Abstract
Accumulated evidence suggests that neurosteroids modulate GABAA receptors through binding interactions with transmembrane domains. To identify these neurosteroid binding sites directly, a neurosteroid-analog photolabeling reagent, (3α,5β)-6-azi-pregnanolone (6-AziP), was used to photolabel membranes from Sf9 cells expressing high-density, recombinant, His8-β3 homomeric GABAA receptors. 6-AziP inhibited 35S-labeled t-butylbicyclophosphorothionate binding to the His8-β3 homomeric GABAA receptors in a concentration-dependent manner (IC50 = 9 ± 1 μM), with a pattern consistent with a single class of neurosteroid binding sites. [3H]6-AziP photolabeled proteins of 30, 55, 110, and 150 kDa, in a concentration-dependent manner. The 55-, 110-, and 150-kDa proteins were identified as His8-β3 subunits through immunoblotting and through enrichment on a nickel affinity column. Photolabeling of the β3 subunits was stereoselective, with [3H]6-AziP producing substantially greater labeling than an equal concentration of its diastereomer [3H](3β,5β)-6-AziP. High-resolution mass spectrometric analysis of affinity-purified, 6-AziP-labeled His8-β3 subunits identified a single photolabeled peptide, ALLEYAF-6-AziP, in the third transmembrane domain. The identity of this peptide and the site of incorporation on Phe301 were confirmed through high-resolution tandem mass spectrometry. No other sites of photoincorporation were observed despite 90% sequence coverage of the whole β3 subunit protein, including 84% of the transmembrane domains. This study identifies a novel neurosteroid binding site and demonstrates the feasibility of identifying neurosteroid photolabeling sites by using mass spectrometry.
Introduction
Certain endogenous pregnane steroids, termed neurosteroids, are potent sedative, anxiolytic, and anesthetic agents in vertebrates (Selye, 1941; Atkinson et al., 1965; Gyermek and Soyka, 1975). There is substantial evidence that the neurobiological effects of neurosteroids are mediated through direct binding interactions with GABAA receptors, the major inhibitory neurotransmitter receptors in the nervous system. Neurosteroids modulate GABAA receptor currents in two ways; they potentiate the actions of GABA at low concentrations, whereas they directly activate GABAA receptors at higher concentrations (Lambert et al., 2009). Several lines of indirect evidence, including electrophysiological studies (Akk et al., 2004; Hosie et al., 2006, 2009) and radioligand binding studies (Evers et al., 2010), indicate that these two effects are mediated by two (or possibly more) different classes of binding sites.
A seminal article by Hosie et al. (2006) used site-directed mutagenesis to show that specific mutations in GABAA receptors could selectively prevent neurosteroid potentiation or activation. Mutations in α1 TM1 (Gln241) and TM4 (Tyr410 and Asn407) prevented potentiation and activation, whereas mutations in α1 TM1 (Thr236) and β2 TM3 (Tyr284) selectively prevented direct activation. Hosie et al. (2006) proposed a model in which a potentiating site existed within the α1 subunit, between TM1 and TM4, and an activating site existed in the cleft between the intramembranous regions of the α1 and β2 subunits. Electrophysiological experiments examining the effects of a variety of structurally modified neurosteroids on GABAA receptors with various mutations in the putative binding sites (Akk et al., 2008; Li et al., 2009) suggested that neurosteroid binding is more complex than originally suggested in the model proposed by Hosie et al. (2006). A possible alternative explanation is that the amino acid residues mutated in the study by Hosie et al. (2006) represent sites that transduce neurosteroid binding to functionally important changes in channel conformation.
One method to distinguish between these two possibilities would be to identify the neurosteroid binding sites directly, by using neurosteroid-analog photolabeling reagents. 6-azi-pregnanolone (6-AziP) is a neurosteroid-analog photolabeling reagent that was shown previously to label VDAC and β-tubulin covalently (Darbandi-Tonkabon et al., 2003, 2004; Chen et al., 2012a). Studies that used tritiated 6-AziP to label rat brain membranes and separated the photolabeled proteins with native gel electrophoresis showed that 6-AziP also labeled native GABAA receptors (data not shown).
Previous studies using anesthetic-analog photolabeling to identify drug binding sites photolabeled large quantities (∼10−9 mol) of detergent-solubilized GABAA receptors and used Edman degradation to identify the sites of photoincorporation (Li et al., 2006). This approach may not be applicable to neurosteroid photolabeling, because neurosteroids are thought to diffuse laterally through the lipid bilayer to their binding sites (Akk et al., 2005) and neurosteroid modulation of GABAA receptors is difficult to observe in detergent-solubilized receptors. The current study was aimed at determining the feasibility of identifying sites of 6-AziP photoincorporation in GABAA receptors by using mass spectrometry, an approach that requires smaller quantities of protein, which would enable photolabeling to be performed in membranes before detergent solubilization. We used His8-β3 homomeric GABAA receptors heterologously expressed in Sf9 cells as an abundant noncomplex source of receptors. β3 homomers have one class (rather than two classes) of neurosteroid binding sites, as evidenced by the binding of 35S-labeled t-butylbicyclophosphorothionate (TBPS) (Davies et al., 1997), which simplifies the analysis of mass spectrometric data. The results of this study demonstrate a novel neurosteroid binding site and illustrate both the feasibility and current limitations of using mass spectrometry to identify neurosteroid binding sites.
Materials and Methods
Membrane Preparation.
The generation of recombinant baculovirus expressing rat His8-β3 subunits of GABAA receptors and infection of Sf9 cells were described previously (Kang et al., 2008). The Sf9 cells (100 ml) were harvested through centrifugation at 1000g for 5 min and were stored at −80°C until use. The cell pellet was resuspended and homogenized in 10 ml of 10 mM sodium phosphate buffer, pH 7.5, by using a Teflon pestle in a motor-driven homogenizer. The homogenate was centrifuged at 1000g for 15 min. The supernatant was saved, and the pellet was homogenized again in 10 ml of 10 mM sodium phosphate buffer, pH 7.5. Homogenization and low-speed centrifugation were repeated twice or until the supernatant was clear. The supernatants were pooled and centrifuged at 130,000g for 45 min, to pellet the membranes. The membrane pellets were resuspended at a concentration of 1 mg of protein/ml in 10 mM potassium phosphate buffer, 100 mM KCl, containing a cocktail of protease inhibitors including 4-(2-aminoethyl)benzenesulfonyl fluoride, pepstatin A, N-[N-(l-3-trans-carboxyirane-2-carbonyl)-l-leucyl]-agmatine (E-64), bestatin, leupeptin, and aprotinin (Sigma-Aldrich, St. Louis, MO), and were stored at −80°C until use. Protein concentrations were measured by using a micro-bicinchoninic acid assay (Thermo Fisher Scientific, Waltham, MA).
Rat brain membranes were prepared as described previously (Evers et al., 2010). Transformed human embryonic kidney (TSA) cells were stably transfected with α1FLAG and β2 GABAA receptor subunits by using methods identical to those described for expression of GABAA receptors in QT6 cells (Evers et al., 2010). Cells were grown in suspension culture in Dulbecco's modified Eagle's medium/F12 medium containing 50 mM HEPES and 10% fetal bovine serum. TSA cells were harvested and membranes were prepared by using the protocol described previously for preparation of QT6 cell membranes (Evers et al., 2010).
[35S]TBPS Binding.
[35S]TBPS binding assays were performed by using previously described methods (Evers et al., 2010). Aliquots of membranes (final protein concentration, 20 μg/ml) were resuspended in 100 mM KCl, 10 mM potassium phosphate, pH 7.5, containing 1 to 2 nM [35S]TBPS (60–100 Ci/mmol; PerkinElmer Life and Analytical Sciences, Waltham, MA) and 5-μl aliquots of 6-AziP in dimethylsulfoxide solution (final steroid concentrations, 3 nM–30 μM), in a total assay volume of 1 ml. Determination of Bmax values was performed by using 30 nM [35S]TBPS. Control binding was defined as binding observed in the presence of 0.5% dimethylsulfoxide and the absence of 6-AziP. All assays contained 0.5% dimethylsulfoxide. Nonspecific binding was defined as binding observed in the presence of 200 μM picrotoxin. Assay tubes were incubated for 2 h at room temperature. A Brandel (Gaithersburg, MD) cell harvester was used for filtration of the assay tubes through Whatman glass fiber (GF/C) filter paper. The filter paper was rinsed with 4 ml of ice-cold buffer three times and dissolved in 4 ml of Safety-Solve (Research Products International, Mt. Prospect, IL). Radioactivity bound to the filters was measured with liquid scintillation counting. Each data point was assessed in triplicate, and the average specific binding values for each triplicate determination were used for curve fitting and determination of IC50. The curves describing 6-AziP inhibition of [35S]TBPS binding were fit to the Hill equation, B = Bmax/[1 + ([C]/IC50)n], where B is the amount of TBPS bound in the presence of 6-AziP, Bmax is the control binding level, [C] is the concentration of 6-AziP, IC50 is the half-maximal inhibitory concentration, and n is the Hill coefficient. All fitting was performed by using SigmaPlot 8 (SPSS Inc., Chicago IL).
Membrane Photolabeling.
Membranes (0.4 mg/ml) in 10 mM potassium phosphate buffer with 100 mM KCl were incubated with 15 or 30 μM [3H]6-AziP, 6-AziP, or pregnanolone on ice for 1 h in the dark. The samples were then irradiated in a quartz cuvette for 3 min, by using a photoreactor emitting light at >320 nm (Darbandi-Tonkabon et al., 2003). Membranes were collected through centrifugation at 130,000g for 45 min and were solubilized with SDS sample buffer (62.5 mM Tris-HCl, 2% SDS, 5% β-mercaptoethanol, 10% glycerol, and 0.1% bromphenol blue) or purification lysis buffer [1% (w/v) Triton X-100, 1 M NaCl, 80 mM imidazole, and 50 mM sodium phosphate, pH 7.5].
SDS-PAGE and Gel Slicing.
SDS-PAGE was performed by using 10% precast gels under reducing conditions (Laemmli, 1970). After electrophoresis, the gels were cut in vertical columns and sliced in 1-mm horizontal slices by using a DE 113 manual gel slicer (Hoeffer, San Francisco, CA). Slices were digested for 24 h with 4 ml of tissue solubilization solution containing 3a20TM complete counting scintillation cocktail and TS-2 tissue solubilizers (9:1; Research Products International Corp., Mount Prospect, IL), and the radioactivity in each slice was determined through scintillation counting (Bureau and Olsen, 1993).
Solubilization and Affinity Purification of GABAA Receptors.
After photolabeling, membranes were suspended in 10 mM sodium phosphate buffer, pH 7.5, at a concentration of 4 mg of protein/ml. An equal volume of 2× extraction buffer [1% (w/v) Triton X-100, 1 M NaCl, 80 mM imidazole, 50 mM sodium phosphate, pH 7.5] with deoxyribonuclease I (final concentration, 0.2 mg/ml; Sigma-Aldrich) was added to the resuspended membranes. After incubation in this buffer for 90 min on ice with stirring, the mixture was centrifuged at 130,000g for 1 h. The supernatant was loaded onto a nickel column or nickel beads that had been precleaned with 1× extraction buffer (0.5% Triton X-100, 40 mM imidazole, 500 mM NaCl, 25 mM sodium phosphate, pH 7.5). The column or nickel beads were washed with 10 bed volumes of 1× extraction buffer and washing buffer (0.1% Triton X-100, 100 mM imidazole, 300 mM NaCl, 25 mM sodium phosphate, pH 7.5). The receptors were eluted with 10 aliquots of 500 μl of elution buffer (0.1% Triton X-100, 500 mM imidazole, 25 mM sodium phosphate, 300 mM NaCl, pH 7.5). All procedures were conducted at 4°C in the presence of a protease inhibitor cocktail containing 4-(2-aminoethyl)benzenesulfonyl fluoride, pepstatin A, E-64, bestatin, leupeptin, and aprotinin (Sigma-Aldrich). The eluted fractions were analyzed through SDS-PAGE with staining by using SYPRO Ruby protein stain (Invitrogen, Carlsbad, CA) or immunoblotting with an antibody to the β2/3 subunit of the GABAA receptor (1:400 dilution; Santa Cruz Biotechnology, Santa Cruz, CA). For immunoblotting, the receptors were observed by using a horseradish peroxidase-conjugated rabbit anti-mouse Ig secondary antibody and the ECL Plus detection method (GE Healthcare, Chalfont St. Giles, Buckinghamshire, UK).
In-Solution Protein Digestion and Peptide Preparation.
On the basis of immunoblotting with an antibody to the β2/3 subunit of the GABAA receptor, fractions containing the purified β3 subunit of the GABAA receptor were pooled and concentrated to a volume of 200 μl by using a Centricon concentrator with a nominal molecular mass limit of 30 kDa (Millipore Corp., Billerica, MA). Protein digestion was conducted by using a modification (Chen et al., 2012b) of a previously described protocol (Washburn, 2008). The proteins were precipitated by using an SDS-PAGE cleanup kit (GE Healthcare), according to the manufacturer's protocol. The precipitated proteins were solubilized in 100 μl of 8 M urea, 5% RapiGest (Waters, Milford MA), 100 mM Tris, pH 8.5, for 30 min at 37°C with agitation. The proteins were precipitated again with an SDS-PAGE cleanup kit and were resuspended in 50 μl of 8 M urea, 0.1% RapiGest, 100 mM Tris, pH 8.5. Proteins were reduced through the addition of tris(2-carboxyethyl)phosphine to 5 mM and incubation for 30 min at room temperature and then were alkylated with 10 mM iodoacetamide for 30 min at room temperature in the dark. The tris(2-carboxyethyl)phosphine and iodoacetamide were quenched with 5 mM dithiothreitol for 10 min at room temperature. The proteins were then digested with 1 μg of the endoproteinase Lys-C (sequencing grade; Roche, Indianapolis, IN) overnight at 37°C. After Lys-C incubation, the sample was divided into four parts. One part was diluted with 100 mM Tris, pH 8.5, to reduce the concentration of urea to 2 M and was digested with 4 μg of sequencing-grade trypsin (Sigma-Aldrich) overnight at 37°C. The other three parts were diluted with 100 mM Tris, pH 8.5, to a concentration of 1 M urea, and CaCl2 was added to a final concentration of 10 mM. These samples were digested with 4 μg of the endoproteinase chymotrypsin (sequencing grade; Roche) for 1, 2, or 6 h at room temperature. At the conclusion of the incubations, all samples were acidified with 1% formic acid (FA) for 30 min at 37°C.
The protein digests were sequentially extracted with NuTip C4 and porous graphite carbon (PGC) tips (Glygen, Columbia, MD), by using a BRAVO robot (Agilent Technologies, Santa Clara, CA). Both the C4 and PGC tips had volumes of approximately 25 μl, in 200-μl pipette tips, and were conditioned with 125 μl of 60% acetonitrile (ACN)/1% FA and equilibrated with 125 μl of 1% ACN/1% FA. Peptides from each digestion were loaded on the C4 tips, which were then washed with 125 μl of 1% ACN/1% FA; peptides were eluted with 20 μl of 60% ACN/1% FA. Solid-phase extraction (SPE) with the C4 tips was repeated twice, to maximize peptide recovery. Peptides that did not adhere to the C4 tip were loaded onto a conditioned PGC tip and were eluted with the same protocol as used with the C4 tips. SPE with the PGC tips was also repeated twice for each digest. The triplicate elutions from each SPE step were pooled for subsequent analysis through nano-LC-LTQ-orbitrap mass spectrometry. For the samples photolabeled with [3H]6-AziP, SPE was conducted with manual pipetting, according to the same protocol. Aliquots of the samples from each step were analyzed through scintillation counting.
High-Resolution Mass Spectrometry.
Peptide mixtures were analyzed by using high-resolution nano-LC-MS with a hybrid mass spectrometer consisting of a linear quadrupole ion trap and an orbitrap (LTQ-Orbitrap XL; Thermo Fisher Scientific). Chromatographic separations were performed by using a NanoLC-1D Plus system (Eksigent, Dublin, CA) for gradient delivery and a cHiPLC-Nanoflex system (Eksigent) equipped with a 15-cm × 75-μm C18 column (ChromXP C18-CL, 3 μm, 120 Å; Eksigent). The liquid chromatograph was interfaced with the mass spectrometer with a nanospray source (PicoView PV550; New Objective, Woburn, MA). Mobile-phase components were 1% FA in water (solvent A) and 1% FA in 99% ACN (solvent B). After equilibration of the column in 98% solvent A/2% solvent B, the samples were injected (10 μl) by using an AS2 autosampler (Eksigent), at a flow rate of 750 nl/min. The peptides were separated by using an ACN gradient at 400 nl/min, as follows: isocratic elution at 2% solvent B, 0 to 5 min; 2% solvent B to 40% solvent B, 5 to 135 min; 40% solvent B to 80% solvent B, 135 to 52 min; 80% solvent B to 2% solvent B, 152 to 155 min; isocratic elution at 2% solvent B, 155 to 170 min. The total cycle time, including column equilibration, sample loading, and gradient elution of peptides, was 217 min. The survey scans (ion mass/charge ratio m/z 350–2000) (MS1) were acquired at high resolution (60,000 resolution at m/z 400) in the orbitrap in profile mode, and the product-ion mass spectrometry spectra (MS2) were acquired at 7500 resolution in the orbitrap in profile mode, after collision-induced dissociation in the linear ion trap. The maximal injection times for the MS1 scans in the orbitrap and the LTQ were both 500 ms, and the maximal injection times for the MSn scans in the orbitrap and the LTQ were 800 and 5000 ms, respectively. The automatic gain-control targets for the orbitrap and the LTQ were 5 × 105 and 3 × 104, respectively, for the MS1 scans and 2 × 105 and 1 × 104, respectively, for the MS2 scans. The MS1 scans were followed by three MS2 events in the linear ion trap with collision activation in the ion trap (parent threshold, 10,000; isolation width, 4.0 Da; normalized collision energy, 30%; activation Q, 0.250; activation time, 30 ms). Dynamic exclusion was used to remove selected precursor ions (−0.20/+1.0 Da) for 90 s after MS2 acquisition. A repeat count of 3, a repeat duration of 45 s, and a maximal exclusion list size of 500 were used. The following ion source parameters were used: capillary temperature, 200°C; source voltage, 2.7 kV; source current, 100 μA; tube lens, 79 V. The data were acquired by using Xcalibur 2.0.7 (Thermo Fisher).
Data Processing and Analysis.
The LC-MS data files for both photolabeled and nonphotolabeled samples were processed by using MASCOT Distiller 2.3.0.0 (Matrix Science Inc., Boston, MA), with previously described settings (Chen et al., 2012b). The resulting MS2 centroid-calculated files were used to search (with MASCOT 2.1.6; Matrix Science Inc.) a customized database containing the sequence of the GABAA receptor β3 subunit (Uniprot accession no. 63079). The search was conducted with no enzyme specificity and allowed nine missed cleavages, oxidation of Met, carbamidomethylation of Cys, and a 6-AziP adduct at every amino acid as variable modifications, with a parent ion tolerance of 20 ppm and a fragment ion mass tolerance of 100 milli-mass units. Scaffold 3.00.03 (Proteome Software Inc., Portland, OR) was used to validate MS2-based peptide identifications with the Protein Prophet algorithm module (Nesvizhskii et al., 2003).
The spectra that matched 6-AziP-photolabeled peptides from the MASCOT searches were filtered and interpreted by using the flowchart shown in Supplemental Fig. 1. For qualification of a candidate 6-AziP-photolabeled peptide from the MASCOT search, the first criterion was that the MS1 extracted ion chromatogram (XIC) was not observed in unlabeled (control) samples. To determine this, the unprocessed LC-MS data for both photolabeled and unlabeled samples were imported into Rosetta Elucidator 3.3 (Rosetta Biosoftware, Seattle, WA) for retention time and m/z value alignment of the peptide ion chromatograms, by using previously described parameters (Neubert et al., 2008). The MS1 XIC of the MASCOT-matched peptide was assessed for all of the samples, including the samples from the chymotrypsin time course experiments. The second criterion was that the m/z value for the parent ion of the candidate modified peptide was within the mass tolerance range determined on the basis of data for all β3 peptides from the same LC-MS analysis. To determine the applied mass tolerance, data for the β3 peptides with Peptide Prophet probability values of ≥50% and MASCOT ion scores of ≥30 from individual LC-MS analysis blocks were exported from the Scaffold file (Supplemental Table 1). Manual spectral interpretation was performed with Xcalibur, to exclude spectra not consistent with their peptide identities. The mass accuracy tolerance (mean ± 2 S.D.) for these peptide parent ions was determined through comparison of the theoretical m/z values of these peptides with the observed values. The third criterion was that the b/y ions in the MS2 spectra of the candidate 6-AziP-labeled peptides supported the peptide identities determined on the basis of the peptide ion fragmentation tables generated by using MS-Product (University of California, San Francisco, CA).
To localize the photolabeling site to a specific amino acid residue, manual spectral interpretation was performed. First, data for all β3 peptides with Peptide Prophet probability values of ≥50% and MASCOT ion scores of ≥30 from the LC-MS analyses in which the candidate photolabeled peptides were identified were exported from the Scaffold file (Supplemental Table 1). For peptides supported by more than one spectrum, the spectra with the highest peptide probability values and MASCOT scores were selected for verification (Supplemental Table 2). The fragmentation spectra of these peptides were evaluated as follows. The m/z values for the b/y ions with determinable charge states were compared with the theoretical values. The distribution of the differences (m/ztheoretical − m/zobserved) was plotted, and mean ± 2 S.D. values were used as criteria for accepting b or y ions without assigned charge states. Mean ± 3 S.D. values were used as criteria for validating ions with observed charge states (presence of a 13C isotope signal). Only b and y ions that met these criteria were assigned in the MS2 spectra.
We also searched for photolabeled peptides by using another strategy, which started at the MS1 level with time- and m/z value-aligned peptide ion chromatograms (aligned by using Rosetta Elucidator). The unprocessed data for the photolabeled and unlabeled samples were compared by aligning m/z values and LC retention times. For the unique XICs found only in the photolabeled samples, peptide mass-matching was performed to compare this list of observed, unique, parent ion m/z values with a list of theoretical values generated through in silico enzyme digestion of the β3 subunit with peptide mass increments of one or two 6-AziP moieties. The in silico enzyme digestion was performed without consideration of endoprotease specificity (“no enzyme”) and with 15 missed cleavage sites, oxidation of Met, and carbamidomethylation of Cys as variable or constant modifications (www.mmass.org). The candidate peptides with MS2 scans were verified by using the same methods as used for the candidate peptides from the aforementioned MASCOT search approach. For the peptide masses without MS2 scans from data-dependent, directed product-ion spectral acquisition (DMSA) mass spectrometry was performed by using the method described previously (Chen et al., 2012b). The data were analyzed by using MASCOT, and the candidate peptide spectral matches were interpreted as described above.
GABAA Receptor Structure Modeling.
Comparative structural models of the β3 homomeric GABAA receptor were built by using previously described methods (Akk et al., 2008). The sequence of the mature rat β3 subunit was aligned with the existing GABAA receptor sequence alignments by using the program MUSCLE (Edgar, 2004). Alternatively, the sequence of the mature rat β3 subunit was aligned with the sequence of the Caenorhabditis elegans glutamate-gated chloride channel (Glu-Cl) by using the Expresso web service (Armougom et al., 2006), which implements the T-Coffee method of multiple sequence alignment (Notredame et al., 2000). The resulting alignment was then used as input for the program Modeler (Sali and Blundell, 1993). The structural templates used were the acetylcholine-binding protein of Lymnaea stagnalis (PDB code 1I9B) (Brejc et al., 2001), chain A of the nicotinic acetylcholine receptor of Torpedo marmorata (PDB code 2BG9) (Unwin, 2005), and chain A of the C. elegans Glu-Cl (PDB codes 3RHWand 3RIF) (Hibbs and Gouaux, 2011). To improve the quality of the models, the 72-residue cytoplasmic loop connecting the M3 and M4 domains was replaced with 10 glycine residues. A total of 10 models of the β3 subunit were produced, with molecular dynamics-based refinement for each model. The model quality was then assessed by using the default penalty function and was scored with the discrete optimized protein energy method; the best model was used for further analysis (Shen and Sali, 2006). A crude model of the β3 pentameric receptor was built by superimposing copies of the best β3 model onto the individual chains of the cryo-electron microscopy-determined structure of the nicotinic acetylcholine receptor described by Unwin (2005) or Glu-Cl (PDB code 3RHW).
Results
6-AziP Modulation of [35S]TBPS Binding to His8-β3 Homomeric GABAA Receptors.
The quantity of His8-β3 homomeric GABAA receptors in Sf9 membranes was estimated by using [35S]TBPS binding. Membranes (∼20 mg of protein) were harvested from 100 ml of culture medium from Sf9 cells expressing β3 homomeric GABAA receptors. [35S]TBPS binding assays using a saturating (30 nM) concentration of radioligand yielded a receptor density of 7.8 pmol of [35S]TBPS binding/mg of protein. To confirm that 6-AziP is an allosteric modulator of β3 homomeric GABAA receptors, the ability of 6-AziP to modulate the binding of low (1–2 nM) concentrations of [35S]TBPS was measured. As shown in Fig. 1, 6-AziP inhibited [35S]TBPS binding to β3 homomeric GABAA receptors in a concentration-dependent manner. The curve was well described by a single-component Hill equation (IC50 = 9 ± 1 μM, nH = 2.4), which suggested a single class of neurosteroid binding sites, rather than the two classes (Fig. 1) observed with neurosteroid modulation of [35S]TBPS binding in native brain membranes (EC50 = 1.9 ± 0.3 μM; IC50 = 15 ± 1.2 μM) and expressed α1β2 receptors (EC50 = 1.6 ± 0.9 μM; IC50 = 9 ± 4 μM).
Fig. 1.
6-AziP modulation of [35S]TBPS binding to β3 homomeric GABAA receptors in Sf9 cell membranes, rat brain membranes, and α1β2 GABAA receptors expressed in TSA cells. 6-AziP produced one-component inhibition of [35S]TBPS binding to the recombinant His8-β3 homomeric GABAA receptors (IC50 = 9.0 ± 1 μM), whereas it produced two-component (enhancement and inhibition) modulation with rat brain membranes and α1β2 receptors.
6-AziP Photolabeling of His8-β3 Homomeric GABAA Receptors.
Initial studies were conducted by using [3H]6-AziP to determine whether β3 subunits were photolabeled and, if so, at which ligand concentrations. Membranes were photolabeled with varying concentrations of [3H]6-AziP and were analyzed through SDS-PAGE, with quantitation of radiolabel incorporation in gel slices. No detectable photolabeling of proteins of 38 to 75 kDa was observed with 1 or 3 μM 6-AziP. Membranes photolabeled with 10 μM [3H]6-AziP showed detectable counts at ∼55 kDa in gel slices, and membranes labeled with 30 μM [3H]6-AziP, the highest soluble concentration, demonstrated a prominent peak of label incorporation at 55 kDa (data not shown). These results indicated that 6-AziP concentrations of >10 μM should be used in photolabeling experiments, consistent with the 9 μM IC50 value for 6-AziP observed in radioligand binding studies. In all subsequent studies, His8-β3 homomeric GABAA receptors were photolabeled with 6-AziP concentrations of either 15 or 30 μM.
To confirm that the 55-kDa proteins photolabeled with [3H]6-AziP were β3 subunits, Sf9 cell membranes were photolabeled with 30 μM [3H]6-AziP and the His8-β3 subunits were enriched on a nickel affinity column. As illustrated in Fig. 2A, when the nickel-purified protein was analyzed through SDS-PAGE and probed with an antibody to the β2/3 subunits (bd17; Santa Cruz), major bands were observed as doublets at approximately 55, 100, and 150 kDa, consistent with the predicted molecular masses of monomers, dimers, and trimers, respectively, of His8-β3 subunits. The observation of multiple forms of β3 with the bd17 antibody is consistent with previous observations and may be attributable to preferential recognition of a modified receptor (Kannenberg et al., 1999). The enrichment procedure resulted in a relatively pure preparation of His8-β3 subunits, as illustrated in the SYPRO Ruby-stained gel comparing aliquots of unpurified lysate, flow-through from the affinity purification column, and purified receptor (Fig. 2B). The major proteins observed in the purified sample were at 55, 100, and 150 kDa and higher molecular masses, consistent with multimers of His8-β3 subunits. The [3H]6-AziP-photolabeled, nickel affinity column-purified proteins were loaded onto the same gel as the SYPRO Ruby-stained samples and were analyzed through gel slicing. The gel slices with significant radioactivity counts were located near 55 kDa (Fig. 2C), the same mass as the protein identified through SYPRO Ruby staining of nickel affinity-purified proteins and the higher-molecular mass band observed on the bd17 immunoblot. The lower-molecular mass form of β3 observed with bd17 immunoblotting may also be photolabeled, as suggested by the small peak of radioactivity at 52 kDa in Fig. 2C. These data indicate that [3H]6-AziP photolabels His8-β3 subunits expressed as homomeric GABAA receptors in Sf9 cells. Radioactivity counts were also observed in bands near 27 and 31 kDa. On the basis of our previous studies, these bands may be VDAC and VDAC fragments that copurify with GABAA receptors (Bureau and Olsen, 1993; Darbandi-Tonkabon et al., 2003). The radioactivity counts near the dye front likely represent photolabeled lipids that also copurify with GABAA receptors.
Fig. 2.
6-AziP photolabeling of His8-β3 homomeric GABAA receptors. Sf9 cell membranes (2 mg of protein) expressing His8-β3 homomeric GABAA receptors were photolabeled with 30 μM [3H]6-AziP, purified with a nickel affinity column, and analyzed through SDS-PAGE. A, the purified proteins were immunoblotted with bd17 antibody, which showed that the β3 subunits were purified as monomers, dimers, and trimers. B, the lysate (lane L), the flow-through fraction from the column (lane F), and purified proteins (lane E) were stained with SYPRO Ruby. C, scintillation counting of gel slices showed the protein bands photolabeled with [3H]6-AziP. Stars, major photolabeled form of the β3 subunit.
Stereoselectivity of Photoincorporation of [3H]6-AziP into His8-β3 Homomeric GABAA Receptors.
To determine whether [3H]6-AziP labels a specific binding site on the β3 subunit, we examined the stereoselectivity of photolabeling. The 3α-isomers of neurosteroids are anesthetics and modulators of GABAA receptors, whereas the 3β-isomers either are inactive (Pathirathna et al., 2005) or serve as GABAA receptor blockers that do not compete with 3α-isomers (Wang et al., 2002). Stereoselectivity was used as a criterion for specificity because the low affinity of 6-AziP for modulating and photolabeling β3 subunits, coupled with solubility limits for neurosteroids, constrains the use of competitive inhibition as a criterion for specificity. Equal amounts of membranes expressing the His8-β3 homomeric GABAA receptors were photolabeled with either [3H]6-AziP [(3α,5β)-6-AziP] or its diastereomer [3H](3β,5β)-6-AziP (both at 30 μM). Figure 3A shows a plot of radioactivity in slices of SDS-PAGE gels from membranes labeled with the two photolabeling reagents. A peak of radioactivity was observed near 31 kDa with both photolabeling reagents, which probably represents photolabeled VDAC. Radioactive peaks were also observed at higher molecular mass and at the dye front. An expanded view of data between 32 and 150 kDa (Fig. 3A, inset) shows that the photoincorporation of [3H]6-AziP into the 55-kDa protein band was stereoselective. The 3α-isomer-photolabeled samples demonstrated a peak of radioactivity at 55 kDa. However, no specific photolabeling peak was detected at 55 kDa for samples labeled with 3β-6-AziP. The observation that 3α-6-AziP and 3β-6-AziP exhibited different photolabeling patterns demonstrates that they must be interacting with a binding pocket that recognizes chirality and that the 3-OH group is important in the binding interaction.
Fig. 3.
Stereoselectivity of photoincorporation of [3H]6-AziP into β3 homomeric GABAA receptors. Five hundred micrograms of Sf9 cell membranes expressing His8-β3 homomeric GABAA receptors were photolabeled with 30 μM [3H]6-AziP [(3α,5β)-6-AziP] or its isomer [3H](3β,5β)-6-AziP. A, radioactivity in slices from SDS-PAGE of photolabeled β3 membranes. Inset, the 3α-isomer of 6-AziP labeled a 55-kDa protein, whereas the 3β-isomer did not. B, radioactivity in gel slices from SDS-PAGE of His8-β3 receptors purified on a nickel affinity column after photolabeling with either the 3α- or 3β-isomer of 6-AziP. The 3α-isomer labeled the 55-kDa protein (β3 subunit) twice as effectively did as the 3β-isomer.
Membrane samples photolabeled with either [3H]6-AziP (3α,5β-6-AziP) or its isomer [3H](3β,5β)-6-AziP were also affinity-purified with nickel-agarose beads and analyzed through SDS-PAGE; quantitation of radioactivity in gel slices is shown in Fig. 3B. The major band of radioactivity incorporation was at ∼55 kDa. The 3α-isomer of [3H]6-AziP produced 2 times more photolabeling in the 55-kDa band than did the 3β-isomer, which indicates the stereoselectivity of 6-AziP photolabeling. A small amount of radiolabeled 31-kDa protein copurified with the β3 subunits.
Identification of 6-AziP-Modified Peptide in GABAA Receptor β3 Subunit TM3.
To determine the site of 6-AziP photoincorporation in the β3 subunit of the GABAA receptor, 3 mg of Sf9 cell membranes expressing His8-β3 homomeric GABAA receptors were photolabeled with 15 μM 6-AziP. As a control, the same amount of protein was irradiated with UV light in the presence of pregnanolone, a 3α5β-neurosteroid without a photolabeling moiety. The samples were then purified with a nickel affinity column and prepared for electrospray ionization-LC-MS by using the workflow diagram shown in Fig. 4. The Lys-C/trypsin or Lys-C/chymotrypsin digestion and subsequent C4 or PGC SPE produced eight samples for each condition (6-AziP-photolabeled or control); each sample was analyzed through acquisition of high-resolution MS1 and MS2 spectra by using electrospray ionization-LC-MS with an LTQ-orbitrap mass spectrometer.
Fig. 4.
Flowchart for identification of the 6-AziP photolabeling site through mass spectrometry. Membranes from Sf9 cells expressing His8-β3 homomeric GABAA receptors were photolabeled with 15 μM [3H]6-AziP or irradiated with UV light in the presence of pregnanolone (15 μM). The His8-β3 homomeric GABAA receptors were then purified with a nickel affinity column and subjected to timed, in-solution digestion with several endoproteases. Peptides were recovered through sequential solid-phase extractions with C4 and PGC and were analyzed through high-resolution (HR) nano-LC-MS with electrospray ionization. O/N, overnight.
A database search using high-resolution MS2 spectra and 6-AziP incorporation as a variable modification in MASCOT identified a single peptide from one of the digests of the photolabeled sample (Lys-C/chymotrypsin 1-h digest extracted with PGC), 295-ALLEYAF-301, which was modified with a single 6-AziP moiety. This peptide eluted at 50.53 to 50.78 min and was observed as the doubly charged [M+2H]2+ ion at m/z 571.851 (Fig. 5A, inset). The m/z value of the parent ion was within 17 ppm of the theoretical value (m/z 571.841). The 95% mass accuracy tolerance that was determined for this LC-MS block was 13.8 to 19.3 ppm. To confirm that this peptide was uniquely observed in the photolabeled sample, all of the Lys-C/chymotrypsin digests of the photolabeled and control samples were aligned and XICs were analyzed for all MS1 features of m/z 571.851 and a charge of 2+ at the observed retention time for the modified peptide. As shown in Fig. 5B, an MS1 feature (m/z 571.851) with a chromatographic retention time of ∼50 min was observed in all of the Lys-C/chymotrypsin digests of the 6-AziP-photolabeled samples but not in the control samples.
Fig. 5.
Identification of a 6-AziP-modified peptide in TM3 of the His8-β3 subunit of the GABAA receptor. A, MS2 mass spectrum of ALLEYAF-6-AziP, a Lys-C/chymotrypsin-digested peptide from the TM3 segment of the GABAA receptor β3 subunit with a 6-AziP adduct on Phe301. The MS2 spectrum was acquired from an [M+2H]2+ ion (inset). B, XICs of ALLEYAF-6-AziP (m/z 571.851) from timed Lys-C/chymotrypsin digests of photolabeled and nonphotolabeled β3 subunits aligned according to retention times and m/z values by using Rosetta Elucidator. The XIC of ALLEYAF-6-AziP (m/z 571.851 and retention time of 49–51 min) was observed in all of the chymotrypsin digests from photolabeled samples and in none of the digests from nonphotolabeled samples.
To determine the modified amino acid residue of photolabeling adducts, manual spectral interpretation was performed. As shown in Fig. 5A, b2, b3, b6, y1, y2, y4, and y5 ions were observed within the fragment ion mass tolerance range and confirmed the peptide identity as ALLEYAF-6-AziP. The diagnostic y1 and b6 ions indicated that 6-AziP was specifically incorporated at Phe301.
No Observation of Other Photolabeled Peptides Despite 90% β3 Subunit Sequence Coverage.
In addition to the MS2-based MASCOT search for photolabeled peptides, we used an MS1-based search strategy with the aligned data from Rosetta Elucidator. Approximately 800 isotope groups from MS1 scans were uniquely found in the photolabeled samples. A similar number of unique groups was found in the nonphotolabeled control samples, which indicates that most of the unique features were not related to photolabeled peptides. The MS1 features unique to the photolabeled samples were mass-matched to a list of 6-AziP-photolabeled β3 peptides generated through in silico digestions with the addition of the adduct weight of one or two 6-AziP moieties. The mass-matching yielded 70 candidate features as possible photolabeled peptides. Only one of these features had an MS2 spectrum from the data-dependent acquisition mass spectrometric analysis, which was ALLEYAF-6-AziP. Fifty-five of the MS1 features from the list had intensity values of >5 × 105 and were reanalyzed with DMSA. A MASCOT search of these DMSA analyses indicated that the features either were from unrelated proteins or were β3 subunit-derived peptides that did not have a 6-AziP adduct. The remaining 14 MS1 features unique to the photolabeled samples had MS1 signal/noise ratios that precluded MS2 analysis through DMSA; none of those features was predicted to be a TMD peptide. The combined results of the LC-MS analyses of all of the photolabeled samples identified peptides (on the basis of MS2 spectra) covering 90% of the sequence of the β3 subunit (Fig. 6). The identified peptides are listed in Supplemental Table 3. The only 6-AziP-labeled peptide identified was ALLEYAF-6-AziP.
Fig. 6.
Mass spectrometric sequence coverage of the β3 subunit of the GABAA receptor. Five micrograms of Sf9 cell membranes expressing His8-β3 homomeric GABAA receptors were photolabeled with 15 μM 6-AziP. The receptors were purified with a nickel affinity column and were analyzed through mass spectrometry after Lys-C/trypsin or Lys-C/chymotrypsin digestion. Ninety percent of the peptide sequence was detected, as shown in red. Frames indicate the TMDs of the protein.
Recovery of [3H]6-AziP-Labeled Protein/Peptides during Sample Preparation.
Transmembrane domain peptides modified with a neurosteroid are hydrophobic and may be insoluble in the solvents used for sample processing. To determine the recovery of 6-AziP-modified peptides during sample preparation, Sf9 cell membranes expressing His8-β3 homomeric GABAA receptors were photolabeled with 15 μM [3H]6-AziP, which was followed by affinity purification, endoprotease enzyme digestion, and SPE, as shown in Fig. 4. For simplicity, a single Lys-C/chymotrypsin digestion (4 h) was performed. After elution from the nickel affinity column, ∼60,000 cpm of [3H]6-AziP-labeled material (4 pmol) was purified (Table 1). After delipidation (two steps of protein precipitation), 2.65 pmol of [3H]6-AziP-labeled material remained. The loss of radioactivity likely resulted from the removal of photolabeled lipids surrounding integral membrane proteins. For subsequent analysis, it was assumed that the entire 2.65 pmol of [3H]6-AziP-labeled material represented photolabeled β3 subunits. There was no loss of radioactivity after Lys-C digestion and 15% loss of counts after chymotrypsin digestion. Therefore, 2.2 pmol of labeled peptides (83%) remained soluble in 1 M urea solution after digestion. Solid-phase extraction with C4 recovered 0.8 pmol of [3H]6-AziP-labled peptides, and sequential PGC SPE recovered 0.7 pmol. A total of 1.5 pmol of [3H]6-AziP-labeled peptides was recovered from the sample preparation process, which constituted 57% of the [3H]6-AziP-labeled protein and 68% of the [3H]6-AziP-labeled peptides.
TABLE 1.
Recovery of [3H]6-AziP-labeled protein/peptides during sample preparation
Two milligrams of Sf9 cell membranes expressing His8-β3 homomeric GABAA receptors were photolabeled with 15 μM [3H]6-AziP, which was followed by affinity purification, Lys-C/chymotrypsin digestion (4 h), and SPE. Recovery was calculated on the basis of the radioactivity of [3H]6-AziP in protein/peptides.
| Photolabeled Protein/Peptide | |
|---|---|
| pmol | |
| Purified proteins | 4.00 |
| Protein precipitate 1 | 3.23 |
| Protein precipitate 2 | 2.65 |
| Lys-C digest | 2.93 |
| Chymotrypsin digest (4 h) | 2.24 |
| C4 SPE eluate | 0.80 |
| PGC SPE eluate | 0.72 |
Molecular Modeling.
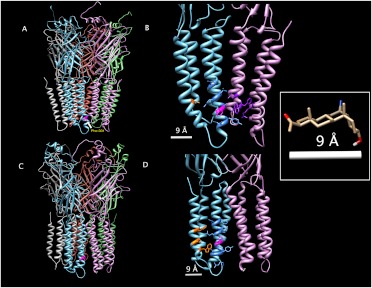
Homology modeling of the homopentameric β3 receptor, using the X-ray structures of C. elegans Glu-Cl as templates, showed Phe301 of the ALLEYAF peptide as being six amino acids removed from the beginning of the TM3-TM4 intracellular loop, facing the membrane directly (Fig. 7, A and B). Molecular structure modeling replacing the Torpedo nicotinic acetylcholine receptor structure with five GABAA receptor β3 subunits showed that Phe301 of the ALLEYAF peptide was seven amino acids removed from the beginning of the TM3-TM4 intracellular loop, facing the membrane (Fig. 7, C and D). To identify other amino acids that might interact with 6-AziP, we screened the structural models for amino acids that were within 9 Å of Phe301 (the distance between the 6′-carbon and the 20′-carbon of 6-AziP) (Fig. 7, inset) and were physically accessible to 6-AziP (e.g., the steroid did not have to pass through an α-helix). This screen yielded 11 amino acids in TM3, TM4, and TM1 and TM4 of the adjacent subunit with the Glu-Cl structure template (Fig. 7B; Table 2) and 12 amino acids in the TM3 and TM4 domains with the nicotinic acetylcholine receptor template (Fig. 7D; Table 2). These lists of candidate neurosteroid-interacting amino acids indicated that the neurosteroid binding pocket could be on the surface of TM3, at the TM3-TM4 interface, or at the interface between TM3 and TM1 or TM4 of an adjacent subunit. These lists provide candidates sites for mutagenesis; mutagenesis of amino acids identified in both models would serve to confirm the functional relevance of Phe301 photolabeling and to provide greater clarity regarding the orientation of the neurosteroid binding pocket.
Fig. 7.
Structural modeling of homomeric β3 GABAA receptors. A and C, β3 GABAA receptor models using the Glu-Cl X-ray-determined structure (A) and the nicotinic acetylcholine receptor cryo-electron microscopy-determined structure (C) as templates. B and D, enlarged view of the transmembrane regions of two adjacent subunits, showing candidate 6-AziP-interacting amino acids with Glu-Cl (B) and nicotinic acetylcholine receptor (D) as templates. In all panels, Phe301 on one subunit is colored magenta; in B and D, all 6-AziP-accessible residues within 9 Å of Phe301 are shown in stick form and residues located on TM4 are colored orange, TM3 blue, and TM1 and TM4 of the adjacent subunit purple. Inset, structure of 6-AziP with only polar hydrogens shown. The distance from C6 to the ketone oxygen is 8.42 Å; this distance was rounded to 9 Å and was used to identify the candidate 6-AziP-accessible residues shown in B and D.
TABLE 2.
6-AziP-accessible amino acids within 9 Å of Phe301
| TM3 |
TM4 |
Adjacent-Subunit TM1 |
Adjacent-Subunit TM4 |
||||
|---|---|---|---|---|---|---|---|
| Glu-Cl | nAchR | Glu-Cl | nAchR | Glu-Cl | nAchR | Glu-Cl | nAchR |
| Phe293 | Trp426 | Val238 | Arg428 | ||||
| Leu294 | Leu294 | Ser427 | Phe240 | ||||
| Leu296 | Ile429 | Trp241 | |||||
| Leu297 | Leu297 | Phe433 | |||||
| Glu298 | Glu298 | ||||||
| Ala300 | Ala300 | ||||||
| Tyr304 | Tyr304 | ||||||
| Ile305 | Ile305 | ||||||
| Arg309 | |||||||
nAchR, nicotinic acetylcholine receptor.
Discussion
Identification of Neurosteroid Photoincorporation Site.
This work was designed to test the feasibility of using mass spectrometry to study neurosteroid photolabeling of GABAA receptors. The study successfully identified a novel site of neurosteroid photoincorporation on Phe301 in TM3 of His8-β3 homomeric GABAA receptors expressed in Sf9 cells. This site seems to be specific, on the basis of stereoselective photoincorporation of 6-AziP into the β3 homomers, and to be functionally significant, on the basis of 6-AziP inhibition of [35S]TBPS binding.
Correlation of Phe301 with Previously Described Neurosteroid Binding Sites.
Evidence from electrophysiological studies indicates the existence of two classes of neurosteroid binding sites on GABAA receptors, one that is occupied at low (nanomolar) concentrations and mediates potentiation of GABA responses and one that is occupied at higher (micromolar) concentrations and mediates direct neurosteroid activation of GABAA currents. It is difficult to assign the Phe301 site to either of these functional effects, because GABA does not activate currents in β3 homomeric receptors (Davies et al., 1997) and occupancy of both classes of neurosteroid sites is thought to be required for direct activation of GABAA currents (Hosie et al., 2006). Results from radioligand binding studies also indicate two classes of neurosteroid binding sites, one that mediates enhancement of [35S]TBPS binding at nanomolar concentrations and one that mediates inhibition of [35S]TBPS binding at micromolar concentrations. As shown in Fig. 1, 6-AziP produces one-component inhibition of TBPS binding in β3 homomers, whereas it produces two-component modulation of TBPS binding in native brain and α1β2 receptors. This is consistent with previous data showing that allopregnanolone causes one-component inhibition of [35S]TBPS binding to β3 homomeric GABAA receptors (Davies et al., 1997) but has a two-component effect on GABAA receptors containing both α and β subunits. The Phe301 site photolabeled by 6-AziP correlates best with the site responsible for neurosteroid inhibition of [35S]TBPS binding. We hypothesized previously that the neurosteroid binding site mediating inhibition of [35S]TBPS binding is synonymous with the site mediating direct GABAA receptor activation, on the basis of the similar neurosteroid concentration dependence of the effects and the identification of a neurosteroid analog that competitively inhibits both neurosteroid-mediated direct current activation and neurosteroid-mediated inhibition of TBPS binding (Evers et al., 2010). Therefore, the Phe301 site identified in this study may be analogous to the direct activation site observed for heteropentameric GABAA receptors.
Two neurosteroid binding sites also have been identified through modeling of the results of site-directed mutagenesis studies with heterologously expressed α1β2 GABAA receptors. Those studies predicted a site mediating potentiation of GABA responses that is contained entirely within an α subunit and a site mediating direct neurosteroid activation that includes residues from both an α subunit and a β subunit (Hosie et al., 2006). The identified residues for the direct activation site were α1 subunit TM1 Thr236 and β2 subunit TM3 Tyr284. A model was proposed in which the neurosteroid binding site mediating direct activation existed in the cleft between the TM1 region of the α1 subunit and the TM3 region of the β2 subunit (Hosie et al., 2006, 2009). β3 homomers do not contain an α subunit, and an analogous binding pocket for neurosteroids would need to include binding residues on adjacent β3 subunits. Therefore, it is not surprising that the TM3 site we identified on the β3 subunit (Phe301) is not identical to the TM3 site on the β2 subunit (Tyr284 site). Phe301 could not have been identified as a neurosteroid interaction site in the previous mutagenesis studies because the homologous amino acid in the β2 subunit is leucine, rather than phenylanine.
Advantages and Limitations of Mass Spectrometric Identification of Photolabeled Peptides.
Detection of photolabeled sites by using mass spectrometry offers several significant advantages, compared with Edman degradation; most notably, the smaller quantity of protein required for MS analysis allows photolabeling to be performed in the native membrane environment, rather than with purified proteins in detergent solution. MS analyses also do not require purification of the photolabeled peptides, because multiple peptides can be identified and sequenced simultaneously with MS. There are several limitations to the use of MS sequencing, however. Because radiolabeled photoligands are not used in MS experiments, tracking of photolabeled peptides from labeling through sequencing is not feasible. Photolabeled peptides might be selectively lost during sample processing. This is a relevant concern because the covalent addition of a steroid molecule to a hydrophobic TMD peptide might render the peptide insoluble in MS-compatible solvents. To assess the recovery of labeled peptides, we used [3H]6-AziP to track radioactivity through purification, digestion, and solid-phase extraction. These data demonstrated high rates of recovery of radiolabeled peptides (68%). Although it is possible that radiolabeled peptides were selectively lost during sample preparation, any lost peptides would be unlikely to represent the major sites of photolabeling, because more than half of the total radioactivity was recovered. It is also possible that photolabeled peptides were irreversibly retained on the reverse-phase liquid chromatography column or were inefficiently ionized. To assess this possibility, we performed LC-MS analyses of a large, synthetic, hydrophobic, TMD peptide (IAFPLLFGIFNLVYWATYLNR, from TM4 of the α1 subunit of the GABAA receptor) photolabeled with 6-AziP. The photolabeled peptide was readily detected (data not shown), which suggests that the loss of photolabeled peptides through precipitation on the reverse-phase column or an inability to be ionized is unlikely, particularly with the smaller chymotryptic peptides generated in our experiments.
ALLEYAF-6-AziP was the only photolabeled peptide identified in our study. Identification of this peptide resulted from a thorough search at both the MS1 and MS2 levels. There are several reasons why possible additional photolabeled peptides might have eluded detection, including 1) unidentified sequences in the protein, 2) fragmentation of the photolabeling moiety during ionization, and 3) inadequate sensitivity or specificity of the method. As shown in Fig. 6, the sequence coverage of the β3 subunit was 90%, with 5 missing sequences. The same sequences were missing from the photolabeled and control (unlabeled) samples, which indicates that the sequence absence is not the result of selective loss of photolabeled peptides. Two of the missing sequences are from the extracellular N-terminal domain [75-SGIPLNL-81-(T) and 146-DEQNC-150-(T)] and contain consensus glycosylation sequons (N-X-S/T). Peptides containing these sequences would be detectable through MS only after deglycosylation (Choudhary and Mann, 2010), an approach that was not pursued because these hydrophilic peptides are unlikely to be sites of neurosteroid binding. Two missing sequences (316-KLAEKTAK-323 and 404-RRRSSQL-410) are from the TM3-TM4 intracellular loop and contain large numbers of Arg and Lys residues. The m/z values of peptides from these sequences are below the detection range of the instrument. The other missing sequence, 227-MPSILITILSWVSFWINY-244, is part of the TM1 domain. This is a potential site of neurosteroid binding, and it is possible that photolabeled peptides from this sequence were missed. It is also possible that 6-AziP undergoes fragmentation during electrospray ionization, which would result in an adduct weight for 6-AziP different from that with simple loss of two nitrogens (Brunner, 1993) and would confound the search protocols. Previous studies with 6-AziP-labeled proteins and synthetic peptides detected peptides containing an intact 6-AziP moiety and failed to detect fragmented 6-AziP adducts (Chen et al., 2012a). Therefore, it is unlikely that we failed to detect photolabeled peptides because of systematic 6-AziP fragmentation during electrospray ionization.
The most important limitations of using LC-MS to identify photolabeled peptides in a complex peptide mixture are the sensitivity and specificity of the search protocols. We identified ALLEYAF-6-AziP by searching MS2 spectra with a list generated through in silico digestion of the β3 sequence, with the added weight of 6-AziP as a variable modification. The success of this method was somewhat serendipitous, because MS2 spectra were collected by using data-dependent acquisition, in which only the highest-intensity MS1 signal (i.e., the most-abundant peptide) at a given retention time on the chromatogram generates an MS2 spectrum. Therefore, generation of an MS2 spectrum requires both an adequate MS1 signal for the labeled peptide and adequate temporal separation from more-abundant peptides. MS1 signals are more sensitive but are limited by the specificity of mass-matching (Wolski et al., 2005). To enhance the detection of possible 6-AziP-labeled peptides, we aligned the MS1 spectra from the photolabeled and control samples according to retention times and m/z values and searched for features that were unique to the photolabeled sample and had reasonable mass-matching to possible 6-AziP-labeled β3 peptides. Analysis of these features by using directed MS2 spectral acquisition identified ALLLEYAF-6-AziP but no additional photolabeled peptides. Enhanced sensitivity and specificity of detection would greatly facilitate the application of the methods used in this study to native biological samples. Both sensitivity and specificity could be enhanced through enrichment of photolabeled peptides, through either chemical or biological affinity purification of the photolabeled peptides (Duncalfe et al., 1996; MacKinnon et al., 2007). The specificity of searches of the MS1 spectra for photolabeled peptides could be greatly enhanced by incorporating stable isotopes into the photolabeling reagent (Sachon et al., 2003).
In summary, this work has identified a novel neurosteroid photoincorporation site (i.e., Phe301) in TM3 of β3 homomeric GABAA receptors expressed in Sf9 cells. The results demonstrate the feasibility, as well as some of the current limitations, of using photoaffinity labeling coupled to mass spectrometry to identify binding sites for neurosteroids, and potentially other receptor-modulating ligands, in native and expressed GABAA receptors.
Supplementary Material
Acknowledgments
We thank Petra Erdmann-Gilmore and Alan E. Davis for expert technical assistance and for performance of the LC-MS analyses and Cheryl Lichti for assistance with data searches. We thank John Braccamontes for preparation of transfected cells.
 The online version of this article (available at http://molpharm.aspetjournals.org) contains supplemental material.
The online version of this article (available at http://molpharm.aspetjournals.org) contains supplemental material.
This work was supported by the National Institutes of Health National Institute of General Medical Sciences [Grant PO1-GM47969] (to D.F.C., J.H.S., A.S.E.), [Grant 8P41-GM103422-35] (to R.R.T); the National Institutes of Health National Center for Research Resources [Grant 5P41RR000954-35] (to R.R.T), the Austrian Ministry of Science and Research; and the European Union Seventh Framework Program [Grant HEALTH-F4-2008-202088] (Neurocypres) (to W.S.).
Article, publication date, and citation information can be found at http://molpharm.aspetjournals.org.
- TM
- transmembrane region
- LC
- liquid chromatography
- MS
- mass spectrometry
- TMD
- transmembrane domain
- FA
- formic acid
- PGC
- porous graphitic carbon
- ACN
- acetonitrile
- PAGE
- polyacrylamide gel electrophoresis
- SPE
- solid-phase extraction
- MS1
- precursor-ion mass spectrometry
- MS2
- product-ion mass spectrometry
- LTQ
- linear quadrupole ion trap
- DMSA
- directed product-ion spectral acquisition
- XIC
- extracted ion chromatogram
- VDAC
- voltage-dependent anion channel
- PDB
- Protein Data Bank
- Glu-Cl
- glutamate-gated chloride channel
- 6-AziP
- 6-azi-pregnanolone
- TBPS
- t-butyl-bicyclophosphorothionate
- E-64
- N-[N-(l-3-trans-carboxyirane-2-carbonyl)-l-leucyl]-agmatine
- TSA
- transformed human embryonic kidney.
Authorship Contributions
Participated in research design: Chen, Townsend, Sieghart, and Evers.
Conducted experiments: Chen and Manion.
Contributed new reagents or analytic tools: Covey, Sieghart, and Fuchs.
Performed data analysis: Chen, Townsend, Reichert, Steinbach, and Evers.
Wrote or contributed to the writing of the manuscript: Chen and Evers.
References
- Akk G, Bracamontes JR, Covey DF, Evers A, Dao T, Steinbach JH. (2004) Neuroactive steroids have multiple actions to potentiate GABAA receptors. J Physiol 558:59–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akk G, Li P, Bracamontes J, Reichert DE, Covey DF, Steinbach JH. (2008) Mutations of the GABA-A receptor α1 subunit M1 domain reveal unexpected complexity for modulation by neuroactive steroids. Mol Pharmacol 74:614–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akk G, Shu HJ, Wang C, Steinbach JH, Zorumski CF, Covey DF, Mennerick S. (2005) Neurosteroid access to the GABAA receptor. J Neurosci 25:11605–11613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armougom F, Moretti S, Poirot O, Audic S, Dumas P, Schaeli B, Keduas V, Notredame C. (2006) Expresso: automatic incorporation of structural information in multiple sequence alignments using 3D-Coffee. Nucleic Acids Res 34:W604–W608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson RM, Davis B, Pratt MA, Sharpe HM, Tomich EG. (1965) Action of some steroids on the central nervous system of the mouse. II. Pharmacology. J Med Chem 8:426–432 [DOI] [PubMed] [Google Scholar]
- Brejc K, van Dijk WJ, Klaassen RV, Schuurmans M, van Der Oost J, Smit AB, Sixma TK. (2001) Crystal structure of an ACh-binding protein reveals the ligand-binding domain of nicotinic receptors. Nature 411:269–276 [DOI] [PubMed] [Google Scholar]
- Brunner J. (1993) New photolabeling and crosslinking methods. Annu Rev Biochem 62:483–514 [DOI] [PubMed] [Google Scholar]
- Bureau MH, Olsen RW. (1993) GABAA receptor subtypes: ligand binding heterogeneity demonstrated by photoaffinity labeling and autoradiography. J Neurochem 61:1479–1491 [DOI] [PubMed] [Google Scholar]
- Chen ZW, Chen LH, Akentieva N, Lichti CF, Darbandi R, Hastings R, Covey DF, Reichert DE, Townsend RR, Evers AS. (2012a) A neurosteroid analogue photolabeling reagent labels the colchicine-binding site on tubulin: a mass spectrometric analysis. Electrophoresis 33:666–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZW, Fuchs K, Sieghart W, Townsend RR, Evers AS. (2012b) Deep amino acid sequencing of native brain GABAA receptors using high-resolution mass spectrometry. Mol Cell Proteomics 11:M111.011445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary C, Mann M. (2010) Decoding signalling networks by mass spectrometry-based proteomics. Nat Rev Mol Cell Biol 11:427–439 [DOI] [PubMed] [Google Scholar]
- Darbandi-Tonkabon R, Hastings WR, Zeng CM, Akk G, Manion BD, Bracamontes JR, Steinbach JH, Mennerick SJ, Covey DF, Evers AS. (2003) Photoaffinity labeling with a neuroactive steroid analogue: 6-azi-pregnanolone labels voltage-dependent anion channel-1 in rat brain. J Biol Chem 278:13196–13206 [DOI] [PubMed] [Google Scholar]
- Darbandi-Tonkabon R, Manion BD, Hastings WR, Craigen WJ, Akk G, Bracamontes JR, He Y, Sheiko TV, Steinbach JH, Mennerick SJ, et al. (2004) Neuroactive steroid interactions with voltage-dependent anion channels: lack of relationship to GABAA receptor modulation and anesthesia. J Pharmacol Exp Ther 308:502–511 [DOI] [PubMed] [Google Scholar]
- Davies PA, Kirkness EF, Hales TG. (1997) Modulation by general anaesthetics of rat GABAA receptors comprised of α1β3 and β3 subunits expressed in human embryonic kidney 293 cells. Br J Pharmacol 120:899–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncalfe LL, Carpenter MR, Smillie LB, Martin IL, Dunn SM. (1996) The major site of photoaffinity labeling of the gamma-aminobutyric acid type A receptor by [3H]flunitrazepam is histidine 102 of the alpha subunit. J Biol Chem 271:9209–9214 [DOI] [PubMed] [Google Scholar]
- Edgar RC. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evers AS, Chen ZW, Manion BD, Han M, Jiang X, Darbandi-Tonkabon R, Kable T, Bracamontes J, Zorumski CF, Mennerick S, et al. (2010) A synthetic 18-norsteroid distinguishes between two neuroactive steroid binding sites on GABAA receptors. J Pharmacol Exp Ther 333:404–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyermek L, Soyka LF. (1975) Steroid anesthetics. Anesthesiology 42:331–344 [DOI] [PubMed] [Google Scholar]
- Hibbs RE, Gouaux E. (2011) Principles of activation and permeation in an anion-selective Cys-loop receptor. Nature 474:54–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosie AM, Clarke L, da Silva H, Smart TG. (2009) Conserved site for neurosteroid modulation of GABAA receptors. Neuropharmacology 56:149–154 [DOI] [PubMed] [Google Scholar]
- Hosie AM, Wilkins ME, da Silva HM, Smart TG. (2006) Endogenous neurosteroids regulate GABAA receptors through two discrete transmembrane sites. Nature 444:486–489 [DOI] [PubMed] [Google Scholar]
- Kang SU, Fuchs K, Sieghart W, Lubec G. (2008) Gel-based mass spectrometric analysis of recombinant GABAA receptor subunits representing strongly hydrophobic transmembrane proteins. J Proteome Res 7:3498–3506 [DOI] [PubMed] [Google Scholar]
- Kannenberg K, Schaerer MT, Fuchs K, Sieghart W, Sigel E. (1999) A novel serine kinase with specificity for β3-subunits is tightly associated with GABAA receptors. J Biol Chem 274:21257–21264 [DOI] [PubMed] [Google Scholar]
- Laemmli UK. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685 [DOI] [PubMed] [Google Scholar]
- Lambert JJ, Cooper MA, Simmons RD, Weir CJ, Belelli D. (2009) Neurosteroids: endogenous allosteric modulators of GABAA receptors. Psychoneuroendocrinology 34 (Suppl 1):S48–S58 [DOI] [PubMed] [Google Scholar]
- Li GD, Chiara DC, Sawyer GW, Husain SS, Olsen RW, Cohen JB. (2006) Identification of a GABAA receptor anesthetic binding site at subunit interfaces by photolabeling with an etomidate analog. J Neurosci 26:11599–11605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Bandyopadhyaya AK, Covey DF, Steinbach JH, Akk G. (2009) Hydrogen bonding between the 17β-substituent of a neurosteroid and the GABAA receptor is not obligatory for channel potentiation. Br J Pharmacol 158:1322–1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKinnon AL, Garrison JL, Hegde RS, Taunton J. (2007) Photo-leucine incorporation reveals the target of a cyclodepsipeptide inhibitor of cotranslational translocation. J Am Chem Soc 129:14560–14561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesvizhskii AI, Keller A, Kolker E, Aebersold R. (2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem 75:4646–4658 [DOI] [PubMed] [Google Scholar]
- Neubert H, Bonnert TP, Rumpel K, Hunt BT, Henle ES, James IT. (2008) Label-free detection of differential protein expression by LC/MALDI mass spectrometry. J Proteome Res 7:2270–2279 [DOI] [PubMed] [Google Scholar]
- Notredame C, Higgins DG, Heringa J. (2000) T-Coffee: a novel method for fast and accurate multiple sequence alignment. J Mol Biol 302:205–217 [DOI] [PubMed] [Google Scholar]
- Pathirathna S, Brimelow BC, Jagodic MM, Krishnan K, Jiang X, Zorumski CF, Mennerick S, Covey DF, Todorovic SM, Jevtovic-Todorovic V. (2005) New evidence that both T-type calcium channels and GABAA channels are responsible for the potent peripheral analgesic effects of 5α-reduced neuroactive steroids. Pain 114:429–443 [DOI] [PubMed] [Google Scholar]
- Sachon E, Tasseau O, Lavielle S, Sagan S, Bolbach G. (2003) Isotope and affinity tags in photoreactive substance P analogues to identify the covalent linkage within the NK-1 receptor by MALDI-TOF analysis. Anal Chem 75:6536–6543 [DOI] [PubMed] [Google Scholar]
- Sali A, Blundell TL. (1993) Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol 234:779–815 [DOI] [PubMed] [Google Scholar]
- Selye H. (1941) Anesthetic effect of steroid hormones. Proc Soc Exp Biol Med 46:116–121 [Google Scholar]
- Shen MY, Sali A. (2006) Statistical potential for assessment and prediction of protein structures. Protein Sci 15:2507–2524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unwin N. (2005) Refined structure of the nicotinic acetylcholine receptor at 4Å resolution. J Mol Biol 346:967–989 [DOI] [PubMed] [Google Scholar]
- Wang M, He Y, Eisenman LN, Fields C, Zeng CM, Mathews J, Benz A, Fu T, Zorumski E, Steinbach JH, et al. (2002) 3β-Hydroxypregnane steroids are pregnenolone sulfate-like GABAA receptor antagonists. J Neurosci 22:3366–3375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washburn MP. (2008) Sample preparation and in-solution protease digestion of proteins for chromatography-based proteomic analysis. Curr Protoc Protein Sci Chapter 23:23.6.1–23.6.11 [DOI] [PubMed] [Google Scholar]
- Wolski WE, Lalowski M, Jungblut P, Reinert K. (2005) Calibration of mass spectrometric peptide mass fingerprint data without specific external or internal calibrants. BMC Bioinformatics 6:203. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.