Abstract
Given the importance of the microbiome for human health, the stability of the microbiome and its response to disturbance are crucial issues. Yet we have an insufficient understanding of them. Early data suggest that there is relative stability in the microbial ecosystem of adults in the absence of gross perturbation, and that long-term stability of human communities is not maintained by inertia, but by the action of restoring forces within a dynamic system. After brief exposures to some antibiotics, there is an immediate and substantial perturbation, and at least a partial recovery of taxonomic composition. Responses to antibiotics are individualized, and influenced by prior experience with the same antibiotic. These findings suggest that the human microbiome has properties of resilience. Besides serving to reveal critical underlying functional attributes, microbial interactions, and keystone species within the indigenous microbiota, responses to disturbance may have value in predicting future instability and disease, and in managing the human microbial ecosystem.
Introduction
More than 300 years after the first description of the human indigenous microbiota, and more than 100 years after the first formal definition of symbiosis as the living together of dissimilar organisms, the human microbiome, or community of microbes and collection of genomes found in and on the human body, is now the subject of renewed, intense study. The emerging perspective of humans as ‘superorganisms’ motivates experimental work and theoretical discussion across the biomedical sciences.1–4 Why the current level of interest in this topic?
Several factors have converged in galvanizing and facilitating scientific inquiry into the human microbiome. First, recent findings suggest that the human indigenous microbial communities explain critical features of human biology, and play a larger role in human health and disease than previously recognized.5,6 Among the benefits to human health, the microbiota contribute to food digestion; nutrition;7 processing and in some cases, detoxification of xenobiotics; regulation of human metabolism; development and terminal differentiation of host mucosa; ‘education’ and regulation of immune system target recognition and responses; 8 integrity of the barrier function of the skin and mucosa; and prevention of colonization and invasion of the host by pathogens. Some of the indigenous microbes derive benefits from this two-way mutualistic relationship as well, including acquisition of nutrients, habitat, and an effective means of dispersal.5 Many of these benefits were only recently revealed and/or their mechanisms clarified. At the same time, a variety of human disease states and other forms of pathology are associated with alterations within the microbiome. These disorders include chronic periodontitis, Crohn’s disease and other forms of inflammatory bowel disease, irritable bowel syndrome, tropical enteropathy, antibiotic-associated diarrhea, and bacterial vaginosis. For each of these forms of host pathology, the concept of (microbial) ‘community as pathogen’ as been proposed, and a distinction drawn between this type of scenario and more traditional medical paradigms for infectious disease in which a single dominant microbial species acts as the pathogenic factor. Stereotypic interactions between community members, such as those of syntrophic partners, may underlie the critical features associated with some of these pathologies.9,10
Revolutionary advances in genomics and associated technologies, and in computational biology, are another factor that explains current attention to the human microbiome.11 Greatly improved capabilities in DNA sequencing have enabled microbial community molecular phylogenetic surveys, and metagenomic and metatranscriptomic analyses of clinical specimens, to far greater depth, at far greater speed, and at far less cost, than was thought to be realistic just one year ago. In contrast to the now common taxonomic surveys of microbial communities which rely on a conserved phylogenetically-reliable molecule, such as the 16S rRNA gene, metagenomics and metabolomics provide a more rich, textured, and multidimensional picture of the community, revealing its genetic potential and metabolic activities.12–14 Tools developed in other fields of science for pattern recognition and identification of predictive features in complex, multi-dimensional data are now being applied to genomic data from the human microbiome.15 Yet, despite these advances, the ability to acquire in-depth assessments of microbial community structure and function from thousands of human microbiomes at many points in time and space, quickly, remains a desired, but as-yet impractical goal. And the ability to manipulate, integrate and explore such large datasets lags even further behind.
A third factor that motivates work on the human microbiome is the convergence of disparate disciplines, and the emergence of transdisciplinarity in this field of study. The latter refers to a research approach in which experts from one discipline adopt the perspective of other disciplines in formulating questions and in designing experimental plans for addressing them. The current study of the human microbiome borrows and exploits concepts from ecology (e.g., the community as the unit of study), environmental microbiology, population biology, and engineering disciplines, to name but a few. Transdisciplinarity creates opportunities to develop novel approaches for diagnosis, prognosis (e.g., see below), therapeutics, and preventive strategies.
Sources of Variation in the Composition and Function of the Human Microbiome
Despite the current research interest and activity focused on the microbiome, there are many critical questions that have not been satisfactorily answered. One of them asks about the most important sources of variation in the composition and function of the human microbiome. One of the dominant sources is habitat, i.e., anatomic site on or within the human body.16 For example, the microbial community in a subgingival crevice differs more from the community in the distal colon from the same adult at the same point in time than it does from the community in a subgingival crevice from a different adult. Although the most abundant taxa have been, or are being characterized from each of the gross human anatomic habitats (e.g., subgingival crevice, distal colon), variation within each of these gross habitats at multiple spatial scales is not so well understood. Another dominant source of variation in composition is the genetic background of the host. The microbiomes of twins are more similar than the microbiomes of a twin and a parent or of non-twin siblings.13 In general, the well-documented differences in the microbiomes of distinct human individuals reflect a combination of multiple factors at play: genetics, and various aspects of life history including antigen, diet, chemical, human, and other animal exposures, and health status. The relative importance of each of these aspects of life history is unclear. Discrete features of the microbiome may be related to very specific exposures. For example, the discovery of porphyranase and agarase genes, whose unusual products are active against the sulfated polysaccharides of marine red algae, in the gut metagenomes of Japanese individuals suggests that their collective, cultural, dietary preference for nutritional seaweeds may have selected for acquisition of these genes by gut bacterial symbionts.17
Time is also a critical factor in explaining variation in microbiome composition and function, especially during the early days of life or the early days of any newly formed habitat (such as the tooth surface). Primary succession of microbial communities in the human body is dramatic, hectic, and stereotyped, with early patterns reflecting differences in mode of delivery.18,19 Subsequent patterns (after the first week or two of life) are reminiscent of a punctuated equilibrium model in which one transient stable state is followed abruptly by another.18,20,21 The major features of an adult-like microbiome are achieved in childhood after 1–3 years of life. However, critical questions remain unanswered here as well, such as the relative importance of deterministic versus stochastic factors in explaining early microbiome features, or how early assembly patterns might dictate microbial community structure and function, as well as host physiology later in childhood and beyond in adulthood.
Given the importance of the microbiome for human health, its stability and response to disturbance would seem to be crucial issues. To what degree and in what ways is the human microbiome altered by disturbance of varying frequency and magnitude? How persistent are these alterations, and what are the consequences for the host? If there is return of the microbiome to its pre-disturbance state, what are the determinants of recovery? These questions are fundamental in the field of ecology.22,23 Although they have not been well-explored in the context of the human microbial ecosystem, they deserve to be. And they are the subject of the remainder of this article.
Stability and Resilience of Ecosystems
An appreciation for the dependency of humans on environmental ecosystem services and the danger of degradation of services to human welfare was apparent to the ancient Greeks. But the formal discussion of ecosystem stability and resilience began in the mid-twentieth century. The definition of “stability” in ecology has varied depending on the user of the term and the context. Elton in 1958 acknowledged the different distinct meanings in referring to population variability, population recovery, ease of invasion, and the consequences of invasion as alternative aspects of stability.24 Pimm defined an ecosystem as “stable” if 1) key variables describing the system return to equilibrium values after displacement; 2) the system is “resilient”, i.e., if there is only a short time before these key system variables returns to their equilibrium value after displacement; 3) there is persistence of key system variables at their set value; 4) the system is resistant to change in the services it provides; or 5) there is limited variability of key system parameters over time.25 He recognized that it is the functions and service output of the ecosystem that matters most in any consideration of stability.
Holling was the first to discuss in depth the meaning of ecosystem resilience.22 He highlighted the importance of defining the range of conditions under which a system is still able to return to its pre-disturbance equilibrium, as a central feature of resilience. He and colleagues wrote that the latter can be defined by the “capacity of a system to absorb disturbance and reorganize while undergoing change so as to still retain essentially the same function, structure, identity, and feedbacks”.26 Resilience can be measured in terms of elasticity, i.e., the rate at which the system returns to its pre-disturbance equilibrium, or the time it takes to return (to either its pre-disturbance equilibrium or some fraction of its maximum displacement). It can also be measured in terms of amplitude, i.e., the magnitude of the maximum displacement the system can absorb and still recover. Traditional conservation ecology is based on assessments of ecosystem resilience in the face of landscape disturbance. Resilience depends in turn on growth rates of community members, interactions between members, and nutrient availability (“ecosystem-level factors”).22 These same principles and definitions of stability and resilience can be applied to our understanding of human health and disease at any of the human habitats where there is a robust indigenous microbial community (see below).
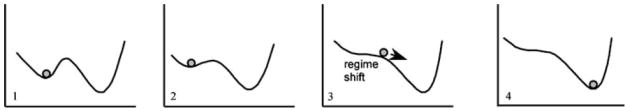
Ecosystem stability and resilience have often been explained in schematic terms with a notional “stability landscape”.22,27 On such a landscape, the community is depicted as a ball sitting on a topographic surface (the ecological landscape) where basins represent alternative equilibrium or stable states, and hilltops represent unstable states (see Figure 1). In this notional construct, gravity causes the ball to seek the lowest points on the landscape, i.e., “basins of attraction”. Holling proposed that some changes in the environment alter the landscape and create new alternative stable states and/or destabilize the position of a community, such that it becomes more attracted to a different basin.22 Using the stability landscape model, he and colleagues in 2004 explained four important attributes of resilience: “latitude” (maximum change permitted in a system before it loses the ability to recover), resistance to change, “precariousness” (proximity to threshold, past which system is more attracted to a different ‘basin’), and panarchy (influence of other equilibrium states and system dynamics on ability of system to recover). Holling acknowledged that models often fail to account for important details of complex systems. But he and others have shown in their models that spatial heterogeneity (mosaics), response time delays (which generate cyclic behavior), non-linear relationships among community components, and random effects—all can lead to enhanced resilience. Importantly, and in ways that might seem counter-intuitive, instability, i.e., large fluctuations, may enhance resilience and persistence.22 Assessments of resilience and stability are spatial and temporal scale-dependent, as well as specific to the particular degree of community complexity and the particular community service being examined.25
Figure 1.
‘Basins of attraction’, or alternative stable states or regimes, in a stability landscape. External forces and internal change can alter the landscape (from scheme 1 to 4) such that an ecosystem (represented as the ball) becomes less resilient and attracted to a new stable state (or regime). Because the landscape topography predicts the near future state of the ecosystem, the topography for a specific human habitat may have clinical utility in patient management. (From ref 27)
Others have discussed the concept of ‘catastrophic regime shift’ that occurs when environmental change occurs of a sufficient degree to degrade the landscape, cause a threshold effect, and tip a community into a new alternative basin of attraction.23,28 This model and concept may be directly relevant to the human microbial ecosystem, as will be discussed later in this paper.
Disturbance Ecology
Disturbance is major source of spatial and temporal heterogeneity in natural communities, and responsible for selection among “life history” variants that arise within these communities.29 As a natural feature of all ecosystems, disturbance has an important effect on system-level processes. Disturbance has traditionally been defined as an event or process (physical or biological) that causes abrupt structural changes to community composition (i.e., die-off in one or more species). In Sousa’s own words, a disturbance is “a discrete, punctuated killing, displacement, or damaging of one or more individuals that…creates an opportunity for other individuals to become established”.29 If it recurs with similar enough intensity and timing, the community will adapt over an evolutionary time scale and reflect this history in its structure and function; if it occurs with erratic features, the community will suffer larger and less predictable changes. The characteristics of the disturbance, e.g., the size of the area disturbed, and the magnitude, frequency, predictability, and duration of each disturbance event, are important in understanding the effect.
Intensity and frequency of disturbance are particularly consequential. Classical ecological studies have shown that intermediate levels of disturbance lead to maximum species diversity in an ecosystem; at intermediate levels, many species have had a chance to colonize but competitive exclusion has not yet intervened.30,31 In classical ecological studies, “pulse” (short duration) disturbances are distinguished from “press” (sustained) disturbances,32 although in the real world, the distinction can be blurred when the relevant time scale is unclear. Nevertheless, the type of disturbance determines which specific organisms and properties in a community are selected over time, and the specific features of adaptation that they display. Of particular note, compounded disturbances, i.e. combinations of disturbances acting together in space and time, are associated with unanticipated consequences (“surprises”) for ecosystems.33
In this context, anthropogenic disturbance is of concern. The activities of humans on the external environment (e.g., in land use) are believed to have caused significantly larger changes to macroscopic species and their ecosystems that are believed to have occurred as a result of non-human associated events (e.g., wind, flooding, fire, drought). Less attention has been directed at the effects of human-mediated disturbance on microbial species, and at the effects of humans, and of modern humans in particular, on the internal (human body) environment. But both, biological disturbance, such as shifts in diet, topical use of detergents, and use of systemic medications, and physical disturbance, such as topical and intra-oral scrubbing, are potentially critical to human health and disease, but poorly understood. Human activity produces disturbances at greater frequency and intensity than that to which most natural ecosystems have had an opportunity to adapt.
Bender has emphasized the value of controlled experiments involving the use of disturbance.32 Such experiments reveal features of a complex ecosystem that would otherwise not be as well appreciated. Species that are less abundant under non-perturbed, equilibrium conditions can become more abundant during or in the wake of disturbance, and more easily detected. Dependencies among community members, and species-species interactions can be identified and characterized. And features of community stability and resilience are revealed in these kinds of experiments. For these reasons and in the context of the above discussion, we have undertaken controlled experiments using antibiotics as a deliberate pulse disturbance in healthy adult human volunteers.
Antibiotics as a Disturbance
The use of antibiotics in massive quantities and concentrations relative to their natural occurrence over millions of years in the environment is a very recent phenomenon of human activity. Most of the focus on adverse impacts of antibiotic use has been on the emergence of resistant strains and species. Relatively few studies have addressed the adverse impact of antibiotics on the community-wide composition and structure of the human indigenous microbiota in individual subjects, using DNA sequencing-based approaches and frequent sampling. Furthermore, studies of antibiotic-treated patients with clinical indications for antibiotic use are plagued by the possible confounding effects of the underlying illness on the microbiota. Jernberg and colleagues have shown that an antibiotic with strong anti-anaerobe activity, clindamycin, causes a decline in Bacteroides diversity after 7 days of use that can persist for at least two years in healthy volunteers.34,35 They have also described effects on the fecal microbiota from the use of a regimen including clarithromycin and metronidazole (the latter of which has broad and strong anti-anaerobe activity) that lasted 4 years.36
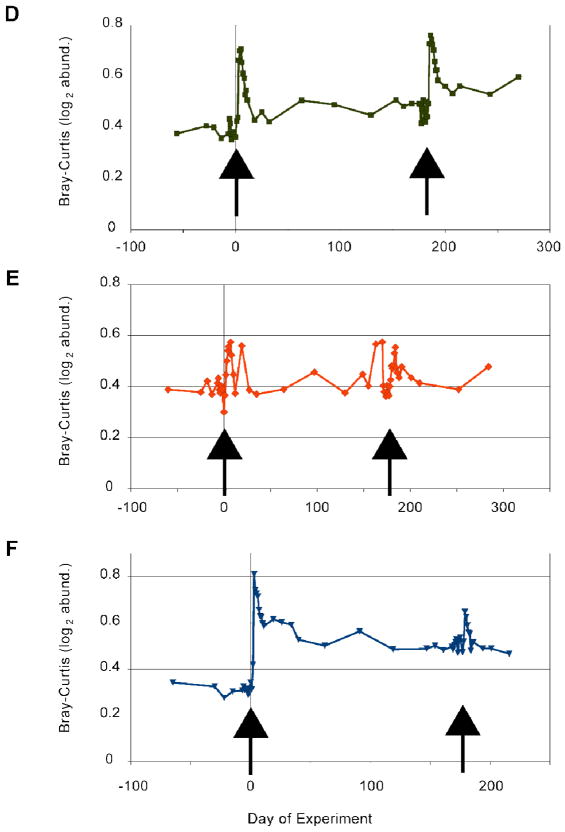
We have examined the effect of a 5-day (pulse) exposure of ciprofloxacin on healthy adult volunteers, with a sampling scheme that included monthly, then weekly, then daily fecal specimens for two months prior to exposure, and then the reverse for six months after exposure.37,38 Ciprofloxacin was chosen because it is thought to have little direct activity against anaerobes, the dominant phenotype of the distal gut microbiota. The 5-day pulse exposure was repeated six months after the first, and subjects then sampled for two additional months. The taxonomic composition of the fecal specimens was assessed by 16S rDNA hypervariable region pyrosequencing. Among the basic findings of these studies, a 5-day exposure to ciprofloxacin causes an abrupt decline in diversity by the end of the exposure period, and a significant change in the relative abundance of approximately one third to one half of all taxa. The taxa most affected included members of Ruminococcaceae and Lachnospiraceae within the phylum Firmicutes, and members of the phylum Bacteroidetes. By 2 weeks following exposure, most taxa had recovered to pre-exposure abundance levels. The second exposure in general caused a similar perturbation, but led to less complete recovery; in addition, each subject showed individualized responses (see Figure 2). In one subject (“D”), taxonomic composition just prior to each of the two exposures was similar, yet the responses were different, suggesting that the ecosystem retained “memory” of its prior perturbation.38 Figure 2 shows results based only on one ecological distance metric, the “Bray-Curtis dissimilarity”.
Figure 2.
Ecological distance (Bray-Curtis metric) between last pre-ciprofloxacin sample and all others from the same subject over time. There is relative stability before each of the two 5-day courses of ciprofloxacin (arrows) and variable bacterial community recovery, but there is a compounded effect of the two antibiotic courses in each of the subjects, D, E and F. (Modified from ref 38)
These data highlight the abrupt nature of the perturbation and the persistence of an effect after some of the ciprofloxacin exposures; however, the Bray-Curtis metric does not take into account the phylogenetic relationships among the taxa, nor their relative abundances. By the end of the study period, the taxonomic composition of the fecal microbiota in each subject had stabilized, but was altered relative to its starting state.37,38 The “compounding” of the disturbance (i.e., two sequential exposures) revealed response features that were not apparent after just one. Analysis of these data for evidence of species-species interactions, and analysis of shotgun metagenomic data from some of these samples for correlations between functional potential and resilience, are ongoing.
A number of important insights may result from such longitudinal studies of the human microbiome, in the same way that insights have been gleaned about the complex dynamics of plant populations in well-studied arid ecosystems.39 Evidence of at least partial recovery of community taxonomic composition from ciprofloxacin disturbance provides an opportunity for identifying the determinants of resilience and homeostasis in the human microbiome, the compensatory capacity of community members, and the possible role of species-species interactions. The results of this and other studies supports the idea that antibiotic “resistance” is the product of direct and indirect mechanisms. The latter include the trans-acting effects of some community subpopulations on the properties of other community members. As an example, heterogeneity in norfloxacin resistance among members of a laboratory-propagated clonal E. coli population has been attributed in part to variation in indole production, and the ‘altruistic’ behavior of rare, highly-resistant cells in producing indole to protect less resistant cells in the population.40 Indole induces expression of molecules and pathways in both producers and nonproducers that are protective against norfloxacin, as well as other antibiotics. The highly-resistant cells carry other mutations associated with antibiotic resistance, and then produce high levels of indole, despite the fitness cost, allowing the other cells to flourish. There are almost certainly many other mechanisms by which antibiotics have indirect effects on members of complex microbial populations, including sequential cascading effects on the microbial food web and alteration of host defenses or other environmental factors. And from a clinical, translational perspective, the ability to predict resilience of the human microbial ecosystem prior to disturbance, or to restore this property to degraded ecosystems, would offer significant potential clinical benefits.
Biodiversity and the insurance hypothesis
Discussions of ecosystem stability early in the twentieth century suggested that stability depends upon there being complexity in the ecosystem.41 Since then, two types of evidence in support of this proposal have been cited: theoretical considerations based on modeling approaches, and experimental data. Classical evidence of the latter type was described by McNaughton and others involving experimental manipulation of the grasslands of the Serengeti-Mara ecosystem in Tanzania and Kenya. These experiments demonstrated that “plant community diversity stabilizes functional properties of the community to environmental perturbations” and reflects “compensating homeostatic interactions among co-occurring species.”42 In a more recent experiment, Naeem and Li constructed several hundred defined, replicate microbial microcosms, containing varying numbers of members of each of two key functional groups for terrestrial and aquatic ecosystems (autotrophs and decomposers), varied nutrients, and then measured biomass and the density of functional group members, as response variables.43 They found that “as the number of species per functional group increased, replicate communities were more consistent in biomass and density measures”, and thus, that “redundancy is a valuable commodity”. This is the basis for the biological “insurance hypothesis”. Other work with experimental microbial microcosms has suggested that the degree of evenness in a community prior to disturbance affects the subsequent response to disturbance.44 (In the described experiments, with richness held constant, the more uneven the community, the less productive it was in the face of a selective stressor.)
Use of a stochastic dynamic model has also provided support for the hypothesis and demonstrated two main effects of species richness: increased richness reduces variability in ecosystem productivity (as one example of an important ecosystem process) over time, and increases mean productivity over time.45 The more asynchronous the response of different species to environmental fluctuations, the larger the effect of richness, i.e., the fewer the number of species required to achieve ecosystem redundancy and maintain the same features of productivity, especially in the face of environmental disturbance. Compensation by one species for loss or decline in another preserves long-term average ecosystem performance and reduces variability in performance, promotes the long-term probability of persistence, and enhances resilience to pulse perturbations.
These findings from traditional ecology have great importance for the human microbial ecosystem and its relationship to health and disease. For example, the features associated with enhanced resilience provide a rudimentary foundation for building and testing a predictive model for resilience in the human microbial ecosystem. Yet, there are critical issues about which we are woefully uninformed. One of these concerns the set of human microbial ecosystem services necessary for maintenance of human health: for each internal human habitat and associated microbial community, what is the complete set of services? How do they vary from one person or state of health to another? And how do we measure these services?
Clinical relevance and applications
Despite long histories of investigation in human health and in ecology, we are just beginning to reap the rewards of transdisciplinary efforts that integrate the approaches of these two disciplines. Several ideas have already been mentioned in the preceding text for translational work arising from the ecological investigations of human microbial communities.
Among the most important questions in need of attention, what are the features of microbial diversity most desired for states of health in humans? How are these features most effectively measured? What disturbance regimes are most desired, and what range of disturbance regimes is tolerated during states of health? What are the microbial ecosystem services of greatest relevance to the wide variety of human states of health? In humans at risk for disease linked to disrupted microbial communities, such as Crohn’s disease, are flares of disease associated with (or due to) loss of microbial community resilience? Can we anticipate flares by detecting early degradation of the stability landscape, or predict treatment failures by identifying a ‘catastrophic regime shift’?
Conclusions
Recent insights into the symbiotic relationships between humans and their indigenous microbial communities, coupled with revolutionary advances in genomic technologies, and the adoption of an ecological perspective in studying these communities and their host, have led to a new-found appreciation for the roles played by the human microbiota in health and disease. One of the next steps will be to address and understand better the functions of the microbiota and the mechanisms associated with these roles. Another will be to understand better stability and resilience in the human microbial ecosystem. Some early data suggest that there is relative stability in the microbial ecosystem of adults in the absence of gross perturbation. Yet, routine, mild fluctuations in community composition in the absence of gross disturbance indicate that long-term stability of human communities is not maintained by inertia, but rather by the action of restoring forces within a dynamic system. With respect to a well-controlled and clinically-relevant disturbance with a brief course of an antibiotic, we find an immediate and substantial perturbation and at least a partial recovery of taxonomic composition. The responses to this antibiotic are individualized, and influenced by prior experience with the same antibiotic. Besides serving to reveal critical underlying functional attributes, microbial interactions, and keystone species within the indigenous microbiota, the response to disturbance may have value in predicting future instability and disease, and in restoring a preferred ecosystem regime.
Acknowledgments
I wish to thank members of the Relman Lab for helpful discussion. DAR is supported by a Distinguished Clinical Scientist Award from the Doris Duke Charitable Trust, by NIH Pioneer Award DP1OD000964, and by the Thomas C. and Joan M. Merigan Endowment at Stanford University.
References
- 1.Lederberg J. Infectious history. Science. 2000 Apr 14;288(5464):287–293. doi: 10.1126/science.288.5464.287. [DOI] [PubMed] [Google Scholar]
- 2.Relman DA, Falkow S. The meaning and impact of the human genome sequence for microbiology. Trends Microbiol. 2001 May;9(5):206–208. doi: 10.1016/s0966-842x(01)02041-8. [DOI] [PubMed] [Google Scholar]
- 3.McFall-Ngai MJ. Unseen forces: the influence of bacteria on animal development. Dev Biol. 2002 Feb 1;242(1):1–14. doi: 10.1006/dbio.2001.0522. [DOI] [PubMed] [Google Scholar]
- 4.Proctor LM. The Human Microbiome Project in 2011 and beyond. Cell Host Microbe. 2011 Oct 20;10(4):287–291. doi: 10.1016/j.chom.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 5.Dethlefsen L, McFall-Ngai M, Relman DA. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature. 2007 Oct 18;449(7164):811–818. doi: 10.1038/nature06245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature. 2007 Oct 18;449(7164):804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turnbaugh PJ, Gordon JI. The core gut microbiome, energy balance and obesity. J Physiol. 2009 Sep 1;587(Pt 17):4153–4158. doi: 10.1113/jphysiol.2009.174136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee YK, Mazmanian SK. Has the microbiota played a critical role in the evolution of the adaptive immune system? Science. 2010 Dec 24;330(6012):1768–1773. doi: 10.1126/science.1195568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lepp PW, Brinig MM, Ouverney CC, Palm K, Armitage GC, Relman DA. Methanogenic Archaea and human periodontal disease. Proc Natl Acad Sci USA. 2004 Apr 20;101(16):6176–6181. doi: 10.1073/pnas.0308766101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vianna ME, Holtgraewe S, Seyfarth I, Conrads G, Horz HP. Quantitative analysis of three hydrogenotrophic microbial groups, methanogenic archaea, sulfate-reducing bacteria, and acetogenic bacteria, within plaque biofilms associated with human periodontal disease. J Bacteriol. 2008 May;190(10):3779–3785. doi: 10.1128/JB.01861-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Relman DA. Microbial genomics and infectious diseases. N Engl J Med. 2011 Jul 28;365(4):347–357. doi: 10.1056/NEJMra1003071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Turnbaugh PJ, Gordon JI. An invitation to the marriage of metagenomics and metabolomics. Cell. 2008 Sep 5;134(5):708–713. doi: 10.1016/j.cell.2008.08.025. [DOI] [PubMed] [Google Scholar]
- 13.Turnbaugh PJ, Hamady M, Yatsunenko T, et al. A core gut microbiome in obese and lean twins. Nature. 2009 Jan 22;457(7228):480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holmes E, Li JV, Athanasiou T, Ashrafian H, Nicholson JK. Understanding the role of gut microbiome-host metabolic signal disruption in health and disease. Trends Microbiol. 2011 Jul;19(7):349–359. doi: 10.1016/j.tim.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 15.Reshef DN, Reshef YA, Finucane HK, et al. Detecting novel associations in large data sets. Science. 2011 Dec 16;334(6062):1518–1524. doi: 10.1126/science.1205438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Bacterial community variation in human body habitats across space and time. Science. 2009 Dec 18;326(5960):1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hehemann JH, Correc G, Barbeyron T, Helbert W, Czjzek M, Michel G. Transfer of carbohydrate-active enzymes from marine bacteria to Japanese gut microbiota. Nature. 2010 Apr 8;464(7290):908–912. doi: 10.1038/nature08937. [DOI] [PubMed] [Google Scholar]
- 18.Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS Biol. 2007 Jul;5(7):e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dominguez-Bello MG, Costello EK, Contreras M, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci USA. 2010 Jun 29;107(26):11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morowitz MJ, Denef VJ, Costello EK, et al. Strain-resolved community genomic analysis of gut microbial colonization in a premature infant. Proc Natl Acad Sci USA. 2011 Jan 18;108(3):1128–1133. doi: 10.1073/pnas.1010992108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koenig JE, Spor A, Scalfone N, et al. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci USA. 2011 Mar 15;108( Suppl 1):4578–4585. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holling C. Resilience and stability of ecological systems. Annual review of ecology and systematics. 1973;4:1–23. [Google Scholar]
- 23.Scheffer M, Carpenter S, Foley JA, Folke C, Walker B. Catastrophic shifts in ecosystems. Nature. 2001 Oct 11;413(6856):591–596. doi: 10.1038/35098000. [DOI] [PubMed] [Google Scholar]
- 24.Elton CS. The ecology of invasions by animals and plants. London: Methuen & Co. Ltd; 1958. [Google Scholar]
- 25.Pimm SL. The balance of nature? : ecological issues in the conservation of species and communities. Chicago: University of Chicago Press; 1991. [Google Scholar]
- 26.Walker B, Holling CS, Carpenter SR, Kinzig A. Resilience, adaptability and transformability in social-ecological systems. Ecol Soc. 2004 Dec;9(2):5. [Google Scholar]
- 27.Folke C, Carpenter S, Walker B, et al. Regime shifts, resilience, and biodiversity in ecosystem management. Annu Rev Ecol Evol S. 2004;35:557–581. [Google Scholar]
- 28.Scheffer M, Carpenter SR. Catastrophic regime shifts in ecosystems: linking theory to observation. Trends Ecol Evol. 2003 Dec;18(12):648–656. [Google Scholar]
- 29.Sousa WP. The Role of Disturbance in Natural Communities. Annual Review of Ecology and Systematics. 1984;15:353–391. [Google Scholar]
- 30.Connell JH. Diversity in tropical rain forests and coral reefs. Science. 1978 Mar 24;199(4335):1302–1310. doi: 10.1126/science.199.4335.1302. [DOI] [PubMed] [Google Scholar]
- 31.Sousa WP. Disturbance in Marine Inter-Tidal Boulder Fields - the Non-Equilibrium Maintenance of Species-Diversity. Ecology. 1979;60(6):1225–1239. [Google Scholar]
- 32.Bender EA, Case TJ, Gilpin ME. Perturbation Experiments in Community Ecology - Theory and Practice. Ecology. 1984;65(1):1–13. [Google Scholar]
- 33.Paine RT, Tegner MJ, Johnson EA. Compounded perturbations yield ecological surprises. Ecosystems. 1998 Nov-Dec;1(6):535–545. [Google Scholar]
- 34.Jernberg C, Lofmark S, Edlund C, Jansson JK. Long-term ecological impacts of antibiotic administration on the human intestinal microbiota. Isme J. 2007 May;1(1):56–66. doi: 10.1038/ismej.2007.3. [DOI] [PubMed] [Google Scholar]
- 35.Jernberg C, Lofmark S, Edlund C, Jansson JK. Long-term impacts of antibiotic exposure on the human intestinal microbiota. Microbiology. 2010 Nov;156(Pt 11):3216–3223. doi: 10.1099/mic.0.040618-0. [DOI] [PubMed] [Google Scholar]
- 36.Jakobsson HE, Jernberg C, Andersson AF, Sjolund-Karlsson M, Jansson JK, Engstrand L. Short-term antibiotic treatment has differing long-term impacts on the human throat and gut microbiome. PLoS One. 2010;5(3):e9836. doi: 10.1371/journal.pone.0009836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008 Nov 18;6(11):e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci USA. 2011 Mar 15;108( Suppl 1):4554–4561. doi: 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brown JH, Whitham TG, Morgan Ernest SK, Gehring CA. Complex species interactions and the dynamics of ecological systems: long-term experiments. Science. 2001 Jul 27;293(5530):643–650. doi: 10.1126/science.293.5530.643. [DOI] [PubMed] [Google Scholar]
- 40.Lee HH, Molla MN, Cantor CR, Collins JJ. Bacterial charity work leads to population-wide resistance. Nature. 2010 Sep 2;467(7311):82–85. doi: 10.1038/nature09354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Macarthur R. Fluctuations of Animal Populations, and a Measure of Community Stability. Ecology. 1955;36(3):533–536. [Google Scholar]
- 42.McNaughton SJ. Diversity and Stability of Ecological Communities - Comment on Role of Empiricism in Ecology. Am Nat. 1977;111(979):515–525. [Google Scholar]
- 43.Naeem S, Li SB. Biodiversity enhances ecosystem reliability. Nature. 1997 Dec 4;390(6659):507–509. [Google Scholar]
- 44.Wittebolle L, Marzorati M, Clement L, et al. Initial community evenness favours functionality under selective stress. Nature. 2009 Apr 2;458(7238):623–626. doi: 10.1038/nature07840. [DOI] [PubMed] [Google Scholar]
- 45.Yachi S, Loreau M. Biodiversity and ecosystem productivity in a fluctuating environment: the insurance hypothesis. Proc Natl Acad Sci USA. 1999 Feb 16;96(4):1463–1468. doi: 10.1073/pnas.96.4.1463. [DOI] [PMC free article] [PubMed] [Google Scholar]