Abstract
We have previously shown that the protein tyrosine phosphatase SHP-1 is highly expressed in CNS glia and is an important modulator of cytokine signaling. As such, mice genetically lacking SHP-1 display constitutive myelin abnormalities, severe virus-induced demyelinating disease, and defects in innate anti-viral responses in the CNS. In this study, we show the differential distribution of the SHP-1 promoter-specific transcripts and demonstrate that several cytokines significantly induce SHP-1 expression in CNS glia. Consistent with these cytokine effects, infection with a neurotropic virus both in vitro and in vivo up-regulates SHP-1 transcripts and protein in CNS cells. Using CNS glial cultures of gene knockout mice, we show that interferons-β and interferons-γ act through STAT-1 and interferon regulatory factor-1 to induce the SHP-1 promoter I transcripts. Conversely, interferons-β and IL-6 act through STAT-3 to induce SHP-1 promoter II transcripts. This study demonstrates that interferons and other cytokines associated with virus infections in the CNS can significantly induce the expression of SHP-1 through STAT-1/3 activity and provides a better understanding of the molecular mechanisms regulating cytokine-induced expression important for multiple homeostatic functions of SHP-1 in the CNS.
Keywords: demyelination, interferons, multiple sclerosis, protein tyrosine phosphatase, signal transducers and activators of transcription, Theiler’s murine encephalomyelitis virus
SHP-1 is an SH2 domain-containing protein tyrosine phosphatase that is a negative regulator of interferon, cytokine, and growth factor signaling primarily via Jak/Stat pathways (David et al. 1995; Haque et al. 1998; Massa and Wu 1998; Paling and Welham 2002; Marsh et al. 2003; Frank et al. 2004). Loss of SHP-1 in the CNS as seen in motheaten (me/me) mice leads to myelin pathology (Massa et al. 2000, 2004) that may be caused by effects of an increased inflammatory milieu and/or specific alterations in cytokine signaling on the myelin-forming oligodendrocytes. Importantly, it was recently shown that leukocytes of MS patients have decreased levels of the phosphatase SHP-1, which results in increased inflammatory gene expression (Christophi et al. 2008). Furthermore, SHP-1 plays a critical role in fighting CNS viral infections such as Theiler’s murine encephalomyelitis virus (TMEV) (Massa et al. 2002). We have recently shown that SHP-1 deficiency leads to higher viral replication as a result of unrestrained STAT-6-dependent arginase I gene expression and reduced nitric oxide production in CNS glia (Massa et al. 2002; Bonaparte et al. 2006).
Two distinct promoters drive the expression of two different SHP-1 transcripts from the SHP-1 gene, both in human (Banville et al. 1995) and mouse (Martin et al. 1999). It was previously shown that the two SHP-1 transcripts are differentially expressed in mouse and human cell lines (Tsui et al. 2002). These distinct transcripts in turn encode two different protein isoforms. The isoform translated from transcript II is two amino acids shorter and has two amino acids in the N-terminus that differ from promoter I derived isoform. In addition, it has been shown that promoter II transcript can generate another longer isoform (SHP-1L) through alternative splicing (Jin et al. 1999). Because the enzymatic activity between the SHP-1 isoforms is negligible (Martin et al. 1999), the alternative promoter usage may be primarily important for homoeostatic regulation of SHP-1 activity under various physiological settings in specific tissues and cell types.
Promoter I, upstream of exon 1, is preferentially active in cells of epithelial origin (Xu et al. 2001). Promoter I activity may be under the control of various transcription factors including nuclear factor-κB (NF-κB), USF, or NF-Y (Xu et al. 2001; Tsui et al. 2002). Promoter II, upstream of exon 2, was shown to be more active in hematopoetic compared to epithelial tissues (Banville et al. 1995). Hematopoietic SHP-1 controls multiple cytokines involved in proliferation and differentiation of these cells. Recently, it was shown that the hematopoietic cell-specific transcription factor PU.1 plays a role in regulating promoter II transcripts in leukemia/lymphoma cell lines (Wlodarski et al. 2007). SHP-1 is a known tumor suppressor in hematopoietic cells and numerous reports document the down-regulation of promoter II transcripts in leukemia/lymphoma, demonstrating the potential importance of epigenetic SHP-1 promoter modulation in human disease (Zhang et al. 2000; Oka et al. 2001;Wu et al. 2003).
We have previously shown that cytokines that act via the JAK/STAT pathway increase the expression of SHP-1 in mouse CNS glia (Massa et al. 2000). These data, along with the observation that SHP-1 is crucial in regulating interferon-induced genes and fighting CNS viral infections led us to hypothesize that the SHP-1 gene may possess cytokine-inducible promoter activity. As such, the antiviral and immunomodulatory activities of interferons (Platanias 2005; van Boxel-Dezaire et al. 2006) may in part be mediated by SHP-1 and may relate to the mechanisms of interferon activity against demyelinating diseases in animals and humans (Teige et al. 2003; Rudick et al. 2004; Ann Marrie and Rudick 2006; Kinkel et al. 2006; Yarilina et al. 2007).
Here we document the differential expression of the two SHP-1 transcripts in several tissues and show that SHP-1 protein and mRNA is induced by cytokines in CNS glia. Importantly, viral infection of CNS cells both in vitro and in vivo significantly induces the expression of SHP-1 protein and SHP-1 mRNA transcripts. Furthermore, we show that virus infection and interferons increase the expression of promoter I transcripts through STAT-1 and interferon regulatory factor-1 (IRF-1) activity in the brain and in cultured CNS glia. Additionally, interferons-β (IFN-β) also induces promoter II transcripts in a STAT-3-dependent manner. Dissecting the specific contribution of each promoter in modulating SHP-1 expression levels in CNS glia will allow a better understanding of the molecular mechanisms regulating the activity of this important protein tyrosine phosphatase in the central nervous and immune systems.
Materials and methods
Animals
Breeding pairs of STAT-1 null mice (129S6/SvEv-Stat1tm1Rds) were purchased from Taconic Farms (Germantown, New York, NY, USA). Breeding pairs of IRF-1 null mice (C57BL/6-irf1 tm1Mak; IRF-1−/−) were obtained from Jackson Laboratories (Bar Harbor, ME, USA). SHP-1-deficient motheaten (me/me) mice on a C3HeB/FeJLe-a/a background were produced from heterozygous breeding pairs obtained from Jackson Laboratories.
Glial cultures
Mixed glial cultures containing astrocytes, oligodendrocytes, and microglia were produced from brains of 8-day-old mice by a modified procedure as previously described (McCarthy and de Vellis 1980; Massa et al. 2000). Briefly, brains were minced in Kreb’s buffer with curved scissors into fine pieces. The minced brain was centrifuged and resuspended in Kreb’s buffer containing 0.25% trypsin and incubated at 37°C for 1 min. After addition of 5% fetal bovine serum and 40 µg/mL Dnase in Kreb’s buffer to stop trypsinization, the tissue was pelleted and resuspended in fresh Kreb’s/FBS/Dnase buffer. The tissue was repeatedly triturated with a fire polished pipette to dissociate cells. The cells were centrifuged, then resuspended in complete culture medium containing Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal bovine serum, 24.5 mM KCl, and 100 µg/mL insulin and plated onto polylysine-coated dishes. Every three days, cells were fed with fresh medium consisting of DMEM with 10% heat-inactivated horse serum (heat-inactivated at 56°C for 1 h), and used at 10 days after plating. Purified cultures of astrocytes, oligodendrocytes, and microglia were produced from 2-week-old mixed glial cultures as described previously (McCarthy and de Vellis 1980; Massa et al. 1986, 2000).
Splenocyte culture
Spleens were removed from 4-week-old mice and ground between the frosted surfaces of two glass slides in Hanks balanced salt solution. Freed cells were left for 2 min on ice, pelleted by centrifugation, and the supernatant discarded. The pellet was resuspended in 4 mL/spleen of red blood cell lysis buffer (155 mM NH3Cl, 0.1 mM EDTA, 12 mM NaHCO2) and incubated on ice for 1 min. Cells were washed twice with Hank’s balanced salt solution and were resuspended in Roswell Park Memorial Institute 1640 medium with 10% fetal bovine serum in a concentration of 106cells/mL.
Cytokine and siRNA treatment
Cells were treated with 100U/mL recombinant murine IFN-β, 10ng/mL of IL-6, 10 ng/mL of recombinant murine IL-1β (R&D Systems, Minneapolis, MN, USA) and 100 U/mL of recombinant murine interferons-γ (IFN-γ) (Genentech INC., San Fransisco, CA, USA). Mouse mixed glial cells were transfected with 1 µg/1 million cells of siRNA against mouse STAT-3 or control scramble siRNA (Santa Cruz, Santa Cruz, CA, USA). The transfection reagent (Dharmafect 3, Dharmacon, Chicago, IL, USA) was used as specified by the manufacturer. Cells were incubated with the transfection medium in serum-free DMEM for 24 h, after which the cells were incubated with serum-containing complete medium for another 48 h before treatment with cytokines or virus. The efficiency of the siRNA to deplete STAT-3 was verified by real-time RT-PCR using primers specific for murine STAT-3 (Santa Cruz).
Virus inoculation
The attenuated strain of TMEV, BeAn 8386 (ATCC, Manassas, VA, USA) was propagated in BHK-21 cells and titrated by plaque assay on BHK-21 cells. Mixed glial cultures were inoculated with 1 × 106 PFU/mL at a multiplicity of infection of 2.0. Mice were inoculated intracerebrally (i.c.) in the right hemisphere with 1 × 106 PFU of BeAn 8386 in a volume of 0.01 mL. At various times after infection, the right cerebral hemispheres of the mice were resuspended in RNA STAT-60 (TEL-TEST, Friendswood, TX, USA) for RNA analysis and the left hemisphere was resuspended in radioimmunoprecipitation assay buffer for protein analysis by western immunoblot assay.
Real-time RT-PCR
Total RNA was isolated using RNA STAT-60 (Tel-test, Friendswood). RNA was quantified spectrophotometrically and 0.5 µg of total RNA was converted into cDNA (Hudson et al. 2006). Briefly, 25 µL volume of the RNA and random primers (Invitrogen, Carlsbad, CA, USA) were incubated at 72°C for 10 min. Reverse transcription was performed using the Superscript II RT enzyme (Invitrogen) and followed the specifications of the manufacturer. cDNA was diluted to 200 µL with water and 2 µL was used for quantitative PCR using SYBR Green kit (Abgene, Rochester, NY, USA). The PCR parameters were 15 min for 95°C and 35 cycles of 95°C for 15 s and 60°C for 1 min in ABI prism 700 (Applied Biosystems, Foster city, CA, USA). The following primers were used at a 10 nM concentration: SHP-1 Promoter I transcript: (5′-GTCGCCCTTTGCCTGTGAC-3′) as the forward and (5′-TCACCCTGGTTCTTGCGG-3′) as the reverse; SHP-1 Promoter II transcript: (5′-CCAGACAAACTGTTCCCTCCAC-3′) as the forward and (5′-GAAACCACCTCACCATCCTG-3′) as the reverse; VP2 region of TMEV: (5′-TGGTCGACTCTGTGGTTACG-3′) as the forward and (5′-GCCGGTCTTGCAAAGATAGT-3′) as the reverse. Serial dilutions of cDNA clones containing a known copy number of each gene were used in each qPCR run to generate a standard curve relating copy number with threshold amplification cycle. Relative gene expression levels were calculated during the logarithmic amplification phase by determining the initial mRNA copy number using the standard curve (Christophi et al. 2004; Scarisbrick et al. 2006). Amplification of each gene-specific fragment was confirmed both by examination of melting peaks and by agarose gel electrophoresis. To control for loading, parallel examination of the housekeeping gene glyceraldehyde phosphate 3-dehydrogenase (GAPDH) was assessed in each RNA sample using (5′-ACCACCATGGAGAAGGC3′) as the forward and (5′GGCATGGACTGTGGTCATGA3′) as the reverse primer.
Western immunoblotting
Whole cell and tissue extracts were prepared as previously described (Massa and Wu 1996b, 1998). Briefly, glial cultures and CNS tissues were rinsed with phosphate-buffered saline, and then lysed with radioimmunoprecipitation assay buffer (50 mM Tris–HCl, pH 7.4, 1% NP-40, 0.25% sodium deoxycholate, 150 mM NaCl, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 1 µg/mL aprotinin, 1 µg/mL leupeptin, 1 µg/mL pepstatin, and 1 mM activated Na3VO4). The protein concentration was quantified by the Lowry protein assay and 100 µg of protein per lane was electrophoresed through a 10% polyacrylamide resolving gel and electroblotted to a polyvinylidene difluoride membrane (Millipore Corporation, Burlington, MA, USA). Membranes were blocked with 5% non-fat dry milk for 1 h, and then incubated with anti-SHP-1 (Upstate, Lake Placid, NY, USA) or anti-IRF-1 (M-20) (Santa Cruz) antibodies followed by horseradish peroxidase conjugated rabbit IgG antibody (DAKO Corporation, Carpinteria, CA, USA). Enhanced chemiluminescence (Amersham Life Sciences, Inc., Cleveland, OH, USA) was used to visualize reactive protein bands on X-ray film.
Statistical analysis
Histograms contain statistical means with the standard error values. The p-values were generated using the unpaired Student’s t-test and a p-value of < 0.05 was chosen to indicate statistical significance between two sample means.
Results
Tissue and cell distribution of SHP-1
The expression of SHP-1 promoter I and promoter II transcripts were quantified in several tissues and cell types derived from C3HeB/FeJLe wild-type mice (Fig. 1). Promoter I transcripts (Fig. 1a) were highly expressed in the brain, liver, and kidney to levels approaching 107 mRNA copies/ng total RNA. In the spinal cord, spleen, and thymus, the expression of promoter I transcripts was 20-fold lower compared to the brain. Mixed glial cultures and microglial cultures had similar levels of promoter I mRNA to the spinal cord. Cultured purified oligodendrocytes and astrocytes had 5 × 104 promoter I transcript copies/ng RNA. The sequence-specific primers that amplify SHP-1 (I), SHP-1(II), and GAPDH were used to amplify 1 × 106 cDNA vectors carrying the SHP-1 (I), SHP-1(II), or GAPDH sequence to illustrate the specificity of the PCR primers (Fig. 1).
Fig. 1.
Tissue and cell distribution of the SHP-1 transcripts. The y-axis represents the copy number of each specific mRNA transcript per ng of total RNA. (a) The epithelial transcript of SHP-1 (I), Accession # NM_001077705, was quantified in several mouse tissues and cell types using sequence-specific primers by real time RT-PCR. (b) The hematopoietic transcript of SHP-1 (II), Accession # NM_013545, was also quantified in the same samples. (c) The housekeeping gene GAPDH was quantified in the same tissues. (The x-axis is labeled: SHP1(I)–106 vectors of the SHP-1 (I) cDNA sequence, SHP1(II)–106 vectors of the SHP-1 (II) cDNA sequence, GAPDH–106 vectors of the GAPDH cDNA sequence, BR-brain, SpCo-Spinal Cord, Glia-mixed glia, Astr-astrocytes, Micgl-microglia, N2a-neuronal cell line, Spl-Spleen, Thy-Thymus, Mast-mast cells, Liv-liver, Kidn-Kidney).
Promoter II transcripts (Fig. 1b) overall showed close to a 100-fold higher expression than promoter I transcripts in all the tissues assayed except in brain, liver, and kidney. The hematopoietic tissues had significantly the highest levels of promoter II transcripts, with spleen expressing 108 copies/ng RNA. The brain and spinal cord had comparable levels of promoter II transcripts (approximately 106 mRNA copies/ng RNA). Cultured glial cells including mixed glia, purified oligodendrocytes, astrocytes, and microglia had a significant 10-fold increase in promoter II copies compared to the brain or spinal cord. The oligodendrocytes had particularly high levels of the promoter II transcripts relative to all other CNS glia in pure culture including microglia and were more than a 100-fold higher than promoter I transcripts. Taken together we concluded that the promoter I and II transcripts are preferentially expressed in epithelial and hematopoetic tissues, respectively. However, unexpectedly, glial cells preferentially expressed the promoter II/hematopoietic transcripts in vitro.
Cytokines regulate SHP-1 protein expression
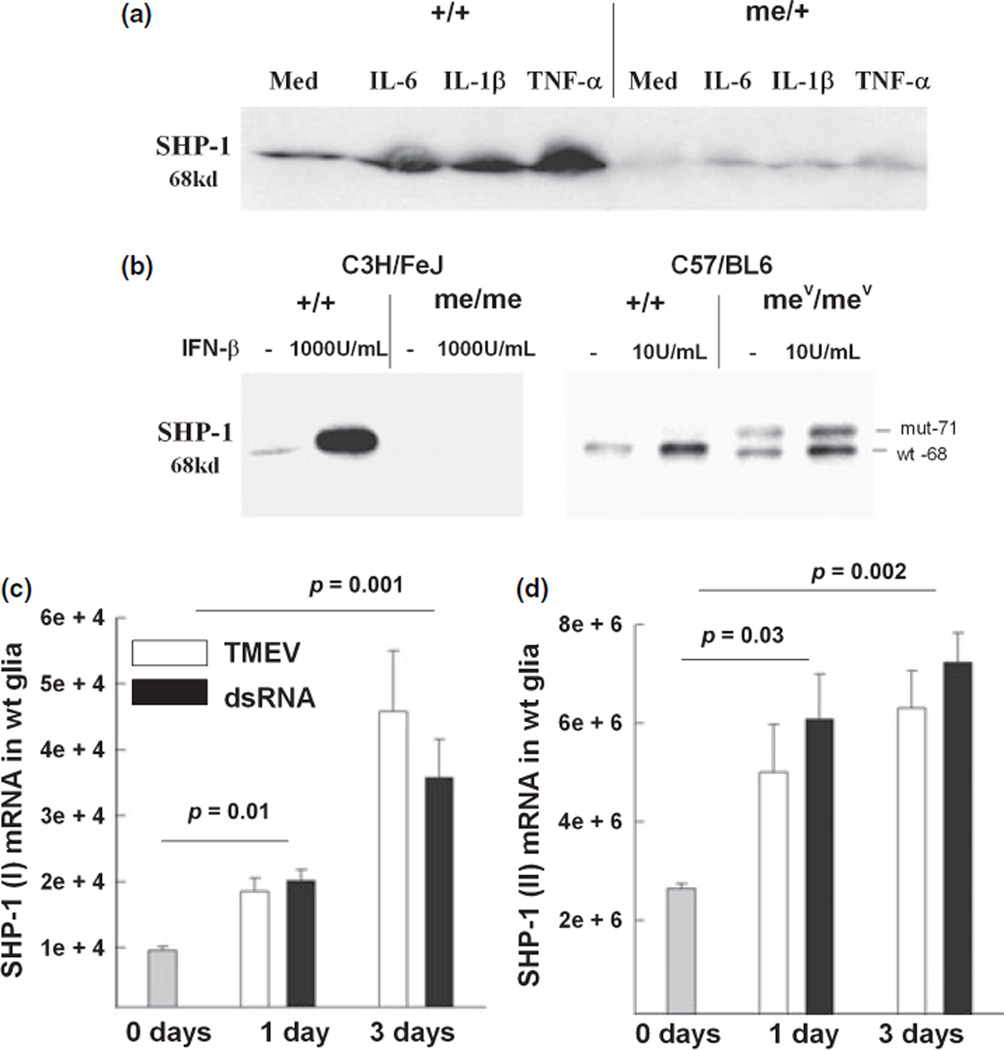
We have previously described that the cytokine IL-6 increases the expression of SHP-1 in CNS glia by a currently unknown pathway (Massa et al. 2000). Glial cultures derived either from wild-type mice or me/+ mice expressed constitutive SHP-1 protein at the expected relative levels and SHP-1 was significantly induced in both genotypes in response to IL-6 (Fig. 2a). Further, IL-1β and tumor necrosis factor-α (TNF-α) considerably increased the expression of SHP-1 protein. Treatment of CNS glia with interferons, IFN-γ (Fig. 5) and IFN-β (Fig. 2b), substantially increased the levels of SHP-1 protein implicating STAT-1 or other STATs in the regulation of SHP-1. As expected, glia from heterozygous (me/+) mice showed induction of SHP-1 following cytokine treatment, but to lower levels compared to +/+ mice. Taken together, these data illustrate that SHP-1 levels are substantially regulated by both gene dosage and in response to a variety of cytokine stimuli.
Fig. 2.
Cytokine treatment and virus infection induce the expression of SHP-1 protein and mRNA in glial cells. (a) Induction of SHP-1 by proinflammatory cytokines. Glial cells from +/+ or me/+ mice were treated with 10 ng/mL of IL-6, 10 ng/mL of IL-1β, and 100 U/mL of TNF-α for 24 h and SHP-1 protein was measured by western immunoblot analysis. Equal amounts of cell protein extract were loaded per lane (100 µg). (b) Glial cells from +/+, me/me, or mev/mev (viable motheaten) mice were treated with 1000 U/mL or 10 U/mL of IFN-β for 24 h and SHP-1 levels were analyzed. (c and d) Wild-type glial cells were infected with TMEV or treated with dsRNA for either 1 day or 3 days and the levels of SHP-1 (I)/epithelial transcripts (c) or SHP-1 (II)/hematopoetic transcripts (d). were measured by real time RT-PCR.
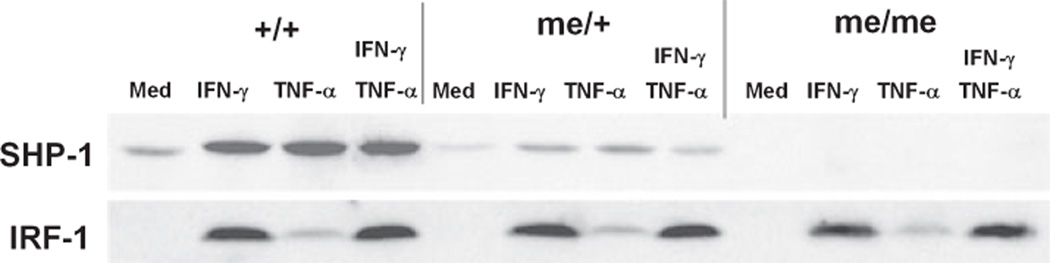
Fig. 5.
Western blot of the induction patterns of SHP-1 and IRF-1 proteins. Astrocytes from +/+, me/+, or me/me mice were treated with either IFN-γ (100 U/mL), TNF-α (1000 U/mL), or IFN-γ and TNF-α combined for 8 h and the levels of SHP-1 and IRF-1 proteins were detected by chemiluminescence. Equal amounts of cell protein extract were loaded per lane (100 µg).
Neurotropic virus infection up-regulates SHP-1 in the CNS
We have previously shown that motheaten mice show increased inflammatory demyelination compared to wild-type littermates when infected with TMEV, implicating SHP-1 in controlling virus-induced inflammation in the CNS. As such, induction of SHP-1 by virus-induced cytokines may represent a negative feedback response to cytokine signaling in the CNS. Therefore, we examined whether SHP-1 levels increased following virus infection of CNS glia in vitro. First, wild-type glial cultures were inoculated with TMEV or viral mimic dsRNA for 24 and 72 h and the levels of SHP-1 transcripts were measured. Promoter I transcripts were significantly increased by 2–4-fold over time after inoculation with either TMEV or treatment with dsRNA compared to untreated controls (Fig. 2c). Similarly, promoter II transcripts were induced by 2–3-fold over time after similar treatment with either TMEV or dsRNA compared to untreated cultures (Fig. 2d).
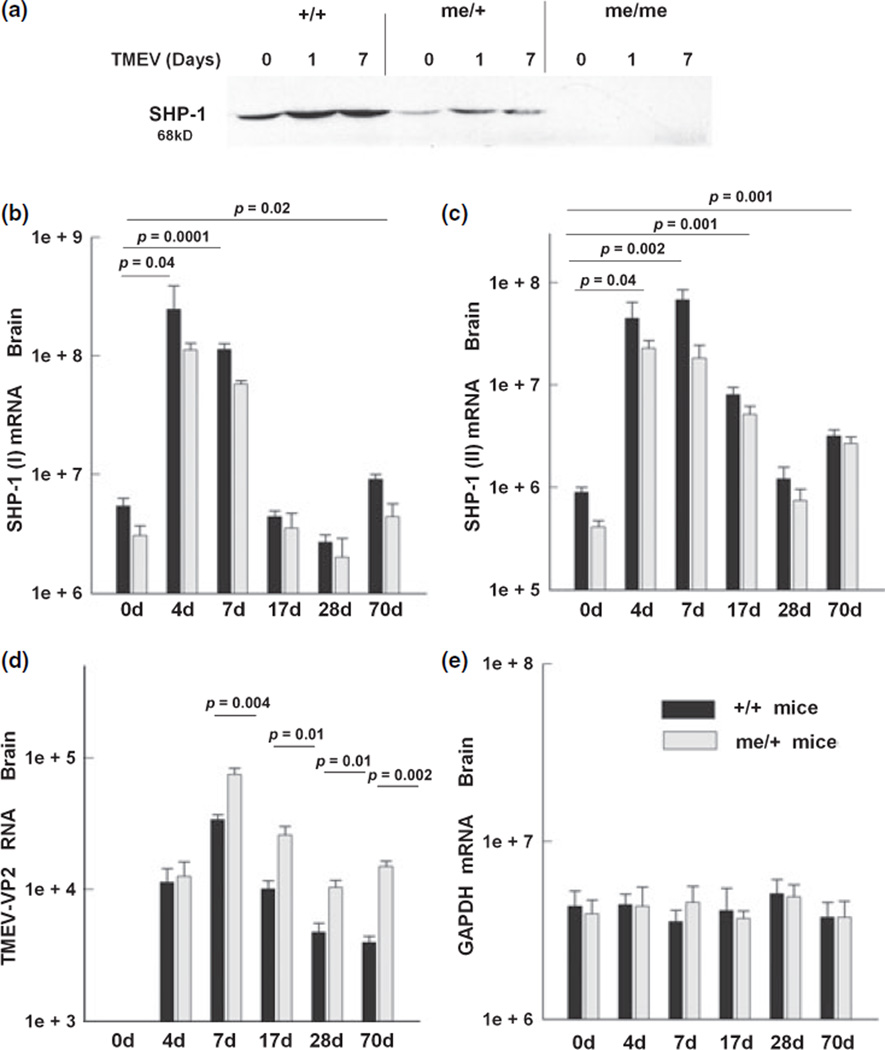
Next, we wanted to determine whether TMEV inoculation in vivo was able to up-regulate the levels of SHP-1. Wild-type (+/+) and me/+ mice were inoculated into the right hemisphere with TMEV and were sacrificed at different time points to examine the expression of SHP-1 in the brain. SHP-1 protein was increased both in the +/+ and me/+ mice at 1 and 7 days post-infection (d.p.i.) (Fig. 3a). In order to delineate the contribution of promoter I and II transcripts in inducing SHP-1, sequence-specific primers were used to quantify each transcript. SHP-1 promoter I transcripts were increased 20-fold after 4 or 7 d. p.i. (Fig. 3b). At 17 and 28 d.p.i., the levels of SHP-1 promoter I transcripts were not significantly different from the levels of SHP-1 promoter I transcripts in the uninfected mice. As expected, the levels of SHP-1 promoter I transcripts were significantly lower in the heterozygous mice and were induced following infection.
Fig. 3.
TMEV infection in vivo induces SHP-1 expression in the CNS. Mice were inoculated intracerebrally in the right hemisphere with 1 × 106 PFU of TMEV. Mice were sacrificed at different time points and the right hemisphere of the mice was used for RNA analysis and the left for protein analysis. (a) Western blot showing SHP-1 expression in +/+, me/+, and me/me mice before and after 1 or 7 days post-infection. Equal amounts of tissue protein extract were loaded per lane (100 µg). (b and c) The SHP-1 (I) and SHP-1 (II) transcript levels in brain of TMEV-infected mice at 0, 4, 7, 17, 28, and 70 days post-infection (d.p.i.) were measured with real time RT-PCR. (d) Primers recognizing the viral protein 2 (VP2) of TMEV were used to quantify the viral RNA loads in the +/+ and me/+ mice. (e) GAPDH mRNA levels from the same samples were quantified to control for RNA loading.
Similarly, SHP-1 promoter II transcripts were up-regulated in the brains of mice that were inoculated with TMEV (Fig. 3c). Early during infection at 4 or 7 d.p.i., the SHP-1 promoter II transcripts were increased 50-fold compared to uninfected mice. As the infection progressed, the levels of SHP-1 promoter II transcripts remained higher compared to the uninfected mice, but were significantly lower compared to the early stage of infection. At 17 d.p.i., SHP-1 promoter II transcripts were nine-fold higher than uninfected mice and at 70 d.p.i. SHP-1 transcripts were 3.5-fold higher than in uninfected mice. The levels of the viral RNA and the mRNA expression of the housekeeping gene GAPDH are also shown to both ascertain the presence of persisting virus during the time course of analysis and determine specificity of SHP-1 induction in response to infection. These data demonstrate that viral infection in the CNS, most likely via virus-induced cytokines, up-regulates SHP-1 transcripts and protein both in vitro and in vivo.
STAT-1 controls SHP-1 promoter I transcript expression
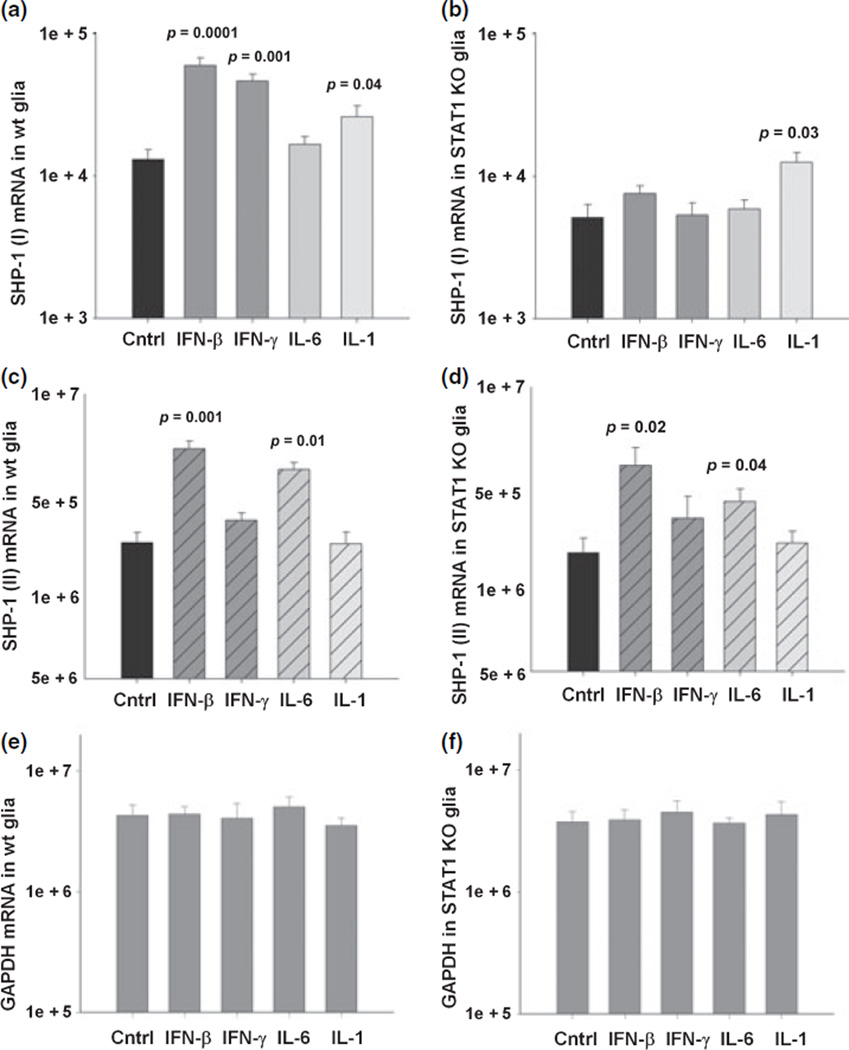
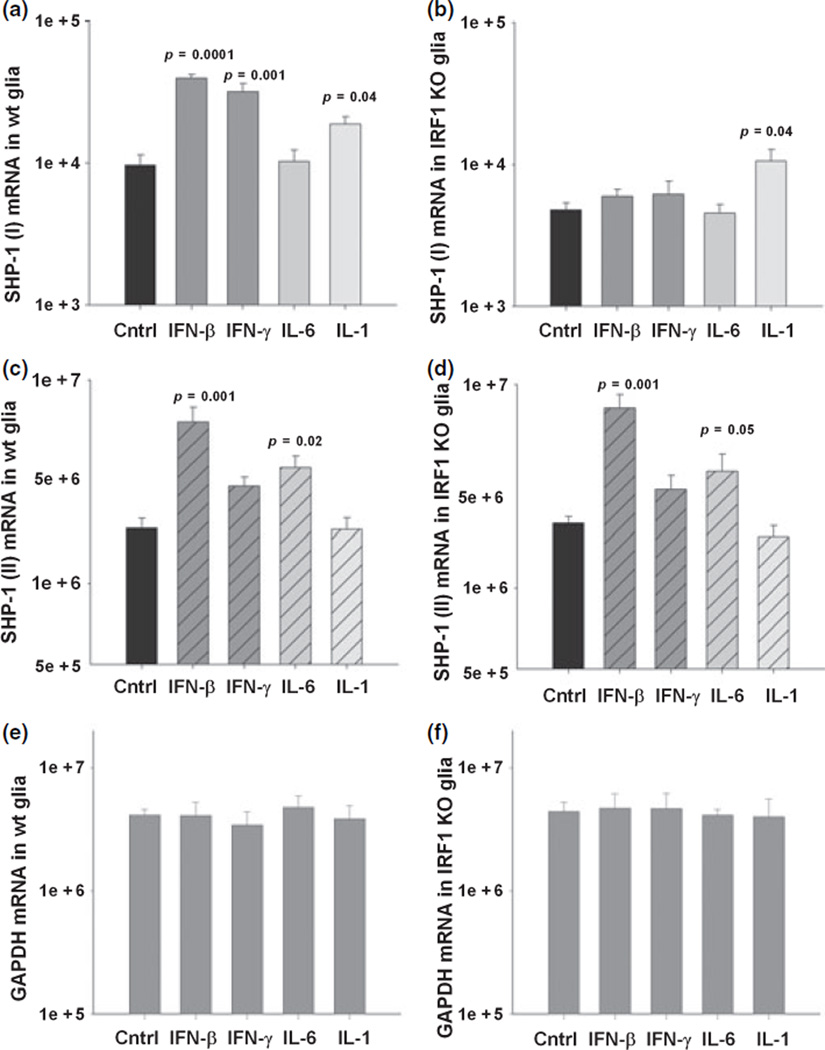
Since SHP-1 was induced by viral infection in vivo and by interferons in cultured glia and interferons signal primarily via STAT-1, it was of interest to determine whether STAT-1 was required for SHP-1 induction. Mixed glial cultures prepared from newborn 129S6/SvEv wild-type and STAT-1 null mice were treated with IFN-β or IFN-γ for 24 h and total RNA was isolated. As controls, cells were also treated with IL-6 and IL-1β, which signal primarily via STAT-3 and NF-κB, respectively. The levels of the promoter I transcripts in wild-type glia showed a significant five-fold increase following treatment with IFN-β, four-fold with IFN-γ treatment, and 2.5-fold with IL-1β treatment (Fig. 4). Treatment with IL-6 did not affect levels of promoter 1 transcripts. Importantly, constitutive levels of promoter I transcripts were four-fold less in STAT-1 null mouse glial cells compared to wild-type cells. Moreover, when STAT-1 null glia were treated with IFN-β or IFN-γ, the increase in the promoter I transcripts seen in wild-type cells was abolished (Fig. 4b). On the other hand, IL-1β treatment significantly increased the SHP-1 promoter transcript in STAT-1 null cells, which is consistent with previous observations that NF-κB activation leads to up-regulation of promoter I transcripts (Tsui et al. 2002). The levels of the SHP-1 promoter II transcripts in wild-type glia (Fig. 4c) also showed a significant four-fold increase with IFN-β treatment. Furthermore, IL-6 treatment significantly increased the promoter II transcripts by two-fold, but treatment with either IL-1β or IFN-γ failed to increase the levels of the promoter II transcripts. The loss of STAT-1 in glial cells did not affect either the constitutive levels of promoter II transcripts or IFN-β-/IL-6-induced levels of the promoter II transcripts.
Fig. 4.
SHP-1 expression in glia of STAT-1 null mice. Expression of SHP-1 transcripts in mixed glial cultures of wild-type (wt) and STAT-1 null (STAT-1 KO) mice following 24 h treatment with medim alone, 100 U/mL of IFN-β, 10 U/mL of IFN-γ, 10 ng/mL of IL-6, and 10 ng/mL of IL-1β measured with real time RT-PCR (mRNA copy number/1.0 ng total RNA). (a) SHP-1 (I) transcript mRNA in +/+ glia, (b) SHP-1 (I) transcript mRNA in STAT-1 null glia, (c) SHP-1 (II) transcript mRNA in +/+ glia, (d) SHP-1 (II) transcript mRNA in STAT-1 null glia, (e and f) GAPDH levels in glia of +/+ and STAT-1 null mice.
IRF-1 controls SHP-1 promoter I transcript expression
Because interferon regulatory factor-1 indirectly mediates multiple STAT-1-dependent responses to interferons and virus infections, we wanted to examine the role of IRF-1 on SHP-1 expression in glia treated with interferon. IFN-γ treatment concomitantly induced the expression of both IRF-1 and SHP-1 protein in wild-type glial cultures (Fig. 5). When me/+ glia were treated with IFN-γ the same amount of IRF-1 was induced compared to +/+ glia, however, as expected, approximately half of the SHP-1 protein was induced. Also, glial cells treated with either TNF-α, which activates NF-κB, or IFN-γ raised the levels of SHP-1 to the same degree, although TNF-α alone showed a very slight induction of IRF-1. The latter may indicate that TNF-α acts via a known κB site in promoter I while IFN-γ acts on promoter 1 in a distinct STAT-1/IRF-1 dependent manner.
In order to directly demonstrate the role of IRF-1 in the regulation of SHP-1, mixed glia cultures from IRF-1−/− mice were compared to wild-type glial cultures for the expression of the two SHP-1 transcripts (Fig. 6). IFN-β and IFN-γ increased the expression of promoter I transcripts by approximately four-fold and IL-1β by two-fold in wild-type 129S6/SvEv mouse glia. However, glial cells from IRF-1−/− mice on the same background showed half the constitutive expression of promoter I transcripts compared to wild-type glial cells (Fig. 6b). Moreover, glial cells from IRF-1−/− showed no induction of promoter I transcripts following treatment with IFN-β or IFN-γ, but showed the same two-fold induction after IL-1β treatment as seen in wild-type cells. On the contrary, there were no differences observed in the constitutive or induced levels of promoter II transcript between glial cells from IRF-1−/− mice and wild-type mice (Fig. 6c and d). Furthermore, promoter II transcripts showed a four-fold induction in glia cells of both wild-type and IRF-1−/− mice following IFN-β treatment. IL-6 showed a two-fold induction of promoter II transcripts in glial cells from both wild-type and IRF-1−/− mice, while IFN-γ and IL-1β failed to significantly raise promoter II transcript levels. Taken together, these data indicate that STAT-1 and IRF-1 contribute to both constitutive and interferon-induced expression of promoter I but not promoter II transcripts in CNS glia.
Fig. 6.
SHP-1 expression in glia of IRF-1 null mice. Expression of the SHP-1 transcripts in mixed glia cultures of wild-type (wt) and IRF-1 null (IRF-1 KO) mice following 24 h treatment with medium alone, 100 U/mL of IFN-β, 10 U/mL of IFN-γ, 10 ng/mL of IL-6, and 10 ng/mL of IL-1β was measured with real time RT-PCR and the mRNA copy number/1.0 ng total RNA is shown. (a) SHP-1 (I) transcript mRNA in +/+ glia. (b) SHP-1 (I) transcript mRNA in IRF-1 null glia. (c) SHP-1 (II) transcript mRNA in +/+ mice. (d) SHP-1 (II) transcript mRNA in IRF-1 null mice. (e and f) GAPDH levels in glia of +/+ and IRF-1 null mice.
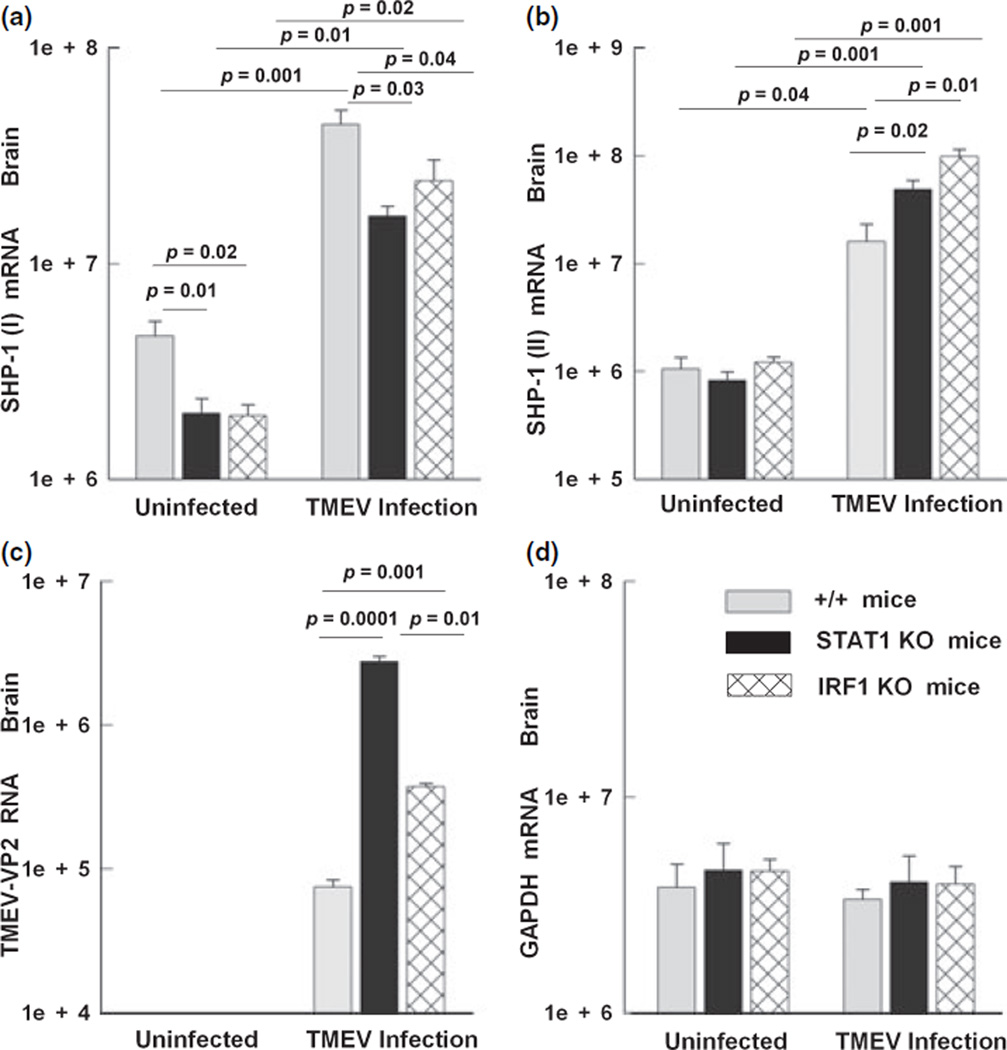
SHP-1 (I) is regulated in vivo by STAT-1 and IRF-1
We wanted to determine whether SHP-1 transcripts of STAT-1 and IRF-1 null mice are lower in brain compared to wild-type mice and whether the lack of STAT-1 or IRF-1 would prevent the induction of the SHP-1 in the CNS of mice infected with TMEV. Wild-type, STAT-1 null, and IRF-1 null mice were inoculated in the right hemisphere with an attenuated strain of TMEV. Since infection of STAT-1 null mice is lethal within 1 week, mice were sacrificed at 5 d.p.i. to examine the expression of SHP-1 in the brain (Fig. 7). SHP-1 promoter I transcripts were constitutively lower in the brain of STAT-1 or IRF-1 KO mice compared to +/+ mice, which is in agreement with the results obtained from glial cultures. When mice were infected with TMEV, the levels of SHP-1 promoter I transcripts rose 10-fold (Fig. 7a). Although SHP-1 promoter I transcripts were considerably induced in STAT-1 and IRF-1 KO mice, this induction was significantly lower than in wild-type control mice. The induction of SHP-1 promoter I transcripts could have been caused by the increased TNF-α levels seen in TMEV infection (Gerhauser et al. 2007), acting via the known NF-κB site (Tsui et al. 2002).
Fig. 7.
SHP-1 expression in the CNS of STAT-1 and IRF-1 null mice following TMEV infection. Wild-type (+/+), STAT-1 null (STAT-1 KO), and IRF-1 (IRF-1 KO) null mice were inoculated in the right hemisphere with an attenuated stain of TMEV. Mice were sacrificed at 5 days p.i. to examine the expression levels of SHP-1 in the brain using real-time RT-PCR (mRNA copy number/1.0 ng total RNA). (a) Promoter I transcript copy numbers per 1.0 ng of total RNA in the right hemisphere of +/+, STAT-1 null, and IRF-1 null mice before and after TMEV infection. (b) SHP-1 (II) transcript levels. (c) TMEV RNA levels, and (d) GAPDH RNA levels.
On the other hand, there were no constitutive differences in the levels of SHP-1 promoter II transcripts between wild-type and STAT-1 or IRF-1 null mice (Fig. 7b). Interestingly, when mice were infected with TMEV, the levels of SHP-1 promoter II transcripts in +/+ wild-type mice increased 20-fold, while STAT-1 and IRF-1 null mice showed significantly higher induction compared to +/+ mice. As expected, STAT-1 null mice had significantly 30-fold higher viral loads and IRF-1 null mice had 10-fold higher viral loads compared to +/+ mice. Therefore, the higher induction of SHP-1 promoter II levels in STAT-1/IRF-1 null mice could be attributed to the higher viral stimulus on cytokine expression seen in those mice. Taken together, these data indicated that STAT-1/IRF-1 increased promoter I activity in response to CNS virus infection, but had little effect on the promoter II activity.
STAT-3 controls SHP-1 promoter II transcript expression
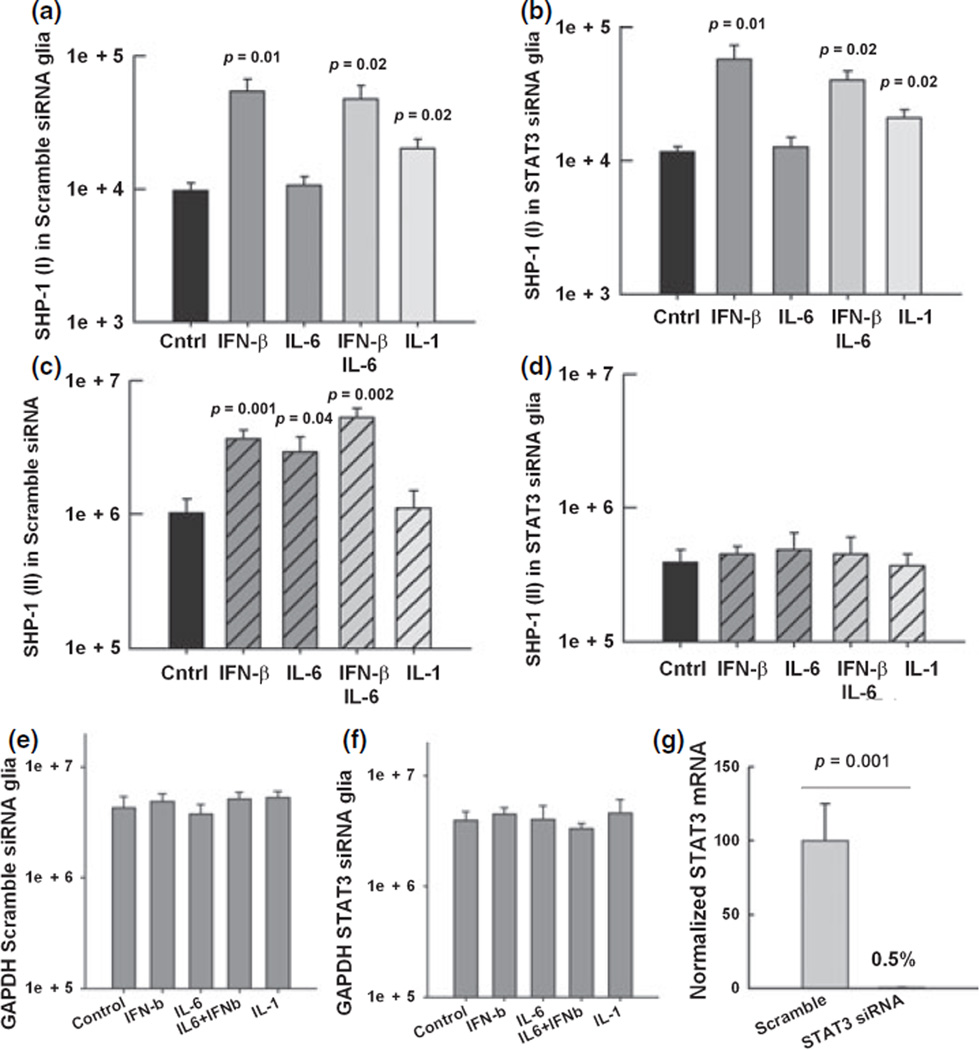
It has previously been shown that STAT-3 binds to and plays a role in constitutive activity of SHP-1 promoter II in human lymphocytic tumor cell lines (Zhang et al. 2005). However, in these cells, STAT-3 appeared to negatively regulate promoter activity. We have previously shown that IL-6 both activated STAT-3 and induced the expression of SHP-1 in CNS glia (Massa et al. 2000) and others have shown functional STAT-3 binding elements in promoter II suggesting that STAT-3 signaling may be involved in regulation of the SHP-1 gene. In order to investigate whether either of the two SHP-1 transcripts were regulated through STAT-3, and considering that targeted disruption of the mouse Stat3 gene leads to early embryonic lethality (Takeda et al. 1997), we used siRNA to deplete STAT-3 expression and analyzed the resulting expression of SHP-1 in wild-type glia. Glial cells from wild-type mice were pre-treated for 72 h with either siRNA against STAT-3 mRNA or siRNA with scrambled sequence as control. Pre-treated glial cells were further treated with medium alone, IFN-β, IFN-γ, IL-1β and IL-6 for 24 h and then RNA was isolated. The efficiency of the siRNA to deplete STAT-3 was verified by measuring the levels of STAT-3 mRNA. We found that STAT-3 expression was 200-fold lower in glia treated with STAT-3 siRNA compared to glia treated with scramble siRNA (Fig. 8g). The constitutive levels of the promoter I transcripts were the same in glial cells that received either no treatment (Fig. 4a), scrambled siRNA, or siRNA against STAT-3 (Fig. 8a and b). Additionally, pre-treatment with either scrambled siRNA or STAT-3 siRNA did not affect the five-fold increase in promoter I transcripts induced by IFN-β or the two-fold increase induced by IL-1β. In contrast, glial cells that were pre-treated with STAT-3 siRNA (Fig. 8d) had three-fold lower constitutive levels of promoter II transcripts compared to cells that received scrambled siRNA. Moreover, in glia pre-treated with siRNA against STAT-3 the increase of the promoter II transcripts following treatment with either IFN-β or IL-6 was abolished. Combined treatment with IFN-β and IL-6 showed a five-fold increase of the promoter II transcripts compared to untreated cultures and STAT-3 siRNA abolished this induction. In summary, induction of promoter II transcripts by SHP-1 by either Il-6 or IFN-β is specifically dependent on STAT-3 in CNS glia.
Fig. 8.
SHP-1 promoter I and II expression in STAT-3 siRNA-treated glia. Real-time PCR was used to quantify the expression of the SHP-1 transcripts (mRNA copy number/1.0 ng total RNA) in mixed glia cultures from wild-type mice pre-treated for 72 h with either siRNA agaist STAT-3 or scrambled sequence siRNA. Then, glia were treated for 24 h with media alone, 100 U/mL of IFN-β, 10 ng/mL of IL-6, and 10 ng/mL of IL-1β. (a) SHP-1 (I) in wild-type glial cells pre-treated with scramble siRNA (b). SHP-1 (I) in glia pre-treated with siRNA against STAT-3. (c) SHP-1 (II) in wild-type glia cells pre-treated with scramble siRNA (d) SHP-1 (II) in glia pre-treated with siRNA against STAT-3. (e and f) The levels of GAPDH were quantified in the same samples. (g) The efficiency of the siRNA to deplete STAT-3 mRNA was verified by real time RT-PCR.
SHP-1 regulation in splenocytes
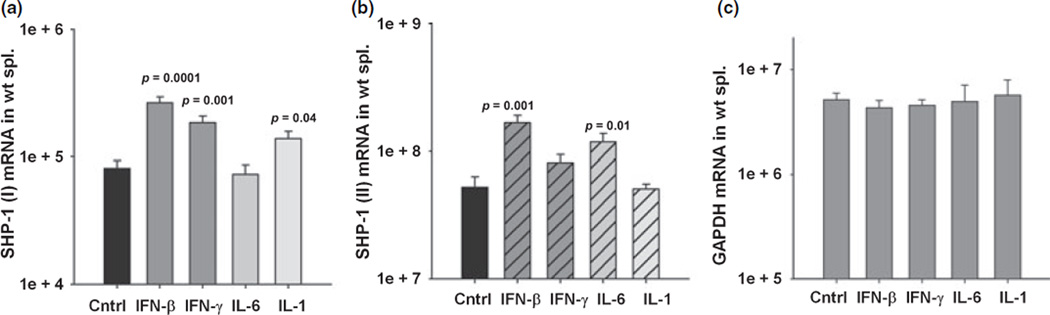
To determine whether SHP-1 is regulated in a similar way in hematopoetic cells as in CNS cells we cultured mouse splenocytes and examined the expression of both SHP-1 transcripts following several treatments. Promoter I transcripts showed a significant three-fold induction following 24 h treatment with either IFN-β or γ compared to untreated controls (Fig. 9a). IL-1β treatment increased promoter I transcripts by two-fold and IL-6 did not induce promoter I compared to control. Promoter II transcript levels showed a significant three-fold increase when splenocytes were treated with IFN-β compared to control (Fig. 9b). IL-6 also significantly increased the promoter II transcript levels two-fold. Thus, inducible promoter I and promoter II activities appear to be similarly regulated in CNS and hematopoetic tissues.
Fig. 9.
SHP-1 transcript expression in splenocyte cultures following cytokine stimulation. Splenocytes from wild-type mice were cultured for 24 h with media alone, 100 U/mL of IFN-β, 10 U/mL of IFN-γ, 10 ng/mL of IL-6, and 10 ng/mL of IL-1β. Total RNA was isolated and analyzed by real time RT-PCR (mRNA copy number/1.0 ng total RNA) to measure the levels of (a) SHP-1 (I) transcript mRNA. (b) SHP-1 (II) transcript mRNA. (c) GAPDH mRNA.
Discussion
In this study, we show the differential expression of the two SHP-1 transcripts in several tissues of mice. Although the designation of epithelial and hematopoetic transcripts generally reflect the greater abundance of the SHP-1 (II)/hematopoetic transcript in hematopoetic cell lines and the SHP-1 (I)/epithelial transcript in epithelial cell lines, here we demonstrate that both promoters are active in tissues of both epithelial and hematopoetic origin. Furthermore, we show that SHP-1 protein and mRNA is induced by a variety of cytokines in CNS glia and peripheral hematopoetic cells in a promoter-specific manner.
Importantly, we demonstrate for the first time that virus infection of CNS cells both in vitro and in vivo significantly induces the expression of SHP-1 mRNA transcripts and protein, which is in accordance with the fact that virus-induced interferons and pro-inflammatory cytokines including IL-6 and TNF-α substantially induce SHP-1 levels. The intermediary role of cytokines in virus-induced SHP-1 was supported by the loss of SHP-1 induction in vivo in STAT-1-and IRF-1-null mice. In agreement with previous observations that complete loss of SHP-1 leads to augmented viral loads and inflammation following TMEV infection (Massa et al. 2002), an increase in SHP-1 activity would be expected to counteract viral replication partly through increased iNOS activity as recently reported by us (Bonaparte et al. 2006). Moreover, the role of iNOS and NO in inhibiting NF-κB activity and tissue inflammation in tissues including the CNS has been demonstrated (Arnett et al. 2002; Ckless et al. 2007).
We have shown that loss of SHP-1 in the CNS as seen in motheaten mice leads to myelin pathology by currently unknown mechanisms (Massa et al. 2000, 2004). However, recent evidence indicates that chronic constitutive inflammation and especially increased production of reactive oxygen species in the CNS of SHP-1-deficient mice during the period of active myelination may be responsible for the observed effects on myelin. Additionally, when motheaten mice are infected with TMEV, me/me mice show severe inflammatory demyelination compared to +/+ littermates (Massa et al. 2002). Similarly, mice deficient in SHP-1 developed a more severe course of experimental allergic encephalomyelitis and a more pronounced inflammatory profile compared to wild-type mice (Deng et al. 2002). These studies point to an important role of SHP-1 in controlling inflammatory demyelinating processes.
Since viral infections and associated cytokines of innate immunity increase the levels of SHP-1 protein by inducing the expression of both SHP-1 promoter I and promoter II transcripts, we elucidated the responsible gene regulatory factors that may be involved. Here we showed that promoter I drives high expression levels of SHP-1 (I) transcripts in the brain in an IRF-1-dependent manner. Our data are also compatible with previous observations that SHP-1 (I) transcripts are under the control of NF-κB (Tsui et al. 2002) because the NF-κB inducer IL-1β induces promoter I transcript expression in IRF-1 null glia. On the other hand, STAT-3 appeared to act exclusively to increase the levels of SHP-1 (II) transcripts via promoter II in response to multiple cytokines. In agreement with these data, analysis of the SHP-1 promoter revealed the presence of IRF-1 and STAT-3 cis regulatory elements in promoter I and promoter II respectively, which could be responsible for the promoter-specific expression of SHP-1 (Fig. 10). Whether these potential regulatory elements are responsible for IRF-1 and STAT-3 effects on promoter activity requires further studies on promoter function in CNS glia. Promoter-specific effects of cytokines and downstream transcription factors may allow modulation of SHP-1 expression in a precise way for tissue-specific homeostatic or therapeutic purposes.
Fig. 10.
Schematic representation of the two SHP-1 transcripts transcribed via promoter I and II. The gray boxes represent the exons that can form the SH-2 binding domains or the catalytic domain. Both transcripts translate into identical isoforms except at the N-terminus where the isoform driven from promoter I, SHP-1 (I), has the amino acids MLSG and the isoform driven by promoter II, SHP-1 (II) has the amino acids MVR. Promoter I is upstream of exon I and contains consensus sequences that can bind NF-κB, IRF-1, and SP-1. Promoter II is found between exon 1 and 2 and has several STAT-3 binding sites. The table lists the nucleotide sequences of the cis-regulatory elements found either in Promoter I or in Promoter II both in human and mouse.
Several studies demonstrate that human MS lesions and peripheral blood mononuclear cells from MS patients contain activated transcription factors like NF-κB, STAT-1, STAT-3, and STAT-6 (Gobin et al. 2001; Cannella and Raine 2004; Frisullo et al. 2006; Zeis et al. 2007; Eggert et al. 2008), suggesting that regulation of inflammatory signaling may be altered in MS and may be responsible for inflammatory demyelination. Furthermore, SHP-1 is a negative regulator of these transcription factors in both the CNS and immune systems (David et al. 1995; Massa and Wu 1996a, 1998; Hanson et al. 2003; Bonaparte et al. 2006). In accordance with that data, we have recently shown that peripheral leukocytes of MS patients have significantly lower levels of SHP-1 mRNA and protein expression and that MS patients have elevated levels of inflammatory gene expression controlled by SHP-1 (Christophi et al. 2008).
Therefore, it is of great interest to determine whether IFN-β, a current effective treatment for the human demyelinating disease multiple sclerosis (Ann Marrie and Rudick 2006; Javed and Reder 2006), acts to elevate the levels of SHP-1 in both CNS resident cells and peripheral leukocytes. Preliminary studies in our laboratory, using peripheral blood mononuclear cells of normal subjects and MS patients have shown that IFN-β increases SHP-1 protein and mRNA expression. Increased levels of SHP-1 would be expected to decrease activation of several pro-inflammatory cytokines and associated transcription factors and attenuate inflammatory gene expression (Fig. 11). Hence, it becomes essential to delineate the tissue-specific molecular mechanisms controlling the transcriptional regulation of SHP-1 and to further study the functional effects of increased SHP-1 levels on CNS inflammatory disease.
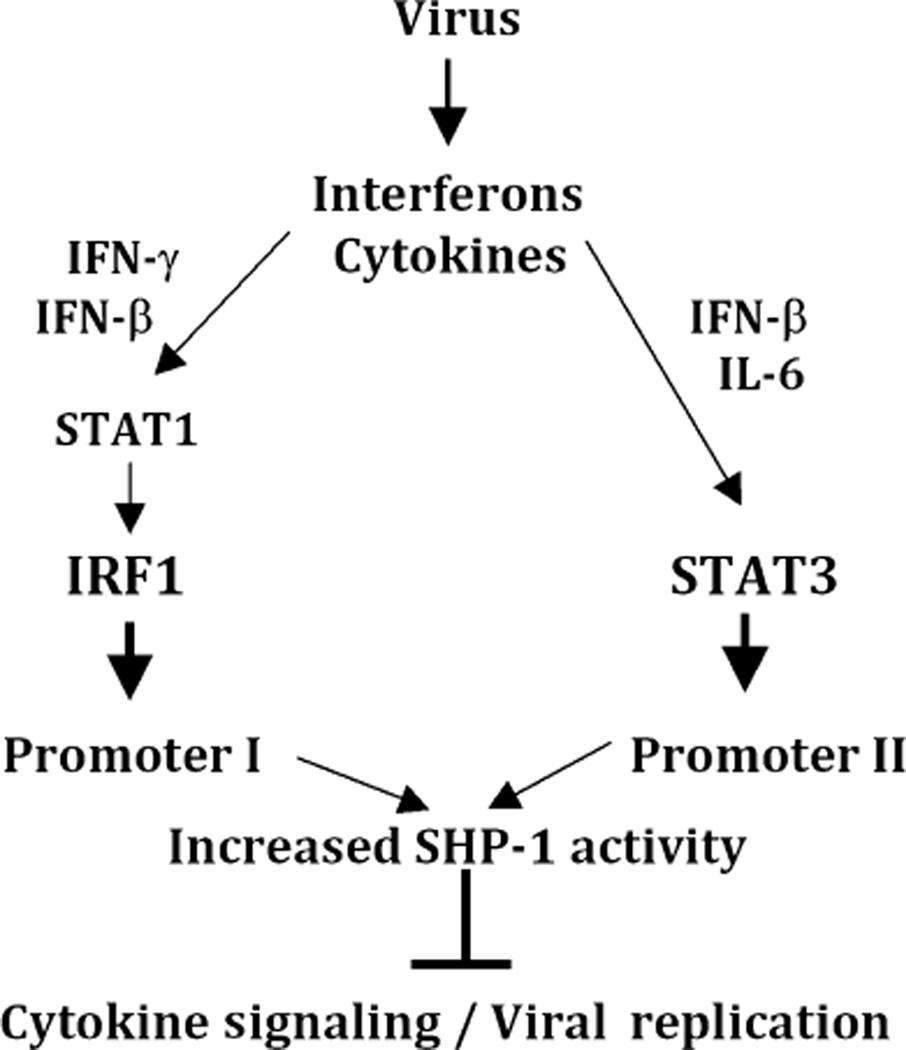
Fig. 11.
Schematic diagram of the signaling pathways contributing to SHP-1 induction. Interferons and cytokines that result from viral infections can activate either promoter I or II of the SHP-1 gene. In particular, IFN-γ and IFN-β can activate STAT-1, resulting in the induction of IRF-1 and subsequent induction of the SHP-1 promoter I transcripts. In contrast, IFN-β and IL-6 can activate STAT-3, resulting in the induction of promoter II transcripts. Consequently, the increased SHP-1 activity is expected to counteract viral replication and inhibit cytokine signaling.
Acknowledgments
This work was supported by grants from the National Multiple Sclerosis Society (RG 2569-C-5) and the National Institutes of Health (R01 NS041593). We would like to thank Dr Isobel Scarisbrick at Mayo Clinic for providing cDNA plamids used in performing real time RT-PCR.
Abbreviations used
- DMEM
Dulbecco’s modified Eagle’s medium
- IFN-β
interferons-β
- IFN-γ
interferons-γ
- IRF-1
interferon regulatory factor-1
- TMEV
Theiler’s murine encephalomyelitis virus
References
- Ann Marrie R, Rudick RA. Drug insight: interferon treatment in multiple sclerosis. Nat. Clin. Pract. Neurol. 2006;2:34–44. doi: 10.1038/ncpneuro0088. [DOI] [PubMed] [Google Scholar]
- Arnett HA, Hellendall RP, Matsushima GK, Suzuki K, Laubach VE, Sherman P, Ting JP. The protective role of nitric oxide in a neurotoxicant-induced demyelinating model. J. Immunol. 2002;168:427–433. doi: 10.4049/jimmunol.168.1.427. [DOI] [PubMed] [Google Scholar]
- Banville D, Stocco R, Shen SH. Human protein tyrosine phosphatase 1C (PTPN6) gene structure: alternate promoter usage and exon skipping generate multiple transcripts. Genomics. 1995;27:165–173. doi: 10.1006/geno.1995.1020. [DOI] [PubMed] [Google Scholar]
- Bonaparte KL, Hudson CA, Wu C, Massa PT. Inverse regulation of inducible nitric oxide synthase (iNOS) and arginase I by the protein tyrosine phosphatase SHP-1 in CNS glia. Glia. 2006;53:827–835. doi: 10.1002/glia.20344. [DOI] [PubMed] [Google Scholar]
- van Boxel-Dezaire AH, Rani MR, Stark GR. Complex modulation of cell type-specific signaling in response to type I interferons. Immunity. 2006;25:361–372. doi: 10.1016/j.immuni.2006.08.014. [DOI] [PubMed] [Google Scholar]
- Cannella B, Raine CS. Multiple sclerosis: cytokine receptors on oligodendrocytes predict innate regulation. Ann. Neurol. 2004;55:46–57. doi: 10.1002/ana.10764. [DOI] [PubMed] [Google Scholar]
- Christophi GP, Isackson PJ, Blaber S, Blaber M, Rodriguez M, Scarisbrick IA. Distinct promoters regulate tissue-specific and differential expression of kallikrein 6 in CNS demyelinating disease. J. Neurochem. 2004;91:1439–1449. doi: 10.1111/j.1471-4159.2004.02826.x. [DOI] [PubMed] [Google Scholar]
- Christophi GP, Hudson CA, Gruber RC, Christophi CP, Mihai C, Mejico LJ, Jubelt B, Massa PT. SHP-1 deficiency and increased inflammatory gene expression in PBMCs of multiple sclerosis patients. Lab. Invest. 2008;88:243–255. doi: 10.1038/labinvest.3700720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ckless K, van der Vliet A, Janssen-Heininger Y. Oxidative-nitrosative stress and post-translational protein modifications: implications to lung structure-function relations. Arginase modulates NF-kappaB activity via a nitric oxide-dependent mechanism. Am. J. Respir. Cell Mol. Biol. 2007;36:645–653. doi: 10.1165/rcmb.2006-0329SM. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David M, Chen HE, Goelz S, Larner AC, Neel BG. Differential regulation of the alpha/beta interferon-stimulated Jak/Stat pathway by the SH2 domain-containing tyrosine phosphatase SHPTP1. Mol. Cell. Biol. 1995;15:7050–7058. doi: 10.1128/mcb.15.12.7050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng C, Minguela A, Hussain RZ, Lovett-Racke AE, Radu C, Ward ES, Racke MK. Expression of the tyrosine phosphatase SRC homology 2 domain-containing protein tyrosine phosphatase 1 determines T cell activation threshold and severity of experimental autoimmune encephalomyelitis. J. Immunol. 2002;168:4511–4518. doi: 10.4049/jimmunol.168.9.4511. [DOI] [PubMed] [Google Scholar]
- Eggert M, Goertsches R, Seeck U, Dilk S, Neeck G, Zettl UK. Changes in the activation level of NF-kappa B in lymphocytes of MS patients during glucocorticoid pulse therapy. J. Neurol. Sci. 2008;264:145–150. doi: 10.1016/j.jns.2007.08.026. [DOI] [PubMed] [Google Scholar]
- Frank C, Burkhardt C, Imhof D, Ringel J, Zschornig O, Wieligmann K, Zacharias M, Bohmer FD. Effective dephosphorylation of Src substrates by SHP-1. J. Biol. Chem. 2004;279:11375–11383. doi: 10.1074/jbc.M309096200. [DOI] [PubMed] [Google Scholar]
- Frisullo G, Angelucci F, Caggiula M, Nociti V, Iorio R, Patanella AK, Sancricca C, Mirabella M, Tonali PA, Batocchi AP. pSTAT1, pSTAT3, and T-bet expression in peripheral blood mononuclear cells from relapsing-remitting multiple sclerosis patients correlates with disease activity. J. Neurosci. Res. 2006;84:1027–1036. doi: 10.1002/jnr.20995. [DOI] [PubMed] [Google Scholar]
- Gerhauser I, Ulrich R, Alldinger S, Baumgartner W. Induction of activator protein-1 and nuclear factor-kappaB as a prerequisite for disease development in susceptible SJL/J mice after theiler murine encephalomyelitis. J. Neuropathol. Exp. Neurol. 2007;66:809–818. doi: 10.1097/nen.0b013e3181461f31. [DOI] [PubMed] [Google Scholar]
- Gobin SJ, Montagne L, Van Zutphen M, Van Der Valk P, Van Den Elsen PJ, De Groot CJ. Upregulation of transcription factors controlling MHC expression in multiple sclerosis lesions. Glia. 2001;36:68–77. doi: 10.1002/glia.1096. [DOI] [PubMed] [Google Scholar]
- Hanson EM, Dickensheets H, Qu CK, Donnelly RP, Keegan AD. Regulation of the dephosphorylation of Stat6. Participation of Tyr-713 in the interleukin-4 receptor alpha, the tyrosine phosphatase SHP-1, and the proteasome. J. Biol. Chem. 2003;278:3903–3911. doi: 10.1074/jbc.M211747200. [DOI] [PubMed] [Google Scholar]
- Haque SJ, Harbor P, Tabrizi M, Yi T, Williams BR. Protein-tyrosine phosphatase Shp-1 is a negative regulator of IL-4- and IL-13-dependent signal transduction. J. Biol. Chem. 1998;273:33893–33896. doi: 10.1074/jbc.273.51.33893. [DOI] [PubMed] [Google Scholar]
- Hudson CA, Christophi GP, Cao L, Gruber RC, Massa PT. Regulation of avoidant behaviours and pain by the anti-inflammatory tyrosine phosphatase SHP-1. Neuron Glia Biol. 2006;2:235–246. doi: 10.1017/S1740925X07000476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javed A, Reder AT. Therapeutic role of beta-interferons in multiple sclerosis. Pharmacol. Ther. 2006;110:35–56. doi: 10.1016/j.pharmthera.2005.08.011. [DOI] [PubMed] [Google Scholar]
- Jin YJ, Yu CL, Burakoff SJ. Human 70-kDa SHP-1L differs from 68-kDa SHP-1 in its C-terminal structure and catalytic activity. J. Biol. Chem. 1999;274:28301–28307. doi: 10.1074/jbc.274.40.28301. [DOI] [PubMed] [Google Scholar]
- Kinkel RP, Kollman C, O’Connor P, et al. IM interferon beta-1a delays definite multiple sclerosis 5 years after a first demyelinating event. Neurology. 2006;66:678–684. doi: 10.1212/01.wnl.0000200778.65597.ae. [DOI] [PubMed] [Google Scholar]
- Marsh HN, Dubreuil CI, Quevedo C, et al. SHP-1 negatively regulates neuronal survival by functioning as a TrkA phosphatase. J. Cell Biol. 2003;163:999–1010. doi: 10.1083/jcb.200309036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A, Tsui HW, Shulman MJ, Isenman D, Tsui FW. Murine SHP-1 splice variants with altered Src homology 2 (SH2) domains. Implications for the SH2-mediated intramolecular regulation of SHP-1. J. Biol. Chem. 1999;274:21725–21734. doi: 10.1074/jbc.274.31.21725. [DOI] [PubMed] [Google Scholar]
- Massa PT, Wu C. The role of protein tyrosine phosphatase SHP-1 in the regulation of IFN-gamma signaling in neural cells. J. Immunol. 1996a;157:5139–5144. [PubMed] [Google Scholar]
- Massa PT, Wu C. Modulation of major histocompatibility complex class I genes by interferon-gamma and ganglioside GT 1b in astrocytes: involvement of protein tyrosine phosphatases. J. Neurochem. 1996b;67:1831–1839. doi: 10.1046/j.1471-4159.1996.67051831.x. [DOI] [PubMed] [Google Scholar]
- Massa PT, Wu C. Increased inducible activation of NF-kappaB and responsive genes in astrocytes deficient in the protein tyrosine phosphatase SHP-1. J. Interferon Cytokine Res. 1998;18:499–507. doi: 10.1089/jir.1998.18.499. [DOI] [PubMed] [Google Scholar]
- Massa PT, Dorries R, ter Meulen V. Viral particles induce Ia antigen expression on astrocytes. Nature. 1986;320:543–546. doi: 10.1038/320543a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massa PT, Saha S, Wu C, Jarosinski KW. Expression and function of the protein tyrosine phosphatase SHP-1 in oligodendrocytes. Glia. 2000;29:376–385. [PubMed] [Google Scholar]
- Massa PT, Ropka SL, Saha S, Fecenko KL, Beuler KL. Critical role for protein tyrosine phosphatase SHP-1 in controlling infection of central nervous system glia and demyelination by Theiler’s murine encephalomyelitis virus. J. Virol. 2002;76:8335–8346. doi: 10.1128/JVI.76.16.8335-8346.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massa PT, Wu C, Fecenko-Tacka K. Dysmyelination and reduced myelin basic protein gene expression by oligodendrocytes of SHP-1-deficient mice. J. Neurosci. Res. 2004;77:15–25. doi: 10.1002/jnr.20155. [DOI] [PubMed] [Google Scholar]
- McCarthy M, de Vellis J. Preparation of separate astroglial and oligodendroglial cell cultures from rat cerebral tissue. The Journal of Cell Biology. 1980;85:890–902. doi: 10.1083/jcb.85.3.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka T, Yoshino T, Hayashi K, et al. Reduction of hematopoietic cell-specific tyrosine phosphatase SHP-1 gene expression in natural killer cell lymphoma and various types of lymphomas/leukemias : combination analysis with cDNA expression array and tissue microarray. Am. J. Pathol. 2001;159:1495–1505. doi: 10.1016/S0002-9440(10)62535-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paling NR, Welham MJ. Role of the protein tyrosine phosphatase SHP-1 (Src homology phosphatase-1) in the regulation of interleukin-3-induced survival, proliferation and signalling. Biochem. J. 2002;368:885–894. doi: 10.1042/BJ20021054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platanias LC. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat. Rev. Immunol. 2005;5:375–386. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- Rudick RA, Lee JC, Simon J, Ransohoff RM, Fisher E. Defining interferon beta response status in multiple sclerosis patients. Ann. Neurol. 2004;56:548–555. doi: 10.1002/ana.20224. [DOI] [PubMed] [Google Scholar]
- Scarisbrick IA, Blaber SI, Tingling JT, Rodriguez M, Blaber M, Christophi GP. Potential scope of action of tissue kallikreins in CNS immune-mediated disease. J. Neuroimmunol. 2006;178:167–176. doi: 10.1016/j.jneuroim.2006.05.022. [DOI] [PubMed] [Google Scholar]
- Takeda K, Noguchi K, Shi W, Tanaka T, Matsumoto M, Yoshida N, Kishimoto T, Akira S. Targeted disruption of the mouse Stat3 gene leads to early embryonic lethality. Proc. Natl Acad. Sci. USA. 1997;94:3801–3804. doi: 10.1073/pnas.94.8.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teige I, Treschow A, Teige A, Mattsson R, Navikas V, Leanderson T, Holmdahl R, Issazadeh-Navikas S. IFN-beta gene deletion leads to augmented and chronic demyelinating experimental autoimmune encephalomyelitis. J. Immunol. 2003;170:4776–4784. doi: 10.4049/jimmunol.170.9.4776. [DOI] [PubMed] [Google Scholar]
- Tsui HW, Hasselblatt K, Martin A, Mok SC, Tsui FW. Molecular mechanisms underlying SHP-1 gene expression. Eur. J. Biochem. 2002;269:3057–3064. doi: 10.1046/j.1432-1033.2002.02986.x. [DOI] [PubMed] [Google Scholar]
- Wlodarski P, Zhang Q, Liu X, Kasprzycka M, Marzec M, Wasik MA. PU.1 activates transcription of SHP-1 gene in hematopoietic cells. J. Biol. Chem. 2007;282:6316–6323. doi: 10.1074/jbc.M607526200. [DOI] [PubMed] [Google Scholar]
- Wu C, Sun M, Liu L, Zhou GW. The function of the protein tyrosine phosphatase SHP-1 in cancer. Gene. 2003;306:1–12. doi: 10.1016/s0378-1119(03)00400-1. [DOI] [PubMed] [Google Scholar]
- Xu Y, Banville D, Zhao HF, Zhao X, Shen SH. Transcriptional activity of the SHP-1 gene in MCF7 cells is differentially regulated by binding of NF-Y factor to two distinct CCAAT-elements. Gene. 2001;269:141–153. doi: 10.1016/s0378-1119(01)00445-0. [DOI] [PubMed] [Google Scholar]
- Yarilina A, DiCarlo E, Ivashkiv LB. Suppression of the effector phase of inflammatory arthritis by double-stranded RNA is mediated by type I IFNs. J. Immunol. 2007;178:2204–2211. doi: 10.4049/jimmunol.178.4.2204. [DOI] [PubMed] [Google Scholar]
- Zeis T, Graumann U, Reynolds R, Schaeren-Wiemers N. Normal-appearing white matter in multiple sclerosis is in a subtle balance between inflammation and neuroprotection. Brain. 2007;131:288–303. doi: 10.1093/brain/awm291. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Raghunath PN, Vonderheid E, Odum N, Wasik MA. Lack of phosphotyrosine phosphatase SHP-1 expression in malignant T-cell lymphoma cells results from methylation of the SHP-1 promoter. Am. J. Pathol. 2000;157:1137–1146. doi: 10.1016/S0002-9440(10)64629-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Wang HY, Marzec M, Raghunath PN, Nagasawa T, Wasik MA. STAT3- and DNA methyltransferase 1-mediated epigenetic silencing of SHP-1 tyrosine phosphatase tumor suppressor gene in malignant T lymphocytes. Proc. Natl Acad. Sci. USA. 2005;102:6948–6953. doi: 10.1073/pnas.0501959102. [DOI] [PMC free article] [PubMed] [Google Scholar]