Abstract
Regulator of G protein signaling 2 (RGS2), a Gq-specific GTPase-activating protein, is strongly implicated in cardiovascular function. RGS2(−/−) mice are hypertensive and prone to heart failure, and several rare human mutations that accelerate RGS2 degradation have been identified among patients with hypertension. Therefore, pharmacological up-regulation of RGS2 protein levels might be beneficial. We used a β-galactosidase complementation method to screen several thousand compounds with known pharmacological functions for those that increased RGS2 protein levels. Several cardiotonic steroids (CTSs), including ouabain and digoxin, increased RGS2 but not RGS4 protein levels. CTSs increased RGS2 protein levels through a post-transcriptional mechanism, by slowing protein degradation. RGS2 mRNA levels in primary vascular smooth muscle cells were unaffected by CTS treatment, whereas protein levels were increased 2- to 3-fold. Na+/K+-ATPase was required for the increase in RGS2 protein levels, because the effect was lost in Na+/K+-ATPase-knockdown cells. Furthermore, we demonstrated that CTS-induced increases in RGS2 levels were functional and reduced receptor-stimulated, Gq-dependent, extracellular signal-regulated kinase phosphorylation. Finally, we showed that in vivo treatment with digoxin led to increased RGS2 protein levels in heart and kidney. CTS-induced increases in RGS2 protein levels and function might modify several deleterious mechanisms in hypertension and heart failure. This novel CTS mechanism might contribute to the beneficial actions of low-dose digoxin treatment in heart failure. Our results support the concept of small-molecule modulation of RGS2 protein levels as a new strategy for cardiovascular therapy.
Introduction
Regulator of G protein signaling (RGS) proteins are GTPase-accelerating proteins that deactivate GTP-bound Gα subunits, thereby reducing the amplitude and duration of G protein-coupled receptor (GPCR) signaling (Ross and Wilkie, 2000; Sjögren et al., 2010). They also represent novel potential drug targets in numerous diseases, including hypertension, heart failure, Parkinson's disease, and schizophrenia (Chowdari et al., 2002; Heximer et al., 2003; Blundell et al., 2008; Blazer and Neubig, 2009). Modulation of RGS proteins might alter GPCR signaling in a pathway- and tissue-specific manner (Blazer and Neubig, 2009). Although RGS proteins are considered to be difficult drug targets (Tesmer et al., 1997), we and others have made significant progress in developing RGS protein inhibitors (Roman et al., 2007; Blazer et al., 2010, 2011; Turner et al., 2012). In other cases, however, increasing RGS protein function may be therapeutically beneficial.
Heart failure and hypertension are major clinical problems, and treatments are only partially effective (Shah et al., 2011). Inhibition of neurohumoral signaling by catecholamines and the renin-angiotensin-aldosterone system is widely used, but mortality rates for heart failure remain high (Shah et al., 2011). Improved understanding of heart failure mechanisms and new treatments are needed.
RGS2 plays key roles in hypertension and heart failure. RGS2(−/−) mice are hypertensive (Heximer et al., 2003), show enhanced catecholamine-induced cardiomyocyte hypertrophy (Nunn et al., 2010), and exhibit worsened heart failure responses to pressure overload in vivo (Takimoto et al., 2009). RGS2 is a relatively selective GTPase-activating protein for Gq-family proteins (Heximer et al., 1997), which mediate phospholipase C and Ca2+ signaling, including that of many vasoconstrictor agents such as norepinephrine, angiotensin II, and endothelin-1. Consequently, hypertension in RGS2(−/−) mice (Heximer et al., 2003; Takimoto et al., 2009) and in humans with low RGS2 expression levels (Yang et al., 2005; Bodenstein et al., 2007; Semplicini et al., 2010) might be attributable to increased GPCR-mediated vasoconstriction. RGS2 expression is increased in patients with Bartter's/Gitelman's syndrome, a disorder characterized by low blood pressure (Calò et al., 2004).
RGS2 protein levels are selectively reduced in ventricular cardiomyocytes early in pressure-overload hypertrophy in mice, and interfering RNA-mediated knockdown of RGS2 leads to increased phenylephrine- and endothelin-induced myocyte hypertrophy (Zhang et al., 2006). Overexpression of RGS2 inhibits Gαq/11-mediated cardiac myocyte hypertrophy (Zou et al., 2006). Endogenous RGS2 also restrains angiotensin-induced cardiac fibroblast signaling, collagen synthesis, and proliferation (Zhang et al., 2011). Furthermore, RGS2 is required for the vasodilatory (Sun et al., 2005) and cardiac antihypertrophic (Takimoto et al., 2009) effects of nitric oxide and cGMP.
An important mechanism of control of RGS proteins involves ubiquitin-mediated proteasomal degradation (Lee et al., 2005, 2011; Bodenstein et al., 2007). RGS4 is degraded through the N-end rule pathway, which detects specific destabilizing amino-terminal residues (Lee et al., 2005; Bodenstein et al., 2007). RGS2 is also degraded through proteasomes in transfected HEK-293T cells (Bodenstein et al., 2007), but the specific pathway has not yet been elucidated. This rapid degradation of RGS2 and other RGS proteins through proteasomal mechanisms provides a useful control point for pharmacological modulation of RGS levels and function (Sjögren and Neubig, 2010). Given the regulatory role of RGS2 in the control of cardiovascular function and the correlations between low RGS2 protein levels and hypertension and heart failure, pharmacologically increasing RGS2 protein expression might be a novel approach to treating cardiovascular disease.
In this study, we tested a collection of known drugs to identify small molecules that increase RGS2 expression. Ouabain, digoxin, and other cardiotonic steroids (CTSs) acted at nanomolar concentrations to increase RGS2 protein levels in vitro. We also demonstrated enhanced function of RGS2 in suppressing Gq-mediated downstream signaling. Digitalis glycosides, including digoxin, are the oldest established treatments for heart failure. The precise mechanisms through which CTSs increase contractility and enhance cardiac function are not known. There has been increasing interest (Ahmed and Waagstein, 2009; Gheorghiade and Braunwald, 2009), however, in the utility of low-dose digoxin in improving clinical outcomes in heart failure. The novel effects of digoxin to increase RGS2 protein levels, as elucidated here, may contribute to improved understanding of CTS function and a new cardiovascular therapeutic strategy.
Materials and Methods
Materials.
N-(Benzyloxycarbonyl)leucinylleucinylleucinal (MG-132) was obtained from Calbiochem (San Diego, CA). Ouabain was obtained from Tocris Bioscience (Ellisville, MO). Digoxin, lactacystin, and cycloheximide were obtained from Sigma-Aldrich (St. Louis, MO). Unless indicated otherwise, all chemicals were obtained from Sigma-Aldrich and all tissue culture supplies from Invitrogen (Carlsbad, CA).
Mammalian Expression Constructs.
A 4-kDa fragment of β-galactosidase [ProLabel (PL); DiscoveRx, Freemont, CA] was added to the C terminus of human RGS2 and RGS4 (i.e., RGS2-PL and RGS4-PL). Human RGS2 and RGS4 were amplified through PCR from the pcDNA3.1 RGS2-HA and RGS4-HA vectors described previously (Bodenstein et al., 2007). Full-length RGS open reading frames, without the HA tag, were cloned into the pCMV-ProLabel-C3 vector (DiscoveRx) in-frame with the C-terminal ProLabel tag. Restriction sites for XhoI (5′) and BamHI (3′) were added to the RGS2 and RGS4 PCR primers to facilitate insertion into the pCMV-ProLabel-C3 vector. The primers for amplification of RGS2 were 5′-CGCTCGAGATGCAAAGTGCTATGTTCTTGGC-3′ (sense) and 5′- CCGCTCGAGATGCAAAGTGCTATGTTCTTGGC-3′ (antisense). The primers for amplification of RGS4 were 5′-CCGCTCGAGATGTGCAAAGGGCTTGCAGGTCTGCC-3′ (sense) and 5′-CGCGGATCCGGCACACTGAGGGACCAGGG-3′ (antisense).
Cell Culture and Transfections.
Human embryonic kidney (HEK) 293 or HEK-293T cells were maintained in a humidified incubator at 37°C with 5% CO2 and were grown to 95% confluence in Dulbecco's modified Eagle's medium (DMEM; Invitrogen) containing 4.5 g/liter glucose, 2 mM l-glutamine, 25 mM HEPES, 10% fetal bovine serum (FBS), 100 U/ml penicillin, and 100 μg/ml streptomycin. Cells were transfected by using Lipofectamine 2000 (Invitrogen) at 5 μl/μg of plasmid DNA, according to the manufacturer's recommended protocol. All transfections were performed under serum-free conditions in Opti-MEM (Invitrogen). Transfections were allowed to proceed for 4 to 5 h before the medium was changed to DMEM with 10% FBS. For transient transfections, experiments were performed 48 h after transfection.
To develop stable HEK-293 cell lines expressing RGS2-PL or RGS4-PL, 400 μg/ml G-418 (Geneticin; Invitrogen) was added to the cells 48 h after transfection. The cells were cultured until nontransfected cells died. For selection of individual clones, cells were sorted into 96-well plates (at 1 cell/well) through flow cytometry, and cells were allowed to grow in selection medium until colonies formed. Individual clones were expanded to 12-well plates and were tested for responses to MG-132 by using the PathHunter ProLabel assay.
Porcine proximal tubular epithelial (LLC-PK) cells with normal (AAC-19) or severely reduced (PY-17; Na+/K+-ATPase-null) expression of Na+/K+-ATPase (α1) were described previously (Liang et al., 2006). Cells were maintained in DMEM with 10% FBS and were allowed to grow to 95% confluence before assays. Puromycin (1 μg/ml) was included in the culture medium to maintain shRNA expression.
Preparation of Rat Aortic Vascular Smooth Muscle Cell Cultures.
All animal studies were reviewed and approved by the University of Michigan Committee on the Use and Care of Animals. Primary rat aortic vascular smooth muscle cells (VSMCs) were prepared as described previously (Atkins et al., 2009). Cells were used in passages 3 to 8.
PathHunter ProLabel β-Galactosidase Complementation Assay.
The 4-kDa PL tag on the C terminus of RGS2 and RGS4 permitted rapid quantitative assessment of protein expression. HEK-293 cells expressing RGS2-PL or RGS4-PL were trypsinized and resuspended in DMEM without phenol red (Invitrogen) containing 4.5 g/liter glucose, 2 mM l-glutamine, 25 mM HEPES, and 0.1% bovine serum albumin and were counted by using a Countess automatic cell counter (Invitrogen). Cells were diluted to 5 × 105 cells/ml and were plated at 15 × 103 cells/well in a white 384-well plate (Corning Life Sciences, Lowell, MA), in 30 μl of DMEM without phenol red containing 0.1% bovine serum albumin. Cells were allowed to attach for at least 3 h before treatment with compounds (10 μM for the screen or as indicated). At the end of treatment, the medium was removed by using an ELx406 plate washer (BioTek Instruments, Winooski, VT), and CellTiter-Fluor viability reagent (5 μl/well; Promega, Madison, WI) was added. The plate was shaken at 400 rpm for 2 min and incubated at 37°C for 30 min before fluorescence (excitation, 390 nm; emission, 505 nm) was measured with a PHERAstar plate reader (BMG Labtech, Cary, NC).
The PathHunter ProLabel protein expression assay (DiscoveRx) was performed immediately after the viability assay, according to the manufacturer's general protocol. Chemiluminescence/lysis reagent was prepared by combining 1 part Galacton-Star substrate, 5 parts Emerald II enhancer, 19 parts chemiluminescence substrate, and 25 parts lysis buffer. Chemiluminescence/lysis reagent was added to each well at 10 μl/well, and the plate was shaken at 400 rpm for 2 min. Plates were incubated at room temperature for an additional 5 min to allow complete cell lysis. Five microliters of enzyme acceptor (incomplete β-galactosidase) were added, and the plate was shaken at 400 rpm for 2 min and then incubated in the dark at room temperature for 3 h. Chemiluminescence corresponding to relative RGS2 or RGS4 protein expression was detected with a PHERAstar plate reader.
Preparation of Cell Lysates.
HEK-293T cells transiently transfected with C-terminally HA-tagged RGS2 (Bodenstein et al., 2007) were plated in 12-well plates in DMEM with 10% FBS and were allowed to grow to 95% confluence before treatment with compound in DMEM with 0.5% FBS. Cells were harvested and lysed on ice through removal of the medium and addition of 100 μl of radioimmunoprecipitation assay buffer containing protease inhibitors [20 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1 mM β-glycerophosphate, 1% Triton X-100, and 0.1% SDS, with complete protease inhibitor cocktail (Roche, Pleasanton, CA)]. For studies of agonist-induced p42/44-ERK1/2 phosphorylation, 2 mM sodium orthovanadate was added to the lysis buffer to inhibit phosphatases. The plate was shaken for 5 min at 4°C, and lysates were transferred to plastic centrifuge tubes. Samples were sonicated for 10 min at 4°C in a bath sonicator and centrifuged at 500g for 3 min, and the supernatant was used for SDS-PAGE and immunoblotting.
For detection of endogenous RGS2 protein expression in VSMCs, AAC-19 cells, or PY-17 cells, cells were plated in six-well plates in DMEM with 0.5% FBS and were allowed to grow to 95% confluence before treatment with the indicated compounds. Cells were trypsinized and harvested in 500 μl of phosphate-buffered saline on ice. Cells were pelleted through centrifugation at 500g for 5 min, and the pellets were lysed through addition of 80 μl of radioimmunoprecipitation assay buffer. All samples for Western blotting were sonicated for 10 min at 4°C in a bath sonicator and centrifuged at 500g for 3 min, and the supernatant was used for SDS-PAGE and immunoblotting.
SDS-PAGE and Immunoblotting.
Protein concentrations in the cell lysates were determined by using the bicinchoninic acid assay (Thermo Fisher Scientific, Waltham, MA) and were adjusted with an appropriate volume of Laemmli buffer (Bio-Rad Laboratories, Hercules, CA). Equal amounts of protein in each lane were resolved for 1 h at 160 V on a 12% SDS-polyacrylamide gel. Samples were transferred to an Immobilon-P membrane (Millipore Corp., Billerica, MA) for 1 h at 100 V and 400 mA, on ice, and were subjected to Western immunoblotting analysis.
The membrane was blocked for 30 min at room temperature in Tris-buffered saline-Tween 20 (TBS-T) (10 mM Tris, pH 8.0, 150 mM NaCl, and 0.1% Tween 20) with 5% (w/v) nonfat dry milk, on an orbital shaker. The membrane was probed overnight at 4°C with primary antibody diluted in TBS-T with 5% (w/v) nonfat dry milk. Rat anti-HA antibody was obtained from Roche (1:1000), rabbit anti-RGS2 antibody was a gift from Dr. David Siderovski (1:2000), rabbit anti-RGS4 antibody was a kind gift from Susan Mumby (1:5000) (Krumins et al., 2004), anti-p42/44-ERK1/2 antibody was obtained from Cell Signaling Technology (Danvers, MA; 1:1000), and total ERK1/2-specific antibody was obtained from Santa Cruz Biotechnology (Santa Cruz, CA; 1:1000). Rabbit anti-glyceraldehyde-3-phosphate dehydrogenase antibody was obtained from Cell Signaling Technology (1:5000).
The membrane was washed four times with TBS-T and was probed for 1 h at room temperature with horseradish peroxidase-conjugated secondary antibody diluted in TBS-T with 5% (w/v) nonfat dry milk. Rabbit anti-rat Ig (1:10,000) and goat anti-rabbit Ig (1:10,000) antibodies were obtained from Sigma-Aldrich, and goat anti-mouse Ig antibody (1:20,000) was obtained from Santa Cruz Biotechnology. Horseradish peroxidase-conjugated anti-actin antibody (1:10,000; Santa Cruz Biotechnology) was used as a loading control. After four washes with TBS-T, the protein bands were observed on autoradiography film by using Super Signal West Pico chemiluminescence substrate (Thermo Fisher Scientific); images were scanned and quantified by using ImageJ software (National Institutes of Health, Bethesda, MD).
Quantitative Real-Time PCR Assays.
RNA was extracted from VSMCs with an RNeasy Mini kit (QIAGEN, Valencia, CA), according to the manufacturer's instructions. After treatment with DNase, 1 μg of total RNA was reverse-transcribed into cDNA with random hexamers by using a cDNA reverse transcription kit (TaqMan; Applied Biosystems, Foster City, CA). Quantitative real-time PCR was performed in 20-μl reactions containing 1 μl of the cDNA sample and 0.3 μM forward and reverse primers with the RT2 SYBR Green qPCR Master Mix (SABiosciences, Frederick, MD). Primers for RGS2 (Doupnik et al., 2001) and β-actin (Schoenfeld et al., 1998) were described previously. Reaction mixtures were incubated at 95°C for 10 min, followed by 40 cycles of 95°C for 30 s, 60°C for 1 min, and 72°C for 30 s. No-template and no-reverse transcriptase control samples were included in each experiment, to detect RNA and/or DNA contamination. Quantification of relative RGS2 mRNA expression levels was performed with the threshold-cycle difference method (Livak and Schmittgen, 2001), with β-actin as an endogenous control.
In Vivo Treatment with Digoxin and RGS2 Expression in Heart.
All animal protocols were consistent with guidelines from the National Institutes of Health and the University of Michigan Committee on the Use and Care of Animals, which approved all procedures. Male C57BL/6 mice (8–13 weeks of age) were housed in cages under specific pathogen-free conditions, maintained in a temperature-controlled room with a 12-h light/dark cycle, and provided with standard chow and water ad libitum.
Osmotic minipumps (model 2002; Alzet, Cupertino, CA) with 0.9% saline solution containing dimethylsulfoxide (0.04%; control) or digoxin (2 μg/kg per day) were incubated overnight at 37°C in sterile saline solution before implantation. Animals were anesthetized with a mixture of ketamine (100 mg/kg) and xylazine (7.5 mg/kg) and then received a small midline incision at the base of the scapula. The skin was retracted, and a small subcutaneous pocket was prepared for osmotic minipump placement. All pumps were inserted with the flow moderator pointed posteriorly away from the surgical site. After 7 days of treatment, the animals were euthanized through intraperitoneal injection of pentobarbital (100 mg/kg), and tissues were removed.
Data Analysis.
All data were analyzed by using Prism 5.0 (GraphPad Software Inc., San Diego, CA). Dose-response curves were fit by using nonlinear regression. Data sets with three or more groups were analyzed by using one-way analysis of variance with Bonferroni's post hoc test for multiple comparisons, and data sets with two groups were analyzed by using Student's t test. Data are presented as mean ± S.D., and p values of <0.05 were considered significant.
Results
RGS2 and RGS4 Were Degraded through Proteasomes.
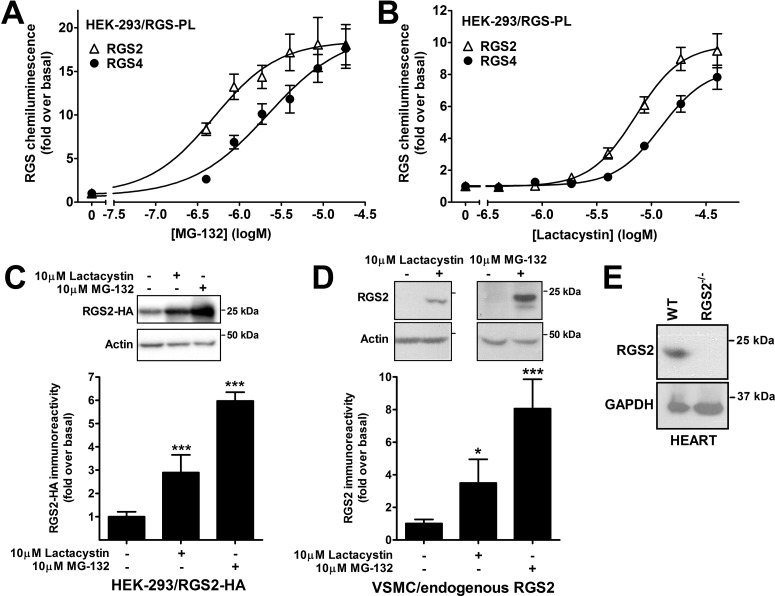
Pharmacologically increasing RGS2 protein expression (and thereby function) might represent a novel strategy for the treatment of hypertension and heart failure. To identify small molecules that selectively increase RGS2 protein expression, we used the PathHunter ProLabel β-galactosidase complementation assay. HEK-293 cell clones stably expressing human RGS2 or RGS4 C-terminally tagged with a 4-kDa part of β-galactosidase (i.e., RGS2-PL and RGS4-PL) were developed. Because both RGS2 and RGS4 were shown previously to be degraded through proteasomes (Bodenstein et al., 2007), the cell lines were validated by using the proteasome inhibitors MG-132 (Fig. 1A) and lactacystin (Fig. 1B). Both compounds increased the chemiluminescence signal corresponding to RGS2 and RGS4 proteins, in a concentration-dependent (Fig. 1) and time-dependent (Supplemental Fig. 1) manner. The increased levels of RGS2 protein expression after proteasome inhibitor treatment were confirmed by using HEK-293T cells transiently transfected with HA-tagged human RGS2 (Fig. 1C). Furthermore, we demonstrated for the first time that endogenous RGS2 protein levels in VSMCs were increased by MG-132 and lactacystin (Fig. 1D).
Fig. 1.
RGS2 and RGS4 were degraded by proteasomes. A and B, HEK-293 cells transfected with either RGS2 or RGS4 C-terminally tagged with the 4-kDa ProLabel tag were treated with the proteasome inhibitors MG-132 (A) or lactacystin (B) overnight at 37°C. Both inhibitors increased RGS2 and RGS4 chemiluminescence in a dose-dependent manner. C and D, these results were confirmed with Western blotting of HEK-293 cells transfected with HA-tagged RGS2 (C) and vascular smooth muscle cells with endogenously expressed RGS2 protein (D). E, the specificity of the RGS2 antibody was confirmed with Western blotting with heart tissue from wild-type and RGS2(−/−) mice. This antibody was described previously (Takimoto et al., 2009). WT, wild-type; GAPDH, glyceraldehyde-3-phosphate dehydrogenase. *, p < 0.05; ***, p < 0.001, using one-way analysis of variance with Bonferroni's post hoc test for multiple comparisons.
Small-Molecule Enhancers of RGS2 Protein Expression Were Identified through High-Throughput Screening.
The stable HEK-293 cell lines expressing RGS2-PL and RGS4-PL were tested in a small-molecule screen with the PathHunter ProLabel assay. To assess and to correct for toxicity of the compounds, the assay was multiplexed with a fluorescence viability assay (CellTiter-Fluor; Promega) in the same wells. MG-132 was used as a positive control for RGS up-regulation, and two compound libraries, namely, Microsource Spectrum 2000 and the Biofocus National Institutes of Health Clinical Collection (collection of drugs approved by the U.S. Food and Drug Administration), were screened at a concentration of 10 μM (∼2900 compounds). Both libraries contain compounds with previously defined pharmacological properties.
Hits in the screen were defined as compounds that increased RGS protein expression >3 S.D. above baseline values (approximately 20% or greater). Only one of the 2900 compounds (thimerosal) affected the expression of both RGS2 and RGS4, which demonstrates distinct mechanisms of regulation and the strong specificity of the assay. Compounds shown to increase the chemiluminescence signal corresponding to RGS2 protein levels in the primary screen were analyzed and divided into groups on the basis of their known pharmacological features (Supplemental Table 1). One group of compounds identified was the CTSs (i.e., digitalis glycosides). These compounds are known for their ability to inhibit the pump function of Na+/K+-ATPase and are commonly used for the treatment of atrial fibrillation and congestive heart failure. Given the known role for RGS2 in the cardiovascular system, these compounds were chosen for further analysis.
Ouabain and Digoxin Selectively Increased RGS2 Protein Expression.
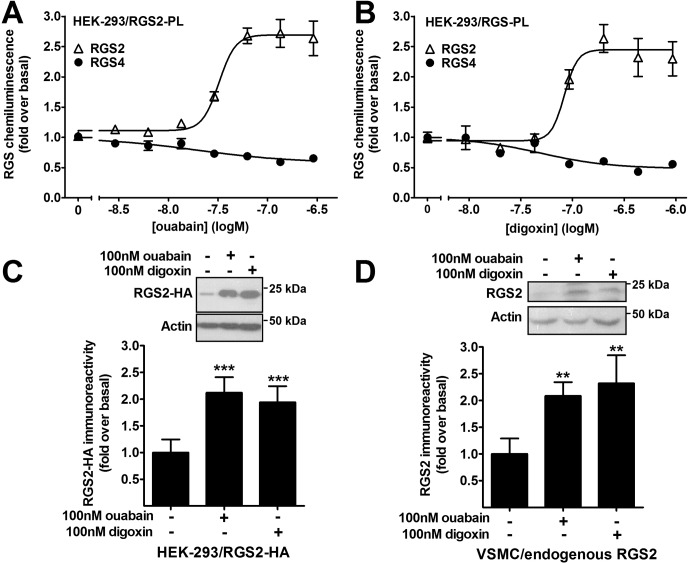
Two CTSs (i.e., ouabain and digoxin) were chosen for follow-up studies, and we initially ascertained the selectivity of these compounds by using the PathHunter ProLabel assay. Both ouabain and digoxin increased RGS2 protein chemiluminescence in a concentration-dependent manner (Fig. 2). The EC50 values for increasing RGS2 protein levels were 32 ± 8 nM and 83 ± 36 nM for ouabain and digoxin, respectively. Neither ouabain nor digoxin increased RGS4 expression (Fig. 2, A and B). The effects of the compounds on RGS2 protein expression were confirmed through Western blotting with HEK-293T cells transiently transfected with RGS2-HA (Fig. 2C). The compounds also increased endogenous RGS2 protein levels in rat VSMCs (Fig. 2D).
Fig. 2.
Ouabain and digoxin selectively increased RGS2 protein expression. A and B, ouabain (A) and digoxin (B) increased RGS2 protein expression in stably transfected HEK-293 cells, in a concentration-dependent manner, as demonstrated with the PathHunter ProLabel assay. The effect was selective over closely related RGS4, because neither compound increased RGS4 protein expression in the same assay system. C, Western blotting showed increased RGS2 protein expression with ouabain and digoxin in transiently transfected HEK-293T cells. D, ouabain and digoxin increased endogenous RGS2 protein expression in rat primary aortic VSMCs, as demonstrated through Western blotting. All treatments were overnight. **, p < 0.01; ***, p < 0.001, using one-way analysis of variance with Bonferroni's post hoc test for multiple comparisons.
In contrast to proteasome inhibitors, ouabain and digoxin did not increase RGS2 protein levels with short-term treatment. The CTS-induced effect on RGS2 protein levels was detected only after overnight treatment (Supplemental Fig. 2), which indicates an indirect mechanism for the effects of these compounds on RGS2 protein expression.
CTSs Exhibited Post-Transcriptional Actions on RGS2.
On the basis of previous reports (Bodenstein et al., 2007) and our own data (Fig. 1), we hypothesized that ouabain and digoxin increased RGS2 protein expression through a post-translational mechanism. To examine this model, we considered whether increased transcription might be a factor. It is unlikely that these compounds activated the promoter for RGS2 transcription, because RGS4 protein expression was not enhanced in our HEK-293 cell lines, in which it was under the regulation of the same cytomegalovirus promoter as RGS2. In VSMCs, in which the endogenous RGS2 promoter drives RGS2 expression, there was no effect of either ouabain or digoxin (both at 100 nM) on RGS2 mRNA levels, as demonstrated with real-time PCR assays (Table 1), despite the significant increase in protein levels (Fig. 2D). The effects of the proteasome inhibitors MG-132 and lactacystin on RGS2 mRNA levels were divergent. Lactacystin (10 μM), which is a more-specific proteasome inhibitor (Fenteany et al., 1995), did not significantly increase RGS2 mRNA levels, whereas overnight treatment with MG-132 (10 μM) resulted in a 10-fold increase in RGS2 mRNA levels (Table 1). This increase could explain the difference in the amount by which the two proteasome inhibitors increased RGS2 protein levels (MG-132, 20-fold; lactacystin, 10-fold) (Fig. 1). Overall, these data support a post-transcriptional mechanism for CTS-induced up-regulation of RGS2 protein expression.
TABLE 1.
RGS2 mRNA expression in vascular smooth muscle cells
Endogenously expressed RGS2 mRNA levels were determined by using real-time PCR assays. Only overnight treatment with 10 μM MG-132 had significant effects on RGS2 mRNA expression. Fold values are expressed relative to control values.
| Treatment | RGS2 mRNA Level |
|
|---|---|---|
| 3 h | Overnight | |
| fold | ||
| 100 nM Digoxin | 1.31 ± 0.71 | |
| 100 nM Ouabain | 0.91 ± 0.17 | |
| 10 μM Lactacystin | 0.80 ± 0.06 | 1.94 ± 1.92 |
| 10 μM MG-132 | 0.56 ± 0.15 | 10.07 ± 5.12*** |
p < 0.001, using one-way analysis of variance with Bonferroni's post hoc test for multiple comparisons.
RGS2 Protein Was Stabilized by CTSs.
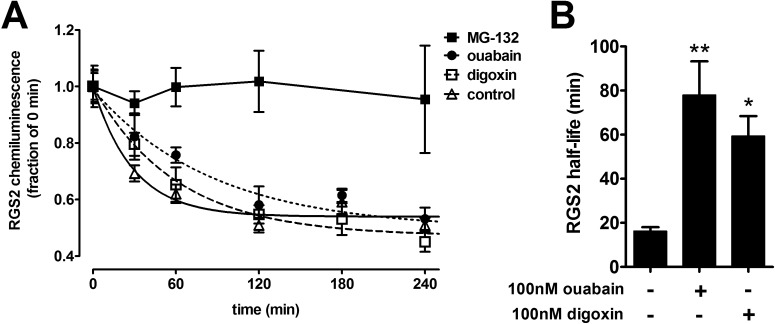
To elucidate further the means by which ouabain and digoxin increased RGS2 protein levels, we investigated the effects of these compounds on the cellular half-life of RGS2 protein. RGS2-PL cells were exposed to the protein translation inhibitor cycloheximide, with or without pretreatment with ouabain, digoxin, or MG-132, and RGS2 levels were measured at different time points. Under control conditions, RGS2 was rapidly degraded (t1/2 = 17 ± 6 min). MG-132 (10 μM) completely blocked the degradation of RGS2 (Fig. 3A), which is consistent with the known proteasomal pathway for RGS2 degradation (Bodenstein et al., 2007) (Fig. 1). Ouabain and digoxin treatment (both at 100 nM) significantly increased the half-life of RGS2 (to 78 ± 43 and 54 ± 26 min, respectively), with a magnitude similar to that of the increase in protein expression levels in both HEK-293 cells and VSMCs (Fig. 3). The increases in the RGS2 protein half-life, together with the lack of effects on RGS2 mRNA levels, support a model in which CTSs increase RGS2 expression through post-translational protein stabilization.
Fig. 3.
RGS2 protein was stabilized by ouabain and digoxin. HEK-293 cells stably transfected with RGS2-PL were treated overnight with 100 nM ouabain, 100 nM digoxin, or 10 μM MG-132. After the treatment, cycloheximide (10 μg/ml) was added to block protein translation at different time points, to determine the RGS2 protein half-life. A, chemiluminescence corresponding to RGS2 protein levels was detected with the PathHunter ProLabel assay. B, quantification of RGS2 protein half-life (n = 3). The half-life of RGS2 under control conditions was 17.5 ± 5.8 min. Ouabain and digoxin increased the half-life of RGS2 to 77.9 ± 43.4 and 54.4 ± 26.1 min, respectively. Treatment with MG-132 completely stabilized RGS2 expression. *, p < 0.05; **, p < 0.01, using one-way analysis of variance with Bonferroni's post hoc test for multiple comparisons.
CTSs Induced Increased RGS2 Protein Expression through Actions on Na+/K+-ATPase.
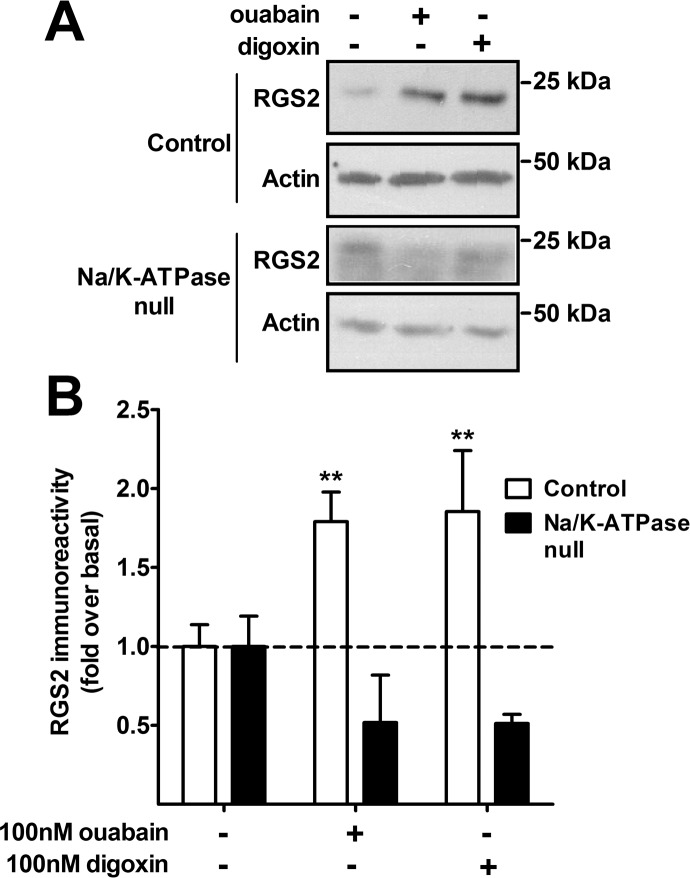
Ouabain and digoxin act on Na+/K+-ATPase, which is the major housekeeping ion pump in many cell types (Skou, 1957), but they may have actions on other targets as well (Yeh et al., 2001). To determine the role of Na+/K+-ATPase in the effects of ouabain and digoxin on RGS2 protein expression, we used LLC-PK cells with normal expression of Na+/K+-ATPase (AAC-19 line) or with Na+/K+-ATPase (α1) levels reduced by 90% with stable shRNA (PY-17 line). These cell lines were used previously to study the mechanisms of Na+/K+-ATPase function (Liang et al., 2006). In AAC-19 cells, both ouabain and digoxin increased endogenous RGS2 protein expression (Fig. 4). Consistent with a mechanism mediated through the Na+/K+-ATPase, no such increase was observed in the PY-17 cells, with reduced Na+/K+-ATPase expression (Fig. 4). Therefore, RGS2 protein expression was stabilized by CTSs through a mechanism that was dependent on the Na+/K+-ATPase.
Fig. 4.
CTSs increased RGS2 protein expression through actions on Na+/K+-ATPase. Endogenous RGS2 protein expression was detected in LLC-PK cells after overnight treatment with 100 nM ouabain or 100 nM digoxin. The ouabain/digoxin-induced increase in RGS2 protein expression was maintained in cells with normal expression of Na+/K+-ATPase (AAC-19). This increase was lost in cells in which Na+/K+-ATPase levels were reduced with shRNA (PY-17; Na+/K+-ATPase-null). A, representative Western blots from four experiments. B, RGS2 levels normalized to actin loading control levels. **, p < 0.01, using two-way analysis of variance with Bonferroni's post hoc test for multiple comparisons.
Molecular Mechanisms of CTS-Mediated RGS2 Up-Regulation Were Investigated.
To determine whether ouabain- and digoxin-mediated stabilization of RGS2 protein was mediated by blockade of the Na+/K+-ATPase pump function itself or was related to actions of the Na+/K+-ATPase to turn on intracellular signaling cascades such as PI-3-kinase, Src, or downstream Ras/Raf/mitogen-activated protein kinase pathways (Xie and Cai, 2003), we used inhibitors of Src and PI-3-kinase to study effects on RGS2 protein levels. Activation of signal transduction cascades was shown to occur at lower (10–100 nM) concentrations of digoxin, similar to those that caused RGS2 up-regulation. Neither the Src inhibitor 4-amino-5-(4-chlorophenyl)-7-(dimethylethyl)pyrazolo[3,4-d]pyrimidine (PP2) nor the PI-3-kinase inhibitor 2-morpholin-4-yl-8-phenylchromen-4-one (LY294002) had any effect on basal RGS2 protein levels in HEK-293 cells. Inhibition of either Src or PI-3-kinase had no effect on CTS-induced increases in RGS2 protein levels (Supplemental Fig. 3). Blocking of the effects of CTSs on Na+/K+-ATPase pump function through decreases in extracellular K+ levels also showed no effects on the RGS2 protein up-regulation induced by ouabain and digoxin (data not shown). At this point, the molecular mechanism of CTS-induced up-regulation of RGS2 remains unclear.
Increased RGS2 Protein Levels Had Functional Effects on GPCR Signaling.
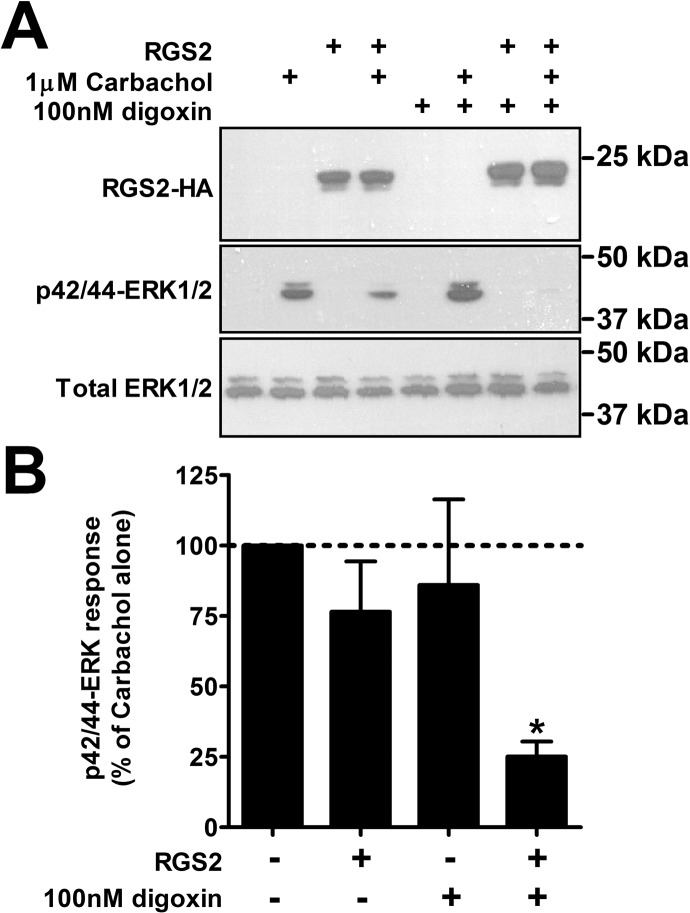
To investigate whether digoxin-mediated increases in RGS2 protein expression had functional effects on signaling through GPCRs, HEK-293T cells were transiently transfected with the Gq-coupled M3 muscarinic receptor alone or in combination with RGS2-HA. Cells were treated with 1 μM levels of the muscarinic agonist carbachol, and agonist-induced ERK1/2 phosphorylation was analyzed through Western blotting (Fig. 5). Carbachol stimulation resulted in a strong increase in phosphorylated ERK1/2 levels, which was slightly (but not statistically significantly) suppressed in the presence of RGS2. Digoxin (100 nM) treatment had no effect on M3 receptor signaling in the absence of RGS2. In the combined presence of RGS2 and digoxin, however, carbachol-induced ERK1/2 phosphorylation was significantly reduced, as expected on the basis of the increase in RGS2 protein levels (Fig. 5). Therefore, increasing RGS2 protein levels by means of pharmacological treatment with digoxin had functional effects on signaling through a Gq-linked GPCR.
Fig. 5.
Increased RGS2 protein expression had functional effects on GPCR signaling. HEK-293T cells were transiently transfected with the M3 muscarinic receptor alone or together with RGS2-HA; 48 h after transfection, carbachol-induced ERK1/2 phosphorylation was measured through Western blotting, with or without overnight pretreatment with digoxin. Carbachol induced strong phosphorylation of ERK1/2 after 10 min. There was a trend toward suppression of the signal in the presence of RGS2-HA, which was significantly enhanced in the presence of 100 nM digoxin (*, p < 0.05, using one-way analysis of variance with Bonferroni's post hoc test for multiple comparisons). In cells with no RGS2-HA, digoxin had no effect on carbachol-induced ERK1/2 phosphorylation. A, representative Western blots. B, quantification of results from three experiments run in triplicate.
Digoxin Increased RGS2 Protein Levels In Vivo.
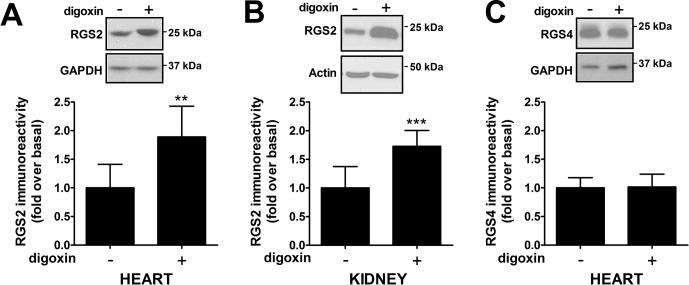
To assess whether pharmacological treatment with digoxin would increase RGS2 protein levels in vivo, we treated mice with 2 μg/kg digoxin daily for 7 days and measured RGS2 protein expression in the heart and other tissues. RGS2 protein levels in the heart were significantly increased, compared with the loading control values (Fig. 6A). The effect on RGS2 mRNA levels was minimal and not significant (control, 100 ± 34.5%; digoxin, 141 ± 50.9%), which confirmed a post-transcriptional mechanism for digoxin-mediated up-regulation of RGS2 protein levels. RGS4 protein levels in the heart were not significantly changed after digoxin treatment, which confirmed that CTSs selectively stabilized RGS2 over RGS4 (Fig. 6C). RGS2 protein levels were also increased in the kidney (Fig. 6B), where RGS2 has been proposed to exert effects on blood pressure homeostasis (Gurley et al., 2010).
Fig. 6.
Digoxin up-regulated RGS2 protein levels in vivo. Male C57BL/6 mice were treated with either vehicle or 2 μg/kg digoxin for 7 days with osmotic minipumps. Levels of RGS2 protein expression in tissue were then analyzed through Western blotting. Treatment with digoxin induced significant increases in RGS2 protein immunoreactivity in both heart (A) and kidney (B), compared with control values. In contrast, closely related RGS4 showed no increase in levels in heart after digoxin treatment (C). Representative blots and quantification of data from eight animals in each group are shown. GAPDH, glyceraldehyde-3-phosphate dehydrogenase. **, p < 0.01; ***, p < 0.001, using Student's unpaired t test.
Discussion
High-throughput chemical screens are used to identify new drugs but also can provide mechanistic insights regarding existing drugs. They can reveal entirely new therapeutic indications for old drugs, in a process termed “repurposing.” In the present study, we used a cell-based, high-throughput, chemical screen for a new cardiovascular target, the RGS proteins. We tested several thousand known drugs and bioactive molecules for potential roles in enhancing RGS2 and/or RGS4 expression and function, and we identified a novel mechanism for digoxin and other cardiotonic steroids. A notable observation from the primary screen was the lack of overlap between the hits for the two RGS proteins screened. Among almost 3000 compounds, only one increased expression of both RGS2 and RGS4. This was unexpected, because of the large number of possible mechanisms that could lead to increased protein expression, including proteasome inhibition and promoter activation. This finding demonstrates a high degree of specificity in the manner in which these two proteins are regulated.
G protein-coupled receptors play key roles in the physiological and pathophysiological processes of cardiovascular disease and heart failure (Salazar et al., 2007; Hendriks-Balk et al., 2008; Brinks and Eckhart, 2010). Consequently, we (Zhong and Neubig, 2001; Sjögren et al., 2010) and others (Doggrell, 2004; Riddle et al., 2005; da Costa Goncalves et al., 2008) suggested that RGS proteins, including RGS2, might represent novel cardiovascular therapeutic targets. Although significant progress has been made in identifying RGS inhibitors (Blazer et al., 2011; Turner et al., 2012), enhancing RGS action is more challenging (Sjögren and Neubig, 2010).
Proteasomal degradation of RGS2 and other RGS proteins might be a useful control point at which to enhance RGS2 expression levels and function (Sjögren and Neubig, 2010). Degradation of the closely related RGS4 and RGS5 is mediated by the N-end rule pathway (Lee et al., 2005; Bodenstein et al., 2007), and the specific E1, E2, and E3 ubiquitination enzymes involved in degradation were recently characterized (Lee et al., 2011). For RGS2, this pathway is not yet characterized. More-detailed information on the specific enzymes involved in RGS2 protein degradation would be useful for the rational development of drugs that aim to increase RGS2 protein expression selectively. In the current study, we showed for the first time in vascular smooth muscle cells that proteasome inhibition, as well as ouabain and digoxin, markedly enhanced endogenous RGS2 protein expression. It is striking that the action of digoxin was selective for RGS2 over RGS4 even in vivo, which indicates that the action is not related to gross inhibition of proteasomal function but the drug somehow targets specific mechanisms related to RGS2 degradation.
The digoxin-induced increase in RGS2 expression was accompanied by a RGS2-dependent reduction in Gq-mediated ERK phosphorylation responses in transfected HEK-293 cells. If this proves to be true also for endogenous RGS2 (i.e., in VSMCs and in vivo), then suppression of signaling by many Gq-coupled receptors, such as angiotensin, vasopressin, thromboxane A2, thrombin, α1-adrenergic, and endothelin receptors, would be expected. Furthermore, RGS2 has non–G protein-mediated effects such as direct inhibition of cAMP production by the cardiac type V and VI adenylyl cyclase isoforms (Salim et al., 2003; Gu et al., 2008) and inhibition of protein translation (Nguyen et al., 2009). These effects might reduce detrimental β-adrenergic receptor signaling-related, proapoptotic, and hypertrophic mechanisms in heart failure. The combination of broad Gq-dependent and -independent mechanisms provides several potentially beneficial effects of increased RGS2 protein expression. At this point, we cannot state whether pharmacologically enhanced RGS2 protein expression would have functional effects in vivo. However, previous studies showed that even small changes in RGS2 protein levels [i.e., heterozygous RGS2(+/−) mice] had effects on endogenous GPCR signaling (Heximer et al., 2003). We predict that the digoxin-mediated increases in RGS2 protein levels observed in heart and kidney might affect canonical and noncanonical RGS2 protein function.
The effects on other components affected by digoxin also need to be investigated. Chronic digoxin treatment, and subsequent increases in RGS2 protein levels, might lead to altered expression of Na+/K+-ATPase and changes in the relative expression and/or function of proteins involved in G protein-mediated signaling. These possible scenarios should be investigated but are beyond the scope of our current study.
Our data showed that ouabain and digoxin stabilized RGS2 protein through a Na+/K+-ATPase-dependent mechanism. At this point, we cannot say whether the effects were mediated through inhibition of Na+/K+-ATPase pump function or induction of signaling cascades. It is unclear how these compounds mediate increases in RGS2 protein levels. However, the selectivity over the closely related RGS4 both in vitro and in vivo suggests that the effects are not related to a global cellular mechanism but represent specific RGS2-related effects, possibly through decreased negative regulation of RGS2 protein.
Because adverse effects related to excessive pump inhibition may contribute to the loss of survival benefits at higher plasma concentrations (and doses) of digoxin in the treatment of heart failure (Ahmed and Waagstein, 2009), it will be important to understand the relative structure-activity relationships for blocking ion transport, inducing signaling pathways, and modulating RGS2 expression. A divergence in structure-activity relationships among those effects might lead to the discovery of less-toxic compounds that have selective actions to induce RGS2 protein increases. A CTS-analog inhibitor, rostafuroxin, is in clinical trials for treatment of hypertension (Lanzani et al., 2010).
It has long been known that rodent Na+/K+-ATPase displays far lower sensitivity to CTSs than its human counterpart (Detweiler, 1967). In light of this observation, it might be surprising that equal concentrations of ouabain and digoxin produced similar effects on RGS2 protein levels in HEK-293 cells and rat VSMCs. However, several studies demonstrated effects on rodent Na+/K+-ATPase at very low concentrations of CTSs. Abramowitz et al. (2003) showed that 100 nM ouabain induced a significant increase in rat VSMC proliferation. Furthermore, VSMCs express both high- and low-affinity Na+/K+-ATPase, and very low concentrations of ouabain (<1 nM) could modulate aortic vascular smooth muscle contraction (Weiss et al., 1993). It is not clear whether these effects are a direct result of ouabain inhibiting Na+/K+-ATPase pump function or inducing signal transduction cascades. It was suggested that acute effects (5–10 min) of CTSs require much higher concentrations (micromolar) than the long-term effects (hours to days; nanomolar) observed in the present study (Quintas et al., 2010).
CTS effects to increase RGS2 expression may influence other aspects of cardiovascular and noncardiovascular function. RGS2(−/−) mice show enhanced M3 receptor responses in the atrium and are more susceptible to atrial arrhythmias (Tuomi et al., 2010). We found that digoxin caused RGS2-dependent suppression of M3 receptor signaling (Fig. 5), which might have relevance in atrial fibrillation, the most well accepted clinical indication for the use of digoxin. The situation regarding hypertension is more complex. Because low RGS2 levels in mice cause hypertension (Heximer et al., 2003) and heterozygous destabilizing mutations in RGS2 are found selectively in hypertensive humans (Yang et al., 2005; Bodenstein et al., 2007), increased RGS2 protein levels resulting from CTS treatment might be predicted to ameliorate high blood pressure. However, endogenous ouabain has long been identified as a prohypertensive factor (Bagrov et al., 2009), although this is controversial (Nicholls et al., 2009). Genetic alterations in Na+/K+-ATPase have opposite effects on blood pressure in different models (Hou et al., 2009; Rindler et al., 2011). The actions of ouabain and other CTSs on blood pressure are probably multifactorial, with contributions from pump inhibition, effector signaling, and the increases in RGS2 expression described here. More studies will be needed for a full understanding of this topic. Outside the cardiovascular system, CTSs are being considered as novel cancer therapies. RGS2 protein levels are down-regulated in androgen-independent prostate cancer cells and in clinical specimens (Cao et al., 2006). Overexpression of RGS2 in LNCaP cells suppressed cell growth, whereas silencing of RGS2 had the opposite effect (Cao et al., 2006). In light of our present data, there may be a role for RGS2 in these effects of CTSs in prostate cancer models.
In summary, we have developed an approach to identify compounds or treatments that enhance RGS protein expression and function. Furthermore, we report the novel observation that digoxin and other CTSs selectively increase RGS2 protein expression in vitro and in vivo. This may represent an element of the mechanism of beneficial effects of CTS treatment in heart failure. Clear assessment of the relative in vivo contributions of RGS2 regulation versus other actions of CTSs will be needed in future work. Our data provide a proof of principle for the concept of increasing RGS2 activity as an approach to the treatment of heart failure and hypertension. Digoxin has been shown to have clinical benefits in heart failure even in the context of ongoing angiotensin-converting enzyme inhibitor treatment (Ahmed and Waagstein, 2009); it is plausible that increased RGS2 protein expression might be a factor. More-detailed in vivo studies will be needed to investigate this possibility.
Supplementary Material
Acknowledgments
We thank the Center for Chemical Genomics, especially Steven Swaney, for providing compound libraries and assistance with high-throughput screening. We also thank Dr. David Siderovski for the anti-RGS2 antibody.
 The online version of this article (available at http://molpharm.aspetjournals.org) contains supplemental material.
The online version of this article (available at http://molpharm.aspetjournals.org) contains supplemental material.
This work was supported by the National Institutes of Health National Institute on Drug Abuse [Grant DA023252]; the Swedish Heart and Lung Foundation [Grant 20110193]; and Bristol-Meyers-Squibb (to R.R.N.).
Article, publication date, and citation information can be found at http://molpharm.aspetjournals.org.
- CTS
- cardiotonic steroid
- GPCR
- G protein-coupled receptor
- RGS
- regulator of G protein signaling
- VSMC
- vascular smooth muscle cell
- PI
- phosphoinositide
- ERK
- extracellular signal-regulated kinase
- PAGE
- polyacrylamide gel electrophoresis
- DMEM
- Dulbecco's modified Eagle's medium
- FBS
- fetal bovine serum
- HEK
- human embryonic kidney
- TBS-T
- Tris-buffered saline-Tween 20
- shRNA
- short hairpin RNA
- HA
- hemagglutinin
- PCR
- polymerase chain reaction
- PL
- ProLabel
- MG-132
- N-(benzyloxycarbonyl)leucinylleucinylleucinal
- PP2
- 4-amino-5-(4-chlorophenyl)-7-(dimethylethyl)pyrazolo[3,4-d]pyrimidine
- LY294002
- 2-morpholin-4-yl-8-phenylchromen-4-one.
Authorship Contributions
Participated in research design: Sjögren, Parra, Heath, Xie, and Neubig.
Conducted experiments: Sjögren, Parra, and Heath.
Contributed new reagents or analytic tools: Atkins and Xie.
Performed data analysis: Sjögren, Parra, and Heath.
Wrote or contributed to the writing of the manuscript: Sjögren, Parra, Atkins, Xie, and Neubig.
References
- Abramowitz J, Dai C, Hirschi KK, Dmitrieva RI, Doris PA, Liu L, Allen JC. (2003) Ouabain- and marinobufagenin-induced proliferation of human umbilical vein smooth muscle cells and a rat vascular smooth muscle cell line, A7r5. Circulation 108:3048–3053 [DOI] [PubMed] [Google Scholar]
- Ahmed A, Waagstein F. (2009) Low-dose digoxin and reduction in mortality and morbidity in heart failure. Int J Cardiol 136:91–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkins KB, Irey B, Xiang N, Brosius FC., 3rd (2009) A rapid, PPAR-gamma-dependent effect of pioglitazone on the phosphorylation of MYPT. Am J Physiol Cell Physiol 296:C1151–C1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagrov AY, Shapiro JI, Fedorova OV. (2009) Endogenous cardiotonic steroids: physiology, pharmacology, and novel therapeutic targets. Pharmacol Rev 61:9–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazer LL, Neubig RR. (2009) Small molecule protein-protein interaction inhibitors as CNS therapeutic agents: current progress and future hurdles. Neuropsychopharmacology 34:126–141 [DOI] [PubMed] [Google Scholar]
- Blazer LL, Roman DL, Chung A, Larsen MJ, Greedy BM, Husbands SM, Neubig RR. (2010) Reversible, allosteric small-molecule inhibitors of regulator of G protein signaling proteins. Mol Pharmacol 78:524–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazer LL, Zhang H, Casey EM, Husbands SM, Neubig RR. (2011) A nanomolar-potency small molecule inhibitor of regulator of G-protein signaling proteins. Biochemistry 50:3181–3192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blundell J, Hoang CV, Potts B, Gold SJ, Powell CM. (2008) Motor coordination deficits in mice lacking RGS9. Brain Res 1190:78–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodenstein J, Sunahara RK, Neubig RR. (2007) N-terminal residues control proteasomal degradation of RGS2, RGS4, and RGS5 in human embryonic kidney 293 cells. Mol Pharmacol 71:1040–1050 [DOI] [PubMed] [Google Scholar]
- Brinks HL, Eckhart AD. (2010) Regulation of GPCR signaling in hypertension. Biochim Biophys Acta 1802:1268–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calò LA, Pagnin E, Davis PA, Sartori M, Ceolotto G, Pessina AC, Semplicini A. (2004) Increased expression of regulator of G protein signaling-2 (RGS-2) in Bartter's/Gitelman's syndrome. a role in the control of vascular tone and implication for hypertension. J Clin Endocrinol Metab 89:4153–4157 [DOI] [PubMed] [Google Scholar]
- Cao X, Qin J, Xie Y, Khan O, Dowd F, Scofield M, Lin MF, Tu Y. (2006) Regulator of G-protein signaling 2 (RGS2) inhibits androgen-independent activation of androgen receptor in prostate cancer cells. Oncogene 25:3719–3734 [DOI] [PubMed] [Google Scholar]
- Chowdari KV, Mirnics K, Semwal P, Wood J, Lawrence E, Bhatia T, Deshpande SN, B K T, Ferrell RE, Middleton FA, et al. (2002) Association and linkage analyses of RGS4 polymorphisms in schizophrenia. Hum Mol Genet 11:1373–1380 [DOI] [PubMed] [Google Scholar]
- da Costa Goncalves ACh, Luft FC, Gross V. (2008) Fine tuning of blood pressure by the regulator of G protein signaling (RGS) 2. J Am Soc Hypertens 2:403–409 [DOI] [PubMed] [Google Scholar]
- Detweiler DK. (1967) Comparative pharmacology of cardiac glycosides. Fed Proc 26:1119–1124 [PubMed] [Google Scholar]
- Doggrell SA. (2004) Is RGS-2 a new drug development target in cardiovascular disease? Expert Opin Ther Targets 8:355–358 [DOI] [PubMed] [Google Scholar]
- Doupnik CA, Xu T, Shinaman JM. (2001) Profile of RGS expression in single rat atrial myocytes. Biochim Biophys Acta 1522:97–107 [DOI] [PubMed] [Google Scholar]
- Fenteany G, Standaert RF, Lane WS, Choi S, Corey EJ, Schreiber SL. (1995) Inhibition of proteasome activities and subunit-specific amino-terminal threonine modification by lactacystin. Science 268:726–731 [DOI] [PubMed] [Google Scholar]
- Gheorghiade M, Braunwald E. (2009) Reconsidering the role for digoxin in the management of acute heart failure syndromes. JAMA 302:2146–2147 [DOI] [PubMed] [Google Scholar]
- Gu S, Anton A, Salim S, Blumer KJ, Dessauer CW, Heximer SP. (2008) Alternative translation initiation of human regulators of G-protein signaling-2 yields a set of functionally distinct proteins. Mol Pharmacol 73:1–11 [DOI] [PubMed] [Google Scholar]
- Gurley SB, Griffiths RC, Mendelsohn ME, Karas RH, Coffman TM. (2010) Renal actions of RGS2 control blood pressure. J Am Soc Nephrol 21:1847–1851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendriks-Balk MC, Peters SL, Michel MC, Alewijnse AE. (2008) Regulation of G protein-coupled receptor signalling: focus on the cardiovascular system and regulator of G protein signalling proteins. Eur J Pharmacol 585:278–291 [DOI] [PubMed] [Google Scholar]
- Heximer SP, Knutsen RH, Sun X, Kaltenbronn KM, Rhee MH, Peng N, Oliveira-dos-Santos A, Penninger JM, Muslin AJ, Steinberg TH, et al. (2003) Hypertension and prolonged vasoconstrictor signaling in RGS2-deficient mice. J Clin Invest 111:445–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heximer SP, Watson N, Linder ME, Blumer KJ, Hepler JR. (1997) RGS2/G0S8 is a selective inhibitor of Gqα function. Proc Natl Acad Sci USA 94:14389–14393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X, Theriault SF, Dostanic-Larson I, Moseley AE, Lingrel JB, Wu H, Dean S, Van Huysse JW. (2009) Enhanced pressor response to increased CSF sodium concentration and to central ANG I in heterozygous α2 Na+-K+-ATPase knockout mice. Am J Physiol Regul Integr Comp Physiol 296:R1427–R1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumins AM, Barker SA, Huang C, Sunahara RK, Yu K, Wilkie TM, Gold SJ, Mumby SM. (2004) Differentially regulated expression of endogenous RGS4 and RGS7. J Biol Chem 279:2593–2599 [DOI] [PubMed] [Google Scholar]
- Lanzani C, Citterio L, Glorioso N, Manunta P, Tripodi G, Salvi E, Carpini SD, Ferrandi M, Messaggio E, Staessen JA, et al. (2010) Adducin- and ouabain-related gene variants predict the antihypertensive activity of rostafuroxin, part 2: clinical studies. Sci Transl Med 2:59ra87. [DOI] [PubMed] [Google Scholar]
- Lee MJ, Tasaki T, Moroi K, An JY, Kimura S, Davydov IV, Kwon YT. (2005) RGS4 and RGS5 are in vivo substrates of the N-end rule pathway. Proc Natl Acad Sci USA 102:15030–15035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PC, Sowa ME, Gygi SP, Harper JW. (2011) Alternative ubiquitin activation/conjugation cascades interact with N-end rule ubiquitin ligases to control degradation of RGS proteins. Mol Cell 43:392–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang M, Cai T, Tian J, Qu W, Xie ZJ. (2006) Functional characterization of Src-interacting Na/K-ATPase using RNA interference assay. J Biol Chem 281:19709–19719 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔCT) method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- Nguyen CH, Ming H, Zhao P, Hugendubler L, Gros R, Kimball SR, Chidiac P. (2009) Translational control by RGS2. J Cell Biol 186:755–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls MG, Lewis LK, Yandle TG, Lord G, McKinnon W, Hilton PJ. (2009) Ouabain, a circulating hormone secreted by the adrenals, is pivotal in cardiovascular disease: fact or fantasy? J Hypertens 27:3–8 [DOI] [PubMed] [Google Scholar]
- Nunn C, Zou MX, Sobiesiak AJ, Roy AA, Kirshenbaum LA, Chidiac P. (2010) RGS2 inhibits beta-adrenergic receptor-induced cardiomyocyte hypertrophy. Cell Signal 22:1231–1239 [DOI] [PubMed] [Google Scholar]
- Quintas LE, Pierre SV, Liu L, Bai Y, Liu X, Xie ZJ. (2010) Alterations of Na+/K+-ATPase function in caveolin-1 knockout cardiac fibroblasts. J Mol Cell Cardiol 49:525–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle EL, Schwartzman RA, Bond M, Insel PA. (2005) Multi-tasking RGS proteins in the heart: the next therapeutic target? Circ Res 96:401–411 [DOI] [PubMed] [Google Scholar]
- Rindler TN, Dostanic I, Lasko VM, Nieman ML, Neumann JC, Lorenz JN, Lingrel JB. (2011) Knockout of the Na,K-ATPase α2-isoform in the cardiovascular system does not alter basal blood pressure but prevents ACTH-induced hypertension. Am J Physiol Heart Circ Physiol 301:H1396–H1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman DL, Talbot JN, Roof RA, Sunahara RK, Traynor JR, Neubig RR. (2007) Identification of small-molecule inhibitors of RGS4 using a high-throughput flow cytometry protein interaction assay. Mol Pharmacol 71:169–175 [DOI] [PubMed] [Google Scholar]
- Ross EM, Wilkie TM. (2000) GTPase-activating proteins for heterotrimeric G proteins: regulators of G protein signaling (RGS) and RGS-like proteins. Annu Rev Biochem 69:795–827 [DOI] [PubMed] [Google Scholar]
- Salazar NC, Chen J, Rockman HA. (2007) Cardiac GPCRs: GPCR signaling in healthy and failing hearts. Biochim Biophys Acta 1768:1006–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salim S, Sinnarajah S, Kehrl JH, Dessauer CW. (2003) Identification of RGS2 and type V adenylyl cyclase interaction sites. J Biol Chem 278:15842–15849 [DOI] [PubMed] [Google Scholar]
- Schoenfeld JR, Vasser M, Jhurani P, Ng P, Hunter JJ, Ross J, Jr, Chien KR, Lowe DG. (1998) Distinct molecular phenotypes in murine cardiac muscle development, growth, and hypertrophy. J Mol Cell Cardiol 30:2269–2280 [DOI] [PubMed] [Google Scholar]
- Semplicini A, Strapazzon G, Papparella I, Sartori M, Realdi A, Macchini L, Calò LA, Ceolotto G. (2010) RGS2 expression and aldosterone:renin ratio modulate response to drug therapy in hypertensive patients. J Hypertens 28:1104–1108 [DOI] [PubMed] [Google Scholar]
- Shah NK, Choksi R, Epstein BJ. (2011) Less RAAS is more, or not. Expert Rev Cardiovasc Ther 9:1363–1365 [DOI] [PubMed] [Google Scholar]
- Sjögren B, Blazer LL, Neubig RR. (2010) Regulators of G protein signaling proteins as targets for drug discovery. Prog Mol Biol Transl Sci 91:81–119 [DOI] [PubMed] [Google Scholar]
- Sjögren B, Neubig RR. (2010) Thinking outside of the “RGS box”: new approaches to therapeutic targeting of regulators of G protein signaling. Mol Pharmacol 78:550–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skou JC. (1957) The influence of some cations on an adenosine triphosphatase from peripheral nerves. Biochim Biophys Acta 23:394–401 [DOI] [PubMed] [Google Scholar]
- Sun X, Kaltenbronn KM, Steinberg TH, Blumer KJ. (2005) RGS2 is a mediator of nitric oxide action on blood pressure and vasoconstrictor signaling. Mol Pharmacol 67:631–639 [DOI] [PubMed] [Google Scholar]
- Takimoto E, Koitabashi N, Hsu S, Ketner EA, Zhang M, Nagayama T, Bedja D, Gabrielson KL, Blanton R, Siderovski DP, et al. (2009) Regulator of G protein signaling 2 mediates cardiac compensation to pressure overload and antihypertrophic effects of PDE5 inhibition in mice. J Clin Invest 119:408–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesmer JJ, Berman DM, Gilman AG, Sprang SR. (1997) Structure of RGS4 bound to AlF4−-activated Giα1: stabilization of the transition state for GTP hydrolysis. Cell 89:251–261 [DOI] [PubMed] [Google Scholar]
- Tuomi JM, Chidiac P, Jones DL. (2010) Evidence for enhanced M3 muscarinic receptor function and sensitivity to atrial arrhythmia in the RGS2-deficient mouse. Am J Physiol Heart Circ Physiol 298:H554–H561 [DOI] [PubMed] [Google Scholar]
- Turner EM, Blazer LL, Neubig RR, Husbands SM. (2012) Small molecule inhibitors of regulator of G protein signalling (RGS) proteins. ACS Med Chem Lett 3:146–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss DN, Podberesky DJ, Heidrich J, Blaustein MP. (1993) Nanomolar ouabain augments caffeine-evoked contractions in rat arteries. Am J Physiol 265:C1443–C1448 [DOI] [PubMed] [Google Scholar]
- Xie Z, Cai T. (2003) Na+-K+–ATPase-mediated signal transduction: from protein interaction to cellular function. Mol Interv 3:157–168 [DOI] [PubMed] [Google Scholar]
- Yang J, Kamide K, Kokubo Y, Takiuchi S, Tanaka C, Banno M, Miwa Y, Yoshii M, Horio T, Okayama A, et al. (2005) Genetic variations of regulator of G-protein signaling 2 in hypertensive patients and in the general population. J Hypertens 23:1497–1505 [DOI] [PubMed] [Google Scholar]
- Yeh JY, Huang WJ, Kan SF, Wang PS. (2001) Inhibitory effects of digitalis on the proliferation of androgen dependent and independent prostate cancer cells. J Urol 166:1937–1942 [PubMed] [Google Scholar]
- Zhang P, Su J, King ME, Maldonado AE, Park C, Mende U. (2011) Regulator of G protein signaling 2 is a functionally important negative regulator of angiotensin II-induced cardiac fibroblast responses. Am J Physiol Heart Circ Physiol 301:H147–H156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Anger T, Su J, Hao J, Xu X, Zhu M, Gach A, Cui L, Liao R, Mende U. (2006) Selective loss of fine tuning of Gq/11 signaling by RGS2 protein exacerbates cardiomyocyte hypertrophy. J Biol Chem 281:5811–5820 [DOI] [PubMed] [Google Scholar]
- Zhong H, Neubig RR. (2001) Regulator of G protein signaling proteins: novel multifunctional drug targets. J Pharmacol Exp Ther 297:837–845 [PubMed] [Google Scholar]
- Zou MX, Roy AA, Zhao Q, Kirshenbaum LA, Karmazyn M, Chidiac P. (2006) RGS2 is upregulated by and attenuates the hypertrophic effect of α1-adrenergic activation in cultured ventricular myocytes. Cell Signal 18:1655–1663 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.