Abstract
The DOK1 gene is a putative tumour suppressor gene located on the human chromosome 2p13 which is frequently rearranged in leukemia and other human tumours. We previously reported that the DOK1 gene can be mutated and its expression down-regulated in human malignancies. However, the mechanism underlying DOK1 silencing remains largely unknown. We show here that unscheduled silencing of DOK1 expression through aberrant hypermethylation is a frequent event in a variety of human malignancies. DOK1 was found to be silenced in nine head and neck cancer (HNC) cell lines studied and DOK1 CpG hypermethylation correlated with loss of gene expression in these cells. DOK1 expression could be restored via demethylating treatment using 5-aza-2′deoxycytidine. In addition, transduction of cancer cell lines with DOK1 impaired their proliferation, consistent with the critical role of epigenetic silencing of DOK1 in the development and maintenance of malignant cells. We further observed that DOK1 hypermethylation occurs frequently in a variety of primary human neoplasm including solid tumours (93% in HNC, 81% in lung cancer) and hematopoietic malignancy (64% in Burkitt’s lymphoma). Control blood samples and exfoliated mouth epithelial cells from healthy individuals showed a low level of DOK1 methylation, suggesting that DOK1 hypermethylation is a tumour specific event. Finally, an inverse correlation was observed between the level of DOK1 gene methylation and its expression in tumour and adjacent non tumour tissues. Thus, hypermethylation of DOK1 is a potentially critical event in human carcinogenesis, and may be a potential cancer biomarker and an attractive target for epigenetic-based therapy.
Keywords: DOK1, DNA hypermethylation, gene silencing, tumour suppressor, cancer
Introduction
Cancer is thought to arise through accumulation of genetic alterations of at least two sets of cellular regulatory genes, i.e. proto-oncogenes and tumour suppressor genes. In general, gene mutations are the hallmarks of these alterations, which may result in activation of proto-oncogenes or inactivation of tumour suppressor genes. Increasing evidence argues that inactivation of tumour suppressor genes by epigenetic silencing also plays a major role in human carcinogenesis.1, 2 DNA methylation is one of the most common epigenetic modifications in mammalian genome involved in the regulation of gene expression.3, 4 Importantly, aberrant DNA methylation is closely connected to a wide variety of human cancers.1, 2 The discovery of cancer-associated genes silenced through hypermethylation during tumour development may represent an attractive target for therapeutic and preventive strategies.1-4
Recent work from us and other groups led to the characterization of a putative tumour suppressor gene, DOK1 (downstream of tyrosine kinases 1). DOK1 was first identified as an abundant tyrosine-hyperphosphorylated protein in chronic myelogenous leukemia cells (CML).5, 6 DOK1 is also constitutively tyrosine phosphorylated in a number of other transformed cells,5-7 suggesting that this event may regulate its tumour suppression functions. DOK1 inhibits MAP kinase activity, and displays anti-proliferative activities.8-14 Genetic disruption of DOK1 and its related member DOK2 in mice increases their susceptibility to leukemia development.8, 9 Furthermore, DOK1 locus in human chromosome 2p13, is frequently rearranged in various human tumours.10 We have reported that, consistent with its potential role as a tumour suppressor, DOK1 expression is altered in a series of Burkitt lymphoma cell lines and in chronic lymphocytic leukemia (CLL).11, 12 Moreover, we reported a frameshift mutation of the DOK1 gene in CLL,12 although DOK1 does not seem to play a major role in familial CLL cases.13 The suppressive effects of DOK1 appear to correlate with its subcellular localisation. Indeed, cytoplasmic wild-type DOK1-mediated cell proliferation inhibition is impaired in the nuclear DOK1 mutant found in CLL12 and in DOK1 mutated in its nuclear exclusion site.14 These studies indicate that DOK1 has the properties of a tumour suppressor gene in human leukemia, although its role in other human malignancies remains unknown.
In this study, we examined DOK1 gene expression in various human tumour cell lines and primary tumour specimens. We found that DOK1 gene is frequently silenced in a variety of human malignancies through epigenetic mechanism, highlighting the importance of the deregulation of this putative tumour suppressor gene in cancer.
MATERIALS AND METHODS
Cell lines and primary tumour tissue samples
Head and neck cancer (HNC) cell lines were kindly provided Dr C. Herold-Mende (University of Heidelberg, Heidelberg, Germany).15 HNC-97, HNC-124, HNC-199, HNC-212 were derived from the oral cavity; HNC-41, HNC-206, HNC-211, from the tonsils; PNS-136, from para-nasal sinus and do not belong to the group of head and neck cancer; HNC-150, from larynx; and HNC-180, from hypopharynx. The colon cancer cell lines TC7 and Lovo were obtained from A. Puisieux (Centre Leon Berard, Lyon, France). The Burkitt’s lymphoma cell lines (BL) (N=44) were established at the International Agency for Research on Cancer (IARC) from primary tumours.11 Primary tumour samples of head and neck (N=120) were embedded surgical materials in paraffin obtained from archive materials. Of these, 98 were collected as part of a multicenter case–control study coordinated by IARC and obtained from three centers in Brazil (43 from Rio de Janeiro, 34 from Sao Paulo and 11 from Porto Alegre). The 10 remaining samples were obtained from Argentina. Topography of tumour and patient information are presented in Table 2. 22 head and neck cancer samples consisted exclusively of larynx tumours collected from Otorhinolaryngology department of Hospital San Juan de Dios of Santiago. 34 additional head and neck tumours samples and their corresponding distant non tumour tissue isolated from the same patients from the European Institute of Oncology (Milan, Italy) were also included. Lung cancer samples (N=84), were obtained as part of a case–control study on lung cancer conducted at the Cancer Research Centre, Moscow (Russia), in a larger multicenter case–control study coordinated by IARC.16 The control samples consisting of lymphocytes (N=96) from 50 lung cancer patients, 46 healthy individuals from the same multicenter case–control study and 45 samples of mouth exfoliated epithelial cells from healthy individuals.
Table 2.
Median of DNA methylation levels of DOK1 in head and neck tumours, stratified by sex, age, topography, tobacco consumption, and alcohol intake
| Patients | Median | |
|---|---|---|
| Sex | ||
| Men | 65 | 49.5 |
| Women | 33 | 41.5 |
| Age | ||
| ≤40 | 6 | 53.9 |
| 41-50 | 24 | 43.1 |
| 51-60 | 35 | 53.5 |
| 61-70 | 20 | 54.4 |
| >70 | 13 | 34.3 |
| Alcohol consumption (g/day) | ||
| 0-138 | 22 | 34.9 |
| 139-889 | 21 | 52.9 |
| 890-3119 | 26 | 51.4 |
| 3120+ | 28 | 53.7 |
| Unknown | 1 | 62.1 |
| Tobacco pack/years | ||
| Never | 6 | 46.3 |
| 0-20 | 24 | 61.0 |
| 20-40 | 29 | 50.2 |
| 40-60 | 23 | 43.1 |
| 60+ | 16 | 45.8 |
| Topography | ||
| Oral cavity - tongue - floor of the mouth |
23 | 46.9 |
| Oropharynx | 37 | 49.3 |
| Hypopharynx | 17 | 53.7 |
| Larynx | 21 | 34.3 |
Colony formation assay
Cells were transduced with empty pBabe-puro retrovirus vector and pBabe-puro-Dok1. 24 hours after infection, cells were split for selection and plated at different dilution 1:100, 1:1000, 1:10000. Cells were grown for 10-15 days, until visible colonies appeared. Cells were then stained with crystal violet in 20% of methanol. The number of colonies from 5 random fields was determined.
Antibodies, reagents and immunoblotting
The antibodies used were the following: rabbit anti-Dok1 antibody (R. Kobayashi, University of Texas M.D. Anderson Cancer Center, Houston, USA); mouse anti-Flag (M5) monoclonal antibody, mouse anti-β-actin (Sigma). Immunoblotting was performed as previously described.14
Reverse transcription, RT-PCR and qRT-PCR
Total RNA was extracted and reverse transcribed using Absolutely RNA kit (Stratagene) and ReverAid Minus First Strand cDNA Synthesis Kit (Fermentas) followed by semi-quantitative PCR.11 qRT-PCR was conducted using MesaGreen qPCR MasterMix plus for SYBR assay kit (Eurogentec, USA). GAPDH mRNA was used to normalize RNA inputs. The sequence of the primers is presented in Supplementary Table 1. Relative expression level of DOK1 gene was calculated using the comparative CT method (2−ddCt).17
DNA extraction and pyrosequencing assay
DNA was extracted from tissue sections embedded in paraffin blocks and quantified as previously described.16 DNA were treated with bisulfite and subjected to pyrosequencing as described previously.18 The primer sequences are shown in Supplementary Table 1. The percentage of methylation evaluated as the mean of all CpG analysed at a given gene promoter and the methylation levels at the promoter of CDKN2A and RASSF1A, were analysed as previously described.16
Generation of DOK1 gene promoter in pGL3
A DNA fragment of 2.8 kbp of the promoter region spanning the promoter region 5′ upstream of the ATG of DOK1 (−2 000 to + 800) and including all the amplicons (Fig. 2a) (Gene bank number NC 000002), was amplified from genomic DNA and cloned in pGL3 luciferase reporter plasmid. The primers used were 5′-CGGGTACCAGACAACAGGAGAGAA AGAGCCC-3′ (Forward) and 5′-GGAAGATCT AGCATCGAGAAACCCGTAATTTC-3′ (Reverse) with the sequence for KpnI and BglII sites for cloning in the corresponding sites of pGL3 to obtain pGL3-DOK1-pro). The nucleotide sequence was verified by sequencing.
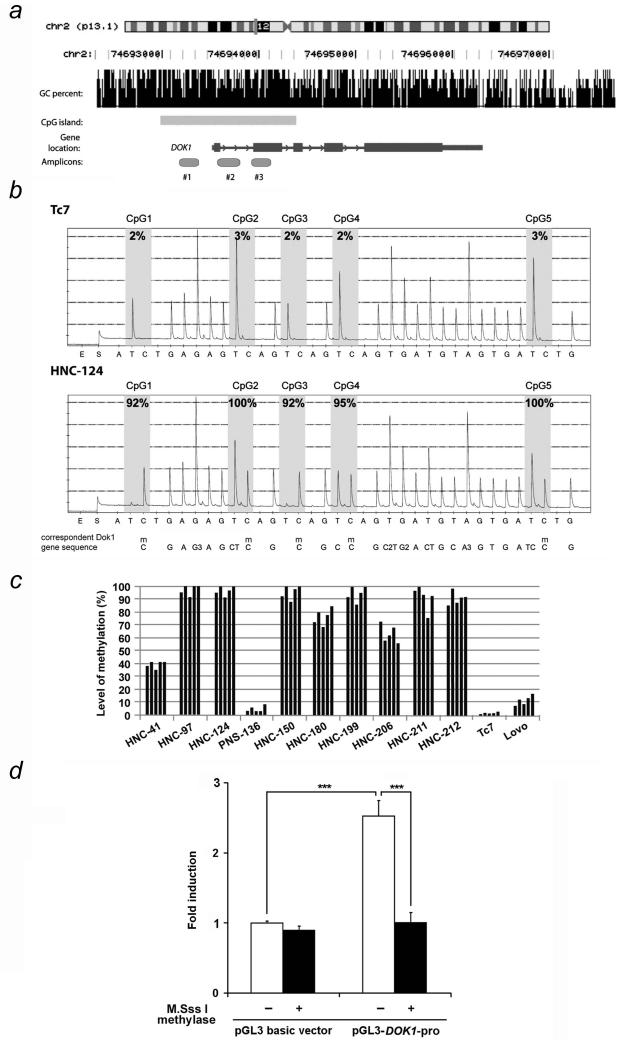
Figure 2.
DNA methylation analysis of the DOK1 in HNC cell lines. (a) Schematic representation of three different parts of DOK1 (amplicons) analysed for their DNA methylation status. GC percent and CpG island are represented based on the software prediction (http://genome.ucsc.edu). (b) Quantitative DNA methylation of CpGs sites in DOK1 for amplicon #3. Pyrosequencing data were obtained from Tc7 control expressing DOK1 and the non-expressing HNC-124. Analysed cytosines are indicated by the gray area. The ratio between T/C allows quantification of the number of alleles which are methylated compared to the unmethylated ones.29-31 Note that when the sequence has single or repeats of Ts and/or Cs preceding the methylated C (mC) (such as CpG2, CpG4 and CpG5), the software uses the reference peaks (peaks of the rest of the DNA sequence, excluding CpG sites.) to substract from the height of the peak.29-31 “m” means methylated. “G3, C2, G2, A3” mean consecutive same nucleotides. (c) DNA methylation levels from HNC cell lines and two control cell lines Lovo and Tc7 with the amplicon #3. Each bar represents the results obtained for individual CpG site. (d) Equal amount of unmethylated and methylated empty pGL3 and DOK1 promoter construct (pGL3-DOK1-pro) was transfected into HEK 293 cells. 48 h post-transfection, cell extracts were prepared and the firefly as well as the Renilla luciferase activity was measured. The relative fold induction of luciferase is shown with ± standard deviation. ***, p<0.001.
In vitro methylation and luciferase assay
pGL3 basic vector and pGL3-DOK1-pro were not treated (mock methylated) or treated with CpG methylase M.SssI (methylated) (New England Biolabs, USA) according to the manufacturer’s instructions. The unmethylated and methylated plasmids were transfected into HEK 293 cells in the presence of Renilla as the internal standard for reporter assay, according to the manufacturer’s instructions (Promega, USA). Quantification of luminescent signal was done by luminometer (Mgm Instruments Optocomp I, USA).
5′-Aza-2′-deoxycytidine treatment
Cells were incubated in culture medium containing 30 μM of 5′-Aza-2′-deoxycytidine (5′-AzadC) (Sigma, St. Louis, MO) or DMSO for four days with medium change every day. DOK1 expression was monitored by RT-PCR.
Statistical analysis
All methylation data were generated without knowledge of the case–control status of the subjects nor the histological features of the samples analysed. Wilcoxon rank-sum test was used to compare methylation levels in tumour samples and normal samples. p value <0.05 considered statistically significant. To assess DNA hypermethylation frequency, we calculated the percentage of tumour samples with methylation above 95% of the levels in blood samples.
Results
DOK1 expression in head and neck cancer cell lines
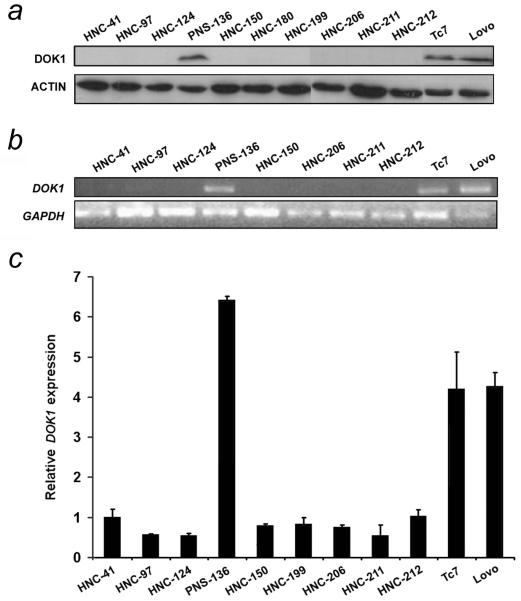
We have previously reported that the DOK1 gene was mutated and its expression downregulated in CLL and in BL cell lines.11, 12 To further evaluate the involvement of DOK1 inactivation in human carcinogenesis, we analysed DOK1 protein expression in 9 HNC cell lines derived from various sites including tonsil, oral cavity, larynx, and hypopharynx. One cell line from para-nasal sinus cancer was also included (see Materials and Methods). Immunoblotting analysis showed that DOK1 protein was not detected in 9 of the 10 (90%) cancer cell lines analysed (Fig. 1a). The only DOK1-expressing cell line was PNS-136 that was derived from a para-nasal sinus cancer. DOK1 protein was detected in two colon cancer lines (Tc7 and Lovo) used as controls (Fig. 1a). In order to determine whether loss of DOK1 protein correlates with an absence of mRNA transcription, a single 362 bp region covering exons 1 and 2 of the DOK1 gene was amplified using the RT-PCR approach. As shown in Fig. 1b, we found high DOK1 mRNA levels only in PNS-136 and control cell lines, and no detectable DOK1 mRNA in any of nine other HNC cell lines investigated. Real time quantitative RT-PCR approach confirmed the inhibition of DOK1 gene expression in HNC cell lines (Fig. 1c). Thus, a tight correlation exists between DOK1 protein and transcription levels in the different HNC cell lines analysed. Genomic amplification using primers flanking the first and last exons (1 to 5) showed that the gene encoding DOK1 is present in all tested cell lines with no detectable deletion or rearrangements (data not shown). Thus, loss of DOK1 expression is likely due to a defect at the transcriptional level.
Figure 1.
DOK1 expression is downregulated in HNC cell lines. (a) Protein extracts from indicated HNC cell lines and two colon cancer cell lines (Lovo and Tc7) used as control were analysed by immunoblotting. Actin was used as a loading control. Semi-quantitative PCR (b) and qRT-PCR (c) of DOK1 mRNA expression using primers DE-F1/DE-R1 are presented.
Hypermethylation of DOK1 gene correlates with the inhibition of its expression in HNC cell lines
Aberrant methylation of CpG islands in the promoter regions is one of the major mechanisms for the silencing of tumour suppressor genes. Analysis of the DOK1 gene locus revealed the presence of a typical sequence (1408 bp) matching the CpG island that covers the promoter region, exon 1 and 2 and intron 1 and 2, of the DOK1 gene (Fig. 2a). To evaluate the methylation status of the region in HNC cell lines, three sets of PCR primers covering different regions of the CpG island were designed for pyrosequencing assays (Supplementary Table 1). The region chosen for analysis spans the areas of greatest CpG density 5′ and 3′ to the transcription start site.
Our initial screening revealed that among three amplicons analysed in the DOK1 CpG island, the amplicon #3, harbouring five CpG sites, showed the highest methylation status in most HNC cell lines analysed, whereas DOK1-expressing control cell lines exhibited virtually no methylation at any of CpG sites covered by the 3 regions (Fig. 2b, and Supplementary Fig. 1). Therefore, the amplicon #3 which shows the highest level of methylation in HNC cells (Supplementary Fig. 1), was used for DNA methylation analysis for the rest of the study. We found a robust methylation of CpG sites present in all the HNC lines lacking DOK1 expression (Fig. 2c and Fig. 1). In contrast, the DOK1 CpG sites were virtually unmethylated in the control cells and in the PNS-136 cells that expressed a high level of DOK1 mRNA (Fig. 2c). Levels of methylation observed were similar across all five CpG sites in the amplicon #3 in all HNC cell lines analysed (Fig. 2c). To further confirm that the hypermethylation of the promoter region of DOK1 is responsible for the inhibition of its expression, we cloned the CpG islands-containing 2.8 kbp sequence including the 2 kbp DOK1 promoter region and the 0.8 kbp gene sequence covering the three amplicons analysed in pyrosequencing, in the pGL3 luciferase reporter plasmid to obtain pGL3-DOK1-pro. pGL3-DOK1-pro construct was in vitro methylated using SS1 CpG methylase, or left unmethylated, and transfected in HEK 293 cells and the luciferase activity was determined. As shown in Fig. 2d, unmethylated DOK1 promoter construct induced a significantly higher level of luciferase activity compared to the methylated plasmid. Thus, hypermethylation of DOK1 gene in HNC cell lines correlates with the inhibition of its promoter activity and low expression of DOK1 both at protein and mRNA levels (Fig. 1).
5′-Aza-2′-deoxycytidine treatment restores DOK1 gene expression in HNC cell lines
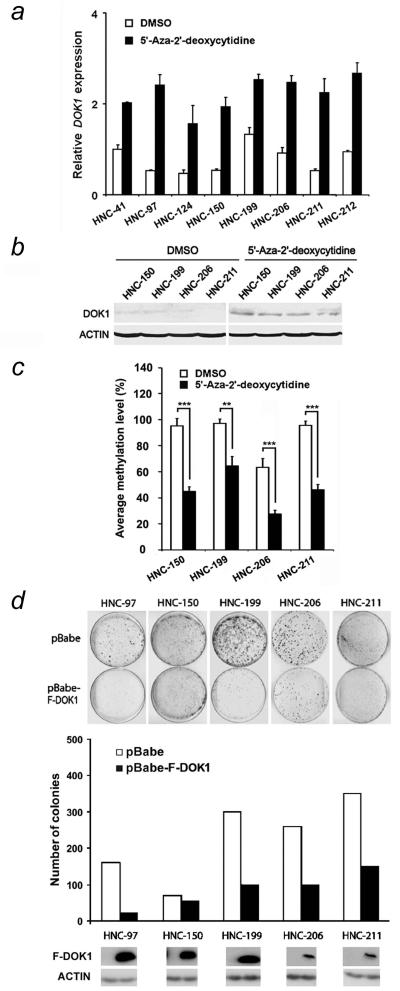
To further investigate the role of DNA methylation in DOK1 gene silencing, we tested whether treatment with a DNA methylating inhibitor agent could restore expression of DOK1 in cancer cells. HNC cell lines and control cells were treated with 5′-AzadC, an inhibitor of DNA methylation, and the expression of DOK1 mRNA was monitored by qRT-PCR. 5′-AzadC treatment resulted in a reactivation and increased expression of DOK1 mRNA in all HNC cell lines tested, with a relative low expression in mock treated cells (Fig. 3a). Moreover, the induction of mRNA by 5′-AzadC correlated with the DOK1 protein expression (Fig. 3b) and a significant decrease in the level of DOK1 methylation (Fig. 3c). These results indicate that aberrant hypermethylation was responsible for silencing of DOK1 gene expression in head and neck cancer cells.
Figure 3.
Treatment of HNC cell lines with 5′-AzadC restores DOK1 expression and re-expression of DOK1 inhibits colony formation efficiency of cells. (a) HNC cells were exposed to the solvent DMSO or to a demethylating agent 5′-AzadC for four days, and DOK1 expression was monitored by q-RT-PCR. (b) Four HNC cell lines were treated as in (a) and DOK1 protein expression was monitored by immunoblotting. (c) Cells were treated as in (b) and the level of DOK1 methylation is presented. (d) Cells were transduced either with empty retrovirus (pBabe) or with retrovirus expressing Flag-DOK1 (pBabe-F-DOK1). Two days after infection, cells were put under selection with pyromycin, and colony formation was monitored (upper panel) with the quantification of the data (lower panel) and the corresponding expression level of DOK1 (below). Data are representative of two independent experiments carried out in triplicate for the quantification.
Effects of re-expression of DOK1 in HNC cell lines
As 5′-AzadC treatment restores DOK1 expression in HNC cells, we sought to evaluate the importance of unscheduled silencing of DOK1 in cancer cells and the impact of its re-expression on cellular functions. We therefore investigated the effects of exogenous expression of DOK1 in cancer cell lines harbouring the DOK1 silenced by promoter hypermethylation. To this end, HNC cell lines were transduced with a retrovirus expressing DOK1 or control empty vector and their proliferation capacity was monitored. As shown in Fig. 3d, all five DOK1-transduced HNC cell lines expressed DOK1 protein (the third bottom panel). Excepted for one cell line (HNC-150), the DOK1 protein-expressing cells exhibited a significant reduction in colony formation capacity, with a different magnitude of growth reduction, in comparison to those transduced with empty vector-expressing retrovirus. Ectopic DOK1 expression resulted in the inhibition of the colony formation in DOK1-expressing cell line PNS-136 (a non-HNC cell line) (data not shown). This event may be explained by the elevated DOK1 levels due to its exogenous and endogenous expression. Alternatively, PNS-136 may harbour other mutations which may compensate for the loss of DOK1 expression. Overall, the restoration of DOK1 expression in cancer cells inhibits their proliferation, and suggests that epigenetic silencing of DOK1 may promote proliferation of transformed cells.
Hypermethylation of DOK1 in primary head and neck tumours
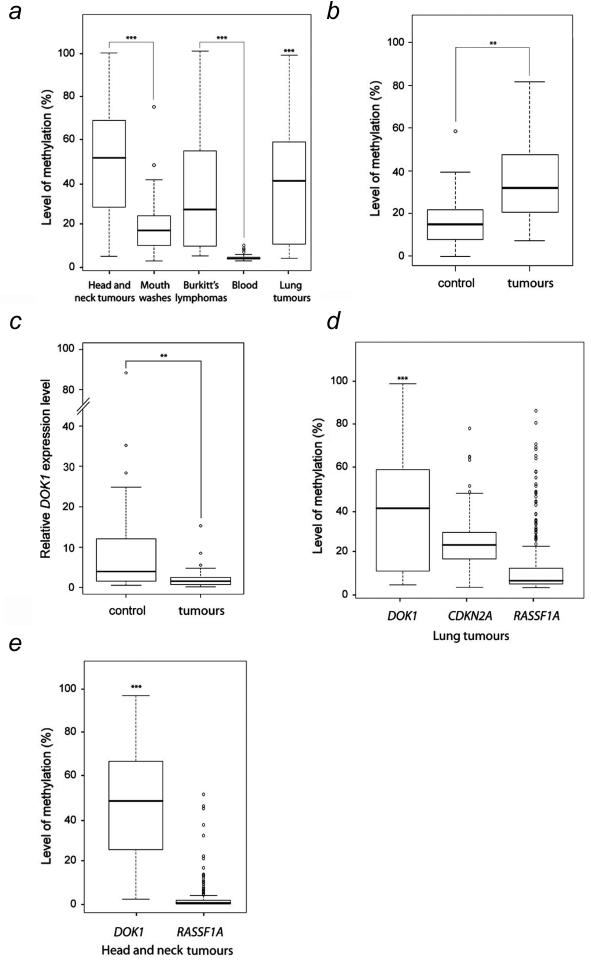
To extend our findings on DOK1 hypermethylation found in HNC cell lines to primary cancer, we evaluated the status of DOK1 methylation in primary head and neck cancers. We found that DOK1 is heavily methylated in primary HNC (Fig. 4a). We found that 93% (112 of 120) of primary cancer samples showed a significant increase of DNA methylation (> 40%) at all the 5 CpG sites in the DOK1 CpG island analysed from various sites of the head and neck (Supplementary Fig. 2a). DNA methylation levels in control blood samples (lymphocytes) or exfoliated epithelial mouth cells from healthy individuals showed that DOK1 DNA methylation was virtually absent and significantly reduced in all samples analysed (Fig. 4a and Supplementary Fig. 2b, c and d). To further correlate the hypermethylation of DOK1 gene with the tumour status, we compared the level of DOK1 promoter methylation in 34 tumours samples and the corresponding control non-tumour adjacent tissue from the same patients. As shown in Fig. 4b, a relative high level of hypermethylation occurred in the tumour samples in comparison to the non-tumour samples. Interestingly, the hypermethylation status inversely correlates with the expression level of DOK1 mRNA (Fig. 4c). Thus, DOK1 gene is hypomethylated and expressed in non-tumour samples, while it is highly methylated and its expression down-regulated in tumour. In the samples analysed, we found no significant difference in the levels of DOK1 methylation in tumour samples from different anatomical sites, although a slight increased was noticed in samples from the oral cavity (Table 2). Overall the DOK1 methylation levels were significantly higher in tumour cells than those in blood and exfoliated epithelia non-tumour control samples (Fig. 4a and b, and Supplementary Fig. 2). Together, these results suggest a tumour-specific hypermethylation of DOK1 and are consistent with the results obtained in the HNC cell lines (Figs. 2 and 3).
Figure 4.
Methylation of DOK1 in human tumour samples. (a). Summary of data obtained from the analysis of mean levels of methylation of the five CpG sites in DOK1 is presented for HNC (n=120); mouth washes (n=45); BL (n=44), lymphocytes (non-tumorigenic tissue) (n=96) and lung tumours (n=84). The raw methylation values from all five CpG sites of each sample were first averaged to achieve the mean values. Then, all the mean values were used to draw the box plot chart. (b). Methylation of DOK1 in selected head and neck tumours and corresponding control distant non-tumour samples from the same patients (n=34 pair samples; p-value ** (p<0.01). (c). DOK1 expression monitored by qRT-PCR in samples analysed in (b) (n=34 pair samples, p-value ** (p<0.01). (d). Comparison of the levels of DNA methylation of DOK1, CDKN2A and RASSF1A in lung. (e) Comparison of the levels of DNA methylation of DOK1 and RASSF1A in head and neck tumours. The raw methylation values from all five CpG sites of each sample were first averaged to achieve the mean values correspondent to samples detected. Then, all the mean values were used to draw the box plot chart. The level of statistical significance for different methylation levels is given by the p-value *** (p<0.001) was obtained using the Wilcoxon test.
Hypermethylation of DOK1 in other human cancers
To evaluate whether DOK1 hypermethylation is a common event in human carcinogenesis, the methylation status of DOK1 gene was monitored in samples from other cancer types, including lung cancer and Burkitt’s lymphoma. We found that DOK1 gene is heavily methylated in 81% (68 of 84) of primary lung cancers (Fig. 4a, Supplementary Fig. 2e and Table 1), suggesting that DOK1 hypermethylation occurs in solid tumours other than HNC. Interestingly, we also found a significant methylation of DOK1 gene in 64% (28 out of 44) of BL cell lines (Fig. 4a, Supplementary Fig. 2f and Table 1), whereas all 5 DOK1 CpG sites in control blood samples were virtually unmethylated (<5%; Supplementary Fig. 2d, e). Comparison of mean methylation levels of all CpG sites in head and neck tumours and blood samples revealed a highly significant increase in methylation levels in all tumours compared to blood samples and mouth epithelial cells from healthy individuals (p<0.01, Fig. 4). Comparison of hypermethylation levels and frequencies of DOK1 with CDKN2A and RASSF1A, two tumour suppressor genes known to be among the most frequent targets of hypermethylation in cancer, revealed that DOK1 hypermethylation occurs at similar (in lung) or higher (in HNC) frequency than of CDKN2A and RASSF1A (Fig. 4d and e, and Table 1). Together, these results demonstrate a high frequency of DOK1 hypermethylation in a variety of human cancers, and define DOK1 as one of the most frequent targets of aberrant hypermethylation, thus strongly supporting the key role of epigenetic silencing of this gene in human carcinogenesis.
Table 1.
Comparison of hypermethylation frequency of DOK1, RASSF1A and CDKN2A in different tumour types
| Tumours | Genes | Hypermethylated samples* |
Samples analysed |
Hypermethylated tumour samples |
|---|---|---|---|---|
| Head & neck tumours |
DOK1
RASSF1A |
112 36 |
120 185 |
93% 19% |
| Lung tumours |
DOK1
CDKN2A RASSF1A |
68 97 64 |
84 106 178 |
81% 92% 36% |
| Burkitt lymphomas |
DOK1 | 28 | 44 | 64% |
Samples with methylation levels above the quantity representing the upper 95% of methylation in blood samples (3.5 % of methylation)
Association between DOK1 methylation and clinicopathologic features of head and neck cancer and risk factors exposure
We next analysed associations between the methylation of DOK1 and available epidemiological and clinical information including alcohol intake, smoking status, sex, age and tumour site. Table 2 shows the association between methylation levels of DOK1 (measured as median of DOK1 methylation levels in individual groups) in 98 head and neck cancer patients and their clinical features and risk factor exposures. With the exception of alcohol consumption, no association was found between methylation levels of the DOK1 gene promoter in tumours and any risk factor included in the analysis (Table 2). In contrast, head and neck tumours from alcohol drinkers exhibited relatively higher methylation levels of DOK1 than those from non-drinkers. The relative increase of DOK1 gene methylation correlates with the quantity of the daily consumption of alcohol (Table 2) (compare the median of 34.9 of low drinkers to 53.7 of heavy drinkers). The analysis also indicated that laryngeal tumours showed a relatively lower methylation levels than those from other anatomic regions of head and neck (Table 2). These results indicate that hypermethylation of the promoter region of DOK1 and subsequent silencing of the gene in head and neck tumours may be associated with alcohol intake.
Discussion
We and others have previously reported that DOK1 displays tumour suppressive activities, including its inhibitory effect on cellular proliferation and tumour formation, and MAP kinase activity.11, 12, 14, 19-22 Consistent with a tumour suppressor role of DOK1, the mice harbouring inactivated DOK1 gene exhibits high susceptibility to cancer development.8, 9, 19 In addition, DOK1 expression was found to be downregulated or inactivated in human malignancies, notably leukemia and lymphomas, 11, 12 through an unknown mechanism. In the present study, we show for the first time that silencing of DOK1 expression also occurs frequently in a variety of human malignancies, including solid tumours and that aberrant hypermethylation may be an underlying mechanism. Indeed, pyrosequencing assays revealed that DOK1 promoter region is frequently hypermethylated in cell lines derived from head and neck cancers, as well as primary tumour samples. DOK1 hypermethylation correlated with loss of gene expression in cancer cell lines and primary cancer cells, and could be readily restored by inhibiting DNA methylation with 5′-AzadC. The possibility however that 5′-AzadC may also induce DOK1 expression by demethylating other genes which regulate the expression of DOK1 cannot be excluded. We further found a high incidence of methylation of the DOK1 promoter in different primary human cancers including solid tumours (93% in head and neck cancers, 81% in lung cancer) and hematopoietic malignancy (64% in BL). These observations are consistent with our recent study showing that hypermethylation of DOK1 also occurred in hepatocellular carcinoma.23 Interestingly, DOK1 hypermethylation appears to occur at similar or higher frequency than in other cancer-associated genes known to be among the most frequent targets of hypermethylation in human malignancies, such as CDKN2A and RASSF1A.16 While further studies are required to precisely identify the role of DOK1 gene silencing during the multi-step process of carcinogenesis, our results argue that DOK1 may be among the most frequent targets of aberrant methylation and unscheduled silencing in human neoplasia. It is well established that DNA methylation of the CpG island in the promoter region is causally involved in gene silencing,2, 24 therefore a tight correlation between DOK1 CpG island hypermethylation in cancer cell lines and loss of gene expression in these cells provides an explanation for the loss or inactivation of DOK1 previously reported in different human neoplasia.11, 12
Frequent hypermethylation of DOK1 in a variety of human neoplasia suggests that this event contributes to the development of malignant cells and cancer phenotype; however, the cellular processes affected by unscheduled hypermethylation and consequent silencing of DOK1 remain largely unknown. The DOK1 gene product functions as a key negative regulator downstream of several receptor and non-receptor tyrosine kinase cascades and mediates activin-induced apoptosis20; therefore, epigenetic silencing of DOK1 may impair apoptotic competence and promote emergence of transformed cells. Since DOK1 inhibits MAP kinase ERKs by interacting with and activating Ras-GTPase activity,5, 7, 22 loss of DOK1 expression can promote cell proliferation and transformation through sustained activation of ERK MAP kinases as observed in DOK1 knockout cells.8, 9 In addition, DOK1 may inhibit other cellular pathways implicated in tumour formation and progression. While DOK1 has been shown to be silenced in human leukemia,12 our results demonstrating hypermethylation of DOK1 in a variety of human malignancies argue that DOK1 may play a role in the tissues other than hematopoietic lineages, and that its silencing through DNA hypermethylation may promote a wide range of human neoplasia.
Interestingly, the level of DOK1 methylation is similar, across all the HNC cell lines analysed (Fig. 2c), except for one cell line from tonsil (HNC-41) that showed relatively low levels of methylation. In general, tumours of the oral cavity (HNC-97, HNC-124, HNC-199 and HNC-212) exhibit high levels of DOK1 methylation. These observations are consistent with data obtained from primary tumours where tumours from the oral cavity appear to have a relative high level of DOK1 methylation (Table 2). In contrast with other cell lines studied, the cell line derived from tumour of the para-nasal sinus (PNS-136) showed low levels of DOK1 CpG methylation and an substantial level of DOK1 expression (Figs. 1 and 2C). Considering para-nasal sinus cancer to be a distinct entity from classical HNC and the overall variation of methylation pattern, these findings suggest that specific DOK1 methylation patterns may reflect a different molecular basis of tumours from different anatomical regions of the head and neck, or different exposures to risk factors including alcohol consumption, tobacco smoking or human papillomavirus infections. Interestingly, with a limited number of samples analysed, alcohol consumption, a major risk factor for head and neck cancer, appears to be associated with the methylation levels of DOK1 (Table 2). Hypermethylation of specific tumour suppressor genes associated with high alcohol intake has been reported in colon and gastric cancers.25, 26 The mechanisms underlying alcohol-induced DNA methylation are unclear. This epigenetic modification is attributed to ethanol metabolic stress (EMESS) that is generated by oxidative (acetylaldehyde, ROS) and non-oxidative (phosphatidylethanol, fatty acylethyl) ethanol metabolism.27 Reduction in folate level and deregulation of methionine metabolism triggered by exaggerated alcohol consumption are also assumed to affect DNA methylation.26, 27 No significant association was found between DOK1 gene methylation and a specific anatomic site of the tumour, although tumours from the larynx showed relatively low level of DOK1 methylation in comparison to other anatomical regions (Table 2). From a clinical standpoint, our findings that 5′-AzadC treatment is capable of restoring DOK1 expression in cancer cell lines and that overexpression of exogenous DOK1 impairs proliferation of cancer cells suggest that DNA hypermethylation of DOK1 may represent an attractive target for intervention strategies.
DOK1 inactivation is not restricted to head and neck neoplasia. Indeed, silencing of DOK1 gene also occurred in a proportion of lung and lymphoid cancers (Fig. 4), suggesting the broad range of inactivation of DOK1 in various human cancer types. During the preparation of our manuscript, Berger et al.28 identified DOK1 and DOK2 as tumour suppressors for lung cancer in animal models. In addition they reported that the expression of DOK2 was frequently down-regulated in human lung cancer in comparison to DOK1. This difference may be linked to the variability of the samples analysed, but also the types of tumours. While most of the lung cancer samples analyzed by Berger et al. are from lung adenocarcinoma, the majority of the samples analyzed in our study are from squamous cell carcinoma (n=66) with few adenocarcinoma (n=13) and other origins (n=5). Nevertheless, these independent studies reveal the important role of the DOK1 family members in human carcinogenesis.
In summary, we report a frequent loss of DOK1 gene expression caused by promoter hypermethylation in a variety of human malignancies and histological subtypes which reinforces the notion that DOK1 may function as an important tumour suppressor gene. A high frequency of DOK1 hypermethylation in a wide range of human cancers may prove a valuable biomarker for early cancer detection and could represent an attractive target for clinical intervention.
Supplementary Material
Acknowledgments
We thank R. Kobayashi for reagents, N. Lyandrat for technical assistance and John Daniel for editing. TV was supported by a PhD fellowship from La Ligue Nationale contre le Cancer (France), and RS by the IARC postdoctoral Fellowship Program, IF by the PhD fellowship from la “Bourse de la Coopération Française”. This work was partially supported by grants from La Ligue Régionale de la Lutte Contre le Cancer du Rhône et de la Drôme (to B.S.S), and the National Institutes of Health/National Cancer Institute (NIH/NCI, Grant No: R03 CA122396-02), USA, the Association pour la Recherche sur le Cancer (ARC, France), la Ligue Nationale Française Contre le Cancer and the Swiss Bridge Award (to Z.H.).
References
- 1.Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer. 2004;4:143–53. doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]
- 2.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–28. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 3.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 4.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33(Suppl):245–54. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 5.Carpino N, Wisniewski D, Strife A, Marshak D, Kobayashi R, Stillman B, Clarkson B. p62(dok): a constitutively tyrosine-phosphorylated, GAP-associated protein in chronic myelogenous leukemia progenitor cells. Cell. 1997;88:197–204. doi: 10.1016/s0092-8674(00)81840-1. [DOI] [PubMed] [Google Scholar]
- 6.Yamanashi Y, Baltimore D. Identification of the Abl- and rasGAP-associated 62 kDa protein as a docking protein, Dok. Cell. 1997;88:205–11. doi: 10.1016/s0092-8674(00)81841-3. [DOI] [PubMed] [Google Scholar]
- 7.Neet K, Hunter T. The nonreceptor protein-tyrosine kinase CSK complexes directly with the GTPase-activating protein-associated p62 protein in cells expressing v-Src or activated c-Src. Mol Cell Biol. 1995;15:4908–20. doi: 10.1128/mcb.15.9.4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Niki M, Di Cristofano A, Zhao M, Honda H, Hirai H, Van Aelst L, Cordon-Cardo C, Pandolfi PP. Role of Dok-1 and Dok-2 in leukemia suppression. J Exp Med. 2004;200:1689–95. doi: 10.1084/jem.20041306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yasuda T, Shirakata M, Iwama A, Ishii A, Ebihara Y, Osawa M, Honda K, Shinohara H, Sudo K, Tsuji K, Nakauchi H, Iwakura Y, et al. Role of Dok-1 and Dok-2 in myeloid homeostasis and suppression of leukemia. J Exp Med. 2004;200:1681–7. doi: 10.1084/jem.20041247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nelms K, Snow AJ, Noben-Trauth K. Dok1 encoding p62(dok) maps to mouse chromosome 6 and human chromosome 2 in a region of translocation in chronic lymphocytic leukemia. Genomics. 1998;53:243–5. doi: 10.1006/geno.1998.5514. [DOI] [PubMed] [Google Scholar]
- 11.Lee S, Huang H, Niu Y, Tommasino M, Lenoir G, Sylla BS. Dok1 expression and mutation in Burkitt’s lymphoma cell lines. Cancer Lett. 2007;245:44–50. doi: 10.1016/j.canlet.2005.10.045. [DOI] [PubMed] [Google Scholar]
- 12.Lee S, Roy F, Galmarini CM, Accardi R, Michelon J, Viller A, Cros E, Dumontet C, Sylla BS. Frameshift mutation in the Dok1 gene in chronic lymphocytic leukemia. Oncogene. 2004;23:2287–97. doi: 10.1038/sj.onc.1207385. [DOI] [PubMed] [Google Scholar]
- 13.Sellick GS, Coleman RJ, Talaban RV, Fleischmann C, Rudd MF, Allinson R, Catovsky D, Houlston RS. Germline mutations in Dok1 do not predispose to chronic lymphocytic leukemia. Leuk Res. 2005;29:59–61. doi: 10.1016/j.leukres.2004.05.022. [DOI] [PubMed] [Google Scholar]
- 14.Niu Y, Roy F, Saltel F, Andrieu-Soler C, Dong W, Chantegrel AL, Accardi R, Thepot A, Foiselle N, Tommasino M, Jurdic P, Sylla BS. A nuclear export signal and phosphorylation regulate Dok1 subcellular localization and functions. Mol Cell Biol. 2006;26:4288–301. doi: 10.1128/MCB.01817-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ninck S, Reisser C, Dyckhoff G, Helmke B, Bauer H, Herold-Mende C. Expression profiles of angiogenic growth factors in squamous cell carcinomas of the head and neck. Int J Cancer. 2003;106:34–44. doi: 10.1002/ijc.11188. [DOI] [PubMed] [Google Scholar]
- 16.Vaissiere T, Hung RJ, Zaridze D, Moukeria A, Cuenin C, Fasolo V, Ferro G, Paliwal A, Hainaut P, Brennan P, Tost J, Boffetta P, et al. Quantitative analysis of DNA methylation profiles in lung cancer identifies aberrant DNA methylation of specific genes and its association with gender and cancer risk factors. Cancer Res. 2009;69:243–52. doi: 10.1158/0008-5472.CAN-08-2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 18.Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, Molloy PL, Paul CL. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci U S A. 1992;89:1827–31. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Di Cristofano A, Niki M, Zhao M, Karnell FG, Clarkson B, Pear WS, Van Aelst L, Pandolfi PP. p62(dok), a negative regulator of Ras and mitogen-activated protein kinase (MAPK) activity, opposes leukemogenesis by p210(bcr-abl) J Exp Med. 2001;194:275–84. doi: 10.1084/jem.194.3.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamakawa N, Tsuchida K, Sugino H. The rasGAP-binding protein, Dok-1, mediates activin signaling via serine/threonine kinase receptors. EMBO J. 2002;21:1684–94. doi: 10.1093/emboj/21.7.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kashiwada M, Cattoretti G, McKeag L, Rouse T, Showalter BM, Al-Alem U, Niki M, Pandolfi PP, Field EH, Rothman PB. Downstream of tyrosine kinases-1 and Src homology 2-containing inositol 5′-phosphatase are required for regulation of CD4+CD25+ T cell development. J Immunol. 2006;176:3958–65. doi: 10.4049/jimmunol.176.7.3958. [DOI] [PubMed] [Google Scholar]
- 22.Zhao M, Janas JA, Niki M, Pandolfi PP, Van Aelst L. Dok-1 independently attenuates Ras/mitogen-activated protein kinase and Src/c-myc pathways to inhibit platelet-derived growth factor-induced mitogenesis. Mol Cell Biol. 2006;26:2479–89. doi: 10.1128/MCB.26.7.2479-2489.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lambert MP, Paliwal A, Vaissiere T, Chemin I, Zoulim F, Tommasino M, Hainaut P, Sylla B, Scoazec JY, Tost J, Herceg Z. Aberrant DNA methylation distinguishes hepatocellular carcinoma associated with HBV and HCV infection and alcohol intake. J Hepatol. 2010;54:705–15. doi: 10.1016/j.jhep.2010.07.027. [DOI] [PubMed] [Google Scholar]
- 24.Lin JC, Jeong S, Liang G, Takai D, Fatemi M, Tsai YC, Egger G, Gal-Yam EN, Jones PA. Role of nucleosomal occupancy in the epigenetic silencing of the MLH1 CpG island. Cancer Cell. 2007;12:432–44. doi: 10.1016/j.ccr.2007.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nan HM, Song YJ, Yun HY, Park JS, Kim H. Effects of dietary intake and genetic factors on hypermethylation of the hMLH1 gene promoter in gastric cancer. World J Gastroenterol. 2005;11:3834–41. doi: 10.3748/wjg.v11.i25.3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Engeland M, Weijenberg MP, Roemen GM, Brink M, de Bruine AP, Goldbohm RA, van den Brandt PA, Baylin SB, de Goeij AF, Herman JG. Effects of dietary folate and alcohol intake on promoter methylation in sporadic colorectal cancer: the Netherlands cohort study on diet and cancer. Cancer Res. 2003;63:3133–7. [PubMed] [Google Scholar]
- 27.Shukla SD, Aroor AR. Epigenetic effects of ethanol on liver and gastrointestinal injury. World J Gastroenterol. 2006;12:5265–71. doi: 10.3748/wjg.v12.i33.5265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berger AH, Niki M, Morotti A, Taylor BS, Socci ND, Viale A, Brennan C, Szoke J, Motoi N, Rothman PB, Teruya-Feldstein J, Gerald WL, et al. Identification of DOK genes as lung tumor suppressors. Nat Genet. 2010;42:216–23. doi: 10.1038/ng.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Colella S, Shen L, Baggerly KA, Issa JP, Krahe R. Sensitive and quantitative universal Pyrosequencing methylation analysis of CpG sites. Biotechniques. 2003;35:146–50. doi: 10.2144/03351md01. [DOI] [PubMed] [Google Scholar]
- 30.Tost J, Dunker J, Gut IG. Analysis and quantification of multiple methylation variable positions in CpG islands by Pyrosequencing. Biotechniques. 2003;35:152–6. doi: 10.2144/03351md02. [DOI] [PubMed] [Google Scholar]
- 31.Yang AS, Estecio MR, Doshi K, Kondo Y, Tajara EH, Issa JP. A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements. Nucleic Acids Res. 2004;32:e38. doi: 10.1093/nar/gnh032. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.