SUMMARY
Myc oncoproteins directly regulate transcription by binding to target genes, yet this only explains a fraction of the genes affected by Myc. mRNA turnover is controlled via AU-binding proteins (AUBPs) that recognize AU-rich elements (AREs) found within many transcripts. Analyses of precancerous and malignant Myc-expressing B cells revealed that Myc regulates hundreds of ARE-containing (ARED) genes and select AUBPs. Notably, Myc directly suppresses transcription of Tristetraprolin (TTP/ZFP36), an mRNA-destabilizing AUBP, and this circuit is also operational during B lymphopoiesis and IL7 signaling. Importantly, TTP suppression is a hallmark of cancers with MYC involvement, and restoring TTP impairs Myc-induced lymphomagenesis and abolishes maintenance of the malignant state. Further, there is a selection for TTP loss in malignancy; thus, TTP functions as a tumor suppressor. Finally, Myc/TTP-directed control of select cancer-associated ARED genes is disabled during lymphomagenesis. Thus, Myc targets AUBPs to regulate ARED genes that control tumorigenesis.
INTRODUCTION
Myc family oncoproteins (c-Myc, N-Myc, and L-Myc) are activated in over half of human cancers where they regulate critical pathways that contribute to tumorigenesis (Meyer and Penn, 2008). c-Myc (hereafter Myc) expression is activated by MYC/Immunoglobulin (Ig) chromosomal translocations in human Burkitt lymphoma (BL) (Dalla-Favera et al., 1982; Taub et al., 1982). This event is sufficient to promote tumorigenesis as Eμ-Myc transgenics, a mouse model of human BL where c-Myc is under the control of the Ig Eμ enhancer, develop aggressive B cell lymphoma (Adams et al., 1985). The Eμ-Myc model has proven remarkably useful as a platform for discovery of pathways activated by Myc and for defining checkpoints bypassed during tumorigenesis, for example the Arf-p53 tumor suppressor apoptotic pathway (Eischen et al., 1999), the Myc-to-p27Kip1 proliferative circuit (Keller et al., 2007) and the DNA damage response (Gorrini et al., 2007).
Myc oncoproteins function as basic-helix-loop-helix leucine zipper (bHLH-Zip) transcription factors that control the expression of a large cast (>1,600) of genes (Zeller et al., 2003) by binding to specific E-box sequences (CAC/AGTG). Binding of Myc to these elements requires dimerization with Max, a bHLH-Zip family member and binding of Myc:Max complexes recruits transcriptional coactivators to induce transcription (Dang et al., 2006). Further, Myc:Max heterodimers repress transcription by binding to and inhibiting the functions of the Miz-1 transcription factor at Initiator (Inr) elements found at some transcription start sites (Seoane et al., 2001; Staller et al., 2001). However, Myc binding does not always connote direct regulation of a target (Zeller et al., 2006) and Myc can indirectly affect gene expression via its regulation of other mediators or by its effects on cell growth, survival or transformation (Dang, 1999).
The regulation of mRNA turnover is a critical node for controlling gene expression, and many short-lived transcripts harbor AU-rich elements (AREs), usually an AUUUA sequence, within their 3’ untranslated regions (3’UTRs). Indeed, using computational analysis an ARE database (ARED) has shown that at least 11% of human genes contain AREs (Halees et al., 2008). A set of RNA binding proteins coined AU-binding proteins (AUBPs) specifically bind to AREs, serving to either stabilize or promote destruction of mRNAs (Chen and Shyu, 1995). For example, the AUBPs HuR, HuB, HuC, HuD, Auf1, Auf2 and Nucleolin (Ncl), typically stabilize ARE-containing mRNAs (Brennan and Steitz, 2001; Dean et al., 2002; Sengupta et al., 2004). In contrast, others such as Tristetraprolin (TTP/Tis11/Zfp36) and its family members Tis11b (Brf1/Zfp36l1) and Tis11d (Brf2/Zfp36l2) bind to ARE-containing mRNAs, marking them for delivery to processing bodies (P-bodies) where transcripts are deadenylated and degraded by mRNA decay enzymes (Blackshear, 2002; Franks and Lykke-Andersen, 2007).
The ability of AUBPs to control gene expression through mRNA stability has been suggested to play roles in tumorigenesis. For example, HuR binds to the COX-2 mRNA ARE in colon cancer cells, increasing levels of this proinflammatory protein (Dixon et al., 2001). Further, the NPM-ALK oncoprotein phosphorylates AUF1, augmenting its ability to stabilize some mRNAs (Fawal et al., 2006). In addition, β-actin promoter-driven expression of the p37 isoform of AUF1 can trigger sarcoma in transgenic mice (Gouble et al., 2002). Conversely, TTP levels are reduced in aggressive prostate and breast cancer and connote poor outcome (Brennan et al., 2009), and inactivation of both Tis11b and Tis11d in mouse T cells can lead to leukemia (Hodson et al., 2010). Thus, although largely anecdotal, these studies suggest that at least some AUBPs play roles in cancer.
Since mRNA stability is a common mechanism for controlling transcript levels, we hypothesized that Myc indirectly regulates ARE-containing mRNAs via the agency of AUBPs, and that this pathway is important for tumorigenesis. Here we show Myc regulates the expression of hundreds of ARED genes and several AUBPs. In particular, Myc directly suppresses the transcription of TTP, which regulates the levels of cancer-associated ARED genes, and this control is disabled during tumorigenesis. Notably, repression of TTP is a hallmark of malignancies with MYC involvement and enforced expression of TTP impairs lymphoma development and abolishes maintenance of the malignant state. Thus, TTP functions as a tumor suppressor.
RESULTS
Myc Regulates the Expression of ARE-Containing mRNAs
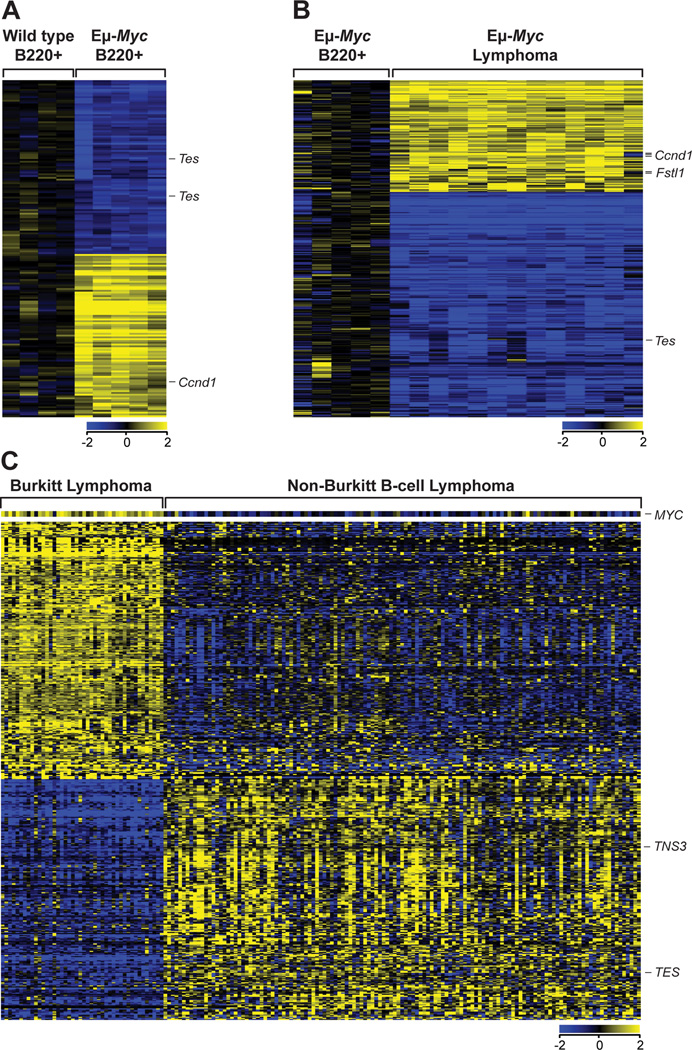
To assess effects of Myc on the expression of ARED genes (Halees et al., 2008), we performed expression profiling analyses of B220+ B cells from 4–6 week old wild type versus Eμ-Myc littermates, and of lymphomas from several Eμ-Myc mice. 153 ARED genes were significantly altered in precancerous Eμ-Myc B cells (74 induced and 79 repressed), or 7.8% of ARED genes expressed (Figure 1A, Table S1). Most of these were similarly regulated in lymphomas, although the magnitude of this response was often greater in tumors (Figure S1A). Further, a comparison of precancerous and malignant Eμ-Myc B cells showed 344 ARED genes (16.5% of ARED genes expressed) were significantly altered by neoplastic conversion (Figure 1B, Table S2). Collectively, the expression of nearly 20% of all ARED genes expressed in B cells is altered during Myc-driven lymphomagenesis.
Figure 1. Myc Alters the Expression of Hundreds of Genes Containing AREs.
(A and B) Gene expression profiling of arrays showing genes from the ARED Organism database whose expression is specifically altered in B220+ B cells from premalignant Eμ-Myc (n=5) and wild type (n=4) mice (A) and in Eμ-Myc lymphomas (n=13) compared to premalignant Eμ-Myc B cells (B). All probe sets shown have a >2.0-fold change and are significantly altered by unpaired t-test analysis (p<0.05).
(C) Gene expression profiling comparing ARED genes differentially expressed between 44 human BL and 129 human non-BL samples from GSE4475. All probe sets shown have >2.0-fold change and are significantly altered by unpaired t-test analysis (p<0.05). See also Figure S1 and Tables S1–S3.
To assess if MYC alters ARED gene expression in human B lymphoma, we queried expression datasets of primary human BL samples that bear MYC/Ig translocations versus other human B lymphoma subtypes (non-BL) (Hummel et al., 2006). Indeed, 366 ARED genes (15.2% of ARED genes expressed) are significantly different in human BL (Figure 1C, Table S3). Altered ARED gene regulation is also manifest in neuroblastoma, where analyses of expression datasets (Wang et al., 2006) demonstrated 129 ARED genes significantly differ in MYCN-amplified versus non-MYCN-amplified tumors (Figure S1B). Finally, there is a significant (24%) overlap in the expression of ARED genes altered in BL and MYCN-amplified neuroblastoma. Thus, Myc affects expression of a large cast of ARED genes in different tumor types and in B cells.
The Expression of AUBPs is Altered During Myc-Driven Lymphomagenesis
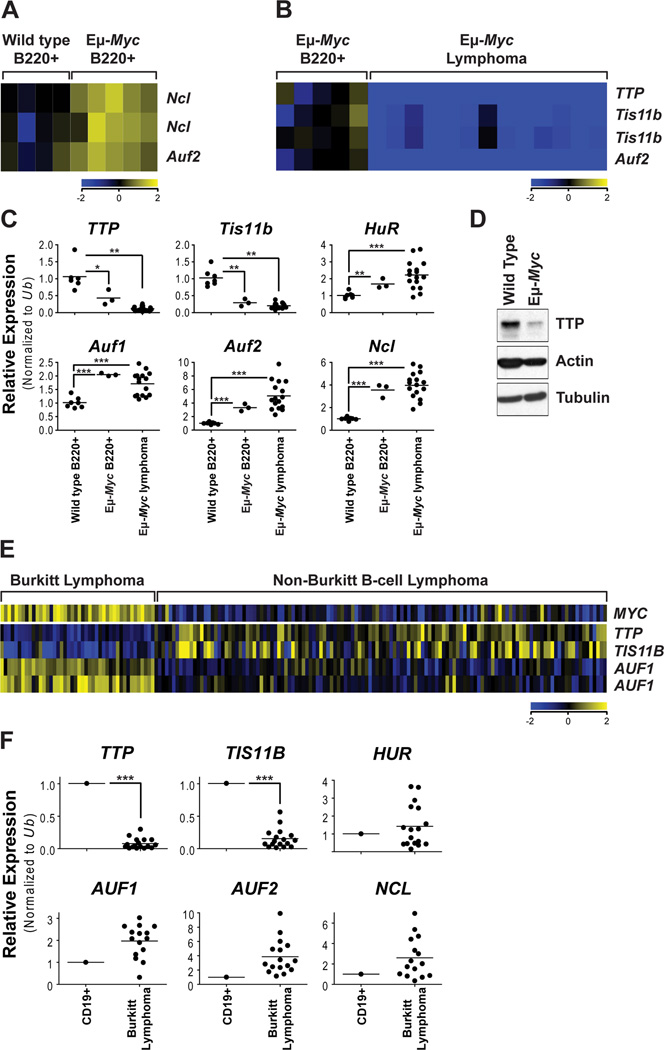
Given the effects of Myc on ARED genes, expression datasets were mined to determine if Myc alters the expression of AUBPs. The expression of Ncl and Auf2 was significantly increased in premalignant Eμ-Myc versus wild type B220+ B cells (Figure 2A). Further, levels of TTP, Tis11b and Auf2 were significantly reduced in malignant versus precancerous Eμ-Myc B cells (Figure 2B). The differences in Auf2 expression are likely due to different probe sets, one recognizing both spliced variants, while the other only recognizes the shorter isoform. Quantitative real time PCR (qRT-PCR) analyses of wild type and premalignant Eμ-Myc B cells, and of Eμ-Myc lymphomas, demonstrated that TTP and Tis11b transcripts were markedly reduced in precancerous and malignant Eμ-Myc B cells (Figure 2C). TTP protein levels were also drastically reduced in premalignant Eμ-Myc B cells (Figure 2D). In contrast, mRNAs encoding HuR, Auf1, Auf2 and Ncl were elevated in precancerous and malignant Eμ-Myc B cells (Figure 2C). Thus, the expression of select AUBPs is altered during Myc-driven lymphomagenesis.
Figure 2. Myc Alters the Expression of AUBPs.
(A–B and E) Gene expression profiling of arrays showing differentially expressed AUBP genes in B220+ B cells from premalignant Eμ-Myc versus wild type littermates (A), Eμ-Myc lymphomas compared to premalignant Eμ-Myc B cells (B) and human BL and non-BL samples (E) from GSE4475. All probe sets shown have >2.0-fold change and are significantly altered by unpaired t-test analysis (p<0.05).
(C and F) qRT-PCR analysis of total RNA isolated from B220+ B cells of wild type and premalignant Eμ-Myc mice and lymphomas that arose in independent Eμ-Myc transgenics (C), and of RNA of CD19+ B cells from a healthy individual compared to human BL samples (F). The relative expression was determined for TTP (ZFP36), TIS11B (ZFP36L1), HUR, AUF1, AUF2 and NCL (Nucleolin). Individual samples are indicated by circles and the mean for each group is represented by a line. Results were normalized to the expression of Ubiquitin (Ub).
(D) Immunoblots comparing the levels of TTP, Actin, and Tubulin in B220+ B cells of wild type and premalignant Eμ-Myc littermates.
Student’s t-test (* p<0.05, ** p<0.01, *** p<0.001)
See also Figure S2.
Suppression of TTP Family Members is a Hallmark of Malignancies with MYC Involvement
To assess AUBP gene expression in human tumors with MYC involvement, we determined their expression in human BL versus non-BL lymphomas. TTP and TIS11B expression is markedly repressed in human BL (Figure 2E and 2F). By contrast, levels of HUR, AUF1, AUF2 and NCL mRNA were not significantly altered. Notably, expression analyses of other cancers and tumor subtypes established that repression of TTP, or of its family member TIS11D, are hallmarks of Myc-expressing malignancies. Strikingly, TTP expression was the inverse of MYC in human tumors, where TTP levels were low in MYC-expressing breast, colorectal and metastatic prostate cancer (Figure S2B–S2E). Further, TIS11D is repressed in MYCN-amplified neuroblastoma and in colorectal cancers with elevated MYC levels (Figure S2A and S2C).
A Myc-TTP Circuit is also Operational in B Lymphocytes
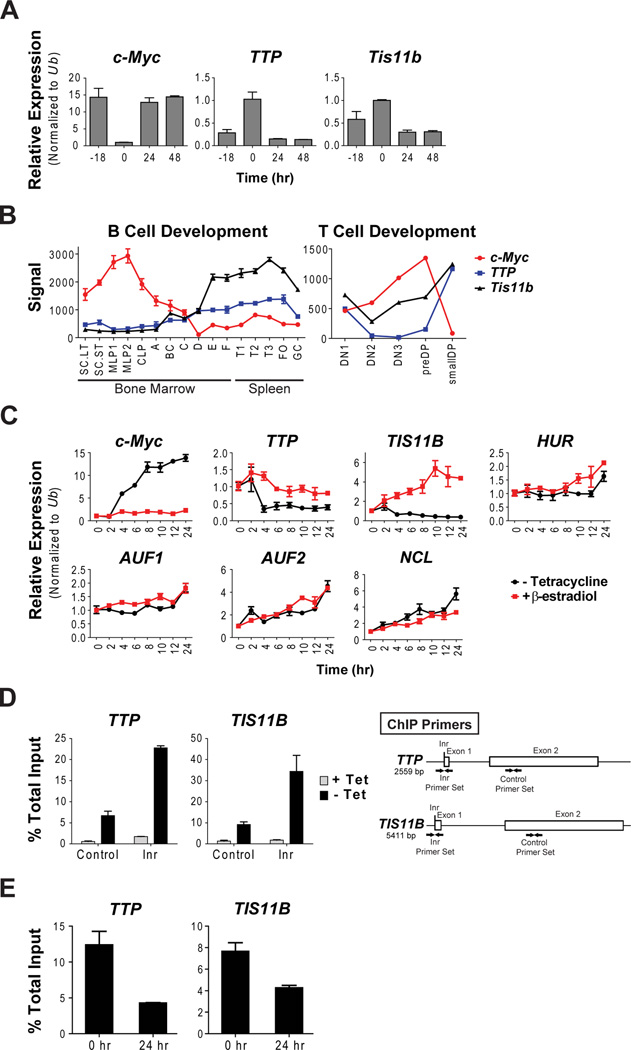
The finding that TTP levels inversely correlated with MYC in tumors versus normal tissues, suggested that Myc and TTP expression might be coordinately controlled during development, and by mitogenic signaling. In progenitor B cells Myc is regulated by IL7 (Morrow et al., 1992). To address control of TTP and Tis11b in this context, primary mouse progenitor B cells grown in IL7 medium were deprived and then re-stimulated with ligand. As expected Myc mRNA levels were dependent upon ligand (Figure 3A). Notably, control of TTP and to a lesser extent Tis11b, were the inverse of Myc, where TTP and Tis11b mRNA levels were high in ligand-deprived cells yet suppressed following IL7 stimulation. Further, analyses of both B and T lymphocyte developmental expression (Painter et al., 2011; Tabrizifard et al., 2004) revealed that TTP and Tis11b levels were the inverse of those of Myc, where they are induced when Myc is shut off in more mature, non-proliferating lymphocytes (Figure 3B). Thus, TTP and Tis11b expression is regulated by physiological signals that control Myc.
Figure 3. Myc Directly Represses TTP and TIS11B Transcription.
(A) qRT-PCR analysis of total RNA isolated from primary wild type B220+ B cells grown ex vivo in the presence of IL7. IL7 was removed at −18 hr and re-introduced at 0 hr and cells were collected at the indicated intervals. The relative expression was determined for c-Myc, TTP and Tis11b. Results were normalized to Ubiquitin (Ub) expression.
(B) Expression of c-Myc, TTP, and Tis11b throughout mouse B cell (GSE15907) and T cell (GSE30631) development.
(C) Total RNA was isolated from P493-6 B cells treated with Tet for 72 hr to repress c-Myc expression. Cells were either washed to remove Tet (black lines with circles), allowing Myc activation, or were treated with β-E2 (red lines with squares), allowing proliferation via ER-EBNA2 activation. Cells were collected at the indicated intervals. qRT-PCR analysis was performed to determine the relative expression of c-Myc, TTP, TIS11B, HUR, AUF1, AUF2, and NCL, and results were normalized to Ubiquitin (Ub) expression.
(D–E) ChIP analyses establish that Myc binds to the initiator element (Inr) found in the promoters of TTP and TIS11B and that this is associated with reduced binding of RNA Pol II. Chromatin was immunoprecipitated with an α-c-Myc antibody from P493-6 cells with repressed c-Myc (+Tet) or following 8 hr of c-Myc activation (−Tet) (D), or with an α-RNA Pol II antibody from P493-6 cells when c-Myc was repressed by Tet (0 hr) or following c-Myc activation by removal of Tet (24 hr) (E). Bound chromatin was evaluated by qRT-PCR and compared to the total amount of chromatin. Results show the mean with error bars indicating ± SEM.
See also Figure S3.
Myc Suppresses TTP and TIS11B Transcription
P493-6 lymphoblastoid B cells harbor a tetracycline (Tet)-repressed c-Myc transgene that allows Myc to be turned on and off (Pajic et al., 2000). To test if Myc regulates the expression of AUBPs, these cells were treated with Tet to turn off Myc. Tet was then removed and cells harvested for expression analysis. As c-Myc mRNA and protein were induced (Figure 3C, black line, data not shown) AUF2 and NCL mRNA levels also increased, whereas TTP and TIS11B mRNA levels dropped. In contrast, HUR and AUF1 mRNA levels do not increase until 24 hr after c-Myc induction, suggesting they are indirect targets of Myc.
P493-6 cells also harbor an estrogen-regulated ER-EBNA2 fusion transgene that can drive cell proliferation in the presence of β-estradiol (β-E2) even when c-Myc is repressed by Tet, allowing one to discriminate direct versus proliferative effects of Myc. Notably, β-E2 had little effect on TTP and actually induced TIS11B in the presence of Tet (Figure 3C, red line), suggesting these are direct targets repressed by Myc. In contrast, β-E2 induced HUR, AUF1, AUF2 and NCL mRNA levels; thus, these AUBP genes may be induced via Myc’s proliferative effects.
Myc:Max complexes can repress transcription by binding to Inr elements. TTP harbors an Inr downstream of the TATA box (Lai et al., 1995) and a putative Inr is present in TIS11B near the TATA box. To test if Myc bound to these Inr elements, chromatin immunoprecipitation (ChIP) assays were performed using c-Myc antibody in P493-6 cells +/− Tet. There was an inducible enrichment (of ~3.5-fold) of Myc binding to the Inr elements of TTP and TIS11B (Figure 3D and S3A). Further, genome-wide Myc ChIP analyses of the ENCODE project (Rosenbloom et al., 2010) showed that Myc binds to the Inr regions of both TTP and TIS11B in several tumor and normal cell lines (Figure S3B).
To assess if Myc-mediated repression of TTP and TIS11B was transcriptional, we performed primary transcript (i.e., unspliced) qRT-PCR analyses and RNA Pol II ChIP analyses in P493-6 cells +/− Tet. Myc induction was associated with repression of nascent TTP transcripts (Figure S3C) and with a marked reduction in binding of RNA Pol II to the transcription start regions of both TTP and TIS11B (Figure 3E). Thus, Myc binding to the Inr elements of TTP and TIS11B is associated with their reduced transcription.
TTP Impairs Myc-Induced Lymphomagenesis
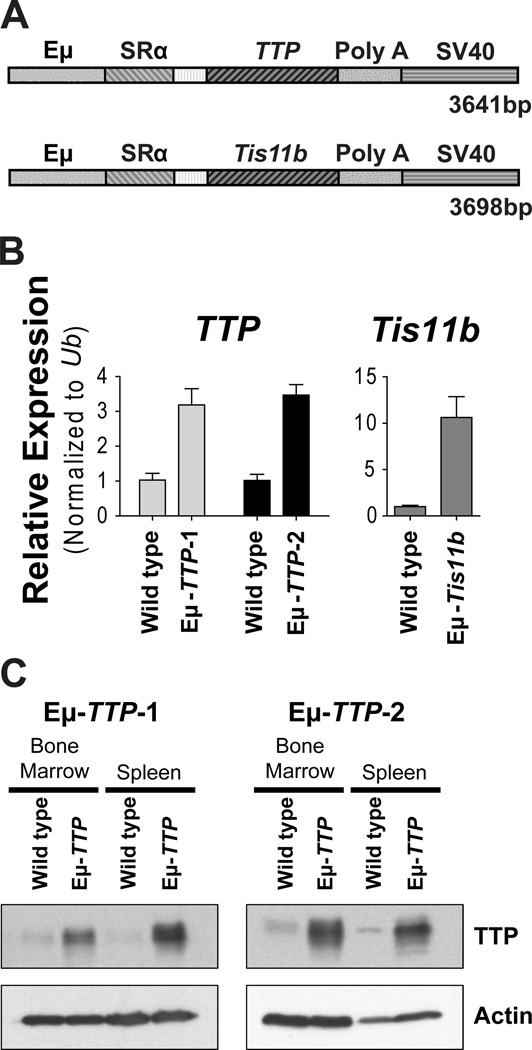
Given the effects of Myc on TTP and TIS11B expression and their suppression in Myc-driven lymphoma, we hypothesized that Myc-directed repression of TTP and/or Tis11b contributes to lymphomagenesis. To test this, Eμ-TTP and Eμ-Tis11b transgene vectors were generated (Figure 4A) that contained complete coding regions for mouse TTP or Tis11b (but lacking their 5’ and 3’UTRs) cloned into the pEμSR vector (Bodrug et al., 1994). These constructs were used to generate Eμ-TTP and Eμ-Tis11b transgenic coined Eμ-TTP-1, Eμ-TTP-2, and Eμ-Tis11b; all expressed elevated levels of TTP or Tis11b mRNA in B cells (Figure 4B) and the TTP transgenics had high levels of TTP protein in splenic and BM B220+ B cells (Figure 4C). Eμ-TTP and Eμ-Tis11b transgenics mice grow normally and have no apparent phenotype. Further, analyses of B cell development established that overexpression of TTP or Tis11b does not alter B cell numbers or immunophenotypes (Figure 5C, data not shown). Indeed, expression analysis showed only 16 genes were significantly altered by elevated TTP expression in Eμ-TTP B cells versus wild type B cells that express endogenous TTP (and only six of these are ARED genes, Figure S4). Thus, TTP overexpression alone has very minor effects on B cell gene expression programs.
Figure 4. Generation of Eμ-TTP and Eμ-Tis11b Transgenic Mice.
(A) Schematic of Eμ-TTP and Eμ-Tis11b transgenes. The complete coding region for mouse TTP or Tis11b, was cloned into the pEμSR plasmid downstream of the SRα promoter. The transgenes also include the mouse Igh 5’ enhancer (Eμ), the rabbit globin poly A and an SV40 tag region.
(B) qRT-PCR analysis of TTP or Tis11b expression in total RNA isolated from B220+ B cells from wild type and Eμ-TTP littermates from two different founders (Line 1 and Line 2) or from Eμ-Tis11b littermates. Results were normalized to Ubiquitin (Ub) expression. (C) Immunoblots analyses of TTP and Actin levels in bone marrow and splenic B220+ B cells of wild type and Eμ-TTP littermates.
Results show the mean with error bars indicating ± SEM.
See also Figure S4.
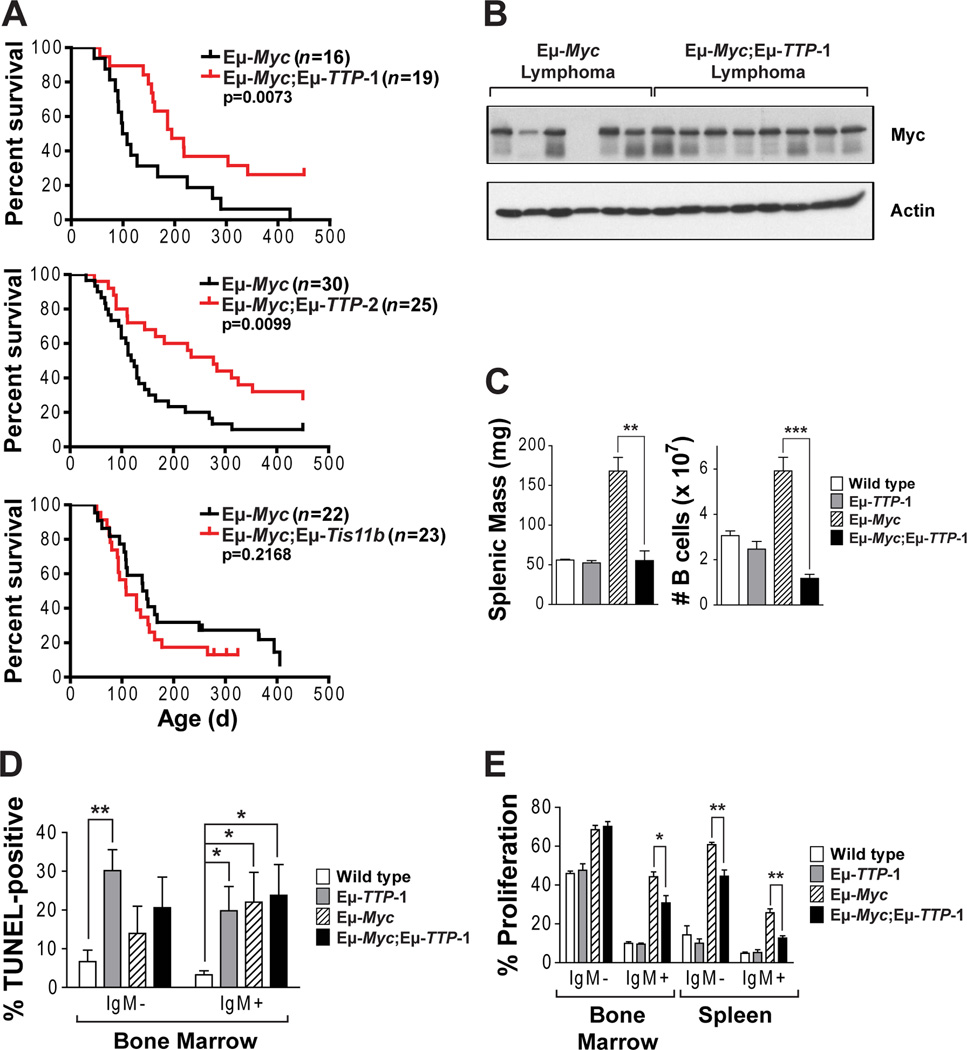
Figure 5. TTP Suppresses Myc-driven Lymphomagenesis.
(A) Survival curves of Eμ-Myc;Eμ-TTP-1 (n=19), Eμ-Myc;Eμ-TTP-2 (n=25), and Eμ-Myc;Eμ-Tis11b (n=23) transgenic mice was compared to Eμ-Myc littermates (n=16, 30 or 22, respectively). Tick marks above survival curve indicate mice that were still alive when the analysis was completed. P-values were determined by using Mantel-Cox logrank test.
(B) Immunoblots analyses of Myc and Actin levels in Eμ-Myc versus Eμ-Myc;Eμ-TTP-1 lymphomas.
(C) Splenic mass and numbers of B220+ B cells in the spleens of 5-week-old wild type, Eμ-TTP-1, Eμ-Myc, and Eμ-Myc;Eμ-TTP-1 littermates.
(D–E) Analysis of TUNEL-positive (D) and BrdU+ (E) B220+ cells, separated based upon their IgM expression, from BM and spleen of 5-week-old wild type, Eμ-TTP-1, Eμ-Myc, and Eμ-Myc;Eμ-TTP-1 mice.
Results show the mean with error bars indicating ± SEM.
Student’s t-test (* p<0.05, ** p<0.01, *** p<0.001)
See also Figure S5.
To test roles of TTP or Tis11b in lymphomagenesis, Eμ-TTP-1, Eμ-TTP-2 or Eμ-Tis11b transgenics were bred to Eμ-Myc mice, and single and double transgenics littermates were assessed for their premalignant state and tumor-free survival. As expected, Eμ-TTP-1, Eμ-TTP-2 and Eμ-Tis11b mice never developed disease and Eμ-Myc transgenics died at 3–4 months of age (median survival 103.5 and 121 days, top and middle panels, Figure 5A). Enforced TTP expression markedly extended the lifespan of Eμ-Myc transgenics (median survival, 194 and 277 days for Eμ-Myc;Eμ-TTP-1 and Eμ-Myc;Eμ-TTP-2, Figure 5A). In contrast, enforced Tis11b expression did not impair disease (Figure 5A, bottom panel). Thus, TTP, but not Tis11b, harnesses Myc-induced lymphomagenesis.
Tumors that ultimately developed in Eμ-Myc;Eμ-TTP transgenics had phenotypes typical of Eμ-Myc lymphomas. Silencing of the Eμ-TTP or Eμ-Myc transgenes was not observed, as tumors expressed high levels of TTP and Myc (Figure 5B and S5A–B). In addition, c-Myc transgene expression is similar in premalignant bone marrow B cells of Eμ-Myc;Eμ-TTP-1 and Eμ-Myc mice, and there are no differences in the levels of endogenous c-Myc transcripts in Eμ-TTP and wild type B cells (Figure S5C). Further, a comparison of the 1697 genes in the Myc target gene database (http://myc-cancer-gene.org/) demonstrated that only three Myc targets differed in Eμ-Myc versus Eμ-Myc;Eμ-TTP-1 lymphomas (Hlcs, Ngfrap1 and Asns) and none of these are ARED genes. Thus, enforced TTP expression neither affects the expression nor function of the Myc transgene. Rather, improved survival of Eμ-Myc;Eμ-TTP transgenics was due to a protracted premalignant state where splenomegaly and increased B cell numbers, hallmarks of eminent disease in Eμ-Myc mice, were manifest in 5-week old Eμ-Myc but not Eμ-Myc;Eμ-TTP-1 littermates (Figure 5C).
TTP Disables Myc’s Proliferative Response
Premalignant Eμ-Myc B cells have high rates of proliferation that are offset by increased apoptosis. TUNEL-FACS analyses revealed no significant differences in the apoptotic index of Eμ-Myc and Eμ-Myc;Eμ-TTP-1 B cells (Figure 5D). Myc triggers apoptosis via induction of Arf and activation of p53; accordingly, >70% of Eμ-Myc lymphomas bear inactivating mutations in Arf or p53 (Eischen et al., 1999). However, there were no differences in Arf or p53 expression, or in the frequency of alterations in Arf or p53, in Eμ-Myc;Eμ-TTP-1 versus Eμ-Myc lymphomas (Figure S5D and S5E) indicating that TTP does not affect Myc-induced apoptosis. Surprisingly, Eμ-TTP-1 BM B cells had an increased apoptotic index. Only one of the 16 genes in BM B cells affected by the TTP transgene (Figure S4), Lims1, has a role in apoptosis. Lims1 encodes Pinch1, which controls destruction of the pro-apoptotic BH3-only protein Bim (Chen et al., 2008). Indeed, Bim levels were elevated in Eμ-TTP-1 B cells (Figure S5F). Regardless, this TTP-directed response is not manifest in the context of Myc, and it has no obvious effects on B cell development.
To assess if Myc-induced proliferation was affected by TTP, wild type and transgenic littermates were injected with BrdU, and B220+ cells were collected from BM and spleen and analyzed by FACS. As expected, Eμ-Myc B cells had a high proliferative index, whereas Eμ-TTP-1 B cells had proliferative rates similar to wild type B cells; thus, TTP alone does not affect B cell growth (Figure 5E). Notably, the proliferative index of Eμ-Myc;Eμ-TTP-1 B cells was significantly reduced compared to Eμ-Myc B cells, with the exception of IgM-B220+ BM B cells. These data indicate that TTP impairs lymphoma development primarily by disabling Myc’s proliferative response.
TTP Functions as a Tumor Suppressor that Abolishes the Malignant State
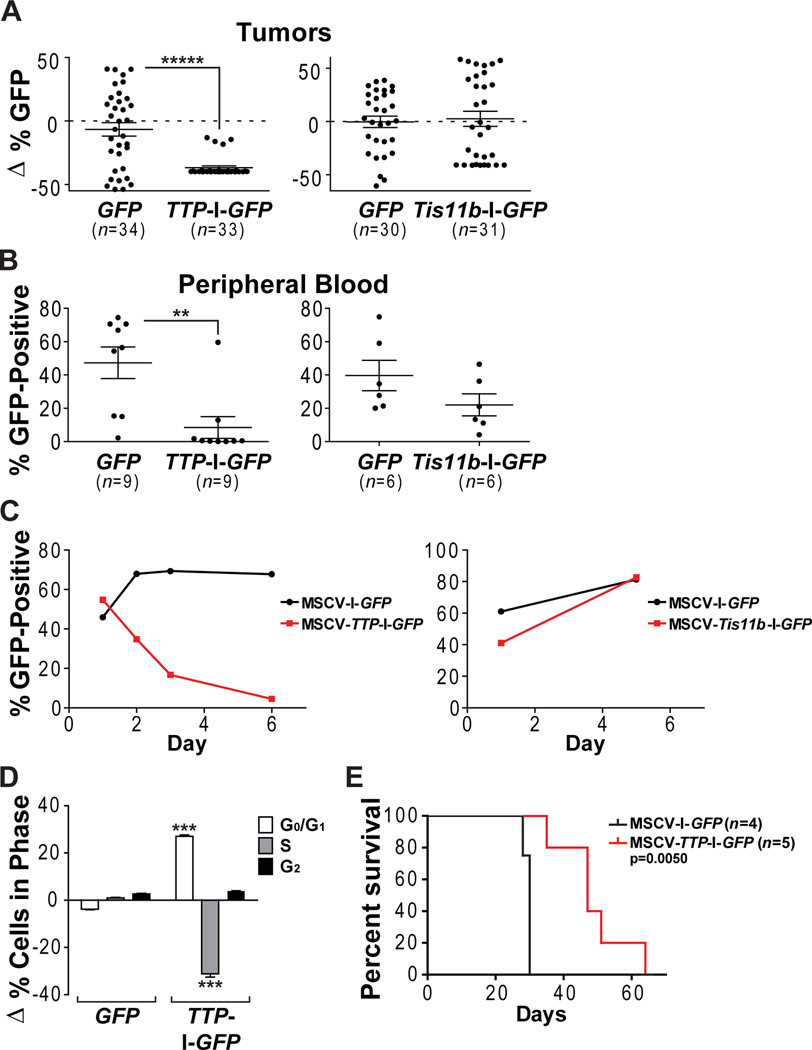
All Eμ-Myc lymphomas express little if any TTP and Tis11b (Figure 2). Thus, we hypothesized that TTP and/or Tis11b suppression was necessary for maintenance of the malignant state. To test this, an Eμ-Myc lymphoma was harvested and infected with MSCV-I-GFP (GFP-only) versus MSCV-TTP-I-GFP (TTP-GFP) retroviruses, or with GFP only versus MSCV-Tis11b-I-GFP (Tis11b-GFP) retroviruses. Transduction efficiencies of lymphoma cells were similar for GFP-only virus in the two experiments (54% and 61%), and were comparable to those for TTP-GFP (40%) and Tis11b-GFP (41%) viruses. Unsorted transduced Eμ-Myc lymphoma cells were injected i.v. into Nude mice and followed for tumor development. In both experiments recipients developed tumors at a similar rate and frequency, two to eight tumors per mouse. Tumors and peripheral blood were collected and GFP expression was assessed. Comparing the percentage of GFP+ lymphoma cells in the GFP-only versus TTP-GFP cohort revealed a striking selection against TTP-expressing tumor cells - a 13-fold drop in lymphoma cells expressing TTP compared to donor input versus a 1.1-fold drop in GFP+ lymphoma cells in the GFP-only cohort (Figure 6A). In contrast, there was less than a 3% change in GFP+ lymphoma cells in both the GFP-only and Tis11b-GFP cohort versus donor input (Figure 6A). Thus, there is a strong selection against TTP, but not Tis11b, expression in Eμ-Myc lymphoma.
Figure 6. TTP Impairs Maintenance of Eμ-Myc Lymphoma.
(A–B) Nude mice were injected with unsorted Eμ-Myc lymphoma cells infected with MSCV-I-GFP versus MSCV-TTP-I-GFP retrovirus (left panel) or with Eμ-Myc lymphoma cells infected with MSCV-I-GFP versus MSCV-Tis11b-I-GFP retrovirus (right panel). Shown is analysis of the change in the percentage of GFP+ B cells found in tumors compared to the percentage of GFP+ lymphoma cells that were injected (A) and the percentage of GFP+ B cells found in peripheral blood (B) at the time of sacrifice. Individual samples are indicated by circles and the mean for each group is represented by a line.
(C) Percentage of GFP+ unsorted ex vivo Eμ-Myc lymphoma cells infected with MSCVI-GFP versus MSCV-TTP-I-GFP retrovirus (left panel) or with MSCV-I-GFP versus MSCV-Tis11b-I-GFP retrovirus (right panel) was determined at the indicated intervals.
(D) Changes in cell cycle distribution of unsorted ex vivo Eμ-Myc lymphoma cells infected with MSCV-I-GFP versus MSCV-TTP-I-GFP retrovirus based upon the presence or absence of GFP within the lymphoma cells (i.e., GFP+ cells minus GFP-negative cells). Cells were labeled with BrdU, harvested and analyzed by FACS. The graph represents the average of three individually infected plates of Eμ-Myc lymphoma cells for each retrovirus.
(E) Survival curves of Nude mice injected with Eμ-Myc lymphoma cells sorted for GFP+ following infection with MSCV-I-GFP or MSCV-TTP-I-GFP retrovirus. P-values were determined by using Mantel-Cox log-rank test.
Results show the mean with error bars indicating ± SEM.
Student’s t-test (**p<0.01, ***p<0.001, *****p<0.00001)
See also Figure S6.
Analysis of peripheral blood B cells from recipients with lymphomas expressing GFP-only virus showed an average of 47% and 40% GFP+ cells (Figure 6B), while those of recipients with Tis11b-GFP virus averaged 22%. Again, in TTP-GFP virus-infected recipients there was a marked selection against TTP-expressing lymphoma cells, where on average only 8.4% of B cells were GFP+ (Figure 6B). Further, seven of these nine recipients had <2% GFP+ B cells; thus, few, if any, lymphoma cells expressing TTP remained.
To determine if TTP-GFP virus was present in lymphomas lacking GFP, genomic qPCR copy number and immunoblot analyses were performed. All tumors from the TTP-GFP cohort having modest levels of GFP+ cells expressed TTP, whereas those with no GFP+ cells lacked TTP (Figure S6B). In addition, genomic qPCR analyses showed that most TTP-GFP tumors have little to no TTP-GFP virus; thus, TTP-expressing lymphoma cells are out-competed (Figure S6A). There was one exception where the TTP-GFP virus was present but silenced, indicating this is another mode of circumventing TTP. In contrast, in the GFP-only tumors all contain at least some GFP virus. As expected, Myc expression was sustained in all lymphomas (Figure S6B). Thus, there is a selection for TTP loss in malignancy.
To assess if TTP affected lymphoma cell survival or proliferation, unsorted Eμ-Myc lymphoma cells infected with GFP-only, TTP-GFP, or Tis11b-GFP viruses were analyzed ex vivo. There were no differences in the apoptotic index of GFP+ versus GFP-negative cells in any of the lymphomas (n=4, data not shown). However, there was a rapid reduction in the percent of GFP+ cells in TTP-GFP virus-infected, but not in GFP-only or Tis11b-GFP virus-infected, lymphomas (n=4). Indeed, by 6 days there were virtually no GFP+ cells in TTP-GFP virus-infected lymphoma cultures (Figure 6C). This correlated with striking reductions in the percent of GFP+ (i.e., TTP-expressing) cells in S phase and corresponding increases in GFP+ cells in G0/G1 phase (Figure 6D). Thus, TTP abolishes the malignant state by disabling Myc’s proliferative response.
These data suggested lymphoma cells with low TTP levels have an advantage in vivo. To test this, we purified GFP+ cells from GFP-only and TTP-GFP virus-infected lymphomas and injected these into Nude mice. Again, TTP-expressing lymphoma cells took significantly longer to develop into tumors than GFP-only expressing lymphoma (p=0.005, Figure 6E). Thus, tumorigenic potential is augmented by suppression of TTP.
TTP-Dependent Control of its Targets is Bypassed During Tumorigenesis
To gain insights into the mechanism of TTP tumor suppression, expression analysis was performed on premalignant BM B220+ B cells isolated from wild type, Eμ-TTP-1, Eμ-Myc and Eμ-Myc;Eμ-TTP-1 littermates. This analysis revealed 115 genes significantly differ between Eμ-Myc and Eμ-Myc;Eμ-TTP-1 B cells (Table S4). 36 of these are ARED genes, where those repressed may represent direct TTP targets and those induced may reflect downstream genes indirectly affected by TTP (Figure 7A). Strikingly, the effects of TTP in premalignant B cells were very selective, as there were no changes on other aspects of mRNA metabolism or upon other biological processes operational in cancer (e.g., angiogenesis, metabolism, etc., Table S5). Notably, ten of these ARED genes have roles in cancer (Table S6) and qRT-PCR analyses confirmed six were differentially expressed in Eμ-Myc and Eμ-Myc;Eμ-TTP-1 B cells. Ccnd1 (Cyclin D1), which regulates cell cycle and proliferation (Peters, 1994), is significantly elevated in Eμ-Myc B cells yet reduced in Eμ-Myc;Eμ-TTP-1 B cells (Figure 7B). Further, Fstl1, a proinflammatory cytokine (Miyamae et al., 2006), is repressed in Eμ-Myc;Eμ-TTP-1 versus Eμ-Myc B cells. In contrast, the expression of ARED genes involved in autophagy (Gabarapl1, Hemelaar et al., 2003) or apoptosis (Uaca, Sakai et al., 2004), were elevated in Eμ-Myc;Eμ-TTP-1 B cells. Additionally, Tes, a putative tumor suppressor (Tobias et al., 2001), and Tns3, a metastasis suppressor (Katz et al., 2007), are reduced in Eμ-Myc B cells yet restored to wild type levels in Eμ-Myc;Eμ-TTP-1 B cells. Moreover, these ARED genes are induced (Ccnd1 and Fstl1) or repressed (TES, TNS3 and GABARAPL1) in Eμ-Myc lymphoma and BL, and in MYCN-amplified neuroblastoma (Figure 1 and Figure S1). Finally, these TTP-dependent targets are also regulated by IL7 signaling in primary B cells (Figure S7A), indicating they are physiological targets of the Myc/TTP circuit.
Figure 7. TTP-dependent Myc Targets Associated with Tumorigenesis.
(A and E) Gene expression profiling of arrays showing ARED genes whose expression is specifically altered in B220+ BM B cells from premalignant Eμ-Myc versus Eμ-Myc;Eμ-TTP-1 mice (A) or all genes whose expression is specifically altered in lymphomas from Eμ-Myc versus Eμ-Myc;Eμ-TTP-1 mice (E). Genes labeled in (A) have known roles in cancer. All probe sets shown have >2.0-fold change and are significantly altered by unpaired t-test analysis (p<0.05).
(B and C) qRT-PCR analysis of total RNA isolated from B220+ BM B cells of wild type and Eμ-TTP-1 transgenics and from premalignant Eμ-Myc and Eμ-Myc;Eμ-TTP-1 mice (B) or from wild type B220+ splenic B cells and from Eμ-Myc and Eμ-Myc;Eμ-TTP-1 lymphomas (C). The relative expression of Ccnd1, Fstl1, Gabarapl1, Tes, Tns3 and Uaca is shown. Results were normalized to Ubiquitin (Ub) expression.
(D) qRT-PCR (top left panel) and immunoblot (top right panel) analyses show the mRNA and protein levels of TTP-Flag, Cyclin D1 and Actin in HeLa Tet-Off/TTP-Flag cells that were grown +Dox (repressed TTP-Flag) or −Dox (induced TTP-Flag) for 48 hr. For RNP-IP analyses (bottom panel), RNPs were immunoprecipitated with α-Flag antibody from control or TTP-Flag-expressing HeLa cells and used for qPCR detection of cyclin D1 mRNA.
Results show the mean with error bars indicating ± SEM.
Student’s t-test (* p<0.05, ** p<0.01, *** p<0.001)
A feature of malignant conversion is bypass of regulatory circuits. Analyses of Eμ-Myc and Eμ-Myc;Eμ-TTP-1 lymphomas confirmed bypass of five out of six TTP-dependent cancer-associated ARED genes (Figure 7C). Specifically, TTP-directed control of Ccnd1, Gabarapl1, Tes, Tns3 and Uaca were all lost during the conversion to frank malignancy. Thus, in the presence of TTP there is a selection for loss of control of its targets during tumorigenesis.
The most profound effects of TTP were on proliferation and cyclin D1. Thus, we tested if, as suggested (Marderosian et al., 2006), Ccnd1 was a direct target of TTP. Dox-dependent control of expression of Flag-tagged TTP in HeLa cells led to marked reductions in endogenous cyclin D1 mRNA and protein (Figure 7D). Further, ribonucleoprotein immunoprecipitation (RNP-IP) of HeLa cells transfected with a TTP-Flag vector established that TTP binding to the 3’UTR of CCND1 mRNA was enriched by over 250-fold compared to control transfected cells (Figure 7D).
To test if TTP-directed suppression of cyclin D1 is necessary for the anti-proliferative response provoked by TTP and for the selection against TTP-expression in Eμ-Myc lymphoma, we first infected Eμ-Myc lymphomas with MSCV-IRES-dsRed2(RFP-only) or MSCV-D1a-IRES-dsRed2 (D1a-RFP) retroviruses (this D1a transgene lacks the 3’UTR harboring TTP recognition elements). dsRed2-expressing cells were purified and then infected with GFP-only or TTP-GFP virus. Notably, enforced expression of D1a was not sufficient to override the selection against TTP-GFP-expressing lymphoma cells or upon cell proliferation (n=3, Figure S7B and S7C). In accord with analyses of Eμ-Myc;Eμ-TTP lymphoma (Figure 7C), TTP did not suppress endogenous Ccnd1 in Eμ-Myc lymphoma and also did not affect the expression of exogenous D1a (Figure S7D). Thus, the ability of TTP to target Ccnd1 transcript destruction is bypassed in Eμ-Myc lymphoma and enforced D1a expression is not sufficient to override the anti-proliferative effects of TTP. These findings are in accord with the profound and broad effects of TTP on the transcriptome manifest in Eμ-Myc;Eμ-TTP versus Eμ-Myc lymphoma (Figure 7E and Table S7, ~200 genes altered) and following acute TTP expression in Eμ-Myc lymphoma cells infected with TTP-GFP virus (Figure S7E, ~1,100 genes altered in GFP+ versus GFP- lymphoma cells).
DISCUSSION
The data presented herein show that Myc indirectly affects the expression of hundreds of ARED genes, and that this is linked to transcriptional regulation of AUBPs that control turnover of their mRNA substrates. The ability of Myc to repress TTP and TIS11B transcription via binding to Inr elements suggests that Myc controls some ARE-containing mRNAs via AUBPs. Notably, the physiological relevance of the Myc-to-AUBP-to-ARED gene response is clear, where the Myc-to-TTP pathway is operational during B cell development and is controlled by IL7 signaling, and where TTP functions as a tumor suppressor that impairs the development and maintenance of Myc-driven lymphoma (Figure S7F). Importantly, this pathway is a hallmark of malignancies with MYC involvement, suggesting that it might be exploited by targeted therapies. Given our findings, it is likely that oncogene-to-AUBP-to-ARED responses are a general feature of cancer and that they serve as checkpoints or effectors that control many features of malignancy.
Previous studies suggested that TTP might play roles in tumorigenesis, as its substrates include mRNAs encoding the LATS2 tumor suppressor and the E6-AP ligase that directs p53 destruction (Lee et al., 2010; Sanduja et al., 2009). Further, TTP can block the tumorigenic potential of immortal v-H-Ras transformed mast cells (Stoecklin et al., 2003). Finally, low TTP expression connotes poor outcome in breast cancer (Brennan et al., 2009).
The most convincing evidence for a role for TTP in cancer comes from the current studies, where overriding Myc-directed repression of TTP more than doubles the lifespan of Eμ-Myc mice, and where TTP compromises tumor maintenance by disabling Myc’s proliferative response. The tumor suppressor roles of TTP are underscored by the marked selection against TTP expression in Myc-driven lymphomas. While in our studies the dominant phenotype of TTP is disruption of Myc’s proliferative response, the ability of TTP to induce apoptosis might be important for its tumor suppressor functions in other contexts and/or in harnessing transformation by other oncogenes.
A prediction of our findings is that repression of TTP, or one of its family members, is necessary for Myc-induced tumorigenesis. In accord with this, TTP suppression is a hallmark of many tumors with MYC involvement, and TIS11D is repressed in others. These connections do not necessarily connote roles in cancer, where for example both TTP and Tis11b are targets repressed by Myc, yet only TTP functions as a tumor suppressor. However, this selectivity in the Eμ-Myc model does not exclude possible tumor suppressor roles for TIS11B or TIS11D in other contexts.
TTP knockout mice rapidly develop an autoimmune disease characterized by cachexia, arthritis and dermatitis (Taylor et al., 1996), which are provoked by marked increases in the TTP target TNF-α (Carballo et al., 1998). Thus, crossing TTP knockout mice to Eμ-Myc transgenics was not informative. Loss of Tnfr1 and Tnfr2 abrogates nearly all aspects of the TTP deficiency (Carballo and Blackshear, 2001); thus, we also generated Eμ-Myc;TTP−/−;Tnfr1−/−;Tnfr2−/− transgenics. Notably, TTP−/−;Tnfr1−/−;Tnfr2−/− mice do not develop tumors and the course of lymphoma onset and survival was similar between these Eμ-Myc;TTP+/+ and Eμ-Myc;TTP−/− cohorts (data not shown). We conclude that TTP loss alone is not sufficient to provoke tumorigenesis, and that Myc-directed suppression of TTP effectively cancels its tumor suppressor functions.
The Myc/TTP circuit appears operational during lymphopoiesis and is controlled by IL7 signaling. However, B cell development and proliferation are essentially unaffected by enforced TTP expression, where phenotypes are not evident even in aged Eμ-TTP transgenics. Thus, while TTP alone has limited effects on B cell physiology, it has profound and selective effects on the development and maintenance of Myc-driven tumors, supporting the notion that agents targeting TTP may be effective therapeutics.
Six ARED genes having known roles in cancer were TTP dependent in precancerous Myc-expressing B cells and TTP-dependent control of these targets was bypassed in lymphomas, suggesting important roles for these targets in the conversion to the malignant state. However, as established by our expression analyses of Eμ-Myc;Eμ-TTP tumors and of Eμ-Myc lymphoma transduced with TTP-expressing retrovirus, there are broad and profound effects of TTP on the cancer transcriptome. Thus, agents that specifically reactivate TTP expression and/or augment its activity may be superior to those that affect the control of any one of its individual targets. Notably, such TTP-targeting agents would represent attractive therapeutics for the treatment of malignancies with MYC involvement.
EXPERIMENTAL PROCEDURES
Mice and Tumor Analyses
The coding region for mouse TTP or Tis11b was cloned into the pEμSR plasmid (Bodrug et al., 1994). The University of Michigan Transgenic Animal Model Core microinjected the Eμ-TTP or Eμ-Tis11b transgenes into fertilized mouse eggs that were then implanted into pseudopregnant females. Transgenic founders were identified by PCR.
Eμ-Myc transgenic mice were bred to Eμ-TTP-1, Eμ-TTP-2, or Eμ-Tis11b transgenic mice. Mice were monitored for illness and tumor development. Sick animals were sacrificed and tumors were collected. Nude mice were used for Eμ-Myc lymphoma transplant experiments. All animal studies were approved by the Scripps Florida IACUC.
RNA Preparation and Analyses
RNA from B cells, tumor samples and P493-6 cells was prepared using the NucleoSpin RNA II kit (Macherey-Nagel). RNA was extracted from 17 BL tumors using the RNA/DNA kit (Qiagen) with institutional review board approval and after informed consent. RNA was used to prepare cDNA and qRT-PCR was performed. Data analyses used the ΔΔCt method, where ubiquitin served as the internal control. Primers are listed in the Extended Experimental Procedures.
Microarray Expression Profiling Analyses
Biotin-labeled cRNA, prepared from total RNA, was fragmented and hybridized to Affymetrix GeneChip Mouse Genome 430A microarray (Eμ-Myc lymphomas) or Mouse Genome 430 2.0 microarray (precancerous Eμ-Myc and Eμ-Myc;Eμ-TTP-1 BM B cells, along with littermate controls, and Eμ-Myc and Eμ-Myc;Eμ-TTP-1 lymphomas). Microarrays were washed, stained and scanned. Microarray data is available from the NCBI GEO database under Accession GSE32239 and GSE37792. Affymetrix data were normalized based on GCRMA algorithm and analyzed to generate heatmaps using GeneSpring GX11 (GS GX11, Agilent). Yellow indicates up-regulation and blue denotes down-regulation on all heatmaps.
For B cell development expression analysis, data from the Immunological Genome Project (GSE15907) was imported into GS GX11 and analyzed using ExonRMA16 algorithm (Painter et al., 2011). For T cell development analysis, the GEO database GSE30631 (Tabrizifard et al., 2004) was used and MAS5 signals were derived using Gene Expression Console software (Affymetrix).
Western Blot Analyses
B cells or tumor samples were lysed and protein concentration was determined. Protein was separated on SDS-PAGE, transferred to PVDF membranes and blotted for specific antibodies. Antibodies are listed in the Extended Experimental Procedures.
Cell Culture
Before harvesting, P493-6 cells (Pajic et al., 2000) were cultured in the presence of Tet (0.1µg/ml) for 72 hr and replated in media either without Tet or with β-E2 (1µM). Eμ-Myc lymphomas were harvested, homogenized in PBS with 2% FBS, filtered through a 100µm strainer and cultured as a single cell suspension. For IL7 stimulation, BM-derived B cells were washed and replated in media without IL7 for 18 hr before re-addition of IL7. HeLa Tet-OFF/TTP-Flag cells were maintained in 2 µg/mL doxycycline (Dox) and in the absence of Dox for 48 hr to induce TTP-Flag expression.
Chromatin Immunoprecipitation
P493-6 B cells were cultured in media with Tet, and then without Tet, fixed and harvested. Sonicated chromatin was immunoprecipitated with antibodies specific for Myc (sc-764, Santa Cruz) or RNA Pol II (sc-47701, Santa Cruz) or isotype matched IgG (GenScript) using magnetic protein G beads (Active Motif). qRT-PCR was run on the chromatin and the percent of chromatin bound by Myc, RNA Pol II or IgG was determined. Primers are listed in the Supplemental Experimental Procedures.
Flow Cytometry
Cell cycle analysis was performed using a BrdU Flow Kit (BD Biosciences). Mice were injected intraperitoneally with 1 mg of 5-bromo-2-deoxyuridine (BrdU) and sacrificed after 16 hr. Cells were blocked with FcBlock and stained with CD45R/B220 and IgM (BD Biosciences). Cultured Eμ-Myc lymphoma cells were pulse-labeled in 10 µM BrdU and harvested. Cells were prepared according to protocol and stained with DAPI and either α-BrdU FITC or α-BrdU APC.
Apoptotic analysis was performed using an APO-BrdU TUNEL kit (Phoenix Flow Systems). Cells were blocked with FcBlock and stained with CD45R/B220 and IgM antibodies. Cells were fixed and BrdU-labeled with terminal deoxynucleotidyl transferase (Tdt) and stained with anti-BrdU-FITC mAb.
White blood cells were isolated from spleens and cells were treated with FcBlock and stained with CD45R/B220 antibody. For cell cycle, apoptotic and B cell number analysis, stained cells were analyzed using a BD LSRII and BD FACSDiva. Doublets were excluded. Total B cell number was determined by the number of cells counted multiplied by the percentage of B cells in each sample.
Peripheral blood and tumors were collected from Nude mice transplanted with Eμ-Myc lymphoma cells infected with MSCV-I-GFP, MSCV-TTP-I-GFP or MSCV-Tis11b-I-GFP retrovirus. Cells were treated with FcBlock and stained with an Alexa Fluor 647 B220 antibody (BD). Stained cells were analyzed using a BD FACSCanto II; doublets were excluded and single cells were gated based on GFP versus Alexa Fluor 647 (B220+).
Lymphoma Cell Infection and Transplantation
293T cells were CaPO4 transfected with MSCV-I-GFP or MSCV-TTP-I-GFP, pMD1-old-gag pol and pCAG-Eco added to 2x HEPES Buffered Saline. Retrovirus was collected and filtered. Eμ-Myc lymphoma cells were added to fresh retrovirus with polybrene and centrifuged. 6 hr post-infection, media was replaced with fresh retrovirus with polybrene. 24 hr after infection Eμ-Myc lymphoma cells were resuspended in PBS and 3×106 unsorted cells or 1×106 GFP+ sorted cells were injected via tail vein into Nude recipients. Peripheral blood and tumors were collected from sick mice.
Ribonucleoprotein Immunoprecipitation (RNP-IP)
HeLa cells were transfected with pcDNA3-Flag-TTP or pcDNA3 vector using Lipofectamine Plus (Invitrogen). Cells were lysed and cytoplasmic extracts incubated with α-Flag mAb or mouse IgG precoated to protein A/G PLUS agarose beads (Santa Cruz) overnight. Total RNA was isolated from beads using Trizol and used for cDNA synthesis. qPCR reactions were performed and data normalized to the IgG controls. Primers are listed in the Extended Experimental Procedures.
Supplementary Material
Highlights.
Myc controls levels of ARE-containing mRNAs via select AU-binding proteins (AUBPs)
Repression of tristetraprolin (TTP) is a hallmark of tumors with MYC involvement
TTP is a tumor suppressor that impairs development and maintenance of lymphoma
Myc/TTP-directed control of select cancer genes is disabled during lymphomagenesis
ACKNOWLEDGEMENTS
We thank Jerry Adams for providing the pEμSRα plasmid; Mihaela Onciu and John Sandlund for providing Burkitt lymphoma samples; J. Alan Diehl for providing MSCV-cyclin D1a retrovirus; Thomas Saunders and the Transgenic Animal Model Core of the University of Michigan’s Biomedical Research Core Facilities for generating the Eμ-TTP and Eμ-Tis11b transgenics Wi Lai and Deborah Stumpo for technical assistance; Shannon Sunday of the Scripps Florida ARC for assistance; Brandon Young and Brad Long of the Scripps Florida Genomics Core; Bivian Torres and Kim Lowe of the Scripps Florida Flow Cytometry Core; Frank C. Dorsey for helpful discussions; and Marika Kernick for editing. Supported by NIH grants (DK44158 and CA167093 to J.L.C., F32-CA115075 to R.R. and CA134609 to D.A.D.), by monies from the ThinkPink Kids Foundation, and by monies from the State of Florida to TSRI. Dr. Blackshear is supported by the Intramural Research Program of the NIH, NIEHS. R.R. also received support from the National City Postdoctoral Fellowship, the Glenn W. Bailey Postdoctoral Fellowship and the PGA National Women’s Cancer Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental information includes Extended Experimental Procedures, seven figures, seven tables and Supplementary References and can be found with this article online.
REFERENCES
- Adams JM, Harris AW, Pinkert CA, Corcoran LM, Alexander WS, Cory S, Palmiter RD, Brinster RL. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature. 1985;318:533–538. doi: 10.1038/318533a0. [DOI] [PubMed] [Google Scholar]
- Blackshear PJ. Tristetraprolin and other CCCH tandem zinc-finger proteins in the regulation of mRNA turnover. Biochem Soc Trans. 2002;30:945–952. doi: 10.1042/bst0300945. [DOI] [PubMed] [Google Scholar]
- Bodrug SE, Warner BJ, Bath ML, Lindeman GJ, Harris AW, Adams JM. Cyclin D1 transgene impedes lymphocyte maturation and collaborates in lymphomagenesis with the myc gene. EMBO J. 1994;13:2124–2130. doi: 10.1002/j.1460-2075.1994.tb06488.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan CM, Steitz JA. HuR and mRNA stability. Cell Mol Life Sci. 2001;58:266–277. doi: 10.1007/PL00000854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan SE, Kuwano Y, Alkharouf N, Blackshear PJ, Gorospe M, Wilson GM. The mRNA-destabilizing protein tristetraprolin is suppressed in many cancers, altering tumorigenic phenotypes and patient prognosis. Cancer Res. 2009;69:5168–5176. doi: 10.1158/0008-5472.CAN-08-4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carballo E, Lai WS, Blackshear PJ. Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science. 1998;281:1001–1005. doi: 10.1126/science.281.5379.1001. [DOI] [PubMed] [Google Scholar]
- Carballo E, Blackshear PJ. Roles of tumor necrosis factor-alpha receptor subtypes in the pathogenesis of the tristetraprolin-deficiency syndrome. Blood. 2001;98:2389–2395. doi: 10.1182/blood.v98.8.2389. [DOI] [PubMed] [Google Scholar]
- Chen CY, Shyu AB. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- Chen K, Tu Y, Zhang Y, Blair HC, Zhang L, Wu C. PINCH-1 regulates the ERK-Bim pathway and contributes to apoptosis resistance in cancer cells. J Biol Chem. 2008;283:2508–2517. doi: 10.1074/jbc.M707307200. [DOI] [PubMed] [Google Scholar]
- Dalla-Favera R, Bregni M, Erikson J, Patterson D, Gallo RC, Croce CM. Human cmyc onc gene is located on the region of chromosome 8 that is translocated in Burkitt lymphoma cells. Proc Natl Acad Sci U S A. 1982;79:7824–7827. doi: 10.1073/pnas.79.24.7824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CV. c-Myc target genes involved in cell growth, apoptosis and metabolism. Mol Cell Biol. 1999;19:1–11. doi: 10.1128/mcb.19.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CV, O'Donnell KA, Zeller KI, Nguyen T, Osthus RC, Li F. The c-Myc target gene network. Semin Cancer Biol. 2006;16:253–264. doi: 10.1016/j.semcancer.2006.07.014. [DOI] [PubMed] [Google Scholar]
- Dean JL, Sully G, Wait R, Rawlinson L, Clark AR, Saklatvala J. Identification of a novel AU-rich-element-binding protein which is related to AUF1. Biochem J. 2002;366:709–719. doi: 10.1042/BJ20020402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon DA, Tolley ND, King PH, Nabors LB, McIntyre TM, Zimmerman GA, Prescott SM. Altered expression of the mRNA stability factor HuR promotes cyclooxygenase-2 expression in colon cancer cells. J Clin Invest. 2001;108:1657–1665. doi: 10.1172/JCI12973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eischen CM, Weber JD, Roussel MF, Sherr CJ, Cleveland JL. Disruption of the ARF-Mdm2-p53 tumor suppressor pathway in Myc-induced lymphomagenesis. Genes Dev. 1999;13:2658–2669. doi: 10.1101/gad.13.20.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawal M, Armstrong F, Ollier S, Dupont H, Touriol C, Monsarrat B, Delsol G, Payrastre B, Morello D. A "liaison dangereuse" between AUF1/hnRNPD and the oncogenic tyrosine kinase NPM-ALK. Blood. 2006;108:2780–2788. doi: 10.1182/blood-2006-04-014902. [DOI] [PubMed] [Google Scholar]
- Franks TM, Lykke-Andersen J. TTP and BRF proteins nucleate processing body formation to silence mRNAs with AU-rich elements. Genes Dev. 2007;21:719–735. doi: 10.1101/gad.1494707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorrini C, Squatrito M, Luise C, Syed N, Perna D, Wark L, Martinato F, Sardella D, Verrecchia A, Bennett S, et al. Tip60 is a haplo-insufficient tumour suppressor required for an oncogeneinduced DNA damage response. Nature. 2007;448:1063–1067. doi: 10.1038/nature06055. [DOI] [PubMed] [Google Scholar]
- Gouble A, Grazide S, Meggetto F, Mercier P, Delsol G, Morello D. A new player in oncogenesis: AUF1/hnRNPD overexpression leads to tumorigenesis in transgenic mice. Cancer Res. 2002;62:1489–1495. [PubMed] [Google Scholar]
- Halees AS, El-Badrawi R, Khabar KS. ARED Organism: expansion of ARED reveals AUrich element cluster variations between human and mouse. Nucleic Acids Res. 2008;36:D137–D140. doi: 10.1093/nar/gkm959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemelaar J, Lelyveld VS, Kessler BM, Ploegh HL. A single protease, Apg4B, is specific for the autophagy-related ubiquitin-like proteins GATE-16, MAP1-LC3, GABARAP, and Apg8L. J Biol Chem. 2003;278:51841–51850. doi: 10.1074/jbc.M308762200. [DOI] [PubMed] [Google Scholar]
- Hodson DJ, Janas ML, Galloway A, Bell SE, Andrews S, Li CM, Pannell R, Siebel CW, MacDonald HR, De Keersmaecker K, et al. Deletion of the RNA-binding proteins ZFP36L1 and ZFP36L2 leads to perturbed thymic development and T lymphoblastic leukemia. Nat Immunol. 2010;11:717–724. doi: 10.1038/ni.1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummel M, Bentink S, Berger H, Klapper W, Wessendorf S, Barth TF, Bernd HW, Cogliatti SB, Dierlamm J, Feller AC, et al. A biologic definition of Burkitt's lymphoma from transcriptional and genomic profiling. N Engl J Med. 2006;354:2419–2430. doi: 10.1056/NEJMoa055351. [DOI] [PubMed] [Google Scholar]
- Katz M, Amit I, Citri A, Shay T, Carvalho S, Lavi S, Milanezi F, Lyass L, Amariglio N, Jacob-Hirsch J, et al. A reciprocal tensin-3-cten switch mediates EGF-driven mammary cell migration. Nat Cell Biol. 2007;9:961–969. doi: 10.1038/ncb1622. [DOI] [PubMed] [Google Scholar]
- Keller UB, Old JB, Dorsey FC, Nilsson JA, Nilsson L, MacLean KH, Chung L, Yang C, Spruck C, Boyd K, et al. Myc targets Cks1 to provoke the suppression of p27Kip1, proliferation and lymphomagenesis. EMBO J. 2007;26:2562–2574. doi: 10.1038/sj.emboj.7601691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai WS, Thompson MJ, Taylor GA, Liu Y, Blackshear PJ. Promoter analysis of Zfp- 36, the mitogen-inducible gene encoding the zinc finger protein tristetraprolin. J Biol Chem. 1995;270:25266–25272. doi: 10.1074/jbc.270.42.25266. [DOI] [PubMed] [Google Scholar]
- Lee HH, Vo MT, Kim HJ, Lee UH, Kim CW, Kim HK, Ko MS, Lee WH, Cha SJ, Min YJ, et al. Stability of the LATS2 tumor suppressor gene is regulated by tristetraprolin. J Biol Chem. 2010;285:17329–17337. doi: 10.1074/jbc.M109.094235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marderosian M, Sharma A, Funk AP, Vartanian R, Masri J, Jo OD, Gera JF. Tristetraprolin regulates Cyclin D1 and c-Myc mRNA stability in response to rapamycin in an Aktdependent manner via p38 MAPK signaling. Oncogene. 2006;25:6277–6290. doi: 10.1038/sj.onc.1209645. [DOI] [PubMed] [Google Scholar]
- Meyer N, Penn LZ. Reflecting on 25 years with MYC. Nat Rev Cancer. 2008;8:976–990. doi: 10.1038/nrc2231. [DOI] [PubMed] [Google Scholar]
- Miyamae T, Marinov AD, Sowders D, Wilson DC, Devlin J, Boudreau R, Robbins P, Hirsch R. Follistatin-like protein-1 is a novel proinflammatory molecule. J Immunol. 2006;177:4758–4762. doi: 10.4049/jimmunol.177.7.4758. [DOI] [PubMed] [Google Scholar]
- Morrow MA, Lee G, Gillis S, Yancopoulos GD, Alt FW. Interleukin-7 induces N-myc and c-myc expression in normal precursor B lymphocytes. Genes Dev. 1992;6:61–70. doi: 10.1101/gad.6.1.61. [DOI] [PubMed] [Google Scholar]
- Nilsson JA, Keller UB, Baudino TA, Yang C, Norton S, Old JA, Nilsson LM, Neale G, Kramer DL, Porter CW, et al. Targeting ornithine decarboxylase in Myc-induced lymphomagenesis prevents tumor formation. Cancer Cell. 2005;7:433–444. doi: 10.1016/j.ccr.2005.03.036. [DOI] [PubMed] [Google Scholar]
- Painter MW, Davis S, Hardy RR, Mathis D, Benoist C Immunological Genome Project Consortium. Transcriptomes of the B and T lineages compared by multiplatform microarray profiling. J Immunol. 2011;186:3047–3057. doi: 10.4049/jimmunol.1002695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajic A, Spitkovsky D, Christoph B, Kempkes B, Schuhmacher M, Staege MS, Brielmeier M, Ellwart J, Kohlhuber F, Bornkamm GW, et al. Cell cycle activation by c-myc in a burkitt lymphoma model cell line. Int J Cancer. 2000;87:787–793. doi: 10.1002/1097-0215(20000915)87:6<787::aid-ijc4>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- Peters G. The D-type cyclins and their role in tumorigenesis. J Cell Sci Suppl. 1994;18:89–96. doi: 10.1242/jcs.1994.supplement_18.13. [DOI] [PubMed] [Google Scholar]
- Rosenbloom KR, Dreszer TR, Pheasant M, Barber GP, Meyer LR, Pohl A, Raney BJ, Wang T, Hinrichs AS, Zweig AS, et al. ENCODE whole-genome data in the UCSC Genome Browser. Nucleic Acids Res. 2010;38:D620–D625. doi: 10.1093/nar/gkp961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai T, Liu L, Teng X, Mukai-Sakai R, Shimada H, Kaji R, Mitani T, Matsumoto M, Toida K, Ishimura K, et al. Nucling recruits Apaf-1/pro-caspase-9 complex for the induction of stressinduced apoptosis. J Biol Chem. 2004;279:41131–41140. doi: 10.1074/jbc.M402902200. [DOI] [PubMed] [Google Scholar]
- Sanduja S, Kaza V, Dixon DA. The mRNA decay factor tristetraprolin (TTP) induces senescence in human papillomavirus-transformed cervical cancer cells by targeting E6-AP ubiquitin ligase. Aging (Albany NY) 2009;1:803–817. doi: 10.18632/aging.100086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta TK, Bandyopadhyay S, Fernandes DJ, Spicer EK. Identification of nucleolin as an AU-rich element binding protein involved in bcl-2 mRNA stabilization. J Biol Chem. 2004;279:10855–10863. doi: 10.1074/jbc.M309111200. [DOI] [PubMed] [Google Scholar]
- Seoane J, Pouponnot C, Staller P, Schader M, Eilers M, Massague J. TGFbeta influences Myc, Miz-1 and Smad to control the CDK inhibitor p15INK4b. Nat Cell Biol. 2001;3:400–408. doi: 10.1038/35070086. [DOI] [PubMed] [Google Scholar]
- Staller P, Peukert K, Kiermaier A, Seoane J, Lukas J, Karsunky H, Moroy T, Bartek J, Massague J, Hanel F, et al. Repression of p15INK4b expression by Myc through association with Miz-1. Nat Cell Biol. 2001;3:392–399. doi: 10.1038/35070076. [DOI] [PubMed] [Google Scholar]
- Stoecklin G, Gross B, Ming XF, Moroni C. A novel mechanism of tumor suppression by destabilizing AU-rich growth factor mRNA. Oncogene. 2003;22:3554–3561. doi: 10.1038/sj.onc.1206418. [DOI] [PubMed] [Google Scholar]
- Tabrizifard S, Olaru A, Plotkin J, Fallahi-Sichani M, Livak F, Petrie HT. Analysis of transcription factor expression during discrete stages of postnatal thymocyte differentiation. J Immunol. 2004;173:1094–1102. doi: 10.4049/jimmunol.173.2.1094. [DOI] [PubMed] [Google Scholar]
- Taub R, Kirsch I, Morton C, Lenoir G, Swan D, Tronick S, Aaronson S, Leder P. Translocation of the c-myc gene into the immunoglobulin heavy chain locus in human Burkitt lymphoma and murine plasmacytoma cells. Proc Natl Acad Sci U S A. 1982;79:7837–7841. doi: 10.1073/pnas.79.24.7837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor GA, Carballo E, Lee DM, Lai WS, Thompson MJ, Patel DD, Schenkman DI, Gilkeson GS, Broxmeyer HE, Haynes BF, et al. A pathogenetic role for TNF alpha in the syndrome of cachexia, arthritis, and autoimmunity resulting from tristetraprolin (TTP) deficiency. Immunity. 1996;4:445–454. doi: 10.1016/s1074-7613(00)80411-2. [DOI] [PubMed] [Google Scholar]
- Tobias ES, Hurlstone AF, MacKenzie E, McFarlane R, Black DM. The TES gene at 7q31.1 is methylated in tumours and encodes a novel growth-suppressing LIM domain protein. Oncogene. 2001;20:2844–2853. doi: 10.1038/sj.onc.1204433. [DOI] [PubMed] [Google Scholar]
- Wang Q, Diskin S, Rappaport E, Attiyeh E, Mosse Y, Shue D, Seiser E, Jagannathan J, Shusterman S, Bansal M, et al. Integrative genomics identifies distinct molecular classes of neuroblastoma and shows that multiple genes are targeted by regional alterations in DNA copy number. Cancer Res. 2006;66:6050–6062. doi: 10.1158/0008-5472.CAN-05-4618. [DOI] [PubMed] [Google Scholar]
- Zeller KI, Jegga AG, Aronow BJ, O'Donnell KA, Dang CV. An integrated database of genes responsive to the Myc oncogenic transcription factor: identification of direct genomic targets. Genome Biol. 2003;4:R69. doi: 10.1186/gb-2003-4-10-r69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeller KI, Zhao X, Lee CW, Chiu KP, Yao F, Yustein JT, Ooi HS, Orlov YL, Shahab A, Yong HC, et al. Global mapping of c-Myc binding sites and target gene networks in human B cells. Proc Natl Acad Sci U S A. 2006;103:17834–17839. doi: 10.1073/pnas.0604129103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.