Abstract
Objective
Coreference resolution of concepts, although a very active area in the natural language processing community, has not yet been widely applied to clinical documents. Accordingly, the 2011 i2b2 competition focusing on this area is a timely and useful challenge. The objective of this research was to collate coreferent chains of concepts from a corpus of clinical documents. These concepts are in the categories of person, problems, treatments, and tests.
Design
A machine learning approach based on graphical models was employed to cluster coreferent concepts. Features selected were divided into domain independent and domain specific sets. Training was done with the i2b2 provided training set of 489 documents with 6949 chains. Testing was done on 322 documents.
Results
The learning engine, using the un-weighted average of three different measurement schemes, resulted in an F measure of 0.8423 where no domain specific features were included and 0.8483 where the feature set included both domain independent and domain specific features.
Conclusion
Our machine learning approach is a promising solution for recognizing coreferent concepts, which in turn is useful for practical applications such as the assembly of problem and medication lists from clinical documents.
Keywords: Coreference resolution, machine learning, NLP, natural language processing, text mining, quality measures, CDI
Introduction
The Health Information Technology for Economic and Clinical Health (HITECH) act passed in February 2009 as part of the American Reinvestment and Recovery Act (ARRA) sets aside significant funds as incentives for the adoption of electronic medical records (EMR). In particular, the act calls for providers to demonstrate ‘meaningful use’ of a certified EMR to qualify for financial incentives. Evidence of meaningful use has been defined, in part, as the capturing of structured elements in an EMR such as problem lists, medications, procedures, allergies, and quality measures. Identifying coreferent concepts is an essential part of capturing these elements.
The 2011 natural language processing (NLP) challenge's focus on coreference (different terms in a document that refer to the same concept) gives it a high practical relevance in the marketplace. In particular, the relationship of problems and treatments in transcribed medical documents can be used to assemble complete, yet precise, problem and medication lists. For instance, the terms: ‘congestive heart failure,’ ‘CHF,’ ‘systolic heart failure,’ and ‘heart failure’ may be coreferent—that is, they may all describe the same condition in the same patient. The objective of the challenge is to assemble all such relationships into chains of coreferent problems, treatments, tests, or persons.
This year, i2b2 provided two sets of annotations using two different guidelines for the challenge: ODIE and i2b2. The ODIE guidelines were more elaborate and detailed than the i2b2 guidelines. We entered track 1c, with i2b2 annotations, to capitalize on our experience with this annotation set from last year's competition. The training document set had the following set of annotations: Test, Problem, Procedure, Person, and Pronoun. The annotation chains (coreferent chains) were of the following categories: Test, Problem, Procedure, and Person.
The effort discussed in this report details our solution to the coreference challenge using the i2b2 guidelines.
Methods
Background
There has been significant recent research effort in the NLP community to address the problem of coreference resolution. The BART system1 uses a classifier that relies on a feature set that can be tuned to different languages. Facets of the feature set described in that system include: gender agreement (he/she), number agreement (singular/plural), animacy agreement (him/it, them/that), string match (exact or partial match), distance between concepts (physical separation—number of characters, words between mentions), and aliases (synonyms). The authors also describe a semantic tree compatibility, in which a frame of slot-value pairs (that include the above features) is associated with each concept and the frames are compared for compatibility. Most approaches to coreference resolution rely on supervised-learning techniques. However, the method used by Raghunathan and coworkers2 uses a completely different approach. They order the feature sets to resolve coreference from most precise to least precise and apply them successively to collate coreferent chains. A cluster-ranking approach, where coreference resolution is recast as a problem of finding the best preceding cluster to link a particular mention, is discussed in the paper by Rahman and Ng.3
Overview of procedure
Our approach in this effort builds upon the methods described by Culotta et al.4 It uses a learning engine and a feature set that is fine-tuned to the clinical domain. The core learning engine is implemented using the Scala programming language.
Machine learning approach
We used the ‘Factorie’ toolkit5 to support the learning task. The toolkit is used to implement factor graphs. In the factor graph, the mentions are represented as nodes. Mentions which are coreferent in a given configuration are connected by edges, which we call pairwise-affinity factors. As the system considers different possible configurations, it constructs a factor graph to represent each configuration.
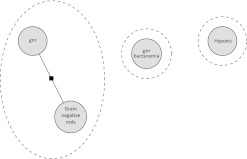
For example, figure 1 shows an incorrect configuration with four mentions divided into three chains. The mentions ‘gnr’ and ‘gram negative rods’ are chained with each other, but the mention ‘gnr bacteremia’ is incorrectly omitted from the chain.
Figure 1.
Four mentions configured as three chains.
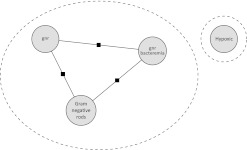
The system will consider adding ‘gnr bacteremia’ to the correct chain as in figure 2.
Figure 2.
Four mentions configured correctly as two chains.
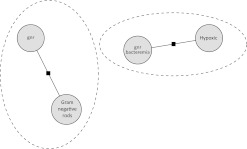
It will also consider adding ‘gnr bacteremia’ to the chain containing the mention ‘hypoxic’ as in figure 3. The features for the pairwise-affinity factors are of two types. Some of the features attempt to capture the relationship of the two mentions. This uses a fairly standard hand-constructed list including: distance metrics, gender agreement (he/she), laterality agreement (left/right), number agreement (singular/plural), overlap, synonyms in SNOMED, hypernyms (broader concepts) in SNOMED, string equality, etc. For the second set of pair-wise features, we take the cross-product of certain mention-wise features. Mention-wise features include words, bi-grams, four character prefixes, and enclosing section type. We also used chain-wise factors, one per chain, to capture information about the chain as a whole. As an example, a chain which included both ‘Mr.’ and ‘she’ would be noted as having a gender inconsistency. The graph was trained using a maximum entropy model with adaptive regularization of weight vectors (AROW) updates.6 Sampling was from the plausible permutations generated for the mentions.
Figure 3.
Four mentions in alternate configuration.
In the next section, we discuss feature selection in greater detail.
Feature selection
Regardless of whether the features discussed above are used in a pair-wise fashion or as an aspect of a single mention or a whole chain, we can classify the feature selection as being domain independent or domain dependent.
Domain independent features
These include four and five character prefixes, words, bi-grams, string match, gender match, and number match. We also considered headword (root/stem matches) and animacy match approaches but lacked the development time to implement these prior to the contest deadlines. Although we had access to parts-of-speech tagging based on the cTAKES system,7 we did not use them. We anticipate trialing these as time permits in the future.
Clinical domain specific features
Here we implemented a variety of features, as follows:
Laterality compatible—determines if the concept refers to the left side or the right side: for example, left knee meniscus tear versus right hip fracture.
Site—identifies whether the body location site is compatible. Clearly meniscus tear (site: knee) and hip fracture (site: hip) are not. (Note: This feature was implemented in the end but there was not enough time to train with this feature before the competition test date).
Section type—clinical documents sections are generally well structured and a mention's location within the document conveys significant information. For instance, a section labeled, ‘past medical history,’ clearly conveys information about the patient's past and any mention there may not be coreferant to similar mentions in other sections.
Aliases—general implementation of coreference resolution tends to use WordNet for gathering aliases. However, for the clinical domain, such aliases are better assembled using SNOMED or other clinical vocabularies. We used the Apelon TermWorks engine to search for aliases for concepts.
Parents of mentions—we also used the SNOMED hierarchy to determine the parents and grandparents of problem concepts.
Acute or chronic—chronic conditions are generally coreferent as they refer to patient conditions that persist over time. For example, hypertension and diabetes are chronic conditions. Acute conditions on the other hand may or may not be coreferent in the same document. So we introduced a feature to recognize acute conditions. Using a data mining infrastructure, we determined that terms such as the following co-occur with acute conditions: acute, attack, stroke, accident, infarction, exacerbation, meningitis, trauma, hemorrhage, rupture, pneumonia, infiltrate, epilepticus, etc.
Surgery—as for chronic/acute disambiguation for problems, one can make a similar distinction between treatments that are surgeries versus those that are medications versus those that are device based.
Sign or symptom—this feature attempts to categorize whether a problem concept is a sign or a symptom.
Diagnostic procedure—this feature categorizes a treatment or a test as to whether it is a diagnostic procedure based on the SNOMED list of diagnostic procedures.
Screening procedure—this feature categorizes a treatment or a test as to whether it is a screening procedure based on the SNOMED list of screening procedures.
Body part mention—this feature analyzes a mention based on the SNOMED body parts list.
-
Related terms—we used the Microsoft Bing search engine to search the web for terms related to the mentions. To carry out this search, we used Bing to search the ‘eMedicine’ (now called Medscape) and ‘webMD’ websites and took the intersection of terms (words) from the two searches to determine related terms. Example are shown below:
meniscal tear eMedicine: [, drez's, disk, sports, good, expendable, ga, that's, surgery, center, medical, refer, tear, howard, meniscal, medicine, miller, knee, drez, information, connection, cartilage…]
meniscal tear webMD: [, discomfort, replacement, vertical, lateral, via, organizedwisdom, treatment, hip, more,, jointreplacement, type, surgery, prior, doctor, disc, medical, webmd:, risks,, tear, depends, knee, joint, tear…]
Intersection of eMedicine and webMD: [, medical, meniscus, tear, knee, treatments, tears]
Although this approach identified ‘knee’ as a related term of ‘meniscal tear,’ we found that using SNOMED vocabulary afforded better results in general. However, the use of web searches does appear interesting and promising and merits further investigation. Of note, Google did not allow programmatic use of their search engine, while Microsoft Bing provided useful software sources to help with such searches.
Temporal features—this mostly identified whether a term is current or past. If, for example, a concept occurred in the ‘past medical history’ section, it would be tagged as in the past. We considered, but did not implement, future tense assessment and tagging.
Technologies and support systems used
The whole NLP framework was developed using the Scala programming language which supports both object-oriented computing and functional programming. Implementations of the language are available for Java and .NET platform. Our NLP platform was built using the Java environment. We also relied on the Apelon terminology environment to determine relevant SNOMED-based aliases.
Evaluation metrics used
The evaluation metrics were supplied by the i2b2 contest organizers. Four evaluation metrics have been specified. For an excellent comparison of the metrics chosen by the i2b2 contest see Recasens and Hovy.8 Cai and Strube also have discussions surrounding some of these metrics.9 The various measures are:
B3—measures10 the number of mentions in the response set (R) that are in common with the gold key set (K). The precision and recall are computed as shown below:
where is the response chain (system response) for the ith mention and is the gold standard. These are then summed over the entire set.
B3—measures are overly sensitive to a large number of singleton mentions, a fact that we verified by assigning every mention as its own coreference chain. This resulted in a B3 value of 0.955, higher than any result we obtained in the actual runs.
MUC—This is a link-based scoring scheme, where a link is the coreference relationship between two mentions.11 The measure fundamentally evaluates how many links are in common between the sets R and K. Recall errors are equated to missing links and precision errors linked to superfluous links. The MUC measure ignores singletons and can be fooled by assigning all mentions to be one big chain.
CEAF—computes a similarity metric between key and response.12 In essence, to score a coreference task it attempts to find the best one to one mapping between the ground truth and the system output. This measure also is susceptible to providing optimistic results in the presence of a large number of singleton mentions.
BLANC—BiLateral Assessment of Noun-phrase Coreference—developed by Recasens and Hovy,8 is an attempt to address the limitations of the above-mentioned measures. The metric capitalizes on the fact that every mention falls into two categories: one that is part of a coreference chain or one that is part of a non-coreference chain. Precision and recall metrics are calculated independently for both categories and the result is then averaged. Singleton mentions will only contribute 50% of the measure and hence will not overwhelm the metric when large numbers of such mentions exist.
The BLANC measure, although computed, was not used by the i2b2 organizers and is not included in our results.
Results and discussion
Training phase—results
Table 1 shows the results from training using only the domain independent features set. On our computing hardware, the training phase required approximately 18 h to run on the 489 documents in the contest training set.
Table 1.
Results for domain independent feature training
| Category | Statistical measures | Recall | F measure | Precision |
| Test | B3 | 0.979 | 0.945 | 0.913 |
| MUC | 0.146 | 0.215 | 0.405 | |
| CEAF | 0.87 | 0.885 | 0.901 | |
| Person | B3 | 0.755 | 0.829 | 0.918 |
| MUC | 0.919 | 0.834 | 0.763 | |
| CEAF | 0.809 | 0.535 | 0.399 | |
| Problem | B3 | 0.961 | 0.948 | 0.936 |
| MUC | 0.418 | 0.491 | 0.596 | |
| CEAF | 0.828 | 0.848 | 0.87 | |
| Treatment | B3 | 0.966 | 0.935 | 0.905 |
| MUC | 0.474 | 0.571 | 0.717 | |
| CEAF | 0.863 | 0.867 | 0.87 | |
| Total | B3 | 0.961 | 0.951 | 0.942 |
| MUC | 0.715 | 0.729 | 0.745 | |
| CEAF | 0.81 | 0.819 | 0.828 | |
| Average total F measure | 0.833 | |||
Table 2 shows the results from training using only the domain independent and domain dependent features set that extensively used the SNOMED vocabularies as discussed in the previous sections. The training phase took approximately 40 h to run.
Table 2.
Results for domain independent and dependent feature training
| Category | Statistical measures | Recall | F measure | Precision |
| Test | B3 | 0.979 | 0.946 | 0.916 |
| MUC | 0.155 | 0.225 | 0.405 | |
| CEAF | 0.892 | 0.897 | 0.903 | |
| Person | B3 | 0.774 | 0.839 | 0.915 |
| MUC | 0.928 | 0.848 | 0.78 | |
| CEAF | 0.819 | 0.556 | 0.42 | |
| Problem | B3 | 0.966 | 0.946 | 0.926 |
| MUC | 0.414 | 0.509 | 0.66 | |
| CEAF | 0.817 | 0.846 | 0.877 | |
| Treatment | B3 | 0.968 | 0.94 | 0.913 |
| MUC | 0.502 | 0.594 | 0.727 | |
| CEAF | 0.852 | 0.865 | 0.878 | |
| Total | B3 | 0.964 | 0.952 | 0.941 |
| MUC | 0.72 | 0.742 | 0.764 | |
| CEAF | 0.811 | 0.825 | 0.839 | |
| Average total F measure | 0.8486 | |||
In general, we did less well on the MUC metric, and scored particularly poorly in the ‘tests’ category by MUC analysis, perhaps because the training documents had fewer instances of ‘tests’ than the other categories, offering less material for our learning engine to build upon.
Testing phase—results
Table 3 shows the result of testing/evaluation carried out on 322 documents. The test required only 10 min to run. It used the model created from training on the domain independent features set.
Table 3.
Results for domain independent testing run
| Category | Statistical measures | Recall | F measure | Precision |
| Test | B3 | 0.964 | 0.93 | 0.898 |
| MUC | 0.174 | 0.24 | 0.385 | |
| CEAF | 0.849 | 0.858 | 0.868 | |
| Person | B3 | 0.812 | 0.86 | 0.914 |
| MUC | 0.917 | 0.866 | 0.82 | |
| CEAF | 0.8 | 0.592 | 0.47 | |
| Problem | B3 | 0.938 | 0.939 | 0.94 |
| MUC | 0.651 | 0.629 | 0.609 | |
| CEAF | 0.877 | 0.856 | 0.836 | |
| Treatment | B3 | 0.941 | 0.917 | 0.895 |
| MUC | 0.566 | 0.62 | 0.68 | |
| CEAF | 0.846 | 0.832 | 0.819 | |
| Total | B3 | 0.951 | 0.945 | 0.94 |
| MUC | 0.76 | 0.764 | 0.768 | |
| CEAF | 0.815 | 0.818 | 0.821 | |
| Average total F measure | 0.8423 | |||
Table 4 shows the result for the test run, based on the model created from using domain independent and domain dependent features. This test run was completed in about 10 min.
Table 4.
Results for domain independent and dependent testing run
| Category | Statistical measures | Recall | F measure | Precision |
| Test | B3 | 0.963 | 0.934 | 0.907 |
| MUC | 0.191 | 0.254 | 0.379 | |
| CEAF | 0.858 | 0.866 | 0.875 | |
| Person | B3 | 0.827 | 0.86 | 0.895 |
| MUC | 0.921 | 0.879 | 0.84 | |
| CEAF | 0.808 | 0.615 | 0.496 | |
| Problem | B3 | 0.942 | 0.936 | 0.93 |
| MUC | 0.62 | 0.633 | 0.647 | |
| CEAF | 0.86 | 0.855 | 0.843 | |
| Treatment | B3 | 0.94 | 0.922 | 0.905 |
| MUC | 0.583 | 0.625 | 0.673 | |
| CEAF | 0.851 | 0.836 | 0.822 | |
| Total | B3 | 0.953 | 0.946 | 0.939 |
| MUC | 0.767 | 0.775 | 0.784 | |
| CEAF | 0.817 | 0.824 | 0.83 | |
| Average total F measure | 0.8483 | |||
Table 5 compares the results across all four runs.
Table 5.
Results comparison across the four runs
| Domain independent features only | Domain independent+dependent features | |
| Training | 0.833 | 0.8486 |
| Testing | 0.8423 | 0.8483 |
Discussion
General observations
The striking finding is that the addition of the domain dependent features, which extensively used SNOMED concepts, did not provide as much benefit in the scoring. Our assumption when approaching the task was exactly the opposite, a notion supported by the literature.13 Multiple factors appear at play in our results. The training samples were fairly extensive (for some of the categories) and the testing samples were drawn from the same corpus. Routine aliases were already captured in the training set and therefore the benefit of the SNOMED vocabulary was less powerful. In addition, our domain dependent features were not targeted at pronoun resolutions which formed the greater part of the recognition task for the challenge.
Our machine learning technique performed poorly at recognizing tests, most likely because of the fewer number of tests in the training documents to engage the learning process. We also performed relatively weakly at pronouns recognition, primarily due to lack of attention to this facet of the challenge by us, the investigators. Disambiguating pronouns are of less practical significance to a commercial entity such as ours, than collating a precise collection of problems or medications.
Tables 1, 2 and 5 also show that the machine learning algorithm did not over-fit. In fact, the results from the test run were quite comparable to the training run.
Error analysis
We looked at the errors made by the learning engine and placed them in the following general classes:
Synonymy failure—failure to recognize two terms representing the same concept. Examples here include: ‘cardiac dz’ and ‘coronary artery disease,’ ‘Transesophageal Echocardiogram’ and ‘The TEE,’ ‘SBP’ and ‘his blood pressure,’ ‘Oxycodone’ and ‘Oxycontin.’ Failure to recognize synonyms typically leads to a number of false negatives.
Temporality failure—failure to recognize the temporality of a concept. Example: ‘surveillance cultures’ versus ‘Plan to have surveillance cultures’ in the future. Although the concept is clearly the same, it is not referring to the same event.
Contextual cues missed. In one example: Patient Name: Mrs. YYY. Later the patient is referred to as ‘the patient’ and with various pronouns. In another example, ‘chronic back pain’ is falsely linked to ‘chronic pain’ and ‘pain’ where the later two references to pain refer to other types of pain understandable within the context of the note.
Our efforts to use SNOMED vocabulary were clearly targeted at addressing errors that arose from failure to recognize synonymous terms. However, that effort has not yet borne fruit. Temporal features were not implemented, which might have eliminated some of the errors. Recognizing when a concept is coreferent and when it is not remains a challenge for NLP systems.
Conclusion
The coreference challenge has focused attention on an area that has sometimes been ignored in clinical document analysis. Our machine learning approach is a promising solution to the task of automating the assembly of problem and medication lists from clinical documents. Our results also suggest that having a high-quality set of annotated training documents is the key—and domain independent features are sufficient for obtaining reasonable results. In real world deployment, however, we consider that it will be critical to employ well formulated domain specific features to provide for a more robust engine that will work across different document types and sources. Of course, problem and medication lists generated through this method will need to be manually reviewed and corrected, but the techniques explored here will capably render an excellent first draft for a human validator.
Acknowledgments
We would like to thank our M*Modal management for supporting i2b2 and our participation in this challenge.
Footnotes
Funding: The 2011 i2b2/VA challenge and the workshop are funded in part by grant number 2U54LM008748 on Informatics for Integrating Biology to the Bedside from the National Library of Medicine. This challenge and workshop are also supported by the resources and facilities of the VA Salt Lake City Health Care System with funding support from the Consortium for Healthcare Informatics Research (CHIR), VA HSR HIR 08-374 and the VA Informatics and Computing Infrastructure (VINCI), VA HSR HIR 08-204, and the National Institutes of Health, National Library of Medicine under grant number R13LM010743-01.
Competing interests: None.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data sharing statement: The i2b2 organizers are making the data used for the challenge available to research institutions.
References
- 1.Broscheit S, Poesio M, Ponzetto SP, et al. BART: a multilingual anaphora resolution system. Proceedings of the 5th International Workshop on Semantic Evaluation. PA, USA: Association for Computational Linguistics Stroudsburg, 2010:104–7 [Google Scholar]
- 2.Raghunathan K, Lee H, Rangarajan S, et al. A multi-pass sieve for coreference resolution. Conference on Empirical Methods in Natural Language Processing. PA, USA: Association for Computational Linguistics Stroudsburg, 2010:492–501 [Google Scholar]
- 3.Rahman A, Ng V. Supervised models for coreference resolution. Conference on Empirical Methods in Natural Language Processing. PA, USA: Association for Computational Linguistics Stroudsburg, 2009:968–77 [Google Scholar]
- 4.Culotta A, Wick M, Hall R, et al. First-order probablistic models for coreference resolution. Proceedings of NAACL HLT. PA, USA: Association for Computational Linguistics Stroudsburg, 2007:81–8 [Google Scholar]
- 5.McCallum A, Schultz K, Singh S. FACTORIE: probabilistic programming via imperatively defined factor graphs. In: Bengio Y, Schuurmans D, Lafferty J, et al., eds. Advances in Neural Information Processing Systems. Cambridge, MA, USA: MIT Press, 2009;22:1249–57 [Google Scholar]
- 6.Crammer K, Kulesza A, Dredze M. Adaptive regularization of weight vectors. In: Bengio Y, Schuurmans D, Lafferty J, et al., eds. Advances in Neural Information Processing Systems. Cambridge, MA, USA: MIT Press, 2009;22:414–22 [Google Scholar]
- 7.Savova GK, Masanz JJ, Ogren PV, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTakes): architecture, component evaluation and applications. J Am Med Inform Assoc 2010;17:507–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Recasens M, Hovy E. BLANC: Implementing the Rand index for coreference evaluation. Nat Lang Eng 2011;17:1–26 [Google Scholar]
- 9.Cai J, Strube M. Evaluation Metrics For End-to-End Coreference Resolution Systems. Stroudsburg, PA, USA: Association for Computational Linguistics, SIGDIAL, 2010:28–36 [Google Scholar]
- 10.Bagga A, Baldwin B. Algorithms for scoring coreference chains. The First International Conference on Language Resources and Evaluation Workshop on Linguistics Coreference. Stroudsburg, PA, USA: Association for Computational Linguistics, 1998:563–6 [Google Scholar]
- 11.Vilain M, Burger J, Aberdeen J, et al. A model-theoretic coreference scoring scheme. Proceedings of the 6th Conference on Message Understanding. Stroudsburg, PA, USA: Association for Computational Linguistics, 1995:45–52 [Google Scholar]
- 12.Luo X. On coreference resolution performance metrics. Proceedings of the conference on Human Language Technology and Empirical Methods in Natural Language Processing. Stroudsburg, PA, USA: Association for Computational Linguistics, 2005:25–32 [Google Scholar]
- 13.Tetrault JR. A corpus-based evaluation of centring and pronoun resolution. Comput Ling 2001;27:507–20 [Google Scholar]