Abstract
Cancer stem cells (CSCs) have been identified in an ever-increasing number of human malignancies based on the ability to recapitulate tumors in the ectopic setting and maintain long-term tumorigenic potential. In pancreatic adenocarcinoma, CSCs may also display additional properties, such as relative drug resistance and enhanced invasive and migratory potential that implicate a role in disease pathogenesis spanning initial tumor formation to metastatic disease progression. Importantly, these findings also suggest that the development of novel therapeutic strategies capable of inhibiting or eliminating CSCs will improve clinical outcomes. Preclinical studies have already described a wide array of potential targeting approaches that target CSC-specific surface antigens and cellular pathways involved in cell survival, adhesion, self-renewal and differentiation. Furthermore, progress in this area should continue to move forward as the unique biology of CSCs is better understood. All preclinical studies to date have focused on targeting specific and phenotypically defined CSCs, but multiple cell populations with the ability to form tumors and self-renew have been identified in pancreatic carcinoma. Since the clinical efficacy of CSC-directed therapies will depend on the inhibition of all sources of tumor self-renewal, better understanding how specific CSC populations are related to one another and whether each possesses specific functional properties will be critical. In this review, we will discuss the potential relationships between different pancreatic CSC populations and strategies to identify novel targeting approaches.
Keywords: Pancreatic cancer, cancer stem cells, tumor heterogeneity, CD44, ALDH
Introduction
Pancreatic ductal adenocarcinoma (PDAC) carries one of the worst prognoses of any malignancy and is the fourth leading cause of cancer related deaths in the United States (1). Despite advances in better understanding the basic biology of PDAC, survival rates have not significantly improved over the past 30 years, and less than five percent of patients remain alive 5-years after diagnosis. Therefore, new treatments are needed for PDAC, and cancer stem cells (CSCs) have emerged as potential targets.
CSCs represent phenotypically distinct cells that possess enhanced tumor-initiating potential, self-renewal, and the ability to recapitulate the cellular heterogeneity of the original tumor (2). Since these initial findings, additional features including their rarity, relative chemoresistance, and metastatic potential have been described, and these properties have allowed them to be referred to by more precise terms, such as tumor initiating cells. However, due to the heterogeneous properties exhibited by CSCs, it has been difficult to provide a label capable of encompassing all of these attributes. Therefore, we will refer to these specialized cell populations by the general term CSC throughout this review. Although the identification of CSCs was limited to myeloid leukemias in the 1990s (3, 4), they have been described in an increasing number of solid tumors over the last decade, including multiple reports in PDAC (5–7). Several aspects of the CSC hypothesis have been hotly debated (8–10), but most relevant is their clinical significance. In PDAC, early data have suggested that the identification of CSCs in primary tumors is associated with shorter overall survival (6), and it is likely that additional functional properties including relative resistance to the standard cytotoxic agent gemcitabine and enhanced metastatic potential are in part responsible for these findings (7, 11).
The identification and characterization of CSCs has generated novel hypotheses regarding the mechanisms involved in PDAC growth and dissemination, but several critical questions remain. We will initially review studies identifying pancreatic CSCs and speculate how these distinct cell populations may be related to one another. We will then discuss potential strategies to target pancreatic CSCs.
Identification of Pancreatic CSCs
At the most basic level, the CSC hypothesis links phenotypically defined tumor cells with specific functional properties, and CSCs have been stringently defined by their ability to differentiate and self-renew (12). The differentiation of CSCs gives rise to the full range of malignant cell types and histological recapitulation of the original tumor whereas self-renewal is responsible for maintaining long-term growth potential. In most diseases, the ability of putative CSCs to form tumors has been evaluated using immunodeficient mice (e.g., NOD/Scid and NSG) followed by histological examination and serial transplantation to demonstrate self-renewal (13). Although these mouse models remain the gold standard to evaluate CSCs, in vitro assays have also been developed to assess the clonogenic potential of CSCs including colony formation in semi-solid media or tumor sphere formation in liquid culture. Moreover, these in vitro assays may quantify the number of cells with self-renewal and long-term growth potential through serial rounds of plating.
Candidate CSC markers have largely consisted of differentially expressed cell surface antigens or drug resistance pathways. One approach to identify novel CSC populations has been the use of surface antigens expressed by normal stem cells in the tissue of origin, such as CD34 in myeloid leukemias (3, 14). Alternatively, antigens or enzymes capable of identifying normal stem cells in multiple tissues, such as CD133 and Aldehyde dehydrogenase (ALDH), have also been used to isolate CSCs in several diseases (15–19). Finally, specific antigens associated with poor prognosis, such as CD44 or c-Met, have also served as CSC markers (5, 20–22).
The initial identification of pancreatic CSCs extended ground-breaking work in breast cancer and investigated the expression of CD44, CD24, and epithelial specific antigen (ESA) (Table 1) (5). Relative to unsorted cells, CD44+CD24+ESA+ cells isolated from low-passage PDAC xenografts were highly tumorigenic and recapitulated the histology and cellular heterogeneity of the original tumor. Furthermore, the functional differences between CD44+CD24+ESA+ and CD44−CD24−ESA− cells were maintained following subcutaneous or orthotopic injection suggesting that tumorigenic potential was cell autonomous and independent of local environmental factors. A second report demonstrated that CD133 could also identify pancreatic CSCs (7). In addition to being highly tumorigenic, CD133+ pancreatic cancer cells were found to be relatively resistant to gemcitabine treatment compared to CD133− cells.
Table 1.
Phenotype and functional properties of pancreatic CSC populations.
| Study | Phenotype | Properties |
|---|---|---|
| Li et al.(5) | CD44+CD24+ESA+ | Increased Sonic Hedgehog expression. |
| Hermann et al.(7) | CD133+ | Chemoresistant. CD133+CXCR4+ cells responsible for metastasis. |
| Rasheed et al.(6) Ishizawa et al.(25) |
ALDH+ | CSCs associated with overall survival. CSCs exhibit mesenchymal feature and are frequently found in metastatic lesions. ALDH+ and CD44+CD24+ cells are equally tumorigenic. |
| Li et al.(22) | CD44+c-Met+ | Highly metastatic. |
Cellular markers associated with drug resistance have also been used to identify CSCs. ALDH, specifically ALDH1A1, is required for the synthesis of all-trans-retinoic and high enzyme activity marks normal mouse pancreatic progenitor cells and normal human stem cells in several organ systems (23, 24). ALDH may also play a role in drug resistance as it can metabolize and neutralize cytotoxic alkylators, such as cyclophosphamide. We studied ALDH in PDAC and found that ALDH+ cells are highly tumorigenic compared to bulk tumor cells (6, 25). Moreover, ALDH+ cells appear to be relatively resistant to gemcitabine in vivo and have increased invasive potential suggesting a role in disease progression (6, 11).
Despite the importance of CD44, CD133, and ALDH in identifying pancreatic CSCs, it is unclear whether these antigens are involved in regulating CSC function or merely serve as phenotypic markers. However, other pancreatic CSC markers have been identified that may be functionally relevant. For example, CXCR4 serves as the chemokine receptor for Stromal cell-derived factor-1 (SDF-1, CXCL12) and is expressed by a subset of CD133+ CSCs that have enhanced metastatic capacity (7). Recent studies have also demonstrated that c-Met can identify and regulate pancreatic CSCs similar to findings in glioblastoma (22, 26). Thus, several strategies have been used to identify pancreatic CSCs, and some of these may provide insights into regulatory factors and potential targeting strategies.
The relationship between distinct pancreatic CSC populations
In most normal organ systems, such as the blood, CNS, and skin, cells are functionally and phenotypically organized according to a strict cellular hierarchy in which self-renewing stem cells give rise to short lived progenitors then terminally differentiated effector cells. The earliest studies in acute myeloid leukemia (AML) demonstrated that tumor cells resembling normal hematopoietic stem cells can self-renew and give rise to relatively differentiated and non-tumorigenic blasts (4). Therefore, it has been generally assumed that cancers are organized in a hierarchical manner similar to normal tissues. However, several CSC populations have been identified in PDAC, and it is not clear how each of these fits into a specific hierarchy or are related to one another. One possibility is that all of the current markers recognize the same cell, but the vast majority of ALDH+ pancreatic tumor cells appear to lack CD44 and CD133. Therefore, it is likely that these antigens identify at least two, or even three, unique cell populations (6, 27). Alternatively, since each putative CSC marker enriches for cells with increased tumorigenic potential but fails to isolate pure populations of CSCs (i.e., every cell expressing a specific marker is not tumorigenic), it is possible that combining antigens will greatly increase CSC purity. However, this does not appear to be the case as the tumor initiating cell frequency of rare PDAC cells co-expressing CD44, CD24, and ALDH is not significantly greater than either ALDH+ or CD44+CD24+ cells (25). Moreover, c-Met is expressed, albeit at variable levels, on CD44+, CD133+, or ALDH+ cells, but increased tumorigenic potential is limited to CD44+c-Methigh cells (22).
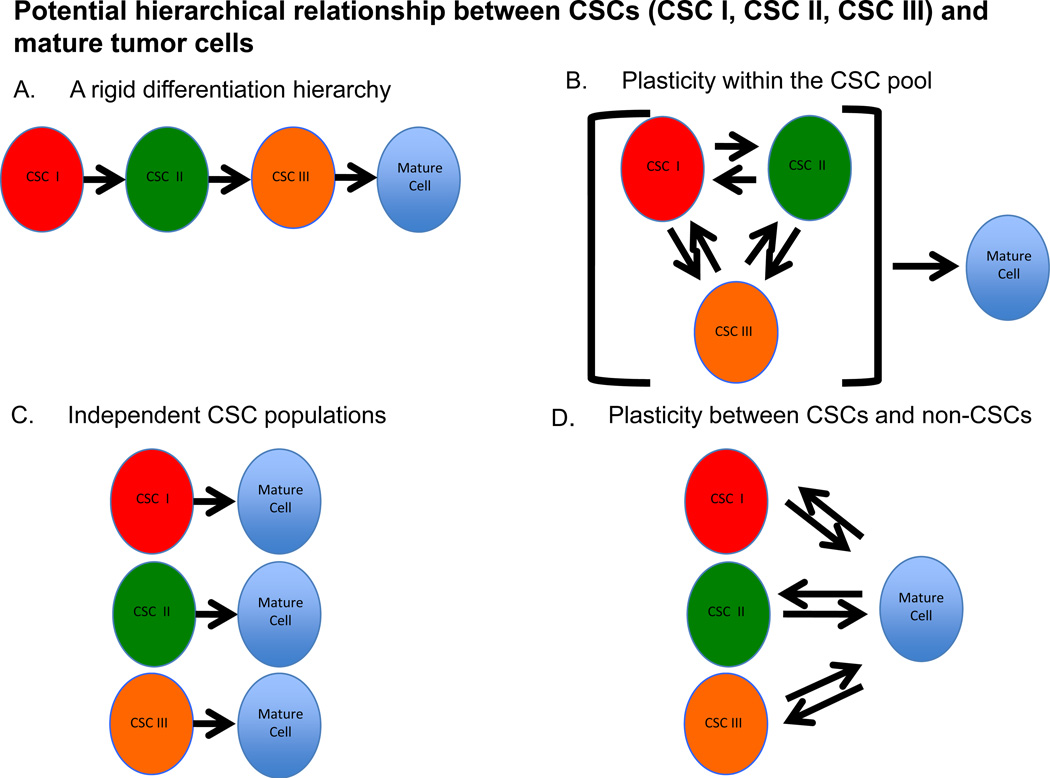
The significance of the various pancreatic CSC markers and the cells they identify clearly requires further clarification. If multiple CSC populations actually exist, an understanding of how they are related to one another will be important since clinically effective targeting likely requires the elimination of all self-renewing cells within the tumor. One possibility is that PDAC cells are organized in a hierarchical and linear manner with a single, phenotypically distinct CSC at the apex giving rise to the other CSC populations and ultimately non-clonogenic mature tumor cells (Figure 1). It is also possible that each phenotypically distinct CSC population represents a specific cellular state of the same clonogenic cell that gives rise to mature tumor cells. Another possibility is that each CSC population is unrelated to another and parallel lines of mature tumor cell production exist. Finally, it is conceivable that a rigid hierarchy of unidirectional differentiation does not exist, but that the system is plastic with non-clonogenic cells giving rise to tumorigenic CSCs displaying a variety of phenotypes. In order to better understand how different CSCs are related to one another, studies examining the overlap between putative CSC populations and the cell types that arise from each specific CSC are needed.
Figure 1. Potential relationships between CSCs and mature tumor cells.
(A) A linear organization with a single phenotypically distinct CSC giving rise to the other CSC populations and ultimately non-clonogenic mature cells. (B) Each phenotypic CSC represents a distinct state of the same clonogenic cell that gives rise to the mature tumor cell. (C) Each CSC population is unrelated to another and parallel lines of mature tumor cell production exist. (D) A plastic system in which non-clonogenic mature cells give rise to CSC displaying a variety of phenotypes.
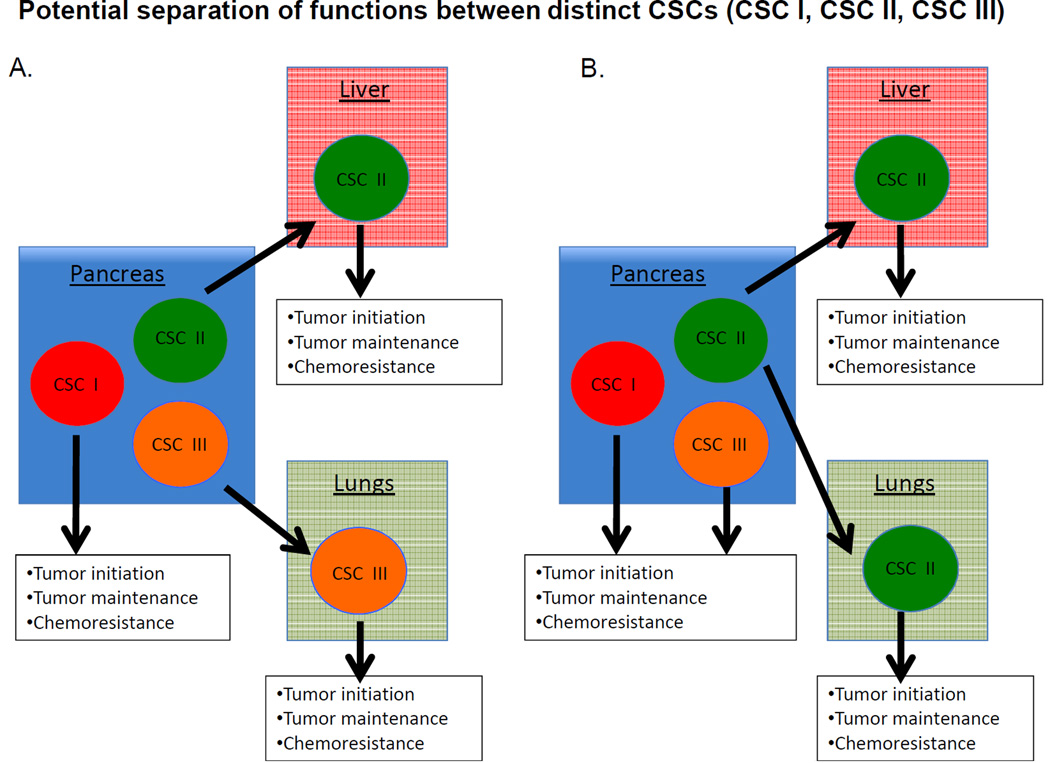
Beyond the organization of phenotypically defined CSC populations, it is also unclear whether the various CSCs are functionally similar or distinct. Although tumor formation, histologic recapitulation, and self-renewal define CSCs, other properties, including relative drug resistance, invasion, migration, and metastatic potential have been ascribed to CSCs and may contribute to their clinical impact (28). It is possible that certain CSC populations could be primarily responsible for tumor initiation and maintenance at the primary site of disease, whereas others could be responsible for tumor dissemination and growth at metastatic sites, such as the subpopulation of CD133+ CSCs expressing CXCR4 (7). It is also possible different organs, such as the liver and lung, harbor different microenvironments with distinct endothelial or stromal cell types or extracellular matrix components that promote or inhibit tumor growth (Figure 2) (29). Therefore, if metastatic dissemination depends on the interaction of CSCs with a particular niche, then different niches might call for unique CSCs. An evaluation of the tumor forming potential of specific CSCs at orthotropic and different metastatic sites may determine whether certain populations are better suited to grow within particular locations.
Figure 2. Potential functional relationships between CSCs.
(A) Distinct CSCs may give rise to macroscopic tumors in distinct anatomical locations. Each population is responsible for tumor growth and resistance to therapy in different organs. (B) A primary CSC population is responsible for tumor initiation and growth at the primary site. Additional populations are responsible for initiation and maintenance at metastatic sites and for resistance to chemotherapy.
It is also possible that the phenotypes exhibited by CSCs are dictated by the external microenvironment. For example, pancreatic tumors are characterized by desmoplasia and dense fibrosis that may expose cells to relative hypoxia, and the hypoxic state has been found to alter the expression of the CSC marker CD133 in brain tumors (30). In addition, several markers used to identify CSCs, such as ALDH and the side population assay, are indicative of drug resistance mechanisms and it is possible that their expression is induced in response to cellular damage. Finally, it is possible that the adaptive metabolic changes undertaken by tumor cells also modifies the expression of CSC makers, although such findings have yet to be reported (31).
Recent studies have demonstrated a clear link between CSCs and the epithelial-to-mesenchymal transition (EMT) in solid tumors. Therefore, it is possible that CSCs represent a specific cellular state expressing multiple phenotypes. Reports using breast cancer models have demonstrated that the induction of EMT by TGF-β or the modulation of specific gene expression (e.g., induction of Twist or repression of E-Cadherin) results in increased expression of CD44 and tumorigenic potential (32, 33). In pancreatic cancer, ALDH+ cells appear have a gene expression profile consistent with EMT and increased invasive and migratory potential compared to bulk tumor cells and CD44+CD24+ cells (6). Moreover, studies examining ZEB1, an inducer of EMT, in pancreatic cancer cells have identified a direct link between EMT, increased tumorigenicity, and drug resistance (34). Therefore, it is possible that a more “epithelial” or “mesenchymal” state is important in determining the functional properties of CSCs. The specific functional properties of different pancreatic CSCs are unclear, and the quantification of tumor formation, metastatic potential, and drug resistance is needed.
Inter- and intra-patient diversity of pancreatic CSCs
Inter-patient heterogeneity may also contribute to the existence of multiple pancreatic CSCs. Recurrent genetic alterations are a hallmark of cancer, and mutations in KRas are present in the vast majority of PDAC (35, 36). On the other hand, mutations in other genes, such as p53 and Smad4/DPC4, can be identified in some, but not all tumors (37, 38). Therefore, pancreatic cancers are not genetically homogeneous, but vary from patient to patient (39). If alterations in specific genes are prognostic and CSCs truly dictate the natural history of PDAC given their potential roles in tumor formation, drug resistance, and metastatic progression, it is likely that specific mutations influence both the phenotype and function of CSCs. Currently, it is unclear whether phenotypically identical CSCs from different patients have the same functional attributes and contribute to disease progression in similar ways. However, such a finding would imply that personalized and individualized CSC targeting therapies are needed. In order to examine inter-patient diversity, the functional properties of different CSCs derived from human tumors with distinct genotypes will need to be determined. In addition, the examination of CSC phenotypes and functional properties in tumors derived from transgenic animal models of pancreatic cancer may be particularly helpful since specific genetic lesions can be modulated in these systems (40).
To further complicate matters, increasing evidence suggest that human cancers can be genetically heterogeneous within the same individual (41–44). Therefore, intra-patient genetic heterogeneity may also drive the phenotypic and functional diversification CSCs. In many cancers, including PDAC, specific genetic alternations may accumulate in an orderly fashion during disease progression (45, 46), thus, it is also possible that different CSCs are responsible for relapse and progression over the course of the disease. In PDAC, metastatic lesions may be genetically distinct from one another and the primary tumor (47). Moreover, primary tumors are composed of geographically and genetically distinct subclones. The role of genetic evolution and diversification in the emergence of distinct CSCs, or conversely, the impact of CSCs on the clonal composition of an individual tumor is not entirely clear, but it is likely that these two processes interact at some level. A systematic investigation of genetic lesions within CSCs, their phenotypes, and functional properties, such as tumorigenic potential, metastasis, and drug resistance, within primary tumors and metastatic lesions derived from the same patient may address this possibility.
Targeting of Pancreatic CSCs
The self-renewal potential and resistance to traditional cytotoxic agents suggest that successful CSC targeting strategies will improve clinical outcomes. One potential approach is targeting the cell surface antigens that characterize pancreatic CSCs using monoclonal antibodies. For example, a bi-specific antibody recognizing both ESA and CD3 has been found to eliminate pancreatic CSCs by redirecting cytotoxic T-lymphocytes (48). CD44 is another surface protein expressed by CSCs in multiple diseases (49), and a specific monoclonal antibody against CD44 can eliminate AML stem cells by inducing terminal differentiation (50). The functional activities of specific pancreatic CSC markers may also serve as potential targets (Table 2). The hepatocyte growth factor (HGF) receptor c-Met identifies highly tumorigenic CSCs in combination with CD44, and the pharmacological inhibition of its activity has been found to inhibit tumor growth and metastasis (22). Another functionally relevant marker is CXCR4 that plays an important role in the homing of hematopoietic stem cells to the bone marrow. CXCR4 has been identified on a subset of CD133+ pancreatic CSCs with enhanced metastatic capacity, and CXCR4 antagonists may prevent tumor dissemination (7). Another potential cell surface target is Death receptor 5 (DR5) that induces apoptosis following binding to TRAIL. A recent study found that ALDH+ and CD44+CD24+ pancreatic CSCs express relatively increased levels of DR5, and receptor engagement using an agonistic monoclonal antibody markedly reduced CSC frequency and tumor growth in vivo (51).
Table 2.
Pancreatic CSC specific targeting strategies and agents
| Study | Target receptor/pathway |
Target population | Agent |
|---|---|---|---|
| Li et al.(22) | c-Met | c-MethighCD44+ CD44+CD24+ESA+ |
XL184 |
| Rajeshkumar et al.(51) | DR5 | ALDH+ CD44+CD24+ESA+ |
DR5 Agonistic monoclonal antibody |
| Lonardo et al.(52) | ALK4 | CD133+ | SB431542 |
| Jimeno et al.(11) Feldmann et al.(54) Feldmann et al.(53) |
Hedgehog | ALDH+ CD44+CD24+ESA+ |
Cyclopamine, IPI269609 |
| Mullendore et al.(57) | Notch | ALDH+ | GSI-18 |
| Zhang et al.(58) | EMT | CD133+ | Salinomycin |
Several cellular signaling pathways have been identified that regulate the self-renewal of normal stem cells and may serve as targets against CSCs. These include pathways required for normal embryonic development, and the Hedgehog (Hh), Notch, and Nodal/Activin pathways may be active in pancreatic CSCs. Nodal and Activin are ligands of the TGF-β superfamily, and a recent study demonstrated that these ligands and their receptor ALK4 are overexpressed in pancreatic CSCs (52). The pharmacological inhibition or knockdown of ALK4 abrogated self-renewal and tumorigenicity as well as sensitized CSCs to gemcitabine. Another series of studies has examined the Hh signaling pathway in pancreatic CSCs and found that pharmacological pathway inhibition reduced the frequency of CSCs and decreased tumor formation and metastasis (11, 53, 54). Of note, a recent phase 2 clinical trial compared gemcitabine alone or in combination with the novel Hh inhibitor saridegib (IPI-926) in patients with metastatic pancreatic cancer based on preclinical data demonstrating enhanced responses to cytotoxic chemotherapy (55). A higher rate of progressive disease was observed at an interim analysis in patients receiving saridegib (56). Although the precise reasons for these results are unclear, it is possible that the Hh pathway regulates the development, rather than maintenance, of metastatic lesions and other ongoing trials of Hh inhibitors in the neoadjuvant setting may provide a better scenario to detect these potential anti-CSC effects. Finally, the inhibition of Notch signaling has been found to inhibit EMT and cellular invasion as well as decrease the frequency of ALDH+ CSCs (57). Thus, cellular pathways involved in regulating the self-renewal of normal stem cells may represent pancreatic CSC targets.
The association of EMT and CSCs may also form the basis for identifying novel targeting agents. High-throughput strategies to screen for novel anti-CSC compounds have been difficult to carry out because of the lack of pure CSC populations and the complex nature of the assays used to assess their functions, but several methods may induce EMT and increase the frequency of CSCs. This approach was ingeniously used by Gupta et al. who genetically engineered human breast cancer cell lines to induce EMT and screened for compounds that could induce cell death (32). The ionophore salinomycin was identified as a potential CSC targeting agent then subsequently found to block tumor formation and metastasis in vivo. Shortly thereafter, salinomycin was shown to inhibit the growth of pancreatic CSCs, indicating that it may represent a potential CSC targeting agent in multiple malignancies (58). Therefore, similar strategies based on EMT may identify novel agents that inhibit pancreatic CSCs.
Conclusion
The CSC hypothesis may provide novel insights into the pathogenesis of PDAC and the mechanisms that regulate clinical chemoresistance and the propensity to develop metastatic disease. Moreover, it may lead to a better understanding of self-renewal that allows tumors to persist over time and the development of novel therapeutic strategies targeting the regulatory pathways involved. With the increased recognition of cellular heterogeneity in PDAC and identification of tumor cells with enhanced tumorigenic potential and self-renewal, questions have emerged regarding the diversity of pancreatic CSCs and their relationships to one another that will need to be addressed. However, these findings are likely to provide a framework to better understand advancements in many of the other fields that are the subject of this Series, including alterations in genetics, tumor metabolism, and the microenvironment. Ultimately, merging these distinct aspects of PDAC biology may provide the basis for truly novel and effective therapies that positively impact clinical outcomes.
Acknowledgments
Financial support: Financial support was provided by research grants from the NIH (R01 CA150142, R01 CA113669, R01 CA134767, P01CA134292) and the Lustgarten Foundation. Z.A.R. is a recipient of the Pancreatic Cancer Action Network-AACR Career Development Award.
Footnotes
Conflict of interest: The authors report no conflicts of interest pertaining to this publication.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 3.Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3:730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- 4.Lapidot T, Sirard C, Vormoor J, Murdoch B, Hoang T, Caceres-Cortes J, et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367:645–648. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- 5.Li C, Heidt DG, Dalerba P, Burant CF, Zhang L, Adsay V, et al. Identification of pancreatic cancer stem cells. Cancer Res. 2007;67:1030–1037. doi: 10.1158/0008-5472.CAN-06-2030. [DOI] [PubMed] [Google Scholar]
- 6.Rasheed ZA, Yang J, Wang Q, Kowalski J, Freed I, Murter C, et al. Prognostic significance of tumorigenic cells with mesenchymal features in pancreatic adenocarcinoma. J Natl Cancer Inst. 2010;102:340–351. doi: 10.1093/jnci/djp535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hermann PC, Huber SL, Herrler T, Aicher A, Ellwart JW, Guba M, et al. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell. 2007;1:313–323. doi: 10.1016/j.stem.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 8.Kern SE, Shibata D. The Fuzzy Math of Solid Tumor Stem Cells: A Perspective. Cancer Res. 2007;67:8985–8988. doi: 10.1158/0008-5472.CAN-07-1971. [DOI] [PubMed] [Google Scholar]
- 9.Adams JM, Strasser A. Is Tumor Growth Sustained by Rare Cancer Stem Cells or Dominant Clones? Cancer Res. 2008;68:4018–4021. doi: 10.1158/0008-5472.CAN-07-6334. [DOI] [PubMed] [Google Scholar]
- 10.Tomasson MH. Cancer stem cells: A guide for skeptics. J Cell Biochem. 2009;106:745–749. doi: 10.1002/jcb.22050. [DOI] [PubMed] [Google Scholar]
- 11.Jimeno A, Feldmann G, Suárez-Gauthier A, Rasheed Z, Solomon A, Zou G-M. A direct pancreatic cancer xenograft model as a platform for cancer stem cell therapeutic development. Mol Cancer Ther. 2009;8:310–314. doi: 10.1158/1535-7163.MCT-08-0924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clarke MF, Dick JE, Dirks PB, Eaves CJ, Jamieson CHM, Jones DL, et al. Cancer stem cells--perspectives on current status and future directions: AACR Workshop on cancer stem cells. Cancer Res. 2006;66:9339–9344. doi: 10.1158/0008-5472.CAN-06-3126. [DOI] [PubMed] [Google Scholar]
- 13.Brennan S, Matsui W. Cancer stem cells: controversies in multiple myeloma. J Mol Med. 2009 doi: 10.1007/s00109-009-0531-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lapidot T, Fajerman Y, Kollet O. Immune-deficient SCID and NOD/SCID mice models as functional assays for studying normal and malignant human hematopoiesis. Journal of Molecular Medicine. 1997;75:664–673. doi: 10.1007/s001090050150. [DOI] [PubMed] [Google Scholar]
- 15.Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, et al. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 16.O'Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445:106–110. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- 17.Matsui W, Wang Q, Barber JP, Brennan S, Smith BD, Borrello I, et al. Clonogenic multiple myeloma progenitors, stem cell properties, and drug resistance. Cancer Res. 2008;68:190–197. doi: 10.1158/0008-5472.CAN-07-3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ginestier C, Hur MH, Charafe-Jauffret E, Monville F, Dutcher J, Brown M, et al. ALDH1 Is a Marker of Normal and Malignant Human Mammary Stem Cells and a Predictor of Poor Clinical Outcome. Cell Stem Cell. 2007;1:555–567. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones RJ, Gocke CD, Kasamon YL, Miller CB, Perkins B, Barber JP, et al. Circulating clonotypic B cells in classic Hodgkin lymphoma. Blood. 2009;113:5920–5926. doi: 10.1182/blood-2008-11-189688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dalerba P, Dylla SJ, Park I-K, Liu R, Wang X, Cho RW, et al. Phenotypic characterization of human colorectal cancer stem cells. Proc Natl Acad Sci USA. 2007;104:10158–10163. doi: 10.1073/pnas.0703478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li C, Wu JÄ, Hynes M, Dosch J, Sarkar B, Welling TH, et al. c-Met Is a Marker of Pancreatic Cancer Stem Cells and Therapeutic Target. Gastroenterology. 2011;141:2218.e5–2227.e5. doi: 10.1053/j.gastro.2011.08.009. [DOI] [PubMed] [Google Scholar]
- 23.Emadi A, Jones RJ, Brodsky RA. Cyclophosphamide and cancer: golden anniversary. Nat Rev Clin Oncol. 2009;6:638–647. doi: 10.1038/nrclinonc.2009.146. [DOI] [PubMed] [Google Scholar]
- 24.Rovira M, Scott S-G, Liss AS, Jensen J, Thayer SP, Leach SD. Isolation and characterization of centroacinar/terminal ductal progenitor cells in adult mouse pancreas. Proc Natl Acad Sci USA. 2009 doi: 10.1073/pnas.0912589107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ishizawa K, Rasheed ZA, Karisch R, Wang Q, Kowalski J, Susky E, et al. Tumor-initiating cells are rare in many human tumors. Cell Stem Cell. 2010;7:279–282. doi: 10.1016/j.stem.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Y, Li A, Glas M, Lal B, Ying M, Sang Y, et al. c-Met signaling induces a reprogramming network and supports the glioblastoma stem-like phenotype. Proceedings of the National Academy of Sciences. 2011;108:9951–9956. doi: 10.1073/pnas.1016912108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim MP, Fleming JB, Wang H, Abbruzzese JL, Choi W, Kopetz S, et al. ALDH Activity Selectively Defines an Enhanced Tumor-Initiating Cell Population Relative to CD133 Expression in Human Pancreatic Adenocarcinoma. PloS one. 2011;6:e20636. doi: 10.1371/journal.pone.0020636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rasheed ZA, Kowalski J, Smith BD, Matsui W. Concise Review: Emerging Concepts in Clinical Targeting of Cancer Stem Cells. Stem Cells. 2011;29:883–887. doi: 10.1002/stem.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feig C, Gopinathan A, Neesse A, Chan D, Cook N, Tuveson DA. The pancreas cancer microenvironment. Clin Cancer Res. 2012;18 doi: 10.1158/1078-0432.CCR-11-3114. XX-XX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bar EE, Lin A, Mahairaki V, Matsui W, Eberhart CG. Hypoxia increases the expression of stem-cell markers and promotes clonogenicity in glioblastoma neurospheres. American Journal Of Pathology. 2010 doi: 10.2353/ajpath.2010.091021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Le A, Rajeshkumar N, Maitra A, Dang CV. Conceptual framework for cutting the pancreatic cancer fuel supply. Clin Cancer Res. 2012;18 doi: 10.1158/1078-0432.CCR-12-0041. XX-XX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gupta PB, Onder TT, Jiang G, Tao K, Kuperwasser C, Weinberg RA, et al. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell. 2009;138:645–659. doi: 10.1016/j.cell.2009.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mani S, Evans K, Hollier B, Guo W, Weinberg R. Generation of Stem-like Cells via EMT: A New Twist in Cancer Initiation and Progression. AACR Education Book. 2009;2009:173. [Google Scholar]
- 34.Wellner U, Schubert J, Burk UC, Schmalhofer O, Zhu F, Sonntag A, et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol. 2009;11:1487–1495. doi: 10.1038/ncb1998. [DOI] [PubMed] [Google Scholar]
- 35.Maitra A, Adsay NV, Argani P, Iacobuzio-Donahue C, De MA, Cameron JL, et al. Multicomponent analysis of the pancreatic adenocarcinoma progression model using a pancreatic intraepithelial neoplasia tissue microarray. ModPathol. 2003;16:902–912. doi: 10.1097/01.MP.0000086072.56290.FB. [DOI] [PubMed] [Google Scholar]
- 36.Hruban RH, van Mansfeld AD, Offerhaus GJ, van Weering DH, Allison DC, Goodman SN, et al. K-ras oncogene activation in adenocarcinoma of the human pancreas. A study of 82 carcinomas using a combination of mutant-enriched polymerase chain reaction analysis and allele-specific oligonucleotide hybridization. American Journal of Pathology. 1993;143:545–554. [PMC free article] [PubMed] [Google Scholar]
- 37.Redston MS, Caldas C, Seymour AB, Hruban RH, da Costa L, Yeo CJ, et al. p53 Mutations in Pancreatic Carcinoma and Evidence of Common Involvement of Homocopolymer Tracts in DNA Microdeletions. Cancer Res. 1994;54:3025–3033. [PubMed] [Google Scholar]
- 38.Hahn SA, Schutte M, Hoque ATMS, Moskaluk CA, da Costa LT, Rozenblum E, et al. DPC4, A Candidate Tumor Suppressor Gene at Human Chromosome 18q21.1. Science. 1996;271:350–353. doi: 10.1126/science.271.5247.350. [DOI] [PubMed] [Google Scholar]
- 39.Jones S, Zhang X, Parsons DW, Lin JC-H, Leary RJ, Angenendt P, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tuveson DA, Hingorani SR. Ductal Pancreatic Cancer in Humans and Mice. Cold Spring Harbor Symp Quant Biol. 2005;70:65–72. doi: 10.1101/sqb.2005.70.040. [DOI] [PubMed] [Google Scholar]
- 41.Anderson K, Lutz C, van Delft FW, Bateman CM, Guo Y, Colman SM, et al. Genetic variegation of clonal architecture and propagating cells in leukaemia. Nature. 2011;469:356–361. doi: 10.1038/nature09650. [DOI] [PubMed] [Google Scholar]
- 42.Notta F, Mullighan CG, Wang JCY, Poeppl A, Doulatov S, Phillips LA, et al. Evolution of human BCR-ABL1 lymphoblastic leukaemia-initiating cells. Nature. 2011;469:362–367. doi: 10.1038/nature09733. [DOI] [PubMed] [Google Scholar]
- 43.Yachida S, Jones S, Bozic I, Antal T, Leary R, Fu B, et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467:1114–1117. doi: 10.1038/nature09515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, et al. Intratumor Heterogeneity and Branched Evolution Revealed by Multiregion Sequencing. New Engl J Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maitra A, Kern SE, Hruban RH. Molecular pathogenesis of pancreatic cancer. Best PractRes Clin Gastroenterol. 2006;20:211–226. doi: 10.1016/j.bpg.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 46.Greaves M, Maley CC. Clonal evolution in cancer. Nature. 2012;481:306–313. doi: 10.1038/nature10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Iacobuzio-Donahue C, Velculescu V, Wolfgang C, Hruban RH. The genetic basis of pancreas cancer development and progression: insights from whole-exome and whole-genome sequencing. Clin Cancer Res. 2012;18 doi: 10.1158/1078-0432.CCR-12-0315. XX-XX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cioffi M, Dorado J, Baeuerle PA, Heeschen C. EpCAM/CD3-Bispecific T-cell Engaging Antibody MT110 Eliminates Primary Human Pancreatic Cancer Stem Cells. Clin Cancer Res. 2012;18:465–474. doi: 10.1158/1078-0432.CCR-11-1270. [DOI] [PubMed] [Google Scholar]
- 49.Dalerba P, Cho RW, Clarke MF. Cancer stem cells: models and concepts. Annu Rev Med. 2007;58:267–284. doi: 10.1146/annurev.med.58.062105.204854. [DOI] [PubMed] [Google Scholar]
- 50.Jin L, Hope KJ, Zhai Q, Smadja-Joffe F, Dick JE. Targeting of CD44 eradicates human acute myeloid leukemic stem cells. Nat Med. 2006;12:1167–1174. doi: 10.1038/nm1483. [DOI] [PubMed] [Google Scholar]
- 51.Rajeshkumar NV, Rasheed ZA, Garcia-Garcia E, Lopez-Rios F, Fujiwara K, Matsui WH, et al. A combination of DR5 agonistic monoclonal antibody with gemcitabine targets pancreatic cancer stem cells and results in long-term disease control in human pancreatic cancer model. Mol Cancer Ther. 2010;9:2582–2592. doi: 10.1158/1535-7163.MCT-10-0370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lonardo E, Hermann PC, Mueller M-T, Huber S, Balic A, Miranda-Lorenzo I, et al. Nodal/Activin Signaling Drives Self-Renewal and Tumorigenicity of Pancreatic Cancer Stem Cells and Provides a Target for Combined Drug Therapy. Cell Stem Cell. 2011;9:433–446. doi: 10.1016/j.stem.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 53.Feldmann G, Fendrich V, McGovern K, Bedja D, Bisht S, Alvarez H, et al. An orally bioavailable small-molecule inhibitor of Hedgehog signaling inhibits tumor initiation and metastasis in pancreatic cancer. Mol Cancer Ther. 2008;7:2725–2735. doi: 10.1158/1535-7163.MCT-08-0573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Feldmann G, Dhara S, Fendrich V, Bedja D, Beaty R, Mullendore M, et al. Blockade of hedgehog signaling inhibits pancreatic cancer invasion and metastases: a new paradigm for combination therapy in solid cancers. Cancer Res. 2007;67:2187–2196. doi: 10.1158/0008-5472.CAN-06-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Olive KP, Jacobetz MA, Davidson CJ, Gopinathan A, McIntyre D, Honess D, et al. Inhibition of Hedgehog signaling enhances delivery of chemotherapy in a mouse model of pancreatic cancer. Science. 2009;324:1457–1461. doi: 10.1126/science.1171362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. http://phx.corporate-ir.net/phoenix.zhtml?c=121941&p=irol-newsArticle&ID=1653550&highlight=.
- 57.Mullendore ME, Koorstra J-B, Li Y-M, Offerhaus GJ, Fan X, Henderson CM, et al. Ligand-dependent Notch signaling is involved in tumor initiation and tumor maintenance in pancreatic cancer. Clin Cancer Res. 2009;15:2291–2301. doi: 10.1158/1078-0432.CCR-08-2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang G-N, Liang Y, Zhou L-J, Chen S-P, Chen G, Zhang T-P, et al. Combination of salinomycin and gemcitabine eliminates pancreatic cancer cells. Cancer Lett. 2011;313:137–144. doi: 10.1016/j.canlet.2011.05.030. [DOI] [PubMed] [Google Scholar]