Abstract
Because breast tissue composition partially predicts breast cancer risk, classification of mammography reports by breast tissue composition is important from both a scientific and clinical perspective. A method is presented for using the unstructured text of mammography reports to classify them into BI-RADS breast tissue composition categories. An algorithm that uses regular expressions to automatically determine BI-RADS breast tissue composition classes for unstructured mammography reports was developed. The algorithm assigns each report to a single BI-RADS composition class: ‘fatty’, ‘fibroglandular’, ‘heterogeneously dense’, ‘dense’, or ‘unspecified’. We evaluated its performance on mammography reports from two different institutions. The method achieves >99% classification accuracy on a test set of reports from the Marshfield Clinic (Wisconsin) and Stanford University. Since large-scale studies of breast cancer rely heavily on breast tissue composition information, this method could facilitate this research by helping mine large datasets to correlate breast composition with other covariates.
Keywords: Mammography, natural language processing, data mining, radiology, breast, text mining, machine learning, pharmacogenomics, breast imaging, quality, outcomes research
Background
Tissue composition and breast cancer
Breast tissue composition is an important component of the radiological evaluation of the breast for two reasons. First, dense fibroglandular tissue is a risk factor for breast cancer.1–3 Second, this dense tissue decreases mammographic sensitivity in detecting breast cancer.4 For these reasons, mammography reports typically contain a description of the overall tissue composition of the breast.
The BI-RADS system
The American College of Radiology Breast Imaging Reporting and Data System (BI-RADS) divides breast composition into four categories: 1 (predominantly fatty), 2 (scattered fibroglandular densities), 3 (heterogeneously dense), and 4 (extremely dense).5 These standardized categories help to minimize ambiguity in mammography reporting. They also enable radiologists to qualify their observations by discussing how a patient's breast composition may limit mammographic sensitivity. Finally, they help stratify patients at the time of imaging, with additional screening recommended for women with dense, fibroglandular breast tissue, which can obscure small masses. Reliable, standardized information on breast composition can facilitate large-scale studies of breast cancer, and may also play an important role in the development of classification systems for the early detection of malignancy.
Limitations of the current system
One limitation of applying the BI-RADS system is that breast composition information is typically not reported in coded form; descriptions of breast composition occur as narrative text within the surrounding text of a mammography report. Although the corresponding BI-RADS category may be obvious to a radiologist reading the report, such inference would prove challenging for a computer. For example, radiologists use characteristic phrases like ‘scattered fibroglandular’, ‘mostly fatty’, and ‘focally dense’ when describing breast composition, but no one textual pattern could be used to obtain this information with 100% sensitivity and specificity. This fact thwarts large research studies, which often require breast tissue composition information from thousands of reports. Manual curation of this information is not feasible, being both time-consuming and error-prone.
An automated classification method
These limitations led us to explore automated approaches for obtaining breast composition information based on principles from natural language processing. The use of natural language processing is not new to radiology or clinical medicine in general; for a comprehensive review of its important role in radiology, see Lacson and Khorasani.6 7 Informatics researchers have already explored ways to extract meaningful textual features from radiology reports,8 classify reports automatically by body location or disease,9 10 automatically produce structured reports from free text reports,11 12 and assess variability in and deficiencies of radiology reporting using text mining.13
We present here the first automated method for addressing one classification problem that has not yet been solved: classifying mammography reports automatically by breast composition. Previous authors have investigated the use of BI-RADS descriptors in mammography reports,14 15 but have not addressed the extraction of breast composition data. Our method classifies each report according to its BI-RADS tissue composition category accurately, efficiently, and automatically, which we hope will aid researchers, clinicians, and policy analysts who need access to large-scale mammography data.
Methods
Data
We used mammography corpora from three different institutions to develop and test our algorithm. Our data included 34 489 reports from Stanford's RADTF (RADiology Teaching File) database,16 and a further 146 972 reports from the University of California, San Francisco (UCSF) Medical Center, which we used to construct a set of textual patterns indicative of each breast composition class. We also built an independent test set comprised of 500 reports from the Stanford corpus (which were held out during the rule-construction phase) and 100 reports from the Marshfield Clinic in Wisconsin.
The reports were independently annotated by a board-certified radiologist with 1 year of fellowship training in breast imaging. Our radiologist annotator was blinded to the automatically assigned breast composition classes when assessing the reports.
Rule construction
Using unstructured mammography reports as its input, our algorithm classifies each report into one of five classes: predominantly fat (class 1), scattered fibroglandular densities (class 2), heterogeneously dense (class 3), extremely dense (class 4), or ‘unspecified’. These classes correspond to the four BI-RADS breast composition classes and one additional category for reports that do not include breast composition information.
We constructed our classifier by modifying the BI-RADS feature extraction approach presented in Nassif et al17 to retrieve breast composition information. We began by mapping all of the key terms and phrases from the full BI-RADS lexicon5 to specific breast composition classes. For example, the key phrase ‘extremely dense’ was mapped to breast composition class 4. We then augmented this lexicon using expert knowledge, adding other breast composition class descriptors frequently used in clinical practice, such as ‘breast is dense’. We worked with multiple radiologists throughout this process to develop an understanding of the different ways radiologists describe different breast composition classes.
Once we had established which BI-RADS key terms/phrases corresponded to each BI- RADS breast composition class, we mined the UCSF and Stanford datasets for all words occurring in close proximity to the key terms. We then established how far (and in what direction) these ‘neighbor’ words could be from the keywords before they ceased to be informative. This was accomplished via an iterative process in which we examined the number of incorrect classifications obtained as the key term and neighbor were moved further and further apart. We then chose the maximum value of separation that corresponded to the lowest overall classification error. For example, the stem scatter could occur up to three words before the key term fibroglandular before the number of false positive class 2 errors began to increase (see figure 1). This process established a set of rules for automatically classifying reports into BI-RADS breast composition classes.
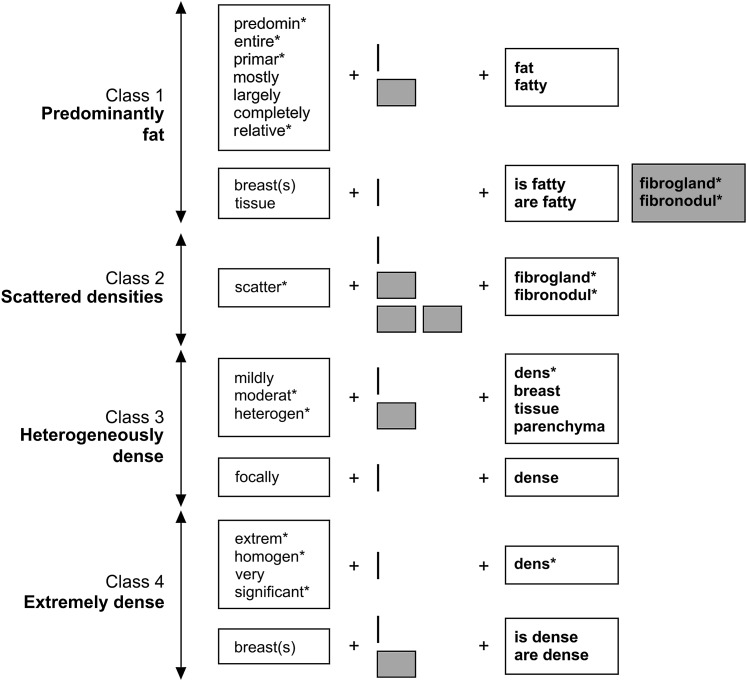
Figure 1.
A diagrammatic explanation of the rules used to assign reports to different BI-RADS tissue composition classes. Each row represents a pattern unique to the class shown at the left. White rectangles represent sets of words or word stems that must be present at a given location to fulfill the rule. Gray rectangles represent words/stems that cannot be present at a location for the rule to be fulfilled. The small gray boxes represent unspecified words. The asterisk (*) is used to denote multiple possible word endings. So, for example, a report would be assigned to class 2 if it contained the stem scatter followed by 0, 1, or 2 other words, and then the stem fibrogland or fibronodul. Similarly, a report would be assigned to class 1 if it contained the word breast(s) or tissue followed immediately by the phrase is/are fatty, but the stem fibrogland or fibronodul did not occur immediately after fatty.
Algorithm development
Figure 1 shows the final set of rules used to assign reports to each BI-RADS breast composition class. A report was classified as BI-RADS composition class 1 if it contained the word fat or fatty preceded (within two words) by a modifier from the following set: predomin*, primar*, largely, relative*, entire*, mostly, completely. The symbol * means that we specified the word stem but not its ending, so any word containing the given stem was accepted. If a report contained the phrase is fatty or are fatty preceded (within two words) by the term breast(s) or tissue, it was also assigned to class 1 provided that fatty was not followed by the stem fibrogland* or fibronodul*. A report was assigned to class 2 if it contained the word fibrogland* or fibronodul*, preceded (within three words) by a modifier of the form scatter*. It was assigned to class 3 if it contained one of the terms dens*, breast, tissue, or parenchyma, preceded (within two words) by a modifier from the set: mildly, moderat*, heterogen*. Reports containing the specific phrase focally dense were also assigned to class 3. Finally, a report was assigned to class 4 if it contained the stem dens* immediately preceded by a modifier from the list extrem*, homogen*, very, significant*, or if it contained the phrase is dense or are dense preceded (within two words) by the term breast or breasts.
Evaluation
Using our algorithm, we classified each mammography report in the test set as BI-RADS breast composition class 1–4, or ‘unspecified’. We then compared the algorithm's results to our radiologist annotator's classifications of the same reports.
Results
Table 1 contains a list of the descriptors found in the three datasets, along with their associated frequencies. A greater variety of descriptors were used to describe class 1 (predominantly fat) mammograms than any other class; class 2 (scattered fibroglandular) mammography reports were the most consistent, always using the phrase scattered fibroglandular or scattered fibronodular.
Table 1.
A summary of the descriptors used to report the different breast composition classes
| Class | Rule | Number of occurrences | |||
| Stanford (training) | UCSF (training) | Stanford (test) | Marshfield (test) | ||
| 1 | predomin* fat(ty) | 163 | 79 | 1 | 0 |
| entire* fat(ty) | 52 | 16 953 | 3 | 0 | |
| primar* fat(ty) | 7 | 219 | 0 | 0 | |
| mostly fat(ty) | 846 | 422 | 1 | 0 | |
| largely fat(ty) | 5149 | 3 | 112 | 1 | |
| completely fat(ty) | 0 | 5 | 0 | 0 | |
| relative* fat(ty) | 0 | 8 | 0 | 0 | |
| breast(s)/tissue is/are fat(ty) | 180 | 39 | 8 | 0 | |
| 2 | scatter* fibro(gland/nodul)* | 13 947 | 51 358 | 123 | 35 |
| 3 | mildly (dens/breast/tissue/parenchyma)* | 1 | 14 | 0 | 6 |
| moderat* (dens/breast/tissue/parenchyma)* | 3 | 282 | 0 | 9 | |
| heterogen* (dens/breast/tissue/parenchym)* | 11 006 | 49 106 | 128 | 19 | |
| focally dense | 2 | 123 | 0 | 0 | |
| 4 | extrem* dens* | 1220 | 11 080 | 9 | 2 |
| homogen* dens* | 5 | 16 | 0 | 0 | |
| very dens* | 2041 | 40 | 45 | 1 | |
| significant* dense | 0 | 0 | 0 | 1 | |
| breast(s) is/are dense | 17 | 60 | 66 | 0 | |
The asterisk (*) is used to denote multiple possible word endings. Note that a single mammography report may contain multiple rule occurrences.
Our algorithm's performance relative to the radiologist's gold standard is shown in table 2. Our algorithm correctly classified 499/500 (99.8%) reports from the Stanford dataset and 99/100 (99%) reports from the Marshfield Clinic dataset. On the Stanford data, the only incorrectly classified report contained the description ‘bilateral breasts redemonstrate dense glandular tissue’, which the radiologist assigned to class 4 and the algorithm assigned to the ‘unspecified’ class. On the Marshfield Clinic test set, the radiologist assigned the description ‘the right breast shows fibroglandular tissue which is finely nodular and strandlike’ to class 2, but the algorithm assigned it to ‘unspecified’.
Table 2.
System performance results on the Stanford and Marshfield testing sets
| Dataset | Records with descriptors present | Records with no descriptors | Correctly classified records | Total |
| Stanford | 497 | 3 | 499 | 500 |
| Marshfield | 73 | 27 | 99 | 100 |
The first two columns contain the number of records that were classifiable and the number that were not (some did not include any BI-RADS tissue composition descriptors whatsoever). The third column contains the number of records that were classified correctly (either assigned to the correct BI- RADS composition class or classified as ‘no descriptors’ when that assessment was correct).
Discussion
Breast tissue composition has consistently been associated with breast cancer and other proliferative breast lesions.1 18–22 For example, breast cancer risk increases by 4–5 times in women with very dense breasts relative to women with little or no dense breast tissue.1–3 It is difficult to diagnose early-stage breast cancer in women with dense breasts, which contributes to the increased risk of breast cancer for these women; rates of interval breast cancers (cancers discovered between yearly screening mammograms) are much higher in women with dense breast tissue than in those with predominantly fatty breasts.23–25
Automated classification of free-text radiology reports into tissue composition classes therefore has important clinical, research, and policy implications. In the clinical arena, the algorithm may enable hospital systems and other healthcare delivery organizations to predict and prepare for potential increases in referrals for and utilization of screening breast ultrasound and breast MRI. For example, the Connecticut state legislature recently passed a bill requiring that women undergoing mammography be counseled about their breast composition and that insurance companies pay for additional screening for women with dense breast tissue.26 Similar bills are currently up for discussion in the Texas and California state legislatures. We might expect such legislation to lead to an increase in the use of these alternative screening methods.
In the research and policy arenas, our algorithm may facilitate large-scale population-based studies of breast tissue composition and other covariates of breast cancer risk, and enable improvement in risk prediction models by allowing them to better incorporate information on breast composition. Breast composition has a strong genetic component,3 27–31 so population-based studies of breast cancer, especially those investigating patterns of occurrence within families and non-genetic causes of breast cancer, must control for it. The difficulty associated with manually extracting breast composition information from thousands of unstructured mammography reports for research purposes was what originally led us to develop an automated method for performing this task. To our knowledge, our methods are the first attempt at automated classification of mammography reports into breast tissue composition classes.
We built and validated our algorithm using reports from three different institutions: UCSF and Stanford for algorithm development, and Stanford and Marshfield for testing. By including reports from multiple institutions during the development process, and by searching thousands of reports to detect variations in how different breast composition classes were described, we hoped to avoid creating an algorithm that was too institution- or radiologist-specific.
Despite its high accuracy, our approach still has a few limitations. Empirical observations of reports from UCSF and Stanford revealed that radiologists at different institutions tend to describe breast composition in characteristic, sometimes divergent ways. For example, most of the class 1 reports at Stanford used the phrase ‘largely fatty’, while UCSF radiologists favored the phrase ‘entirely fatty’ and almost never used ‘largely fatty’. This could be due to the use of institution-specific templates in mammography reporting, which could indicate that we need to include reports from several more institutions to develop a truly robust algorithm. Future studies with larger and more diverse datasets should be performed to confirm the accuracy and generalizability of our algorithm to reports obtained from other institutions. Although our use of regular expressions was highly accurate for classifying breast tissue composition based on the text of unstructured mammography reports, the method is highly domain-specific and might not be generalizable to other applications within the field of radiology.
Conclusion
In conclusion, we have created an algorithm that automatically processes unstructured, free-text mammography reports and reliably classifies them into BI-RADS breast composition classes. Our algorithm achieves extremely high accuracy (>99%) during testing. This method could facilitate research and policy analysis by enabling investigators to efficiently mine large collections of mammography reports.
Acknowledgments
The authors would like to thank Jacqueline Bohne for her help obtaining the Marshfield Clinic data.
Footnotes
Funding: This work was supported by grants from the National Cancer Institute, National Institutes of Health (grant numbers U01-CA-142555, K07-CA114181, and R01-CA127379), and by a training grant from the National Library of Medicine (grant number 5T15LM007033-27). None of the funding sources had any involvement in the preparation of the manuscript.
Competing interests: None.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Boyd NF, Martin LJ, Bronskill M, et al. Breast tissue composition and susceptibility to breast cancer. J Nat Cancer Inst 2010;102:1224–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boyd NF, Rommens JM, Vogt K, et al. Mammographic breast density as an intermediate phenotype for breast cancer. Lancet Oncol 2005;6:798–808 [DOI] [PubMed] [Google Scholar]
- 3.Martin LJ, Melnichouk O, Guo H, et al. Family history, mammographic density, and risk of breast cancer. Cancer Epidemiol Biomarkers Prev 2010;19:456–63 [DOI] [PubMed] [Google Scholar]
- 4.Carney PA, Miglioretti DL, Yankaskas BC, et al. Individual and combined effects of age, breast density, and hormone replacement therapy use on the accuracy of screening mammography. Ann Intern Med 2003;138:168–75 [DOI] [PubMed] [Google Scholar]
- 5.American College of Radiology (ACR) Breast Imaging Reporting and Data System (BI-RADS). 3rd edn Reston, VA: American College of Radiology, 2003 [Google Scholar]
- 6.Lacson R, Khorasani R. Natural language processing: the basics (part 1). J Am Coll Radiol 2011;8:436–7 [DOI] [PubMed] [Google Scholar]
- 7.Lacson R, Khorasani R. Natural language processing for radiology (part 2). J Am Coll Radiol 2011;8:583–4 [DOI] [PubMed] [Google Scholar]
- 8.Garla V, Re VL, Dorey-Stein Z, et al. The Yale cTAKES extensions for document classification: architecture and application. J Am Med Inform Assoc 2011;18:614–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sevenster M, van Ommering R, Qian Y. Automatically correlating clinical findings and body locations in radiology reports using MedLEE. J Digital Imaging. Published Online First: 8 July 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Solti I, Cooke CR, Xia F, et al. Automated classification of radiology reports for acute lung injury: comparison of keyword and machine learning based natural language processing approaches. Proceedings (IEEE Int Conf Bioinformatics Biomed) 2009;2009:314–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Apostolova E, Channin DS, Demner-Fushman D, et al. Automatic segmentation of clinical texts. Conf Proc IEEE Eng Med Biol Soc 2009;2009:5905–8 [DOI] [PubMed] [Google Scholar]
- 12.Hasegawa Y, Matsumura Y, Mihara N, et al. Development of a system that generates structured reports for chest x-ray radiography. Methods Inf Med 2010;49:360–70 [DOI] [PubMed] [Google Scholar]
- 13.Reiner B. Uncovering and improving upon the inherent deficiencies of radiology reporting through data mining. J Digit Imaging 2010;23:109–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Starren J, Johnson SM. Notations for high efficiency data presentation in mammography. Proc AMIA Annu Fall Symp 1996:557–61 [PMC free article] [PubMed] [Google Scholar]
- 15.Starren J, Johnson SM. Expressiveness of the breast imaging reporting and database system (BI-RADS). Proc AMIA Annu Fall Symp 1997:655–9 [PMC free article] [PubMed] [Google Scholar]
- 16.Do BH, Wu A, Biswal S, et al. RADTF: a semantic search-enabled, natural language processor-generated radiology teaching file. RadioGraphics 2010;30:2039–48 [DOI] [PubMed] [Google Scholar]
- 17.Nassif H, Woods R, Burnside E, et al. Information extraction for clinical data mining: a mammography case study. 9th IEEE International Conference on Data Mining Workshops (ICDMW′09). Miami, pp. 37–42, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kerlikowske K, Ichikawa L, Miglioretti DL, et al. Longitudinal measurement of clinical mammographic breast density to improve estimation of breast cancer risk. J Natl Cancer Inst 2007;99:386–95 [DOI] [PubMed] [Google Scholar]
- 19.Nagao Y, Kawaguchi Y, Sugiyama Y, et al. Relationship between mammographic density and the risk of breast cancer in Japanese women: a case-control study. Breast Cancer 2003;10:228–33 [DOI] [PubMed] [Google Scholar]
- 20.Ursin G, Ma H, Wu AH, et al. Mammographic density and breast cancer in three ethnic groups. Cancer Epidemiol Biomarkers Prev 2003;12:332–8 [PubMed] [Google Scholar]
- 21.Nagata C, Matsubara T, Fujita H, et al. Mammographic density and the risk of breast cancer in Japanese women. Br J Cancer 2005;92:2102–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heusinger K, Loehberg CR, Haeberle L, et al. Mammographic density as a risk factor for breast cancer in a German case-control study. Eur J Cancer Prev 2011;20:1–8 [DOI] [PubMed] [Google Scholar]
- 23.Mandelson MT, Oestreicher N, Porter PL, et al. Breast density as a predictor of mammographic detection: comparison of interval- and screen-detected cancers. J Natl Cancer Inst 2000;92:1081–7 [DOI] [PubMed] [Google Scholar]
- 24.Chiarelli AM, Kirsh VA, Klar NS, et al. Influence of patterns of hormone replacement therapy use and mammographic density on breast cancer detection. Cancer Epidemiol Biomarkers Prev 2006;15:1856–62 [DOI] [PubMed] [Google Scholar]
- 25.Kavanagh AM, Byrnes GB, Nickson C, et al. Using mammographic density to improve breast cancer screening outcomes. Cancer Epidemiol Biomarkers Prev 2008;17:2818–24 [DOI] [PubMed] [Google Scholar]
- 26.Connecticut Bill No. 458, Public Act No. 09-41 An Act Requiring Communication of Mammographic Breast Density Information to Patients. http://www.cga.ct.gov/2009/ACT/PA/2009PA-00041-R00SB-00458-PA.htm (accessed 17 May 2011).
- 27.Boyd NF, Dite GS, Stone J, et al. Heritability of mammographic density, a risk factor for breast cancer. N Engl J Med 2002;347:886–94 [DOI] [PubMed] [Google Scholar]
- 28.Boyd NF, Martin LJ, Rommens JM, et al. Mammographic density: a heritable risk factor for breast cancer. Methods Mol Biol 2009;472:343–60 [DOI] [PubMed] [Google Scholar]
- 29.Stone J, Gurrin LC, Byrnes GB, et al. Mammographic density and candidate gene variants: a twins and sisters study. Cancer Epidemiol Biomarkers Prev 2007;16:1479–84 [DOI] [PubMed] [Google Scholar]
- 30.Boyd NF, Martin LJ, Chavez S, et al. Breast-tissue composition and other risk factors for breast cancer in young women: a cross-sectional study. Lancet Oncol 2009;10:569–80 [DOI] [PubMed] [Google Scholar]
- 31.Ziv E, Shepherd J, Smith-Bindman R, et al. Mammographic breast density and family history of breast cancer. J Nat Cancer Inst 2003;95:556–8 [DOI] [PubMed] [Google Scholar]