Abstract
We have developed and validated a sandwich chemiluminescence immunoassay (SCIA) which measures polycyclic aromatic hydrocarbon (PAH)–DNA adducts combining high throughput and adequate sensitivity, appropriate for evaluation of adduct levels in human population studies. Fragmented DNA is incubated with rabbit antiserum elicited against DNA modified with r7,t8-dihydroxy-t-9,10-epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene (BPDE) and subsequently trapped by goat anti-rabbit IgG bound to a solid surface. Anti-single-stranded (ss) DNA antibodies binds in a quantity proportional to the adduct levels and is detected by chemiluminescence. The BPDE–DNA SCIA has a limit of detection of 3 adducts per 109 nucleotides with 5 μg DNA per well. We have validated the BPDE–DNA SCIA using DNA modified in vitro, DNA from benzo[a]pyrene (BP)-exposed cultured cells and mice. The levels of adduct measured by SCIA were lower (30–60%) than levels of bulky DNA adducts measured in the same samples by 32P-postlabelling. The BPDE–DNA SCIA also detected adducts produced in vivo by PAHs other than BP. When blood DNA samples from maternal/infant pairs were assayed by BPDE–DNA SCIA, the adduct levels obtained were significantly correlated. However, there was no correlation between 32P-postlabelling and SCIA values for the same samples. The SCIA can be extended to any DNA adduct and is expected to provide, when fully automated, a valuable high-throughput approach in large-scale population studies.
Introduction
Polycyclic aromatic hydrocarbons (PAHs), a group of chemicals released into the environment primarily as a result of incomplete fuel combustion, comprise a number of carcinogenic components. For this reason and because of their universal presence in the environment, the impact of PAHs on human health has been of worldwide concern (1–5).
Carcinogenic PAHs are metabolised to electrophilic species that react with DNA. The resulting DNA adducts initiate processes that result in cancer development. The prototypical PAH, benzo[a]pyrene (BP), gives rise to multiple DNA adducts of guanine and adenine. However, one major adduct formed in vivo, the r7,t8, t9-trihydroxy-c-10-(N2deoxyguanosyl)-7,8,9,10-tetrahydro-benzo[a]pyrene (BPdG), is thought to make critical contributions to cellular events leading to carcinogenesis (6,7). PAH–DNA adducts may serve as integrated biomarkers of PAH exposure, reflecting the overall impact of chronic exposure, individual metabolic capacity and DNA repair (8–12). In addition, they can be biomarkers of cancer risk (13–17).
The ability to measure PAH–DNA adducts in large-scale human studies would seem critical to exposure evaluation and for human cancer risk analysis. The two most commonly used assay methods, 32P-postlabelling (18) and competitive immunoassays utilising antisera elicited against DNA modified with BP (19), have advantages but also disadvantages that hamper their use in such studies. 32P-postlabelling is highly sensitive, able to detect a few adducts in 1010 nucleotides using 1 μg of DNA, but for the analysis of DNA from environmentally exposed humans has very low chemical specificity. The procedure is labour intensive and uses 32P radioisotope. Competitive enzyme-linked immunoabsorbent assays (ELISA), on the other hand, recognise only PAH–DNA adducts and has the potential for high throughput. However, competitive ELISA-based immunoassays typically require 50–150 μg of total DNA per sample, an amount that may not be easily available in many studies.
Here, we report the development and validation of a new direct ELISA with a chemiluminescence end point, the BPDE–DNA sandwich chemiluminescence immunoassay (BPDE–DNA SCIA). For the detection of PAH–DNA adducts in human samples, this assay has all the advantages of the BPDE–DNA chemiluminescence immunoassay (CIA) (20) and, in addition, requires only 11 μg of DNA for duplicate analysis (including 1 μg which compensates inaccuracies during pipetting), indicating suitability for large-scale population studies. An adaptation of a recently reported analogous assay for O6-methylguanine (O6-meG) (21), the BPDE–DNA SCIA employs DNA fragments (Mspl restriction) containing ≤1 PAH–DNA adduct residue. A rabbit antiserum elicited against DNA modified with BPDE (22) selectively transfers the adduct-containing fragments from the bulk of the DNA to a solid surface. For the final detection, monoclonal anti-single-stranded (ss) DNA antibody is utilised in combination with a chemiluminescence end point. The high ratio of normal nucleotides to adducts in each fragment serves to enhance the signal and sensitivity while maintaining proportionality with the PAH–DNA adducts.
It has been previously shown that the antiserum cross reacts with DNA adducted with other PAHs (23) and hence, when analysing human DNA samples, the signal obtained reflects a composite exposure to a mixture of PAHs and therefore the presence of adducts of a variety of PAHs (24).
In this series of experiments, we have validated the new BPDE–DNA SCIA using a variety of approaches. We have established reproducibility for both the standard curve and the repeated assay of a single sample. We have shown dosimetry for BP bound to DNA in HepG2 cells and dosimetry and cross reactivity for benzo[a]pyrene (BP), as well as dibenzo[a,h]anthracene (DB[a,h]A) and benzo[b]fluoranthene (B[b]F) bound to DNA in mouse liver. Comparison between the BPDE–DNA SCIA and 32P-postlabelling correlated well for values obtained using HepG2 cells and mouse liver. Human blood DNA samples were largely positive in both assays, but there was no correlation in magnitude between values obtained by the two methods.
Materials and methods
Biochemicals
The anti-BPDE–DNA rabbit antiserum was from rabbit #33 bleed 8/2/1978 and was elicited against DNA modified in vitro with BPDE, where the only measurable adduct was the BPdG (22). BPDE was custom synthesised by Dr Albrecht Seidel at Biochemisches Institut für Umweltcarcinogene, Prof. Dr Gernot Grimmer-Stiftung, Grosshansdorf, Germany. All other biochemicals were from Sigma Chemical Co. (St Louis, MO, USA) unless otherwise stated.
Purification of DNA
The purification of DNA from HepG2 cells, human blood mononuclear cells and mouse tissues was performed with Qiagen Blood & Cell Culture DNA purification kits, according to the manufacturer’s instructions with minor modifications (18,21). DNA purified with this method had an average length of 35 000–50 000 bp and an A260/A280 ratio within the range of 1.75–1.85. DNA from MCF-7 cells was purified by three methods: Qiagen columns, traditional phenol/chloroform/isoamyl alcohol extraction (25) and the salting out method for protein precipitation and DNA isolation (26).
Cell culture
HepG2 cells were grown in Dulbecco’s minimal essential medium (Gibco) (containing glucose 4.5 g/l, L-glutamine and pyruvate) supplemented with 10% fetal bovine serum (Gibco) and 1% penicillin/streptomycin (Gibco), at 37°C in a humidified incubator with 5% CO2. Cells were treated with 0.02–2.00 μM BP and harvested after 24 h for DNA isolation. Pellets of MCF-7 cells, which had been treated in aliquots of 2 × 107 cells with 1 μM BP at 37°C for 24 h, were kindly provided by Dr D.H. Phillips (see Acknowledgments).
Mouse exposures
Male CD2F1 mice, 10 weeks old and weighing 25–30 g (purchased from Charles River Laboratories Italia S. P. A., Italy), were administered single intraperitoneal injections of BP (100 and 200 mg/kg), B[b]F (200 and 400 mg/kg) and DB[a,h]A (2.5, 5 and 10 mg/kg), in tricaprylin. The animals were sacrificed 14 days after the treatment, the liver tissue was removed and used for DNA extraction by Qiagen mini columns, as described above and subsequent adduct analysis In addition, aliquots of liver DNA’s from several mice, treated with BP, were pooled and served as a quality control standard (QC) in all the ELISA assays.
Standard BPDE–modified DNA (BPDE–DNA)
Samples of DNA modified to different levels with BPDE were prepared by incubating HeLa cell DNA (20 mg; 1 mg/ml in 0.1 M Tris–HCl, pH 7.4) overnight in the dark at 37°C with BPDE dissolved in tetrahydrofuran. The final concentrations of BPDE were 100, 25, 10 and 2.5 μg/ml. After repeated extractions with diethyl ether (eight times) and isoamyl alcohol (four times), both saturated with water, the aqueous phase was collected and the DNA was precipitated by adding 2.5 volumes cold ethanol, washed twice with 70% ethanol, dried and redissolved in distilled water. The levels of BPdG adducts were determined by spectrophotometric analysis, using an extinction coefficient for the BPdG adduct at 350 nm of 29 000. The concentrations of the adducted double-stranded DNA samples were calculated assuming that one absorbance unit at 260 nm equals 50 μg/ml. The modification levels of the DNA samples obtained for the concentrations of BPDE mentioned above were 7.00 (0.7%), 1.31 (0.13%), 0.64 (0.064%) and 0.16 (0.016%) adducts per 1000 nucleotides, respectively. The modified DNA samples were sent to the University of Leicester (see Acknowledgments), where they were analysed for BPdG adducts using liquid chromatography-mass spectrometry (LC-MS/MS), and the results obtained were in excellent agreement with the spectrophotometric measurements. The DNA with the lowest level of modification (∼1 adduct per 6000 nucleotides) was used for the SCIA standard curves.
PAH–DNA adduct analysis with the BPDE–DNA SCIA
The new immunoassay was first developed for O6-meG, and details have been described in Georgiadis et al. (21). In general, the same assay conditions were used for the BPDE–DNA SCIA, but the use of the BPDE–DNA antiserum and the additional steps required optimisation. For the BPDE–DNA SCIA, a 96-well high-binding microtiter plate (Greiner Labortechnik, Germany) was coated with 100 μl goat anti-rabbit IgG, species absorbed for minimum cross reactivity (Millipore Inc., Billerica, MA, USA), diluted 1:300-fold in coating buffer (Na2CO3 0.015 M, NaHCO3 0.035 M; pH: 9.6) and incubated overnight at 4°C. After washing three times with water, non-specific binding was blocked by adding to the wells 310 μl 0.25% casein (I-block; Tropix, Bedford, MA, USA) in phosphate-buffered saline with 0.5% Tween (PBST) for 1.5 h at 37°C, followed by two cycles of washing with PBST.
Standard adducted DNA, containing 0.16 BPdG adducts per 1000 nucleotides, and the unknown DNA samples (duplicates of 5.5 μg each; 11 μg in total) were exhaustively digested with MspI (10 units/μg DNA) (New England Biolabs Inc.) by overnight incubation at 37°C in a total volume of 55 μl in 50 mM NaCl, 10 mM Tris–HCl, 10 mM MgCl2, 1 mM dithiothreitol; pH 7.9. The DNA was subsequently denatured by heating to 95°C for 10 min, snap frozen in liquid nitrogen and then allowed to thaw on ice. Subsequently, an equal volume of anti-BPDE–DNA rabbit antiserum (diluted 1:12 500 in 2×PBST, 0.5% casein) was added to the DNA and the mixture (110 μl in total) was incubated at 37°C for 1.5 h in a shaking water bath. One hundred microlitres of each DNA plus antiserum mixture (containing 10 μg of DNA) was then transferred into triplicate microtiter plate wells and incubated for 1.5 h at room temperature, followed by five cycles of washing with PBST. Two sequential steps then followed: (i) addition of 100 μl mouse anti-ss DNA antibodies, semi-purified from culture cell supernatant (Millipore Inc.) diluted 100-fold with PBST, 0.25% casein and (ii) addition of purified goat anti-mouse-IgG antiserum conjugated with alkaline phosphatase, with minimum species cross reactivity, (Biolegend, San Diego, CA, USA) diluted (1:500) in PBST containing 0.25% casein. Each step was followed by five cycles of washing with PBST. Finally, two additional washing cycles with Tris buffer (20 mM Tris–1 mM MgCl2, pH 9.5) were performed. One hundred microlitres CDP-Star containing Emerald II enhancer (Tropix, Bedford, MA, USA) was then added to the wells, the plates were incubated at room temperature for 30 min and luminescence was read using a Safire II Microplate Luminometer (TECAN) at 542 nm. The levels of PAH–DNA adducts in the unknown samples were calculated by comparison with a BPDE–DNA standard curve (common range of standard: 0.1–2 fmol BPdG adducts per well), constructed for each microwell plate employed on each assay occasion. In addition, a sample of BP-treated mouse liver DNA of known adduct content (measured also by LC–MS–MS) was used in triplicate with each microtiter plate as a quality control standard.
Determination of bulky DNA adducts by 32P-postlabelling
The levels of bulky DNA adducts were determined by the regular 32P-postlabelling procedure, as described in detail previously (18).
Samples from mother–infant pairs
Samples of blood mononuclear cells were collected from subjects (40 mothers and paired 40 newborns) participating in a mother–newborn child biobank in Copenhagen (27), a pilot study within the Newborns and genotoxic exposure risks (NewGeneris) project (28). Ethical clearance for the study was provided by the Danish Ethics Committee, and the project was reviewed and approved prior to initiation by the Ethics Committee of the Capital Region of Denmark and the Danish Data Protection Agency (J. Nr. H-KF-01-327603; J. Nr. 2007-41-0415). Venous peripheral blood from the mother and cord blood of her newborn child was collected at delivery into heparinised vacutainer tubes. Samples were processed within 1 h to obtain mononuclear cells, which were stored at −80°C until transfer to the assay laboratory.
Statistical methods
Tests used for statistical analysis included: (i) analysis of variance for comparisons of DNA adduct levels between DNA samples purified with different protocols; (ii) independent and paired Student’s t-tests to evaluate the difference of the adduct levels measured by SCIA and 32P-postlabelling; (iii) the Mann–Whitney and two-tailed Wilcoxon tests for assessing the difference of DNA adduct levels between maternal and cord blood and, (iv) Spearman correlation tests for calculating the correlation between DNA adduct levels measured in maternal and cord blood or samples analysed by different methods. For calculations of the levels of DNA adducts measured by SCIA in the human samples, samples with non-detectable adducts were assigned a value half way between zero and the limit of detection.
Results
Standard curves and assay precision
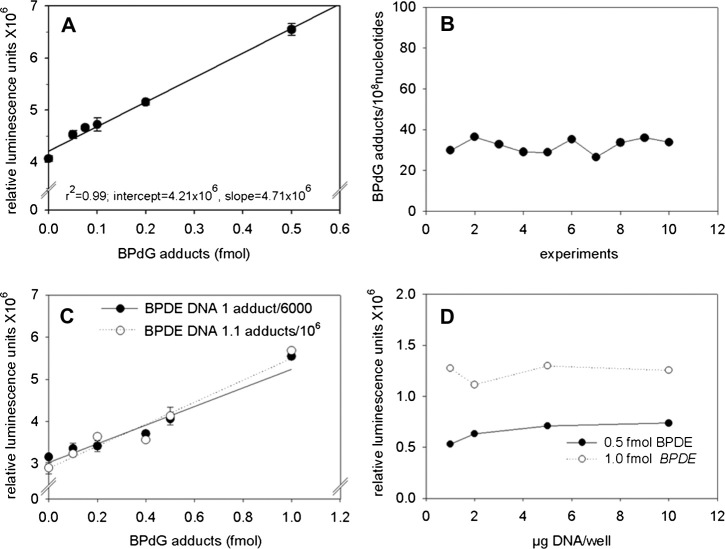
In order to minimise the between-assay variation, each microtiter plate contained a standard curve (see Materials and methods) constructed by mixing different amounts of standard DNA of known BPdG adduct content with adduct-free HeLa DNA. The level of modification of the standard DNA (∼1 adduct per 6000 normal nucleotides) ensured that the proportion of MspI digestion fragments with more than one adduct would be negligible. The signal-to-noise ratio of the assay continued to increase throughout the range of decreasing mouse anti-ss DNA antiserum dilutions tested (results not shown), and for this reason, final dilutions were adopted taking into consideration material availability and cost. As shown in Figure 1A, the luminescence signal obtained was linearly related (r 2 = 0.99) to the amount of BPDE–DNA adducts added (range: 0.05–0.5 fmol) and the coefficient of variance (CV) was 2.5%. Linearity is retained up to 2 or 3 fmols and the curve deviates from linearity (negatively) only above this concentration due to the instrument upper limit for signal detection (results not shown). Despite this excellent reproducibility, the absolute levels of the chemiluminescence signals showed significant day-to-day variation, which necessitated the construction of a standard curve for each experiment. The mean slope-to-intercept ratio of 10 standard curves, constructed over a period of 1 month, was 0.82 ± 0.22 (SD) fmol−1, demonstrating consistent and stable assay response. The mean limit of detection (3× SD of the signal of unmodified carrier DNA) of the above-mentioned standard curves was 42.0 ± 17.8 (SD) attomole. Most importantly, a QC sample of BP-treated DNA (see Materials and methods), which was included in all the above analyses, gave a mean adduct level of 32.2 adducts per 108 normal nucleotides (Figure 1B) with a CV of 10.5%. The same sample was also analysed by LC–MS–MS and the BPdG adduct levels were found to be 36.2 adducts per 108 nucleotides, in excellent agreement with the mean value determined by SCIA.
Fig. 1.
Validation of the direct sandwich ELISA assay (SCIA) for the detection of BPdG–DNA adduct levels, using 10 μg DNA per well and performing all reactions in triplicate. Rabbit polyclonal antiserum elicited against BPDE–DNA was diluted 1:25 000-fold. (A)‘High sensitivity’ standard curve (linear regression analysis) obtained using 0–0.5 fmol BPdG–DNA adducts. (B) Inter-day variability shows repetitive analyses of a quality control DNA sample containing 36.2 adducts per 108 nucleotides (assayed by LC–MS–MS). Ten different analyses, performed on different microtiter plates, on different days and utilising different standard curves gave a mean adduct level of 32.2 adducts per 108 normal nucleotides with a CV 10.5%. (C) Standard curves obtained using a DNA containing 1 BPdG per 6000 nucleotides (by spectrophotometry) and a DNA containing 1.1 BPdG per 106 nucleotides (assayed by LC–MS–MS). (D) Effect of the amount of DNA added per well on the chemiluminescence signal obtained with 0.5 and 1 fmol BPDE–DNA. Reactions were performed in triplicate and the background was subtracted.
In traditional competitive ELISAs, the accuracy of the measurement of the adduct levels is greatly dependent on the density of the adducts in the standard DNA, something that necessitates the latter being adducted at a similar level to the test samples (29). In the SCIA, the digestion of the DNA to fragments containing no more than one adduct is anticipated to overcome this necessity. In order to test this hypothesis and further validate the assay, we constructed two standard curves on the same plate, one using our standard DNA, which contained 1 adduct per 6000 nucleotides and a second one using DNA modified at a level of 1.1 adducts per 106 nucleotides as measured by LC–MS–MS (generous gift of Dr Fred Beland, see Acknowledgements). An excellent agreement between the two standard curves was obtained (Figure 1C), with their intercepts being 3.02 and 2.88 × 106 relative luminescence units (RLU) and the slopes 2.32 and 2.60 × 106 RLU.fmol−1, respectively. The impact of the total amount of DNA per well on assay sensitivity was evaluated by examining the variation of the intensity of the signal generated by a constant amount of BPdG (0.5 or 1.0 fmol) in the presence of 1–10 μg DNA per well. No significant variation in luminescence signal between 1 and 10 μg DNA was found (Figure 1D) and subsequently, when using 5 or 10 μg of DNA per well, the limit of detection was estimated as ∼3 or 1.5 adducts per 109 nucleotides, respectively.
Assay validation: dynamic range, influence of DNA isolation method comparison with 32P-postlabelling
Using DNA isolated from HepG2 cells treated for 24 h with 0.02–2.0 μM BP, the BPdG adducts measured by SCIA were found to be linearly related to the BP concentrations (Figure 2), indicating that the dynamic range of the assay is at least two orders of magnitude. The highest dose of BP gave rise to 22 BPdG adducts per 108 nucleotides while the lowest dose to 0.27 BPdG per 108 nucleotides.
Fig. 2.
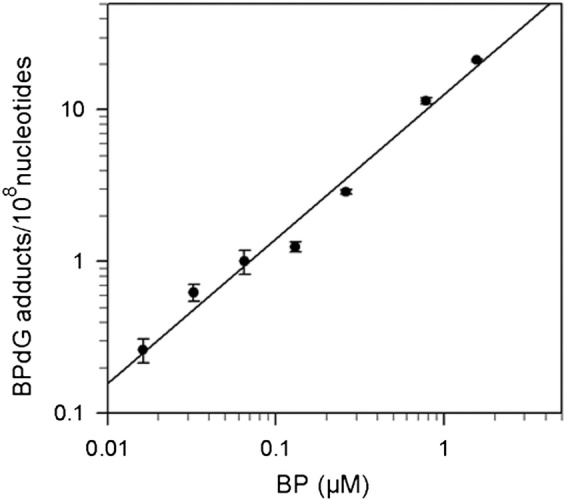
Log–log plot of DNA adduct levels, measured by BPDE–DNA SCIA, in HepG2 cells exposed for 24 h to 0.02–2.0 μM BP.
Experiments were conducted to evaluate the accuracy and reproducibility of the new assay, as well as the influence of the DNA isolation method on the measured adduct levels, and to compare the adduct levels measured in the same DNA by BPDE–SCIA and 32P-postlabelling (Table I). For this purpose, MCF-7 cells were treated with 1 μM BP for 24 h, harvested and separated into aliquots which were used to isolate DNA using three different methods: Qiagen columns, phenol–chloroform extraction and salting out. For each DNA isolation method, three different DNA samples were analysed twice (SCIA) or three times (32P-postlabelling) on consecutive days. SCIA showed better reproducibility than 32P-postlabelling, and the DNA adduct measurements by SCIA showed good reproducibility for samples analysed multiple times on the same day or on different days (Table I). In addition, unlike 32P-postlabelling, SCIA allowed us to analyse up to 70 samples/week per person and was proved to be a high-throughput method, when fully automated.
Table I.
Comparison of the BPdG levels measured in DNA from BP-treated MCF-7 cells by BPDE–DNA SCIA and 32P-postlabelling
| Purification methods | SCIAa, mean ± SD adducts per 108 | Mean % CV1 (within-day) | Mean % CV2 (inter-day) | 32P-postlabellingb, mean ± SD adducts per 108 | Mean % CV1 (within-day) | Mean % CV2 (Inter-day) | Ratioc | P1 | P2 |
| Phenol extraction | 1162 ± 256 | 20.7 | 8.9 | 1682 ± 496 | 27.0 | 26.3 | 0.7 | 0.11 | 0.27 |
| Qiagen columns | 871 ± 92 | 10.25 | 6.5 | 2048 ± 340 | 40.3 | 15.7 | 0.4 | <0.01 | <0.01 |
| Salting out | 799 ± 104 | 11.95 | 9.3 | 1712 ± 308 | 16.3 | 12.8 | 0.5 | <0.01 | 0.05 |
%CV1 refers to the within-day coefficient of variance for adduct levels measured by either method in independently extracted DNA samples, while %CV2 refers to the inter-day coefficient of variance of the same samples analysed twice or three times on different days; P1: P value for independent t-test for the inter-day mean adducts levels obtained by the two methods; P2: P value for paired t-test.
Triplicate DNA samples were extracted by the three methods and assayed independently twice.
Triplicate DNA samples were extracted by the three methods and assayed independently three times.
Ratio of adduct levels measured by SCIA to those measured by 32P-postlabelling.
There was no significant difference in DNA adduct value by SCIA when the DNA was prepared by any of the three methods. The levels of BPdG adducts measured by SCIA were 30–60% (depending on the DNA extraction method) lower than those measured by 32P-postlabelling. However, the mean SCIA adduct values measured on different days in the DNA samples prepared by Qiagen and salting out were highly correlated with those obtained by 32P-postlabelling, either when examined independently or when examined pairwise (P values in Table I). Although the SCIA gave consistently lower adducts than the 32P-postlabelling assay, it is probable that the major adduct in these cells was BPdG, and this is likely why the two assays showed a good correlation.
Assay specificity
The anti-BPDE–DNA antiserum employed in our assay was elicited against DNA modified with BP, but it has been shown to recognise and cross react with DNA modified with a series of carcinogenic PAHs in addition to BP (23). Therefore, because humans are exposed to mixtures of environmental PAHs, values for human samples, assayed using this antiserum with a BPDE–DNA standard curve, will be semi-quantitative. In addition, it is important to know as much as possible about the cross reactivity of the antiserum and two common PAHs which were not previously examined for anti-BPDE–DNA cross reactivity are B[b]F and DB[a,h]A. Therefore, in this experiment, mice were given single injections of BP, B[b]F and DB[a,h]A, and livers were obtained 14 days after exposure. DNA was extracted and analysed by BPDE–DNA SCIA and 32P-postlabelling (Figure 3). Although repair of DNA adducts may have occurred, as the mouse protocol was conducted in the context of a different study, the BPDE–DNA SCIA detected an essentially linear increase in DNA adducts with dose, not only for the BP but also for the other two PAHs (not shown). The levels of adducts measured by BPDE–DNA SCIA and 32P-postlabelling were proportional to each other (Figure 3), confirming the anticipated cross reactivity, although the numbers of adducts measured by BPDE–DNA SCIA were 3.3% for B[b]F, and 20% for BP and DB[a,h]A, of those measured by 32P-postlabelling. The observation that BP-induced mouse liver DNA adducts measured by BPDE–DNA SCIA were ∼20% of those measured by 32P-postlabelling, as compared to 30–60% reported above for adducts produced in MCF7 cells, may be explained on the basis of the known ability of the liver to form multiple types of DNA-binding products that may not cause similar responses in SCIA.
Fig. 3.
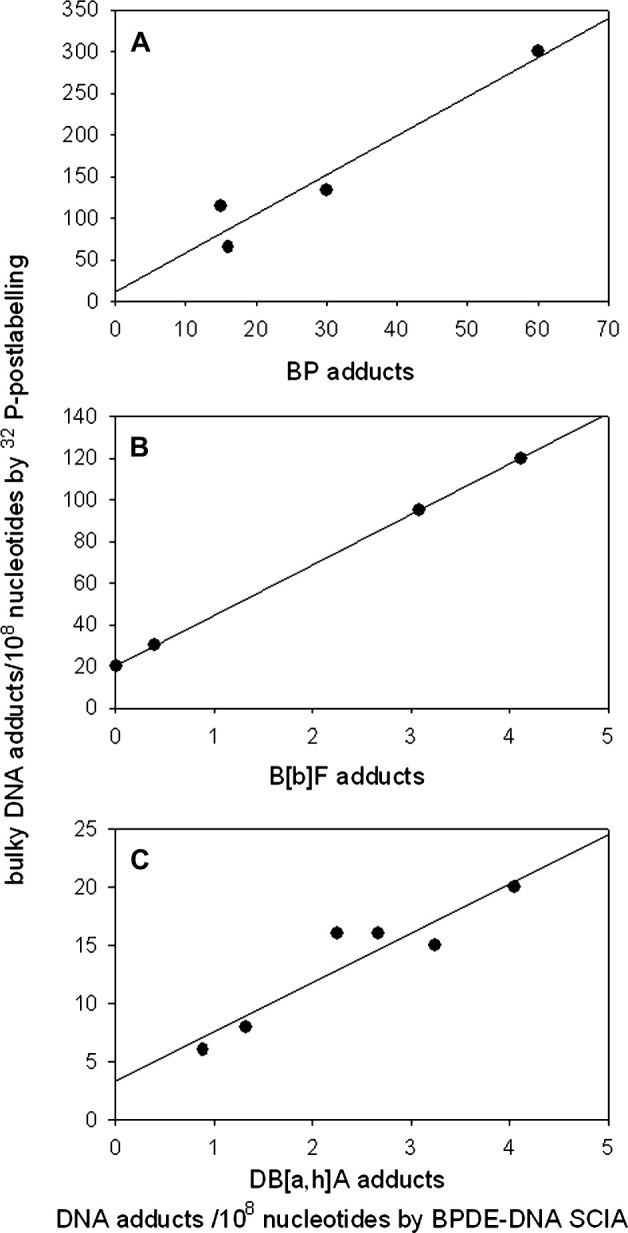
Comparison of DNA adduct levels detected by SCIA and 32P-postlabelling in liver DNA from mice treated in duplicate with: (A) 100 and 200 mg/kg body weight BP, (B) 200 and 400 mg/kg body weight B[b]F and (C) 2.5, 5 and 10.0 mg/kg body weight DB[a,h]A. Samples were taken 14 days after exposure.
DNA adducts determined in maternal peripheral and infant cord blood mononuclear cells
Mononuclear cell DNA extracted from peripheral blood of mothers and cord blood from their infants (n = 40 matched pairs) was analysed for DNA adducts. The standard curves were constructed using 5 μg DNA containing 0.1–2 fmol BPdG adducts per well since most samples (5 μg per well) had detectable adduct levels within that range. By BPDE–DNA SCIA, the mean maternal and cord blood adduct levels were 1.41 ± 0.93 (mean ± SD) and 1.13 ± 0.99 adducts per 108 nucleotides, respectively (Figure 4A), while the corresponding median values were 1.32 and 0.93 adducts per 108 nucleotides, respectively. The mean maternal adduct levels were statistically significantly higher than those of cord blood (P = 0.04; two-tailed Wilcoxon test). Three maternal and eight cord blood DNA samples, which fell below the detection limit (3 adducts per 109 nucleotides; 5 μg DNA/well), were assigned a value of 1.5 adducts per 109 nucleotides (half way between zero and the limit of detection).
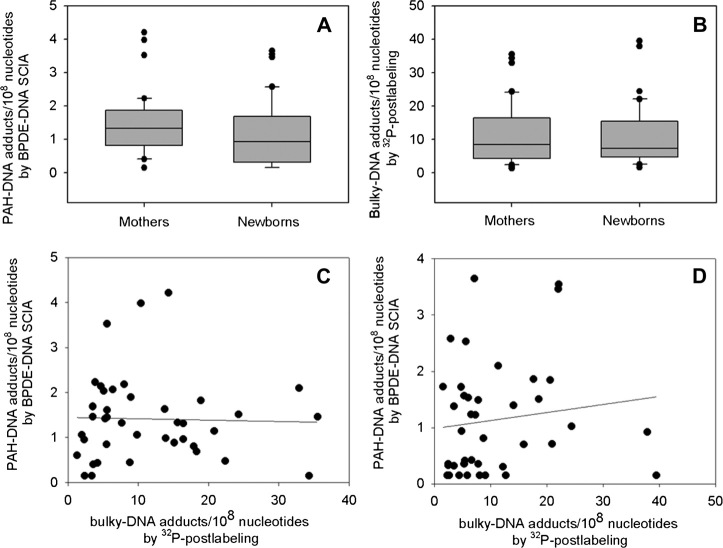
Fig. 4.
Human maternal peripheral and cord blood mononuclear cell DNA assayed by SCIA and 32P-postlabelling. Box and whisker plots illustrate the distribution of DNA adduct levels (per 108 nucleotides) obtained by SCIA (A) and 32P-postlabelling (B). The horizontal lines inside the boxes indicate the median values, the box boundaries the 25th and 75th percentiles and the capped bars show the 10th and 90th percentiles. Outliers are shown as individual symbols. (C and D), scatter plots, illustrate the correlation between PAH–DNA adducts measured by SCIA, and bulky DNA adducts measured by 32P-postlabelling, in maternal (C) and newborn (D) DNA samples.
The levels of bulky DNA adducts measured by 32P-postlabelling were ∼10-fold higher than those measured by BPDE–DNA SCIA for both maternal and cord blood samples (Figure 4B). Mean maternal and cord blood bulky DNA adduct values were 11.5 ± 9.0 (mean ± SD) and 10.7 ± 9.1 adducts per 108 nucleotides, respectively (P > 0.05), while the corresponding median values were 8.4 and 7.2 adducts per 108 nucleotides, respectively. There was no correlation between the DNA adduct levels measured by the two methods for either the maternal or the cord DNA samples (Figure 4C and D). However, a comparison of maternal and cord blood samples measured by the same assay showed significant correlations between maternal and cord blood adducts measured by each of the two methods (Figure 5A and B; Spearman correlation coefficient = 0.66 and 0.60, respectively; P < 0.001 for both).
Fig. 5.
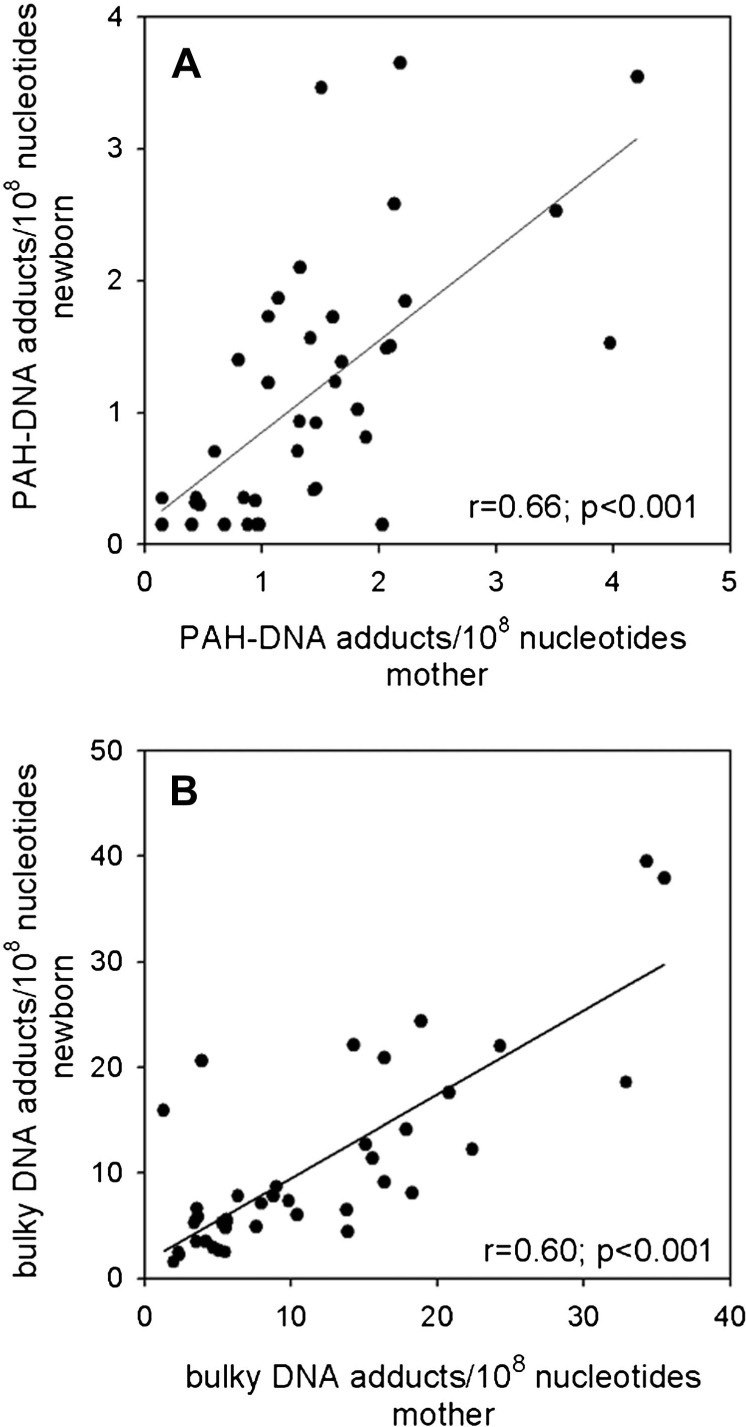
Relationship between adduct levels in maternal and newborn blood DNA measured by SCIA (A) and 32P-postlabelling (B). The ‘r’ and ‘P’ values were determined by Spearman correlation test.
Discussion
Numerous methods have been developed for the detection and measurement of PAH-derived DNA adducts, including high pressure liquid chromatography (HPLC) (30,31), and MS-based methods, which allow for chemical-specific adduct identification and quantitation (32,33). Typically, structure-specific methods require so much DNA that they cannot be applied to population studies involving general environmental exposures (e.g. HPLC–MS–MS requires 100 μg of DNA for a single analysis). For this reason, 32P-postlabelling (34), immunoassays (19) and immunohistochemistry (24) have been employed more widely in such studies. 32P-postlabelling for PAH adducts analysis is very sensitive (the limit of detection is commonly 1 adduct per 109 nucleotides using 10 μg DNA per reaction), but it lacks chemical specificity and the majority of the adducts detected are largely unknown. In addition, the procedure is time consuming, labour intensive and has low resolution. The ‘classic’ competitive ELISA assay for PAH–DNA adducts was reported to have a sensitivity of ∼4 adducts per 108 nucleotides using 25–35 μg of DNA per well. However, >200 μg of DNA are required per sample, due to the high background and variability of the signal. The related competitive dissociation-enhanced lanthanide fluoro-immunoassayassay (DELFIA) increased the sensitivity to <1 adduct per 108 nucleotides without reducing however the total quantity of DNA required. A further increase in the sensitivity—approaching that of 32P-postlabelling—was achieved by the CIA (20), but the requirements for DNA remained substantial (∼50 μg per sample). Therefore, there is still a need for DNA adduct assays of improved sensitivity and throughput, which can be employed in large-scale population studies, and for which only limited amounts of DNA are available. We have recently developed a direct sandwich ELISA for the detection and quantification of O6-methylguanine (O6-meG SCIA) in human samples. This type of assay combines high sensitivity and high throughput (21), exhibits a linear dose–response relationship over a wide range of adduct levels and can, in principle, be adapted for the quantification of different DNA adducts if suitable anti-adduct antisera and appropriate standard adducted DNA are available.
Here, we have described the development, validation and pilot application of a similar assay for PAH–DNA adducts. The BPDE–DNA SCIA provides substantially improved sensitivity as compared to the BPDE–DNA competitive ELISAs and improved throughput as compared to 32P-postlabelling and enables the detection of BPdG or PAH–DNA adducts in the majority of samples from environmentally exposed subjects. Duplicate analyses can be obtained with 11 μg DNA (allowing for 1 μg to be consumed during sample processing). The method is based on exhaustive restriction digestion of DNA to fragments <6000 bp in length, a size expected to contain no more than a single adduct residue. The adduct-containing fragments are bound to a solid surface with BPDE–DNA antibodies, and the plate-immobilised DNA fragments are detected by incubation with anti-ss DNA secondary antisera with alkaline phosphatase and a chemiluminescence end point.
The BPDE–DNA SCIA has several advantages. After optimisation, an assay limit of detection of ∼0.05 fmol per well was achieved, which corresponds to 3 adducts per 109 nucleotides when using 5 μg DNA per well. Up to 10 μg DNA per well does not affect assay performance, and sensitivity can be increased with higher concentrations of anti-DNA antiserum. The assay exhibits satisfactory accuracy and reproducibility characteristics. Analysis of DNA samples from cells treated with different doses of BP gave results in good agreement with those previously reported by others (35) and indicated that the dynamic range of the assay covers at least two orders of magnitude. Standard curves obtained with BPdG–DNA containing 1 adduct per 6000 nucleotides, as measured by spectrophotometry, and 1.1 adduct per 106 nucleotides as measured by LC–MS–MS were almost superimposable, confirming our hypothesis that restriction digestion of the DNA to fragments containing no more than a single adduct residue should eliminate the requirement, common for competitive ELISAs, for a standard DNA modified at the same level as the test samples (29).
The influence of the method of DNA isolation as well as the intra- and between-day variability of the assay was evaluated. Comparable results were obtained with DNA isolated by phenol extraction, Qiagen columns or salting out. The within-day reproducibility of the assay for independently extracted DNAs was higher, and the inter-day variability significantly lower than reported values for previously employed competitive immunoassays (20). The adduct levels as measured by SCIA in MCF-7 cells treated with BP were compared with those obtained by 32P-postlabelling and found to be 30–60% lower (depending on the DNA extraction method). It is possible that some adducts detected by 32P-postlabelling are not recognised by the BPDE–DNA SCIA (36). Nevertheless, a very strong correlation was observed between the adduct levels obtained by the two methods.
The chemical specificity of immunoassays is limited by the recognition characteristics of the antisera employed. The anti-adduct antiserum was raised against BPDE-modified DNA and is known to also recognise adducts produced by other carcinogenic PAHs (22). In order to further characterise the specificity of this antiserum, we analysed DNA samples isolated from the livers of mice exposed to two additional PAHs, DB[a,h]A and B[b]F and found strong cross reactivity with the BPDE–DNA antiserum in the SCIA. In addition, we analysed the same samples by 32P-postlabelling and found that the absolute levels measured by SCIA were substantially lower than those measured by 32P-postlabelling. The reduced capacity of SCIA to detect DNA adducts of DB[a,h]A and B[b]F reflects the reduced avidity of the anti-BPDE–DNA antiserum for these PAH–DNA adducts. This finding confirms the broad compound class specificity of the BPDE–DNA SCIA and emphasises the fact that DNA samples from environmentally exposed humans should be considered to provide a semi-quantitative aggregate measure of PAH–DNA adduct formation.
The new assay is currently being employed in the NewGeneris project (28). Here, we report measurements of the levels of PAH–DNA adducts by the new method and of bulky DNA adducts by 32P-postlabelling from buffy coats of 40 pairs of maternal and cord blood samples. The donors were non-smoking women living in Denmark and were likely to have experienced exposure to very low levels of PAHs. Using SCIA, the adduct levels observed were of the order of 1 adduct per 108 nucleotides comparable to those previously reported with the use of other immunoassays (13,20,37). On the other hand, the adduct levels measured in the same samples by 32P-postlabelling were of the order of 10 adducts per 108 nucleotides, close to those reported in some studies (38) but significantly higher (∼10-fold) than those of other studies which involved exposure to high levels of air pollution (39,40). In a study utilising both methods for adduct detection, the levels obtained by ELISA were 2- to 4-fold lower than those obtained by postlabelling (41), although other studies reported comparable values for either assays (42). It should be noted that the majority of previous human studies utilised samples of DNA purified by phenol extraction, unlike our study where they were purified by Qiagen columns. Kovács et al. (18) used both phenol extraction and Qiagen columns for DNA purification prior to the measurement of bulky DNA adducts from lymphocytes of environmentally exposed individuals and MCF-7 cells treated with BPDE. They found slightly higher bulky adduct levels in MCF-7 cells and several fold higher levels in the human samples when the Qiagen columns were used for DNA purification. These results indicate that our knowledge of the nature of the various bulky DNA adducts and of their stability during the different DNA isolation procedures is limited and underline the need for fully standardised methods to obtain comparable results.
The adduct levels measured with either assay in maternal and the respective cord blood DNA were quantitatively comparable and closely correlated. On the other hand, there was no correlation between the DNA adduct levels determined by SCIA and 32P-postlabelling, with SCIA yielding substantially lower values. Lack of correlation in the levels of BPDE–DNA adducts in PAH-exposed study populations, measured by ELISA, DELFIA or CIA immunoassays, and 32P-postlabelling have also been reported in the past (43,44,38). Most recently, Pratt et al. (45) utilised a similar to ours anti-adduct antiserum to determine in placentas PAH–DNA adduct levels semi-quantitatively by immunohistochemistry and bulky DNA adducts by 32P-postlabelling and found no correlation between the levels of the two types of adducts. The lack of correlation between adduct levels measured by immunochemical assays and 32P-postlabelling is in concordance with our findings and probably reflects (i) the variable ability of antisera raised against a particular type of PAH-DNA adduct to recognise adducts of other PAHs and (ii) the fact that probably only some of the bulky hydrophobic DNA adducts detected by 32P-postlabelling are related to PAHs.
In summary, this report demonstrates the practical utility of a new direct sandwich ELISA for PAH–DNA adducts characterised by increased sensitivity, capable of detecting this type of DNA damage in a large proportion of DNA samples of humans suffering low environmental exposures. The small amount of material required (just >10 μg DNA for duplicate analysis) and its throughput (70 samples/week per person) makes this assay suitable for use in the context of studies utilising samples from biorepositories.
Funding
This work was supported by contracts from the European Union (to S.A.K): Network of Excellence ECNIS — ‘ Environmental cancer risk, nutrition and individual susceptibility’ and Integrated Project NewGeneris — ‘ Newborns and genotoxic exposure risks ’ (contract numbers FOOD-CT-2005-513943, FOOD-CT-2005-016320). The work was also supported in part by the intramural program of the Center for Cancer Research, National Cancer Institute, National Institutes of Health, Bethesda, MD, USA (to M.C.P.).
Acknowledgments
We are grateful to Dr Fred Beland (National Center for Toxicological Research, Jefferson, USA) for his generous gift of BPdG modified DNA and to Drs Peter Farmer and Rajinder Singh (University of Leicester, Cancer Biomarkers and Prevention Group Leicester, UK) for measuring the BPdG containing DNA samples by LC–MS/MS. We also acknowledge the technical assistance of Mr George Koukouves.
Conflict of interest statement: None declared.
References
- 1.International Agency for Research on Cancer. Vol. 92. Lyon, France: IARC Press; 2010. Some non-heterocyclic polycyclic aromatic hydrocarbons and some related exposures. IARC Monographs on the Evaluation of Carcinogenic Risks to Humans . [PMC free article] [PubMed] [Google Scholar]
- 2.International Programme on Chemical Safety Environmental Health Criteria 202. Selected Non-heterocyclic Policyclic Aromatic Hydrocarbons. Geneva, Switzerland: World Health Organization; 1998. [Google Scholar]
- 3.Bostrom CE, Gerde P, Hanberg A, et al. Cancer risk assessment, indicators, and guidelines for polycyclic aromatic hydrocarbons in the ambient air. Environ. Health Perspect. 2002;110(Suppl. 3):451–488. doi: 10.1289/ehp.110-1241197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Panel on Contaminants in the Food Chain of the European Food Safety Agency. Scientific opinion on a request from the European Commission on polycyclic aromatic hydrocarbons in food. EFSA J. 2008;724:1–114. [Google Scholar]
- 5.Roth MJ, Strickland KL, Wang GQ, Rothman N, Greenberg A, Dawsey SM. High levels of carcinogenic polycyclic aromatic hydrocarbons present within food from Linxian, China may contribute to that region’s high incidence of oesophageal cancer. Eur. J. Cancer. 1998;34:757–758. doi: 10.1016/s0959-8049(97)10071-5. [DOI] [PubMed] [Google Scholar]
- 6.Denissenko MF, Pao A, Tang M, Pfeifer GP. Preferential formation of benzo[a]pyrene adducts at lung cancer mutational hotspots in P53. Science. 1996;274:430–432. doi: 10.1126/science.274.5286.430. [DOI] [PubMed] [Google Scholar]
- 7.Ross JA, Nesnow S. Polycyclic aromatic hydrocarbons: correlations between DNA adducts and ras oncogene mutations. Mutat. Res. 1999;424:155–166. doi: 10.1016/s0027-5107(99)00016-0. [DOI] [PubMed] [Google Scholar]
- 8.Jedrychowski WA, Perera FP, Maugeri U, et al. Intrauterine exposure to polycyclic aromatic hydrocarbons, fine particulate matter and early wheeze. Prospective birth cohort study in 4-year olds. Pediatr. Allergy Immunol. 2010;21:e723–e732. doi: 10.1111/j.1399-3038.2010.01034.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi H, Perera F, Pac A, et al. Estimating individual-level exposure to airborne polycyclic aromatic hydrocarbons throughout the gestational period based on personal, indoor, and outdoor monitoring. Environ. Health Perspect. 2008;116:1509–1518. doi: 10.1289/ehp.10972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Godschalk RW, Van Schooten FJ, Bartsch H. A critical evaluation of DNA adducts as biological markers for human exposure to polycyclic aromatic compounds. J. Biochem. Mol. Biol. 2003;36:1–11. doi: 10.5483/bmbrep.2003.36.1.001. [DOI] [PubMed] [Google Scholar]
- 11.Rothman N, Poirier MC, Haas RA, Correa-Villasenor A, Ford P, Hansen JA, O'Toole T, Strickland PT. Association of PAH-DNA adducts in peripheral white blood cells with dietary exposure to polyaromatic hydrocarbons. Environ. Health Perspect. 1993;99:265–267. doi: 10.1289/ehp.9399265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rothman N, Poirier MC, Baser ME, Hansen JA, Gentile C, Bowman ED, Strickland PT. Formation of polycyclic aromatic hydrocarbon-DNA adducts in peripheral white blood cells during consumption of charcoal-broiled beef. Carcinogenesis. 1990;11:1241–1243. doi: 10.1093/carcin/11.7.1241. [DOI] [PubMed] [Google Scholar]
- 13.Gunter MJ, Divi RL, Kulldorff M, et al. Leukocyte polycyclic aromatic hydrocarbon-DNA adduct formation and colorectal adenoma. Carcinogenesis. 2007;28:1426–1429. doi: 10.1093/carcin/bgm022. [DOI] [PubMed] [Google Scholar]
- 14.Kyrtopoulos SA. Biomarkers in environmental carcinogenesis research: striving for a new momentum. Toxicol. Lett. 2006;162:3–15. doi: 10.1016/j.toxlet.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 15.Rundle A, Tang D, Hibshoosh H, Schnabel F, Kelly A, Levine R, Zhou J, Link B, Perera F. Molecular epidemiologic studies of polycyclic aromatic hydrocarbon-DNA adducts and breast cancer. Environ. Mol. Mutagen. 2002;39:201–207. doi: 10.1002/em.10048. [DOI] [PubMed] [Google Scholar]
- 16.Rundle A, Tang D, Hibshoosh H, Estabrook A, Schnabel F, Cao W, Grumet S, Perera FP. The relationship between genetic damage from polycyclic aromatic hydrocarbons in breast tissue and breast cancer. Carcinogenesis. 2000;21:1281–1289. [PubMed] [Google Scholar]
- 17.Phillips DH. DNA adducts as markers of exposure and risk. Mutat. Res. 2005;577:284–292. doi: 10.1016/j.mrfmmm.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 18.Kovács K, Anna L, Rudnai P, Schoket B. Recovery of bulky DNA adducts by the regular and a modified 32P-postlabelling assay; influence of the DNA-isolation method. Mutat. Res. 2011;721:95–100. doi: 10.1016/j.mrgentox.2010.12.012. [DOI] [PubMed] [Google Scholar]
- 19.Santella RM, Zhang YJ. Immunologic detection of benzo(a)pyrene-DNA adducts. Methods Mol. Biol. 2011;682:271–278. doi: 10.1007/978-1-60327-409-8_19. [DOI] [PubMed] [Google Scholar]
- 20.Divi RL, Beland FA, Fu PP, et al. Highly sensitive chemiluminescence immunoassay for benzo[a]pyrene-DNA adducts: validation by comparison with other methods, and use in human biomonitoring. Carcinogenesis. 2002;23:2043–2049. doi: 10.1093/carcin/23.12.2043. [DOI] [PubMed] [Google Scholar]
- 21.Georgiadis P, Kaila S, Makedonopoulou P, Fthenou E, Chatzi L, Pletsa V. Development and validation of a new, sensitive immunochemical assay for O6-methylguanine in DNA and its application in a population study. Cancer Epidemiol. Biomarkers Prev. 2011;2:82–90. doi: 10.1158/1055-9965.EPI-10-0788. [DOI] [PubMed] [Google Scholar]
- 22.Poirier MC, Santella R, Weinstein IB, Grunberger D, Yuspa SH. Quantitation of benzo(a)pyrene-deoxyguanosine adducts by radioimmunoassay. Cancer Res. 1980;40:412–416. [PubMed] [Google Scholar]
- 23.Weston A, Manchester DK, Poirier MC, Choi JS, Trivers GE, Mann DL, Harris CC. Derivative fluorescence spectral analysis of polycyclic aromatic hydrocarbon-DNA adducts in human placenta. Chem. Res. Toxicol. 1989;2:104–108. doi: 10.1021/tx00008a006. [DOI] [PubMed] [Google Scholar]
- 24.Pratt MM, John K, MacLean AB, Afework S, Phillips DH, Poirier MC. Polycyclic aromatic hydrocarbon (PAH) exposure and DNA adduct semi-quantitation in archived human tissues. Int. J. Environ. Res. Public Health. 2011;8:2675–2691. doi: 10.3390/ijerph8072675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schoket B, Horkay I, Kosa A, Paldeak L, Hewer A, Grover PL, Phillips DH. Formation of DNA adducts in the skin of psoriasis patients, in human skin in organ culture, and in mouse skin and lung following topical application of coal-tar and juniper tar. J. Invest. Dermatol. 1990;94:241–246. doi: 10.1111/1523-1747.ep12874576. [DOI] [PubMed] [Google Scholar]
- 26.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pedersen M, Wichmann J, Autrup H, et al. Increased micronuclei and bulky DNA adducts in cord blood after maternal exposures to traffic-related air pollution. Environ. Res. 2009;109:1012–1020. doi: 10.1016/j.envres.2009.08.011. [DOI] [PubMed] [Google Scholar]
- 28.Merlo DF, Wild CP, Kogevinas M, Kyrtopoulos S, Kleinjans J. NewGeneris: a European study on maternal diet during pregnancy and child health. Cancer Epidemiol. Biomarkers Prev. 2009;18:5–10. doi: 10.1158/1055-9965.EPI-08-0876. [DOI] [PubMed] [Google Scholar]
- 29.Santella RM, Weston A, Perera FP, Trivers GT, Harris CC, Young TL, Nguyen D, Lee BM, Poirier MC. Interlaboratory comparison of antisera and immunoassays for benzo[a]pyrene-diol-epoxide-I-modified DNA. Carcinogenesis. 1988;9:1265–1269. doi: 10.1093/carcin/9.7.1265. [DOI] [PubMed] [Google Scholar]
- 30.Pavanello S, Favretto D, Brugnone F, Mastrangelo G, Dal PG, Clonfero E. HPLC/fluorescence determination of anti-BPDE-DNA adducts in mononuclear white blood cells from PAH-exposed humans. Carcinogenesis. 1999;20:431–435. doi: 10.1093/carcin/20.3.431. [DOI] [PubMed] [Google Scholar]
- 31.Manchester DK, Weston A, Choi JS, Trivers GE, Fennessey PV, Quintana E, Farmer PB, Mann DL, Harris CC. Detection of benzo[a]pyrene diol epoxide-DNA adducts in human placenta. Proc. Natl. Acad. Sci. U. S. A. 1988;85:9243–9247. doi: 10.1073/pnas.85.23.9243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koc H, Swenberg JA. Applications of mass spectrometry for quantitation of DNA adducts. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2002;778:323–343. doi: 10.1016/s1570-0232(02)00135-6. [DOI] [PubMed] [Google Scholar]
- 33.Boysen G, Hecht SS. Analysis of DNA and protein adducts of benzo[a]pyrene in human tissues using structure-specific methods. Mutat. Res. 2003;543:17–30. doi: 10.1016/s1383-5742(02)00068-6. [DOI] [PubMed] [Google Scholar]
- 34.de Kok TM, Moonen HJ, van DJ, Van Schooten FJ. Methodologies for bulky DNA adduct analysis and biomonitoring of environmental and occupational exposures. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2002;778:345–355. doi: 10.1016/s0378-4347(01)00543-6. [DOI] [PubMed] [Google Scholar]
- 35.Schoket B, Doty WA, Vincze I, Strickland PT, Ferri GM, Assennato G, Poirier MC. Increased sensitivity for determination of polycyclic aromatic hydrocarbon-DNA adducts in human DNA samples by dissociation-enhanced lanthanide fluoroimmunoassay (DELFIA) Cancer Epidemiol. Biomarkers Prev. 1993;2:349–353. [PubMed] [Google Scholar]
- 36.Sayer JM, Chadha A, Agarwal SK, Yeh HJC, Yagi H, Jerina DM. Covalent nucleoside adducts of benzo[a]pyrene 7,8-diol 9,10-epoxide: structural reinvestigation and characterization of a novel adenosine adduct on the ribose moiety. J. Org. Chem. 1991;56:20–29. [Google Scholar]
- 37.Whyatt RM, Santella RM, Jedrychowski W, et al. Relationship between ambient air pollution and DNA damage in Polish mothers and newborns. Environ. Health Perspect. 1998;106(Suppl. 3):821–826. doi: 10.1289/ehp.98106821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gyorffy E, Anna L, Gyori Z, et al. DNA adducts in tumour, normal peripheral lung and bronchus, and peripheral blood lymphocytes from smoking and non-smoking lung cancer patients: correlations between tissues and detection by 32P-postlabelling and immunoassay. Carcinogenesis. 2004;25:1201–1209. doi: 10.1093/carcin/bgh131. [DOI] [PubMed] [Google Scholar]
- 39.Topinka J, Milcova A, Libalova H, Novakova Z, Rossner P, Jr, Balascak I, Sram RJ. Biomarkers of exposure to tobacco smoke and environmental pollutants in mothers and their transplacental transfer to the foetus. Part I: bulky DNA adducts. Mutat. Res. 2009;669:13–19. doi: 10.1016/j.mrfmmm.2009.04.011. [DOI] [PubMed] [Google Scholar]
- 40.Binkova B, Chvatalova I, Lnenickova Z, Milcova A, Tulupova E, Farmer PB, Sram RJ. PAH-DNA Adducts in environmentally exposed population in relation to metabolic and DNA repair gene polymorphisms. Mutat. Res. 2007;620:49–61. doi: 10.1016/j.mrfmmm.2007.02.022. [DOI] [PubMed] [Google Scholar]
- 41.Cheng YW, Hsieh LL, Lin PP, Chen CP, Chen CY, Lin TS, Su JM, Lee H. Gender difference in DNA adduct levels among nonsmoking lung cancer patients. Environ. Mol. Mutagen. 2001;37:304–310. doi: 10.1002/em.1037. [DOI] [PubMed] [Google Scholar]
- 42.Schoket B. DNA damage in humans exposed to environmental and dietary polycyclic aromatic hydrocarbons. Mutat. Res. 1999;424:143–153. doi: 10.1016/s0027-5107(99)00015-9. [DOI] [PubMed] [Google Scholar]
- 43.Poirier MC, Weston A, Schoket B, et al. Biomonitoring of United States Army soldiers serving in Kuwait in 1991. Cancer Epidemiol. Biomarkers Prev. 1998;7:545–551. [PubMed] [Google Scholar]
- 44.Schoket B, Phillips DH, Poirier MC, Vincze I. DNA adducts in peripheral blood lymphocytes from aluminum production plant workers determined by 32P-postlabelling and enzyme-linked immunosorbent assay. Environ. Health Perspect. 1993;99:307–309. doi: 10.1289/ehp.9399307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pratt MM, King LC, Adams LD, et al. Assessment of multiple types of DNA damage in human placentas from smoking and nonsmoking women in the Czech Republic. Environ. Mol. Mutagen. 2011;52:58–68. doi: 10.1002/em.20581. [DOI] [PMC free article] [PubMed] [Google Scholar]