Since 2006, he has been managing the Nucleic Acid Synthetic Biology Research Team at the Systems and Structural Biology Center, RIKEN, as the team leader. In 2007, he founded the venture company ‘TagCyx Biotechnologies’ with Dr. Shigeyuki Yokoyama, to provide unnatural base pair technologies toward the expansion of the genetic alphabet of DNA.
Michiko Kimoto (Hirao) was born in Kanagawa, Japan, in 1974. She graduated from the University of Tokyo, Faculty of Science, Department of Biophysics and Biochemistry in 1997. In the laboratory of Prof. Shigeyuki Yokoyama, in the Department of Biophysics and Biochemistry, Graduate School of Science, The University of Tokyo, she worked on the generation of RNA aptamers capable of regulating protein–protein interactions, and the site-specific introduction of functional components into the aptamers by using an unnatural base pair transcription system, and received her Ph.D. in 2002. She became a research associate at the RIKEN Genomic Sciences Center, and joined Dr. Ichiro Hirao’s laboratory. She has been continuing her work on developing unnatural base pair systems that function in replication, transcription, and translation. She established methods to evaluate the fidelity of unnatural base pairs in replication and transcription, by taking advantage of the properties of the unnatural bases. She is currently a research scientist at RIKEN (2006–) and a visiting scientist at TagCyx Biotechnologies (2008–).
References
- 1).Watson J.D., Crick F.H. (1953) Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature 171, 737–738 [DOI] [PubMed] [Google Scholar]
- 2).Nirenberg M.W., Matthaei J.H. (1961) The dependence of cell-free protein synthesis in E. coli upon naturally occurring or synthetic polyribonucleotides. Proc. Natl. Acad. Sci. U.S.A. 47, 1588–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3).Nirenberg M., Leder P. (1964) RNA codewords and protein synthesis. The effect of trinucleotides upon the binding of sRNA to ribosomes. Science 145, 1399–1407 [DOI] [PubMed] [Google Scholar]
- 4).Nishimura S., Jones D.S., Khorana H.G. (1965) Studies on polynucleotides. 48. The in vitro synthesis of a co-polypeptide containing two amino acids in alternating sequence dependent upon a DNA-like polymer containing two nucleotides in alternating sequence. J. Mol. Biol. 13, 302–324 [DOI] [PubMed] [Google Scholar]
- 5).Rich, A. (1962) Problems of evolution and biochemical information transfer. In Horizons in Biochemistry (eds. Kasha, M. and Pullman, B.). Academic Press, New York, pp. 103–126. [Google Scholar]
- 6).Krueger A.T., Kool E.T. (2009) Redesigning the architecture of the base pair: toward biochemical and biological function of new genetic sets. Chem. Biol. 16, 242–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7).Szathmary E. (2003) Why are there four letters in the genetic alphabet? Nat. Rev. Genet. 4, 995–1001 [DOI] [PubMed] [Google Scholar]
- 8).Service R.F. (2000) Molecular biology. Creation’s seventh day. Science 289, 232–235 [DOI] [PubMed] [Google Scholar]
- 9).Benner S.A., Sismour A.M. (2005) Synthetic biology. Nat. Rev. Genet. 6, 533–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10).Hirao I., Kimoto M., Yamashige R. (2012) Natural versus artificial creation of base pairs in DNA: Origin of nucleobases from the perspectives of unnatural base pair studies. Acc. Chem. Res. (in press) [DOI] [PubMed] [Google Scholar]
- 11).Kimoto M., Cox R.S., 3rd, Hirao I. (2011) Unnatural base pair systems for sensing and diagnostic applications. Expert Rev. Mol. Diagn. 11, 321–331 [DOI] [PubMed] [Google Scholar]
- 12).Switzer C., Moroney S.E., Benner S.A. (1989) Enzymatic incorporation of a new base pair into DNA and RNA. J. Am. Chem. Soc. 111, 8322–8323 [Google Scholar]
- 13).Piccirilli J.A., Krauch T., Moroney S.E., Benner S.A. (1990) Enzymatic incorporation of a new base pair into DNA and RNA extends the genetic alphabet. Nature 343, 33–37 [DOI] [PubMed] [Google Scholar]
- 14).Bain J.D., Switzer C., Chamberlin A.R., Benner S.A. (1992) Ribosome-mediated incorporation of a non-standard amino acid into a peptide through expansion of the genetic code. Nature 356, 537–539 [DOI] [PubMed] [Google Scholar]
- 15).Sherrill C.B., Marshall D.J., Moser M.J., Larsen C.A., Daude-Snow L., Jurczyk S., Shapiro G., Prudent J.R. (2004) Nucleic acid analysis using an expanded genetic alphabet to quench fluorescence. J. Am. Chem. Soc. 126, 4550–4556 [DOI] [PubMed] [Google Scholar]
- 16).Switzer C.Y., Moroney S.E., Benner S.A. (1993) Enzymatic recognition of the base pair between isocytidine and isoguanosine. Biochemistry 32, 10489–10496 [DOI] [PubMed] [Google Scholar]
- 17).Horlacher J., Hottiger M., Podust V.N., Hubscher U., Benner S.A. (1995) Recognition by viral and cellular DNA polymerases of nucleosides bearing bases with nonstandard hydrogen bonding patterns. Proc. Natl. Acad. Sci. U.S.A. 92, 6329–6333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18).Benner S.A. (2004) Understanding nucleic acids using synthetic chemistry. Acc. Chem. Res. 37, 784–797 [DOI] [PubMed] [Google Scholar]
- 19).Martinot T.A., Benner S.A. (2004) Artificial genetic systems: exploiting the “aromaticity” formalism to improve the tautomeric ratio for isoguanosine derivatives. J. Org. Chem. 69, 3972–3975 [DOI] [PubMed] [Google Scholar]
- 20).Guckian K.M., Krugh T.R., Kool E.T. (1998) Solution structure of a DNA duplex containing a replicable difluorotoluene-adenine pair. Nat. Struct. Biol. 5, 954–959 [DOI] [PubMed] [Google Scholar]
- 21).Morales J.C., Kool E.T. (1998) Efficient replication between non-hydrogen-bonded nucleoside shape analogs. Nat. Struct. Biol. 5, 950–954 [DOI] [PubMed] [Google Scholar]
- 22).Khakshoor O., Wheeler S.E., Houk K.N., Kool E.T. (2012) Measurement and theory of hydrogen bonding contribution to isosteric DNA base pairs. J. Am. Chem. Soc. 134, 3154–3163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23).Kool E.T., Sintim H.O. (2006) The difluorotoluene debate – a decade later. Chem. Commun. (Camb.), 3665–3675 [DOI] [PubMed] [Google Scholar]
- 24).Zhang X., Lee I., Berdis A.J. (2005) The use of nonnatural nucleotides to probe the contributions of shape complementarity and π-electron surface area during DNA polymerization. Biochemistry 44, 13101–13110 [DOI] [PubMed] [Google Scholar]
- 25).Kincaid K., Beckman J., Zivkovic A., Halcomb R.L., Engels J.W., Kuchta R.D. (2005) Exploration of factors driving incorporation of unnatural dNTPS into DNA by Klenow fragment (DNA polymerase I) and DNA polymerase α. Nucleic Acids Res. 33, 2620–2628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26).Frey J.A., Leist R., Leutwyler S. (2006) Hydrogen bonding of the nucleobase mimic 2-pyridone to fluorobenzenes: an ab initio investigation. J. Phys. Chem. A 110, 4188–4195 [DOI] [PubMed] [Google Scholar]
- 27).Henry A.A., Romesberg F.E. (2003) Beyond A, C, G and T: augmenting nature’s alphabet. Curr. Opin. Chem. Biol. 7, 727–733 [DOI] [PubMed] [Google Scholar]
- 28).Hirao I. (2006) Unnatural base pair systems for DNA/RNA-based biotechnology. Curr. Opin. Chem. Biol. 10, 622–627 [DOI] [PubMed] [Google Scholar]
- 29).Bergstrom, D.E. (2009) Unnatural nucleosides with unusual base pairing properties. Curr. Protoc. Nucleic Acid Chem. Chapter 1, Unit 1.4. [DOI] [PubMed] [Google Scholar]
- 30).Yang Z., Sismour A.M., Sheng P., Puskar N.L., Benner S.A. (2007) Enzymatic incorporation of a third nucleobase pair. Nucleic Acids Res. 35, 4238–4249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31).Yang Z., Chen F., Alvarado J.B., Benner S.A. (2011) Amplification, mutation, and sequencing of a six-letter synthetic genetic system. J. Am. Chem. Soc. 133, 15105–15112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32).Malyshev D.A., Seo Y.J., Ordoukhanian P., Romesberg F.E. (2009) PCR with an expanded genetic alphabet. J. Am. Chem. Soc. 131, 14620–14621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33).Malyshev D.A., Pfaff D.A., Ippoliti S.I., Hwang G.T., Dwyer T.J., Romesberg F.E. (2010) Solution structure, mechanism of replication, and optimization of an unnatural base pair. Chem. Eur. J. 16, 12650–12659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34).Hirao I., Kimoto M., Mitsui T., Fujiwara T., Kawai R., Sato A., Harada Y., Yokoyama S. (2006) An unnatural hydrophobic base pair system: site-specific incorporation of nucleotide analogs into DNA and RNA. Nat. Methods 3, 729–735 [DOI] [PubMed] [Google Scholar]
- 35).Hirao I., Mitsui T., Kimoto M., Yokoyama S. (2007) An efficient unnatural base pair for PCR amplification. J. Am. Chem. Soc. 129, 15549–15555 [DOI] [PubMed] [Google Scholar]
- 36).Kimoto M., Kawai R., Mitsui T., Yokoyama S., Hirao I. (2009) An unnatural base pair system for efficient PCR amplification and functionalization of DNA molecules. Nucleic Acids Res. 37, e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37).Yamashige R., Kimoto M., Takezawa Y., Sato A., Mitsui T., Yokoyama S., Hirao I. (2012) Highly specific unnatural base pair systems as a third base pair for PCR amplification. Nucleic Acids Res. 40, 2793–2806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38).Hirao I., Kimoto M., Yamakage S., Ishikawa M., Kikuchi J., Yokoyama S. (2002) A unique unnatural base pair between a C analogue, pseudoisocytosine, and an A analogue, 6-methoxypurine, in replication. Bioorg. Med. Chem. Lett. 12, 1391–1393 [DOI] [PubMed] [Google Scholar]
- 39).Kiefer J.R., Mao C., Braman J.C., Beese L.S. (1998) Visualizing DNA replication in a catalytically active Bacillus DNA polymerase crystal. Nature 391, 304–307 [DOI] [PubMed] [Google Scholar]
- 40).Doublie S., Tabor S., Long A.M., Richardson C.C., Ellenberger T. (1998) Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 A resolution. Nature 391, 251–258 [DOI] [PubMed] [Google Scholar]
- 41).Guo M.J., Hildbrand S., Leumann C.J., McLaughlin L.W., Waring M.J. (1998) Inhibition of DNA polymerase reactions by pyrimidine nucleotide analogues lacking the 2-keto group. Nucleic Acids Res. 26, 1863–1869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42).Morales J.C., Kool E.T. (1999) Minor groove interactions between polymerase and DNA: more essential to replication than Watson-Crick hydrogen bonds? J. Am. Chem. Soc. 121, 2323–2324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43).Eom S.H., Wang J., Steitz T.A. (1996) Structure of Taq polymerase with DNA at the polymerase active site. Nature 382, 278–281 [DOI] [PubMed] [Google Scholar]
- 44).Tahirov T.H., Temiakov D., Anikin M., Patlan V., McAllister W.T., Vassylyev D.G., Yokoyama S. (2002) Structure of a T7 RNA polymerase elongation complex at 2.9 Å resolution. Nature 420, 43–50 [DOI] [PubMed] [Google Scholar]
- 45).Ludwig J., Eckstein F. (1989) Rapid and efficient synthesis of nucleoside 5′-O-(1-thiotriphosphates), 5′-triphosphates and 2′,3′-cyclophosphorothioates using 2-chloro-4H-1,3,2-benzodioxaphosphorin-4-one. J. Org. Chem. 54, 631–635 [Google Scholar]
- 46).Yoshikawa M., Kato T., Takenishi T. (1967) A novel method for phosphorylation of nucleosides to 5′-nucleotides. Tetrahedron Lett. 50, 5065–5068 [DOI] [PubMed] [Google Scholar]
- 47).Petruska J., Goodman M.F., Boosalis M.S., Sowers L.C., Cheong C., Tinoco I., Jr. (1988) Comparison between DNA melting thermodynamics and DNA polymerase fidelity. Proc. Natl. Acad. Sci. U.S.A. 85, 6252–6256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48).Goodman M.F., Creighton S., Bloom L.B., Petruska J. (1993) Biochemical basis of DNA replication fidelity. Crit. Rev. Biochem. Mol. Biol. 28, 83–126 [DOI] [PubMed] [Google Scholar]
- 49).Moran S., Ren R.X., Rumney S., Kool E.T. (1997) Difluorotoluene, a nonpolar isostere for thymine, codes specifically and efficiently for adenine in DNA replication. J. Am. Chem. Soc. 119, 2056–2057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50).Kimoto M., Yokoyama S., Hirao I. (2004) A quantitative, non-radioactive single-nucleotide insertion assay for analysis of DNA replication fidelity by using an automated DNA sequencer. Biotechnol. Lett. 26, 999–1005 [DOI] [PubMed] [Google Scholar]
- 51).Roberts C., Bandaru R., Switzer C. (1997) Theoretical and experimental study of isoguanine and isocytosine: Base pairing in an expanded genetic system. J. Am. Chem. Soc. 119, 4640–4649 [Google Scholar]
- 52).Sepiol J., Kazimierczuk Z., Shugar D. (1976) Tautomerism of isoguanosine and solvent-induced keto-enol equilibrium. Z. Naturforsch. C 31, 361–370 [DOI] [PubMed] [Google Scholar]
- 53).Maciejewska A.M., Lichota K.D., Kusmierek J.T. (2003) Neighbouring bases in template influence base-pairing of isoguanine. Biochem. J. 369, 611–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54).Ohtsuki T., Kimoto M., Ishikawa M., Mitsui T., Hirao I., Yokoyama S. (2001) Unnatural base pairs for specific transcription. Proc. Natl. Acad. Sci. U.S.A. 98, 4922–4925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55).Ishikawa M., Hirao I., Yokoyama S. (2000) Synthesis of 3-(2-deoxy-β-D-ribofuranosyl)pyridin-2-one and 2-amino-6-(N,N-dimethylamino)-9-(2-deoxy-β-D-ribofuranosyl)purine derivatives for an unnatural base pair. Tetrahedron Lett. 41, 3931–3934 [Google Scholar]
- 56).Fujiwara T., Kimoto M., Sugiyama H., Hirao I., Yokoyama S. (2001) Synthesis of 6-(2-thienyl)purine nucleoside derivatives that form unnatural base pairs with pyridin-2-one nucleosides. Bioorg. Med. Chem. Lett. 11, 2221–2223 [DOI] [PubMed] [Google Scholar]
- 57).Hirao I., Ohtsuki T., Fujiwara T., Mitsui T., Yokogawa T., Okuni T., Nakayama H., Takio K., Yabuki T., Kigawa T., Kodama K., Nishikawa K., Yokoyama S. (2002) An unnatural base pair for incorporating amino acid analogs into proteins. Nat. Biotechnol. 20, 177–182 [DOI] [PubMed] [Google Scholar]
- 58).Mitsui T., Kimoto M., Harada Y., Yokoyama S., Hirao I. (2005) An efficient unnatural base pair for a base-pair-expanded transcription system. J. Am. Chem. Soc. 127, 8652–8658 [DOI] [PubMed] [Google Scholar]
- 59).Mitsui T., Kimoto M., Kawai R., Yokoyama S., Hirao I. (2007) Characterization of fluorescent, unnatural base pairs. Tetrahedron 63, 3528–3537 [Google Scholar]
- 60).Ohno S., Yokogawa T., Nishikawa K. (2001) Changing the amino acid specificity of yeast tyrosyl-tRNA synthetase by genetic engineering. J. Biochem. 130, 417–423 [DOI] [PubMed] [Google Scholar]
- 61).Chatterji D., Gopal V. (1996) Fluorescence spectroscopy analysis of active and regulatory sites of RNA polymerase. Methods Enzymol. 274, 456–478 [DOI] [PubMed] [Google Scholar]
- 62).Qin P.Z., Pyle A.M. (1999) Site-specific labeling of RNA with fluorophores and other structural probes. Methods 18, 60–70 [DOI] [PubMed] [Google Scholar]
- 63).Huang F., Wang G., Coleman T., Li N. (2003) Synthesis of adenosine derivatives as transcription initiators and preparation of 5′ fluorescein- and biotin-labeled RNA through one-step in vitro transcription. RNA 9, 1562–1570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64).Draganescu A., Hodawadekar S.C., Gee K.R., Brenner C. (2000) Fhit-nucleotide specificity probed with novel fluorescent and fluorogenic substrates. J. Biol. Chem. 275, 4555–4560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65).Seetharaman S., Zivarts M., Sudarsan N., Breaker R.R. (2001) Immobilized RNA switches for the analysis of complex chemical and biological mixtures. Nat. Biotechnol. 19, 336–341 [DOI] [PubMed] [Google Scholar]
- 66).Zhang L., Sun L., Cui Z., Gottlieb R.L., Zhang B. (2001) 5′-Sulfhydryl-modified RNA: initiator synthesis, in vitro transcription, and enzymatic incorporation. Bioconjug. Chem. 12, 939–948 [DOI] [PubMed] [Google Scholar]
- 67).Kimoto M., Endo M., Mitsui T., Okuni T., Hirao I., Yokoyama S. (2004) Site-specific incorporation of a photo-crosslinking component into RNA by T7 transcription mediated by unnatural base pairs. Chem. Biol. 11, 47–55 [DOI] [PubMed] [Google Scholar]
- 68).Endo M., Mitsui T., Okuni T., Kimoto M., Hirao I., Yokoyama S. (2004) Unnatural base pairs mediate the site-specific incorporation of an unnatural hydrophobic component into RNA transcripts. Bioorg. Med. Chem. Lett. 14, 2593–2596 [DOI] [PubMed] [Google Scholar]
- 69).Kawai R., Kimoto M., Ikeda S., Mitsui T., Endo M., Yokoyama S., Hirao I. (2005) Site-specific fluorescent labeling of RNA molecules by specific transcription using unnatural base pairs. J. Am. Chem. Soc. 127, 17286–17295 [DOI] [PubMed] [Google Scholar]
- 70).Moriyama K., Kimoto M., Mitsui T., Yokoyama S., Hirao I. (2005) Site-specific biotinylation of RNA molecules by transcription using unnatural base pairs. Nucleic Acids Res. 33, e129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71).Hirao I. (2006) Placing extra components into RNA by specific transcription using unnatural base pair systems. Biotechniques 40, 711–715 [DOI] [PubMed] [Google Scholar]
- 72).Petach H., Gold L. (2002) Dimensionality is the issue: use of photoaptamers in protein microarrays. Curr. Opin. Biotechnol. 13, 309–314 [DOI] [PubMed] [Google Scholar]
- 73).Brody E.N., Willis M.C., Smith J.D., Jayasena S., Zichi D., Gold L. (1999) The use of aptamers in large arrays for molecular diagnostics. Mol. Diagn. 4, 381–388 [DOI] [PubMed] [Google Scholar]
- 74).Kimoto M., Shirouzu M., Mizutani S., Koide H., Kaziro Y., Hirao I., Yokoyama S. (2002) Anti-(Raf-1) RNA aptamers that inhibit Ras-induced Raf-1 activation. Eur. J. Biochem. 269, 697–704 [DOI] [PubMed] [Google Scholar]
- 75).Hirao I., Harada Y., Kimoto M., Mitsui T., Fujiwara T., Yokoyama S. (2004) A two-unnatural-base-pair system toward the expansion of the genetic code. J. Am. Chem. Soc. 126, 13298–13305 [DOI] [PubMed] [Google Scholar]
- 76).Kool E.T. (1998) Replication of non-hydrogen bonded bases by DNA polymerases: a mechanism for steric matching. Biopolymers 48, 3–17 [DOI] [PubMed] [Google Scholar]
- 77).Hirao I., Ohtsuki T., Mitsui T., Yokoyama S. (2000) Dual specificity of the pyrimidine analogue, 4-methylpyridin-2-one, in DNA replication. J. Am. Chem. Soc. 122, 6118–6119 [Google Scholar]
- 78).McMinn D.L., Ogawa A.K., Wu Y., Liu J., Schultz P.G., Romesberg F.E. (1999) Efforts toward expansion of the genetic alphabet: DNA polymerase recognition of a highly stable, self-pairing hydrophobic base. J. Am. Chem. Soc. 121, 11585–11586 [Google Scholar]
- 79).Mitsui T., Kitamura A., Kimoto M., To T., Sato A., Hirao I., Yokoyama S. (2003) An unnatural hydrophobic base pair with shape complementarity between pyrrole-2-carbaldehyde and 9-methylimidazo[(4,5)-b]pyridine. J. Am. Chem. Soc. 125, 5298–5307 [DOI] [PubMed] [Google Scholar]
- 80).Sismour A.M., Benner S.A. (2005) The use of thymidine analogs to improve the replication of an extra DNA base pair: a synthetic biological system. Nucleic Acids Res. 33, 5640–5646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81).Kimoto M., Mitsui T., Harada Y., Sato A., Yokoyama S., Hirao I. (2007) Fluorescent probing for RNA molecules by an unnatural base-pair system. Nucleic Acids Res. 35, 5360–5369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82).Hikida Y., Kimoto M., Yokoyama S., Hirao I. (2010) Site-specific fluorescent probing of RNA molecules by unnatural base-pair transcription for local structural conformation analysis. Nat. Protoc. 5, 1312–1323 [DOI] [PubMed] [Google Scholar]
- 83).Xia G., Chen L., Sera T., Fa M., Schultz P.G., Romesberg F.E. (2002) Directed evolution of novel polymerase activities: mutation of a DNA polymerase into an efficient RNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 99, 6597–6602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84).Fa M., Radeghieri A., Henry A.A., Romesberg F.E. (2004) Expanding the substrate repertoire of a DNA polymerase by directed evolution. J. Am. Chem. Soc. 126, 1748–1754 [DOI] [PubMed] [Google Scholar]
- 85).Mulder B.A., Anaya S., Yu P., Lee K.W., Nguyen A., Murphy J., Willson R., Briggs J.M., Gao X., Hardin S.H. (2005) Nucleotide modification at the γ-phosphate leads to the improved fidelity of HIV-1 reverse transcriptase. Nucleic Acids Res. 33, 4865–4873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86).Morohashi N., Kimoto M., Sato A., Kawai R., Hirao I. (2012) Site-specific incorporation of functional components into RNA by an unnatural base pair transcription system. Molecules 17, 2855–2876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87).Kimoto M., Hirao I. (2010) Site-specific incorporation of extra components into RNA by transcription using unnatural base pair systems. Methods Mol. Biol. 634, 355–369 [DOI] [PubMed] [Google Scholar]
- 88).Kimoto M., Mitsui T., Yokoyama S., Hirao I. (2010) A unique fluorescent base analogue for the expansion of the genetic alphabet. J. Am. Chem. Soc. 132, 4988–4989 [DOI] [PubMed] [Google Scholar]
- 89).Kimoto M., Mitsui T., Yamashige R., Sato A., Yokoyama S., Hirao I. (2010) A new unnatural base pair system between fluorophore and quencher base analogues for nucleic acid-based imaging technology. J. Am. Chem. Soc. 132, 15418–15426 [DOI] [PubMed] [Google Scholar]
- 90).Yamashige R., Kimoto M., Mitsui T., Yokoyama S., Hirao I. (2011) Monitoring the site-specific incorporation of dual fluorophore-quencher base analogues for target DNA detection by an unnatural base pair system. Org. Biomol. Chem. 9, 7504–7509 [DOI] [PubMed] [Google Scholar]