Summary
Posttranslational histone modifications are important for gene regulation, yet the mode of propagation and the contribution to heritable gene expression states remains controversial. To address these questions, we developed a Chromatin in vivo (CiA) Assay system employing chemically-induced proximity to initiate and terminate chromatin modifications in living cells. We selectively recruited HP1α to induce H3K9me3-dependent gene silencing and describe the kinetics and extent of chromatin modifications at the Oct4 locus in fibroblasts and pluripotent cells. H3K9me3 propagated symmetrically and continuously at rates of ~0.18 nucleosomes/hr to produce domains of up to 10kb. After removal of the HP1α stimulus, heterochromatic domains were heritably transmitted, undiminished through multiple cell generations. Our data enabled quantitative modeling of reaction kinetics, which revealed that dynamic competition between histone marking and turnover determines the boundaries and stability of H3K9me3 domains. Applying this framework, we were able to predict the steady-state dynamics and spatial features of the majority of euchromatic H3K9me3 domains.
Introduction
In multicellular organisms, cellular identity is defined by distinct profiles of gene expression that are faithfully transmitted through cell division. There are multiple mechanisms that convey heritable transcriptional information independent of DNA sequence. These epigenetic mechanisms are self-sustaining in the absence of the initial stimulus (Bonasio et al., 2010; Ptashne, 2007). Chemical modifications of DNA and nucleosomal histones have been implicated in contributing to epigenetic programs. However, to date, only DNA methylation has been shown to mediate gene repression that is conserved through cell generations (Goll and Bestor, 2005; Wigler et al., 1981). Some posttranslational histone modifications exhibit strong correlations with transcriptional states (Kouzarides, 2007), and mechanisms for their propagation have been proposed (Margueron et al., 2009; Nakayama et al., 2001; Talbert and Henikoff, 2006). However, genetic approaches have not been able to address the cellular dynamics of chromatin regulation and biochemical approaches have been limited by the inability to faithfully reconstitute chromatin in vitro. Hence, new experimental techniques are required to develop a kinetic understanding of chromatin regulation in living cells.
Position Effect Variegation (PEV) has been a classical paradigm to study the role of histone modifications in inheritance of transcriptional patterns. In flies, PEV describes a mottled eye phenotype, caused by random silencing of the white gene when translocated into the proximity of a heterochromatic region (Muller, 1930). Patches of red and white cell clones are maintained through cell divisions in the developing fly eye indicating that silencing of the white gene product is clonally inherited. Genetic screens for PEV modifiers in several organisms have identified multiple proteins implicated in methylation of histone H3 at lysine 9 (H3K9me) (Fodor et al., 2010). In mammalian cells, H3K9 trimethylation (H3K9me3) is a hallmark of heterochromatin (Peters et al., 2002), and is also required for transcriptional silencing of genes and retroviral elements (Magklara et al., 2011; Matsui et al., 2010; Nielsen et al., 2001). Transcriptional repression involves Heterochromatin Protein 1 (HP1), which specifically binds to methylated H3K9 (Bannister et al., 2001; Lachner et al., 2001; Nakayama et al., 2001). HP1 can form oligomers, which are thought to bridge neighboring nucleosomes and mediate chromatin condensation (Canzio et al., 2011; Verschure et al., 2005). HP1 also directly interacts with and recruits H3K9-specific histone methyltransferases (HMTs) Suv39h1/2 and SETDB1 (Fritsch et al., 2010; Peters et al., 2003; Rea et al., 2000; Schultz et al., 2002). HMT interactions could facilitate self-propagation and sequential methylation of adjacent nucleosomes consistent with a model of linear spreading (Hall et al., 2002; Schotta et al., 2002). Alternatively, it has been suggested that H3K9 methylation could propagate along the chromosome discontinuously through a mechanism of skipping (Talbert and Henikoff, 2006). In Drosophila, spreading of heterochromatin depends on the activity and dosage of heterochromatin components and can be antagonized by euchromatic factors (Ebert et al., 2004; Schotta et al., 2002). While this competitive balance appears to determine the boundaries of pericentric heterochromatin, it is unclear if H3K9me3 repressed target genes are also dosage sensitive and subject to dynamic regulation. Moreover, it is unknown whether at these genes persistence of the mark through cell division is a result of self-propagation or renewed targeting of HMTs through recruitment by cis-regulatory factors after each replication cycle (Moazed, 2011).
We sought to study the formation of heterochromatin at a well-defined euchromatic target promoter in living cells. In embryonic stem (ES) cells, Oct4 (Pou5f1) is highly expressed, encoding a transcription factor that is critical for pluripotency and self-renewal. Upon cellular differentiation, Oct4 expression is rapidly and completely silenced through a series of events including histone H3K9 methylation, HP1 binding and DNA methylation (Feldman et al., 2006). Interestingly, in differentiated tissues Oct4 repression can be overcome by ectopic expression of pluripotency transcription factors (including OCT4 itself), which leads to the formation of pluripotent cells (Takahashi and Yamanaka, 2006). However, cellular reprogramming is highly inefficient, possibly due to repressive chromatin structure that presents a barrier to transcription factor binding.
To investigate the kinetics of chromatin modification and the transmission of epigenetic information, we have generated a murine strain that allows rapid addition and removal of chromatin regulatory activities to a genetically modified Oct4 allele in any cell type using small molecule-mediated recruitment. Selective targeting of HP1α induced H3K9me3 at the Oct4 reporter locus and subsequent linear spreading in cis over a distance of 10 kbp to form a heterochromatic domain with features of PEV. Removal of HP1α from the locus allowed us to study the epigenetic properties of the histone mark, clearly demonstrating that the H3K9me3 domain was inherited through cell divisions in the absence of the initial stimulus. Transcriptional activators could oppose the maintenance of heterochromatin suggesting that the steady-state of H3K9me3 is governed by antagonizing activities of histone marking and turnover. Mathematical modeling based on competitive dynamics enabled us to describe the spatial features of heterochromatic domains and to calculate the rates of histone H3K9 methylation and turnover at the CiA:Oct4 locus in ES cells and fibroblasts. Finally, when applied to genomic data sets, our quantitative framework predicts the steady-state dynamics of the vast majority of all non-centromeric H3K9me3 domains in the mouse genome.
Results
Generation of the murine Chromatin in vivo Assay (CiA) system at Oct4
We envisioned an approach where Chemically Induced Proximity (CIP) enables selective addition and removal of different chromatin and transcriptional activities to an endogenous chromosomal locus in vivo (Figure 1). CIP uses bi-functional small molecules that are membrane-permeable and can cause rapid association of two different peptide tags fused to proteins of interest inside cells. Parallel or sequential addition of orthogonal small-molecule ligands has been successfully employed to dissect the mechanism and order of events of various biochemical processes (Graef et al., 1997; Gruber et al., 2006; Ho et al., 1996; Spencer et al., 1993). Importantly, induction of protein proximity is readily reversible in cells or animals by washout or specific displacement using one-sided molecules that bind to only one peptide tag.
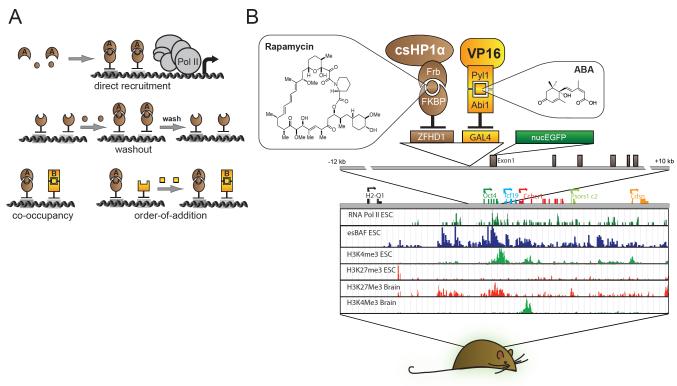
Figure 1. Design of Chromatin in vivo Assay at Oct4 (CiA:Oct4) ES cell line and mouse.
(A) CIP allows direct recruitment, washout, co-occupancy, and order-of-addition experiments. (B) The CIA:Oct4 mouse contains one modified Oct4 allele harboring two arrays of DNA binding sites (12XZFHD1 and 5XGal4) in the promoter region upstream of an in-frame EGFP reporter. Distribution of histone modifications at the Oct4 locus in murine ES cells and brain tissue (Mikkelsen et al., 2007) reveals the distinct chromatin substrates for CiA modulation.
Since Oct4 gene dosage is haplosufficient (Nichols et al., 1998), we genetically modified one Oct4 allele to recruit chromatin regulators by CIP and to study the effects on gene expression and chromatin structure. We introduced two arrays of different DNA binding sites (12xZFHD1 and 5xGAL4) upstream of the Oct4 promoter and an in-frame nuclear EGFP reporter replacing the first exon of Oct4 (Figure 1). Targeted ES cells retained good morphology (Figure S1A-B) and provided real-time fluorescence-based readout of gene expression at single cell resolution. Protein tethering using the CIP system involves expression of two sets of chimeric proteins designed to bind different sides of a CIP molecule. The first set is composed of the respective DNA binding domain (GAL4 or ZFHD1) fused to a CIP anchor partner (e.g. FKBP12). The second set of protein chimeras contains the protein of interest fused to the CIP recruitment partner (e.g. FRB). Addition of bifunctional small molecules (e.g. rapamycin) induces the CIP anchor to reversibly bind the CIP recruitment partner, tethering any given protein to the modified Oct4 allele. We call this system the Chromatin in-vivo Assay (CiA) system and the allele harboring the recruitment domains and reporter at Oct4, CiA:Oct4.
Nucleosome modifications at the Oct4 promoter have been well defined in a variety of cell types including mouse ES cells (Figure 1B), where the active gene is modified with histone H3 acetylation (H3ac) and histone H3K4 trimethylation (H3K4me3) (Mikkelsen et al., 2007). In contrast, Oct4 is transcriptionally repressed in differentiated tissues and packaged into nucleosomes marked by H3K9me3 and H3K27me3. We induced cellular differentiation of CiA ES cells by removal of LIF and treatment with retinoic acid (RA) (Athanasiadou et al., 2010; Sato et al., 2006). Differentiation of CiA ES cells reduced expression of both the endogenous OCT4 protein and the GFP reporter protein (Figure S1C). Subsequent chromatin-immunoprecipitation (ChIP) analysis showed that GFP repression was paralleled with a loss of H3K4me3 and gain of repressive H3K9me3 and H3K27me3 (Figure S1D). We concluded that the CiA:Oct4 allele faithfully reflected physiological regulation of wild-type Oct4.
Kinetic analysis of heterochromatin induced by chemical-mediated recruitment
Initiation and maintenance of Oct4 repression during cellular differentiation involves substantial changes in histone modifications and gain of DNA methylation. To determine the kinetics of H3K9me3-dependent gene repression, we sought to recruit HP1 to the CiA:Oct4 promoter in ES cells. HP1α is composed of a chromo-domain (CD), which confers specific binding to methylated H3 lysine 9, and a chromo-shadow domain (CSD), which directly interacts with H3K9-specific histone methylases, including SetDB1 and Suv39h1/2 (Hiragami and Festenstein, 2005; Schultz et al., 2002). We infected CiA ES cells with a lentiviral construct of the DNA binding domain of GAL4 (GAL4) fused to either full-length murine HP1α or a truncated form of HP1α containing only the CSD (csHP1α). While infection with GAL4 alone did not alter GFP, both full-length HP1α and csHP1α fusion proteins induced complete silencing of gene expression as measured by flow cytometry (Figure S1E). To minimize potential non-specific effects of ectopic HP1α expression, we used csHP1α in subsequent studies.
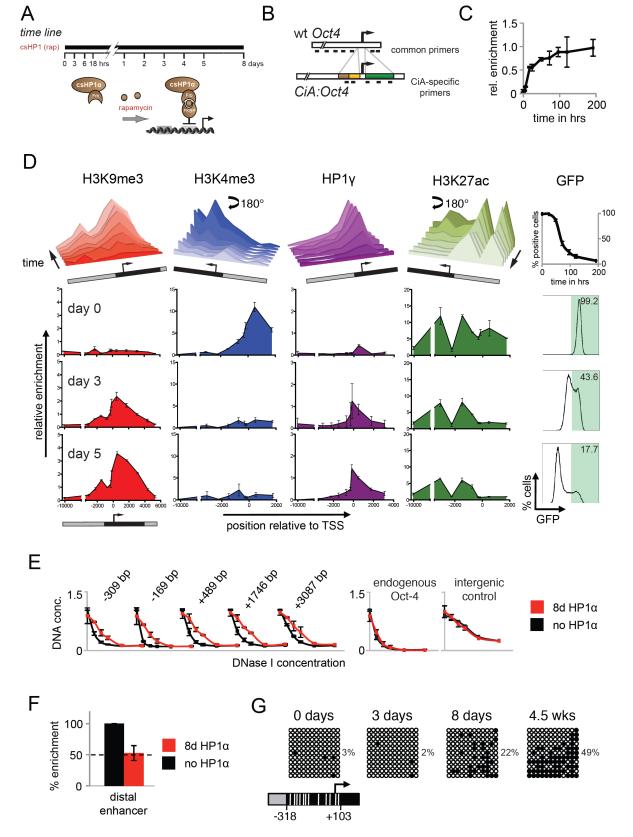
To gain better temporal resolution of the dynamic processes involved in HP1α-mediated repression, we used the CIP system to recruit csHP1α. We infected CiA ES cells with two lentiviral constructs, one containing GAL4 fused to the CIP anchor, FKBP12, and the other encoding the csHP1α fragment fused to two repeats of the 98 aa FKBP12-rapamycin binding (FRB) domain of mTor with a V5 epitope tag for detection (Figure 2A). After addition of rapamycin, we monitored changes in GFP expression and chromatin structure by flow cytometry and ChIP, respectively. ChIP against the V5-tag revealed significant recruitment of csHP1α within 6 hours and nearly saturated binding within 24 hours after rapamycin addition (Figure 2C). csHP1α targeting led to complete repression of the CiA:Oct4 reporter within 5 days (Figure 2D). Interestingly, CiA ES cells did not display a gradual decrease in GFP expression but instead segregated into a bimodal population of GFP-positive and GFP-negative cells (Figure 2D, right panel). The distribution and enrichment of histone modifications was determined by ChIP and real-time PCR using sets of common and reporter allele-specific primers, which cover the distal and proximal regulatory regions upstream and the gene body downstream of the CiA:Oct4 promoter (Figure 2B). Prior to csHP1α targeting (day 0), H3K27ac was broadly enriched at promoter-distal and -proximal sites, while H3K4me3 was only enriched downstream of the active transcription start site (TSS). At this time, H3K9me3 was absent and only basal levels of endogenous HP1γ were detectable (Figure 2D). After 18 hours of csHP1α tethering, we started to observe H3K9me3, which coincided with recruitment of endogenous HP1γ at the CiA:Oct4 promoter. For the next four days, H3K9me3 and HP1γ increased and spread upstream and downstream of the GAL4 binding site. After 5 days of csHP1α recruitment, H3K9me3 had formed a large domain of approximately 10 kbp, which peaked adjacent to the DNA binding site and gradually decreased for 5 kbp to either side (Figure 2D). Interestingly, consistent with the bimodal expression pattern, GFP-negative sorted cells displayed a fully established heterochromatic domain after only three days of rapamycin, while GFP-positive cells lacked H3K9me3 (Figure S2). This suggests that csHP1α-dependent repression initiates stochastically in an all-or-nothing fashion in individual cells of the population. Gain of H3K9me3 appeared to involve recruitment of SETDB1 (Figure S3A). Loss of active marks and establishment of the H3K9me3 domain did not significantly affect nucleosome occupancy at the reporter allele, as measured by total histone H3 ChIP, nor did it alter chromatin structure or expression of the endogenous Oct4 allele (Figures S3B and S3D).
Figure 2. Kinetics of heterochromatin formation following HP1α recruitment in ES cells.
A) Experimental design: rapamycin addition recruits HP1α chromoshadow fragment (csHP1α) to the CiA:Oct4 promoter. B) Schematic representation of wild-type and CiA alleles depicts location of allele-specific and common real-time PCR primers C) ChIP analysis shows rapamycin-mediated csHP1α recruitment over time. D) ChIP analysis reveals dynamic changes of active (H3K4me3, H3K27ac) and repressive (H3K9me3, HP1 gamma) chromatin modifications at the CiA:Oct4 locus. Upper panel summarizes time course of chromatin remodeling at 0h, 24h, 48h, 72h, 96h, 120h, and 192h. Data rotated 180° as indicated to display loss of active marks. Lower panels display ChIP analysis of histone modifications (y-axis) across the CiA:Oct4 locus (x-axis) at selected time points. GFP expression was measured by flow cytometry at each time point. Schematic of the reporter allele indicates CiA:Oct4-specific primer pairs in black. E) Dnase I sensitivity across the CiA:Oct4 locus before and after csHP1α recruitment. F) ChIP analysis of OCT4 transcription factor binding at Oct4 enhancer before and after csHP1α recruitment. G) Bisulfite sequencing analysis of DNA methylation changes at the CiA:Oct4 promoter following csHP1α targeting, with percentage methylated CpGs. White lines in schematic below mark relative positions of CpG dinucleotides.
To test how changes in histone modifications related to chromatin compaction, we analyzed the sensitivity of the CiA:Oct4 locus to endonuclease digestion. DNase I sensitivity is a hallmark of active promoters, while inactive genes and heterochromatic regions are more resistant to digestion (Groudine and Weintraub, 1982; Kerem et al., 1984). We found that the CiA:Oct4 locus was significantly more resistant to DNase I digestion after 8 days of csHP1α recruitment, while DNase I sensitivity remained unaffected at the endogenous Oct4 promoter as well as at a control locus (Figure 2E). Reduced accessibility to nuclease digestion was also observed at the OCT4 transcription factor binding site located at the distal enhancer 2 kbp upstream of the promoter. PCR primers at this region are outside of the knock-in sequence, and thus do not allow discrimination between the endogenous and reporter alleles (Figure 2B). We found that OCT4 binding was reduced by approximately two-fold (Figure 2F). Since OCT4 expression from the wild-type allele is critical for ES cell maintenance, this decline reflected a near-complete eviction of OCT4 protein from the distal enhancer of the CiA:Oct4 allele.
Upon RA treatment and in differentiated tissues, the Oct4 promoter is subject to DNA methylation, which has been proposed to stabilize maintenance of gene repression through cell division (Athanasiadou et al., 2010; Feldman et al., 2006). We analyzed levels of CpG methylation at the CiA:Oct4 promoter during the course of gene silencing. Bisulfite-sequencing revealed low levels of DNA methylation in CiA ES cells prior to csHP1α recruitment. Unlike H3K9me3, DNA methylation was only slightly increased after 8 days when transcription at the locus was fully repressed (Figure 2G). However, promoter methylation continued to gradually increase and was significantly higher at 4.5 weeks, similar to levels found in differentiated cells.
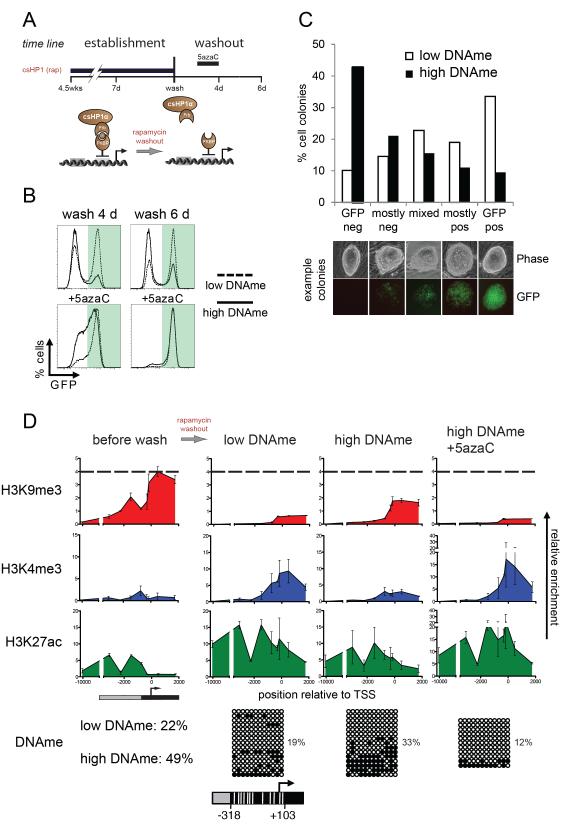
Maintenance of heterochromatin at the CiA:Oct4 locus in ES cells
We determined the stability of induced heterochromatin and its transmission through cell generations by the timed removal of rapamycin. The CiA:Oct4 allele was silenced by rapamycin-mediated recruitment of csHP1α for either 7 days or 4.5 weeks followed by rapamycin by washout (Figure 3A). We observed comparable levels of H3K9me3 after 7 days or 4.5 weeks (Figure S3F), yet the shorter pulse of csHP1α recruitment induced only low levels of DNA methylation while the extended pulse resulted in high levels of methylation at the CiA:Oct4 promoter. While we cannot exclude additional differences, for simplicity, we will refer to the short pulse as “low DNAme” (Figure 2G) and the long pulse as “high DNAme” (Figure 2G). Homogenous populations of GFP-negative cells were obtained by fluorescence-activated cell sorting (FACS) after short (7 days) or long (4.5 weeks) csHP1α treatment. Cells were then passaged in the absence of rapamycin to release csHP1α from the CiA:Oct4 promoter (Figure 3A). To our surprise, a significant fraction of CiA ES cells began to re-express GFP. CiA ES cells with low DNAme displayed GFP reactivation in 50.7% of the population four days after washout and in 63.2% six days after washout (Figure 3B). In contrast, only 18.9% of the CiA ES cells with high DNAme displayed GFP reactivation four days after washout, and 30.7% after six days of washout. To address whether maintenance of GFP repression was controlled by DNA methylation, we treated cells with DNA methyltransferase inhibitor 5-azacytidine (5azaC) (Yoo and Jones, 2006). After a 5azaC pulse for two days during washout (days 2-4), GFP repression was rapidly lost regardless of the duration of csHP1α targeting (Figure 3B, lower panel). To test if increased DNA methylation enhanced heritable repression from one cell generation to the next or whether reactivation of CiA:Oct4 was stochastic within the population, we followed GFP expression in individual colonies founded by single parental CiA ES cells. GFP-negative cells sorted from low or high DNAme samples were sparsely plated and cultured in the absence of rapamycin for four days to form colonies. Colonies were imaged and scored for GFP expression (Figure 3C). Colonies founded by parental cells with low DNAme were 53% GFP-positive or mostly positive, while only 10% of colonies were completely GFP-negative. We also observed that many of these colonies contained a mixed population of GFP-positive and -negative cells, indicating that stochastic reactivation occurred in cells with low DNAme. In contrast, colonies formed from high DNAme parental ES cells showed less GFP reactivation. Only 21% of the colonies were GFP-positive while 43% were completely GFP-negative and 21% of the colonies were mostly GFP-negative. These results argue that DNA methylation of the CiA:Oct4 promoter enhanced maintenance and heritable transmission of gene repression by suppressing spontaneous reactivation in the absence of the HP1 stimulus.
Figure 3. Maintenance of heterochromatin and heritable Oct4 repression in ES cells.
A) Experimental design: rapamycin was added for either 7 days or 4.5 weeks and then washed-out with or without Dnmt-inhibitor 5azaC. B) Flow cytometry analysis after release from csHP1α after 7 days (low promoter DNAme), or after 4.5 weeks (high promoter DNAme). C) Colony growth assay tests heritability of CiA:Oct4 repression. GFP expression of individual colonies was quantified by microscopy (7 days, n=158; 4.5 weeks, n=199). D) ChIP analysis of histone modifications after rapamycin washout. The bottom panels depict bisulfite sequencing analysis of DNA methylation at CiA:Oct4 promoter after csHP1α washout for 4 days. White lines in schematic below mark relative positions of CpG dinucleotides.
To examine whether the stability of the heterochromatic domain was dependent on DNA methylation, we measured H3K9me3 after four days of rapamycin washout and in the presence or absence of 5azaC. Consistent with rapid gain of GFP expression (Figure 3B), cells with low DNAme exhibited a dramatic reduction of H3K9me3 concomitant with an increase in active histone modifications (Figure 3D). CiA ES cells with high DNAme also displayed reduced levels of H3K9me3 and increased H3K4me3. However, these changes were less extensive than in cells with low DNAme. H3K9me3 was rapidly and uniformly lost upon addition of DNA methylation inhibitors 5azaC (Figure 3D).
Activation of the CiA:Oct4 locus in fibroblasts and dynamics of re-establishment of heterochromatin
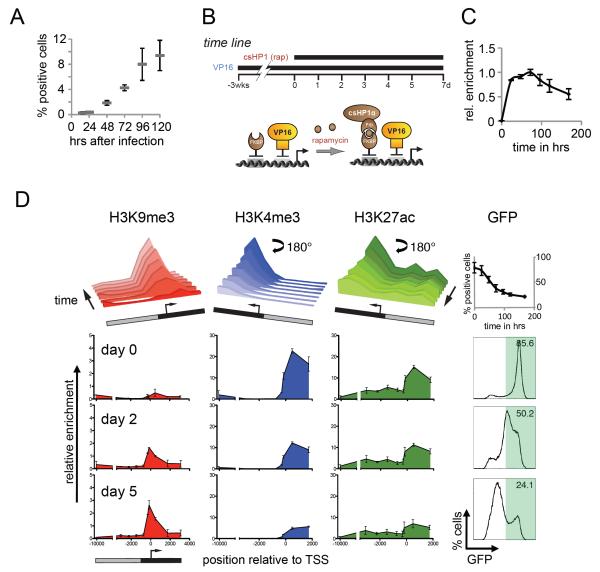
To determine if transcriptional opposition led to H3K9me3 instability, we needed to examine the maintenance of heterochromatin in a tissue lacking ES cell pluripotency factors. We generated a mouse from CiA ES cells and prepared mouse embryonic fibroblasts (MEFs). In these cells, the CiA:Oct4 allele was transcriptionally silent and embedded in repressive chromatin with high levels of H3K9me3 and H3K27me3 (Figure S4). To erase these repressive marks, we recruited transcriptional activator VP16 to the CiA:Oct4 promoter. Based on studies of reprogramming, we expected Oct4 reactivation to require 14-16 days, or not to be possible at all. Surprisingly, 24 hours after infection we detected a small fraction of reactivated cells by flow cytometry, and by 48 hours we observed GFP reactivation in approximately 2% of CiA MEFs (Figure 4A). Five days following infection with GAL4-VP16, nearly 10% of the cell population stably expressed the CiA:Oct4 reporter. This is an especially high proportion considering that the artificially reactivated reporter allele conferred no competitive growth advantage. These results demonstrate that directed recruitment of a strong transcriptional activator was capable of rapidly reactivating Oct4 in differentiated cells.
Figure 4. CiA:Oct4 activation and kinetics of heterochromatin formation in MEFs.
(A) CiA E14.5 p4 MEFs were infected with lentiviral constructs of either GAL4 alone or a GAL4-VP16 fusion. Puromycin was added at 48 hrs and cells analyzed by flow cytometry. (B) Experimental design: the CiA:Oct4 allele was reactivated by GAL4-VP16 in transformed CiA MEFs. GFP-positive cells were enriched by FACS and treated with rapamycin to induce csHP1α targeting. C) ChIP analysis of rapamycin-mediated csHP1α recruitment over time. D) ChIP analysis reveals dynamic changes of active (H3K4me3, H3K27ac) and repressive (H3K9me3) histone modifications at the CiA:Oct4 locus. Upper panel summarizes time course of chromatin remodeling at 0h, 24h, 48h, 72h, 96h, 120h, and 192h. Data rotated 180° as indicated to display loss of active marks. Lower panels display loss of active and gain of repressive histone modifications (y-axis) across the CiA:Oct4 locus (x-axis) at selected time points. GFP expression was measured by flow cytometry analysis at each time point.
We generated a uniform cell line by transforming CiA MEFs with simian virus 40 large T antigen. We sorted a homogenous population of GFP-positive MEFs and examined the chromatin structure at the CiA:Oct4 locus. Similar to CiA ES cells, sorted VP16-activated MEFs displayed H3K4me3 and H3K27ac and lacked significant levels of H3K9me3 (Figures S4). The ability to reactivate the CiA:Oct4 locus allowed us to examine initiation and maintenance of csHP1α-dependent gene repression and compare the dynamics of heterochromatin formation between CiA fibroblasts and ES cells. VP16-activated MEFs were infected with ZFHD1-FKBP12 and csHP1α-2xFRB fusion constructs (Figure 4B). As in CiA ES cells, addition of rapamycin led to rapid csHP1α recruitment within 24 hours (Figure 4C). GFP expression was readily repressed in reactivated CiA MEFs, resembling the overall rate of silencing observed in CiA ES cells (Figures 4D and 2D). However, both the mode of GFP reduction and the formation of heterochromatin were different. In CiA ES cells, csHP1α targeting induced a bimodal transition, while a gradual reduction of GFP signal was induced in MEFs (Figure 2D and 4D). During the course of GFP silencing, H3K4me3 and H3K27ac were reduced and H3K9me3 was established (Figures 4D). To examine if nucleosome displacement during DNA replication was required for the transition from the active to repressed chromatin state, we halted cell division in CiA MEFs by serum starvation. Similar to cycling cells; non-dividing cells rapidly silenced GFP expression and established a heterochromatic domain within 5 days of csHP1α tethering (Figure S5). Hence, we conclude that replication-dependent histone exchange was not required for chromatin reprogramming, which may involve replication-independent histone variant incorporation or active demethylation instead. In contrast to CiA ES cells, where the heterochromatic domain covered approximately 10 kbp, the extent of H3K9me3 in both, cycling and starved MEF cells, was much more constricted, expanding only about 2 kbp at the CiA:Oct4 promoter (Figures 4D and S5). These data demonstrated that csHP1α targeting could overcome strong transcriptional stimulation by VP16 and induce H3K9me3-dependent repression in MEFs despite forming a smaller heterochromatic domain compared to CiA ES cells.
Heterochromatin is stable in the absence of transcription at Oct4
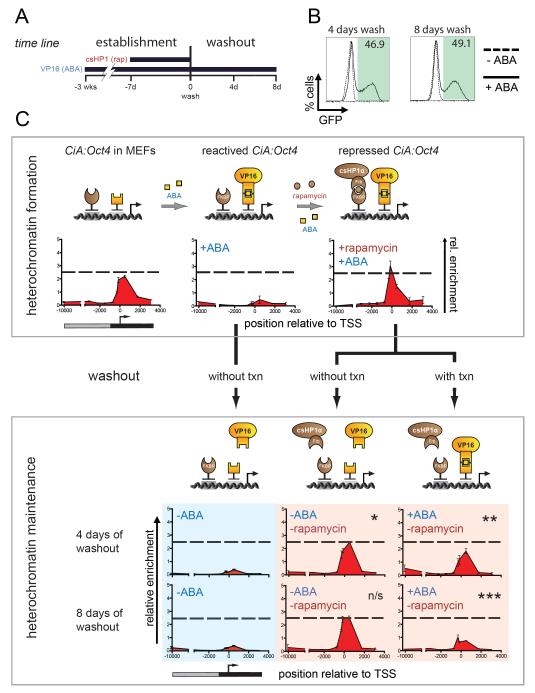
We sought to address the interplay between transcription and H3K9me3 maintenance by sequential recruitment and removal of the transcriptional activator VP16 in CiA MEFs. We made use of an orthogonal pair of CIP partners, PYL1 and ABI1, which dimerize upon binding of the plant hormone abscisic acid (ABA) (Liang et al., 2011). CiA MEFs were infected with lentiviral constructs containing GAL4-ABI1 and PYL1-VP16 (Fig 5A). Upon addition of ABA, VP16 was recruited to the CiA locus, resulting in removal of repressive histone modifications and GFP reactivation to a similar extent as the direct fusion of GAL4-VP16 (Figure 4D, Figure 5C). GFP-positive cells were enriched by FACS and maintained consistent GFP expression in the presence of ABA for weeks without defects in growth or morphology. However, within four days following ABA removal, more than 99% lost detectable GFP expression and H3K4me3 (Figure S4). Importantly, ChIP analysis did not reveal reemergence of a H3K9me3 domain, indicating that heterochromatin does not form spontaneously without transcription (Figure 5C, blue panel). In this context, the chromatin state of the CiA:Oct4 locus could be considered as “neutral” for further experimentation on H3K9me3 stability.
Figure 5. Maintenance of heterochromatin and dependence on transcription.
A) Experimental design: the CiA:Oct4 allele was reactivated in transformed CiA MEFs by abscisic acid (ABA)-mediated recruitment of VP16. GFP-positive reactivated cells were enriched by FACS. Rapamycin was added for 7 days to recruit csHP1α. GFP-negative cells were sorted by FACS. Finally, rapamycin was washed out in the presence or absence of ABA-recruited VP16. Cells were analyzed four and eight days later. B) Flow cytometry analysis after removal of csHP1α in the presence and absence of ABA-recruited VP16. C) Cartoon depicts recruitment strategy to form heterochromatin and test its maintenance. ChIP analysis of H3K9me3 along the CiA:Oct4 allele during heterochromatin formation and after csHP1α removal with or without ABA-mediated VP16 recruitment for 4 and 8 days. H3K9me3 is maintained after rapamycin washout when not opposed by ABA-mediated transcription (P-values: * p=0.052, n/s=not significant, ** p=0.007, *** p=0.004).
Heterochromatin was induced by rapamycin-mediated recruitment of csHP1α for 7 days in VP16-activated CiA MEFs. We then tested H3K9me3 stability through cell division in the absence of the initiating csHP1α stimulus. Rapamycin was removed and cells with or without ABA (recruiting VP16) were analyzed after four and eight days of washout (Figure 5). Unlike in CiA ES cells, H3K9me3 was stably maintained through cell division for at least eight days following rapamycin washout in absence of ABA (Figure 5C, red panels). H3K9me3 levels remained unchanged even when maintenance of DNA methylation was inhibited with 5azaC (Figure S6). In contrast, upon VP16 recruitment, GFP expression was significantly increased (Figure 5B) and coincided with a reduction of H3K9me3 close to basal levels after eight days of rapamycin washout. This result indicated that strong transcriptional activation could overcome repressive chromatin structure in the absence of csHP1α tethering similar to our results in CiA ES cells (Figure 5C, red panel). Most importantly however, these results demonstrated that H3K9me3 could be transmitted undiminished through numerous cell divisions in the absence of the initial stimulus.
Model of heterochromatin dynamics reveals in vivo rates of histone modification
We sought a quantitative model for our observations to rationalize the observed kinetics and spatial distribution of these marks, and to define the rates of heterochromatin formation in vivo. One model of histone marking has provided insight into metastable H3K9me switching in the yeast mate-type locus (Dodd et al., 2007). However, this model does not accurately account for the localized peaks and soft borders of the heterochromatic islands observed in our experiments (Figure S7).
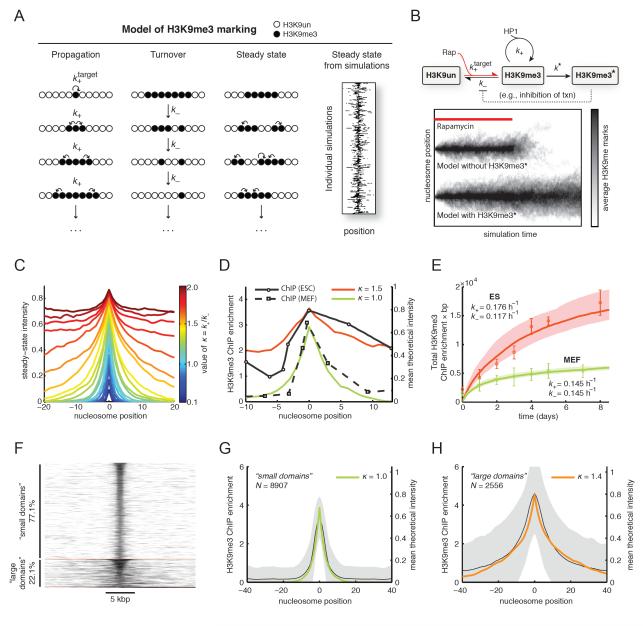
We therefore developed a novel and generalizable steady-state kinetic scheme that can be validated by Monte Carlo simulation. We considered the CiA:Oct4 locus as a one-dimensional lattice, with each lattice position corresponding to an individual nucleosome (full details in Supplemental text). We reasoned that H3K9me3 dynamics at a given locus would be governed by processes involved in either addition or removal of the mark. Our general kinetic scheme integrates all processes that propagate H3K9me3 into a single net propagation rate (k+), and integrates all processes that result in H3K9me3 removal into a separate turnover rate (k−, see Figure 6A). In our model, k+ describes the net rate of H3K9me3 addition at nucleosomes adjacent to H3K9me3-marked sites. In this model, H3K9me3 spreading along the chromosome occurs exclusively through linear propagation to neighboring nucleosomes, consistent with our experimental results and the proposed propagation model via HP1 oligomerization (Bannister et al., 2001; Canzio et al., 2011; Hall et al., 2002; Lachner et al., 2001; Schotta et al., 2002). Unlike propagation, we reasoned that turnover of H3K9me3 is equally likely everywhere, thus k− describes the stochastic turnover of H3K9me3 at any marked nucleosome. In our model, the origin corresponds to the site of csHP1α recruitment; H3K9me3 marks are nucleated at this unique site at rate k target+ in the presence of the CIP (i.e., rapamycin).
Figure 6. Kinetic model of H3K9me3 dynamics.
A) We consider chromatin as a one-dimensional beads-on-a-string lattice. Three processes: nucleation, propagation and turnover, yield a bounded steady-state island of marks. Nucleation occurs at the target site at rate k+. Propagation of the mark continues at sites immediately adjacent to marked sites, at rate k+. Turnover of the mark is equally likely everywhere at rate k−. When these processes are allowed to occur at the same time, a stochastic, bounded island of H3K9me3 marks is established at steady state. Sample output of the model with H3K9me3 domains at steady-state (right panel; each horizontal line represents a single simulation). B) Simplified kinetic scheme of H3K9me3 dynamics. Without a feedback mechanism to reinforce placement of H3K9me3 marks, the domain collapses in the absence of continued nucleation (lower panel). In the presence of a feedback mechanism that stabilizes H3K9 methylation (denoted by H3K9me3*) the domain persists. C) The profile of the steady-state island varies with κ (defined in main text). Larger values of κ increase the size of the island until κ > 1.5; above this value, the island grows without bounds. D) Fits of the experimental H3K9me3 ChIP data shown in Figure 2 to the kinetic model. Data from ES cells are best described by κ=1.5, while the data from MEFs are described by κ=1.0. E) Specific values of k+ and k− were obtained by fitting the simulations to a time course of integrated H3K9me3 ChIP enrichment at the locus (see Figure 2). Resulting values of k+ and k− are shown next to the data for each cell type. Our estimated uncertainty in these values is 35% (shaded regions). F) Clustering of genomic H3K9me3 domains in ES cells (Bilodeau et al., 2009) by k-means, with k=3. Clustering identified two predominant groups ("small" and "large" H3K9me3 domains), and a very small number of aberrant domains. G) Small H3K9me3 domains (mean ± SD) are described well by our model with κ=1.0. H) Large H3K9me3 domains (mean ± SD) are described well by our model with κ=1.4.
Our kinetic model thus describes a single nucleation site, from which H3K9me3 can be placed at rate k target+, and propagated to neighboring sites at rate k+. This propagation is opposed by random turnover at rate k−. According to this scheme, a stochastic, bounded steady-state domain of H3K9me3 centered on the csHP1α nucleation site was established (Figure 6A, right panel). This simple scheme led to the formation of a stable heterochromatic domain, which peaked at the nucleation site and displayed soft continuous borders, consistent with our observations in CiA ES cells and MEFs. Moreover, under these conditions the model predicted a rapid collapse of H3K9me3 upon withdrawal of the initiating stimulus (Figure 6B, lower panel). This was similar to the lack of H3K9me3 maintenance observed in CiA ES cells. Thus, additional mechanisms might be required to support H3K9me3 stability and epigenetic memory. We extended our dynamic competition model of H3K9me3 marking to include an additional transition that stabilized the H3K9me3 state, which we call H3K9me3*. We reasoned that this stabilization process occurs at a slow rate k*, and only at sites that are already H3K9me3 modified. In our simulations, this parameter led to sparse transitions to the H3K9me3* state, which prevented H3K9me3 turnover (Figure 6B, upper panel). Importantly, with this feedback mechanism, the H3K9me3 domain was stably maintained even upon withdrawal of the initial stimulus (Figure 6B). We conclude that besides HP1α, additional heritable feedback mechanisms (e.g. lack of transcriptional activators, DNA methylation, or chromatin compaction of the locus) are necessary to ensure epigenetic memory of H3K9me3 silencing.
Next, we determined the cellular modification and turnover rates by comparing our empirical measurements to our model of H3K9me3 marking. Our kinetic model indicated that a restricted domain of H3K9me3 naturally arises from the dynamic competition between marking and turnover when the ratio κ=k+/k− ≤ 1.5. At values of κ > 1.5, the domain of H3K9me3 marks spreads without bounds (Figure 6C). Despite the simplicity of the kinetic scheme, the profiles of the H3K9me3 domains established in simulations under these constraints fit well to our data (Figure 6D). The profile of the H3K9me3 domain in CiA ES cells is best described by κ≈1.5, and in CiA MEFs by κ≈1.0. We conclude that slight differences in the relative rates of propagation or turnover can explain the observation that the H3K9me3 domain is characteristically larger in CiA ES cells.
We then obtained specific values for nucleosome modification and turnover (k+ and k−) in each cell type by comparing the empirical rates of H3K9me3 accumulation measured by ChIP. As described above, the ratio κ was obtained by fitting the simulation profile to the in vivo steady-state H3K9me3 domain (Figure 6D). This ratio constrains the relationship between k+ and k−, requiring us only to align the timescale of the simulations to the experimental data to obtain the real rates. To perform this fit, we considered that each simulated nucleosome plus internucleosomal DNA covers a length of 200 bp and integrated our ChIP enrichment values within 3 kbp of the transcription start site. We also used the same relationship between ChIP enrichment and theoretical intensity shown in Figure 6D. We found that the rates k+ and k− in ES cells are respectively 0.176 h−1 and 0.117 h−1, and in MEFs are respectively 0.145 h−1 and 0.145 h−1 (Figure 6E). We estimated the uncertainty in these values to be ~35%. Thus, we conclude that H3K9me3 marks propagate along the chromosome, marking neighboring nucleosomes on average every ~5.7 h in ES cells and every ~6.9 h in MEFs.
Kinetic model predicts shapes of genomic H3K9me3 domains
We next compared the predictions of our H3K9me3 model with previously published H3K9me3 ChIP-seq data from mouse ES cells (Bilodeau et al., 2009). Using k-means clustering, we clustered 11,556 H3K9me3 domains into three groups (Figure 6F; full details in Supplement). This clustering separated the H3K9me3 data into similarly sized “small domains” (8,907; 77.1% of the data), “large domains” (2,556; 22.1%), and a small number of “aberrant domains” (93; 0.8%), which did not fit well in either group.
Within the “large” and “small” H3K9me3 domains, we compared the class averages of H3K9me3 ChIP intensity to our model. The average profiles of both large and small domains displayed localized H3K9me3 peaks with shapes that could be readily fitted to our model. Small H3K9me3 domains were described by κ=1.0 (Figure 6G), and large domains fit well with κ=1.4 (Figure 6H). Unlike H3K9me3 peaks, H3K36me3 is broadly distributed over active gene bodies forming plateau-shaped domains (Mikkelsen et al., 2007). Genomic H3K36me3 profiles did not cluster well into groups consistent with our model (not shown). We conclude that the general model described above is consistent with the profiles of 99.2% of non-centromeric H3K9me3 domains.
Discussion
Kinetics and bounding of heterochromatic domains
The temporal control achieved with the CiA system provided the motivation to develop a mathematical model of the kinetics and spreading of H3K9me3. We reasoned that at steady-state, H3K9me3 is governed by dynamic competition of opposing activities with a rate of addition (k+) and a rate of H3K9me3 turnover (k−). Thus, the ratio of these two rates weighs the relative contributions of various cellular processes, including csHP1α recruitment, H3K9 methylation, demethylation and nucleosomal turnover. Remarkably, by modeling different ratios of these two rates (κ), we identified good fits to the spatial distributions of H3K9me3 in both ES cells and fibroblasts. Based on these results, we were able to define the net rates of H3K9 methylation as well as the turnover rates of modified histones at the CiA:Oct4 locus in ES cells and fibroblasts. The estimated net turnover rate k− approximates previously measured rates of global histone turnover in HeLa S3 cells (Zee et al., 2010), and rates of histone H3 displacement from chromatin in Drosophila S2 cultures (Deal et al., 2010) supporting the validity of our model (Figure S7). Furthermore, our model suggests that heterochromatic boundaries need not necessarily be limited by local insulator elements. Instead, transitions between heterochromatin and euchromatin might reflect a gradual shift in the balance of activities that add and remove histone modifications. In CiA cells, H3K9me3 marking gradually decreases to either side of the csHP1α recruitment site. According to the model, this is consistent with a reduction in the maintenance of heterochromatin with increasing distance from the initiation site.
In addition to heterochromatin formation at the CiA:Oct4 locus, our model accounts for endogenous steady-state distribution profiles of H3K9me3 domains throughout the mouse genome. Indeed, an overwhelming majority (99.2%) of non-centromeric H3K9me3 domains can be described well by our model with a single variable parameter. This general parameter, κ, describes the rate of H3K9me3 propagation relative to mark turnover at a given nucleosomal position. Although these individual rates may vary considerably throughout the genome, we find that the value of κ falls within a narrow range for nearly all H3K9me3 domains. This may reflect that H3K9me3 domains are compacted and less accessible, and therefore the turnover rates within the domains are expected to be quite slow. In this case, even modest variation in the propagation rate will result in larger or smaller H3K9me3 domains.
Memory and propagation of H3K9me3 through cell generations
The ability to initiate heterochromatin formation and then to terminate csHP1α recruitment allowed us to examine the epigenetic properties of H3K9me3 through cell division. We found that the stability of H3K9 methylation domains differed between cell types and varied in the context of transcription and DNA methylation. Chemically-induced H3K9me3 was stably maintained through cell divisions upon removal of the csHP1α stimulus in CiA MEFs. In the absence of induced transcription, this maintenance did not rely on DNA methylation, as enrichment and extent of H3K9me3 was retained at low levels of promoter methylation and also after 5azaC treatment. However, upon washout of the csHP1α stimulus and recruitment of a potent transcriptional activator, H3K9me3 retention was compromised. The collapse of heterochromatin upon transcriptional activator recruitment resembled the rapid loss of H3K9me3 after washout in CiA ES cells with low DNAme or after treatment with 5azaC. While GFP reactivation was also detected in ES cells with high DNAme, this population was significantly smaller, suggesting that high levels of DNA methylation could enhance heterochromatin stability in these cells. We conclude that H3K9me3 is indeed an epigenetic mark in the strict sense in that it persists after removal of the initial stimulus. However, recruitment of transcriptional activators could disrupt heritable maintenance indicating the reversible nature of this modification.
In our model of dynamic competition, maintenance of the heterochromatic domain after rapamycin withdrawal required the establishment of a stabilized state, which we refer to as H3K9me3*. One could imagine that loss of transcriptional activators, gain of DNA methylation, changes in nuclear localization, and/or higher-order chromatin structure may contribute to the increased stability of the H3K9me3* state. Our experiments in MEFs indicated that the absence of transcriptional opposition is an important component of the H3K9me3* state. Along these lines, enhanced H3K9me3 stability in ES cells with high DNAme likely reflects the ability of CpG methylation to interfere with transcriptional activation at the Oct4 promoter.
Implications for artificial modulation of mammalian gene expression programs
Our model faithfully captures the steady-state dynamics of heterochromatin with the ratio κ=k+/k−, where the opposing effects of histone marking and mark turnover are sufficient to establish an inherently bounded H3K9me3 domain. Synthetic recruitment of the transcriptional activator VP16 to the CiA:Oct4 locus in MEFs illustrates how disruption of the steady-state conditions can compromise the maintenance of chromatin and expression state. In keeping with our model, we speculate that this rapid reactivation reflects a sudden shift in the dynamic balance in favor of k− due to recruitment of transcriptional machinery. Even in terminally differentiated primary mouse fibroblasts, we observed GFP reactivation within 24 to 48-hours of direct recruitment of VP16 to the CiA:Oct4 locus. This demonstrates that strong transcription alone can rapidly override the multiple epigenetic mechanisms involved in silencing a single allele.
Synthetic approaches to productively regulate gene expression in development and cellular reprogramming will require a better understanding of the dynamics and interactions of various chromatin modifications involved in establishment and maintenance of stable gene expression. We demonstrate that the tight temporal control of the CiA system enables quantitative studies, which in the future could be extended to different genomic loci. These approaches will facilitate the generation of integrative models of chromatin dynamics in living cells to understand how gene regulation is achieved through the regulation of chromatin structure.
Experimental Procedures
Construction and culture of Chromatin in vivo Assay at Oct4 (CiA:Oct4) ES cells
A BAC containing the mouse Pou5f1 locus was manipulated through recombineering (see details in Extended Experimental Procedures). Cells were cultured using standard conditions, see Extended Experimental Procedures.
Generation and culture of Chromatin in vivo Assay mouse embryo fibroblasts (MEF)s
CiA ES cells were injected into Bl/6 derived blastocysts and implanted into surrogate mothers. CiA:Oct4 allele presence was confirmed by agouti coat color and Southern Blot. MEFs were produced and cultured using standard conditions. See Extended Experimental Procedures for full details.
Construct design and chemical induction of proximity
All constructs were created in a modified lentivirus backbone with EF1-a promoter driving the gene of interest and a second PGK promoter driving production of a gene resistant to selection. All essential plasmids can be obtained from Addgene. Details on lentiviral production and chemical induction technique in Extended Experimental Procedures.
Retinoic acid (RA) assay and Western blots
CiA ES cells were treated with Retinoic Acid at 5uM for indicated time. Lysates were collected in RIPA buffer (150mM NaCL, 1% triton, 0.5% sodium deoxycholate, 0.1% SDS, 50mM Tris pH 8.0). 30ug/lane total protein was run on 4-12% Bis-tris gel, transferred to PVDF membranes and imaged by Infrared fluorescence (Li-Cor Biosciences) with the following antibodies: Oct-3/4 (Santa Cruz Biotech, #SCBT-9081), GFP (Clontech, #632375), GAPDH (Santa Cruz Biotech, #SCBT-32233 or #SCBT-25778).
Flow cytometry analysis and sorting
All Flow cytometry analysis was performed on an LSR II (BD Biosciences) and analyzed with FlowJo software, individual cells were gated based on forward and side scatter, auto-fluorescent cells were omitted, and remaining cells were then analyzed for GFP levels. Cells were sorted using an Aria or Aria II (BD Biosciences).
Chromatin immunoprecipitation (ChIP) Analysis
ChIP was essentially performed as described previously (Mohn et al., 2008), complete details in Extended Experimental Procedures. Antibodies used for ChIP are as follows: H3K4me3 (Millipore, #05-745R), H3K27me3 (Millipore, #07-449), H3K9me3 (Abcam, #ab8898), H3K27ac (Abcam, #ab4729), HP1 gamma (Millipore, #05-690), V5 (Invitrogen, #46-0705), GAL4 (Santa Cruz Biotechnology, sc510), Oct-3/4 (Santa Cruz Biotech, #SCBT-9081). Primers used for real-time PCR listed in Supplemental table 1.
Bisulfite sequencing analysis of DNA methylation
1 μg genomic DNA was bisulfite converted with the EpiTec Bisulfite Kit (QIAGEN). Endogenous and knock-in-specific Oct4 promoter sequences were amplified by PCR and PCR products were cloned using TOPOTA cloning kit (Invitrogen) followed by sequencing. Methylation profiles were analyzed using BiQ Analyzer software (Bock et al., 2005). Primers for PCR amplification are listed in Supplemental table 1.
DNase I sensitivity assay
DNase I sensitivity assay was carried out as previously described (Lu and Richardson, 2004), see Extended Experimental Procedures.
Supplementary Material
Highlights.
New method allows first kinetic measurements of chromatin modification in vivo
Mathematical model defines propagation and extent of heterochromatic domains
Oct4 can be rapidly activated by directed transcription within 48 hrs in fibroblasts
Heterochromatic islands are stably propagated in the absence of initial stimulus
Acknowledgements
We thank D. Schübeler and members of the Crabtree lab, in particular J.L. Ronan and A.S. Koh for insightful comments in the preparation of the manuscript. We gratefully acknowledge technical assistance by L. Chen, A. Kuo, and the Stanford FACS facility. We thank A. Sun, FS. Liang and J. Wysocka for sharing reagents. O.B. is supported by a Human Frontier Science Program fellowship. Research in the G.R.C lab is supported by NIH grants (HD55391, NS046789, AI060037) and the Howard Hughes Medical Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Athanasiadou R, de Sousa D, Myant K, Merusi C, Stancheva I, Bird A. Targeting of de novo DNA methylation throughout the Oct-4 gene regulatory region in differentiating embryonic stem cells. PLoS One. 2010;5:e9937. doi: 10.1371/journal.pone.0009937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- Bilodeau S, Kagey MH, Frampton GM, Rahl PB, Young RA. SetDB1 contributes to repression of genes encoding developmental regulators and maintenance of ES cell state. Genes Dev. 2009;23:2484–2489. doi: 10.1101/gad.1837309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock C, Reither S, Mikeska T, Paulsen M, Walter J, Lengauer T. BiQ Analyzer: visualization and quality control for DNA methylation data from bisulfite sequencing. Bioinformatics. 2005;21:4067–4068. doi: 10.1093/bioinformatics/bti652. [DOI] [PubMed] [Google Scholar]
- Bonasio R, Tu S, Reinberg D. Molecular signals of epigenetic states. Science. 2010;330:612–616. doi: 10.1126/science.1191078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canzio D, Chang EY, Shankar S, Kuchenbecker KM, Simon MD, Madhani HD, Narlikar GJ, Al-Sady B. Chromodomain-mediated oligomerization of HP1 suggests a nucleosome-bridging mechanism for heterochromatin assembly. Mol Cell. 2011;41:67–81. doi: 10.1016/j.molcel.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deal RB, Henikoff JG, Henikoff S. Genome-wide kinetics of nucleosome turnover determined by metabolic labeling of histones. Science. 2010;328:1161–1164. doi: 10.1126/science.1186777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd IB, Micheelsen MA, Sneppen K, Thon G. Theoretical analysis of epigenetic cell memory by nucleosome modification. Cell. 2007;129:813–822. doi: 10.1016/j.cell.2007.02.053. [DOI] [PubMed] [Google Scholar]
- Ebert A, Schotta G, Lein S, Kubicek S, Krauss V, Jenuwein T, Reuter G. Su(var) genes regulate the balance between euchromatin and heterochromatin in Drosophila. Genes Dev. 2004;18:2973–2983. doi: 10.1101/gad.323004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman N, Gerson A, Fang J, Li E, Zhang Y, Shinkai Y, Cedar H, Bergman Y. G9a-mediated irreversible epigenetic inactivation of Oct-3/4 during early embryogenesis. Nat Cell Biol. 2006;8:188–194. doi: 10.1038/ncb1353. [DOI] [PubMed] [Google Scholar]
- Fodor BD, Shukeir N, Reuter G, Jenuwein T. Mammalian Su(var) genes in chromatin control. Annu Rev Cell Dev Biol. 2010;26:471–501. doi: 10.1146/annurev.cellbio.042308.113225. [DOI] [PubMed] [Google Scholar]
- Fritsch L, Robin P, Mathieu JR, Souidi M, Hinaux H, Rougeulle C, Harel-Bellan A, Ameyar-Zazoua M, Ait-Si-Ali S. A subset of the histone H3 lysine 9 methyltransferases Suv39h1, G9a, GLP, and SETDB1 participate in a multimeric complex. Mol Cell. 2010;37:46–56. doi: 10.1016/j.molcel.2009.12.017. [DOI] [PubMed] [Google Scholar]
- Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- Graef IA, Holsinger LJ, Diver S, Schreiber SL, Crabtree GR. Proximity and orientation underlie signaling by the non-receptor tyrosine kinase ZAP70. EMBO J. 1997;16:5618–5628. doi: 10.1093/emboj/16.18.5618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groudine M, Weintraub H. Propagation of globin DNAase I-hypersensitive sites in absence of factors required for induction: a possible mechanism for determination. Cell. 1982;30:131–139. doi: 10.1016/0092-8674(82)90019-8. [DOI] [PubMed] [Google Scholar]
- Gruber S, Arumugam P, Katou Y, Kuglitsch D, Helmhart W, Shirahige K, Nasmyth K. Evidence that loading of cohesin onto chromosomes involves opening of its SMC hinge. Cell. 2006;127:523–537. doi: 10.1016/j.cell.2006.08.048. [DOI] [PubMed] [Google Scholar]
- Hall IM, Shankaranarayana GD, Noma K, Ayoub N, Cohen A, Grewal SI. Establishment and maintenance of a heterochromatin domain. Science. 2002;297:2232–2237. doi: 10.1126/science.1076466. [DOI] [PubMed] [Google Scholar]
- Hiragami K, Festenstein R. Heterochromatin protein 1: a pervasive controlling influence. Cell Mol Life Sci. 2005;62:2711–2726. doi: 10.1007/s00018-005-5287-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho SN, Biggar SR, Spencer DM, Schreiber SL, Crabtree GR. Dimeric ligands define a role for transcriptional activation domains in reinitiation. Nature. 1996;382:822–826. doi: 10.1038/382822a0. [DOI] [PubMed] [Google Scholar]
- Kerem BS, Goitein R, Diamond G, Cedar H, Marcus M. Mapping of DNAase I sensitive regions on mitotic chromosomes. Cell. 1984;38:493–499. doi: 10.1016/0092-8674(84)90504-x. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Lachner M, O’Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- Liang FS, Ho WQ, Crabtree GR. Engineering the ABA plant stress pathway for regulation of induced proximity. Sci Signal. 2011;4:rs2. doi: 10.1126/scisignal.2001449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q, Richardson B. DNaseI hypersensitivity analysis of chromatin structure. Methods Mol Biol. 2004;287:77–86. doi: 10.1385/1-59259-828-5:077. [DOI] [PubMed] [Google Scholar]
- Magklara A, Yen A, Colquitt BM, Clowney EJ, Allen W, Markenscoff-Papadimitriou E, Evans ZA, Kheradpour P, Mountoufaris G, Carey C, et al. An epigenetic signature for monoallelic olfactory receptor expression. Cell. 2011;145:555–570. doi: 10.1016/j.cell.2011.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ, 3rd, Voigt P, Martin SR, Taylor WR, De Marco V, et al. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature. 2009;461:762–767. doi: 10.1038/nature08398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T, Leung D, Miyashita H, Maksakova IA, Miyachi H, Kimura H, Tachibana M, Lorincz MC, Shinkai Y. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature. 2010;464:927–931. doi: 10.1038/nature08858. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D. Mechanisms for the inheritance of chromatin states. Cell. 2011;146:510–518. doi: 10.1016/j.cell.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohn F, Weber M, Rebhan M, Roloff TC, Richter J, Stadler MB, Bibel M, Schubeler D. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol Cell. 2008;30:755–766. doi: 10.1016/j.molcel.2008.05.007. [DOI] [PubMed] [Google Scholar]
- Muller H. Types of visible variations induced by X-rays in<i>Drosophila</i>. Journal of Genetics. 1930;22:299–334. [Google Scholar]
- Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, Scholer H, Smith A. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379–391. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- Nielsen SJ, Schneider R, Bauer UM, Bannister AJ, Morrison A, O’Carroll D, Firestein R, Cleary M, Jenuwein T, Herrera RE, et al. Rb targets histone H3 methylation and HP1 to promoters. Nature. 2001;412:561–565. doi: 10.1038/35087620. [DOI] [PubMed] [Google Scholar]
- Peters AH, Kubicek S, Mechtler K, O’Sullivan RJ, Derijck AA, Perez-Burgos L, Kohlmaier A, Opravil S, Tachibana M, Shinkai Y, et al. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol Cell. 2003;12:1577–1589. doi: 10.1016/s1097-2765(03)00477-5. [DOI] [PubMed] [Google Scholar]
- Peters AH, Mermoud JE, O’Carroll D, Pagani M, Schweizer D, Brockdorff N, Jenuwein T. Histone H3 lysine 9 methylation is an epigenetic imprint of facultative heterochromatin. Nat Genet. 2002;30:77–80. doi: 10.1038/ng789. [DOI] [PubMed] [Google Scholar]
- Ptashne M. On the use of the word ‘epigenetic’. Curr Biol. 2007;17:R233–236. doi: 10.1016/j.cub.2007.02.030. [DOI] [PubMed] [Google Scholar]
- Rea S, Eisenhaber F, O’Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- Sato N, Kondo M, Arai K. The orphan nuclear receptor GCNF recruits DNA methyltransferase for Oct-3/4 silencing. Biochem Biophys Res Commun. 2006;344:845–851. doi: 10.1016/j.bbrc.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Schotta G, Ebert A, Krauss V, Fischer A, Hoffmann J, Rea S, Jenuwein T, Dorn R, Reuter G. Central role of Drosophila SU(VAR)3-9 in histone H3-K9 methylation and heterochromatic gene silencing. EMBO J. 2002;21:1121–1131. doi: 10.1093/emboj/21.5.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ., 3rd SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 2002;16:919–932. doi: 10.1101/gad.973302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer DM, Wandless TJ, Schreiber SL, Crabtree GR. Controlling signal transduction with synthetic ligands. Science. 1993;262:1019–1024. doi: 10.1126/science.7694365. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Talbert PB, Henikoff S. Spreading of silent chromatin: inaction at a distance. Nat Rev Genet. 2006;7:793–803. doi: 10.1038/nrg1920. [DOI] [PubMed] [Google Scholar]
- Verschure PJ, van der Kraan I, de Leeuw W, van der Vlag J, Carpenter AE, Belmont AS, van Driel R. In vivo HP1 targeting causes large-scale chromatin condensation and enhanced histone lysine methylation. Mol Cell Biol. 2005;25:4552–4564. doi: 10.1128/MCB.25.11.4552-4564.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigler M, Levy D, Perucho M. The somatic replication of DNA methylation. Cell. 1981;24:33–40. doi: 10.1016/0092-8674(81)90498-0. [DOI] [PubMed] [Google Scholar]
- Yoo CB, Jones PA. Epigenetic therapy of cancer: past, present and future. Nat Rev Drug Discov. 2006;5:37–50. doi: 10.1038/nrd1930. [DOI] [PubMed] [Google Scholar]
- Zee BM, Levin RS, Dimaggio PA, Garcia BA. Global turnover of histone post-translational modifications and variants in human cells. Epigenetics Chromatin. 2010;3:22. doi: 10.1186/1756-8935-3-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.