Abstract
Objective
Relation extraction in biomedical text mining systems has largely focused on identifying clause-level relations, but increasing sophistication demands the recognition of relations at discourse level. A first step in identifying discourse relations involves the detection of discourse connectives: words or phrases used in text to express discourse relations. In this study supervised machine-learning approaches were developed and evaluated for automatically identifying discourse connectives in biomedical text.
Materials and Methods
Two supervised machine-learning models (support vector machines and conditional random fields) were explored for identifying discourse connectives in biomedical literature. In-domain supervised machine-learning classifiers were trained on the Biomedical Discourse Relation Bank, an annotated corpus of discourse relations over 24 full-text biomedical articles (∼112 000 word tokens), a subset of the GENIA corpus. Novel domain adaptation techniques were also explored to leverage the larger open-domain Penn Discourse Treebank (∼1 million word tokens). The models were evaluated using the standard evaluation metrics of precision, recall and F1 scores.
Results and Conclusion
Supervised machine-learning approaches can automatically identify discourse connectives in biomedical text, and the novel domain adaptation techniques yielded the best performance: 0.761 F1 score. A demonstration version of the fully implemented classifier BioConn is available at: http://bioconn.askhermes.org.
Keywords: Analysis, automated learning, controlled terminologies and vocabularies, discovery, display, image representation, knowledge acquisition and knowledge management, knowledge bases, knowledge representations, machine learning, natural language processing, NLP, ontologies, processing, text and data mining methods
Introduction
The desire for knowledge discovery through text mining of biomedical literature has led to a great deal of research towards the extraction and retrieval of valuable and useful information from biomedical text, through natural language processing (NLP) methods developed for recognizing entities (eg, proteins, genes, drugs, diseases, etc), facts, hypotheses, events, and relations between entities. However, with the exception of some recent work on coreference resolution,1 much of this processing has been restricted to the level of the clause, focusing on identifying entities and relations within a clause, and has ignored the importance of identifying relations expressed at the level of discourse, ie, relations expressed across clauses or sentences. In example 1, for instance, queries regarding the inhibitory effect of IL-10 could be answered more accurately when the ‘concession’ relation between the two sentences is identified, signaled by the word However. Taking the first sentence alone would otherwise lead to the false inference that the IL-10-mediated inhibitory effect is unrestricted.
Example 1:IL-10-mediated inhibition of CD4+ T-cell cytokine production is principally dependent on its inhibition of macrophage antigen-presenting cell function.1 However, this indirect inhibitory effect is thought to be restricted at the site of T-cell activation in RA… (Concession: contra-expectation)
Knowledge of such relations, called ‘discourse relations’, can be very useful in extracting various kinds of biomedical information. In this paper, we present the first investigations towards identifying discourse relations in biomedical literature. We focus on identifying ‘discourse connectives’, which are words or phrases used to indicate the presence of discourse relations, such as the word However in example 1. Following the terms and definitions of the Penn Discourse Treebank (PDTB),2 discourse relations hold between abstract objects, such as eventualities and propositions, which serve as the arguments to the relation. Each discourse relation is assumed to hold between precisely two arguments (named Arg1 and Arg2). Discourse relations are characterized in terms of several semantic (or sense) classes, including ‘contrast’, ‘conjunction’, ‘cause’, ‘condition’, and ‘instantiation’, among others. In example 2, the word but is a discourse connective that indicates the presence of a ‘contrast’ relation between the eventualities expressed by the two sentences. In all the examples in this paper, Arg2, the argument syntactically associated with or bound by the connective is underlined, while Arg1, is shown in italics. The discourse connective is in bold. The semantics (or sense) of the connective is shown in parentheses at the end of the examples.
Example 2: The phosphorylation of signal transducer and activator of transcription 3 was sustained in both blood and synovial tissue CD4+ T cells of RA, but it was not augmented by the presence of 1 ng/ml IL-10. (Contrast)
Identifying the presence of discourse relations can help in the extraction of valuable information from natural language text and also benefit many NLP applications.3–9 For example, identifying causal discourse relations will make it possible to generate repositories of ‘why’ questions from biomedical text.10 In general, question generation systems,7 as well as question-answering systems, stand to benefit greatly from recognizing discourse relations because it will allow for the generation and answering of complex questions about biomedical events and situations.
Discourse relations can also be used to benefit information extraction from clinical narratives. Unique adverse drug event information often appears in narratives of electronic health records. While most biomedical natural language processing (BioNLP) algorithms for adverse drug event extraction are based on co-occurrence of an adverse event and a drug, problems exist with such an approach, as illustrated by examples 3 and 4. In example 3, connective after has a temporal function describing the administration of atenolol. Whereas in example 4, the connective after has a causal interpretation. The ‘bradycardia’ is caused by atenolol.
Example 3: …atenolol should be continued while he is at hospital and after he is discharged. (Temporal: succession)
Example 4: Patient was noted to bradycardia as heart rate fell to low 50s after taking atenolol, (Cause: result)
Therefore, the identification of discourse relations would enable text-mining engines to discover not only entities and events, but also relations between biological or medical events, such as the temporal and causal relations, relations between facts, and relations between experimental evidence and their conclusions. Further illustration is provided in the supplementary material (available online only).
Words that function as discourse connectives in some instances may have non-discourse-related functions in others. Therefore, one cannot identify discourse connectives by simply using a list of connective expressions and applying pattern matching over the texts. For instance, the word so functions as a connective in example 5(a), expressing a result relation, while acting as an intensifier in example 5(b) with no discourse function at all. A similar example of such functional ambiguity is given for ‘briefly’ in the supplementary material (available online only).
Example 5(a): however, CsA also inhibits activation of the JNK pathway following TcR/CD3 and CD28 stimulation,11 12 and so CsA pretreatment may act to prevent early T cell activation of these pathways, thus blocking cytokine production and protecting mice from the effects of subsequent SEB exposure. (Cause: result)
Example 5(b): It is striking that ductal growth is so exquisitely focused in the end buds.
Automatic discourse parsing comprises several sub-tasks, including discourse connective detection, argument detection, discourse connective sense categorization, and discourse structure composition. The first step towards a full-fledged discourse relation detection system and parser is the detection of discourse connectives. In this study, we explore supervised machine-learning approaches to identify discourse connectives automatically in biomedical literature and compare them with simple lexical pattern matching-based approaches. The main contributions of this paper are: (1) we are the first group to identify discourse connectives in the biomedical domain; (2) we explore the use of domain-specific features in addition to the normal syntactic features used in machine learning and (3) we use domain adaptation techniques to leverage larger open-domain datasets and further improve the performance of the discourse connective identification.
Related work
A great deal of work has been performed to explore methods for discourse parsing13–15 and discourse identification in the open domain.16 17 Pitler and Nenkova18 explored supervised machine-learning approaches to identify explicit discourse connectives and disambiguate their sense in the PDTB.
In contrast, work on discourse parsing in the biomedical domain has been limited. BioNLP tasks have traditionally focused on sentence level analysis and information extraction. Studies19–22 have explored approaches to segment biomedical text into sections and topics. Szarvas et al23 created BioScope, a corpus annotated with negative and speculative keywords and their linguistic scope in biomedical text. Agarwal and Yu24 25 subsequently developed a system automatically to identify negation and hedging cues and their scope in biomedical text.
The most closely related work is the development of an annotated corpus of discourse relations called the Biomedical Discourse Relation Bank (BioDRB),26 27 and studies on the sense disambiguation of discourse connectives.26 Studies have also examined certainty,28 and future research direction in biomedical literature29 using discourse structure. Other discourse aspects have been researched in the biomedical domain, such as the annotation of co-reference relations1 11 12 30 and anaphora resolution.31
Previously, we developed a preliminary conditional random fields (CRF)-based classifier32 to identify discourse connectives using the PDTB and BioDRB corpora. The classifier achieved an F1 score of 0.55 for identifying connectives. In this study, using the same corpora we significantly expand the previous work by exploring new features including syntactic and domain-specific semantic features and novel domain adaptation techniques.
Materials and methods
Discourse relations corpora
The two annotated corpora we used in this study are the PDTB 2.02 (http://www.seas.upenn.edu/∼pdtb) and the BioDRB26 (http://biodiscourserelation.org/). The PDTB annotations are done over 2159 texts (over 1 million word tokens) from the Wall Street Journal (WSJ) articles collection of the Penn Treebank.33 The Penn Treebank is an open-domain large-scale annotated corpus of syntactic phrase structure, which has been very widely used by researchers for data-driven parser development. The source WSJ articles have also been annotated for other kinds of linguistic information, including semantic roles34 and co-reference,35 among others. The PDTB was developed to enrich the WSJ annotations further at the level of discourse and provides annotations of explicit and implicit discourse relations their arguments, their senses, and the attributions of discourse relations and each of their two arguments.
The BioDRB is a corpus of discourse relations annotated over 24 full-text articles (∼112 000 word tokens) taken from the GENIA corpus.36 The GENIA articles were selected by querying the PubMed for ‘blood cells’ and ‘transcription factors‘ and were considered representative of scientific articles in this domain by the GENIA research group.37 Discourse relation annotations of the BioDRB largely follow the PDTB guidelines and, like the PDTB, include annotations of explicit and implicit discourse relations, their arguments and their semantics. Unlike the PDTB, however, the BioDRB does not currently annotate attribution. An overall agreement of 85% was reported among annotators of BioDRB.27
The PDTB and BioDRB contain annotations for 18 459 and 2637 total explicit connectives, or 18.5 and 26.4 discourse connectives per 1000 tokens, respectively. After connective stemming (eg, ‘three days after’ stemmed to ‘after’) there are 100 unique explicit discourse connectives in the PDTB and 123 in the BioDRB.
Our analysis shows that 56% of the explicit discourse connectives in the BioDRB occur in the PDTB, including common connectives like and, also, so, and however. Thirty-three per cent of the connectives in BioDRB comprise the class of ‘subordinators’ like followed by, in order to, and due to, which are not annotated as connectives in the PDTB corpus (connectives in the PDTB are defined as belonging to three grammatical classes: subordinating conjunctions, coordinating conjunctions, and discourse adverbials). The final 11% of the connectives in the BioDRB consist of lexical items that do not occur in the PDTB texts and were therefore not classified as connectives. Examples of these include: In outline, As a consequence, and In summary.
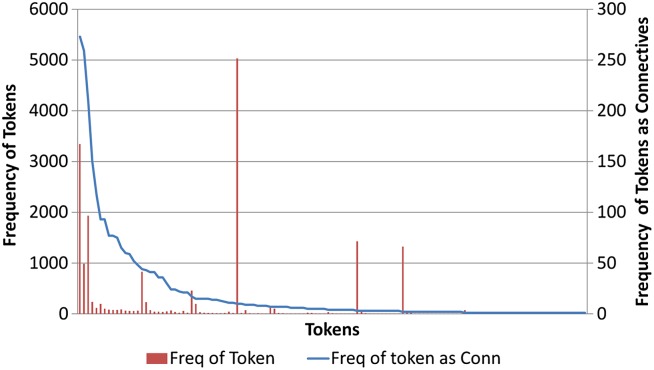
Figure 1 shows the frequency of the tokens in the BioDRB corpus and their frequency as connectives. From our analysis of the BioDRB data we found that 76% of the connectives were functionally ambiguous, in that they also appeared in the text not as part of a discourse relation. We also found that 43.5% of the connectives occurred only once as a connective in the entire corpus.
Figure 1.
Frequency of the tokens in the Biomedical Discourse Relation Bank (BioDRB) corpus and their frequency of as connectives.
As the BioDRB corpus is relatively small, we leveraged the much larger PDTB corpus in order to help deal with data sparseness. Although the PDTB is not from the biomedical domain, we expect the addition of more data to boost the performance of the classifiers.
Domain adaptation approaches
In order to compensate for the relatively small size of the BioDRB (∼112K tokens), and to leverage the much larger open-domain PDTB (∼1 million tokens), we explored domain adaptation approaches to build models trained on both corpora. In domain adaptation, the larger corpus is referred to as the source domain (PDTB in this case) and the smaller one as the target domain (BioDRB in this case). In this study, we explored three supervised domain adaptation techniques:
Instance weighting combines the data from both corpora, but assigns different weights to them during the training phase. The weights are usually inversely proportional to the size of the corpus to compensate for the larger number of training examples and to avoid over fitting to the source domain. The classifier was then trained using this weighted training dataset.
Instance pruning actively removes misleading training instances. For example, if for training example d, we find different labels for d in the source and target domains, then we remove all such instances of d from the source domain training data. To apply instance pruning, we first trained a classifier on the target domain data (BioDRB), and then applied this classifier to the source domain data (PDTB). All the instances in the source domain that were incorrectly classified are pruned from the source training set (∼1% of data was pruned). The final classifier was trained using this pruned source domain dataset.
Feature augmentation is a method in which additional meta-features are added to indicate whether a specific feature came from the source or target dataset. For each training example, the feature vector is expanded to contain not only the original features, but also indicators representing the domain from which each feature was taken. This makes it possible for us to represent the effect of individual features in the source and target domain, respectively, and for the machine-learning algorithms to distinguish between features important to the respective domains. The classifier is then trained on the combined dataset with the additional features. Consider the example, ‘…industry is regulated by commodity futures …’ in the source domain and ‘…resulted in a small overlap in regulated mRNAs at 4 …’ in the target domain. The word ‘regulated’ is used as a verb in source domain where as it is used as an adjective in target domain. In the feature vector for the word ‘regulated’, the source-specific indicator linked to ‘verb’ and the target-specific indicator linked to ‘adjective’ is set.
Supervised machine learning
The two supervised machine-learning approaches we explored were CRF and support vector machines (SVM). Our aim in using these two approaches was to explore whether it was more beneficial to cast the problem of identifying discourse connectives as a sequence-labeling task (with CRF), or as a classification task (with SVM).
CRF are a probabilistic modeling framework38 commonly used for sequence labeling problems. In our experiments, we treated documents as a sequence of words, and the classifier determined whether or not each word in the sequence was part of a connective. We built the CRF classifiers using the ABNER toolkit.39
To test connective identification as a classification task, we built an SVM classifier using Weka. SVM are a well-known statistical machine-learning algorithm and have shown very good performance in many classification tasks.40 41 We used the SVM to classify each word in a sentence as either a discourse connective token or a non-discourse token.
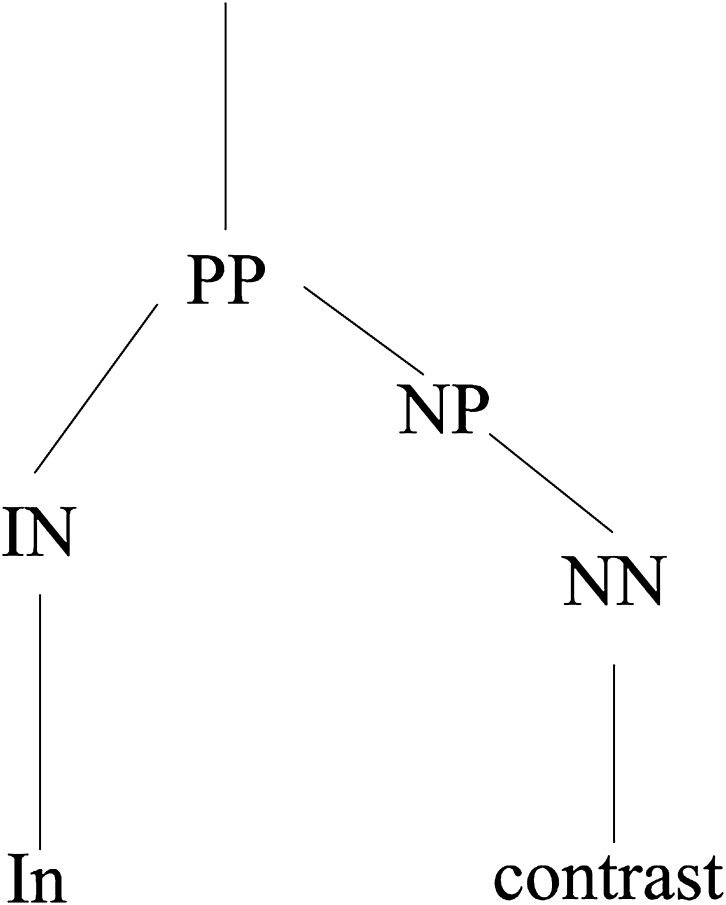
In addition to the default ABNER features, we evaluated syntactic and domain-specific learning features. We explored the syntactic features that have been shown to be important in previous studies,13 16 18 namely part-of-speech (POS) of the token, the label of the immediate parent of the token's POS in a parse tree, and the POS of the left sibling (the token to the left of the current word inside the innermost constituent). Figure 2 shows a sample constituency parse tree, in which the POS tag is always the immediate parent label of a word token at the leaf of the tree. In this example, the POS tag of the word ‘in’ is IN and the POS tag of the word ‘contrast’ is NN. Furthermore, the immediate parent label of the POS for ‘in’ is PP, and of the POS for ‘contrast’ is NP. The left sibling value is NONE assuming it is the start of the sentence. The syntactic features were obtained using the Charniak–Johnson parser trained in the biomedical domain. The parser was evaluated to have the best performance when tested on the GENIA corpus.42 We also explored domain-specific features by applying BANNER43 gene tagger and the LINNAEUS44 species tagger to identify gene and species named entities as well as Metamap to map text elements to the unified medical language system (UMLS)45 semantic types. Mapping free text to concepts or named entities (eg, gene and species) represents a case of back-off smoothing that contributes to improved performance.
Figure 2.
Sample parse tree.
Experiments and systems
We developed several systems to evaluate: the complexity of the task; the impact of different syntactic and domain-specific features; and the impact of different domain adaptation models. As there is a total of 24 articles in the BioDRB, to simplify the task, we used 12-fold cross-validation rather than the common 10-fold so that an article (not a segment of it) was assigned as either a training or a testing article.
Evaluation of in-domain systems
In this experiment, we develop two heuristic baseline systems and compare their performance with our in-domain CRF and SVM-based classifiers.
Baseline systems
The first baseline system, BaseLex, uses a lexical heuristic, creating a lexicon by extracting the connectives annotated in the BioDRB corpus, and then tagging all instances of these words in the text as connectives.
The second baseline system, BaseLexPunct, is a combination of the lexical heuristic from BaseLex and additional heuristics related to observed punctuation patterns associated with connectives. In particular, we observed that connectives were often either preceded or followed by a comma, or appeared as the first word in the sentence. The system first identifies all connective terms from the lexicon in the text, and then filters out the instances that do not match with the manually created punctuation heuristic.
Supervised machine-learning systems
The two baseline systems were compared against our supervised machine-learning systems: In-domainSVM, the SVM classifier and the In-domainCRF, the CRF-based classifier. Both the classifiers were trained and tested on BioDRB, using syntactic features (see Supervised machine learning section).
Measuring the impact of semantic features
In this experiment, we evaluated the impact of different types of features; in particular we wished to determine the relative performance of syntactic versus domain-specific features. For this reason we built variants of the best performing classifier from the first experiment using different features, as follows: The UMLS classifier exclusively uses UMLS features extracted using Metamap; the GeneSpecies classifier exclusively uses the gene and species categories extracted with BANNER and LINNAEUS as features; we then evaluate both of these classifiers after adding the features used in the Evaluation of in-domain systems experiment, which we call UMLS+ and GeneSpecies+, respectively. Finally, we combined all of the features into a classifier, which we will call Semantics+.
Systems to measure impact of domain adaptation
In this experiment, we evaluated the impact of the domain-adaptation approaches described in the Domain adaptation approaches section, for which we compared several classifiers with and without domain adaptation. We used the classifier type and feature sets found to have the best performance in our previous experiments.
Baseline systems
The following systems did not incorporate domain adaptation, and were used as the baseline: the In-domain classifier, trained exclusively on the target domain; the Cross-domain classifier, trained on the source domain; and the Unweighted classifier, trained on the merged source and target domains.
Domain adaptation systems
To test the various domain adaptation techniques, we developed three classifiers: the Instance Weighting classifier, in which source domain data were given a weight 0.1 times that of target domain data (the value of 0.1 was used as an approximation of the relative sizes of the datasets); the Instance Pruning classifier and the FeatAugment classifier, which were trained using the instance weighting, instance pruning and feature augmentation approaches, respectively.
Combined domain adaptation systems
The following systems incorporated combinations of the domain adaptation techniques: the Weighted-Pruning classifier, trained using a combination of instance weighting and instance pruning approaches; the Weighted-FeatAugment classifier, trained using a combination of instance weighting and feature augmentation approaches; the Hybrid classifier, trained using a combination of instance pruning and feature augmentation approaches; and finally, the Weighted-Hybrid classifier, trained using the combination of all three approaches. For the combined methods using instance weighting, the source weight was changed from 0.1 to 0.5 to reflect the effects of the other two adaptation methods.
Evaluation metrics
All the classifiers (including the baseline classifiers) were run at the token level, ie, the word level, marking each token in the evaluation corpus as either connective or not. They were evaluated with 12-fold cross validation, except for the Cross-Domain classifier, which was trained on the source domain and evaluated on the target domain. For systems using the combination of the BioDRB and the PDTB, the training for each fold was always done on the entire PDTB with 11/12s of the BioDRB, and the evaluation done on the remaining BioDRB data. The standard evaluation metrics of recall, precision, and F1 scores were used to measure the performance of all systems.
Results
Table 1 shows the performance evaluation of the in-domain classifiers relative to the baseline systems, as described in the Evaluation of in-domain systems and Measuring the impact of semantic features sections. The heuristic baseline systems BaseLex and BaseLexPunct had an F1 score of 0.33 and 0.272, respectively. The supervised machine-learning classifiers In-domainSVM and In-domainCRF had an F1 score of 0.657 and 0.757, respectively. The supervised machine-learning methods clearly outperform the baseline methods. The CRF-based system had the best performance overall, and was therefore chosen as the system to be adapted for subsequent experiments.
Table 1.
Task complexity: performance (average±Std) of different classifiers for the task complexity measurement. Effect of learning features: performance (average±Std) of in-domain CRF classifiers trained with different learning features
| Classifier type | Overall performance (F1 score) (Precision, Recall) discourse connectives | Overall performance (F1 score) (Precision, Recall) non-discourse connectives |
| Task complexity | ||
| BaseLex | 0.330±0.044 (0.198±0.032, 1.000±0.000) | 0.948±0.005 (1.000±0.000, 0.901±0.010) |
| BaseLexPunct | 0.272±0.058 (0.165±0.041, 0.790±0.072) | 0.946±0.006 (0.994±0.001, 0.901±0.010) |
| In-domainSVM | 0.657±0.061 (0.773±0.066, 0.575±0.07) | 0.945±0.002 (0.998±0.001, 0.897±0.004) |
| In-domainCRF | 0.757±0.059 (0.817±0.058, 0.711±0.086) | 0.994±0.001 (0.992±0.002, 0.996±0.001) |
| Effect of learning features | ||
| UMLS (UMLS Semantic features) | 0.681±0.063 (0.786±0.050, 0.606±0.086) | 0.993±0.001 (0.990±0.003, 0.996±0.001) |
| Gene-Species (Gene + Species features) | 0.686±0.058 (0.797±0.050, 0.608±0.082) | 0.993±0.001 (0.990±0.002, 0.996±0.001) |
| UMLS+ (Syntactic + UMLS Semantic features) | 0.744±0.061 (0.806±0.051, 0.696±0.087) | 0.992±0.001 (0.986±0.003, 0.997±0.001) |
| Gene-Species+ (Syntactic + Gene + Species features) | 0.753±0.052 (0.814±0.045, 0.703±0.075) | 0.994±0.001 (0.992±0.002, 0.996±0.001) |
| In-domain (Syntactic features) | 0.757±0.059 (0.817±0.058, 0.711±0.086) | 0.994±0.001 (0.992±0.002, 0.996±0.001) |
| Semantics+ (All features) | 0.747±0.059 (0.810±0.048, 0.698±0.086) | 0.994±0.001 (0.992±0.002, 0.996±0.001) |
Values in bold indicate the performance of the classifier that had the best performance.
It is clear from the data in table 1 that the addition of domain-specific semantic features did not help improve classifier performance. The In-domain classifier, trained using only the syntactic features, had the best performance, F1 score 0.757, followed by the Gene-Species classifier, F1 0.753. While the difference between the two scores is not statistically significant, the additional features were clearly not providing any benefit. Therefore, in subsequent experiments with domain adaptation, we used only syntactic features to train classifiers. We can also see from table 1 that the identification of non-discourse connectives had good performance in all systems.
Table 2 shows the performance of all CRF classifiers with the impact of different domain adaptation models, as described in the Systems to measure impact of domain adaptation section. Among the simple domain adaptation techniques, the Instance Weighting classifier had the best performance; with an F1 score of 0.730, compared with other individual domain adaptation-based classifiers Instance Pruning and FeatAugment, for which F1 scores were 0.637 and 0.677, respectively.
Table 2.
Performance (average±Std) of different classifiers based on CRF for identifying the discourse connectives using domain adaptation techniques for various categories
| Classifier type | Overall performance (F1 score) (Precision, Recall) discourse connectives | Overall performance (F1 score) (Precision, Recall) non-discourse conn |
| Cross-domain | 0.592±0.066 (0.834±0.061, 0.461±0.065) | 0.992±0.001 (0.986±0.002, 0.998±0.001) |
| UnWeighted | 0.677±0.071 (0.810±0.061, 0.585±0.085) | 0.993±0.001 (0.989±0.002, 0.997±0.001) |
| In-domain | 0.757±0.059 (0.816±0.058, 0.711±0.086) | 0.994±0.001 (0.992±0.002, 0.996±0.001) |
| Weighted | 0.730±0.053 (0.805±0.052, 0.671±0.075) | 0.993±0.001 (0.991±0.002, 0.996±0.001) |
| Pruning | 0.637±0.076 (0.844±0.070, 0.514±0.079) | 0.993±0.001 (0.987±0.002, 0.998±0.001) |
| FeatAugment | 0.695±0.056 (0.760±0.048, 0.647±0.090) | 0.993±0.001 (0.990±0.002, 0.996±0.001) |
| Weighted-Pruning | 0.753±0.057 (0.816±0.051, 0.703±0.083) | 0.994±0.001 (0.992±0.002, 0.996±0.001) |
| Weighted-FeatAugment | 0.757±0.045 (0.809±0.050, 0.716±0.068) | 0.994±0.001 (0.992±0.002, 0.996±0.001) |
| Hybrid | 0.761±0.051 (0.813±0.041, 0.719±0.079) | 0.994±0.001 (0.993±0.002, 0.996±0.001) |
| Weighted-Hybrid | 0.757±0.050 (0.807±0.047, 0.717±0.076) | 0.994±0.001 (0.992±0.002, 0.996±0.001) |
Values in bold indicate the performance of the classifier that had the best performance.
CRF, conditional random fields.
None of the methods, however, performed better than the baseline In-domainCRF classifier. Some classifiers increased recall (Instance Weighting) while others increased precision (Instance Pruning). This indicates that systems combining multiple domain adaptation techniques may be more robust, and therefore produce better F1 scores. Results of these combinations are shown in the last four rows. The Hybrid classifier had the best performance among all classifiers, with an F1 score of 0.761. All the classifiers shown in table 2 were statistically significant (t test, p<0.005) when compared with the Cross-domain classifier.
The performance of classifiers trained using simple domain adaptation methods were statistically significant (t test, p<0.005) when compared with the classifiers trained using combined domain adaptation methods. However, the classifiers trained using combined domain adaptation techniques did not produce statistically significant differences in their results.
Error analysis
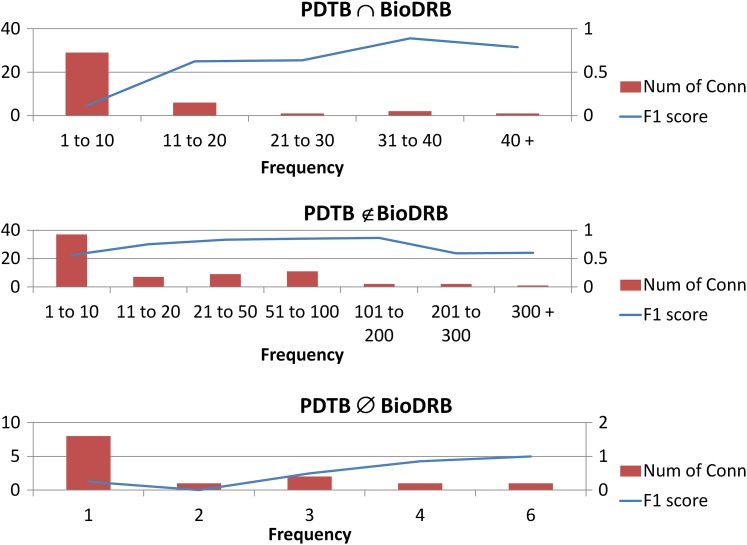
For error analysis, we focused on analyzing the CRF classifiers trained on syntactic features, because they showed the best performance. Error analysis revealed that most of the errors were due to the common problem of data sparseness. In particular, most of the false negatives did not appear in the training set or appeared only once as a connective in the entire corpus. Therefore, we assessed classifier performance while taking these distributions into account. We first categorized the connectives based on their occurrence distributions in the PDTB and BioDRB corpora. There were three categories: connectives that were present and annotated in both corpora (BioDRB ∩ PDTB), present in both but annotated only in BioDRB (BioDRB ∉ PDTB), and finally, present and annotated only in BioDRB (BioDRB ∅ PDTB). We then investigated the performance of each domain-adapted classifier on each of the categories for tokens that appear at least once as connectives in the corpus. Table 3 shows the percentage of connectives identified by the classifier in each category, the classifier's performance in that category for identifying the token as a discourse connective and non-discourse connective. We observed that the weighting technique improved the performance across all three categories.
Table 3.
Performance (F1 score) of the classifiers for identifying the discourse connectives
| BioDRB ∩ PDTB | BioDRB ∉ PDTB | BioDRB ∅ PDTB | |||||||
| % Of conns | Performance as DCONN | Performance as non-DCONN | % Of conns | Performance as DCONN | Performance as Non DCONN | % Of conns | Performance as DCONN | Performance as non-DCONN | |
| Cross-domain | 96.7% | 0.62 | 0.92 | 3.3% | 0.03 | 0.97 | 0% | 0 | 0.86 |
| UnWeighted | 84.3% | 0.70 | 0.93 | 10.5% | 0.21 | 0.98 | 5.2% | 0.55 | 0.91 |
| In-domain | 74% | 0.78 | 0.94 | 19.8% | 0.65 | 0.98 | 6.2% | 0.63 | 0.91 |
| Weighted | 75.7% | 0.75 | 0.94 | 17% | 0.51 | 0.98 | 7.3% | 0.7 | 0.92 |
| Pruning | 93.4% | 0.67 | 0.93 | 3.3% | 0.08 | 0.97 | 3.3% | 0.14 | 0.87 |
| FeatAugment | 72.8% | 0.70 | 0.93 | 21% | 0.58 | 0.98 | 6.2% | 0.5 | 0.9 |
| Weighted-Pruning | 75.3% | 0.77 | 0.94 | 18.5% | 0.60 | 0.98 | 6.2% | 0.63 | 0.91 |
| Weighted-FeatAugment | 72.6% | 0.77 | 0.94 | 20.2% | 0.67 | 0.98 | 7.2% | 0.7 | 0.92 |
| Hybrid | 74.4% | 0.78 | 0.94 | 19.5% | 0.66 | 0.98 | 6.1% | 0.67 | 0.92 |
| Weighted-Hybrid | 73.8% | 0.78 | 0.94 | 20.2% | 0.65 | 0.98 | 6% | 0.67 | 0.92 |
BioDRB, Biomedical Discourse Relation Bank; PDTB, Penn Discourse Treebank.
The impact of the frequency of the connectives on the performance of the classifier was analyzed. Figure 3 shows the graph of the number of connectives and the performance of the top-performing Hybrid classifier against the frequency of connectives in the BioDRB.
Figure 3.
The graph of performance of Hybrid classifier over different distributions of the connectives. BioDRB, Biomedical Discourse Relation Bank; PDTB, Penn Discourse Treebank.
In general, as the frequency of the connectives increased, the performance of the classifier for identifying those connectives increased. This is to be expected, as increased training data resulted in improved classification. The decrease in performance for very frequent connectives can be explained by a small number of very frequent but very ambiguous connectives.
Table 4 below shows the five most common connective forms, the likelihood of each form occurring as a connective, and the F1 scores of the classifiers on these connectives. The Hybrid and In-domain classifiers performed better for frequent connectives (>100 occurrences as connectives). The connective and had an F1 score of approximately 0.7±0.04 for all the classifiers except for the FeatAugment classifier. For the connectives by, to, and after, the table shows that as domain adaptation techniques were applied, the performance increased over Cross-Domain and Unweighted classifiers.
Table 4.
The most common connectives in BioDRB and their F1 scores on the classifiers
| Classifiers | And (8.1%) | By (26.1%) | To (10.8%) | After (52.7%) | However (100%) |
| Cross-domain | 0.72 | 0.03 | 0 | 0.06 | 0.98 |
| UnWeighted | 0.73 | 0.3 | 0.07 | 0.67 | 1 |
| In-domain | 0.7 | 0.64 | 0.66 | 0.74 | 1 |
| Weighted | 0.7 | 0.52 | 0.53 | 0.72 | 1 |
| Pruning | 0.74 | 0.04 | 0 | 0.5 | 0.99 |
| FeatAugment | 0.26 | 0.55 | 0.58 | 0.65 | 0.99 |
| Weighted-Pruning | 0.74 | 0.57 | 0.6 | 0.73 | 1 |
| Weighted-FeatAugment | 0.67 | 0.59 | 0.67 | 0.72 | 1 |
| Hybrid | 0.67 | 0.64 | 0.67 | 0.72 | 1 |
| Weighted-Hybrid | 0.67 | 0.62 | 0.66 | 0.71 | 1 |
Values in bold indicate the performance of the classifier that had the best performance. BioDRB, Biomedical Discourse Relation Bank.
Our results show that a significant percentage of errors was introduced by two of the most frequent connectives, by and to, which were annotated in the BioDRB. We assessed the performance of all the classifiers from the Systems to measure impact of domain adaptation section after removing the connectives by and to. The F1 score of the Hybrid classifier increased from 0.761 to 0.792, which was statistically significant (t test, p<0.001) (see supplementary material, available online only).
Examples
In this section we manually examined the set of classified instances to evaluate the classifier that had the poorest performance (Cross-domain) and the classifier that had the best performance (Hybrid).
Example 6: One day after injection, the swelling of the ears was determined with a gauge (Hahn & Kolb, Stuttgart, Germany). (Temporal: succession)
Example 7: In view of the fact that NF-κB was also activated by anti-CD3/anti-CD28, IL-15 or mitogens in our experiments, it is most likely that the NF-κB pathway is also actively involved in the induction of IL-17 in RA PBMC. (Cause: justification)
Examples 6 and 7 show instances in which both the Cross-domain and Hybrid classifiers failed to identify the connectives. The connectives One day after and In view of the fact that appear only once in the entire BioDRB corpus and do not occur at all in the PDTB corpus. As the classifiers encounter these connectives for the first time during testing, they fail to recognize them as discourse connectives. Example 6 suggests that collecting an exhaustive list of discourse connectives will not be feasible because any number could be inserted into the expression One day after.
Example 8: In order to explain this differential efficacy, several parameters were analyzed. (Purpose: goal)
Example 9: Due to the high level of sensitivity of nested RT-PCR, even low levels of illegitimate transcription in PBMNC can cause false-positive results.2–5 (Cause: reason)
Examples 8 and 9 show instances that were correctly identified by the Hybrid classifier, but were incorrectly classified by the Cross-domain classifier. Both in order to and due to are subordinators that were not annotated as connectives in the PDTB corpus, but were annotated as connectives in the BioDRB corpus. As the Hybrid classifier is trained for the biomedical domain using BioDRB, it identified them as connectives; however, the Cross-domain classifier failed to identify them as connectives as its training set did not contain such instances. In fact, the only connective in the BioDRB ∉ PDTB class that Cross-domain correctly classifies is as an example, which shares words with common connectives in PDTB.
Example 10: We considered this to be an appropriate positive control, as any cell that is detected using the immunobeads should express the EpCAM gene. Tests of the single tumor cell and 100 PBMNC aliquots with EpCAM showed that it was also expressed to a sufficient level to enable detection of the tumor cell in 31/35 (89%) cases after 45 cycles of PCR amplification. (Conjunction)
Example 11: The accelerating effect of the mAb RIB5/2 was reproduced in two additional treatment experiments, and this effect was observed despite a variable onset of AA in the PBS-treated animals (day 9 to 11). (Conjunction)
Examples 10 and 11 show instances that were correctly identified by the Cross-domain classifier, but incorrectly classified by the Hybrid classifier. The connectives also and and occur in both the PDTB and BioDRB corpora. Table 4 shows that the connective and had a better F1 score for the Cross-domain and In-domain classifiers compared with the Hybrid classifier. In addition, the Hybrid classifier incorporates feature augmentation, whose difficulty classifying and is clearly illustrated in table 4.
Discussion
Automatic identification of discourse connectives is a challenging task. We found 76% of connectives to be ambiguous. As such, it is not surprising that using simple lexical features based on a connective-matching system did not perform well (0.33 F1 score as shown in table 1). Our results show that the supervised machine-learning approaches significantly outperformed the simpler pattern-matching approaches, yielding a maximum 0.757 F1 score.
We explored two different machine-learning models: SVM and CRF. We found that the CRF model outperformed the SVM model, yielding 0.757 F1 score, 10% higher than that of the SVM model. Note that the performance of both systems was much lower than in the open domain (0.94 F1 score). For comparison, we trained and tested CRF models on the PDTB with the published feature set.18 The classifier yielded similar results (0.937 F1 score), which demonstrated that our models are state of the art.
Our results have shown that in-domain classifiers outperformed cross-domain classifiers. While the CRF-based in-domain classifier achieved the highest performance of 0.757 F1 score, the best cross-domain classifier yielded only 0.592 F1 score. The results demonstrate that the biomedical domain needs domain-specific models for discourse connective identification. As the PDTB is not taken from the biomedical domain and has different linguistic characteristics, the addition of additional training data from the PDTB does not boost classifier performance.
We explored different learning features. Similar to previous open-domain work,18 we found that syntactic features are important. In contrast, adding domain-specific semantic features (eg, features based on UMLS) did not improve the performance. We speculate that the additional features may have introduced noise that is responsible for decreased performance.
Previous work has demonstrated that domain-adaption approaches can significantly improve the performance of tasks such as semantic role labeling.46 In contrast, our experiments show that different domain adaptation methods have complementary effects on performance and can be combined for further improvement. Our new domain adaptation model Hybrid, which is a CRF model trained with a combination of instance pruning and feature augmentation domain adaptation techniques, outperformed all other models achieving and F1 score of 0.761. The Hybrid classifier used the advantages of both the instance pruning (improved precision) and feature augmentation (improved recall) approaches thus increasing the overall performance.
Data sparseness is a very common problem in statistical NLP. In our study 43.5% of the connective types appeared only once in the entire corpus as connectives. However, our results show that removal of these singleton connectives did not drastically affect system performance. This may be explained by the fact that the singleton connectives accounted for only a small portion (3%) of all discourse connective instances. This suggests that future work should focus on identifying improved features for disambiguating commonly occurring and highly ambiguous (such as by and to) connectives.
Conclusion and future work
We have presented a method to identify discourse connectives automatically in biomedical text. This task is difficult and poses many challenges. The Hybrid classifier based on CRF with a combination of instance pruning and feature augmentation domain adaptation techniques had the best performance (F1 score 0.761) in the biomedical domain, while performance in open domain is still better (F1 score 0.93). This paper explored various supervised machine-learning-based algorithms for automatically identifying explicit discourse connectives and evaluated different domain adaptation techniques to adapt models trained on the PDTB to the biomedical domain with various novel features. Although performance of the Hybrid classifier is not statistically more significant than the In-domain classifier, leveraging the large corpus from another domain makes the classifier trained for biomedical domain more robust when the data are sparse. Future work will explore features to disambiguate the commonly occurring and confounding connectives like by and to. Later, we will extend this work to identify the arguments of explicit discourse connective and perform connective sense categorization, the next step towards developing a discourse parser. We will also explore techniques to identify the presence of implicit discourse relations in the text.
Supplementary Material
Footnotes
Contributors: HY and BPR conceived and designed the experiments. BPR performed the experiments. BPR, HY and TM analyzed the data. BPR, HY, RP, TM and BH wrote the paper.
Funding: Hong Yu received support from the National Library of Medicine, grant number 1R01GM095476. Any opinions, findings, or recommendations are those of the authors and do not necessarily reflect NIH's views.
Competing interests: None.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Zheng J, Chapman WW, Crowley RS, et al. Coreference resolution: a review of general methodologies and applications in the clinical domain. J Biomed Inform 2011;44:1113–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prasad R, Dinesh N, Lee A, et al. The penn discourse treebank 2.0. Proceedings of the 6th International Conference on Language Resources and Evaluation (LREC 2008). Marrakech, Morocco, 2008:2961–8 Organised by ELRA, the European Language Resources Association, with the collaboration of major international institutions and organisations. [Google Scholar]
- 3.Marcu D. Improving summarization through rhetorical parsing tuning. In: The 6th Workshop on Very Large Corpora; Montreal, Canada. New Brunswick, NY: The Association for Computational Linguistics SIGDAT, 1998:206–15 [Google Scholar]
- 4.Hovy EH. Automated discourse generation using discourse structure relations. In: Artificial Intelligence, Volume 63. 1993:341–85 [Google Scholar]
- 5.Hernault H, Piwek P, Prendinger H, et al. Generating dialogues for virtual agents using nested textual coherence relations. Proceedings of the 8th International Conference on Intelligent Virtual Agents; Tokyo, Japan. Tokyo, Japan: International Information Science Foundation, 2008:139–45 [Google Scholar]
- 6.McDonald R, Hannan K, Neylon T, et al. Structured models for fine-to-coarse sentiment analysis. Annual Meeting-Association For Computational Linguistics, Volume 45; Prague, Czech Republic. New Brunswick, NY: The Association for Computational Linguistics, 2007:432 [Google Scholar]
- 7.Mannem P, Prasad R, Joshi A. Question generation from paragraphs at UPenn: QGSTEC system description. Proceedings of QG2010: The Third Workshop on Question Generation. Pittsburgh, Pennsylvania, USA, 2010:84–91 [Google Scholar]
- 8.MacCartney B, Manning CD. Natural logic for textual inference. Proceedings of the ACL-PASCAL Workshop on Textual Entailment and Paraphrasing. Prague, Czech Republic, 2007:193–200 [Google Scholar]
- 9.Mani I, Verhagen M, Wellner B, et al. Machine learning of temporal relations. Proceedings of the 21st International Conference on Computational Linguistics and the 44th annual meeting of the Association for Computational Linguistics; Sydney, Australia. New Brunswick, NY: The Association for Computational Linguistics, 2006:753–60 [Google Scholar]
- 10.Prasad R, Joshi A. A discourse-based approach to generating why-questions from texts. Proceedings of the Workshop on the Question Generation Shared Task and Evaluation Challenge. Arlington, VA: National Science Foundation, 2008 [Google Scholar]
- 11.Coden A, Savova G, Sominsky I, et al. Automatically extracting cancer disease characteristics from pathology reports into a Disease Knowledge Representation Model. J Biomed Inform 2009;42:937–49 [DOI] [PubMed] [Google Scholar]
- 12.Roberts A, Gaizauskas R, Hepple M, et al. Building a semantically annotated corpus of clinical texts. J Biomed Inform 2009;42:950–66 [DOI] [PubMed] [Google Scholar]
- 13.Soricut R, Marcu D. Sentence level discourse parsing using syntactic and lexical information. Proceedings of the 2003 Conference of the North American Chapter of the Association for Computational Linguistics on Human Language Technology-Volume 1. 2003:149–56 [Google Scholar]
- 14.Duverle DA, Prendinger H. A novel discourse parser based on support vector machine classification. Proceedings of the Joint Conference of the 47th Annual Meeting of the ACL and the 4th International Joint Conference on Natural Language Processing of the AFNLP: Volume 2; Singapore. New Brunswick, NY: The Association for Computational Linguistics, 2009:665–73 [Google Scholar]
- 15.Subba R, Di Eugenio B. An effective discourse parser that uses rich linguistic information. Proceedings of Human Language Technologies: The 2009 Annual Conference of the North American Chapter of the Association for Computational Linguistics; Singapore. New Brunswick, NY: The Association for Computational Linguistics, 2009:566–74 [Google Scholar]
- 16.Wellner B, Pustejovsky J. Automatically identifying the arguments of discourse connectives. Proceedings of EMNLP-CoNLL; Prague, Czech Republic. New Brunswick, NY: The Association for Computational Linguistics SIGDAT and The Association for Computational Linguistics SIGNLL, 2007:92–101 [Google Scholar]
- 17.Elwell R, Baldridge J. Discourse connective argument identification with connective specific rankers. The IEEE International Conference on Semantic Computing. Santa Clara, CA, USA, 2008:198–205 [Google Scholar]
- 18.Pitler E, Nenkova A. Using syntax to disambiguate explicit discourse connectives in text. Proceedings of the ACL-IJCNLP 2009 Conference Short Papers; Singapore. New Brunswick, NY: The Association for Computational Linguistics, 2009:13–16 [Google Scholar]
- 19.Light M, Qiu XY, Srinivasan P. The language of bioscience: facts, speculations, and statements in between. Proceedings of BioLink 2004 Workshop on Linking Biological Literature, Ontologies and Databases: Tools for Users. Boston, Massachusetts, 2004:17–24 [Google Scholar]
- 20.Mullen T, Mizuta Y, Collier N. A baseline feature set for learning rhetorical zones using full articles in the biomedical domain. In: ACM SIGKDD Explorations Newsletter, Volume 7. New York, NY, USA: ACM, 2005:52–8 [Google Scholar]
- 21.Teufel S, Siddharthan A, Batchelor C. Towards discipline-independent argumentative zoning: evidence from chemistry and computational linguistics. Proceedings of the 2009 Conference on Empirical Methods in Natural Language Processing: Volume 3; Singapore. New Brunswick, NY: The Association for Computational Linguistics SIGDAT, 2009:1493–502 [Google Scholar]
- 22.Biber D, Jones JK. Merging corpus linguistic and discourse analytic research goals: discourse units in biology research articles. Corpus Linguistics and Linguistic Theory 2005;1:151–82 [Google Scholar]
- 23.Szarvas G, Vincze V, Farkas R, et al. The BioScope corpus: annotation for negation, uncertainty and their scope in biomedical texts. Proceedings of the Workshop on Current Trends in Biomedical Natural Language Processing; Columbus, Ohio. New Brunswick, NY: The Association for Computational Linguistics, 2008:38–45 [Google Scholar]
- 24.Agarwal S, Yu H. Biomedical negation scope detection with conditional random fields. J Am Med Inform Assoc 2010;17:696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Agarwal S, Yu H. Detecting hedge cues and their scope in biomedical literature with conditional random fields. J Biomed Inform 2010;43:953–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prasad R, McRoy S, Frid N, et al. The biomedical discourse relation bank. BMC Bioinformatics 2011;12:188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu H, Frid N, McRoy S, et al. A pilot annotation to investigate discourse connectivity in biomedical text. Proceedings of the Workshop on Current Trends in Biomedical Natural Language Processing; Columbus, Ohio. New Brunswick, NY: The Association for Computational Linguistics, 2008:92–3 [Google Scholar]
- 28.Rizomilioti V. Exploring epistemic modality in academic discourse using corpora. Information technology in languages for specific purposes 2006;7:53–71 [Google Scholar]
- 29.Lisacek F, Chichester C, Kaplan A, et al. Discovering paradigm shift patterns in biomedical abstracts: application to neurodegenerative diseases. In: First International Symposium on Semantic Mining in Biomedicine. Hinxton, Cambridgeshire, UK, 2005:11–13 Organised by the EU Network of Excellence Semantic Mining and the European Bioinformatics Institute (EBI). [Google Scholar]
- 30.Savova GK, Chapman WW, Zheng J, et al. Anaphoric relations in the clinical narrative: corpus creation. J Am Med Inform Assoc 2011;18:459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Castano J, Zhang J, Pustejovsky J. Anaphora resolution in biomedical literature. Proceedings of the International Symposium on Reference Resolution for NLP. Alicante, Spain, 2002 [Google Scholar]
- 32.Ramesh BP, Yu H. Identifying discourse connectives in biomedical text. AMIA Ann Symp Proc 2010;2010:657–61. [PMC free article] [PubMed] [Google Scholar]
- 33.Marcus MP, Marcinkiewicz MA, Santorini B. Building a large annotated corpus of English: the penn treebank. Comput Ling 1993;19:313–30 [Google Scholar]
- 34.Palmer M, Gildea D, Kingsbury P. The proposition bank: an annotated corpus of semantic roles. Comput Ling 2005;31:71–106 [Google Scholar]
- 35.Weischedel R, Palmer M, Marcus M, et al. Ontonotes Release 4.0. Philadelphia: Linguistic Data Consortium, 2011, Technical Report. [Google Scholar]
- 36.Kim JD, Ohta T, Tateisi Y, et al. GENIA corpus—a semantically annotated corpus for bio-textmining. Bioinformatics 2003;19(Suppl 1):i180–2 [DOI] [PubMed] [Google Scholar]
- 37.Verspoor K, Cohen KB, Hunter L. The textual characteristics of traditional and Open Access scientific journals are similar. BMC Bioinformatics 2009;10:183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lafferty J, McCallum A, Pereira F. Conditional random fields: probabilistic models for segmenting and labeling sequence data. ICML '01 Proceedings of the Eighteenth International Conference on Machine Learning. Williamstown, MA, USA, 2001:282–9 [Google Scholar]
- 39.Settles B. ABNER: an open source tool for automatically tagging genes, proteins and other entity names in text. Bioinformatics 2005;21:3191. [DOI] [PubMed] [Google Scholar]
- 40.Tong S, Koller D. Support vector machine active learning with applications to text classification. J Mach Learn Res 2002;2:45–66 [Google Scholar]
- 41.Joachims T. Text categorization with support vector machines: learning with many relevant features. In: Machine Learning: ECML-98. Chemnitz, Germany, 1998:137–42 [Google Scholar]
- 42.McClosky D. Any domain parsing: automatic domain adaptation for natural language parsing. In: Ph. D. thesis, Department of Computer Science, Brown University. Providence, Rhode Island, USA, 2009 [Google Scholar]
- 43.Leaman R, Gonzalez G. BANNER: an executable survey of advances in biomedical named entity recognition. Pac Symp Biocomput 2008;13:652–63 [PubMed] [Google Scholar]
- 44.Gerner M, Nenadic G, Bergman CM. LINNAEUS: a species name identification system for biomedical literature. BMC Bioinformatics 2010;11:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bodenreider O. The unified medical language system (UMLS): integrating biomedical terminology. Nucleic Acids Res 2004;32:D267–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dahlmeier D, Ng HT. Domain adaptation for semantic role labeling in the biomedical domain. Bioinformatics 2010;26:1098–104 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.