Summary
Homeobox domain-containing transcription factors are important regulators of hematopoiesis. Here we report that increased levels of non-clustered H2.0-like homeobox (HLX) lead to loss of functional hematopoietic stem cells and formation of aberrant progenitors with unlimited serial clonogenicity and blocked differentiation. Inhibition of HLX reduces proliferation and clonogenicity of leukemia cells, overcomes the differentiation block, and leads to prolonged survival. HLX regulates a transcriptional program, including PAK1 and BTG1, that controls cellular differentiation and proliferation. HLX is overexpressed in 87% of patients with acute myeloid leukemia (AML) and independently correlates with inferior overall survival (N=601, p=2.3×10−6). Our study identifies HLX as a key regulator in immature hematopoietic and leukemia cells, and a prognostic marker and therapeutic target in AML.
Introduction
Transcription factors are critical for the regulation of normal hematopoiesis as well as leukemogenesis (Friedman, 2007; Laiosa, Stadtfeld and Graf, 2006; Tenen, 2003). Several members of the Hox (class I homeobox genes) family of transcription factors, which contain a conserved homeobox domain and are organized into 4 major gene clusters in humans, have been implicated in the functioning of hematopoietic stem and progenitor cells (HSPC) as well as in leukemic transformation and the generation of leukemia-initiating cells (Argiropoulos and Humphries, 2007; Krumlauf, 1994; Sitwala, Dandekar and Hess, 2008). Less is known about the role of non-clustered (class II) homeobox genes in hematopoiesis and leukemia. Members of the CDX family, for instance, have been found to be overexpressed in acute leukemias and to regulate Hox gene expression (Bansal, Scholl, et al, 2006; Scholl, Bansal, et al, 2007). Transcriptional analysis of purified stem and progenitor populations has recently been utilized as a powerful tool to identify critical regulators of stem and progenitor cell function and transformation to leukemia-initiating cells (Krivtsov, Twomey, et al, 2006; Majeti, Becker, et al, 2009; Passegué, Wagner and Weissman, 2004; Saito, Kitamura, et al, 2010; Somervaille and Cleary, 2006; Steidl, Rosenbauer, et al, 2006; Steidl, Steidl, et al, 2007). Our analysis of pre-leukemic HSPC in a murine model of AML revealed the non-clustered H2.0-like homeobox (Hlx) gene to be 4-fold upregulated compared to wild-type (WT) HSPC ((Steidl, Rosenbauer, et al, 2006); and data not shown), suggesting that Hlx may be involved in malignant transformation. HLX is the highly conserved human/murine homologue of the homeobox gene H2.0, which shows tissue-specific expression throughout development in Drosophila melanogaster (Allen, Lints, et al, 1991; Hentsch, Lyons, et al, 1996). Additional studies two decades ago detected HLX expression in hematopoietic progenitors and in leukemic blasts of patients with AML, and a study of HLX-deficient fetal liver cells suggested a decrease of colony-formation capacity (Deguchi and Kehrl, 1991; Deguchi, Kirschenbaum and Kehrl, 1992). However, the precise function of HLX in HSPC and its role in leukemia have not been studied, which was the objective of the present study. AML is a heterogeneous disease with overall poor clinical outcome (Marcucci, Haferlach and Dohner, 2011). Less than one third of patients with AML achieve durable remission with current treatment regimens. Furthermore, prognostication and risk stratification of individual patients remains very challenging, in particular in favorable and standard risk groups. New targets need to be identified for effective and individualized therapeutic intervention.
Results
HLX overexpression impairs hematopoietic reconstitution and leads to a decrease in long-term hematopoietic stem cells and persistence of a small progenitor population
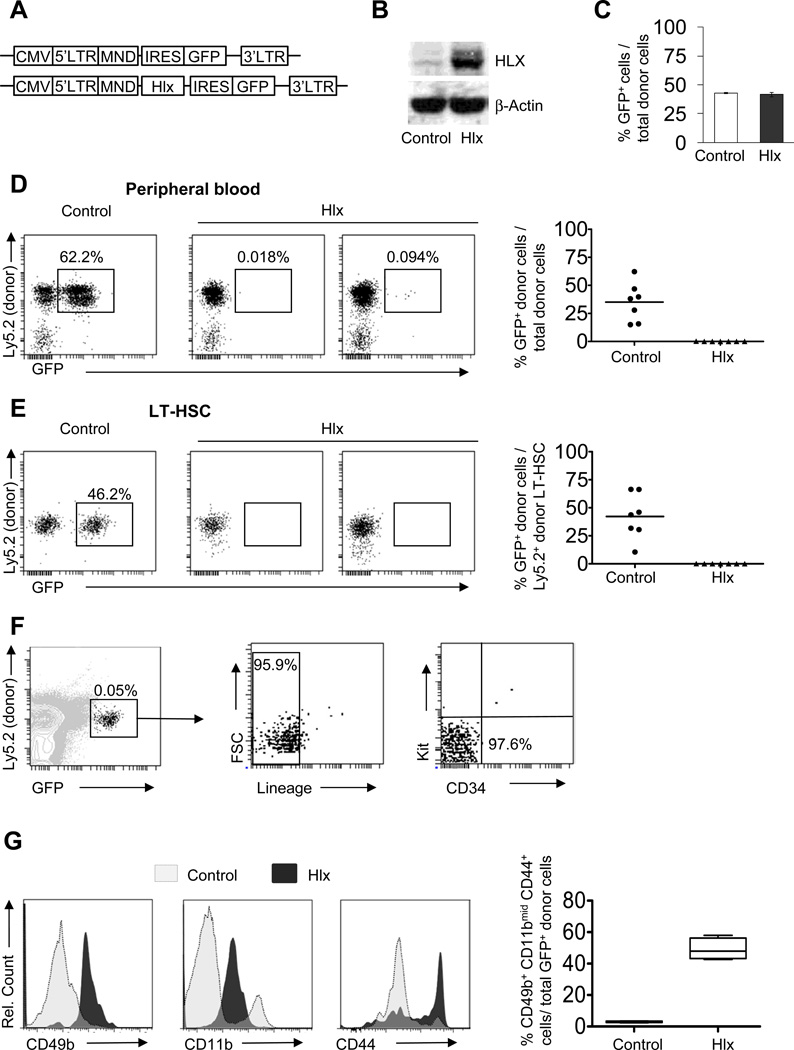
To examine the functional consequences of elevated HLX levels on hematopoiesis, we sorted lineage-negative (Lin−), Kit+ bone marrow (BM) cells from Ly5.2(CD45.2)+ WT mice, transduced them with a lentivirus expressing HLX and GFP, or GFP alone as a control (Fig. 1A+B), and transplanted them into lethally irradiated congenic Ly5.1(CD45.1)+ recipient mice. Transduction efficiency of control lentivirus and Hlx lentivirus was comparable, with both at approximately 50% (Fig. S1A). Twenty-four hours post-transplantation, both control and HLX-overexpressing GFP+ Ly5.2+ donor cells were detected in the BM at similar frequencies (42.8% and 41.6%, respectively) (Fig. 1C), indicating equal homing of the transplanted cells. Twelve weeks after transplantation, we evaluated hematopoietic multilineage reconstitution in the peripheral blood. Both groups engrafted robustly with an average donor chimerism of Ly5.2 cells of 80% (SD: 10%) and 85% (SD: 9%) in the control and Hlx groups, respectively. However, while mice transplanted with control cells showed 35% (SD: 17%) GFP+ cells in the peripheral blood 12 weeks after transplantation, mice transplanted with Hlx-transduced cells displayed drastically less GFP+ cells with only 0.07% (SD: 0.06%), demonstrating a severe defect of HLX-overexpressing cells in hematopoietic reconstitution (Fig. 1D). To determine the cellular compartments in which HLX was effective, we analyzed stem and progenitor cells in recipient BM. No GFP+ long-term hematopoietic stem cells (LT-HSC; Thy1loFlk2−LSK (Lin−Sca1+Kit+)) in mice transplanted with HLX-expressing cells were detected, while control mice displayed on average 42% (SD: 20%) GFP+ LT-HSC (Fig. 1E). Furthermore, in contrast to control animals, we could not find any GFP+ HLX-expressing short-term HSC (ST-HSCs; Thy1loFlk2+LSK), multipotent progenitors (MPP; Thy1− Flk2+LSK), common myeloid progenitors (CMP; Lin−Kit+Sca-1−FcγRII/IIIloCD34lo), granulocyte/monocyte progenitors (GMP; Lin−Kit+Sca-1−FcγRII/III+CD34+) or megakaryocyte/erythrocyte progenitors (MEP; Lin−Kit+Sca-1−FcγRII/III−CD34−), indicating that HLX acts at the level of the earliest hematopoietic stem cells (Fig. S1B). When we analyzed Hlx-GFP-transduced Lin−Kit+ (KL) cells by AnnexinV/DAPI staining, both control and HLX-overexpressing cells displayed the same low percentage of apoptotic/necrotic cells (Fig. S1C), indicating that HLX acts by a mechanism other than induction of apoptosis or necrosis. Further analysis for donor-derived cell populations persisting upon HLX overexpression revealed a small population of GFP+, CD45.2(Ly.5.2)+, Lin−, CD34−, and Kit− cells which were still present in the BM 12 weeks after transplantation (Fig. 1F). Analysis for additional surface markers revealed that these cells were characterized by intermediate expression of CD11b, as well as high expression of CD49b and CD44 (Fig. 1G, left panel), which is consistent with the surface phenotype of myeloid precursor cells slightly past the GMP stage (Novershtern, Subramanian, et al, 2011). This CD45+CD11bmidCD49b+CD44+ cell population was 16-fold expanded upon HLX expression in comparison to control (p=1.1×10−5) (Fig. 1G, right panel).
Figure 1. HLX overexpression impairs hematopoietic reconstitution and leads to a decrease in long-term hematopoietic stem cells and persistence of a small progenitor population.
(A) Schematics of lentiviral vectors. (B) Increased protein expression of HLX in Lin−Kit+ cells after transduction with HLX-expressing lentivirus and sorting of GFP+ cells. (C) Homing is not affected by HLX overexpression. 8×104 lentivirus-transduced Lin−Kit+ cells from WT C57BL/6 mice (Ly5.2) were transplanted into lethally irradiated congenic WT recipients (Ly5.1). Bone marrow mononuclear cells from recipients were analyzed 24 hours after transplantation. The frequency of GFP+ cells in the donor population (Ly5.1− Ly5.2+) was assessed and averages +/− SD are shown (N=3). (D, E) Control- or Hlx-IRES-GFP-transduced Lin−Kit+ cells (Ly5.2) together with spleen cells from congenic WT mice (Ly5.1) were transplanted into lethally irradiated congenic WT recipients (Ly5.1) (N=7) and analyzed 12 weeks after transplantation. Total GFP+ cells in peripheral blood (D), and Lin−Kit+Sca+Thy1loFlk2− LT-HSC in bone marrow (E) are shown. Detailed gating scheme and additional analyses are shown in Fig. S1. Means are indicated by horizontal lines in the panels on the right. (F) Analysis of GFP+ cells in total bone marrow cells from recipients transplanted with Hlx-transduced Lin−Kit+ cells after 12 weeks. Relative percentages of GFP+, Lin−, as well as CD34−Kit− donor cells are indicated. (G) CD49b, CD11b, and CD44 expression on donor Ly5.2+GFP+Lin−Kit− cells from recipients transplanted with Hlx- or control-transduced Lin−Kit+ cells 6 weeks after transplantation. Representative histogram plots are shown on the left. Percent of Ly5.2+GFP+Lin−Kit− cells that co-express CD49b+, CD11bmid, and CD44+ are displayed for the control and Hlx groups in the right panel (N=4/condition).
HLX confers unlimited serial clonogenicity to CD34−Kit− hematopoietic cells
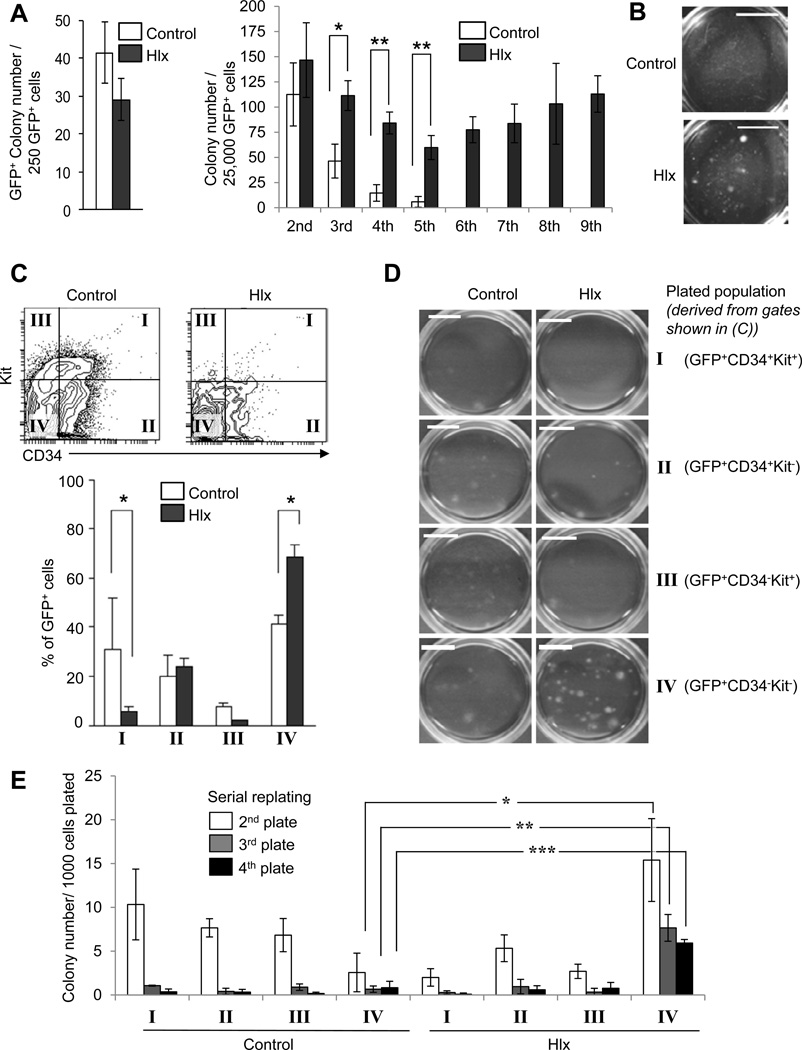
Next, we performed in vitro colony formation assays of transduced LSK cells. Hlx-transduced LSK cells formed slightly fewer and smaller colonies than control-transduced LSK cells in the initial plating (Fig. 2A, left panel, and Fig. S2A). To evaluate long-term clonogenic potential of HLX-overexpressing cells, we performed serial-replating assays. LSK cells overexpressing HLX showed greater clonogenic capacity in the 2nd to 5th plating in comparison to control-transduced cells, and maintained serial replating capacity through the 6th to 9th plating (Fig. 2A, right panel), showing a de facto immortalization of this clonogenic progenitor population by HLX. In addition, colonies were noticeably larger in size after five platings compared to control (Fig. 2B). Analysis of cells isolated from the initial plating revealed that HLX overexpression led to a decrease of Kit+ cells, similar to the in vivo phenotype, and an increased proportion of phenotypically more mature CD34−Kit− cells in comparison to control-transduced cells (Fig. 2C). To further characterize this persisting population, we examined a panel of cell surface markers. While the CD34−Kit− cells were negative for CD11c, CD25, FcγRII/III, CD61, CD115, and CD150 (Fig. S2B; and data not shown), they expressed CD49b and CD44, as well as intermediate levels of CD11b (Fig. S2B), similar to our observations in vivo (Fig. 1G). To determine which cellular subpopulation conferred the increased clonogenic capacity, we sorted equal numbers of CD34+Kit+ cells, CD34+Kit− cells, CD34− Kit+ cells, and CD34−Kit− cells from the first plating (populations I–IV, see Fig. 2C), and subjected each individual population to colony formation assays. Only CD34−Kit− cells derived from HLX-overexpressing cells formed a larger number of colonies in comparison to control cells, while all other populations did not display significant clonogenicity (Fig. 2D). Furthermore, the HLX-overexpressing GFP+CD34−Kit− cells showed serial replating capacity through 4 rounds, while all other populations exhausted significantly earlier (Fig. 2E). Finally, when we injected HLX-overexpressing GFP+CD34− Kit− cells from the fourth, sixth, or eighth plating into irradiated NOD-SCID-IL2Rgamma null (NSG) mice, GFP+ cells were detectable after 7 weeks in the peripheral blood (Fig. S2C; and data not shown). These data indicate that increased levels of HLX confer long-term clonogenicity to a population of CD34−Kit− cells.
Figure 2. HLX overexpression confers serial replating capacity to Lin−CD34−Kit− cells.
(A) Primary colony formation assay (left panel) and serial replating assay (right panel) of Lin−Kit+Sca1+ cells after transduction with control lentivirus or Hlx lentivirus. GFP+ colonies derived from control cells (white bars) and HLX-overexpressing cells (black bars) +/− SD are shown. Statistical significance is indicated (* p<0.05, and ** p<0.005, N=3). (B) Photograph of entire tissue culture dishes after 5th plating shows enlarged size of colonies derived from the Hlx-transduced cells. Scale bar indicates 1cm. (C) HLX overexpression leads to a decrease of CD34+Kit+ cells, and increases the CD34−Kit− population. Representative FACS plots are shown in the upper panel. The lower panel shows the frequency of each population within total GFP+ cells (I=CD34+Kit+, II=CD34+Kit−, III=CD34−Kit+, IV=CD34−Kit−). Means of control cells (white bars) and HLX-overexpressing cells (black bars) +/− SD are shown. Statistical significance is indicated (* p<0.05, N=3). (D) Whole plate photographs of colonies derived from sorted cells from each population (I, II, III, IV). Scale bars indicate 1cm. (E) Serial replating assay of each sorted population (I, II, III, IV). Colony numbers +/− SD after 2nd plating (white bars), 3rd plating (gray bars) and in 4th plating (black bars) are shown (N=3). Statistical significance is indicated (* p=0.013; ** p=0.0015; *** p=0.0004). Additional data on the aberrant clonogenic population is shown in Fig. S2.
HLX induces a myelomonocytic differentiation block
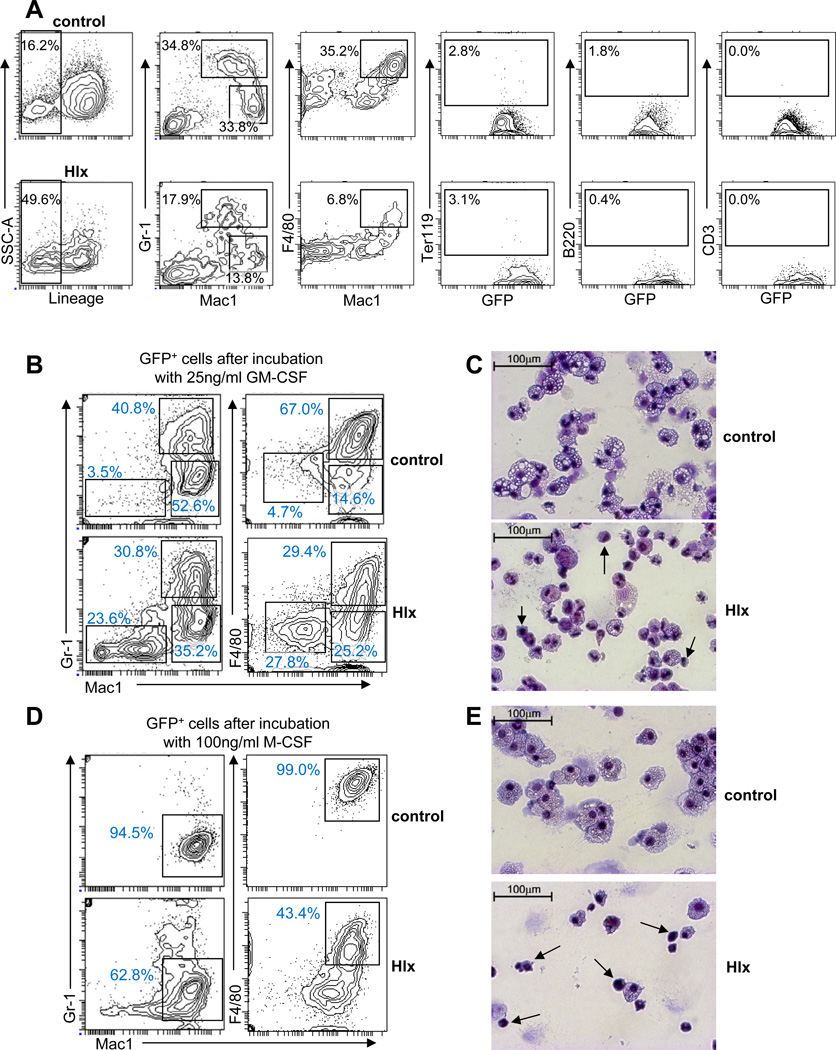
To investigate the effect of HLX overexpression on differentiation capacity, we analyzed the clonogenic GFP+CD34−Kit− cells from the first colony assay for the expression of additional cell surface markers. The proportions of Gr1+Mac1+ and Gr1−Mac1+, as well as F4/80+Mac1+ expressing cells were significantly reduced (Gr1+Mac1+: from 29.7% (control) (SD: 6.6%) to 16.2% (Hlx) (SD: 1.9%), p=0.026; Gr1−Mac1+: from 30.7% (SD: 4.5%) to 11.2% (SD: 2.6%), p=0.003; F4/80+Mac1+: from 31.6% (SD: 4.9%) to 8.9% (SD: 2.8%), p=0.002; N=3), indicative of a defect in myelomonocytic differentiation (Fig. 3A). Expression of erythroid, B-lymphoid, and T-lymphoid markers was unchanged. Furthermore, almost half of the HLX-overexpressing GFP+CD34−Kit− population (47.6%, (SD: 2.8%)) was lineage-negative, in contrast to only 17% (SD: 1.2%) of GFP+CD34−Kit− control-transduced cells (p=0.005) (Fig. 3A). Sorted, HLX-overexpressing GFP+Lin− CD34−Kit− cells also showed a significant increase in clonogenicity compared with control-transduced cells (Fig. S3), indicating that HLX acts at the level of Lin−CD34−Kit− cells. To specifically test myelomonocytic differentiation, we carried out colony-formation assays with GM-CSF or M-CSF stimulation (Fig. 3B–E). Hlx-transduced cells gave rise to lower numbers of Gr1−Mac1+ and F4/80+Mac1+ cells compared to control-transduced cells, upon either GM-CSF or M-CSF stimulation (Fig. 3B, D). Cytomorphological evaluation of cells after stimulation showed an increased percentage of Hlx-transduced cells with immature progenitor morphology, in stark contrast to control-transduced cells which predominantly displayed mature monocytic morphology (Fig. 3C, E). Taken together, our findings show that HLX not only enhances clonogenicity of an increased population of Lin−CD34−Kit− cells, but also confers a partial myelomonocytic differentiation block.
Figure 3. HLX induces a myelo-monocytic differentiation block.
(A) Representative FACS analysis of GFP+CD34−Kit− cells after the primary colony-forming assay. Relative percentages of cells in the indicated gates are given and show a reduction of mature myelomonocytic cells derived from the cells overexpressing HLX. (B) FACS analysis of cells derived from control-transduced or Hlx-transduced LSK cells after culture in methylcellulose with 25ng/ml GM-CSF. Relative percentages of cells are given for each gate and show a lower number of cells expressing mature myelomonocytic markers. (C) Representative morphology of cells from B. Cells with immature morphology can be found in the colonies derived from cells transduced with Hlx (indicated by arrows). (D) FACS analysis of cells derived from control-transduced or Hlx-transduced LSK cells after culture in methylcellulose with 100ng/ml M-CSF. (E) Representative morphology of cells from D, which confirm a monocytic differentiation block. Cells with immature morphology are indicated by arrows. See also Fig. S3 for additional data.
HLX downregulation inhibits AML
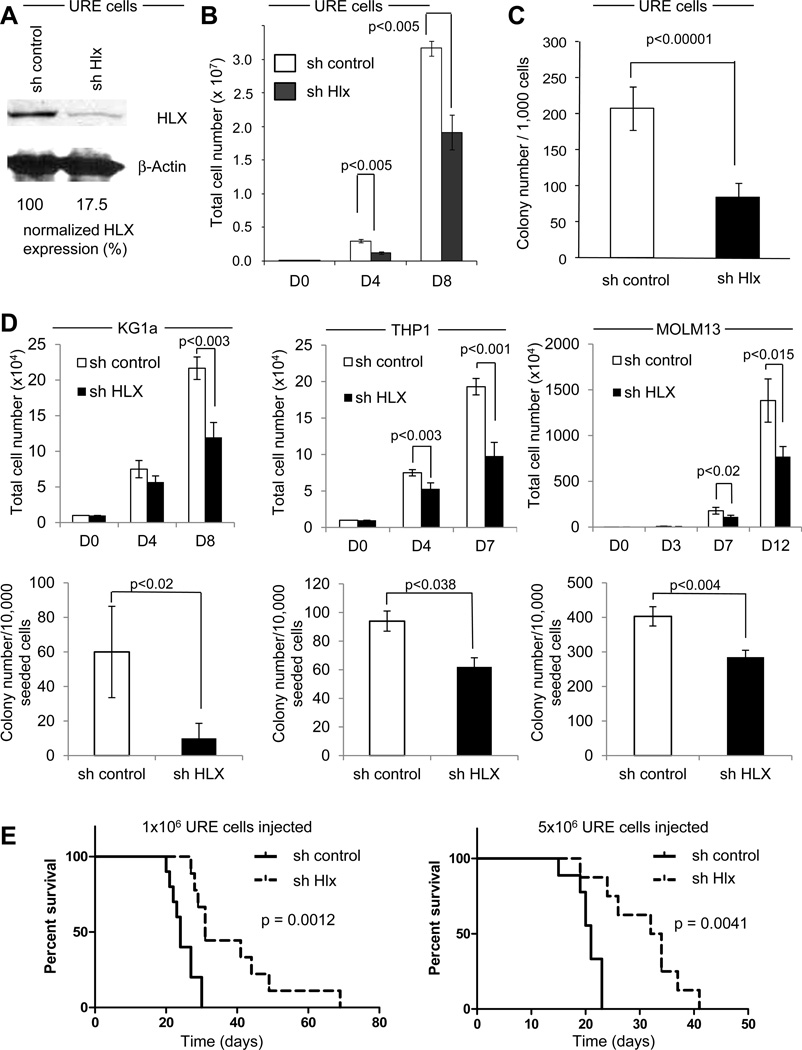
To test whether HLX overexpression is functionally important for AML, we targeted Hlx by RNA interference. We transduced leukemia cells derived from the PU.1 UREΔ/Δ AML model (URE cells) as well as human AML cells with lentiviral constructs expressing either Hlx-directed (sh Hlx) or control shRNA (sh control). Knockdown of HLX by 80% led to significantly reduced cell proliferation in suspension culture, as well as reduced formation of colonies of URE cells in methylcellulose assays in comparison to control cells (p<0.00001) (Fig. 4A, B, C). Likewise, human KG1a, THP1, and MOLM13 AML cells showed 40–60% reduced growth (p=0.003, p=0.001, and p=0.014, respectively) in suspension culture, as well as significantly diminished clonogenicity (p=0.02, p=0.038, and p=0.004, respectively) (Fig. 4D). To test the anti-leukemic effect of HLX suppression in vivo, we performed murine transplantation assays. We found that reduction of HLX levels in transplanted URE cells significantly prolonged recipient survival in comparison to mice transplanted with control shRNA-transduced cells (p=0.0012, and p=0.0041) (Fig. 4E). Taken together, our findings demonstrate that HLX is functionally important for AML cells, and that targeting HLX can inhibit cellular growth, decrease clonogenicity, and lead to improved survival in a murine AML transplantation model. Of note, Hlx heterozygous mice, which show a 50% reduction of HLX protein levels in BM cells, did not show any noticeable effects on normal hematopoiesis, including hematopoietic stem cell functions such as capacity for hematopoietic reconstitution and serial transplantability (Fig. S4).
Figure 4. HLX downregulation inhibits AML.
Lentiviruses expressing short hairpins directed against Hlx (sh Hlx) or a control (sh control) were used to downregulate HLX in mouse AML cells (panels A–C) and human AML cells (sh HLX) (panel D). (A) Western blotting shows a >80% reduction of HLX protein by shRNA. (B) Cell proliferation kinetics were determined by manual cell counts using trypan blue exclusion (N=3) in sh control cells (white bars) and sh Hlx cells (black bars). Different time points (days) are indicated. Error bars indicate SD (C) Clonogenic assay of URE cells treated with sh control or sh Hlx. 1,000 cells each were cultured in methylcellulose and GFP+ colonies were counted. Error bars indicate SD (N=3). (D) Cell proliferation kinetics in suspension culture (top panel) and clonogenicity (bottom panel) of human AML cells (KG1a, left panel; THP1, middle panel; MOLM13, right panel). Time points (days) are indicated. Error bars (SD) and statistical significance is indicated (t test, N=4). (E) Transplantation of URE cells transduced with sh control or sh Hlx into NSG mice. 1 million (left panel) or 5 million cells (right panel) were retroorbitally injected into NSG mice after sublethal irradiation (250cGy). Kaplan-Meier curves of overall survival of recipient mice are displayed. p values (log-rank) are indicated. See also Fig. S4 for additional data.
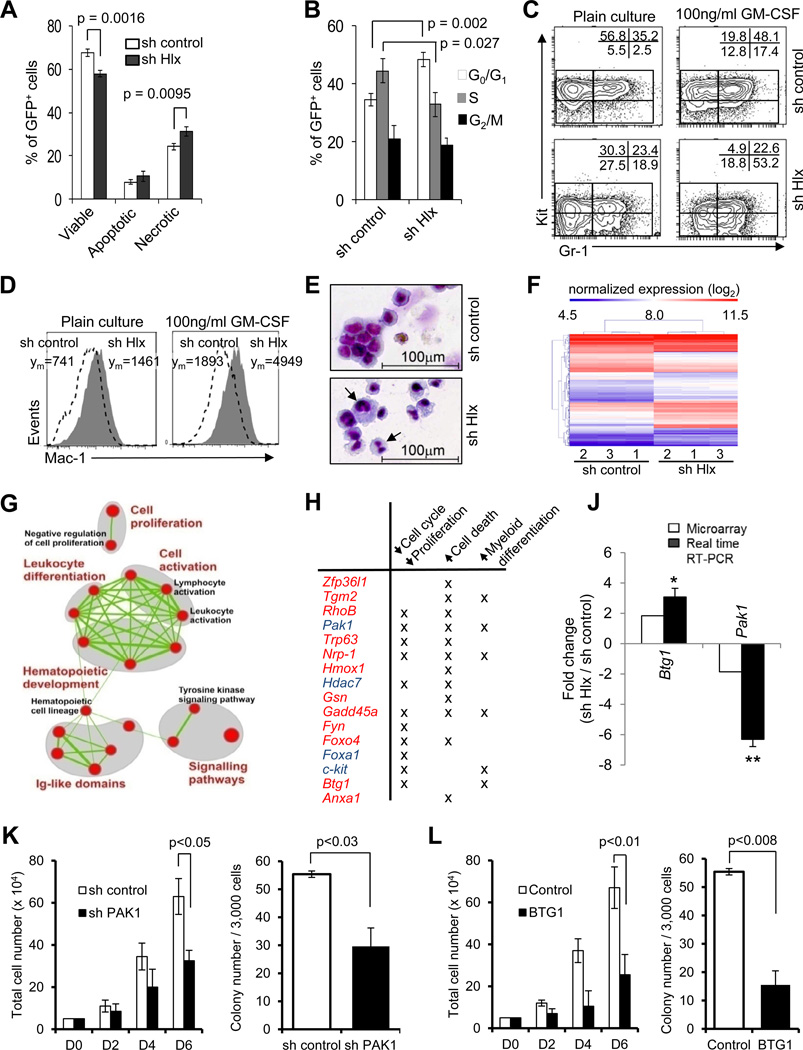
Inhibition of HLX leads to decreased cell cycling, increased cell death, and differentiation of AML cells, and causes significant changes in gene expression
To obtain insight into the mechanism of action of HLX inhibition we studied the cell biological and molecular consequences of knockdown of HLX in leukemia cells. We found that HLX downregulation in URE cells led to a decrease in viable cells, and an increase in necrotic cells (Fig. 5A). This was accompanied by a lower number of cells in S phase (p=0.027), and a higher number of cells in G0/G1 phase of cell cycle (p=0.002) (Fig. 5B). In addition, reduction of HLX led to an increased population of cells expressing lower levels of Kit and higher levels of Gr-1 and Mac1, indicative of myeloid differentiation (Fig. 5C, D). Stimulation with GM-CSF further increased the number of Mac1 and Gr-1 expressing cells, and led to increased differentiation of AML cells upon sh Hlx treatment in comparison to control-treated cells, which retained an immature, leukemic morphology (Fig. 5E). This observation shows that inhibition of HLX can overcome the myeloid differentiation block of AML cells.
Figure 5. Inhibition of HLX leads to decreased cell cycling, increased cell death, and differentiation of AML cells, and causes significant changes in gene expression.
(A) Relative percentages +/− SD of viable cells (DAPI-negative/Annexin V-negative), apoptotic cells (DAPI-negative/AnnexinV-positive) and necrotic cells (entire DAPI-positive) in sh control cells (white bars) and sh Hlx cells (black bars). p values are indicated (N=3). (B) Cell cycle status of sh control and sh Hlx leukemia cells. Percentages of cells in G0/G1 (white bars), S (gray bars), and G2/M (black bars) phase of cell cycle are displayed. Statistical significance is indicated (N=3). (C, D) Cell surface marker analysis after suspension culture for 3 days. Relative percentages of cells in the indicated gates are given, and show a decrease of immature Kit+ cells and an increase of Kit−Gr1+ cells (C), and an increase of Mac1+ cells (D). (E) Morphology of cells after treatment with 100ng/ml recombinant GM-CSF for 3 days. Cells with maturation signs are indicated by arrows. (F) Hierarchical clustering of genes differentially expressed in URE leukemia cells upon HLX knockdown. Expression levels are color-coded. (G) Enrichment map representation of cellular processes perturbed in leukemia cells upon HLX knockdown. Enriched gene sets are represented as nodes (red circles) connected by edges (green links) denoting the degree of gene set overlap. The node size is proportional to the number of genes in the gene set and the edge thickness represents the number of genes that overlap between gene sets. The color intensity of the nodes indicates the statistical significance of enrichment of a particular gene set. Groups of functionally related gene sets are circled in gray and labeled. A more comprehensive enrichment map is shown in Fig. S5. See Table S1 for additional information. (H) Select genes altered by HLX inhibition in URE leukemia cells. Up-regulated genes are shown in red and down-regulated genes are shown in blue. Their involvement in regulation of cell cycle/proliferation, cell death, and myeloid differentiation is indicated. (J) Validation of differential mRNA expression of Btg1 and Pak1. Fold changes +/− SD upon HLX knockdown are shown. (* p<0.05; ** p<0.01, N=3). Additional qRT-PCR validation is shown in Fig. S5. (K) Short hairpin (shPAK1) or a control (sh control) were used to downregulate HLX downstream target PAK1 in human AML cell line KG1a. Proliferation (left panel) and clonogenicity (right panel) are shown. Error bars indicate SD (N=4). (L) Effect of BTG1 overexpression on KG1a proliferation (left panel) and colony formation capacity (right panel). Error bars indicate SD (N=4). Quantification of PAK1 knockdown and BTG1 overexpression are shown in Fig. S5C and S5D.
To gain insight into the molecular effects caused by HLX inhibition, we measured gene expression profiles of sh Hlx-transduced URE cells and control shRNA-transduced cells. Leukemia cells treated with Hlx-directed shRNAs displayed an altered gene expression pattern with 392 genes being significantly differentially expressed (mean difference > 0.5 (log2), p<0.05) (Fig. 5F). Gene set enrichment analysis (GSEA) showed that “leukocyte differentiation”, “cell activation”, and “cell proliferation” were among the most significantly affected cellular functions (Fig. 5G), which is consistent with the leukemia-inhibitory effect of HLX reduction in URE cells. We specifically found that several key genes involved in the regulation of cell cycle and proliferation, cell death, and myeloid differentiation, were significantly changed upon HLX downregulation (Fig. 5H). We confirmed differential expression of several genes, namely Btg1, FoxO4, Fyn, Gadd45a, RhoB, Trp63, Zfp36l1, Hdac7, and Pak1 by qRT-PCR (Fig. 5J and Fig. S5A). We also found enrichment of genes involved in pathways of other cellular functions (Fig. S5B). Further, we utilized GSEA to compare the Hlx knockdown data with the molecular signatures database (MSigDB) (Mootha, Lindgren, et al, 2003; Subramanian, Tamayo, et al, 2005), and found negative enrichment of several known leukemia- and stem cell-related gene signatures (Table S1). Taken together, these data are consistent with a model that HLX overexpression leads to activation of a specific transcriptional program in leukemia cells which affects processes critical for leukemogenesis such as cell differentiation and proliferation, and which can at least partially be reversed by inhibition of HLX.
We focused on two downstream genes modulated upon HLX inhibition in AML cells, Pak1 and Btg1, which have previously been implicated in the regulation of cell cycle and malignant proliferation (Ong, Jubb, et al, 2011; Kuo, Duncavage, et al, 2003). Knockdown of PAK1 in KG1a AML cells led to a significant inhibition of cell proliferation (p<0.05) and clonogenicity (p<0.03) (Fig. 5K, Fig. S5C), mimicking the effects we had observed by knockdown of HLX. Similarly, ectopic expression of BTG1 led to a significant inhibition of cell growth (p<0.01) and colony forming capacity (p<0.008) of KG1a cells (Fig. 5L, Fig. S5D). These findings suggest that downregulation of PAK1 and upregulation of BTG1 are functionally relevant for mediating the leukemia-inhibitory effects of HLX knockdown.
HLX is overexpressed in patients with AML
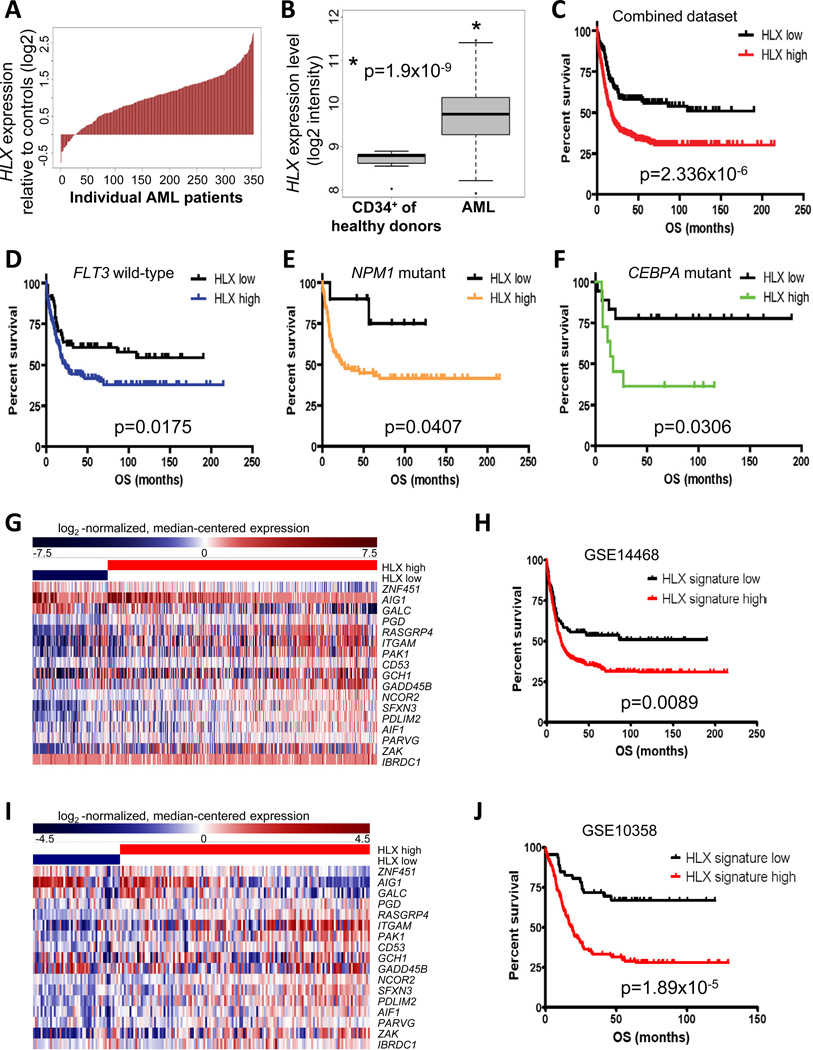
To examine whether HLX overexpression plays a role in human leukemia, we analyzed gene expression data of 354 patients with AML (Figueroa, Lugthart, et al, 2010; Wouters, Lowenberg, et al, 2009). HLX was overexpressed in 87% of patients with AML in comparison to CD34+ cells of healthy donors (Fig. 6A). On average, HLX expression was 2.03-fold higher in AML patients (p=1.9×10−9; Fig. 6B). 54% (190 out of 354) of patients with AML overexpressed HLX more than 2-fold, with the range extending up to 6.8-fold overexpression. These results demonstrate that HLX overexpression is a common feature in patients with AML. HLX overexpression was found across different French-American-British (FAB) classification subsets of AML (Table S2). Of note, high HLX expression was significantly more frequent in AML with myelomonocytic (M4) and monoblastic (M5) morphology, which is consistent with the observed myelomonocytic differentiation block caused by HLX overexpression in vitro.
Figure 6. HLX is overexpressed in patients with AML and correlates with poor overall survival.
(A) Waterfall plot of relative expression (log2) of HLX of 354 patients with AML in comparison to CD34-enriched bone marrow cells from 11 healthy donors. (B) Box plot summary of the HLX expression data shown in A. HLX expression is significantly higher in patients with AML in comparison to CD34+ cells from healthy donors (p=1.9×10−9). The median expression values (bold lines), 25th and 75th percentile (bottom and top of box), and the minimum and maximum (lower and upper whiskers) of both groups are shown. See Table S2 for additional information. (C) Kaplan-Meier survival plots of overall survival (OS) of patients with high versus low HLX expression in a combined dataset of 601 patients with AML (GSE10358, GSE12417 (U133plus2.0) and GSE14468). Patients with high expression of HLX show drastically inferior clinical outcome. The p value (log-rank test) is indicated. Kaplan-Meier plots for all individual datasets are shown in Fig. S6A–F. (D, E, F) Kaplan-Meier survival plots comparing overall survival (OS) of patients with high versus low HLX expression in molecularly defined subsets of AML. High expression of HLX is associated with significantly inferior clinical outcome in all subsets. p values (log-rank test) are given. (D) AML patients with no detectable mutations of the FLT3 gene. (E) AML patients with mutant NPM1. (F) AML patients with mutant CEBPA. (G) Heat map of log2-normalized, median-centered gene expression of a 17-gene “HLX signature” in AML patients of the GSE14468 cohort. HLX expression status is indicated. Relative gene expression is color-coded (red: high, blue: low). (H) Kaplan-Meier plot comparing overall survival (OS) of “HLX signature high” versus “HLX signature low” AML patients of the GSE14468 cohort. The p value (log-rank) is indicated. (I) Heat map of log2-normalized, median-centered gene expression of the “HLX signature” in AML patients of the GSE10358 cohort. (J) Kaplan-Meier survival plot comparing overall survival (OS) of “HLX signature high” versus “HLX signature low” AML patients of the GSE10358 cohort. The p value (log-rank) is indicated. See also Fig. S6 and Table S3 for additional information.
Increased HLX expression correlates with inferior survival
We further sought to examine if HLX expression levels in patients were associated with known clinical or molecular parameters. We analyzed 4 published datasets of patients with AML, of whom gene expression and time-to-event data were available (NCBI GEO accessions GSE10358, GSE12417 (U133A), GSE12417 (U133plus2), and GSE14468 (Metzeler, Hummel, et al, 2008; Tomasson, Xiang, et al, 2008; Wouters, Lowenberg, et al, 2009)). As the lower 25% of patients had HLX expression levels very similar to CD34+ cells of healthy donors (Fig. 6B), we decided to use the 25th percentile to dichotomize patients into “HLX high” and “HLX low” expressers. We compared the overall survival of AML patients with low versus high HLX, and observed that in each of the 4 different data sets, high HLX expression was associated with inferior overall survival (Fig. S6A–D). Overall survival (irrespective of HLX status) in datasets GSE12417 (U133plus2.0), GSE14468 and GSE10358 was very similar, with superimposable survival curves (p=0.4636, log-rank test; Fig. S6E), suggesting that these cohorts and their clinical outcomes could be combined for further analyses. The GSE12417 (U133A) dataset showed a worse overall survival outcome compared with the other datasets (Fig. S6F) and was therefore not included in the combined analysis. Consistent with the analyses of the individual datasets, the evaluation of the combined set of patients from the GSE10358, GSE12417 (U133plus2.0) and GSE14468 datasets (N=601 total) confirmed that high HLX levels are associated with inferior overall survival (p=2.336 × 10−6 (log-rank); hazard ratio (HR)=0.57 (95% CI: 0.046–0.71); median survival: 17.05 months (HLX high), “not reached” (HLX low); 5-yr survival rate: 32.95% (HLX high), 55.85% (HLX low)) (Fig. 6C). To assess whether the impact of HLX expression on overall survival is independent of known prognostic factors for AML, we performed multivariate analysis based on the data of the GSE14468 dataset (Figueroa, Lugthart, et al, 2010; Wouters, Lowenberg, et al, 2009), using a Cox regression model. In this analysis, high HLX status remained an independent prognostic factor (p=0.0416, HR 1.521) along with FLT3 mutation status (p=0.0003, HR 1.925), NPM1 mutation status (p=0.0006, HR 0.518), CEBPA mutation status (p=0.0371, HR 0.693), and cytogenetic risk group (p=0.0109, HR 1.382). Of note, the relative blast percentage in the bone marrow was not correlated with HLX expression or overall survival (Fig. S6G–I). The independent prognostic role of HLX status led us to hypothesize that it may provide additional prognostic information for patients who belong to previously established, molecularly defined subtypes of AML. We indeed observed that among patients with FLT3 WT, NPM1 mutations, or CEBPA mutations, high HLX expression was associated with inferior overall survival (p=0.0175, p=0.0407 and p=0.0306, respectively) (Fig. 6D–F).
An HLX-dependent transcriptional signature is functionally relevant in AML patients
To obtain insight into the molecular consequences of elevated HLX levels we overexpressed HLX in sorted murine LSK cells and performed genome-wide transcriptional analysis. We found that 195 genes were significantly changed, resulting in a clearly distinguishable expression signature induced by HLX overexpression (Fig. S6J). Using GSEA we found enrichment of known leukemia- and stem cell-related gene signatures, consistent with our previous findings (Table S3).
Next, we tested whether this mouse LSK HLX overexpression gene set correlated with HLX expression in human AML patients. Specifically, we compared the human orthologs of the mouse gene set to HLX expression levels of AML patients in the different cohorts using globaltest (Goeman, van de Geer, et al, 2004). We found a highly significant correlation between the mouse gene signature and HLX expression in human AML samples (p=7.43×10−23 for GSE14468, p=2.13×10−08 for GSE10358, p=2.31×10−06 for GSE12417 (U133plus2.0), and p=5.01×10−10 for GSE12417 (U133A)). Further, we intersected differentially expressed genes from the HLX overexpression or inhibition studies with analogously differentially expressed genes in “HLX high” versus “HLX low” patients of the GSE14468 data set, and analyzed these genes for association with survival. Thereby, we were able to define an HLX-dependent core set of 17 genes (referred to as “HLX signature”) correlating with HLX expression status in patients with AML (Fig. 6G). When we dichotomized patients into “HLX signature high” versus “HLX signature low” patients (defined by the genes of the signature, excluding HLX), we found that “HLX signature high” patients had significantly inferior overall survival (p=0.0089 (log-rank); hazard ratio (HR)=0.66 (95% CI: 0.48 to 0.90); median survival: 17.22 months for HLX signature high, “not reached” for HLX signature low; 5-yr survival rate: 34.5% for HLX signature high, 53.9% for HLX signature low) (Fig. 6H).
We subsequently validated the HLX signature in an independent cohort of patients, the GSE10358 data set, and found that the signature correlated strongly (p=7.8×10−11) with “HLX high” versus “HLX low” expression status in these AML patients as well (Fig. 6I). Furthermore, “HLX signature high” patients showed a strikingly inferior overall survival (p=1.89x10−05 (log-rank); hazard ratio (HR)=0.42 (95% CI: 0.28 to 0.62); median survival: 18.3 months (HLX signature high), “not reached” (HLX signature low); 5-yr survival rate: 29.0% (HLX signature high), 67.0% (HLX signature low)) (Fig. 6J). Taken together, these data suggest that elevated HLX levels cause a specific, functionally critical gene expression signature in human AML.
Interestingly, PAK1 was part of the HLX-induced prognostic signature. Given our finding that PAK1 mediates the leukemia-inhibitory effects of HLX knockdown in AML cells (Fig. 5K), we asked whether PAK1 expression levels alone may be functionally relevant in AML patients. We dichotomized AML patients into “PAK1 high” and “PAK1 low” expressers and analyzed clinical outcome. “PAK1 high” patients showed significantly inferior overall survival (p=0.00014 (log-rank)) than “PAK1 low” patients (median: 17.7 months (PAK1 high) vs. 109.1 months (PAK1 low); 5-yr survival rate: 34.0% (PAK1 high) vs. 50.5% (PAK1 low)) (Fig. S6K). Of note, high PAK1 expression was associated with inferior overall survival only in patients of the “HLX high” group (p=0.0005 (log-rank); hazard ratio (HR)= 0.62 (95% CI: 0.48 – 0.81)); median survival: 15.8 months (PAK1 high), 42.0 months (PAK1 low); 5-yr survival rate: 29.7% (PAK1 high), 48.1% (PAK1 low)), but not in “HLX low” patients (p=0.77 (log-rank); hazard ratio (HR)= 1.08 (95% CI: 0.65 – 1.78)); 5-yr survival rate: 55.0% (PAK1 high), 55.0% (PAK1 low)) (Fig. S6L,M). In addition, PAK1 expression levels were on average 1.5-fold higher (p=2.2×10−16) in “HLX high” patients compared to “HLX low” patients (Fig. S6N), and HLX and PAK1 expression levels in individual patients were also significantly correlated (p=8.8x10−15, R=0.31; Fig. S6O). Furthermore, experimental overexpression of HLX in LSK cells led to a significant increase in Pak1 mRNA expression (1.9-fold, p=0.017; Fig. S6P).
Discussion
Utilizing both mouse and human model systems we have shown that the class II homeobox protein HLX affects hematopoietic stem cell function, as well as clonogenicity and differentiation of immature hematopoietic progenitor cells. We found that HLX is significantly overexpressed in the majority of patients with AML, and that high HLX expression levels are independently associated with inferior clinical outcome. Thereby, our study identifies HLX as a class II homeobox gene which is critically involved in the pathogenesis of acute myeloid leukemia. Our finding that increased HLX expression correlates with more aggressive disease, combined with the observation that HLX knockdown results in an inhibition of growth and clonogenicity of leukemia cells further suggest that HLX may be a promising prognostic and therapeutic target. Future studies evaluating HLX in the context of different therapies will be required to determine if HLX also has predictive relevance.
Many clustered (class I, or HOX) homeobox genes have been implicated in normal hematopoiesis as well as leukemia, but much less is known about the role of non-clustered (class II) homeobox transcription factors (for review see (Argiropoulos and Humphries, 2007)). Several HOX genes are expressed at high levels in subtypes of AML (Alcalay, Tiacci, et al, 2005; Ayton and Cleary, 2003; Bullinger, Dohner, et al, 2004; Horton, Grier, et al, 2005). Important roles in leukemic transformation have been demonstrated specifically for several members of the HOX-A and the HOX-B cluster (Fischbach, Rozenfeld, et al, 2005; Krivtsov, Twomey, et al, 2006; Kroon, Krosl, et al, 1998; Sauvageau, Thorsteinsdottir, et al, 1997; Somervaille and Cleary, 2006; Thorsteinsdottir, Sauvageau, et al, 1997). Also, the non-clustered homeobox gene CDX2 was recently reported to be implicated in leukemogenesis (Scholl, Bansal, et al, 2007). However, the clinical significance of these known HOX genes is largely unclear. Here, we report that expression levels of a homeobox gene are strongly associated with inferior overall survival in several large, independent cohorts of patients with AML. Furthermore, the prognostic value of HLX is a broad phenomenon across several molecular subsets of patients, and HLX holds up as an independent prognostic factor in a multivariate model. Gene expression analyses upon experimental overexpression and inhibition of HLX demonstrated that HLX regulates the expression of a specific subset of genes, and that part of this “HLX signature” is also detectable in AML patients and discriminates between patients with poor and favorable clinical outcome. Taken together, these observations suggest that HLX is a key regulator of a gene subset critical for AML pathogenesis, and that it may define a previously unrecognized molecular subtype of AML with distinct biological features and clinical outcome.
We identified PAK1 and BTG1 as two genes downstream of HLX in myeloid cells, and show that PAK1 and BTG1 are functionally relevant in AML cells. Notably, PAK1 is upregulated in human AML patients with high HLX expression, and it is also part of the HLX-induced core signature that independently predicts overall survival in AML patients. Interestingly, high PAK1 expression itself is associated with inferior overall survival, but only in patients with high HLX expression, indicating that there is an interaction of HLX and PAK1 with respect to their impact on overall survival. The correlation of HLX and PAK1 expression levels in the murine HLX knockdown model and in patients with AML, together with the observed induction of Pak1 expression upon HLX overexpression in stem cells strongly suggest that Pak1 is a downstream target of HLX and critically mediates, at least in part, the leukemia-promoting effects of HLX in patients in vivo. Further studies will be required to determine the exact nature of this regulation, and whether it is direct or indirect.
Several HOX genes, such as HOXB4, have been reported to be stimulators of HSC function and expansion (Antonchuk, Sauvageau and Humphries, 2002; Sauvageau, Thorsteinsdottir, et al, 1995). Our data show that HLX suppresses the function of normal immature HSC and progenitors, however leads to an increase of clonogenicity, as well as a differentiation block at the level of phenotypically more mature progenitors. As the loss of HSC does not seem to be mediated by induction of apoptosis or necrosis, one may speculate that HLX exerts this dual role by triggering initial differentiation of HSC and suppression of terminal differentiation at a more committed progenitor level. Further studies will be required to understand the molecular basis of this effect. While overexpression of HLX led to the formation of myeloid progenitors with unlimited serial clonogenicity, we so far did not observe development of leukemia upon transplantation, suggesting that HLX elevation alone may not be sufficient for full transformation. Like other homeobox genes, Hlx may function in concert with co-factors (Moens and Selleri, 2006; Pineault, Abramovich, et al, 2004). Such co-factors could confer cell type specificity to the effects of HLX overexpression, and also contribute to leukemic transformation. It is also possible that increase of HLX plays a role in leukemia maintenance rather than leukemia induction.
Several transcription factors that govern normal hematopoietic differentiation have been implicated in leukemogenesis by blocking differentiation and promoting self-renewal and clonogenicity (for review see (Rosenbauer and Tenen, 2007)). HLX may act similar to those factors by establishing a specific gene expression program in early HSPC, which results in increased long-term clonogenicity and a differentiation arrest, contributing to poor clinical outcome. Thus, HLX expression levels may be utilized to predict clinical outcome and improve risk stratification. Finally, inhibition of HLX may be a promising strategy for treatment of patients with AML.
Experimental Procedures
Mice and cells
FVB/nJ mice (Ly5.1), C57BL/6J (Ly5.2) mice, and B6.SJL-Ptprca Pepcb/BoyJ (Pep boy, Ly5.1), and NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice were used for in vitro assays and in vivo transplantation assays. Mice with targeted disruption of the upstream regulatory element (URE) of the PU.1 gene have been previously described (Rosenbauer, Wagner, et al, 2004). All animal experiments were approved by the Institutional Animal Care and Use Committee of the Albert Einstein College of Medicine (protocol # 20080109). URE cells were established and maintained as described previously (Steidl,Rosenbauer, et al, 2006). Human AML cell lines THP1, MOLM13, and KG1a were cultured under standard conditions.
Flow Cytometric Analysis and Sorting
Mononuclear cells were purified by lysis of erythrocytes. For analysis and sorting we used antibodies directed against CD4[GK1.5], CD8a[53-6.7], CD19[eBio1D3], Gr-1[RB6-8C5], B220[RA3-6B2], F4/80[BM8], c-kit[ACK2], Sca-1[D7], CD34[RAM34], CD16/32[93], Flk-2[A2F10], Mac1[M1/70], Ter119[TER-119], and Thy-1.2[53-2-1]. To distinguish donor from host cells in transplanted mice, cells were additionally stained with anti-CD45.1[A20] and CD45.2[104]. Analysis and sorting were performed using a FACSAria II Special Order System (BD Biosciences, San Jose, CA). See Supplemental Experimental Procedures for more details.
Lentiviral vectors and transduction
For overexpression studies, we introduced the mouse Hlx coding sequence into the EcoRI site of a pCAD-IRES-GFP lentiviral construct (Steidl, Steidl, et al, 2007). For BTG1 overexpression we introduced the human BTG1 coding sequence into the EcoRI site of the pCAD-IRES-GFP construct. For murine knockdown studies, we inserted shRNA template oligonucleotides (target sense strand-loop-target antisense strand-TTTTT) into the pSIH1-H1-copGFP shRNA vector (System Biosciences, Mountain View, CA). For human knockdown studies we utilized the pGIPZ system from Open Biosystems, Huntsville, AL. See Supplemental Experimental Procedures for sequence information. For knockdown of PAK1 we utilized the plko.1-puro vector system (Sigma) with non-silencing control or human PAK1 target. Lentiviral particles were produced utilizing 293T cells, and concentrated by ultracentrifugation. For overexpression studies, we treated sorted Lin−Kit+ cells from WT C57BL/6J (Ly5.2) BM (for in vivo assays) or Lin−Kit+Sca-1+ cells from WT FVB/nJ BM (for in vitro assays) with control virus or Hlx virus. In brief, sorted cells were cultured with lentiviral supernatants in the presence of 8µg/ml polybrene. 24 hours after transduction, cells were washed with PBS and then used for experiments. After 40 hours, transduction efficiency was determined by flow-cytometry. For knockdown studies, cells were incubated with short-hairpin-containing lentivirus for 24 hours. GFP+ cells were sorted using a FACS Aria II sorter (BD Biosciences) and used for experiments.
Quantitative real-time PCR
We extracted total RNA from FACS-sorted cells or cultured cells using RNeasy Micro kit (Qiagen, Valencia, CA) and synthesized cDNA by Superscript II reverse transcriptase (Invitrogen, Carlsbad, CA). We performed real-time PCR using an iQ5 real-time PCR detection system (BIO-RAD, Hercules, CA) with 1 cycle of 50°C (2min) and 95°C (10min) followed by 40 cycles of 95°C (15sec) and 60°C (1min) using Power SYBR Green PCR master mix (AB, Carlsbad, CA) (see Supplemental Experimental Procedures for primer sequences).
Western blotting
Cell proliferation, cell cycle, and apoptosis assays
Colony formation assays and serial replating assays
Assays were performed in MethoCult M3434 (Stem Cell Technologies, Vancouver, BC) containing IL-3, IL-6, SCF, and EPO or in MethoCult M3234 supplemented with M-CSF or GM-CSF as previously described (Higuchi, O'Brien, et al, 2002;Huntly, Shigematsu, et al, 2004;Will, Kawahara, et al, 2009). GFP+ colonies were scored 8–10 days after plating using an AXIOVERT 200M microscope (Zeiss, Maple Grove, MN). After scoring, we resorted GFP+ cells after each round and then proceeded with serial replating assays. Cells were replated and colonies were again scored after 10–14 days.
Transplantation assays
For HLX overexpression studies, 5×104 lentivirus-transduced Lin−Kit+ cells (Ly5.2) together with 2.5×105 spleen cells from congenic WT recipients (Ly5.1) were transplanted into lethally irradiated age-matched congenic WT recipients (Ly5.1) by retroorbital vein injection. Total body irradiation was delivered in a single dose of 950 cGy using a Shepherd 6810 sealed-source 137Cs irradiator.
Micorarray experiments and analysis
RNA was extracted from sorted GFP+ cells utilizing the RNeasy Micro Kit (Qiagen). After evaluation of the quality of RNA with an Agilent2100 Bioanalyzer, total RNA was used for amplification utilizing the Nugen Ovation pico WTA system according to the manufacturer’s instructions. After labeling with the GeneChip WT terminal labeling kit (Affymetrix), labeled cRNA of each individual sample was hybridized to Affymetrix Mouse Gene 1.0ST microarrays (Affymetrix), stained, and scanned by GeneChip Scanner 3000 7G system (Affymetrix) according to standard protocols. For data analysis we utilized Multiple Experiment Viewer v.4 pilot2 (Saeed, Bhagabati, et al, 2006), DAVID bioinformatics tool (Huang da, Sherman and Lempicki, 2009a; Huang da, Sherman and Lempicki, 2009b), Enrichment Map Cytoscape plugin (Cline, Smoot, et al, 2007; Merico, Isserlin, et al, 2010), and Gene Set Enrichment Analysis v2.0 (GSEA) (Mootha, Lindgren, et al, 2003; Subramanian, Tamayo, et al, 2005). See Supplemental Experimental Procedures for details.
Data sets and statistical analysis
We analyzed the publicly available gene expression data sets with accession numbers GSE12417 (training set U133A and U133B; test set U133plus2.0), GSE14468, and GSE10358 (http://www.ncbi.nlm.nih.gov/geo/). Clinical outcome and mutational data for the GSE10358 dataset were obtained from a recent study of the same group (Ley, Ding, et al, 2010). Analyses of the gene expression profiles from GSE14468, GSE12417 training set and GSE10358 were performed based on published (Gentles, Plevritis, et al, 2010) and publicly available MAS5 files (GSE24006) with reanalyzed data. For analysis of the test set of the GSE12417 dataset, CEL files were processed using GenePattern (Broad Institute, Cambridge MA) for normalization (Expression File Creator algorithm). We utilized GraphPad Prism 5.0, R/Bioconductor sva package (R package version 3.2.0)(Leek, Johnson, et al.,2012), and SPSS 18.0 statistical package for further analyses. See Supplemental Experimental Procedures for details.
Signature generation and calculation of signature scores
We utilized dChip (http://biosun1.harvard.edu/complab/dchip/), SAM (significance analysis of microarrays), as well as R/Bioconductor globaltest (Goeman, van de Geer, et al, 2004) for signature identification, followed by calculation of signature scores. See Supplemental Experimental Procedures for details.
Supplementary Material
Highlights.
Elevated Hlx levels lead to loss of HSC and formation of aberrant progenitors with unlimited serial clonogenicity
Decrease of Hlx inhibits leukemic cell growth, overcomes the differentiation block, and prolongs survival in vivo
HLX is overexpressed in the vast majority of AML patients
High levels of HLX independently correlate with inferior survival of AML patients
Significance.
AML is a heterogeneous disease with poor clinical outcome. Less than one third of patients achieve durable remission with current treatment regimens, and prognostication and risk stratification of individual patients is challenging. New molecular targets are desired for more effective therapeutic intervention. We identify the non-clustered homeobox gene Hlx as a key regulator in immature hematopoietic and leukemia cells. HLX is overexpressed in the majority of AML patients, and high levels of HLX correlate with inferior survival. Inhibition of HLX overcomes the differentiation block of AML, and prolongs survival in an in vivo model. As a key factor controlling malignant cell growth and differentiation, HLX may be a prognostic and therapeutic target in AML and possibly other types of cancer.
Acknowledgments
We thank the Einstein Human Stem Cell FACS and Xenotransplantation Facility for expert technical assistance (supported by NYSTEM, C024172). This work was supported by a new investigator award of the Leukemia Research Foundation, NYSTEM grant #CO24350, a Howard Temin Award of the NCI (R00CA131503), and a Medical Research Award of the Gabrielle’s Angel Foundation for Cancer Research (to U.S.). U.S. is the Diane and Arthur B. Belfer Faculty Scholar in Cancer Research of the Albert Einstein College of Medicine.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession number
Complete array data are available in the Gene Expression Omnibus (GEO) of NCBI (Edgar, Domrachev and Lash, 2002), accession number GSE27947.
References
- Alcalay M, Tiacci E, Bergomas R, Bigerna B, Venturini E, Minardi SP, Meani N, Diverio D, Bernard L, Tizzoni L, et al. Acute myeloid leukemia bearing cytoplasmic nucleophosmin (NPMc+ AML) shows a distinct gene expression profile characterized by up-regulation of genes involved in stem-cell maintenance. Blood. 2005;3:899–902. doi: 10.1182/blood-2005-02-0560. [DOI] [PubMed] [Google Scholar]
- Allen JD, Lints T, Jenkins NA, Copeland NG, Strasser A, Harvey RP, Adams JM. Novel murine homeo box gene on chromosome 1 expressed in specific hematopoietic lineages and during embryogenesis. Genes Dev. 1991;4:509–520. doi: 10.1101/gad.5.4.509. [DOI] [PubMed] [Google Scholar]
- Antonchuk J, Sauvageau G, Humphries RK. HOXB4-induced expansion of adult hematopoietic stem cells ex vivo. Cell. 2002;1:39–45. doi: 10.1016/s0092-8674(02)00697-9. [DOI] [PubMed] [Google Scholar]
- Argiropoulos B, Humphries RK. Hox genes in hematopoiesis and leukemogenesis. Oncogene. 2007;47:6766–6776. doi: 10.1038/sj.onc.1210760. [DOI] [PubMed] [Google Scholar]
- Ayton PM, Cleary ML. Transformation of myeloid progenitors by MLL oncoproteins is dependent on Hoxa7 and Hoxa9. Genes Dev. 2003;18:2298–2307. doi: 10.1101/gad.1111603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal D, Scholl C, Frohling S, McDowell E, Lee BH, Dohner K, Ernst P, Davidson AJ, Daley GQ, Zon LI, Gilliland DG, Huntly BJ. Cdx4 dysregulates Hox gene expression and generates acute myeloid leukemia alone and in cooperation with Meis1a in a murine model. Proc. Natl. Acad. Sci. U. S. A. 2006;45:16924–16929. doi: 10.1073/pnas.0604579103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullinger L, Dohner K, Bair E, Frohling S, Schlenk RF, Tibshirani R, Dohner H, Pollack JR. Use of gene-expression profiling to identify prognostic subclasses in adult acute myeloid leukemia. N. Engl. J. Med. 2004;16:1605–1616. doi: 10.1056/NEJMoa031046. [DOI] [PubMed] [Google Scholar]
- Cline MS, Smoot M, Cerami E, Kuchinsky A, Landys N, Workman C, Christmas R, Avila-Campilo I, Creech M, Gross B, et al. Integration of biological networks and gene expression data using Cytoscape. Nat. Protoc. 2007;10:2366–2382. doi: 10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deguchi Y, Kehrl JH. Selective expression of two homeobox genes in CD34-positive cells from human bone marrow. Blood. 1991;2:323–328. [PubMed] [Google Scholar]
- Deguchi Y, Kirschenbaum A, Kehrl JH. A diverged homeobox gene is involved in the proliferation and lineage commitment of human hematopoietic progenitors and highly expressed in acute myelogenous leukemia. Blood. 1992;11:2841–2848. [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;1:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa ME, Lugthart S, Li Y, Erpelinck-Verschueren C, Deng X, Christos PJ, Schifano E, Booth J, van Putten W, Skrabanek L, et al. DNA methylation signatures identify biologically distinct subtypes in acute myeloid leukemia. Cancer. Cell. 2010;1:13–27. doi: 10.1016/j.ccr.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischbach NA, Rozenfeld S, Shen W, Fong S, Chrobak D, Ginzinger D, Kogan SC, Radhakrishnan A, Le Beau MM, Largman C, Lawrence HJ. HOXB6 overexpression in murine bone marrow immortalizes a myelomonocytic precursor in vitro and causes hematopoietic stem cell expansion and acute myeloid leukemia in vivo. Blood. 2005;4:1456–1466. doi: 10.1182/blood-2004-04-1583. [DOI] [PubMed] [Google Scholar]
- Friedman AD. Transcriptional control of granulocyte and monocyte development. Oncogene. 2007;47:6816–6828. doi: 10.1038/sj.onc.1210764. [DOI] [PubMed] [Google Scholar]
- Gentles AJ, Plevritis SK, Majeti R, Alizadeh AA. Association of a leukemic stem cell gene expression signature with clinical outcomes in acute myeloid leukemia. JAMA. 2010;24:2706–2715. doi: 10.1001/jama.2010.1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goeman JJ, van de Geer SA, de Kort F, van Houwelingen HC. A global test for groups of genes: testing association with a clinical outcome. Bioinformatics. 2004;1:93–99. doi: 10.1093/bioinformatics/btg382. [DOI] [PubMed] [Google Scholar]
- Hentsch B, Lyons I, Li R, Hartley L, Lints TJ, Adams JM, Harvey RP. Hlx homeo box gene is essential for an inductive tissue interaction that drives expansion of embryonic liver and gut. Genes Dev. 1996;1:70–79. doi: 10.1101/gad.10.1.70. [DOI] [PubMed] [Google Scholar]
- Higuchi M, O'Brien D, Kumaravelu P, Lenny N, Yeoh EJ, Downing JR. Expression of a conditional AML1-ETO oncogene bypasses embryonic lethality and establishes a murine model of human t(8;21) acute myeloid leukemia. Cancer. Cell. 2002;1:63–74. doi: 10.1016/s1535-6108(02)00016-8. [DOI] [PubMed] [Google Scholar]
- Horton SJ, Grier DG, McGonigle GJ, Thompson A, Morrow M, De Silva I, Moulding DA, Kioussis D, Lappin TR, Brady HJ, Williams O. Continuous MLL-ENL expression is necessary to establish a "Hox Code" and maintain immortalization of hematopoietic progenitor cells. Cancer Res. 2005;20:9245–9252. doi: 10.1158/0008-5472.CAN-05-1691. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009a;1:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009b;1:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Huntly BJ, Shigematsu H, Deguchi K, Lee BH, Mizuno S, Duclos N, Rowan R, Amaral S, Curley D, Williams IR, Akashi K, Gilliland DG. MOZ-TIF2, but not BCR-ABL, confers properties of leukemic stem cells to committed murine hematopoietic progenitors. Cancer. Cell. 2004;6:587–596. doi: 10.1016/j.ccr.2004.10.015. [DOI] [PubMed] [Google Scholar]
- Kawahara M, Hori T, Chonabayashi K, Oka T, Sudol M, Uchiyama T. Kpm/Lats2 is linked to chemosensitivity of leukemic cells through the stabilization of p73. Blood. 2008;9:3856–3866. doi: 10.1182/blood-2007-09-111773. [DOI] [PubMed] [Google Scholar]
- Krivtsov AV, Twomey D, Feng Z, Stubbs MC, Wang Y, Faber J, Levine JE, Wang J, Hahn WC, Gilliland DG, Golub TR, Armstrong SA. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 2006;7104:818–822. doi: 10.1038/nature04980. [DOI] [PubMed] [Google Scholar]
- Kroon E, Krosl J, Thorsteinsdottir U, Baban S, Buchberg AM, Sauvageau G. Hoxa9 transforms primary bone marrow cells through specific collaboration with Meis1a but not Pbx1b. EMBO. J. 1998;13:3714–3725. doi: 10.1093/emboj/17.13.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumlauf R. Hox genes in vertebrate development. Cell. 1994;2:191–201. doi: 10.1016/0092-8674(94)90290-9. [DOI] [PubMed] [Google Scholar]
- Kuo ML, Duncavage EJ, Mathew R, den Besten W, Pei D, Naeve D, Yamamoto T, Cheng C, Sherr CJ, Roussel MF. Arf induces p53-dependent and - independent antiproliferative genes. Cancer Res. 2003;63:1046–1053. [PubMed] [Google Scholar]
- Laiosa CV, Stadtfeld M, Graf T. Determinants of lymphoid-myeloid lineage diversification. Annu. Rev. Immunol. 2006:705–738. doi: 10.1146/annurev.immunol.24.021605.090742. [DOI] [PubMed] [Google Scholar]
- Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28:882–883. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, Larson DE, Kandoth C, Payton JE, Baty J, Welch J, et al. DNMT3A mutations in acute myeloid leukemia. N. Engl. J. Med. 2010;25:2424–2433. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeti R, Becker MW, Tian Q, Lee TL, Yan X, Liu R, Chiang JH, Hood L, Clarke MF, Weissman IL. Dysregulated gene expression networks in human acute myelogenous leukemia stem cells. Proc. Natl. Acad. Sci. U. S. A. 2009;9:3396–3401. doi: 10.1073/pnas.0900089106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcucci G, Haferlach T, Dohner H. Molecular genetics of adult acute myeloid leukemia: prognostic and therapeutic implications. J. Clin. Oncol. 2011;5:475–486. doi: 10.1200/JCO.2010.30.2554. [DOI] [PubMed] [Google Scholar]
- Merico D, Isserlin R, Stueker O, Emili A, Bader GD. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One. 2010;11:e13984. doi: 10.1371/journal.pone.0013984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzeler KH, Hummel M, Bloomfield CD, Spiekermann K, Braess J, Sauerland MC, Heinecke A, Radmacher M, Marcucci G, Whitman SP, et al. An 86-probe-set gene-expression signature predicts survival in cytogenetically normal acute myeloid leukemia. Blood. 2008;10:4193–4201. doi: 10.1182/blood-2008-02-134411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens CB, Selleri L. Hox cofactors in vertebrate development. Dev. Biol. 2006;2:193–206. doi: 10.1016/j.ydbio.2005.10.032. [DOI] [PubMed] [Google Scholar]
- Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003;3:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- Novershtern N, Subramanian A, Lawton LN, Mak RH, Haining WN, McConkey ME, Habib N, Yosef N, Chang CY, Shay T, et al. Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell. 2011;144:296–309. doi: 10.1016/j.cell.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong CC, Jubb AM, Zhou W, Haverty PM, Harris AL, Belvin M, Friedman LS, Koeppen H, Hoeflich KP. p21-activated kinase 1: PAK'ed with potential. Oncotarget. 2011;2:491–496. doi: 10.18632/oncotarget.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passegue E, Wagner EF, Weissman IL. JunB deficiency leads to a myeloproliferative disorder arising from hematopoietic stem cells. Cell. 2004;3:431–443. doi: 10.1016/j.cell.2004.10.010. [DOI] [PubMed] [Google Scholar]
- Pineault N, Abramovich C, Ohta H, Humphries RK. Differential and common leukemogenic potentials of multiple NUP98-Hox fusion proteins alone or with Meis1. Mol. Cell. Biol. 2004;5:1907–1917. doi: 10.1128/MCB.24.5.1907-1917.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbauer F, Tenen DG. Transcription factors in myeloid development: balancing differentiation with transformation. Nat. Rev. Immunol. 2007;2:105–117. doi: 10.1038/nri2024. [DOI] [PubMed] [Google Scholar]
- Rosenbauer F, Wagner K, Kutok JL, Iwasaki H, Le Beau MM, Okuno Y, Akashi K, Fiering S, Tenen DG. Acute myeloid leukemia induced by graded reduction of a lineage-specific transcription factor, PU.1. Nat. Genet. 2004;6:624–630. doi: 10.1038/ng1361. [DOI] [PubMed] [Google Scholar]
- Saeed AI, Bhagabati NK, Braisted JC, Liang W, Sharov V, Howe EA, Li J, Thiagarajan M, White JA, Quackenbush J. TM4 microarray software suite. Methods Enzymol. 2006:134–193. doi: 10.1016/S0076-6879(06)11009-5. [DOI] [PubMed] [Google Scholar]
- Saito Y, Kitamura H, Hijikata A, Tomizawa-Murasawa M, Tanaka S, Takagi S, Uchida N, Suzuki N, Sone A, Najima Y, et al. Identification of therapeutic targets for quiescent, chemotherapy-resistant human leukemia stem cells. Sci. Transl. Med. 2010;17:17ra9. doi: 10.1126/scitranslmed.3000349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauvageau G, Thorsteinsdottir U, Eaves CJ, Lawrence HJ, Largman C, Lansdorp PM, Humphries RK. Overexpression of HOXB4 in hematopoietic cells causes the selective expansion of more primitive populations in vitro and in vivo. Genes Dev. 1995;14:1753–1765. doi: 10.1101/gad.9.14.1753. [DOI] [PubMed] [Google Scholar]
- Sauvageau G, Thorsteinsdottir U, Hough MR, Hugo P, Lawrence HJ, Largman C, Humphries RK. Overexpression of HOXB3 in hematopoietic cells causes defective lymphoid development and progressive myeloproliferation. Immunity. 1997;1:13–22. doi: 10.1016/s1074-7613(00)80238-1. [DOI] [PubMed] [Google Scholar]
- Scholl C, Bansal D, Dohner K, Eiwen K, Huntly BJ, Lee BH, Rucker FG, Schlenk RF, Bullinger L, Dohner H, Gilliland DG, Frohling S. The homeobox gene CDX2 is aberrantly expressed in most cases of acute myeloid leukemia and promotes leukemogenesis. J. Clin. Invest. 2007;4:1037–1048. doi: 10.1172/JCI30182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitwala KV, Dandekar MN, Hess JL. HOX proteins and leukemia. Int. J. Clin. Exp. Pathol. 2008;6:461–474. [PMC free article] [PubMed] [Google Scholar]
- Somervaille TC, Cleary ML. Identification and characterization of leukemia stem cells in murine MLL-AF9 acute myeloid leukemia. Cancer. Cell. 2006;4:257–268. doi: 10.1016/j.ccr.2006.08.020. [DOI] [PubMed] [Google Scholar]
- Steidl U, Rosenbauer F, Verhaak RG, Gu X, Ebralidze A, Otu HH, Klippel S, Steidl C, Bruns I, Costa DB, et al. Essential role of Jun family transcription factors in PU.1 knockdown-induced leukemic stem cells. Nat. Genet. 2006;11:1269–1277. doi: 10.1038/ng1898. [DOI] [PubMed] [Google Scholar]
- Steidl U, Steidl C, Ebralidze A, Chapuy B, Han HJ, Will B, Rosenbauer F, Becker A, Wagner K, Koschmieder S, et al. A distal single nucleotide polymorphism alters long-range regulation of the PU.1 gene in acute myeloid leukemia. J. Clin. Invest. 2007;9:2611–2620. doi: 10.1172/JCI30525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 2005;43:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenen DG. Disruption of differentiation in human cancer: AML shows the way. Nat. Rev. Cancer. 2003;2:89–101. doi: 10.1038/nrc989. [DOI] [PubMed] [Google Scholar]
- Thorsteinsdottir U, Sauvageau G, Hough MR, Dragowska W, Lansdorp PM, Lawrence HJ, Largman C, Humphries RK. Overexpression of HOXA10 in murine hematopoietic cells perturbs both myeloid and lymphoid differentiation and leads to acute myeloid leukemia. Mol. Cell. Biol. 1997;1:495–505. doi: 10.1128/mcb.17.1.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasson MH, Xiang Z, Walgren R, Zhao Y, Kasai Y, Miner T, Ries RE, Lubman O, Fremont DH, McLellan MD, et al. Somatic mutations and germline sequence variants in the expressed tyrosine kinase genes of patients with de novo acute myeloid leukemia. Blood. 2008;9:4797–4808. doi: 10.1182/blood-2007-09-113027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will B, Kawahara M, Luciano JP, Bruns I, Parekh S, Erickson-Miller CL, Aivado MA, Verma A, Steidl U. Effect of the nonpeptide thrombopoietin receptor agonist Eltrombopag on bone marrow cells from patients with acute myeloid leukemia and myelodysplastic syndrome. Blood. 2009;18:3899–3908. doi: 10.1182/blood-2009-04-219493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wouters BJ, Lowenberg B, Erpelinck-Verschueren CA, van Putten WL, Valk PJ, Delwel R. Double CEBPA mutations, but not single CEBPA mutations, define a subgroup of acute myeloid leukemia with a distinctive gene expression profile that is uniquely associated with a favorable outcome. Blood. 2009;13:3088–3091. doi: 10.1182/blood-2008-09-179895. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.