Abstract
Objective
Pharmacogenomics evaluations of variability in drug metabolic processes may be useful for making individual drug response predictions. We present an approach to deriving ‘phenotype scores’ based on existing pharmacogenomics knowledge and a patient's genomics data. Pharmacogenomics plays an important role in the bioactivation of tamoxifen, a prodrug administered to patients for breast cancer treatment. Tamoxifen is therefore considered a model for many drugs requiring bioactivation. We investigate whether this knowledge-based approach can be applied to produce a phenotype score that is predictive of the endoxifen/N-desmethyltamoxifen (NDM) plasma concentration ratio in patients taking tamoxifen.
Materials and methods
We implement a knowledge-based model for calculating phenotype scores from patient-specific genotype data. These data include allelic variants of genes encoding enzymes involved in the bioactivation of tamoxifen. We performed quantile linear regression to evaluate whether six phenotype scoring algorithms are predictive of patient endoxifen/NDM plasma concentration ratio, and validate our scoring methods.
Results
Our model illustrates a knowledge-based approach to predict drug metabolism efficacy given patient genomics data. Results showed that for one phenotype scoring algorithm, scores were weakly correlated with patient endoxifen/NDM plasma concentration ratios. This algorithm performed better than simple metrics for variation in individual and multiple genes.
Discussion
We discuss advantages of the model, challenges to its implementation in a personalized medicine context, and provide example future directions.
Conclusions
We demonstrate the utility of our model in a tamoxifen case study context. We also provide evidence that more complicated polygenic models are needed to represent heterogeneity in clinical outcomes.
Keywords: Pharmacogenetics, pharmacogenomics, pharmacokinetics, knowledge bases, computer reasoning, genotype, genetic variation, predictive genetic testing, cytochrome P-450 enzyme system, tamoxifen, computer reasoning, knowledge bases, genomics, electronic health records, clinical decision support, developing/using computerized provider order entry, classical experimental and quasi-experimental study methods (lab and field), developing/using clinical decision support (other than diagnostic) and guideline systems, other specific ehr applications (results review, medication administration, disease progression, system implementation and management issues, surveys and needs analysis, qualitative/ethnographic field study
Introduction
Pharmacogenomics evaluations are concerned with the effects of several genes on drug disposition and drug response. Drug disposition, or pharmacokinetics (PK), is the process by which drugs are handled within the body following administration. PK processes include drug absorption, distribution, metabolism, and excretion. Drug response, or pharmacodynamics (PD), is the effect of drugs on the body and also describes patient response. Genes encoding enzymes involved in drug metabolism, such as cytochrome P450 enzymes (CYPs), have been investigated in previous studies to determine the impact of genetic variation on enzyme activity. Differences in the enzyme activity of CYPs can affect the rate of drug metabolism and in turn may contribute to the duration and intensity of the pharmacological action of drugs. In theory, knowledge of genetic variation in genes encoding CYPs can allow physicians to prescribe drugs and doses in a way that is better tailored to expected patient responses and to identify individuals at risk for adverse drug reactions (ADRs) or disease reoccurrence. We present a general approach to calculating ‘phenotype scores’ (numeric scores representing an individual's genetic variation in genes encoding enzymes involved in a drug's metabolic pathway) based on existing pharmacogenomics knowledge and a patient's genomics data. Using tamoxifen as a case study, patient genomics data utilized in phenotype score calculations include the allelic variant status of genes encoding CYPs involved in the bioactivation of tamoxifen (eg, CYP2D6*1/*10, *1 and *10 are allelic variants of the cytochrome P450 2D6 gene). Existing pharmacogenomics knowledge includes: (1) curated findings from the primary literature where associations between gene allelic variant and enzyme activity (‘allelic variant’–‘enzyme activity’ associations) are reported (eg, CYP2D6*1/*10, *1 allele is associated with normal enzyme activity and *10 allele is associated with decreased enzyme activity); (2) evidence of associations between genotype and metabolizer activity, or ‘genotype’–‘metabolizer activity’ associations (eg, CYP2D6*1/*10, an allele associated with normal enzyme activity and an allele associated with decreased enzyme activity is associated with overall extensive metabolizer activity); and (3) curated evidence of drug metabolic properties (eg, CYP2D6 is a major enzyme involved in the metabolism of tamoxifen). The primary sources of evidence (evidence sources) utilized in this work include SuperCYP (http://bioinformatics.charite.de/supercyp/),1 a review article that reports ‘genotype’–‘metabolizer activity’ associations based on studies involving tamoxifen,2 and the Pharmacogenomics Knowledge Base (PharmGKB, http://pharmgkb.org).3 SuperCYP is a database that contains information on drugs, CYP–drug interactions, and alleles. Curated evidence of ‘allelic variant’–‘enzyme activity’ associations is made available within SuperCYP. PharmGKB is a comprehensive resource for pharmacogenomics that provides curated knowledge about the impact of genetic variation on drug response. Curated drug metabolic pathway evidence is made available within PharmGKB and is utilized in this work. PharmGKB is also utilized as a source to supplement information on ‘allelic variant’–‘enzyme activity’ associations found in SuperCYP.
We propose a model for reasoning with pharmacogenomics knowledge and clinical (genomics) data, and build a prototype model implementation. Our model incorporates an evidential approach to determine with what pharmacogenomics knowledge reasoning should be performed (involves specifying some belief criteria or minimal evidence requirement). Six phenotype scoring algorithms are investigated in total (see the ‘Reasoning rules and objects’ section).
Pharmacogenomics plays an important role in the bioactivation of tamoxifen, an anti-estrogen agent administered to patients for breast cancer treatment and prevention. Tamoxifen can therefore be considered a model for many drugs requiring bioactivation. Previous work has demonstrated that tamoxifen is a prodrug and CYPs play a role in catalyzing the formation of the anti-estrogenic metabolite endoxifen, with N-desmethyltamoxifen (NDM) as the prominent intermediate metabolite. Here, we explore tamoxifen as a case study for our prototype model implementation and evaluate how well phenotype scores generated by the reasoning system predict the patient endoxifen/NDM plasma concentration ratio (as a marker for drug metabolism efficacy).
In general, anticancer drugs such as tamoxifen provide an interesting test bed for investigating personalized healthcare, specifically pharmacogenomics. Despite the recent increase in existing anticancer drugs, benefits achieved have been less than desired.4 Anticancer drugs are also frequently associated with suspected ADRs.5 6 Lack of efficacy and occurrences of toxicity due to anticancer drugs may be partially due to inter-individual variability in drug metabolism.
Background and significance
We present a model that takes an evidential approach to reasoning across assertions about pharmacogenomics and assigns phenotype scores to breast cancer patients taking tamoxifen. Related work includes creation of the Drug Interaction Knowledge Base (DIKB)7 8 and use of activity scores to predict phenotype in other studies.9–11
Evidential approach to knowledge representation
This work derives assertions about ‘allelic variant’–‘enzyme activity’ and ‘genotype’–‘metabolizer activity’ associations from the published literature. A similar approach was taken to curate and reason across assertions about drug metabolism knowledge. The DIKB7 uses a model for predicting metabolic inhibition interactions that incorporates an evidence base (EB) and a knowledge base (KB). The EB contains assertions (or facts) about a drug's mechanistic properties, while the KB contains assertions included in the EB that meet some belief criteria (the minimal evidence requirement).
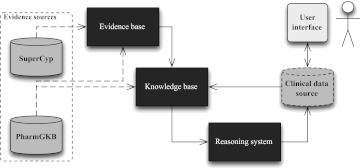
This paper investigates whether a similar approach might be taken with patient genotype data and pharmacogenomics knowledge. We link ‘allelic variant’–‘enzyme activity’ and ‘genotype’–‘metabolizer activity’ assertions to evidence for and against these assertions. Similar to the DIKB approach, our EB contains the full set of assertions and our KB contains a subset of assertions based on a belief criterion. In this case, the EB contains all publication-related assertions including ‘allelic variant’–‘enzyme activity’ association assertions (extracted from SuperCYP, and in some cases PharmGKB). The KB contains a subset of the publication-related assertions, assertions about drug metabolic pathway properties (extracted from PharmGKB), and assertions about patient genomics data (from a clinical data source). An overview of our adapted architecture is shown in figure 1.
Figure 1.
Prototype reasoning system architecture: a prototype implementation of the PEMRIC model. SuperCYP is our primary source for genotype–phenotype association knowledge. PharmGKB is our primary source for pharmacokinetic pathway knowledge. The clinical data source contains patient data. The dashed line linking PharmGKB to the evidence base represents the inclusion of PharmGKB curated publications into the evidence base only for the drug-oriented approach. The user interface element is included for illustrative purposes.
DIKB evidence taxonomy
We utilize the DIKB evidence taxonomy8 to classify evidence that a given gene allelic variant has an effect on enzyme activity. The taxonomy contains 36 evidence types; types relevant to this work are shown in box 1. For our prototype system we utilize a subset of evidence types including in vitro experiment, retrospective study, and clinical trial study.
Box 1. A subset of the Drug Interaction Knowledge Base (DIKB) evidence taxonomy.
Evidence types
Clinical trial types
- A pharmacokinetic clinical trial
- A genotyped pharmacokinetic clinical trial
- A phenotyped pharmacokinetic clinical trial
Retrospective study types
A retrospective population pharmacokinetic study
In vitro experiment types
- A drug metabolism identification experiment
- A CYP450, recombinant, drug metabolism identification experiment with possibly NO probe enzyme inhibitor(s)
- A CYP450, human microsome, drug metabolism identification experiment using chemical inhibitors
Observation-based report
An observation-based adverse drug event (ADE) report (eg, FDA Adverse Event Reporting System)
A published observation-based ADE report
Non-traceable statement types
A non-traceable, but possibly authoritative, statement
A non-traceable drug-label statement
Activity scores for phenotype prediction: tamoxifen case study
There are several studies where gene or enzyme activity scores were assigned to patients for the purpose of predicting their response to drug therapy.9–11 None of these studies take the evidential approach we propose here, but they did provide us with some lessons. The work of Borges et al9 compares the ability of three CYP2D6 gene scoring systems10–12 to predict their CYP2D6 phenotype, where the CYP2D6 phenotype is the endoxifen/NDM plasma ratio in tamoxifen pharmacogenetics trial women participants. Similarly, we evaluate the ability of our scoring system to predict tamoxifen drug metabolism efficacy (via the endoxifen/NDM plasma ratio) in women. We also use a scoring convention similar to those proposed to predict CYP2D6 activity, where zero corresponds to the least amount of activity. In previous work, the endoxifen/NDM plasma ratio was identified as the best predictor of CYP2D6 activity.13 We believe the endoxifen/NDM plasma ratio also serves as a good marker for drug metabolism efficacy in our tamoxifen case study.
In Fuhr's discussion of pharmacogenetic-based clinical scores,14 he outlines reasons why the medical community is reluctant to apply genetic information on drug metabolizing enzymes for treatment personalization. Ambiguous and complex algorithms for interpreting genetic information are unlikely to be utilized. We aim to present a transparent approach to provide a score that characterizes the activity of enzymes involved in the metabolism of a drug.
Methods
The Pharmacogenomics Evidence Mapping for Reasoning with Individualized Clinical Data (PEMRIC) model incorporates an evidential approach to calculating phenotype scores from patient-specific genotype data, in the context of making prescribing decisions. Six different approaches to calculating phenotype scores are described in this manuscript that use the following methods:
Method of selecting evidence: enzyme-oriented or drug-oriented (ie, KB contains all ‘allelic variant’–‘enzyme activity’ association facts reported in the literature, or KB contains only drug-specific ‘allelic variant’–‘enzyme activity’ facts)
Method of calculating phenotype score: enzyme activity or metabolizer activity (ie, calculations incorporate ‘allelic variant’–‘enzyme activity’ facts or ‘genotype’–‘metabolizer activity’ facts, respectively)
Method of calculating phenotype score: unweighted or weighted (ie, weights assigned to enzymes according to their level of involvement in metabolic activities).
Below, we provide an overview of PEMRIC model methods for selecting and reasoning with evidence to provide phenotype scores.
A model for reasoning with pharmacogenomic evidence and patient clinical data
The PEMRIC model builds on the approach used to develop the DIKB that incorporates an EB, a KB, and use of the DIKB evidence taxonomy. The PEMRIC model extends the DIKB model to include evidence sources and clinical data sources (see figure 1). Evidence sources utilized in this study are SuperCYP, PharmGKB, and a review article that reports ‘genotype’–‘metabolizer activity’ associations for CYP enzymes involved in tamoxifen metabolism.2 The clinical data source used in this study was produced by the Specific Estrogen Receptor Modulator Pharmacogenetics (SERM) group (now called the Consortium on Breast Cancer Pharmacogenomics, or COBRA)15 and made publically available through the PharmGKB website. The DIKB model and the PEMRIC model both represent knowledge as frames and take a rule-based strategy for reasoning. With a frame-based approach, objects are represented as classes, classes have attributes (or slots), and slots have assigned values.
We represent the following objects for the EB and KB: patient (patient class), patient genotype data (patient-enz-genotype class), evidence of medication PK mechanism (med-metabolite class), evidence of ‘allelic variant’–‘enzyme activity’ association (enz-allele-activity-publication class), evidence of ‘genotype’–‘metabolizer activity’ association (metabolizer-activity-publication class), and evidence of enzyme contribution in a medication metabolic pathway (enz-contribution class). Table 1 lists examples of each class and describes components of the frame-based representation. Each object has a class name and one or more ‘slots,’ where each slot represents an important attribute of the object. In the PEMRIC model, each object represents an assertion (or fact).
Table 1.
Example Java Expert System Shell (JESS) facts derived from the clinical data source, PharmGKB, SuperCYP, and review article sources
|
Unformatted slots were utilized in all approaches. Bolded slots were only utilized in the drug-oriented approach.
Italicized slots were only utilized with the ‘genotype’–’metabolizer activity’ scoring system. Underlined slots were captured, but not utilized in any approach. IM, intermediate metabolizer.
The PEMRIC model also incorporates rule-based reasoning using forward chaining inference. Rules are if–then statements, where a given set of conditions will lead to a set of results. Assertions are used to determine whether conditions defined in a rule are true. Given the conditions defined in a rule are true, an action will take place. Forward chaining inference specifically starts with a collection of assertions used to infer new assertions until a goal is reached or until nothing new can be inferred. Therefore, the action is often to infer a new assertion, which is then added to the EB or KB. Reasoning concludes when all relevant assertions are considered in calculating a phenotype score. Rules included in the PEMRIC EB and KB are described in table 2.
Table 2.
Pseudocode description of evidence base (EB) and knowledge base (KB) rules
| Rule | Condition (data/knowledge source) | Action | |
| EB | R1 |
|
|
| R2 |
|
|
|
| R3 |
|
|
|
| KB | R4 |
|
|
| R5 |
|
|
|
| R6 |
|
|
|
| R7 |
|
|
|
| R8 |
|
|
|
| R9 |
|
|
|
| R10 |
|
|
|
| R11 |
|
|
|
| R12 |
|
|
|
| R13 |
|
|
|
| R14 |
|
|
|
For each rule (column 2), the bulleted conditions (column 3) are AND statements. The bulleted actions (column 4) are new facts that would be added to the EB or KB if all of the conditions of a rule are satisfied. Rules R1, R2, and R3 specify that EB facts from three evidence types (‘drug metabolism identification experiment,’ ‘retrospective population PK study,’ and ‘pharmacokinetic clinical trial experiment’) would be included in the KB. The order in which rules are fired is undefined and they are thus fired sequentially. Note: pseudocode describes only the enzyme-oriented approach. EB, evidence base; KB, knowledge base; PK, pharmacokinetics.
The PEMRIC model incorporates a subset of DIKB taxonomy evidence types to specify the belief criteria for including evidence of ‘allelic variant’–‘enzyme activity’ associations in the KB. Evidence types specified for publications include in vitro (in vitro experiment evidence type in box 1) and in vivo (retrospective and clinical trial study evidence types in box 1) evidence. A belief criterion distinct from the DIKB evidence taxonomy was specification of the drug of focus in a published study. We took two approaches to implementing the PEMRIC model to facilitate comparison of the predictability of phenotype scores calculated using an enzyme-oriented approach (without the criterion) and using a drug-oriented approach (with the criterion). Taking an enzyme-oriented approach, any publication reporting an ‘allelic variant’–‘enzyme activity’ association for a gene is included in the KB. For the enzyme-oriented approach, the following was one of three rules (a rule for each evidence type) included in the EB specifying whether an ‘allelic variant’-‘enzyme activity’ association is believed to be sufficient evidence for inclusion in the KB: IF publication q reports that allelic variant d has-allele-activity f AND publication q has-evidence-type ‘in vitro experiment’ THEN allelic variant d has-sufficient-evidence-of-allele-activity f. With the drug-oriented approach, evidence provided by a publication is included in the KB if the drug of focus in the study is a drug being taken by the patient. For the drug-oriented approach, the following rule was one of three rules included in the EB: IF publication q reports that allelic variant d has-allele-activity f AND patient p is taking medication m AND publication q has-drug-studied m AND publication q has-evidence-type ‘in vitro experiment’ THEN allelic variant d has-sufficient-evidence-of-allele-activity f.
The model also has flexibility to incorporate other evidence types. A new evidence type can be incorporated into the system by adding a rule to the EB, for example, by adding a rule specifying that an ‘allelic variant’–‘enzyme activity’ assertion of a non-traceable drug-label statement evidence type is believed to be sufficient evidence to be included in the KB: IF publication q reports that allelic variant d has-allele-activity f AND publication q has-evidence-type ‘a non-traceable drug-label statement’ THEN allelic variant d has-sufficient-evidence-of-allele-activity f, where a non-traceable drug-label statement is an assertional statement found in a drug label that does not provide any traceable citations for its evidence support.8
Prototype reasoning system design in a tamoxifen case study context
Our reasoning system is a prototype implementation of the PEMRIC model for reasoning with pharmacogenomics knowledge and clinical data. The initial trigger for the system is retrieval of a medical record number of a patient who is prescribed tamoxifen. Given the patient is being prescribed tamoxifen, the patient's genomics data (clinical source data), and KB facts, our reasoning system calculates a phenotype score. System components are summarized in figure 1.
Pharmacogenomics evidence sources and tamoxifen case study data
Data and knowledge sources from which we derive assertions include two evidence sources, one review article, and one clinical data source. The clinical data source produced by COBRA includes data for 30 subjects who received 20 mg/day tamoxifen. Genotype information utilized in this work includes the results from CYP3A5, CYP2D6, CYP2C9, and CYP2C19 genetic tests. The mode of ascertaining genotypes is specified in the PharmGKB dataset.16
Supplementary phenotypic information was obtained directly from the COBRA group. Specific phenotypic information that was utilized in this work includes measured amounts of endoxifen and NDM at 4 months after initiation of tamoxifen. Two patients were excluded from analyses because they did not have recorded values for endoxifen and NDM plasma levels. We considered metabolite plasma levels at 4 months because tamoxifen serum concentrations reach a steady state by 4 months.17 The primary phenotype we wished to predict using our scoring algorithms was the endoxifen/NDM plasma concentration ratio (as a marker for drug metabolism efficacy).
Evidence sources include PharmGKB and SuperCYP. We derived computable assertions from evidence of the drug PK pathway reported in PharmGKB. Also, with a focus on CYPs, we derive ‘allelic variant’–‘enzyme activity’ association assertions from SuperCYP (and in some cases PharmGKB). Since our evaluation is focused on patient genotype in the context of a drug metabolism, we assume all PK pathway knowledge from PharmGKB to be true. In our evaluation, in vitro experiments, and retrospective and clinical trial (referred to as ‘in vivo’ studies in the SuperCYP database) evidence types are considered acceptable evidence to support a given ‘allelic variant’–‘enzyme activity’ association assertion.
Currently, information needed to define ‘genotype’–‘metabolizer activity’ associations are not captured in PharmGKB or SuperCYP (ie, metabolizer activity levels associated with various genotypes). CYP metabolizer activity levels in patients taking tamoxifen that result from CYP2C9, CYP2C19, and CYP2D6 genotypes are described in a single review article,2 but CYP3A5 is not covered. Therefore, we derived assertions for CYP2C9, -2C19, and -2D6 from the review article, and a conservative approach was taken to define ‘genotype’–‘metabolizer activity’ associations for CYP3A5 (ie, designation as ‘extensive metabolizer’ with at least one wild-type allele, and ‘intermediate metabolizer’ otherwise). Similar to metabolic properties reported in PharmGKB, these assertions are assumed to be true.
Collecting pharmacogenomics evidence for tamoxifen case study
Using tamoxifen as an example, we performed the following manual steps:
Step 1. Define assertions for tamoxifen metabolism properties according to tamoxifen PK pathway details available on the PharmGKB website.3 18 As mentioned previously, these assertions are assumed to be true. Therefore, they are directly included in our KB.
Step 2. Perform a SuperCYP polymorphism search for each enzyme involved in the tamoxifen PK pathway. Results include reports of ‘allelic variant’–‘enzyme activity’ associations and references the PubMed ID of publications containing evidence of each relationship.
Step 3. Define gene ‘allelic variant’–‘enzyme activity’ association assertions and classify evidence types for each publication using the DIKB evidence taxonomy.
Step 4. Enter evidence items into the EB.
Step 5. Define ‘genotype’–‘metabolizer activity’ association assertions according to reports summarized in the review article2 (see table 3). These assertions are assumed to be true and are directly included in our KB.
Table 3.
Genotypes and their associated metabolizer phenotypes
| Metabolizer activity | Gene | Allele activity | Enzyme-oriented approach | Drug-oriented approach |
| Ultrarapid metabolizer | CYP2C9 | Unknown | – | – |
| CYP2C19 | Increased/increased | – | – | |
| CYP2D6 | Wild-type/increased; increased/increased | – | – | |
| CYP3A5 | Unknown | – | – | |
| Extensive metabolizer | CYP2C9 | Wild-type/wild-type | *1/*1,*1/*2,*1/*3 (*1=wild-type, *2=decreased, *3=wild-type) | *1/*1,*1/*2 (*1=wild-type, *2=decreased) |
| CYP2C19 | Wild-type/wild-type | *1/*1,*1/*2 (*1=wild-type, *2=decreased) | *1/*1 (*1=wild-type) | |
| CYP2D6 |
|
*1/*1,*1/*4, *1/*6,*1/*10 (*1=wild-type, *4=decreased, *6=wild-type, *10=decreased) | *1/*1,*1/*4,*1/*6,*1/*10 (*1=wild-type, *4=non-functional, *6=non-functional, *10=non-functional) | |
| CYP3A5 | Unknown | *1/*1,*1/*3 (*1=wild-type, *3=decreased) | *1/*1,*1/*3 (*1=wild-type, *3=non-functional) | |
| Intermediate metabolizer | CYP2C9 | Wild-type/non-functional | *2/*2 (*2=decreased) | *1/*3,*2/*2 (*1=wild-type, *2=decreased, *3=non-functional) |
| CYP2C19 | Wild-type/non-functional | – | *1/*2 (*1=wild-type, *2=non-functional) | |
| CYP2D6 |
|
*4/*4 (*4=decreased) | – | |
| CYP3A5 | Unknown | *3/*3 (*3=decreased) | *3/*3 (*3=non-functional) | |
| Poor metabolizer | CYP2C9 | Non-functional/non-functional | – | – |
| CYP2C19 | Non-functional/non-functional | – | – | |
| CYP2D6 | Non-functional/non-functional | – | *4/*4 (*4=non-functional) | |
| CYP3A5 | Unknown | – | – | |
The ‘Allele activity’ column of this table is adapted from Sheffield and Pillimore2 (review article). ‘increased,’ ‘wild-type,’ ‘decreased,’ and ‘non-functional’ refer to associated allelic activity.
Reasoning rules and objects
Our prototype system performs reasoning over PK pathway knowledge, and ‘allelic variant’–‘enzyme activity’ or ‘genotype’–‘metabolizer activity’ association knowledge to calculate a phenotype score. EB ‘allelic variant’–‘enzyme activity’ association facts are included in the KB if there is at least one primary research article classified as having drug metabolism identification experiment results to support the fact and according to whether an enzyme-oriented or drug-oriented approach is being taken. All PK pathway knowledge facts are included in the KB. We define both facts and rules using the Java Expert System Shell (JESS),19 a Java-based rule engine and scripting environment. When contradictory evidence was observed (eg, CYP2D6*10 has reports indicating non-functional and decreased enzyme activity), we included the most commonly reported value in the KB (eg, CYP2D6*10 leading to decreased enzyme activity). If there was a tie, facts were included in the KB according to the following priority: increased>wild-type>decreased>non-functional. Once reasoning concludes, the system assigns a patient their phenotype score.
We will describe the main approaches taken to select evidence for phenotype score calculations (enzyme-oriented and drug-oriented approaches), to define facts used for calculations (‘allelic variant’–‘enzyme activity’ and ‘genotype’–‘metabolizer activity’ association assertions), and to incorporate into phenotype score calculations weighting values indicating the involvement of enzymes in metabolic activities (weighted and un-weighted approaches).
Six phenotype scoring algorithms are investigated in total: (1) enzyme-oriented, un-weighted, ‘allelic variant’–‘enzyme activity’ scoring algorithm; (2) drug-oriented, un-weighted, ‘allelic variant’–‘enzyme activity’ scoring algorithm; (3) drug-oriented, un-weighted, ‘genotype’–‘metabolizer activity’ scoring algorithm; (4) enzyme-oriented, weighted, ‘allelic variant’–‘enzyme activity’ scoring algorithm; (5) drug-oriented, weighted, ‘allelic variant’–‘enzyme activity’ scoring algorithm; and (6) drug-oriented, un-weighted, ‘genotype’–‘metabolizer activity’ scoring algorithm. An ‘enzyme-oriented, un-weighted, ‘genotype’–‘metabolizer activity’ scoring algorithm’ and a ‘enzyme-oriented, weighted, ‘genotype’–‘metabolizer activity’ scoring algorithm’ are not investigated because the review article2 utilized to determine metabolizer activity is specific to studies involving tamoxifen.
Evidence selection: enzyme-oriented approach
As described above in Step 3, in our evidence collection methods, we derive gene ‘allelic variant’-’enzyme activity’ assertions from publications. Using the enzyme-oriented approach to reasoning, the following objects were captured for each ‘allelic variant’–‘enzyme activity’ fact: the PubMed ID of the publication containing the evidence; gene allelic variant (eg, CYP2C9*2); allele activity (eg, decreased); and evidence type (eg, in vitro) (see pubmed-id, enz-allele, enz-activity, and evidence-type slots for enz-allele-activity-publication object in table 1).
All evidence of gene ‘allelic variant’–’enzyme activity’ was included in the KB as long as the evidence criterion described in the previous section was satisfied. With the drug-oriented approach, information about the drug studied is also incorporated in our reasoning algorithms.
Evidence selection: drug-oriented approach
The drug-oriented approach to reasoning required adding a drug slot to the enz-allele-activity-publication object for each ‘allelic variant’–’enzyme activity’ fact representing the drug studied (see table 1). Evidence was then included in the KB if: (1) defined evidence criteria are satisfied; and (2) the study involves the drug of interest (ie, tamoxifen).
A subset of the phenotype scoring algorithms considered in this work incorporate weighting enzymes in the calculation. A weighted approach incorporates a numeric value indicating the involvement of an enzyme in the metabolic activities of the drug (ie, allelic variants in genes encoding major drug metabolizing enzymes are weighted higher than minor enzymes). All enzymes involved in the metabolism of the drug are considered equal contributors in the un-weighted approach.
Phenotype score calculation: un-weighted approach
| (1) |
| (2) |
The phenotype score is calculated according to equations (1) or (2). pscore is the phenotype score calculated as the sum of allele activity levels (a) or metabolizer activity levels (m), across all genes (g). The ‘allelic variant’–‘enzyme activity’ scoring system and ‘genotype’–‘metabolizer activity’ scoring system are described below. We implemented both un-weighted and weighted approaches because we were interested in whether accounting for the relative contribution of enzymes involved in a drug metabolic pathway would affect our ability to predict drug metabolism efficacy.
Phenotype score calculation: weighted approach
| (3) |
| (4) |
With the weighted approach the phenotype score is calculated according to equation (3) or (4). These differ from equations (1) and (2) in the inclusion of a weight factor (w). Each allelic or genotype activity value is multiplied by the weight factor assigned to that gene. Genes encoding major and minor metabolizing enzymes for a given drug are assigned different weight factors. We added a new object to describe the enzyme contribution in the drug PK pathway (see enzyme-contribution object in table 1). An enzyme was identified as major or minor according to PharmGKB PK pathway evidence. In this case, enzymes CYP2D6 and CYP3A5 were described as major metabolizing enzymes, and CYP2C9 and CYP2C19 as minor metabolizing enzymes. The numeric major/minor values assigned are as follows:
Gene encodes major metabolizing enzyme: 1.0
Gene encodes minor metabolizing enzyme: 0.5
Phenotype scoring algorithms use one of two methods leveraging existing pharmacogenomics knowledge to assigning numeric values to patient genetic variant status. One method assigns numeric values according to ‘allelic variant’–‘enzyme activity’ associations, and the other method assigns numeric values according to ‘genotype’–‘metabolizer activity’ associations.
Phenotype score: ‘allelic variant’–‘enzyme activity’ scoring system
Allelic activities (a) utilized in the phenotype score calculations are assigned according to a convention consistent with other studies10–12:
Increased allele activity: 1.5
Wild-type allele activity: 1.0
Decreased allele activity: 0.5
Non-functional allele activity: 0.0
Phenotype score: ‘genotype’–‘metabolizer activity’ scoring system
Patient genotypes are assigned to metabolizer phenotypes as described in table 3 (see enz-allele-activity-publication object in table 1). Metabolizer activities (m) utilized in the phenotype score calculations are assigned according to a convention similar to the allele activity scoring system:
Ultrarapid metabolizer (UM) metabolizer activity: 1.5
Extensive metabolizer (EM) metabolizer activity: 1.0
Intermediate metabolizer (IM) metabolizer activity: 0.5
Poor metabolizer (PM) metabolizer activity: 0.0
PEMRIC model implementation: reasoning rules
Rules for reasoning with assertions defined in the KB are summarized in table 2. Rules for including facts about ‘allelic activity’–‘enzyme activity’ associations, taking an enzyme-oriented approach, are described. Rules to assign metabolizer activity values based on an individual's genotype (two allelic variant activity values), that is, ‘genotype’–’metabolizer activity’ associations, are not shown. Neither are the rules for applying a drug-oriented approach shown.
PEMRIC model implementation: reasoning facts
We took two main approaches (enzyme-oriented and drug-oriented) to encoding EB facts that were relevant to our tamoxifen case study dataset.
EB facts
Taking the enzyme-oriented approach, our EB included 85 ‘allelic variant’–‘enzyme activity’ facts (from SuperCYP). However, there is currently only one publication reporting results from a tamoxifen study within SuperCYP. Therefore, in order to facilitate our drug-oriented approach to reasoning, we supplemented ‘allelic variant’–‘enzyme activity’ facts derived from SuperCYP, with facts derived from curated publications in PharmGKB. PharmGKB has curated drug–gene relationships as well as curated gene variant annotations. Two authors of this manuscript reviewed publications curated by PharmGKB as having evidence of gene–tamoxifen relationships for CYP2D6, CYP2C9, CYP2C19, and CYP3A5 genes (32 publications). This set of publications was narrowed down to nine publications that are vitro experiment, or retrospective or clinical trial study evidence types, and define activity level for at least one gene allele.
For each PharmGKB publication, the authors read the publication and manually recorded all slot values for enz-allele-activity-publication object facts (example values are shown in table 1). Multiple enz-allele-activity-publication facts can be derived from a single publication (ie, one publication can report results on multiple populations and multiple allelic variants).
KB facts
The facts directly included in the KB are: seven PK pathway facts (PharmGKB), 140 facts from the clinical data source (facts representing the existence of 28 test patients, and facts for the results of five genetic tests for each test patient), and 40 ‘genotype’–‘metabolizer activity’ association facts (one for each combination of increased, wild-type, decreased, and non-functional for two alleles of each gene). Fifteen of the 40 were ‘genotype’–‘metabolizer activity’ facts derived from the review article and 25 were assigned using a conservative approach (see ‘Activity scores for phenotype prediction: tamoxifen case study’ section). Example facts are shown in table 1.
Tamoxifen case study evaluation: statistical methods
We evaluated whether phenotype scores (incorporating information about multiple enzymes) are predictive of differences in the endoxifen/NDM plasma ratio (as a marker for drug metabolism efficacy) for 28 patients. For each of six phenotype scoring algorithms, we performed both linear and quantile linear regression where the independent variable was the phenotype score and the dependent variable was the endoxifen/NDM plasma ratio. We report the more conservative of the two approaches (the quantile regressions). In addition, based on a suggestion from a reviewer, we performed quantile regression with bootstrapped standard errors. The number of bootstrap replicates was set at 2500. We found that one approach to calculating a phenotype score passed tests for significance, compared to three approaches without bootstrapping.
We also investigate how our knowledge-based approach to calculating phenotype scores compares to a simple metric representing the genotype of (1) a single gene, and (2) multiple genes. For the simple metric, for each patient, the genotype of each gene is designated as having two wild-type alleles (Wt/Wt), two variant alleles (Vt/Vt), or one variant and one wild-type allele (Wt/Vt). Simple metrics for individual genes were assigned as follows: Wt/Wt=0, Wt/Vt=1, Vt/Vt=2. Although not discussed in detail in this manuscript, we also evaluated a dominant model (Vt/Vt=0, Wt/Vt=1, Wt/Wt=1) and a recessive model (Vt/Vt=1, Wt/Vt=0, Wt/Wt=0) for representing the genotypes of multiple genes. A simple metric for multiple genes was the sum of these values across all genes. See figure 2 for frequency counts. To evaluate the predictive power of individual genes, quantile linear regression was performed where the independent variable was the simple metric of an individual gene and the dependent variable was the endoxifen/NDM plasma ratio. To evaluate the predictive power of multiple genes, quantile linear regression was performed where the independent variable was the simple metric for multiple genes, and the dependent variable was the endoxifen/NDM plasma ratio.
Figure 2.
Distribution of genotypes: CYP3A5 Wt=*1, Vt=*3,*6; CYP2D6 Wt=*1, Vt=*4,*6; CYP2C9 Wt=*1, Vt=*2,*3; and CYP2C19 Wt=*1, Vt=*2.
The predictive performances of the phenotype scores and of the simple metrics on the endoxifen/NDM plasma ratio were assessed with R2, and their significances are evaluated with p values. All statistical analyses were performed using Stata V.11.2 (StataCorp LP).
Results
Phenotype scores as a predictor of drug metabolism efficacy
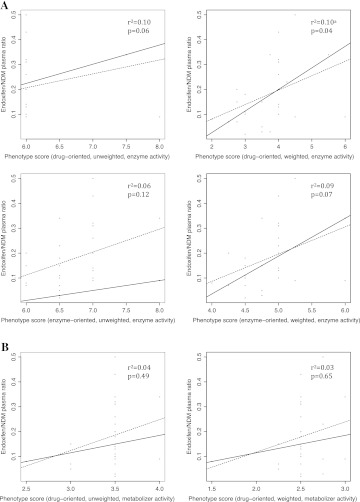
Using tamoxifen as a case study, the predictive performance of six phenotype scoring systems was evaluated. The regression coefficient (difference in medians) for one scoring system was significantly different from zero at the 0.05 level (see figure 3A). This phenotype scoring system was the drug-oriented, weighted, ‘allelic variant’–‘enzyme activity’ scoring algorithm. The scoring system showed a weak correlation with endoxifen/NDM plasma ratio (R2=0.10; p<0.05). There was no statistically significant difference from zero for the regression coefficient for five scoring systems (see figure 3A,B). These phenotype scoring systems included: the drug-oriented, un-weighted, ‘allelic variant’–‘enzyme activity’ scoring algorithm; the enzyme-oriented, un-weighted, ‘allelic variant’–‘enzyme activity’ scoring algorithm; the enzyme-oriented, weighted, ‘allelic variant’–‘enzyme activity’ scoring algorithm; the drug-oriented, un-weighted, ‘genotype’–‘metabolizer activity’ scoring algorithm; and the drug-oriented, weighted, ‘genotype’–‘metabolizer activity’ scoring algorithm. These findings suggest that the phenotype scoring algorithms that assign numeric values according to ‘allelic variant’–‘enzyme activity’ associations are more predictive than those that assign numeric values according to ‘genotype’–’metabolizer activity.’ In addition, drug-oriented approaches to including knowledge about ‘allelic variant’–’enzyme activity’ associations in the KB may be slightly better than enzyme-oriented approaches.
Figure 3.
Scatterplot and quantile regression fit. The plots show scatter plots of the endoxifen/N-desmethyltamoxifen (NDM) ratio versus phenotype scores. Superimposed on the plots are the median fit (solid line) and the least squares estimate of the conditional mean function (dashed line). (A) Phenotype scoring systems that assign numeric values according to ‘allelic variant’–‘enzyme activity’ association assertions. (B) Phenotype scoring systems that assign numeric values according to ‘genotype’–‘metabolizer activity’ association assertions. aA p value of ≤0.05 is considered statistically significant, indicating significant difference in the regression coefficient medians.
Validation of phenotype scoring systems
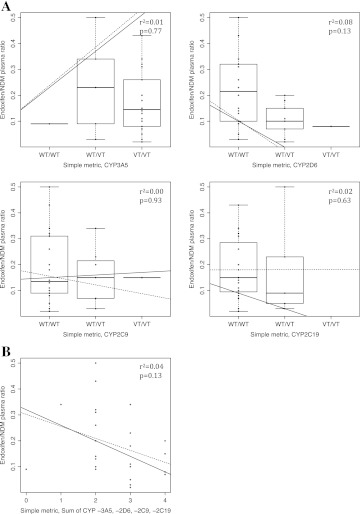
To validate our phenotype scoring system, we compared the predictive performance of the phenotype scoring system to (1) four simple metrics representing the genotypes (Vt/Vt=2, Wt/Vt=1, or Wt/Wt=0) for each gene (CYP3A5, CYP2D6, CYP2C9, and CYP2C19) and (2) a simple metric representing the genotypes of multiple genes (a sum across all genes). We also compared the predictive performance of the phenotype scoring system to a dominant model (Vt/Vt=0, Wt/Vt=1, Wt/Wt=1) and to a recessive model (Vt/Vt=1, Wt/Vt=0, Wt/Wt=0) for representing the genotypes of multiple genes. There was no statistically significant difference from zero for the regression coefficient for the simple metrics representing individual genes (see figure 4A), for the simple metric representing the genotypes of multiple genes (see figure 4B), or for dominant and recessive models for representing the genotypes of multiple genes. Results from investigating dominant and recessive models are not shown in figure 4. The phenotype scoring system is therefore more predictive of patient endoxifen/NDM plasma ratio levels than the simple metrics for individual gene genotypes and the simple metric for the genotypes of all genes.
Figure 4.
(A) Simple metrics for individual genes with genotype values as Vt/Vt=2, Wt/Vt=1, and Wt/Wt=0: CYP3A5 Wt=*1, Vt=*3,*6; CYP2D6 Wt=*1, Vt=*4,*6; CYP2C9 Wt=*1, Vt=*2,*3; and CYP2C19 Wt=*1, Vt=*2. Wt/Wt=0, Wt/Vt=1, Vt/Vt=2. The plots show box-plots of the endoxifen/N-desmethyltamoxifen (NDM) ratio versus genotype values. Superimposed on the plots are the median fit (solid line) and the least squares estimate of the conditional mean function (dashed line). (B) Simple metric for the sum of genotype values for genes CYP3A5, CYP2D6, CYP2C9, and CYP2C19. The lines indicate median bands. The plots show scatter plots of the endoxifen/NDM ratio versus simple metrics. Superimposed on the plots are the median fit (solid line) and the least squares estimate of the conditional mean function (dashed line).
Discussion
Advantages of the PEMRIC model
We suggest an approach that might be incorporated in the electronic health record (EHR) at the point of care. Patient-specific data that need to be captured to facilitate implementation of this model include: the medication being prescribed to a patient, knowledge of what genotype data are currently available for the patient, and the patient's genotype for genes that are involved in the medication drug metabolism pathway. Given a patient's genotype data, the PEMRIC model provides a method for providing a single score that characterizes the combined activity of enzymes involved in drug metabolism.
Future directions
This work investigates how existing knowledge resources could be utilized in a reasoning system. Steps to collect pharmacogenomics evidence have the potential to be automated, although improved download and data access capabilities are needed. For example, with SuperCYP, much of the information needed could be saved as an HTML file that can be parsed; however, a more standardized format would better facilitate automation. Similarly, pathway facts could be derived by parsing downloaded PharmGKB pathway evidence Excel files. Automation of these steps could occur by providing access to evidence file data via the PharmGKB SOAP (Simple Object Access Protocol) interface. Machine learning and artificial intelligence techniques might also be applied to identify relevant passages and extract assertions directly from publications.
The knowledge-based approach we take in this work might be extended to include other forms of clinical data and additional knowledge resources, and an evidential approach to other forms of knowledge could be taken. For example, this approach assumes PK pathway knowledge from PharmGKB to be true. In future work, it could be beneficial to incorporate an evidential approach to defining PK pathway knowledge. In addition, with access to knowledge about allelic variant frequencies in various populations, it could be useful to incorporate patient race/ethnicity into the phenotype score calculation.
Another area for future investigation would be validation of our methods for calculating phenotype scores with a separate dataset. This would facilitate checking for over-fitting and further assessing the potential for the methods to be incorporated into a clinical setting. We were unable to find a separate public dataset with endoxifen plasma concentrations at 4 months and NDM plasma concentrations at 4 months for patients, in addition to genotype data.
Additionally, future work investigating the practical issues of implementing this model in the EHR context would be beneficial. Given the sensitivity of patient-specific data and the evolving nature of evidence in this context, it is important to consider approaches to providing access to, ensuring the security of, connecting, and coordinating disparate data and knowledge sources.
Challenges to PEMRIC model implementation
Using our proof of concept prototype reasoning system, we present several approaches to provide a score that characterizes the combined activity of enzymes involved in the metabolism of a drug. The hope is that rather than evaluating each variable individually, a clinician will be able to review a single score. Even so, such a score would need to be presented in a format and with supportive information that promotes its appropriate use in the making of informed health decisions in a clinical context. There are several technical- and content-related challenges inherent in ensuring that phenotype scores are correct, are current for the patient at hand, and are provided to clinicians in a useful way. In addition, the validity of such a score in the context of predicting response to therapy should be tested in future work.
There are currently mixed results from studies investigating associations between genotype-mediated variation in CYP2D6 enzyme activity and endoxifen concentrations and clinical outcomes. While studies indicate significant associations between CYP2D6 genotype and endoxifen levels,13 20 21 some reports do not indicate an association with clinical outcomes (eg, breast cancer recurrence, incidence of hot flashes).22 23 Therefore, there are currently no recommendations to perform CYP2D6 testing to preferentially select tamoxifen in clinical care. This disagreement in research findings highlights an inability to directly relate CYP2D6 genotype to tamoxifen metabolite levels, and metabolite levels to clinical outcomes. Therefore, before we can facilitate the use of pharmacogenomics data in prescribing decisions, the connection between patient-specific data and clinical outcomes needs to be better understood. Methods incorporating more reasoning steps and evidence have the potential to provide a more complete view of the relationship between patent-specific data (genotypic data and otherwise) and clinical outcomes.
Conclusion
We provided a detailed description of a prototype PEMRIC model implementation in a tamoxifen case study context. Case study findings suggest that phenotype scoring algorithms that assign numeric values according to ‘allelic variant’–‘enzyme activity’ associations have the potential to predict endoxifen/NDM plasma levels (as a marker for drug metabolism efficacy). One scoring system (the drug-oriented, weighted, ‘allelic variant’–‘enzyme activity’ scoring algorithm) showed a weak correlation with endoxifen/NDM plasma ratio (R2=0.10; p<0.05). This approach performed better than simple metrics for variation in individual and multiple genes. Given these results, the PEMRIC model implementation and scoring approaches investigated in this work warrant further investigation. Results also suggest that monogenic models that are classically applied to interpret variants in pharmacogenetic studies may miss important (more complex) associations. More complicated polygenic models, such as those explored in this work, are needed to represent heterogeneity in clinical outcomes.
Acknowledgments
The authors would like to thank the anonymous reviewers for their valuable comments and suggestions for improving this manuscript; Dr Kenneth Thummel (University of Washington Department of Pharmaceutics) for his valuable input; Dr David Flockhart for his willingness to provide the SERM dataset for use in this work; and Drs Isabelle Ragueneau-Majlessi and Houda Hachad (University of Washington Department of Pharmaceutics) for providing guidance on future directions.
Footnotes
Contributors: This study was conceived of by CLO and EBD. CLO collected the data, designed and developed the reasoning system, performed the analyses, interpreted the data, and wrote the manuscript. EBD collected the data, performed the analyses, interpreted the data, and reviewed and edited the manuscript. PTH and IJK reviewed and edited the manuscript.
Funding: This research is supported in part by the UW NIH NLM Informatics Research Training Grant (NIH NLM #T15 LM07442), the regional CTSA – The Institute of Translational Health Sciences (NIH NCRR 1 UL1 RR 025014), a UW Genome Training Grant (NIH NHGRI #T32 HG000053), the Columbia Training in Biomedical Informatics (NIH NLM #T15 LM007079), and a Mentored Clinical Scientist Training Grant (AHRQ 5K08 HS014739, PI: Devine).
Competing interests: None.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Preissner S, Kroll K, Dunkel M, et al. SuperCYP: a comprehensive database on Cytochrome P450 enzymes including a tool for analysis of CYP-drug interactions. Nucleic Acids Res 2010;38(Database issue):D237–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sheffield LJ, Pillimore HE. Clinical use of pharmacogenomic tests in 2009. Clin Biochem Rev 2009;30:55–65 [PMC free article] [PubMed] [Google Scholar]
- 3.Klein TE, Chang JT, Cho MK, et al. Integrating genotype and phenotype information: an overview of the PharmGKB Project. Pharmacogenomics J 2001;1:167–70 [DOI] [PubMed] [Google Scholar]
- 4.Dawood S, Leyland-Jones B. Pharmacology and pharmacogenetics of chemotherapeutic agents. Cancer Invest 2009;27:482–8 [DOI] [PubMed] [Google Scholar]
- 5.Kane-Gill SL, Van Den Bos J, Handler SM. Adverse drug reactions in hospital and ambulatory care settings identified using a large administrative database. Ann Pharmacother 2010;44:983–93 [DOI] [PubMed] [Google Scholar]
- 6.Budnitz DS, Pollock DA, Weidenbach KN, et al. National surveillance of emergency department visits for outpatient adverse drug events. JAMA 2006;296:1858–66 [DOI] [PubMed] [Google Scholar]
- 7.Boyce R, Collins C, Horn J, et al. Computing with evidence Part II: an evidential approach to predicting metabolic drug-drug interactions. J Biomed Inform 2009;42:990–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boyce R, Collins C, Horn J, et al. Computing with evidence Part I: a drug-mechanism evidence taxonomy oriented toward confidence assignment. J Biomed Inform 2009;42:979–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Borges S, Desta Z, Jin Y, et al. Composite functional genetic and comedication CYP2D6 activity score in predicting tamoxifen drug exposure among breast cancer patients. J Clin Pharmacol 2010;50:450–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gaedigk A, Simon SD, Pearce RE, et al. The CYP2D6 activity score: translating genotype information into a qualitative measure of phenotype. Clin Pharmacol Ther 2008;83:234–42 [DOI] [PubMed] [Google Scholar]
- 11.Zineh I, Beitelshees AL, Gaedigk A, et al. Pharmacokinetics and CYP2D6 genotypes do not predict metoprolol adverse events or efficacy in hypertension. Clin Pharmacol Ther 2004;76:536–44 [DOI] [PubMed] [Google Scholar]
- 12.Zanger UM, Raimundo S, Eichelbaum M. Cytochrome P450 2D6: overview and update on pharmacology, genetics, biochemistry. Naunyn Schmiedebergs Arch Pharmacol 2004;369:23–37 [DOI] [PubMed] [Google Scholar]
- 13.Borges S, Desta Z, Li L, et al. Quantitative effect of CYP2D6 genotype and inhibitors on tamoxifen metabolism: implication for optimization of breast cancer treatment. Clin Pharmacol Ther 2006;80:61–74 [DOI] [PubMed] [Google Scholar]
- 14.Fuhr U. Pharmacogenetic-based clinical scores: a useful, simple tool to predict tamoxifen-based CYP2D6 phenotype? J Clin Pharmacol 2010;50:370–2 [DOI] [PubMed] [Google Scholar]
- 15.Consortium on Breast Cancer Pharmacogenomics profile http://www.pharmgkb.org/contributors/pgrnAlumni/cobra_profile.jsp (accessed 5 May 2011).
- 16.Consortium on Breast Cancer Pharmacogenomics (COBRA) Patient responses to tamoxifen. Submission PS202620 [Computer file]. Consortium on breast cancer pharmacogenomics (COBRA) [producer], 2006. PharmGKB [distributor], 2006 [Google Scholar]
- 17.Stearns V, Johnson MD, Rae JM, et al. Active tamoxifen metabolite plasma concentrations after coadministration of tamoxifen and the selective serotonin reuptake inhibitor paroxetine. J Natl Cancer Inst 2003;95:1758–64 [DOI] [PubMed] [Google Scholar]
- 18.Desta Z, Ward BA, Soukhova NV, et al. Comprehensive evaluation of tamoxifen sequential biotransformation by the human cytochrome P450 system in vitro: prominent roles for CYP5A and CYP2D6. J Pharmacol Exp Ther 2004;310:10062–75 [DOI] [PubMed] [Google Scholar]
- 19.Java Expert System Shell. http://www.jessrules.com (accessed 5 May 2011).
- 20.Kiyotani K, Mushiroda T, Imamura CK, et al. Significant effect of polymorphisms in CYP2D6 and ABCC2 on clinical outcomes of adjuvant tamoxifen therapy for breast cancer patients. J Clin Oncol 2010;28:1287–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gjerde J, Hauglid M, Breilid H, et al. Effects of CYP2D6 and SULT1A1 genotypes including SULT1A1 gene copy number on tamoxifen metabolism. Ann Oncol 2008;19:56–61 [DOI] [PubMed] [Google Scholar]
- 22.Rae JM, Drury S, Hayes DF, et al. Lack of correlation between gene variants in tamoxifen metabolizing enzymes with primary endpoints in the ATAC trial. 33rd Annual San Antonio Breast Cancer Symposium. Abstract S1-7. Presented 9 December 2010.
- 23.Leyland-Jones B, Regan MM, Bouzk M, et al. : Outcome according to CYP2D6 genotype among postmenopausal women with endocrine-responsive early invasive breast cancer randomized in the BIG 1-98 trial. 33rd Annual San Antonio Breast Cancer Symposium. Abstract S1-8. Presented 9 December 2010.