Abstract
Members of the caspase family of proteases transmit the events that lead to apoptosis of animal cells. Distinct members of the family are involved in both the initiation and execution phases of cell death, with the initiator caspases being recruited to multicomponent signaling complexes. Initiation of apoptotic events depends on the ability of the signaling complexes to generate an active protease. The mechanism of activation of the caspases that constitute the different apoptosis-signaling complexes can be explained by an unusual property of the caspase zymogens to autoprocess to an active form. This autoprocessing depends on intrinsic activity that resides in the zymogens of the initiator caspases. We review evidence for a hypothesis—the induced-proximity model—that describes how the first proteolytic signal is produced after adapter-mediated clustering of initiator caspase zymogens.
Apoptosis is a mechanism that regulates cell number and is vital throughout the life of all animals. Though several different types of biochemical events have been recognized as important in apoptosis, perhaps the most fundamental is the participation of members of a family of cysteine-dependent, Asp-specific proteases known as the caspases (1–3). Caspases cleave a number of cellular proteins, and the process is one of limited proteolysis in which a small number of cuts, usually only one, are made in interdomain regions. Sometimes cleavage results in activation of the protein, sometimes in inactivation, but never in degradation, because their substrate specificity distinguishes the caspases as among the most restricted of endopeptidases.
Singularly important in this context is that caspase zymogens are themselves substrates for caspases, such that some are able to activate others in a hierarchical relationship (Fig. 1). Thus, pathways exist to transmit signals via sequential caspase activations, and this event has been most extensively examined in apoptosis. It is relatively easy to imagine that the caspases operating at the bottom of the pathway are activated by the ones above. Until recently, the questions of how the first caspase in a pathway became activated and how the first death signal was generated were perplexing issues. Now, several groups have focused on this issue (4–7) and have arrived at a consensus to describe the intriguing operation of the initiation of the proteolytic pathways that execute apoptosis. Though the basic hypothesis is supported, many issues remain to be explained, not the least of which is the nature of the mechanism that governs the process. This paper reviews the support for the hypothesis—the induced-proximity model—and its current limitations.
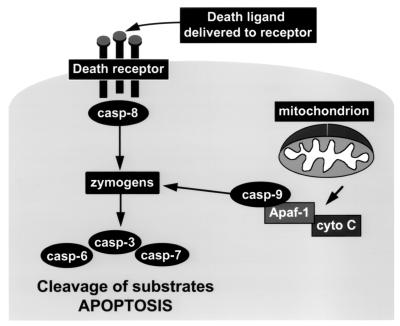
Figure 1.
The framework of apoptosis. Death may be signaled by direct ligand-enforced clustering of receptors at the cell surface, which leads to the activation of the “initiator” caspase-8 (casp-8). This caspase then directly activates the “executioner” caspases 3 and 7 (and possibly 6), which are predominantly responsible for the limited proteolysis that characterizes apoptotic dismantling of the cell. Alternatively, irreparable damage to the genome caused by mutagens, pharmaceuticals that inhibit DNA repair, or ionizing radiation leads to the activation of another initiator, caspase-9 (28). The latter event requires the recruitment of pro-caspase-9 to proteins such as Apaf-1, which requires the proapoptotic factor cytochrome c (cyto C) to be released from mitochondria (29). Though other modulators probably regulate the apoptotic pathway in a cell-specific manner (30), this framework is considered common to most mammalian cells.
Apoptosis Triggered by Death Receptors.
One of the most intensively studied pathways to cell death results from ligation of transmembrane death receptors belonging to the tumor necrosis factor-R1 (TNF-R1) family. After engagement by specific ligands, these receptors transmit a lethal signal that results in classic apoptotic cell death (8, 9). Because simple transfection of death receptors is usually sufficient to sensitize cells to a death ligand, it follows that the components required to transduce this signal reside in many cells. Thus TNF-R1 family members serve as a conduit for the transfer of death signals into the cell’s interior after interaction with their extracellular cognate ligands. The TNF-R1/TNF pair itself presents a rather complex pathway with which to dissect apoptosis initiation, because this receptor/ligand pair can signal either apoptosis or an antagonistic NF-κB-mediated survival pathway, depending on the cellular context. The TNF-R1 homologue Fas (CD95/Apo-1) has been the paradigm of choice, because addition of its cognate ligand, FasL, or even receptor agonist antibodies rapidly signals cell death (10).
Because agonist Fas antibodies can trigger apoptosis, it was possible to use them to isolate the components of the death-inducing signaling complex (DISC) that forms after Fas ligation (4, 11). A combination of yeast two-hybrid and protein-sequence analysis revealed a seemingly simple DISC, comprising Fas itself, the adapter molecule FADD, and caspase-8 (Fig. 1). This discovery revealed a potential solution to the perplexing problem of how the first proteolytic signal was generated during apoptosis, because it implicated a caspase directly in the triggering event. Before this work, receptors were thought to signal either by altering the phosphorylation status of key signaling molecules or by functioning as ion channels. Death receptors, such as Fas, signal by direct recruitment and activation of a protease (caspase-8). How exactly does the recruited zymogen become active? To understand this process as a basis for formulating an adequate hypothesis, one must understand the unusual properties of caspase zymogens that set them apart from most other proteases. Because, unlike most other proteases, simple expression of caspase zymogens in Escherichia coli usually results in their activation (12, 13). This activation results from processing that is a consequence of intrinsic proteolytic activity residing in the caspase zymogens. It is not caused by E. coli proteases, as indicated by the fact that catalytically disabled C285A (caspase-1 numbering convention) mutants fail to undergo processing.
Self-Processing of Caspase Zymogens.
In common with other protease zymogens (14), with notable exceptions (see Table 1), generation of an active caspase usually requires limited proteolysis (Fig. 2). The activating cleavage takes place within a short segment that, in the zymogen, connects the large and small subunits of the catalytic domain with both subunits containing essential components of the catalytic machinery. The location of cleavage within this segment need not be precise in vitro (15); nevertheless, the highly conserved Asp-297 (caspase-1 numbering convention) directs cleavage specificity within this segment in vivo. Proteolytic processing that results in activation usually occurs at this Asp residue, such that most activated caspases can process their own and other caspase zymogens, given sufficient time and a high enough concentration in vitro (16–18). The extent to which this processing occurs in vivo, however, is regulated by the residues surrounding Asp-297. For example, the sequence surrounding Asp-297 in the downstream executioner caspases 3 and 7 fits the extended substrate specificity of the initiator caspases 8 and 9 remarkably well (19). With the notable exception of at least caspase-2 (20), distinctions in substrate specificity within the caspase family fit closely to the S4–S1 subsite preferences deduced from synthetic peptidic substrates (19).
Table 1.
Zymogenicities of some caspases compared with two serine proteases
| Protease | Zymogenicity |
|---|---|
| Caspase-3 | >10,000 |
| Caspase-8 | 100 |
| Caspase-9 | 10 |
| Trypsin | >10,000 |
| tPA | 2–10 |
Zymogenicity is defined as the ratio of the activity of a processed protease to the activity of the zymogen on any given substrate (27). Data for trypsin and tissue plasminogen activator (tPA) are taken from ref. 27. The interesting range of zymogenicity values displayed by members of the caspase family is mirrored by members of the chymotrypsin family, with trypsin and tPA shown for comparison. Presumably, enzmes such as tPA and caspase-9 have down played the requirement for proteolysis as a mechanism of substantially increasing their activities, because allosteric regulators substitute this function: fibrin for tPA and Apaf-1 for caspase-9. In the case of tPA, specific side-chain interactions, absent in other members of the chymotrypsin family, allow activity of the zymogen. However, in the absence of a molecular structure of the caspase-8 and caspase-9 zymogens, little evidence is available to explain the high activity of the unprocessed protein. One clue is suggested by the structure of active caspases 1 and 3, each of which is composed of two catalytic units thought to arise from the dimerization of monomeric zymogens (reiewed in ref. 3). If activation of zymogens of the initiator caspases-8, 9, and CED3 operates by clustering, then the clustering phenomenon may be explained by adapter-driven homodimerization of monomers. However, as detailed in Future Directions, the molecular mechanisms are far from clear.
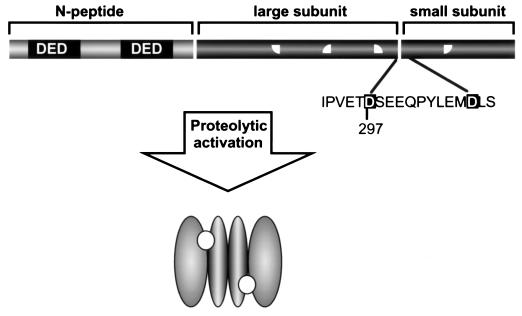
Figure 2.
Caspase activation by proteolysis. Caspases are synthesized as single-chain precursors that await activation within the cell. Activation usually proceeds in all caspases by cleavage at the conserved Asp-297 (caspase-1 numbering convention). After this activation, an as-yet undescribed conformational change is thought to occur, bringing the activity and specificity determinants (quarter circles in the linear precursor) into the correct alignment for catalysis. Frequently an N-terminal peptide is removed; however, the reason for this removal is obscure, because it is apparently not required for zymogen activation. In the example of caspase-8 shown in the figure, the N-peptide (sometimes called the prodomain) contains death-effector domains (DEDs) required for recruitment to the cytosolic face of death receptors. The crystal structures of caspases 1 and 3 reveal a dimer of small and large subunits in the active, processed state, and—it is assumed, though not specifically demonstrated—that this organization is the case for caspases in solution. The active sites in the putative dimer are shown as open circles.
The Induced-Proximity Hypothesis.
Interestingly, depending on expression conditions, one can obtain either processed active caspase or unprocessed zymogen from the same construct, at least for caspases 3, 7, and 9 (15, 21, 22). For example, short induction times (<30 min) yield unprocessed zymogens, but longer ones (>3 hours) yield fully processed enzymes. Significantly, even very short expression times and low inducer concentrations have failed to yield caspase-8 zymogens in our studies (G.S. and H. Stennicke, unpublished work). Caspase-8 processes itself extremely rapidly on heterologous expression in E. coli, suggesting that the zymogen must possess significant intrinsic proteolytic activity, allowing for autoprocessing. These observation are the basis for the induced-proximity hypothesis for the operation of the DISC, the assembly of which forces a locally high concentration of caspase-8 zymogens in a process mediated by recruited FADD (Fig. 3). This clustering of zymogens possessing intrinsic enzymatic activity would allow for processing in trans as well as activation of the first protease in the cascade.
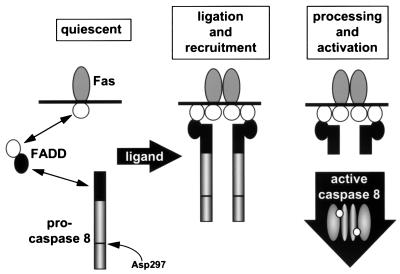
Figure 3.
Model for the operation of the DISC. Assembly of the DISC occurs in a hierarchical manner. On ligation of Fas, its “death domain” (white circle) binds to a homologous domain in the adapter FADD, which in turn recruits the zymogen of caspase-8 by a homophilic interaction requiring the homologous DEDs (black circles). Immediately after recruitment, the zymogen is processed by an adjacent zymogen, resulting in proteolytic activation and origination of active caspase-8 as the initiating death signal. Activation is thought to result from cleavage at Asp-297 (caspase-1 numbering convention). Presumably, the active form of caspase-8 (designated as a dimer as seen in the structures of active caspases 1 and 3) releases itself from the adapter after proteolytic removal of the N-terminal DED, though it is not clear how the endogenous activated enzyme distributes in the cell.
The hypothesis would need to be tested by asking whether the zymogen form of caspase-8 possessed reasonable enzymatic activity. Because such a test could not be made by expressing the wild-type precursor, a nonprocessable mutant was generated by replacing the two Asp cleavage sites within the large/small subunit linker segment with Ala. These replacements enabled the generation of a “frozen” zymogen that could be obtained in quantity after expression in E. coli. Significantly, the frozen zymogen retained the same specificity against caspase inhibitors and synthetic substrates but cleaved these substrates at 1% of the rate of an equivalent concentration of fully processed enzyme. The mechanistic origin of this rate differential is currently unknown, but, significantly, the zymogenicity of caspase-8, the ratio of its activity as a fully active enzyme to the activity of its unprocessed zymogen, was 100 (4). The importance of zymogenicity is detailed in Table 1.
Testing the Hypothesis.
The in vitro observations on the high zymogenicity of caspase-8 suggested that a test of the induced autoprocessing hypothesis was mandated, preferably in vivo. With this mandate in mind, we generated a caspase-8 construct in which the DED domains of the zymogen were replaced by a myristoylation signal, followed by three tandem repeats of a derivative FK506 binding protein (FKBP). The latter had been designed by Schreiber and colleagues (23) to act as an artificial mimic of natural cellular recruitment processes. Artificial oligomerization of proteins carrying the FKBP domains was induced by treatment with the cell penetrant FK1012, a dimeric form of FK506. Ectopic expression of the catalytically active chimera was tolerated fairly well by two human cell lines, even in the presence of monomeric FK506. However, on addition of dimeric FK1012, the cells underwent apoptosis by a mechanism that depended on the catalytic function of the chimeric caspase-8, because replacing the catalytic Cys by Ser failed to elicit the same effect. This technique, later termed the “artificial death switch” (24), has taken a prominent position in the exploration of apoptosis initiation.
These data, the in vitro observations on the zymogenicity of pro-caspase-8, and the artificially induced death of cells harboring the chimeric FKBP-caspase-8 are fully consistent with the induced-proximity model. Indeed, since this original description, the postmitochondrial initiator caspase-9 (7) and the Caenorhabditis elegans caspase CED3 (25) have both been implicated in congruent proximity activation mechanisms. Is this mechanism a common one for the basis of generating biochemical death signals? Possibly. However, a caveat must be added to the caspase-9 issue, because this caspase has a very low zymogenicity (22); in other words, it is almost as active before as it is after proteolytic processing! Thus, in the case of caspase-9, an alternative pathway may be used.
If the single-chain zymogens of caspases 8 and 9 are partly active, why are they not dangerous to healthy cells? They should cause a slow production of active executioner caspases. This question is most readily explained by the presence of endogenous caspase inhibitors, members of the IAP (inhibitor of apoptosis protein) family (31). Members of this family inhibit executioner caspases 3 and 7, and we propose that they present a barrier to caspase activity that must be exceeded before sufficient execution potential can be achieved. Thus, in the presence of IAPs, a little caspase activation is acceptable, because it would be rapidly saturated by the inhibitors. It is only when a sufficient concentration of activated executioner caspases builds up that apoptosis occurs. In this hypothesis, the IAPs regulate the apoptotic threshold.
Future Directions.
Notwithstanding the attractiveness of the induced-proximity model, there remain a number of open questions. For example, although the data support the hypothesis, the molecular mechanisms of the event(s) have not been explained, and there are a number of issues that need to be addressed in the near future. These issues are as follows. (i) Must the processed caspase-8 be released from the DISC to diffuse toward its downstream substrates? (ii) Does activation require dimerization, a consensus for the catalytic form of caspases 1 and 3 at least? (iii) Does processing occur in cis (intramolecular) or in trans (intermolecular)? (iv) Must the zymogens be specifically aligned within the recruitment complex, and how many zymogen molecules constitute an activation locus? (v) Is the minimal operative DISC as simple as the one depicted in Fig. 3, or are other proteins required (26)? These questions cut to the heart of uncertainties surrounding the fundamental activation mechanism of all the caspases, and each is (in principle) answerable by generating specific mutants and by using the artificial death-switch technique. Perhaps it is already possible to settle the issue of cis versus trans processing; in our hands, it is rarely possible to observe activation of caspase zymogens in the nanomolar range, but on artificial concentration toward the micromolar range, one observes processing and activation. This observation would imply a second-order reaction, which is most easily understood in terms of trans processing. Indeed, this proposal makes sense, because it is much easier to regulate zymogen activation in trans than in cis. The answers to these questions will require the molecular structure of at least one caspase zymogen (preferably caspase-8). Their resolution will certainly lead to a better understanding of the molecular mechanism of the DISC, with the attendant possibilities of interfering therapeutically to either initiate or prevent the commitment step in death-receptor-mediated apoptosis.
Acknowledgments
This work was supported by grants from the National Heart, Lung, and Blood Institute, National Institute on Aging, and National Institute of Neurological Disorders and Stroke.
ABBREVIATIONS
- TNF
tumor necrosis factor
- DISC
death-inducing signaling complex
- DED
death-effector domain
References
- 1.Salvesen G S, Dixit V M. Cell. 1997;91:443–446. doi: 10.1016/s0092-8674(00)80430-4. [DOI] [PubMed] [Google Scholar]
- 2.Cohen G M. Biochem J. 1997;326:1–16. doi: 10.1042/bj3260001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thornberry N A, Lazebnik Y. Science. 1998;281:1312–1316. doi: 10.1126/science.281.5381.1312. [DOI] [PubMed] [Google Scholar]
- 4.Muzio M, Stockwell B R, Stennicke H R, Salvesen G S, Dixit V M. J Biol Chem. 1998;273:2926–2930. doi: 10.1074/jbc.273.5.2926. [DOI] [PubMed] [Google Scholar]
- 5.Martin D A, Siegel R M, Zheng L, Lenardo M J. J Biol Chem. 1998;273:4345–4349. doi: 10.1074/jbc.273.8.4345. [DOI] [PubMed] [Google Scholar]
- 6.Yang X, Chang H Y, Baltimore D. Mol Cell. 1998;1:319–325. doi: 10.1016/s1097-2765(00)80032-5. [DOI] [PubMed] [Google Scholar]
- 7.Srinivasula S M, Ahmad M, Fernandes-Alnemri T, Alnemri E S. Mol Cell. 1998;1:949–957. doi: 10.1016/s1097-2765(00)80095-7. [DOI] [PubMed] [Google Scholar]
- 8.Ashkenazi A, Dixit V M. Science. 1998;281:1305–1308. doi: 10.1126/science.281.5381.1305. [DOI] [PubMed] [Google Scholar]
- 9.Ware C F, Santee S, Glass A. The Cytokine Handbook. 3rd Ed. London: Academic; 1998. pp. 549–592. [Google Scholar]
- 10.Nagata S, Goldstein P. Science. 1995;267:1449–1456. doi: 10.1126/science.7533326. [DOI] [PubMed] [Google Scholar]
- 11.Boldin M P, Goncharov T M, Goltsev Y V, Wallach D. Cell. 1996;85:803–815. doi: 10.1016/s0092-8674(00)81265-9. [DOI] [PubMed] [Google Scholar]
- 12.Orth K, O’Rourke K, Salvesen G S, Dixit V M. J Biol Chem. 1996;271:20977–20980. doi: 10.1074/jbc.271.35.20977. [DOI] [PubMed] [Google Scholar]
- 13.Stennicke H R, Salvesen G S. J Biol Chem. 1997;272:25719–25723. doi: 10.1074/jbc.272.41.25719. [DOI] [PubMed] [Google Scholar]
- 14.Neurath H. Trends Biochem Sci. 1989;14:268–271. doi: 10.1016/0968-0004(89)90061-3. [DOI] [PubMed] [Google Scholar]
- 15.Zhou Q, Salvesen G S. Biochem J. 1997;324:361–364. doi: 10.1042/bj3240361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Srinivasula S M, Ahmad M, Fernandes-Alnemri T, Litwack G, Alnemri E S. Proc Natl Acad Sci USA. 1996;93:14486–14491. doi: 10.1073/pnas.93.25.14486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muzio M, Salvesen G S, Dixit V M. J Biol Chem. 1997;272:2952–2956. doi: 10.1074/jbc.272.5.2952. [DOI] [PubMed] [Google Scholar]
- 18.Slee E A, Harte M T, Kluck R M, Wolf B B, Casiano C A, Newmeyer D D, Wang H G, Reed J C, Nicholson D W, Alnemri E S, et al. J Cell Biol. 1999;144:281–292. doi: 10.1083/jcb.144.2.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thornberry N A, Rano T A, Peterson E P, Rasper D M, Timkey T, Garcia-Calvo M, Houtzager V M, Nordstrom P A, Roy S, Vaillancourt J P, et al. J Biol Chem. 1997;272:17907–17911. doi: 10.1074/jbc.272.29.17907. [DOI] [PubMed] [Google Scholar]
- 20.Talanian R V, Quinlan C, Trautz S, Hackett M C, Mankovich J A, Banach D, Ghayur T, Brady K D, Wong W W. J Biol Chem. 1997;272:9677–9682. doi: 10.1074/jbc.272.15.9677. [DOI] [PubMed] [Google Scholar]
- 21.Stennicke H R, Jurgensmeier J M, Shin H, Deveraux Q, Wolf B B, Yang X, Zhou Q, Ellerby H M, Ellerby L M, Bredesen D, et al. J Biol Chem. 1998;273:27084–27090. doi: 10.1074/jbc.273.42.27084. [DOI] [PubMed] [Google Scholar]
- 22.Stennicke H R, Deveraux Q L, Humke E W, Reed J C, Dixit V M, Salvesen G S. J Biol Chem. 1999;274:8359–8362. doi: 10.1074/jbc.274.13.8359. [DOI] [PubMed] [Google Scholar]
- 23.Spencer D M, Belshaw P J, Chen L, Ho S N, Randazzo F, Crabtree G R, Schreiber S L. Curr Biol. 1996;6:839–847. doi: 10.1016/s0960-9822(02)00607-3. [DOI] [PubMed] [Google Scholar]
- 24.MacCorkle R A, Freeman K W, Spencer D M. Proc Natl Acad Sci USA. 1998;95:3655–3660. doi: 10.1073/pnas.95.7.3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang X, Chang H Y, Baltimore D. Science. 1998;281:1355–1357. doi: 10.1126/science.281.5381.1355. [DOI] [PubMed] [Google Scholar]
- 26.Imai Y, Kinura T, Murakami A, Yajima N, Sakamaki K, Yonehara S. Nature (London) 1999;398:777–785. doi: 10.1038/19709. [DOI] [PubMed] [Google Scholar]
- 27.Tachias K, Madison E L. J Biol Chem. 1996;271:28749–28752. doi: 10.1074/jbc.271.46.28749. [DOI] [PubMed] [Google Scholar]
- 28.Li P, Nijhawan D, Budihardjo I, Srinivasula S M, Ahmad M, Alnemri E S, Wang X. Cell. 1997;91:479–489. doi: 10.1016/s0092-8674(00)80434-1. [DOI] [PubMed] [Google Scholar]
- 29.Zou H, Henzel W J, Liu X, Lutschg A, Wang X. Cell. 1997;90:405–413. doi: 10.1016/s0092-8674(00)80501-2. [DOI] [PubMed] [Google Scholar]
- 30.Green D R, Reed J C. Science. 1998;281:1309–1312. doi: 10.1126/science.281.5381.1309. [DOI] [PubMed] [Google Scholar]
- 31.Deveraux Q, Takahashi R, Salvesen G S, Reed J C. Nature (London) 1997;388:300–304. doi: 10.1038/40901. [DOI] [PubMed] [Google Scholar]