Abstract
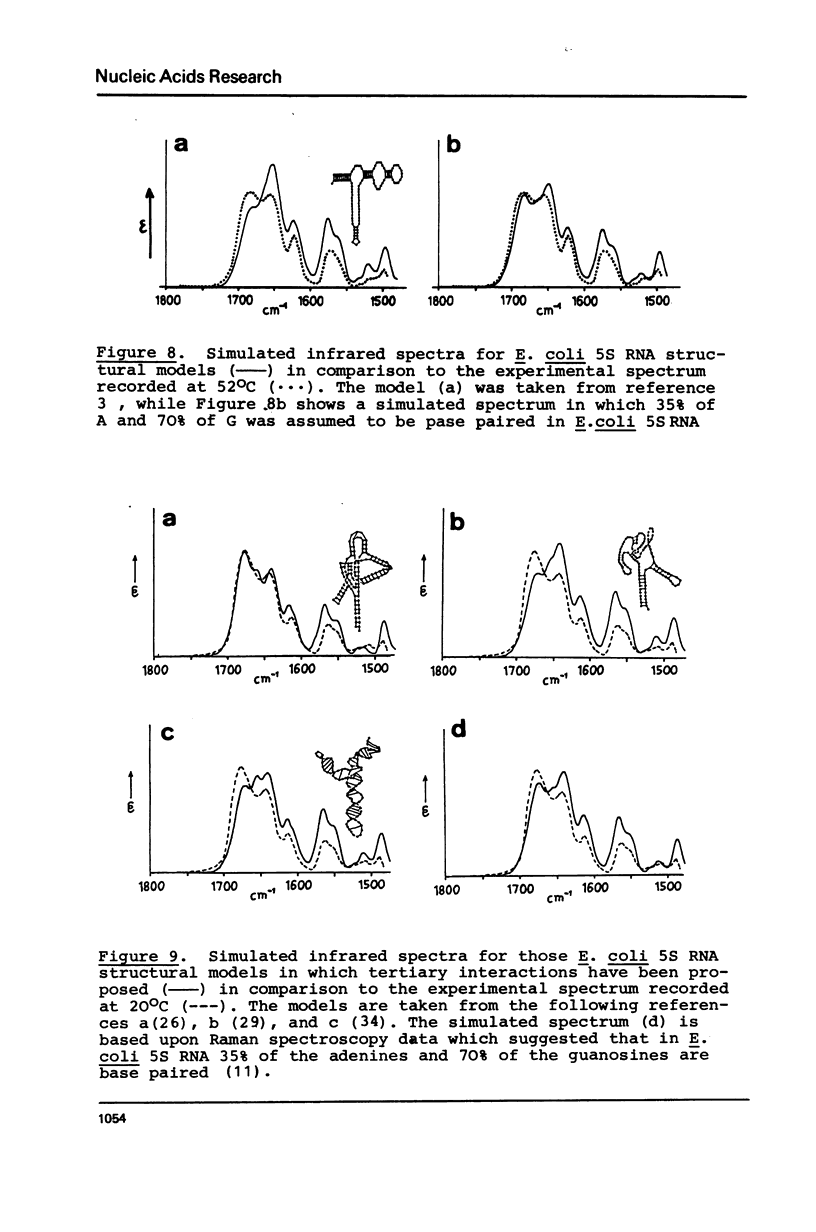
The extent of base pairing in Escherichia coli and Bacillus stearothermophilus 5S RNAs was determined by infrared spectroscopy. From the infrared spectra taken at 20 degrees and 52 degrees C it is concluded that E. coli and B. stearothermophlius 5S RNAs possess a large number of base pairs (Table I). Comparison of our results with those previously published using other methods leads to the conclusion that the structures of prokaryotic 5S RNAs involve a large number of tertiary interactions, in which the base pairing is not necessarily solely of the Watson-Crick type.
Full text
PDF

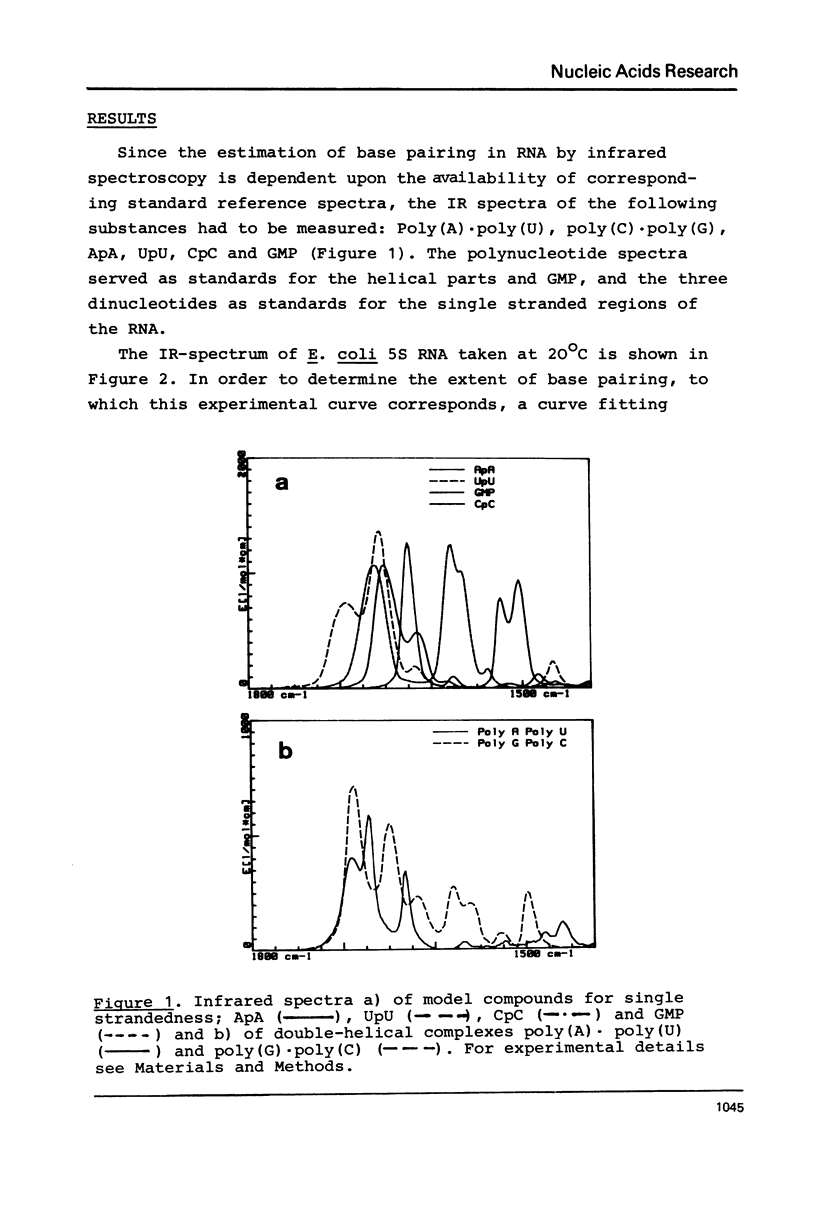
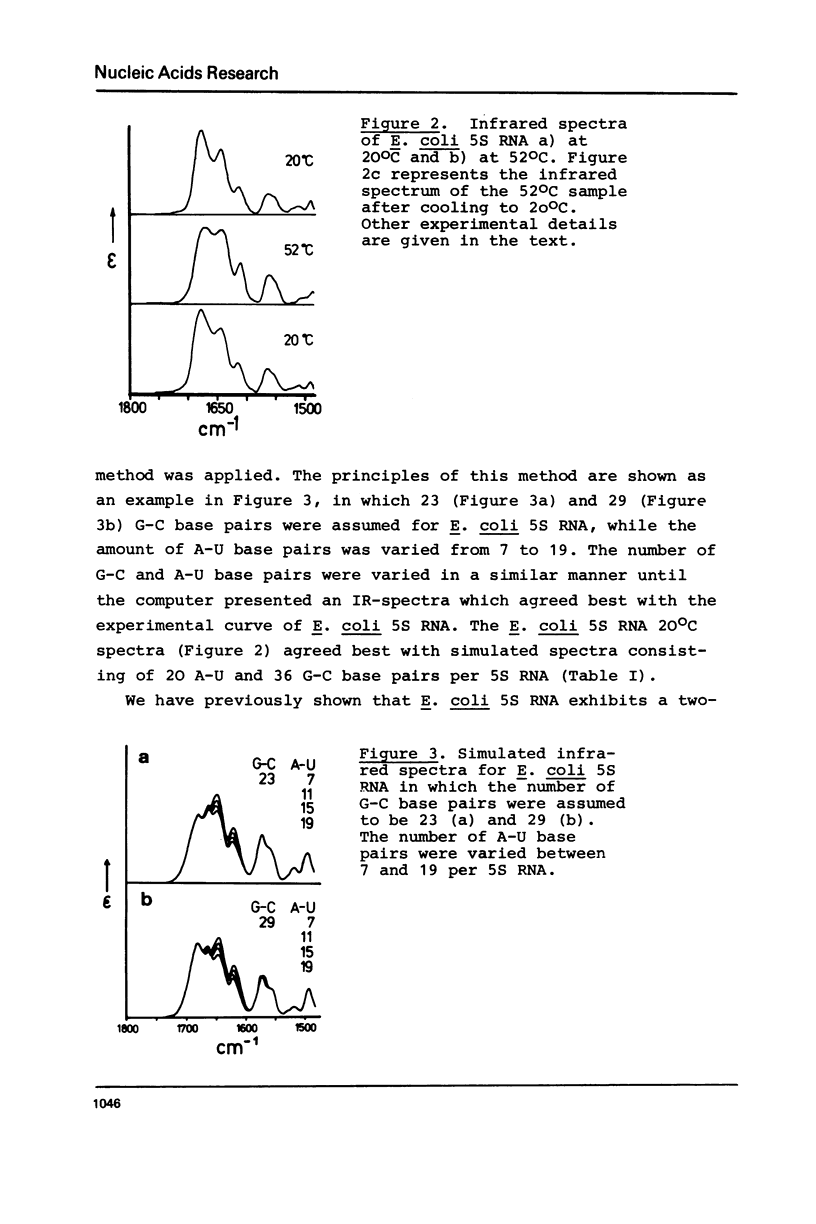
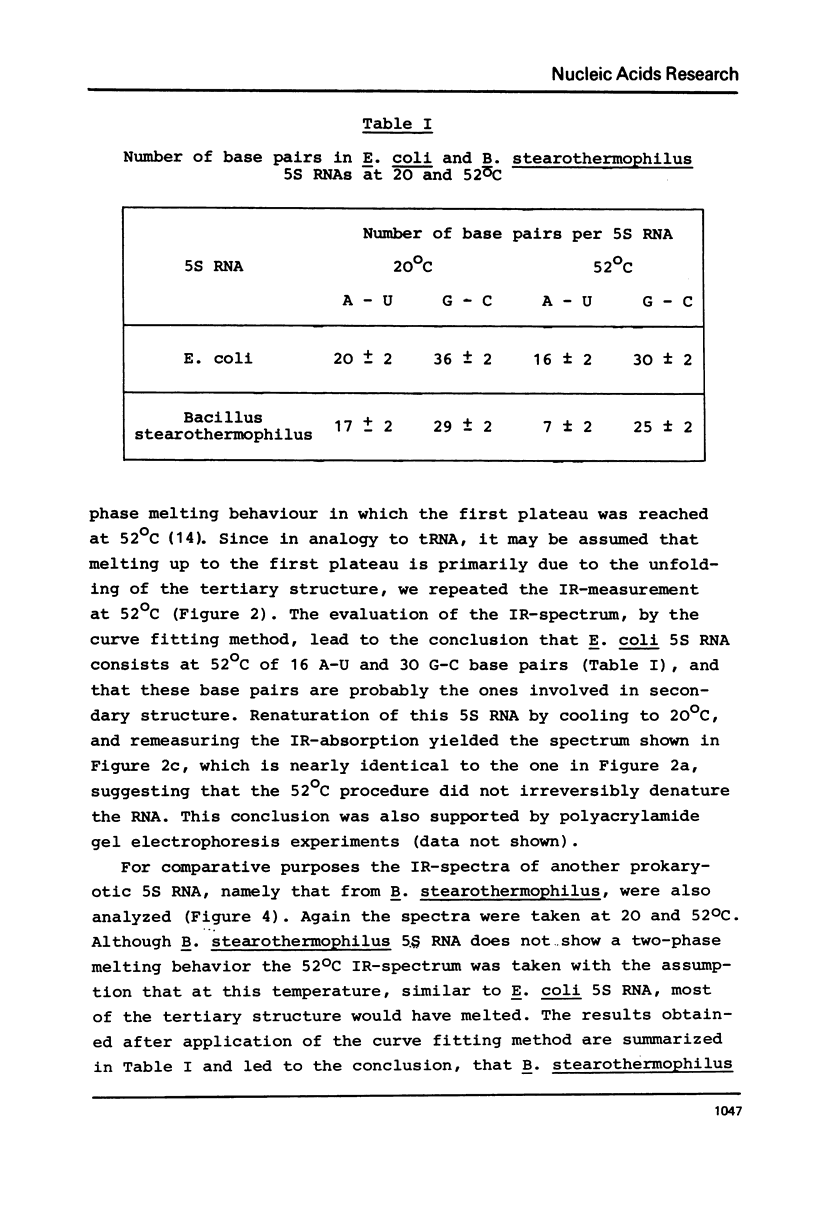
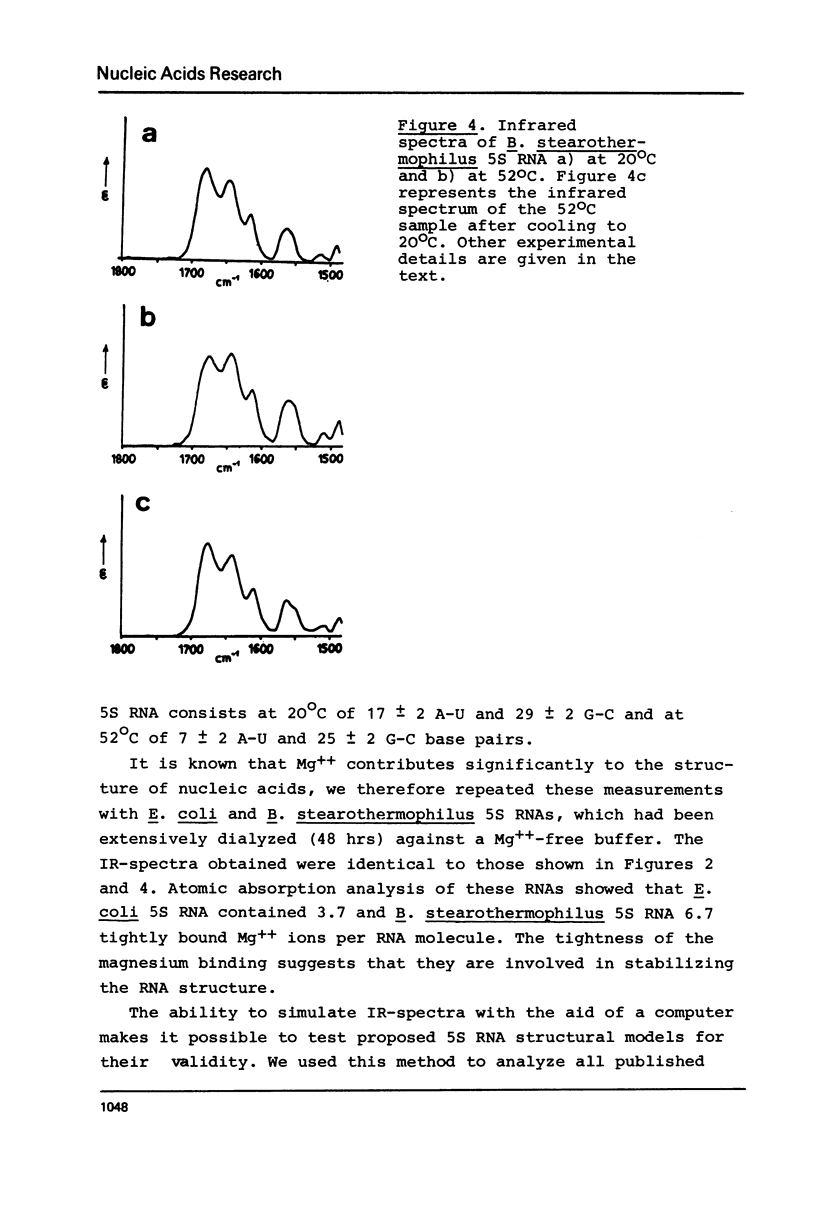

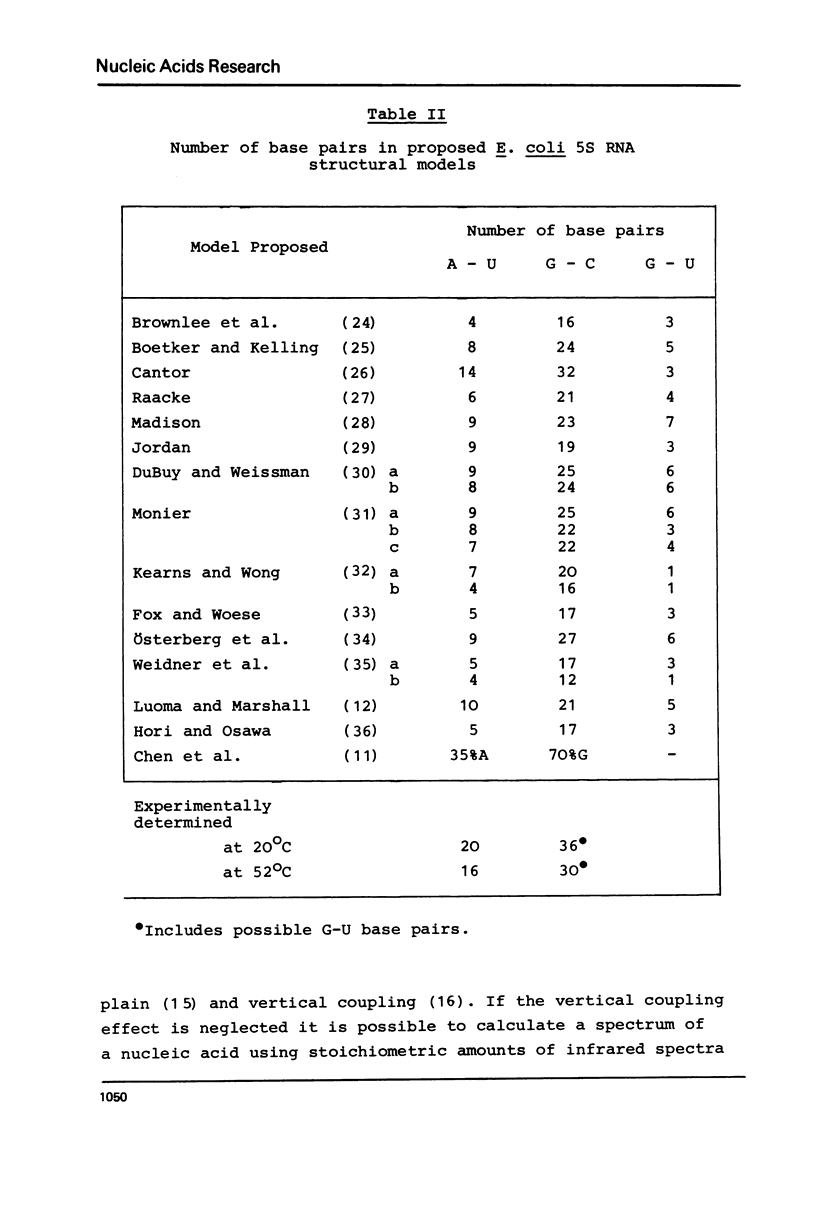
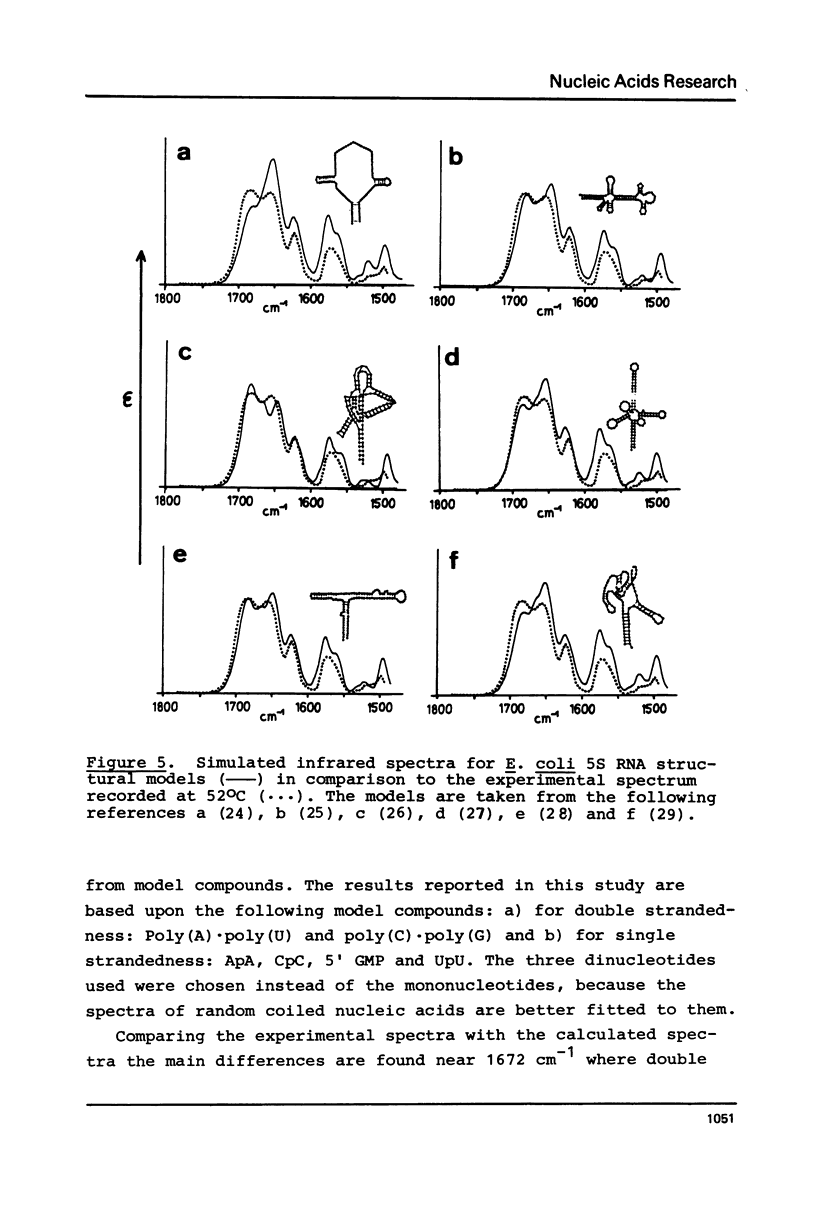
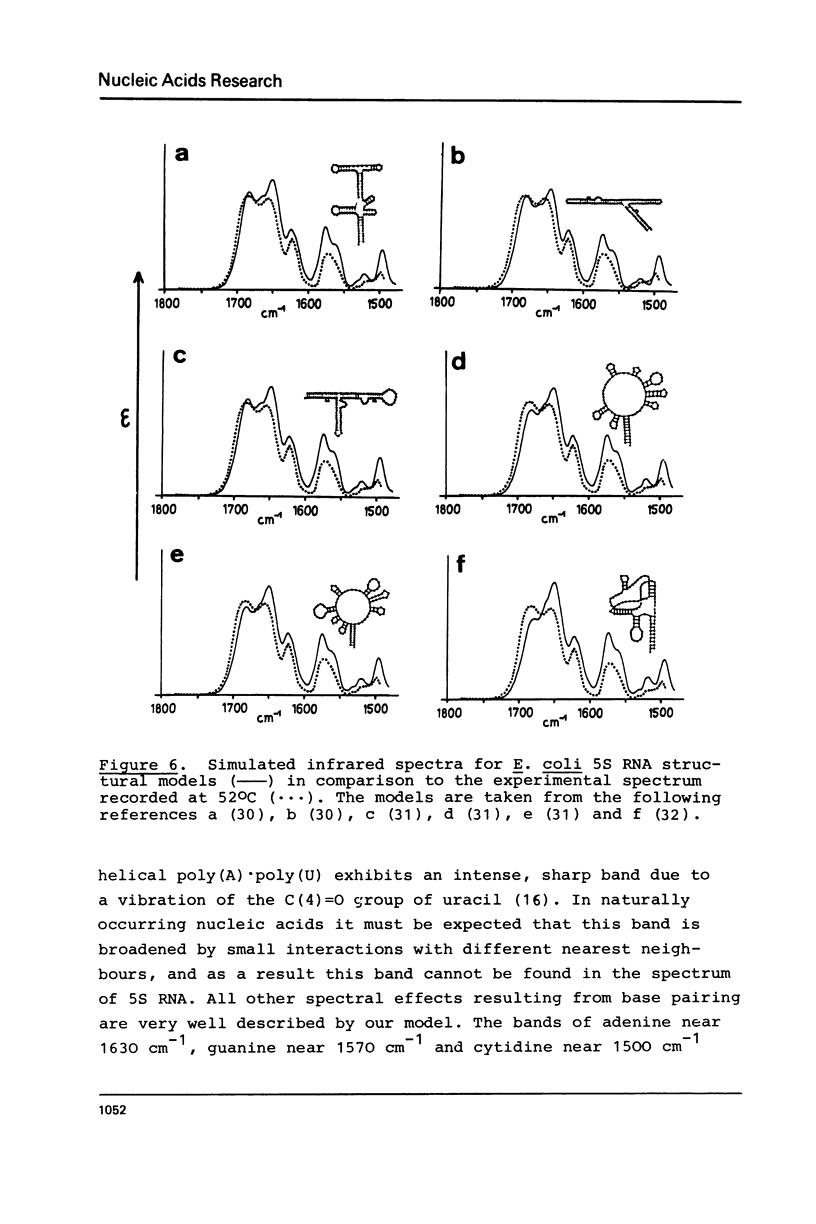
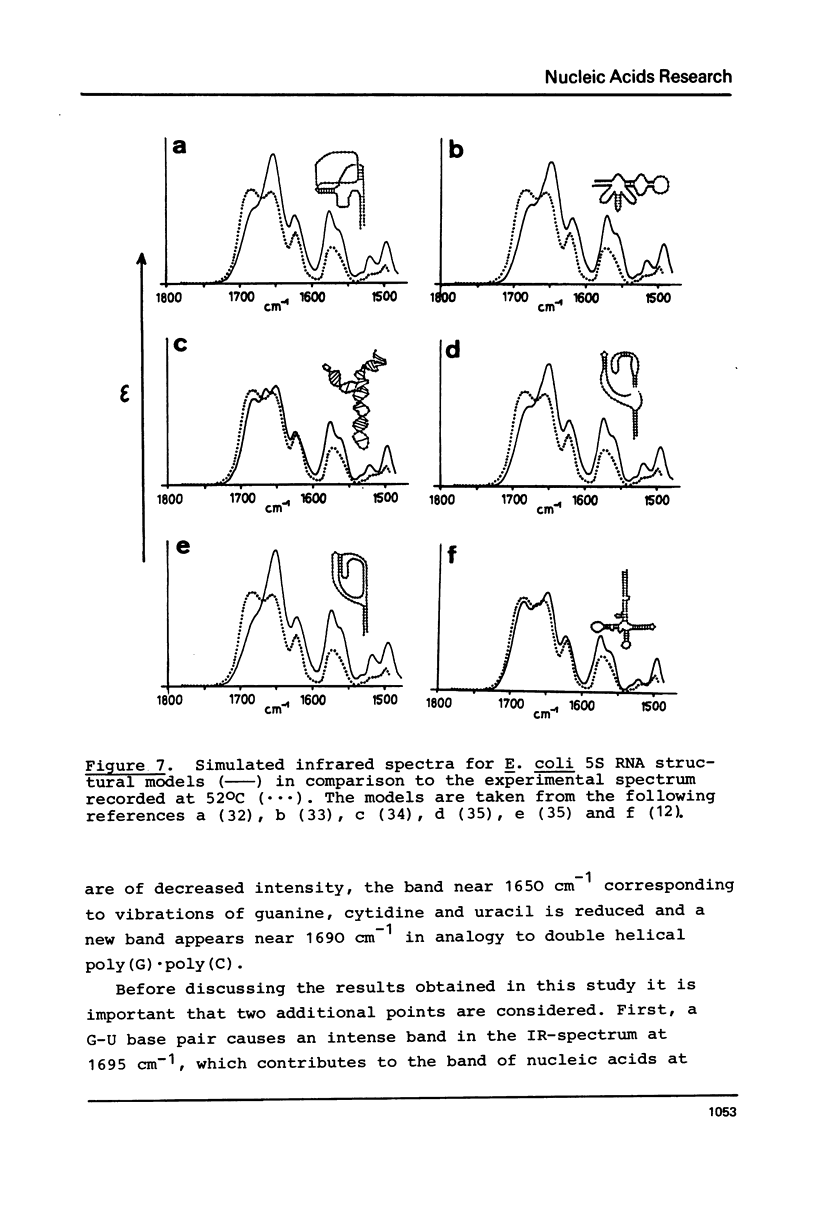



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Boedtker H., Kelling D. G. The ordered structure of 5S RNA. Biochem Biophys Res Commun. 1967 Dec 15;29(5):758–766. doi: 10.1016/0006-291x(67)90283-5. [DOI] [PubMed] [Google Scholar]
- Brownlee G. G., Sanger F., Barrell B. G. Nucleotide sequence of 5S-ribosomal RNA from Escherichia coli. Nature. 1967 Aug 12;215(5102):735–736. doi: 10.1038/215735a0. [DOI] [PubMed] [Google Scholar]
- Cantor C. R. Possible conformations of 5-S ribosomal RNA. Nature. 1967 Nov 4;216(5114):513–514. doi: 10.1038/216513a0. [DOI] [PubMed] [Google Scholar]
- Chen M. C., Giegé R., Lord R. C., Rich A. Raman spectra of ten aqueous transfer RNAs and 5S RNA. Conformational comparison with yeast phenylalanine transfer RNA. Biochemistry. 1978 Jul 25;17(15):3134–3138. doi: 10.1021/bi00608a030. [DOI] [PubMed] [Google Scholar]
- Cramer F., Erdmann V. A. Amount of adenine and uracil base pairs in E. coli 23S, 16S and 5S ribosomal RNA. Nature. 1968 Apr 6;218(5136):92–93. doi: 10.1038/218092a0. [DOI] [PubMed] [Google Scholar]
- DuBuy B., Weissman S. M. Nucleotide sequence of Pseudomonas fluorescens 5 S ribonucleic acid. J Biol Chem. 1971 Feb 10;246(3):747–761. [PubMed] [Google Scholar]
- Erdmann V. A. Collection of published 5S and 5.8S RNA sequences and their precursors. Nucleic Acids Res. 1979 Jan;6(1):r29–r44. doi: 10.1093/nar/6.1.419-c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann V. A., Doberer H. G. Structure and function of 5S RNA: the role of the 3' terminus in 5S RNA function. Mol Gen Genet. 1972;114(2):89–94. doi: 10.1007/BF00332779. [DOI] [PubMed] [Google Scholar]
- Erdmann V. A. Structure and function of 5S and 5.8 S RNA. Prog Nucleic Acid Res Mol Biol. 1976;18:45–90. [PubMed] [Google Scholar]
- Fox G. E., Woese C. R. The architecture of 5S rRNA and its relation to function. J Mol Evol. 1975 Oct 3;6(1):61–76. doi: 10.1007/BF01732674. [DOI] [PubMed] [Google Scholar]
- Hori H., Osawa S. Evolutionary change in 5S RNA secondary structure and a phylogenic tree of 54 5S RNA species. Proc Natl Acad Sci U S A. 1979 Jan;76(1):381–385. doi: 10.1073/pnas.76.1.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard F. B., Frazier J., Miles H. T. Interbase vibrational coupling in G:C polynucleotide helices. Proc Natl Acad Sci U S A. 1969 Oct;64(2):451–458. doi: 10.1073/pnas.64.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan B. R. Computer generation of pairing schemes for RNA molecules. J Theor Biol. 1972 Feb;34(2):363–378. doi: 10.1016/0022-5193(72)90168-3. [DOI] [PubMed] [Google Scholar]
- Katsura T., Morikawa K., Tsuboi M., Kyogoku Y., Seno T. Infrared spectra of transfer ribonucleic acids. 3. Fragments of formyl-methionine tRNA from Escherichia coli. Biopolymers. 1971;10(4):681–698. doi: 10.1002/bip.360100407. [DOI] [PubMed] [Google Scholar]
- Lewis J. B., Doty P. Identification of the single-strand regions in Escherichia coli 5S RNA, native and A forms, by the binding of oligonucleotides. Biochemistry. 1977 Nov 15;16(23):5016–5025. doi: 10.1021/bi00642a012. [DOI] [PubMed] [Google Scholar]
- Luoma G. A., Marshall A. G. Laser Raman evidence for new cloverleaf secondary structures for eukaryotic 5.8S RNA and prokaryotic 5S RNA. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4901–4905. doi: 10.1073/pnas.75.10.4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madison J. T. Primary structure of RNA. Annu Rev Biochem. 1968;37:131–148. doi: 10.1146/annurev.bi.37.070168.001023. [DOI] [PubMed] [Google Scholar]
- Marshall A. G., Smith J. L. Nuclear-spin-labeled nucleic acids. 1 19F nuclear magnetic resonance of Escherchia coli 5-fluorouracil-5S-RNA. J Am Chem Soc. 1977 Jan 19;99(2):635–636. doi: 10.1021/ja00444a066. [DOI] [PubMed] [Google Scholar]
- Miles H. T., Frazier J. Infrared study of helix strandedness in the poly A-poly U system. Biochem Biophys Res Commun. 1964;14:21–28. doi: 10.1016/0006-291x(63)90204-3. [DOI] [PubMed] [Google Scholar]
- Morikawa K., Tsuboi M., Kyogoku Y., Seno T., Nishimura S. Infrared spectrum of an anticodon fragment of a transfer RNA. Nature. 1969 Aug 2;223(5205):537–538. doi: 10.1038/223537a0. [DOI] [PubMed] [Google Scholar]
- Morikawa K., Tsuboi M., Takahashi S., Kyogoku Y., Mitsui Y., Iitaka Y., Thomas G. J., Jr The vibrational spectra and structure of poly(rA-rU)-poly(rA-rU). Biopolymers. 1973 Apr;12(4):799–816. doi: 10.1002/bip.1973.360120409. [DOI] [PubMed] [Google Scholar]
- Osterberg R., Sjöberg B., Garrett R. A. Molecular model for 5-S RNA. A small-angle x-ray scattering study of native, denatured and aggregated 5-S RNA from Escherichia coli ribosomes. Eur J Biochem. 1976 Sep 15;68(2):481–487. doi: 10.1111/j.1432-1033.1976.tb10835.x. [DOI] [PubMed] [Google Scholar]
- Raacke I. D. "Cloverleaf" conformation for 5S RNAs. Biochem Biophys Res Commun. 1968 May 23;31(4):528–533. doi: 10.1016/0006-291x(68)90509-3. [DOI] [PubMed] [Google Scholar]
- Schernau U., Ackermann T. Determination of the base-pairing content of four specific tRNAs by infrared spectroscopy. Biopolymers. 1977 Aug;16(8):1735–1745. doi: 10.1002/bip.1977.360160810. [DOI] [PubMed] [Google Scholar]
- Thomas G. J., Jr Determination of the base pairing content of ribonucleic acids by infrared spectroscopy. Biopolymers. 1969;7(3):325–334. doi: 10.1002/bip.1969.360070305. [DOI] [PubMed] [Google Scholar]
- Tsuboi M., Higuchi S., Kyogoku Y., Nishimura S. Infrared spectra of transfer RNA's. II. Formylmethionine transfer RNA from Escherichia coli in aqueous solution. Biochim Biophys Acta. 1969 Nov 19;195(1):23–28. [PubMed] [Google Scholar]
- Weidner H., Yuan R., Crothers D. M. Does 5S RNA function by a switch between two secondary structures? Nature. 1977 Mar 10;266(5598):193–194. doi: 10.1038/266193a0. [DOI] [PubMed] [Google Scholar]
- Wrede P., Pongs O., Erdmann V. A. Binding oligonucleotides to Escherichia coli and Bacillus stearothermophilus 5 S RNA. J Mol Biol. 1978 Mar 25;120(1):83–96. doi: 10.1016/0022-2836(78)90296-6. [DOI] [PubMed] [Google Scholar]
- Wrede P., Pongs O., Erdmann V. A. Binding oligonucleotides to Escherichia coli and Bacillus stearothermophilus 5 S RNA. J Mol Biol. 1978 Mar 25;120(1):83–96. doi: 10.1016/0022-2836(78)90296-6. [DOI] [PubMed] [Google Scholar]


