Abstract
The bleomycins (BLMs) are used clinically in combination with a number of other agents for the treatment of several types of tumors, and the BLM, etoposide, and cisplatin treatment regimen cures 90–95% of metastatic testicular cancer patients. BLM-induced pneumonitis is the most feared, dose-limiting side effect of BLM in chemotherapy, which can progress into lung fibrosis and affect up to 46% of the total patient population. There have been continued efforts to develop new BLM analogues in the search for anticancer drugs with better clinical efficacy and lower lung toxicity. We have previously cloned and characterized the biosynthetic gene clusters for BLMs from Streptomyces verticillus ATCC15003, tallysomycins from Streptoalloteichus hindustanus E465-94 ATCC31158, and zorbamycin (ZBM) from Streptomyces flavoviridis SB9001. Comparative analysis of the three biosynthetic machineries provided the molecular basis for the formulation of hypotheses to engineer novel analogues. We now report engineered production of three new analogues, 6′-hydroxy-ZBM, BLM Z, and 6′-deoxy-BLM Z and the evaluation of their DNA cleavage activities as a measurement for their potential anticancer activity. Our findings unveiled: (i) the disaccharide moiety plays an important role in the DNA cleavage activity of BLMs and ZBMs, (ii) the ZBM disaccharide significantly enhances the potency of BLM, and (iii) 6′-deoxy-BLM Z represents the most potent BLM analogue known to date. The fact that 6′-deoxy-BLM Z can be produced in reasonable quantities by microbial fermentation should greatly facilitate follow-up mechanistic and preclinical studies to potentially advance this analogue into a clinical drug.
INTRODUCTION
The bleomycins (BLMs), a family of glycopeptide-derived antibiotics produced by several Streptomyces species, are used clinically in combination with a number of other agents for the treatment of several types of tumors, notably germ cell tumors, squamous cell carcinomas, and malignant lymphomas.1–4 The combination BLM, etoposide and cisplatin treatment regimen cures 90–95% of metastatic testicular cancer patients, clearly representing a therapeutic success story.5,6 Almost uniquely among anticancer drugs, BLM exhibits low myelo- and immunosuppression, promoting its wide application in combination chemotherapy.1–4 However, BLM-induced pneumonitis is the most feared, dose-limiting side effect of BLM in chemotherapy, which can progress into lung fibrosis and affect up to 46% of the total patient population.3,5–7 Central to the development of BLM-induced pneumonitis is endothelial damage of lung vasculature due to BLM-induced cytokines and free radicals, and the resulting lung toxicity has been proposed to be independent of the cytotoxicity of BLMs to tumor cells.3,7
BLMs are thought to exert their biological effects through a sequence selective, metal-dependent oxidative cleavage of double stranded DNA in the presence of oxygen.2–4 The BLMs can be dissected into four functional domains (Figure 1). The pyrimidoblamic acid subunit along with the adjacent β-hydroxyl histidine constitutes the metal-binding domain for Fe2+ complexation and molecular oxygen activation responsible for DNA cleavage. The bithiazole and C-terminal amine moiety provide the majority of the BLM-DNA affinity and may contribute to polynucleotide recognition and the DNA cleavage selectivity. The (2S, 3S, 4R)-4-amino-3-hydroxy-2-methylpentanoic acid subunit provides the linker between the metal-binding and DNA-binding sites and plays an important role in the efficiency of DNA cleavage by BLMs. The disaccharide moiety has been speculated to participate in cell recognition by BLMs and possibly in cellular uptake and metal ion coordination. Consequently, there have been extraordinary efforts to develop new BLM analogues by both organic synthesis1–3,8–10 and directed biosynthesis11,12 in the search for anticancer drugs with better clinical efficacy and lower lung toxicity. Numerous BLM analogues have been prepared in the past three decades, helping to define the fundamental functional roles of the individual domains, and thereby lending support to the premise that DNA damage is the major contributor to the cytotoxicity of BLMs to tumor cells.1–3,8–12 However, it can be argued that none of these analogues have improved therapeutic properties, and tinkering at any position within the metal-binding domain, the linker region, or the bithiazole moiety severely reduced the ability of the drugs to mediate DNA cleavage. In contrast, the disaccharide moiety remains largely unexplored, mainly due to the lack of BLM analogues prepared with varying sugars, although it is known that deglyco-BLMs are much less effective at mediating DNA cleavage.1–3,8,9
Figure 1.
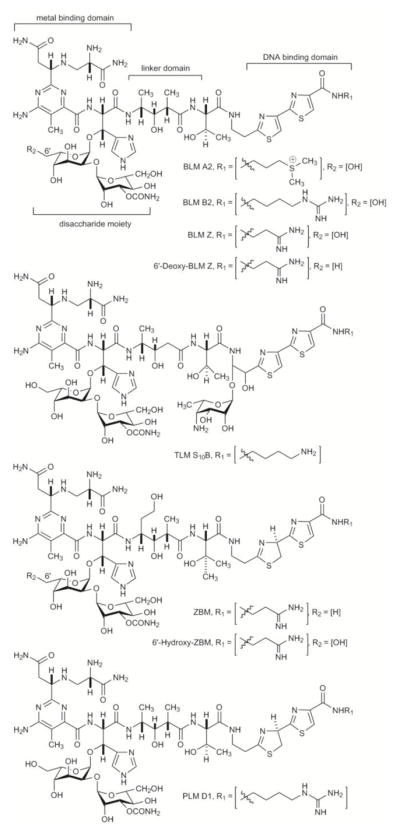
Structures of selected members of the bleomycin (BLM) family of antitumor antibiotics: BLM A2, BLM B2, tallysomycin (TLM) S10B, phleomycin (PLM) D1, and zorbamycin (ZBM), as well as the engineered ZBM analogue, 6′-hydroxy-ZBM, and BLM analogues, BLM Z and 6′-deoxy-BLM Z. See Figures S3 and S6 for the numberings of ZBMs and BLMs, respectively.
The naturally occurring BLMs differ structurally primarily at the C-terminus of the hybrid peptide-polyketide aglycones of the glycopeptides. The commercial product, Blenoxane®, contains BLM A2 and BLM B2 as the principle constituents (Figure 1).
Structurally related to the BLMs are the tallysomycins (TLMs), phleomycins (PLMs), and zorbamycin (ZBM).2,12,13 One characteristic structural difference among these compounds is the presence of a bithiazole unit in the BLM and TLM aglycones and a thiazolinylthiazole unit in the PLM and ZBM aglycones. In addition, while BLMs, PLMs, and TLMs share the same disaccharide moiety and many of the same terminal amine moieties, TLMs feature an additional sugar moiety and ZBM has its unique disaccharide and terminal amine moiety (Figure 1). The PLMs, TLMs, and ZBM exert their biological effect through the same oxidative DNA cleavage mechanism as the BLMs.2–4 Although the BLMs are the only members of this family that have been developed into clinical drugs, the PLMs and ZBM are significantly more potent than the BLMs in mediating DNA cleavage. This raises the question what is the structure and activity relationship of PLMs and ZBM in comparison with the BLMs and can the superior DNA cleavage activity associated with the PLMs and ZBM be engineered into new BLM analogues and exploited for anticancer drug discovery.
Complementary to chemical synthesis, combinatorial biosynthesis – the generation of novel analogues of natural products by genetic engineering of biosynthetic pathways – offers a promising alternative to preparing microbial metabolites and their analogues biosynthetically.12,14 Specific structural alterations in the presence of abundant functional groups can often be achieved by precise rational manipulation of the biosynthetic machinery. A minimum of four requirements must be met before combinatorial biosynthesis can be successfully exploited to generate structural diversity of natural products: (i) availability of the gene clusters encoding the production of a particular natural product or a family of natural products, (ii) genetic and biochemical characterizations of the biosynthetic machinery for the targeted natural products to a degree that the combinatorial biosynthesis principles can be rationally applied to engineer the designer analogues, (iii) expedient genetic systems for in vivo manipulation of genes governing the production of the target molecules in their native producers or heterologous hosts, and (iv) production of the natural products or their engineered analogues to levels that are appropriate for detection, isolation, and structural and biological characterizations.
We have previously cloned, sequenced and characterized the biosynthetic gene clusters for BLMs from Streptomyces verticillus ATCC15003,15–21 TLMs from Streptoalloteichus hindustanus E465-94 ATCC31158,22–24 and ZBM from Streptomyces flavoviridis SB9001,25 a ZBM-overproducer derived from the wild-type S. flavoviridis ATCC21892 strain13 (Figure S1). Comparative analysis of the three biosynthetic gene clusters and pathways provided the molecular basis on which to formulate hypotheses for engineering novel analogues.12 We postulate that (i) the assembly of the hybrid peptide-polyketide aglycones is catalyzed by the hybrid nonribosomal peptide synthetase (NRPS)-polyketide synthase (PKS) megasynthases, in which domain variations can account for the major structural differences among the BLM, TLM, and ZBM aglycones, (ii) in contrast to this, the seemingly identical domain organization of the NRPS modules responsible for the heterocycle formation, produces the distinct structural entities bithiazole and thiazolinylthiazole, for BLM/TLM and ZBM, respectively, and (iii) variations in sugar biosynthesis and glycosylation dictate the final BLM, TLM, and ZBM glycopeptide scaffolds (Figure S2). The feasibility of engineering the biosynthesis of BLM, TLM, and ZBM in their native producers has also been demonstrated by the generation of the ΔblmD mutant strain S. verticillus SB5, which produces decarbamoyl-BLM,21 the ΔtlmH mutant strain S. hindustanus SB8005, which produces destalose TLM,24 and the numerous mutant strains of S. flavoviridis, which help define the precise boundaries of the ZBM gene cluster.12,25 However, in vivo manipulations in S. verticillus and S. hindustanus remain inefficient, despite great effort, precluding their practical use as a preferred host in which to engineer novel BLM analogues. Although an expedient genetic system has been developed for S. flavoviridis, the molecuar mechanism differentiating bithiazole biosynthesis in BLM and TLM from thiazolinylthiazole biosynthesis in ZBM remains unknown, and consequently, manipulation of the ZBM biosynthetic machinery for the production of analogues containing the characteristic BLM bithiazole moiety, in the BLM aglycone, has not been successful.12,25 One way to circumvent both problems is to produce novel BLM analogues by engineering the BLM biosynthetic machinery in S. flavoviridis.
Here we report the engineered production of a ZBM analogue, 6′-hydroxy-ZBM, containing the BLM disaccharide and two BLM analogues, BLM Z and 6′-deoxy-BLM Z, having the ZBM terminal amine moiety and the BLM or ZBM disaccharide, respectively, all in S. flavoviridis. We also evaluated the engineered ZBM and BLM analogues, in comparison with ZBM and BLM A2 as controls, as to their DNA cleavage activities as a measurement for their potential anti-cancer activity. Our findings unveiled: (i) the disaccharide moiety plays an important role in the DNA cleavage activity of BLMs and ZBM, (ii) the ZBM disaccharide significantly enhances the potency of BLM, and (iii) 6′-deoxy-BLM Z represents the most potent BLM analogue known to date. The fact that 6′-deoxy-BLM Z is biosynthesized by a recombinant strain and can be produced in reasonable quantities by scale-up microbial fermentation should alleviate the concern for its availability, thereby setting the stage to carry out the mechanistic and preclinical studies needed to potentially advance this analogue into a clinical drug.
METHODS
The S. flavoviridis recombinant strain SB9013, which produces 6′-hydroxy-ZBM, was constructed from the S. flavoviridis ΔzbmL mutant strain SB900325 by cross-complementation of the ΔzbmL mutation with blmG. The S. flavoviridis recombinant strain SB2026, which produces BLM Z and 6′-deoxy-BLM Z, was constructed from the S. flavoviridis ΔzbmVIII mutant strain SB9025 by expression of the BLM gene cluster introduced on pBS54, the bacterial artificial chromosome (BAC)- based vector containing the entire BLM cluster. The structures of 6′-hydroxy-ZBM, BLM Z, and 6′-deoxy-BLM Z were elucidated by a combination of mass spectrometry (MS) and 1H and 13C NMR spectroscopic analyses, as well as by comparison to the 1H and 13C NMR data of BLM A221,23,24 and ZBM.13 The DNA cleavage activities of 6′-hydroxy-ZBM, BLM Z, and 6′-deoxy-BLM Z were determined in plasmid relaxation assays with pBluescript II SK(+) in comparison with BLM A2 and ZBM.9,21,23 A complete description of materials, methods, and detailed experimental procedures, as well as tables and figures, is available as Supporting Information (SI).
RESULTS
Engineering the S. flavoviridis recombinant strain SB9013 that produces 6′-hydroxy-ZBM
We previously constructed the S. flavoviridis ΔzbmL mutant strain SB9003, in which ZBM production was abolished (Figure 2B, panels I and II). Introduction of a complementation construct, pBS9019, in which the expression of zbmL is under the control of the constitutive promoter ErmE*, into SB9003 afforded a recombinant strain SB9009, whose ZBM production was restored (Figure 2B, panels I, II, and III).12,25 These results demonstrate the feasibility of manipulating disaccharide biosynthesis in the ZBM machinery of S. flavoviridis.
Figure 2.
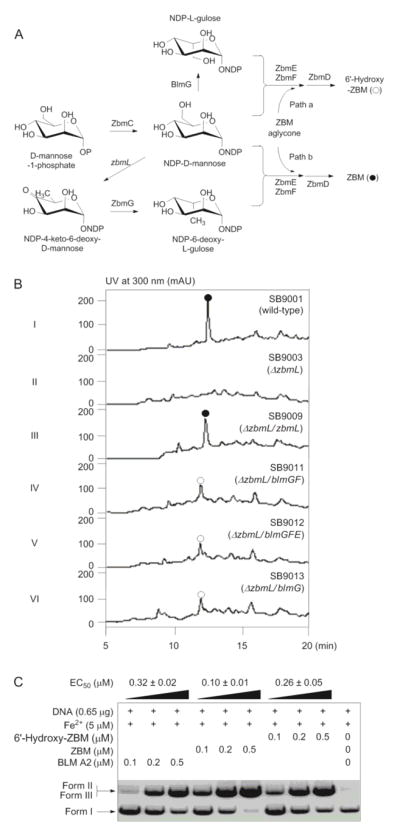
Engineered production of 6′-hydroxy-ZBM and evaluation of its DNA cleavage activity in comparison with BLM A2 and ZBM. (A) Proposed pathways for BLM and ZBM sugar biosynthesis in S. verticillus and S. flavoviridis, respectively, and cross-complementation of the ΔzbmL mutant strain SB9003 with blmG for the production of 6′-hydroxy-ZBM in SB9011, SB9012, and SB9013. (B) HPLC analysis of ZBM (●) production in SB9001, SB9003, and SB9009 and 6′-hydroxy-ZBM (○) production in SB9011, SB9012, and SB9013. (C) DNA cleavage activities of 6′-hydroxy-ZBM as observed in plasmid relaxation assays with pBluescript II SK(+) in comparison with BLM A2 and ZBM. Form I, supercoiled plasmid DNA; form II, open circular plasmid DNA; form III, linearized plasmid DNA; EC50, effective concentration at 50% plasmid DNA relaxation. See Figure S7 for relative ratios of single-strand cleavage (Form II) and double-strand cleavage (Form III) of supercoiled plasmid DNA (Form I).
The ΔzbmL mutant strain SB9003 is thought to accumulate NDP-D-mannose instead of NDP-4-keto-6-deoxy-D-mannose, the native substrate for the ZbmG epimerase.12,25 ZbmG apparently cannot accept NDP-D-mannose as an alternative substrate for catalyzing the analogous epimization at the C-5′ position to afford the corresponding NDP-L-gulose, without which ZBM biosynthesis was apparently abolished (Figure 2A, path b).12,25 In contrast, the BlmG epimerase in BLM biosynthesis catalyzes the epimerization of NDP-D-mannose directly into NDP-L-gulose, which is subsequently incorporated into the disaccharide moiety of BLM by the corresponding BlmE and BlmF glycosyltransferases (Figure S2).12,16,21,22 Cross-complementation of the ΔzbmL mutation with constructs containing either blmG alone or in combinations with blmE, blmF, or both, therefore, represents an attractive strategy for engineering a new ZBM analogue carrying the BLM disaccharide (Figure 2A, path a).
Three integrative plasmids, pBS57, pBS61, and pBS63 were constructured, each containing the constitutive ErmE* promoter to control the expression of blmGF (pBS57), blmGEF (pBS61), or blmG (pBS63), respectively (SI). These plasmids were introduced into SB9003 to yield the ΔzbmL cross-complementation strains SB9011 (SB9003/pBS57), SB9012 (SB9003/pBS61), and SB9013 (SB9003/pBS63) (SI). The recombinant strains SB9011, SB9012, and SB9013, were fermented under conditions described previously,13,25 using the S. flavoviridis SB9001 and the ΔzbmL mutant strain SB9003 as controls (SI). HPLC analysis of the metabolites produced showed that ZBM production was not restored and instead, a new metabolite, named 6′-hydroxy-ZBM, with a distinct retention time in comparison to ZBM, was detected in all three cross-complementation strains (Figure 2B, panels IV, V, VI). The titers of 6′-hydroxy-ZBM in SB9011, SB9012, and SB9013 were similar, estimated to be ~4 mg/L (Figure 2B, panels IV, V, VI), in comparison with that for ZBM (~10 mg/L) from the SB9001 strain (Figure 2B, panel I).13
A scale-up fermentation of SB9013 was carried out to purify 6′-hydroxy-ZBM for structural elucidation (SI). Briefly, the fermentation broth (30 L) was centrifuged, and the supernatant was collected and adjusted to pH 7.0. After sequential column chromatography on Amberlite® IRC-50 and Diaion HP-20, followed by semipreparative HPLC on a C-18 column, the new metabolite was purified as a blue 6′-hydroxy-ZBM•Cu2+ complex (10 mg). Further treatment of the 6′-hydroxy-ZBM•Cu2+ complex with EDTA to remove Cu2+, followed by HPLC on a C-18 column, finally afforded Cu2+-free 6′-hydroxy-ZBM as a white powder (3.6 mg).
Structural elucidation of 6′-hydroxy-ZBM was first carried out by a combination of MS and 1H and 13C NMR spectroscopic analyses, as well as by comparison of the resultant data with ZBM13 and BLM A221,23,24 (SI). Electrospray ionization (ESI)-MS analysis, in positive mode, of the 6′-hydroxy-ZBM•Cu2+ complex afforded an [M + Cu]2+ ion at m/z 745.5 and of Cu2+-free 6′-hydroxy-ZBM afforded two ions, an [M + 2H]2+ ion at m/z 714.8 and an [M + H + Na]2+ ion at m/z 725.8. High resolution (HR) matrix-assisted laser desorption ionization (MALDI)-MS analysis, in positive mode, of Cu2+-free 6′-hydroxy-ZBM yielded an [M + H]+ ion at m/z 1428.5634, establishing the molecular formula of 6′-hydroxy-ZBM as C55H85N19O22S2 (calculated for C55H85N19O22S2 + H+, 1428.5631). The 16 mass unit increase of the new metabolite in comparison with ZBM supported the assignment of the new metabolite as a hydroxylated-ZBM analogue, the hydroxylation regiochemistry of which was assigned on the basis of collision-induced dissociation (CID) tandem MS analysis. The CID tandem MS of the [M + 2H]2+ ion at m/z 714.8 yielded four diagnostic fragments at m/z 1233.2, 1061.2, 612.2, and 531.2, corresponding to the [M − (3-O-carbamoyl-mannose) + H]+, [M − (2-O-(3-O-carbamoyl-mannosyl)-gulose) + H]+, [M − (3-O-carbamoyl-mannose) + 2H]2+, and [M − (2-O-(3-O-carbamoyl-mannosyl)-gulose) + 2H]2+ ions, respectively. Taken together, these results suggested that the new metabolite contained the BLM disaccharide 3-O-carbamoyl-α-D-mannosyl-L-gulose (Figure 1), as would be expected from cross-complementation of the ΔzbmL mutant strain by blmG (Figure 2A).
The structure of 6′-hydroxy-ZBM, a new ZBM analogue containing a BLM disaccharide (Figure 1), was finally established on the basis of comprehensive 1D and 2D NMR analysis (SI, Figures S3, S8–S12 and Tables S1, S2). The 1H and 13C NMR spectra of 6′-hydroxy-ZBM are very similar to those of ZBM13 except for the disaccharide moiety. The signals for the methyl group [δC at 17.3 and δH at 0.92 (d, J = 8.0 Hz) for C-6′ in ZBM] disappeared and instead, signals for the hydroxymethylene group [δC at 63.4 and δH at 3.58 (m) for C-6′ in 6′-hydroxy-ZBM] appeared. The signals for the disaccharide moiety of 6′-hydroxy-ZBM are almost identical to those of BLM A2.13,21,23,24 These analyses also enabled the full 1H and 13C NMR spectroscopic assignments of 6′-hydroxy-ZBM as summarized in Tables S1 and S2 (SI).
Engineering the S. flavoviridis recombinant strain SB9027 that produces ZBM, 6′-hydroxy-ZBM, BLM Z, and 6′-deoxy-BLM Z
We have previously reported a BAC-based strategy for natural product production and engineering in heterologous Streptomyces hosts.26–29 We adopted the BAC-based strategy to circumvent the technical difficulties encountered in manipulating BLM biosynthesis in its native producer S. verticillus by expressing the BLM biosynthetic gene cluster in S. flavoviridis, for which an expedient genetic system has been developed and the feasibility of engineering ZBM biosynthesis has been demonstrated.12,25
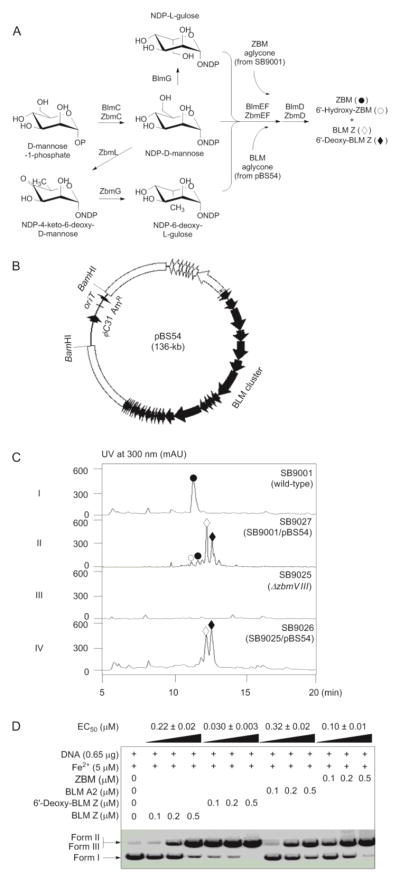
A BAC library of the BLM-producing S. verticillus ATCC15003 strain was constructed according to the published protocol (SI).27 The average insert size of the BAC library was ~80 kb as determined by pulsed-field gel electrophoresis analysis of HindIII digests from 36 randomly selected clones. The S. verticillus library (~2000 clones) was screened using DNA fragments flanking the BLM cluster to isolate BAC clones that harbored the intact BLM cluster (SI). Two positive clones, pBS54 and pBS55, were identified, containing a 120-kb and an 80-kb insert, respectively. End-sequencing of both clones confirmed that pBS54 harbored the entire intact BLM cluster (Figure 3B) (SI).
Figure 3.
Engineered production of BLM Z and 6′-deoxy-BLM Z and evaluation of their DNA cleavage activities in comparison with BLM A2 and ZBM. (A) Proposed pathways for BLM and ZBM sugar biosynthesis in S. verticillus and S. flavoviridis, respectively, and expression of the BLM biosynthetic gene cluster in the ZBM-producing SB9001 for the production of ZBM, 6′-hydroxy-ZBM, BLM Z, and 6′-deoxy-BLM Z by SB9027 and in the ΔzbmVIII mutant strain SB9025 for the production of BLM Z and 6′-deoxy-BLM Z by SB9026. (B) The pBS54 clone with a 120-kb insert from S. verticillus ATCC15003 that harbors the intact BLM biosynthetic gene cluster cloned into the two BamHI sites, along with the apramycin resistance marker (AmR), oriT, and ΦC31. (C) HPLC analysis of ZBM (●) production in SB9001, SB9025, and SB9027, 6′-hydroxy-ZBM (○) production in SB9027, and BLM Z (◇) and 6′-deoxy-BLM Z (◆) production in SB9026 and SB9027. (D) DNA cleavage activities of BLM Z and 6′-deoxy-BLM Z as observed in plasmid relaxation assays with pBluescript II SK(+) in comparison with BLM A2 and ZBM. Form I, supercoiled plasmid DNA; form II, open circular plasmid DNA; form II, linearized plasmid DNA; EC50, effective concentration at 50% plasmid DNA relaxation. See Figure S7 for relative ratios of single-strand cleavage (Form II) and double-strand cleavage (Form III) of supercoiled plasmid DNA (Form I).
pBS54 was first mobilized into selected model Streptomyces hosts,27,29 Streptomyces albus J1074, Streptomyces lividans K4-114, and Streptomyces coelicolor M512, via conjugation, affording the corresponding recombinant strains SB9028 (S. albus J1074/pBS54), SB9029 (S. lividans k4-114/pBS54), and SB9030 (S. coelicolor M512/pBS54), respectively. The three recombinant strains were fermented along with S. verticillus ATCC15003 as a positive control.16 BLM production was not detected under any conditions examined (SI and Figure S4).
pBS54 was next mobilized into S. flavoviridis SB900113 via conjugation to afford the recombinant strain SB9027. Fermentation of SB9027, with SB9001 as a positive control, was carried out under the optimized conditions for ZBM production13 (SI). HPLC analysis of the fermentation showed the production of two new major metabolites, together with several minor metabolites, including ZBM and 6′-hydroxy-ZBM, the structures of which were confirmed by MS analysis and comparison to known standards (Figure 3C, panels I and II) (SI). Production of ZBM and 6′-hydroxy-ZBM in SB9027 indicated that the biosynthetic machineries for both BLM and ZBM disaccharides are functional (Figure 3A).
The two new major products from SB9027 exhibited almost identical UV-Vis spectra as BLMs, and both featured absorbance at 248 nm and 294 nm, which is characteristic for the bithiazole moiety of the BLM aglycone, suggesting that they are most likely BLM analogues.13,21,23,24 ESI-MS analysis in positive mode of the two new products yielded [M + Cu]2+ ions at m/z 722.3 and 714.4, respectively, which differ from any of the BLMs known to date.1–4,8–13,21,24 In an analogy to the structural relationship between ZBM and 6′-hydroxy-ZBM, the apparent 16 mass unit difference between the two compounds suggested that they could be new BLM analogues with disaccharides varying by one hydroxyl group. Production of ZBM, 6′-hydroxy-ZBM, as well as BLM analogues in SB9027 suggested that the biosynthetic machineries for both BLM and ZBM aglycones are functional, and inactivation of the latter could afford a platform for the engineered production of BLM analogues exclusively (Figure 3A).
Engineering the S. flavoviridis recombinant strain SB9026 that produces BLM Z and 6′-deoxy-BLM Z
The zbmVIII gene encodes the PKS that is responsible for the biosynthesis of the polyketide moiety of ZBM (Figures S1 and S2).25 We first deleted a 3.7-kb SbfI internal fragment of zbmVIII in SB9001 to construct an in-frame ΔzbmVIII deletion mutant strain SB9025 (Figure S5). The in-frame deletion strategy was preferred to ensure that the SB9025 mutant strain abolishes ZBM aglycone biosynthesis but remains competent to support ZBM disaccharide biosynthesis. pBS54 was next introduced into SB9025 via conjugation to afford the recombinant strain SB9026. Fermentation of SB9026, together with SB9001, SB9025, and SB9027 as controls, was carried out under the optimized conditions for ZBM production13 (SI). HPLC analysis of the metabolites produced confirmed that SB9025 completely lost ZBM production (Figure 3C, panels I vs. III) and that SB9026 produced only the two predicted BLM analogues (Figure 3C, panels II vs. IV), the exact phenotype that would be expected when expressing the BLM cluster in a ZBM aglycone-null host such as SB9025 (Figure 3A). The two new BLM analogues were produced in approximately a 1:1 ratio (Figure 3C, panel IV), and under the optimized fermentation conditions, their titers were estimated to be ~10 mg/L each.
Isolation of the two new metabolites produced by SB9026 was carried out by following protocols described previously for BLMs and analogues13,21,23,24 (SI). In brief, the fermentation broth (12 L) was centrifuged, and the supernatant was collected and adjusted to pH 7.0. After sequential column chromatography on Amberlite® IRC-50, Diaion HP-20, and Sephadex LH-20, followed by semipreparative HPLC on a C-18 column, the two new metabolites were purified as blue BLM•Cu2+ complexes, named BLM Z•Cu2+ (8.3 mg) and 6′-deoxy-BLM Z•Cu2+ (9.7 mg), respectively. Final treatment of the BLM•Cu2+ complexes with EDTA, followed by HPLC on a C-18 column, afforded Cu2+-free BLM Z (2.1 mg) and 6′-deoxy-BLM Z (2.3 mg) as white powders, respectively.
The structures of BLM Z, a new BLM analogue with the ZBM terminal amine moiety (Figure 1), and 6′-deoxy-BLM Z, a new BLM analogue with both the ZBM terminal amine moiety and the ZBM disaccharide (Figure 1), were elucidated by a combination of MS and 1H and 13C NMR spectroscopic analyses, as well as by comparison to the 1H and 13C NMR data of BLM A221,23,24 and ZBM13 (SI, Figures. S6, S13–S23 and Tables S1 and S2). Thus, HR-MALDI-MS analysis in positive mode of the Cu2+-free BLM Z, yielded an [M + H]+ ion at m/z 1382.5274, establishing the molecular formula of BLM Z as C53H79N19O21S2 (calculated for C53H79N19O21S2 + H+, 1382.5212). Analysis of 1D and 2D NMR spectra of BLM Z, along with comparison to those of BLM A2, indicated the presence of structural moieties characteristic of the BLMs, including the metal-binding domain – the pyrimidoblamic acid subunit along with the adjacent hydroxyl histidine, the (2S,3S,4R)-4-amino-3-hydroxy-2-methylpentanoic acid subunit, the disaccharide moiety, and the bithiazol group.13,21,23,24 Apart from these moieties, immediately identifiable from the NMR spectroscopic data for BLM Z were resonances consistent with the ZBM terminal amine moiety, 3-aminopropionamidine [δC 169.2 (s), 33.0 (t), and 36.9 (t); δH 3.82 (m) and 2.83 (t, J = 6.8 Hz)].13 BLM Z was finally determined to be a new BLM analogue featuring the ZBM terminal amine moiety, whose assignment was unambiguously established on the basis of the key HMBC correlations between H2-37 [δH 3.82 (m)] and the two tertiary carbons C-39 [δC 169.2 (s)] and C-36 [δC 163.7 (s)]. This deduced structure was further confirmed by detailed analysis of 1D and 2D NMR spectra (Figure S6), which also enabled the full 1H and 13C NMR spectroscopic assignments for BLM Z as summarized in Tables S1 and S2 (SI).
HR-MALDI-MS analysis in positive mode of Cu2+-free 6′-deoxy-BLM Z yielded an [M + H]+ ion at m/z 1366.5198, establishing the molecular formula of 6′-deoxy-BLM Z as C53H79N19O21S2 (calculated for C53H79N19O20S2 + H+, 1366.5263). The 16 mass unit decrease in comparison with BLM Z supported the assignment of this new metabolite as a deshydroxyl analogue of BLM Z. Detailed comparison of the 1H and 13C NMR data of this metabolite with those of BLM Z led to the conclusion that the only difference was that the hydroxymethylene group [δC at 60.5 and δH at 3.57 (overlap) and 3.42 (dd, J = 11.8 and 5.5 Hz)] at C-6′ of the disaccharide in BLM Z was replaced by a methyl group [δC at 14.7 and δH at 0.90 (d, J = 6.5 Hz)] in 6′-deoxy-BLM Z (Figgure S6). Taken together, these analyses finally established the new metabolite as 6′-deoxy-BLM Z, whose full 1H and 13C NMR spectroscopic assignments are summarized in Tables S1 and S2 (SI).
Evaluating the DNA cleavage activities of 6′-hydroxy-ZBM, BLM Z, and 6′-deoxy-BLM Z, in comparison to ZBM and BLM A2
6′-Hydroxy-ZBM, BLM Z, and 6′-deoxy-BLM Z were compared to ZBM and BLM A2 for their ability to cleave pBluescript II SK(+) supercoiled plasmid DNA in the presence of Fe2+ as described previously9,21,23 (SI). BLM-mediated single-strand cleavage initially results in the conversion of supercoiled plasmid DNA (form I) to open circular plasmid DNA (form II), and double-strand cleavage subsequently generates linearized plasmid DNA (form III) (Figures 2C, 3D, and S7). In this assay, ZBM was significantly more potent than BLM A2. ZBM at 0.1 μM resulted in nearly 50% plasmid relaxation, whereas 0.2 μM BLM A2 was required for the same amount of plasmid relaxation (Figure 2C and 3D). Replacing the ZBM disaccharide with the BLM disaccharide resulted in an approximately 2-fold reduction in DNA cleavage activity. This was evident when comparing 6′-hydroxy-ZBM with ZBM, which exhibited 50% plasmid relaxation at 0.2 μM and 0.1 μM, respectively (Figure 2C). This finding also suggested that the ZBM disaccharide may be superior to the BLM disaccharide in mediating DNA cleavage. BLM Z and BLM A2 showed almost identical DNA cleavage activity, with 50% plasmid relaxation at 0.2 μM each (Figure 3D). The ZBM terminal amine moiety showed no significant difference in mediating DNA cleavage activity from the terminal amine moiety of BLM A2. The 6′-deoxy-BLM Z was most potent compound tested, exhibiting almost 100% plasmid relaxation at 0.1 μM, whereas 100% plasmid relaxation would require BLM A2, BLM Z, or ZBM minimally at 0.5 μM each (Figure 3D). This finding supports the previous data obtained with 6′-hydroxy-ZBM (Figure 2C) and allows us to conclude that the ZBM disaccharide is preferred and could be exploited to engineer BLM analogues with improved DNA cleavage acitivity (Figures 2 and 3).
The observed DNA cleavage activity was concentration-dependent for all of the BLMs and ZBMs tested. To determine the relative DNA cleavage activities of 6′-hydroxy-ZBM, BLM Z, and 6′-deoxy-BLM Z, in comparison with ZBM and BLM A2, the effective concentrations at 50% plasmid DNA relaxation (EC50) were determined using 0.65 μg of pBluescript SK II (+) plasmid DNA, 10 μM Fe2+, and varying concentrations of 6′-hydroxy-ZBM (from 0.025 to 3.00 μM), BLM Z (from 0.010 to 1.00 μM), 6′-deoxy-BLM Z (from 0.010 to 1.00 μM), ZBM (from 0.02 to 2.00 μM), and BLM A2 (from 0.05 to 1.00 μM). Reactions were incubated at 37 °C for 30 min (Figure S7) (SI). The EC50 values were estimated to be 0.26 ± 0.05 μM for 6′-hydroxy-ZBM, 0.22 ± 0.02 μM for BLM Z, 0.030 ± 0.003 μM for 6′-deoxy-BLM Z, 0.10 ± 0.01 μM for ZBM, and 0.32 ± 0.02 μM for BLM A2 as summarized in Figures 2C and 3D. Again, 6′-deoxy-BLM Z exhibited an EC50 that was at least 10-fold more potent than BLM A2, representing the most potent BLM analogue known to date.1–4,8–13,21,24
DISCUSSION
BLMs and TLMs differ from ZBM in their disaccharide moieties (Figure 1). Comparative analysis of the BLM, TLM, and ZBM biosynthetic machinery revealed that the L-gulose moiety in BLM resulted from direct epimerization of the NDP-D-mannose precursor by BlmG, while the combined action of ZbmL and ZbmG converted the NDP-D-mannose precursor into the 6-deoxy-L-gulose moiety found in ZBM (Figure S2).12 The fact that ZBM production was abolished in the ΔzbmL mutant strain SB9003 and could be restored upon expression of a functional copy of zbmL in trans in recombinant strain SB9009 suggested that ZbmG and BlmG, in spite of their high sequence homology (64% identity/71% similarity), apparently exhibit distinct substrate specificity, ZbmG for NDP-4-keto-6-deoxy-D-mannose, and BlmG for NDP-D-mannose (Figures 2 and 3).12,16,25 ZbmL/ZbmG and BlmG therefore represent ideal candidates to use for engineering designer ZBM and BLM analogues with altered disaccharide moieties, on the assumption that the BlmEF or ZbmEF glycosyltransferases in both BLM and ZBM machineries are forgiving in the choice of the disaccharides to be incorporated into the BLM and ZBM aglycones (Figures 2A, 3A, and S2).
We took advantage of the expedient genetic system developed for S. flavoviridis25 to investigate if novel ZBM analogues could be produced by cross-complementation of the ΔzbmL mutant strain SB9003 with blmGEF (as in SB9012), blmGE (as in SB9011), or blmG (as in SB9013) (Figure 2A). It was rather unexpected that the three recombinant strains afforded the same phenotype (Figure 2B), producing the same new ZBM analogue, which was subsequently characterized as 6′-hydroxy-ZBM featuring the BLM disaccharide (Figure 1). These findings confirmed that BlmG can functionally reconstitute NDP-L-gulose biosynthesis in the ΔzbmL mutant strain SB9003 and that the ZbmEF glycosyltransferase apparently can efficiently incorporate the altered sugar into the designer ZBM analogue. SB9011, SB9012, and SB9013 produced 6′-hydroxy-ZBM in a similar titer (~4 mg/L), which was approximately 40% of that of ZBM (~10 mg/L) by SB9003.13 The structure of 6′-hydroxy-ZBM was established on the basis of extensive MS and NMR spectroscopic data analyses (Figures S3, S8–S12 and Tables S1, S2).
Engineering sugar biosynthetic pathways for BLM in S. verticillus21 and for TLM in S. hindustanus22–24 have all been demonstrated, respectively, resulting in the production of BLM or TLM analogues with altered sugar moieties. We therefore attempted to carry out the reciprocal experiments in the BLM producer S. verticillus, by cross-complementing a ΔblmG mutant with zbmLG, to engineer a BLM analogue with the ZBM disaccharide. This approach was eventually abandoned due to the inability to effectively genetically manipulate S. verticillus.
We opted to express the entire BLM biosynthetic machinery in the ZBM producer S. flavoviridis to explore the feasibility of engineering novel BLM analogues in a genetically amenable heterologous host (Figure 3A).12,25 The recently developed E. coli-Streptomyces BAC shuttle vector26 enabled us to construct a BAC (pBS54) that contained the entire BLM cluster on a 120-kb insert (Figure 3B). The BAC pBS54 was introduced into Streptomyces heterologous hosts by oriT-mediated intergeneric conjugation and incorporated into chromosomes of the hosts via site-specific integration.26–28 Upon introduction of pBS54 into S. flavoviridis SB9001, the resultant recombinant strain SB9027 produced four major metabolites (Figure 3C, panel II). As expected, BLM Z and ZBM were produced, confirming functional expression of the BLM machinery from the introduced BAC clone pBS54 and of the ZBM machinery from the host background SB9001 under the conditions examined. However, two new hybrid metabolites, 6′-deoxy-BLM Z, a BLM analogue with the ZBM disaccharide and 6′-hydroxy-ZBM, a ZBM analogue with the BLM disaccharide were also produced. The production of 6′-deoxy-BLM Z and 6′-hydroxy-ZBM confirmed functional cross-talk between the biosynthetic machineries for the BLM and ZBM aglycones and their respective disaccharide moieties in the recombinant strain SB9027 (Figure 3A).12
We next constructed the ΔzbmVIII PKS mutant strain SB9025 in order to abolish the biosynthesis of the ZBM aglycone and create a cleaner background in the host so that the production of the BLM analogues, upon expression of the BLM biosynthetic machinery from pBS54, could be more readily discerned (Figure 3A). Introduction of pBS54 into the ΔzbmVIII PKS mutant strain SB9025 afforded recombinant strain SB9026, fermentation of which under the optimized conditions for ZBM production indeed produced only the two BLM analogues (Figure 3C, panel IV), each of which carried the BLM and ZBM disaccharide, respectively. SB9026 produced BLM Z and 6′-deoxy-BLM Z in approximately a 1:1 ratio with a titer of ~10 mg/L for each of the metabolites, which was comparable to that of ZBM levels (~10 mg/L) in the parent SB9001 strain.13 The structures of BLM Z and 6′-deoxy-BLM Z were established on the basis of extensive MS and NMR spectroscopic data analyses (Figures S6, S19–S23 and Tables S1, S2).
The anticancer activity of the BLM family of metabolites has been correlated with their in vitro DNA cleavage activity.1–4,8,9,21,23 Evaluation of the in vitro DNA cleavage activity of 6′-hydroxy-ZBM, BLM Z, and 6′-deoxy-BLM Z, compared to ZBM and BLM A2 as controls, shows the remarkable impact of the disaccharide on the activity of BLMs and ZBMs (Figures 2C, 3D, and S7). Among the over 160 BLM analogues that have been prepared and analyzed to date, none has shown much improvement in activity over the parent BLMs and very few have modifications at the disaccharide moiety.1–4,8–13,21,24 Although PLM and ZBM are known to be significantly more potent than the BLMs in mediating DNA cleavage, the BLMs are the only members of this family that have been developed into clinical drugs.1–4 We now show that the ZBM disaccharide contributes, at least in part, to the superior activity of ZBM in mediating DNA cleavage and that the designer BLM analogue 6′-deoxy-BLM Z, which features the ZBM disaccharide, exhibits outstanding DNA cleavage activity in vitro. This was evidenced by its impressive EC50 of 0.030 ± 0.003 μM, which is approximately one order of magnitude smaller than BLM Z with an EC50 of 0.22 ± 0.02 μM and BLM A2 with an EC50 of 0.32 ± 0.02 μM under the conditions examined (Figures 2C, 3D, and S7). To the best of our knowledge, 6′-deoxy-BLM Z represents the most potent BLM known to date.1–4,8–13,21,24 Most importantly, these findings should inspire further effort toward creating additional analogues of BLM, specifically targeting the disaccharide moiety of BLM and screening for even further improved activity. Emerging technologies in “glycorandomization” for natural product structural diversity should greatly facilitate such an endeavor.30
It has been proposed that the dose-limiting, accumulative lung toxicity associated with BLM chemotherapy is independent of the cytotoxicity of BLMs to tumor cells.2,5–7 It has yet to be confirmed by in vivo study that the improved DNA cleavage activity observed in vitro with 6′-deoxy-BLM can be translated into improved anticancer activity with decreased side effects. Still, it is exciting to speculate that the increased potency of 6′-deoxy-BLM could have a great chance to result in new BLM chemotherapy regimens with a reduced dosage, thereby lessening the dose-limiting side effect and the incidence of lung toxicity.5–7 The fact that the 6′-deoxy-BLM is biosynthesized by the recombinant S. flavoviridis strain SB9026 and can be produced in reasonable quantities by scale-up microbial fermentation should greatly facilitate the follow-up mechanistic and preclinical studies as well as its development into a potential clinical drug.
Supplementary Material
Acknowledgments
We thank Dr. Marie F. Coeffet-LeGal, Cubist Pharmaceuticals Inc., Lexington, MA 02421, for providing the pStreptoBAC V vector, the Analytical Instrumentation Center of the School of Pharmacy, UW-Madison for support in obtaining MS and NMR data, and the John Innes Center, Norwich, U.K., for providing the REDIRECT Technology kit. This work was supported in part by the Chinese Ministry of Education 111 Project B08034 (to Y.D.) and the NIH grant CA94426 (to B.S.).
ABBREVIATIONS
- BAC
bacterial artificial chromosome
- BLM
bleomycin
- CID
collision-induced dissociation
- ESI
electrospray ionization
- HR
high resolution
- MALDI
matrix-assisted laser desorption ionization
- MS
mass spectrometry
- NRPS
noribosomal peptide synthetase
- PKS
polyketide synthase
- PLM
phleomycin
- SI
- TLM
tallysomycin
- ZBM
zorbamycin
Footnotes
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript. 6These authors contributed equally.
Supporting Information. A complete description of materials, methods, and detailed experimental procedures. Also included in SI are: (i) Figures S1 and S2 depicting the BLM, TLM, and ZBM biosynthetic gene clusters and the proposed pathways for BLM, TLM, and ZBM sugar biosynthesis and attachment to their respective aglycones; (ii) Tables S1 and S2 summarizing 1H and 13C NMR data of ZBM, 6′-hydroxy-ZBM, BLM Z, and 6′-deoxy-BLM Z; (iii) Figures S3 and S8–S12 of 1H, 13C, 1H-1H gCOSY, gHMBC, and gHMQC NMR spectra of 6′-hydroxy-ZBM; (iv) Figure 4 summarizing the failed attempt to produce BLMs in SB9028, SB9029, and SB9030; (v) Figure 5 depicting the construction of the ΔzbmVIII mutant strain SB9025 and confirmation of its genotype by Southern analysis; (vi) Figures S6 and S13–S18 of 1H, 13C, 1H-1H gCOSY, gHMBC, gHSQC, and HSQC-TOCSY NMR spectra of BLM Z; (vii) Figures S6 and S19–S23 of 1H, 13C, 1H-1H gCOSY, gHMBC, and gHMQC NMR spectra of 6′-deoxy-BLM Z; and (viii) Figure S7 summarizing DNA cleavage activities of 6′-hydroxy-ZBM, BLM Z, and 6′-deoxy-BLM Z in comparison with BLM A2 and ZBM as observed in plasmid relaxation assays with pBlue script II SK(+). This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Boger DL, Cai H. Angew Chem Int Ed. 1999;38:448–476. doi: 10.1002/(SICI)1521-3773(19990215)38:4<448::AID-ANIE448>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 2.Hecht SM. J Nat Prod. 2000;63:158–168. doi: 10.1021/np990549f. [DOI] [PubMed] [Google Scholar]
- 3.Chen J, Stubbe J. Nat Rev Cancer. 2005;5:102–112. doi: 10.1038/nrc1547. [DOI] [PubMed] [Google Scholar]
- 4.Galm U, Hager MH, Van Lanen SG, Ju H, Thorson JS, Shen B. Chem Rev. 2005;105:739–758. doi: 10.1021/cr030117g. [DOI] [PubMed] [Google Scholar]
- 5.Einhorn LH. Proc Natl Acad Sci USA. 2002;99:4592–4595. doi: 10.1073/pnas.072067999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huddart RA, Birtle AJ. Expert Rev Anticancer Ther. 2005;5:123–138. doi: 10.1586/14737140.5.1.123. [DOI] [PubMed] [Google Scholar]
- 7.Sleijfer S. Chest. 2001;120:617–624. doi: 10.1378/chest.120.2.617. [DOI] [PubMed] [Google Scholar]
- 8.Leitheiser CJ, Smith KL, Rishel MJ, Hashimoto S, Konishi K, Thomas CJ, Li C, McCormick MM, Hecht SM. J Am Chem Soc. 2003;125:8218–8227. doi: 10.1021/ja021388w. [DOI] [PubMed] [Google Scholar]
- 9.Ma Q, Xu Z, Schroeder BR, Sun W, Wei F, Hashimoto S, Konishi K, Leitheiser CJ, Hecht SM. J Am Chem Soc. 2007;129:12439–12452. doi: 10.1021/ja0722729. [DOI] [PubMed] [Google Scholar]
- 10.Chapuis J-C, Schmaltz RM, Tsosie KS, Belohlavek M, Hecht SM. J Am Chem Soc. 2009;131:2438–2439. doi: 10.1021/ja8091104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takita T, Muraoka Y. In: Biochemistry of Peptide Antibiotics: Recent Advances in the Biotechnology of β-Lactam and Microbial Peptides. Kleinkauf H, von Dohren H, editors. W de Gruyter; New York: 1990. pp. 289–309. [Google Scholar]
- 12.Galm U, Wendt-Pienkowski E, Wang L, Huang S-X, Unsin C, Tao M, Coughlin JM, Shen B. J Nat Prod. 2011;74:526–536. doi: 10.1021/np1008152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang L, Yun B-S, George NP, Wendt-Pienkowski E, Galm U, Oh T-J, Coughlin JM, Zhang G, Tao M, Shen B. J Nat Prod. 2007;70:402–406. doi: 10.1021/np060592k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van Lanen SG, Shen B. In: Antitumor Agents from Natural Products. 2. Kingston DGI, Cragg GM, Newman DJ, editors. CRC Press; Boca Raton: 2012. pp. 667–694. [Google Scholar]
- 15.Du L, Shen B. Chem Biol. 1999;6:507–517. doi: 10.1016/S1074-5521(99)80083-0. [DOI] [PubMed] [Google Scholar]
- 16.Du L, Sanchez C, Chen M, Edwards DJ, Shen B. Chem Biol. 2000;7:623–642. doi: 10.1016/s1074-5521(00)00011-9. [DOI] [PubMed] [Google Scholar]
- 17.Du L, Chen M, Sanchez C, Shen B. FEMS Microbiol Lett. 2000;189:171–175. doi: 10.1111/j.1574-6968.2000.tb09225.x. [DOI] [PubMed] [Google Scholar]
- 18.Sanchez C, Du L, Edwards DJ, Toney MD, Shen B. Chem Biol. 2001;8:725–738. doi: 10.1016/s1074-5521(01)00047-3. [DOI] [PubMed] [Google Scholar]
- 19.Schneider TL, Shen B, Walsh CT. Biochemistry. 2003;42:9722–9730. doi: 10.1021/bi034792w. [DOI] [PubMed] [Google Scholar]
- 20.Du L, Chen M, Zhang Y, Shen B. Biochemistry. 2003;42:9731–9740. doi: 10.1021/bi034817r. [DOI] [PubMed] [Google Scholar]
- 21.Galm U, Wang L, Wendt-Pienkowski E, Tao M, Coughlin JM, Shen B. J Biol Chem. 2008;283:28236–28245. doi: 10.1074/jbc.M804971200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tao M, Wang L, Wendt-Pienkowski E, George NP, Galm U, Zhang G, Coughlin JM, Shen B. Mol BioSyst. 2007;3:60–74. doi: 10.1039/b615284h. [DOI] [PubMed] [Google Scholar]
- 23.Wang L, Tao M, Wendt-Pienkowski E, Galm U, Coughlin JM, Shen B. J Biol Chem. 2009;284:8256–8264. doi: 10.1074/jbc.M900640200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tao M, Wang L, Wendt-Pienkowski E, Zhang N, Yang D, Galm U, Coughlin JM, Xu Z, Shen B. Mol BioSyst. 2010;6:349–356. doi: 10.1039/b918106g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Galm U, Wendt-Pienkowski E, Wang L, George NP, Oh T-J, Yi F, Tao M, Coughlin JM, Shen B. Mol BioSyst. 2009;4:77–90. doi: 10.1039/b814075h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miao V, Coeffet-LeGal MF, Brian P, Brost R, Penn J, Whiting A, Martin S, Ford R, Parr I, Bouchard M, Silva CJ, Wrigley SK, Baltz RH. Microbiology. 2005;151:1507–1523. doi: 10.1099/mic.0.27757-0. [DOI] [PubMed] [Google Scholar]
- 27.Feng Z, Wang L, Rajski SR, Xu Z, Coeffet-LeGal MF, Shen B. Bioorg Med Chem. 2009;17:2147–2153. doi: 10.1016/j.bmc.2008.10.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang D, Zhu X, Wu X, Feng Z, Huang L, Shen B, Xu Z. Appl Microbiol Biotechnol. 2011;89:1709–1719. doi: 10.1007/s00253-010-3025-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Galm U, Shen B. Exp Opinion Drug Discov. 2006;1:409–437. doi: 10.1517/17460441.1.5.409. [DOI] [PubMed] [Google Scholar]
- 30.Gantt RW, Peltier-Pain P, Thorson JS. Nat Prod Rep. 2011;28:1811–1853. doi: 10.1039/c1np00045d. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.