Abstract
Objective
A system that translates narrative text in the medical domain into structured representation is in great demand. The system performs three sub-tasks: concept extraction, assertion classification, and relation identification.
Design
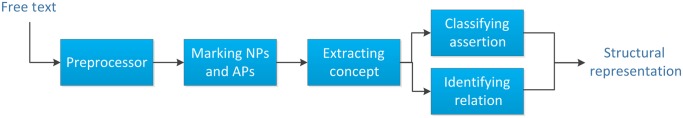
The overall system consists of five steps: (1) pre-processing sentences, (2) marking noun phrases (NPs) and adjective phrases (APs), (3) extracting concepts that use a dosage-unit dictionary to dynamically switch two models based on Conditional Random Fields (CRF), (4) classifying assertions based on voting of five classifiers, and (5) identifying relations using normalized sentences with a set of effective discriminating features.
Measurements
Macro-averaged and micro-averaged precision, recall and F-measure were used to evaluate results.
Results
The performance is competitive with the state-of-the-art systems with micro-averaged F-measure of 0.8489 for concept extraction, 0.9392 for assertion classification and 0.7326 for relation identification.
Conclusions
The system exploits an array of common features and achieves state-of-the-art performance. Prudent feature engineering sets the foundation of our systems. In concept extraction, we demonstrated that switching models, one of which is especially designed for telegraphic sentences, improved extraction of the treatment concept significantly. In assertion classification, a set of features derived from a rule-based classifier were proven to be effective for the classes such as conditional and possible. These classes would suffer from data scarcity in conventional machine-learning methods. In relation identification, we use two-staged architecture, the second of which applies pairwise classifiers to possible candidate classes. This architecture significantly improves performance.
Keywords: xy804280, thu, text processing, natural language processing, medical records
Introduction
Digitization of medical records is a growing and inevitable trend with the increasing adoption of information technology in hospitals. For physicians, it would be extremely useful if relevant information could be extracted from medical records and stored in structured form. With structured representation of medical concepts, clinical assertions, and their relations, physicians could quickly access patients' medical records for clinical decision making.
We built a complete system that performs the three tasks1 defined by the i2b2 2010 challenge: concept extraction, assertion classification, and relation identification. The concepts, called named-entities, consist of medical problem, treatment, and test. Assertions denote the status of a medical problem. A medical problem is to be associated with one of six assertion classes: present, absent, possible, conditional, hypothetical, and not_associated_with_the_patient. A relation exists between a medical problem and a concept. The relations to be identified are TrIP, TrWP, TrCP, TrAP, TrNAP, TeRP, TeCP, and PIP. TrIP indicates that the treatment improves the medical problem, TrWP that the treatment worsens the problem, TrCP that the treatment causes the problem, TrAP that the treatment is administered for the problem, TrNAP that the treatment is not administered because of the problem, TeRP that the test reveals the problem, TeCP that the test is conducted to investigate the problem, and PIP that the first problem indicates the second problem.
An example illustrating the three tasks is shown at the bottom of the page. The phrases ‘metabolic acidosis’ and ‘low bicarb,’ underlined and in red, belong to medical problem, the gray background color means present, and the arrow indicates PIP between ‘metabolic acidosis‘ and ‘low bicarb.’
The system is evaluated using the open published data provided by the i2b2 organizer. The training set includes 349 labeled discharge summaries with 27 837 concepts, 11 968 assertions, 5264 relations, and 827 unlabeled discharge summaries. The test set consists of 477 labeled discharge summaries with 45 009 concepts, 18 550 assertions, and 9070 relations.
Our contribution in this paper is threefold. First, after establishing a processing framework, we have invested significant effort in feature engineering to achieve the best performance for the three tasks. Second, both machine learning and rule-based methods were incorporated in the system. In assertion classification we used, as features for machine learning-based classifiers, carefully designed values that denote the classification result by a rule-based subsystem and its confidence, and thus combined the advantages of the two approaches. Third, we developed a new concept extraction algorithm, which dynamically switches two separate conditional random fields (CRF) models2 by using a dosage-unit dictionary. The two models, Model_medication and Model_other, are applied to different sets of sentences that are easily recognized using the dosage-unit dictionary.
The whole system consists of three sub-systems: (1) a concept extractor that dynamically switches two CRF models,2 Model_medication and Model_other, to extract concepts; (2) an assertion classifier that combines a rule-based classifier and four machine learning classifiers3 to decide a category for assertion; and (3) a relation identifier that combines four machine learning classifiers with discriminating features4 5 to identify relations. The experiment results show that our system demonstrates state-of-the-art performance in all three tasks.
Background
In the medical domain, there has been extensive work on natural language processing (NLP) tools and information extraction over the past 20 years. Spyns6 surveyed medical language processing and listed NLP application systems and tools in the medical domain, such as LSP-MLP7 and MEDLEE.8 Meystre et al9 provided a comprehensive review of information extraction from electronic medical records since 1995. Szolovits10 built a medical lexicon into the Link Grammar Parser (LGP) to obtain more accurate syntax information and applied it to emergency department notes. The recent trend in event/relation identification in the bio-medical domain was surveyed in Ananiadou et al.11
There have been several reports directly related to our work. Sibanda12 presented a category and relationship extractor (CaRE) that extracts concepts, classifies assertions, and identifies relations, and reported that the rule-based classifier outperformed the statistical classifier when the training data available were limited. Jagannathan et al13 evaluated the effectiveness of commercial NLP engines for extracting medication information concerning medication name, strength, route, and dosing frequency. Their work was based on 2000 medical records. Furthermore, in recent years, several challenge tasks using medical records have been devised. In 2007, a shared task on multi-label classification of radiology reports14 was organized, in which 45 ICD-9-CM codes were chosen as the class labels. The i2b2 organizer has held a series of challenges since 2006. The tasks were diverse, such as de-identification15 and smoking assertion16 of discharge summaries (2006), multi-label classification of obesity and co-morbidities from discharge summaries17 (2008), medication extraction of seven labels from discharge summaries18 19 (2009) and concepts, assertions, and relations extraction in clinical text (2010). This paper deals with the 2010 challenge and uses the same dataset for evaluation and comparison. Similarly to Sun et al20 and Minard et al21 in the challenge, we followed the ideas of NegEx22 and ConText23 for our rule-based component in the assertion task. However, the rule-based component is only part of our system. The output of the rule-based system is not used as the final result but is presented to machine learning-based classifiers, with the final judgment made through voting by five classifiers.
Methods
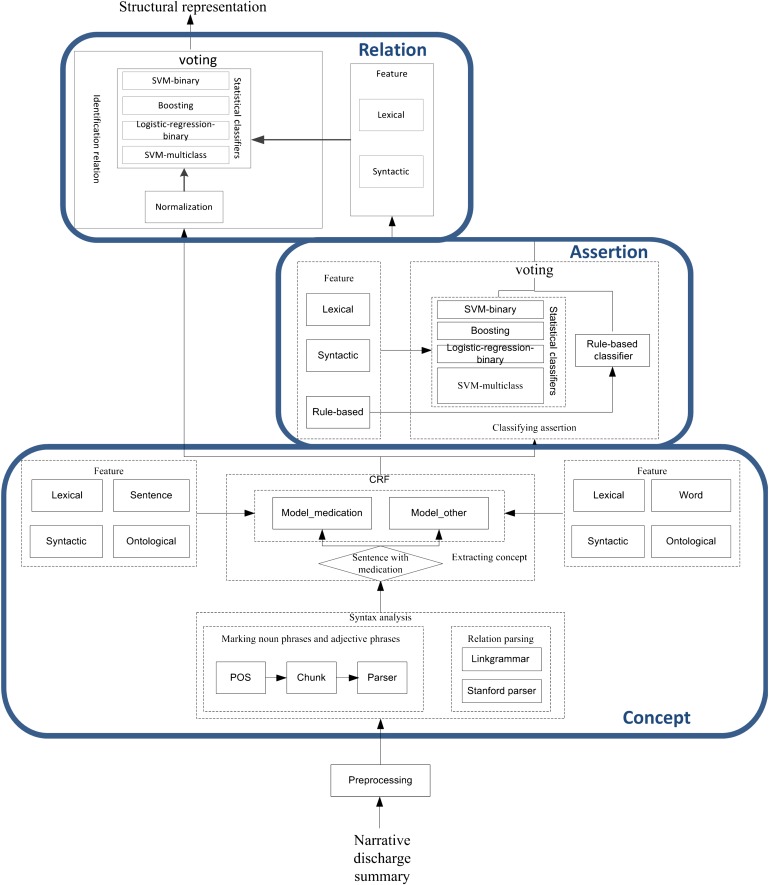
Figure 1 provides an overview of the system. Figure 2 shows both the general and detailed flow diagrams of the overall system. All of the dictionaries used in our system are available at http://research.microsoft.com/en-us/projects/ehuatuo/default.aspx.
Figure 1.
A flow diagram of the overall system.
Figure 2.
A detailed overall flow chart of the system. CRF, conditional random field; POS, part of speech; SVM, support vector machine.
Pre-processing sentences
This step reformats sentences so that standard NLP tools can be applied. This includes removal of itemized symbols (eg, ‘1)’), replacement of symbols with corresponding expressions (eg, ‘r/o’ by ‘rule out’), deletion of dots, etc.
Marking noun phrases and adjective phrases
Because concepts are expressed in the form of NP (noun phrase) or AP (adjective phrase), NPs and APs are marked by an NLP tool (SharpNLP4) as candidates of concepts. SharpNLP is an open source NLP tool originally developed for processing of newspaper articles. Since discharge summaries show peculiar linguistic characteristics not common in newspaper articles, we made several modifications. Furthermore, since only the boundaries of NP/AP phrases are relevant to concept extraction, their internal structures, which are serious causes of ambiguity in other NLP applications, are ignored. Ambiguities between the gerund and the adjectival reading of a verb in the progressive form, for example, are suppressed from being generated. We also mark only the smallest phrases in complex NPs and APs as candidates of concepts. Only exceptions are NPs that contain prepositional phrases that indicate organ/body part.24 25 We also modified SharpNLP to deal with NPs inside peculiar expressions, such as ‘2019-06-14 10:30 AM BLOOD CK-MB - 3 cTropnT - 0.05 *.’ We first detect the date and time expression at the beginning, which is often followed by pairs of NPs (a test and its value).
Concept extractor
Concepts in the i2b2 challenge belong to the three classes, medical problem, treatment, and test. However, careful examination of the training corpus reveals that there is a distinct subclass of treatment, which we call medication. Not only is the frequency of the subclass very high but also a sentence containing the subclass often has a specific style as a sentence. Out of randomly selected 300 treatment concepts in the training set, 197 concepts belong to this subclass. Sentences with medication concepts in them are often telegraphic, and do not follow the ordinary English syntax. Instead, they often have a fixed semantic pattern such as [Medication] [digits] [dosage-unit] [mode]. ‘Folic acid 1 mg p.o. daily’ is a typical example. These telegraphic sentences are interspersed freely with ordinary sentences. Since they are very different from ordinary sentences, we have to provide a concept-extraction module specifically constructed for them.
In order to construct a special extraction model for these sentences, we need training data, which i2b2 does not provide. A simple dictionary-based concept extraction method does not work for medication, since the class of medication has an abundance of lexical variances. For example, term expressions in a medication dictionary (eg, UMLS medication brand name dictionary) are canonical, while their colloquial equivalents frequently appear in discharge summaries. Furthermore, a medication name often has synonyms, abbreviations, trade names, systematic (chemical) names, and various combinations. For example, ‘morphine’ has a systematic name (morphine), two trade names (Mscontin and Oramorph), synonyms (MS Contin, Avinza, Kadian, Oramorph, and Roxanol), an abbreviation (MSIR), and various combinations such as ‘morphine (Oramorph).’ Furthermore, unlike the other classes, new medications appear regularly. It is very difficult, if not impossible, to construct a dictionary that covers all these medication names and keep it up to date.
Instead of expressions in medication, we use expressions of dosage-unit to detect telegraphic sentences. The same set of dosage-units such as ml, gr, m-grams, etc, is used for a wide range of different types of medication and they constitute a closed set. Whenever medication occurs in a telegraphic sentence, it is accompanied by a dosage-unit. In other words, the occurrence of a dosage-unit signals that the sentence is telegraphic, so we can identify a telegraphic sentence by just looking for dosage-unit expressions. These characteristics of dosage-unit expressions and telegraphic sentences allow us to construct a training set of telegraphic sentences. At the actual processing phase, we switch the concept extraction model from the model for ordinary sentences to the model for telegraphic sentences when a dosage-unit expression is found.
While we construct a dictionary for standard dosage-units from UMLS, SNOMED_CT, and the training data, we use simple elastic matching for dictionary look-up instead of exact matching. That is, each word in a sentence, if it literally intersects with a word in the dictionary of dosage-units, is considered as a dosage-unit. This simple elastic matching works very well. Among 8894 expressions in the test data retrieved by this elastic matching, only 130 are not dosage-unit expressions. The exact matching can only identify 6835 expressions for the same test data. We use several dictionaries for concept extraction, other than the dosage-unit dictionary. When an expression matches an entry in one of these dictionaries, we assign specific features that indicate the dictionary in which the match is found. Since each dictionary corresponds to a specific class of concepts, these features are effective cues for machine learning-based classifiers. In the following, we will briefly describe each of the dictionaries, how the dictionaries are constructed, and how the features from the dictionary are used for the two concept extractors, Model_medication (for telegraphic sentences) and Model_other (for ordinary sentences).
Building dictionaries for concept extractors
It is well known that a dictionary can be used to facilitate concept extractors or named-entity recognizers. However, a dictionary alone does not result in high performance, due to the incomplete coverage of a dictionary (low recall) and the existence of ambiguous terms (low precision). Instead, we use information from dictionaries as features for CRF-based extractors to overcome the deficiencies of the dictionary-only approach. Four dictionaries are constructed from the following current resources.
UMLS dictionary
The UMLS dictionary contains medical problem that is subclassified into 14 semantic types, treatment into six types, and test into six types.25
MeSH dictionaries
Every phrase in MeSH belongs to one semantic type, which is denoted by an ID of alpha-numeric characters.24 The MeSH vocabulary branches with identifiers with ‘B’ and ‘C’ as the initial character, those with ‘D’ as the initial character, and those with ‘E’ as the initial character correspond to medical problem, treatment, and test, respectively.
Medication dictionary
The medication dictionary is constructed from UMLS21 and SNOMED_CT.
Head-noun dictionaries
Head-noun dictionaries26 27 are built by extracting the head nouns of the three categories (ie, medical problem, treatment, and test) from UMLS dictionaries and the training data.
Each category in the dictionaries is used as a binary feature in our feature vector representation. When a NP or AP contains a term in a specific category of one of these four dictionaries, the corresponding feature will be set to 1. Otherwise, it is set to 0. In total, we have 10 binary features (ie, UMLS: 3, MeSH: 3, medication: 1, head-noun: 3).
Creating features
Five sets of features are used in our CRF-based28 29 extractors: lexical context, syntactic context, ontological, sentence, and word features. Table 1 shows these five feature sets. Table 2 lists the selected feature sets for both Model_medication and Model_other. Although most of the features are those commonly used in named-entity recognizers and are self-evident, the features that we specifically introduced for our extractors are described below.
Table 1.
List of various features for the concept task
| Category | Features |
| Lexical context features | The target itself (n-gram) |
| Syntactic context features | The POS |
| The phrases of NPs and APs | |
| Ontological features | UMLS-based dictionary matching |
| MeSH-based dictionary matching | |
| Medication dictionary matching | |
| Head-noun dictionary matching | |
| Sentence features | The sentence with a dosage-unit or temporal adverb (eg, twice daily or q4h) |
| The sentence with numerals before dosage-units | |
| The sentence containing a drug name before a numeral | |
| The sentence with characteristics such as [the phrase] [numeral] [dosage] | |
| The sentence with a drug name followed by an alternative drug name in parentheses, eg, fosamax (alendronate) | |
| Word features | Word capitalized |
| Entire word capitalized | |
| Word abbreviation | |
| The phrase before the numerals | |
| Assertion word | |
| Body word | |
| Pattern matching | |
| Word normalization | |
| N-character-prefix-and-suffix | |
| Word clustering |
AP, adjective phrase; NP, noun phrase; POS, part of speech.
Table 2.
Selected feature sets for both Model_medication and Model_other
| Lexical context | Syntactic context | Ontological | Sentence | Word | |
| Model_medication | x | x | x | x | |
| Model_other | x | x | x | x |
x Indicates that the dictionary in each column exists for the corresponding category.
Pattern matching (CF1)
Some items in discharge summaries are expressed in highly stylized ways. For example, in the section describing the status of patients, a sentence such as ‘2018-10-25 11:15 AM BLOOD WBC - 18.3 * RBC - 3.42’ may appear. In this sentence, a pattern of ‘test - digit’ appears twice, and the same pattern appears repeatedly in other sentences as well. These patterns, which are beyond ordinary English syntax, are treated by a pattern matching module. Once a candidate for a NP or AP is recognized as part of such a pattern, the candidate is given a corresponding feature and used by the CRF-based extractors.
Word normalization (CF2)
We normalize the words containing numbers and symbols.30 We replace decimals with *Num*, telephone number like 319-335-9253 with *PhoneNum*, date like 01/01/2011 with *Date*, and time like 10:20am with *Time*. We also add word stems as features.
N-character-prefix-and-suffix (CF3)
In the medical domain, concepts with the same topics often share the same prefix and/or suffix.31 32 For instance, the words ‘pharmacotherapy’ and ‘chemotherapy’ have the same suffix ‘therapy,’ which indicates that they both correspond to treatment. Therefore, we extract all possible N-character (2≤N≤7) prefixes and suffixes as features to be used for concept extraction. For example, from the word ‘angiograph,’ we extract a total of 14 features (prefix: ‘a,’ ‘an,’ ‘ang,’ ‘angi,’ ‘angio,’ ‘angiog,’ and ‘angiogr’; suffix: ‘h,’ ‘ph,’ ‘aph,’ ‘raph,’ ‘graph,’ ‘ograph,’ and ‘lograph’).
Word clustering (CF4)
Jargon (eg, prn), abbreviations (eg, CT), and rare terms (eg, bibasilar) are used in discharge summaries. Clustering words into classes can aggregate words with common properties and alleviate the problems caused by data scarcity. We used an unsupervised word clustering based on distributional and morphological information.33 As morphological information, we used the dictionary of medical suffixes.31 All the data released by the i2b2 organizer are supplied to the algorithm.33 After clustering, every word is associated with a class. We have a feature for the word class in our feature representation. The number of clusters (and thus the number of the values for the word class feature) is predefined in our clustering algorithm. We use various configurations of the cluster number, that is, 50, 100, 200, 400, 600, and 800.
Training CRF models for extracting concepts and matching corresponding types
The linear-chain CRF28 is used as a framework for the two concept extractors. The standard Begin/Inside/Outside (BIO) tags were used to mark concept boundaries.
Assertion classifier
In the i2b2 challenge, for assertion classification, a lot of groups used rule-based methods. Sun et al20 proposed a method based on NegEx22 and ConText23 algorithms, and used the SNOMED_CT dictionary to match self-asserted concepts such as ‘afebrile.’ Minard et al21 proposed a method combined of a rule-based method and machine learning. The rule-based method is an extension of NegEx. The support vector machine technique (SVM) is used for machine learning. We also used the hybrid approach combining one rule-based classifier with four statistical classifiers34 by voting. In statistical features, we used syntax features (2-gram LGP syntax, verb) and section names.
Generating the dictionary for the rule-based classifier
Considering the limited availability of training data and motivated by Sibanda's work,12 we put emphasis on the manual construction of an effective dictionary such as NegEx22 and ConText.23 Careful examination of the training data reveals that the appearance of a particular clue word in the neighborhood of the medical problem concept strongly signifies the assertion status of the concept. The scope of the neighborhood depends on a clue word which should precede or succeed the medical problem in a sentence for it to affect the assertion status of the medical problem. The neighborhood is normally the whole sentence, but the existence of particular words (eg, but) splits the eligible neighborhoods in a sentence. An entry in the keyword dictionary consists of a clue word, the assertion class that the word implies, and the scope of neighborhood (Preceding, Succeeding, or Any). Another list of words that delimit the scope was also provided. An example of description in the dictionaries is shown in table 3. The underlined word is the matched term in the dictionary and the italic word is the medical problem concept in the example column. The number of the entries in the dictionary is shown in table 4. All the dictionaries are available online.
Table 3.
Definitions and examples of dictionaries
| Definition | Example | |
| Keywords | Terms looked up in the sentence with a problem | One of his brothers has had a myocardial infarction. |
| Preceding words/phrases | Terms can be looked up before the problem in a sentence | Percocet, 5/325, 1-2 tabs PO q4-6h prn pain. |
| Succeeding words/phrases | The term looked up after the problem in a sentence | Return to the ER for high fevers, vomiting, uncontrolled pains or other concerns as needed. |
| Termination words | The term signifying the invalidation of signal words | In terms of his liver abnormalities, it was felt that viral hepatitis was in the differential but also was felt that Bactrim could be a cause of these abnormalities. |
| Self-indicative | The problem in a sentence that includes information itself, such as the prefix ‘a-’ | In ‘asymptomatic,’ and ‘non-tender’, the prefixes ‘a-’ and ‘non-’ are self-indicative |
| Personal nouns | Personal nouns qualifying nouns | His pain |
Table 4.
List of dictionaries for each category in assertion
| Category | Keyword | Preceding | Succeeding | Termination | Self-indicative | Personal nouns |
| Present | x | |||||
| Absent | x | x | x | x | ||
| Possible | x | x | x | |||
| Conditional | x | |||||
| Hypothetical | x | x | ||||
| Not_associated_with_the_patient | x |
x Indicates that the dictionary in each column exists for corresponding category.
Classifying assertions by the rule-based classifier
Assertion classification is the task of classifying an occurrence of medical problem into one of the six categories: not_associated_with_the_patient, hypothetical, conditional, possible, absent, and present. When one concept can be classified into more than one category (eg, not_associated_with_the_patient and present), the category with the highest priority should be selected as the final choice. The above six categories are listed in decreasing order of priority. However, the rule-based classifier itself does not make the final judgment, but its output is used by a group of statistical classifiers. It gives a vector of six dimensions as its output, each of which corresponds to a confidence score for the corresponding category.
The current version of the rule-based classifier is very simple. It only checks whether a clue word in the dictionary appears in the legitimate neighborhood of the medical problem, and if it does, it adds xcategory in the following formula to the previous value of xcategory (each of the six xcategory is initially set to 0). In this formula, w is a window size fixed for each category and d is the distance between the occurrence of medical problem and the clue word.
| (1) |
The window size is fixed to 4 for the present experiment. When xcategory for a category becomes greater or equal to 1, the xcategory is set to 1 and the rest of the categories have 0 as their value. Furthermore, the classifier gives 1 to xnot_associated_with_the_patient whenever medical problem appears in either the ‘family history’ section or the ‘social history’ section.
Extracting features for the statistical classifiers
In addition to the output of the rule-based classifier, our statistical classifiers use two sets of features: lexical context12 (3-gram words) and syntactic context12 (2-gram LGP syntax, verb, and section name) to complement the decision by the rule-based classifier.
Classifying assertions by statistical classifiers
Four statistical classifiers developed by MSR Asia3 are used: SVM-binary, boosting, logistic-regression-binary, and SVM-multiclass. We chose them because: (1) SVM-binary has been shown to be effective in many learning scenarios, and is also feasible for a small training set or high-dimensional feature space12; (2) by combining many weak classifiers, boosting can achieve high performance in training data and relatively high performance in testing data29; (3) logistic-regression-binary is used extensively in the medical field, where it achieves high performance even when the features are correlated34; and (4) SVM-multiclass is used because of its ability to directly solve multi-label problems.34
Voting
The final results are decided by voting of the five classifiers, that is, the rule-based classifier and the four statistical classifiers. In a cross-validation procedure, we find that the SVM classifier performs better than the other three classifiers. Therefore, if the vote is a draw (2 vs 2), we choose the output of the SVM classifier as the final result.
Relation identifier
There are three distinct types of relations according to their argument classes: problem-problem, problem-treatment, and problem-test. We provide three modules for these three types. The first classifier classifies a relation into (PIP, Non-PIP). The second and the third modules classify a relation into (TrIP, TrAP, TrCP, TrWP, TrNAP, NonTrP) and (TeRP, TeCP, Non-TeP), respectively. Each of these three modules consists of the same set of four classifiers and its decision is based on the votes of the four classifiers. Classification is performed in the following four steps.
Normalizing sentences
Since concepts in a sentence are already recognized, each concept occurrence is replaced with a placeholder (eg, ‘have dyspnea and tachypnea and required O2’ is normalized to ‘have [this.p] and [that.p] and required [this.tr]’).
Extracting features for the relation classifier
The features used by the relation classifiers are divided into three sets: lexical context features, syntactic context features by the LGP, and a set of the features that we newly introduced for this task. The lexical context features and syntactic context features resemble those used by Sibanda, where they are described in detail.12 The features that we newly introduced are described below.
Concepts co-occurrence (RF1)
In the medical domain, it is common for two concepts to co-occur in the same sentences as a matching pair with high frequency (eg, morphine and pain).35 We select a set of pairs of concepts whose co-occurrence frequency exceeds a threshold. Then we examine if a pair in the set is a good indicator for classification of one of the relation classes (ie, PIP, TrIP, etc). If more than 80% cases in which the pair appears are classified as positive for a class, then the pair is considered as a positive pair for that class. We choose 291 concept pairs with high frequency that are positive for at least one of the relation classes. In the feature representation, these pairs are represented as 291 binary features.
Verb-based rules (RF2)
One of the major cues for relation identification are a set of verbs, such as ‘improve,’ ‘worsen,’ etc. These specific verbs tend to denote single or a few specific relations. After examining the training data, we chose 34 verbs as such relation-indicating verbs and use them as features.
N-gram-sequential patterns (RF3)
Some n-gram-sequential patterns (n=2, 3, 4, 5) were identified as useful features by examining the training data. For example, when classifying PIP/Non-PIP, we first extract the uni-gram pattern, bi-gram pattern, … and five-gram pattern. Some of them are indicative of PIP. For instance, ‘associated with [this.p]’ is a positive pattern. For each pattern P and a relation R, PositiveP/R is the number of times that P appears in examples positive for R, and NegativeP/R is the number of times P appears in examples negative for R. We only consider n-gram patterns that appear more than 60 times for a given relation. Then, we choose n-gram patterns, Ps, which satisfy either (2) or (3) as effective n-gram patterns for classification of R.
| (2) |
| (3) |
Assertion status (RF4)
The assertion status is also useful for relation identification and is used as a feature expressed by having one of six possible values set to 1 and the rest set to 0 in a six-dimensional binary feature vector.
Stanford dependency path (RF5)
Previous work36 37 indicates syntactic features from multiple parsers improve relation extraction. Besides LGP, we used the Stanford dependency parser. The parser takes a sentence as input and outputs a dependency graph that demonstrates dependency relations between words in the sentence. For each pair of placeholder words (eg, [this.p], etc) to be classified, we identify a path in the dependency graph that connects them. We use all syntactic labels on the path as features. Features are represented as binary features, each of which corresponds to a syntactic label used in the Stanford parser.
Tense information (RF6)
Tense information is an important cue for certain relations. Suppose we have a test, and we conduct the test to examine if the patient has a medical problem (TeCP). Then it is likely that the sentence is in the future tense. In our implementation, we apply a simple rule to identify the tense of a sentence. We use SharpNLP to find the tense of a verb that appears in the rightmost position in the sentence.
Word clustering (RF7)
Word clustering was applied to target words (ie, head nouns) and verbs to avoid the difficulties caused by data scarcity. Every target word and verb is given a class label, which denotes the cluster it belongs to.
Identifying relations by statistical classifiers
The same four statistical classifiers as used for assertion classification are employed. The second classification (TrIP, TrAP, TrCP, TrWP, TrNAP, Non-TrP) and the third classification (TeRP, TeCP, Non-TeP) are not binary. We first apply a set of binary classifiers, each of which is to determine whether a given relation belongs to the relation. When more than one classifier returns positive results, pairwise classifiers determine the winner. A pairwise classifier helps to classify the categories that are difficult to distinguish, for instance TrIP and TrCP, TrAP and TrNAP, etc.
Voting
The final results are determined by the voting of the four classifiers. As in the assertion task, we choose the SVM output when the vote was a draw.
Results
Performance was measured by the three standard measures: precision (P), recall (R), and F measure (F). The results were micro-averaged and macro-averaged for each of the three tasks.
In the 2010 i2b2 challenge, the F measures of concept extraction, assertion classification, and relation identification of the top performing systems are 0.8523, 0.9362, and 0.7365, respectively.38 Table 5 lists the results of our system for the three tasks. The results demonstrate that our system achieved state-of-the-art results. In particular, for small labeled training data, the rule-based method is shown to improve performance.
Table 5.
Macro-averaged and micro-averaged results for the three tasks
| Macro-averaged | Micro-averaged | |||||
| Precision | Recall | F measure | Precision | Recall | F measure | |
| Concept | 0.8653 | 0.8319 | 0.8482 | 0.8652 | 0.8334 | 0.8489 |
| Assertion | 0.8235 | 0.8071 | 0.8196 | 0.9392 | 0.9392 | 0.9392 |
| Relation | 0.6400 | 0.5547 | 0.5943 | 0.7400 | 0.7252 | 0.7326 |
Concept task
Table 6 summarizes the improvement resulting from the addition of individual discriminating features. The baseline system used all the features except CF1, CF2, CF3, and CF4. We then listed the results by adding the features of CF1, CF2, CF3, and CF4. CF4 improved the F measure most.
Table 6.
Micro-averaged results from feature contributions for the concept task
| Methods for the concept task | Precision | Recall | F measure | Increment |
| (b)+Baseline | 0.8461 | 0.7866 | 0.8153 | |
| (c)+CF1 | 0.8574 | 0.7982 | 0.8267 | 1.14% |
| (d)+CF2 | 0.8530 | 0.8091 | 0.8305 | 0.38% |
| (e)+CF3 | 0.8539 | 0.8170 | 0.8351 | 0.46% |
| (f)+CF4 | 0.8652 | 0.8334 | 0.8489 | 1.38% |
CF1, pattern matching; CF2, word normalization; CF3, N-character-prefix-and-suffix; CF4, word clustering.
Table 7 shows the performances of four different systems, that is, (1) Model_medication (OM), (2) Model_other (OO), (3) the combined model of Model_medication and Model_other, dynamically switched by elastic matching of the dosage-unit dictionary (CMOA), and (4) the combined model of Model_medication and Model_other, dynamically switched by exact matching of the dosage dictionary (CMOS). The two combined models significantly outperform the two single models. The difference between CMOA and CMOS (0.3%) shows that the elastic matching works effectively. We used the z test on the F measure for testing the significance in (CMOA vs OM) and (CMOA vs OO). We divided 477 test records into 48 collections, each containing 9 or 10 records. As an F measure is calculated for each collection and model, respectively, we obtain 48 F scores for each of the models (ie, OM, OO, and CMOA). For the OM model, the SD of the 48 F scores is 0.019, and for the OO model, it is 0.017. The p value of the z test between the OM and CMOA models is 0.0397, and that between the OO and CMOA models is 0.0251. Since they both satisfy α≤0.05, the CMOA model significantly outperforms the two single models (OM and OO).
Table 7.
F measure results from every concept for the concept task
| Methods | Problem | Treatment | Test | Micro-average |
| OM | 0.8374 | 0.8490 | 0.8026 | 0.8421 |
| OO | 0.8580 | 0.8195 | 0.8404 | 0.8422 |
| CMOA | 0.8524 | 0.8441 | 0.8106 | 0.8489 |
| CMOS | 0.8500 | 0.8398 | 0.8102 | 0.8459 |
CMOA, combined Model_medication and Model_other with Adaptive dictionary; CMOS, combined Model_medication and Model_other with Standard dictionary; OM, only Model_medication; OO, only Model_other.
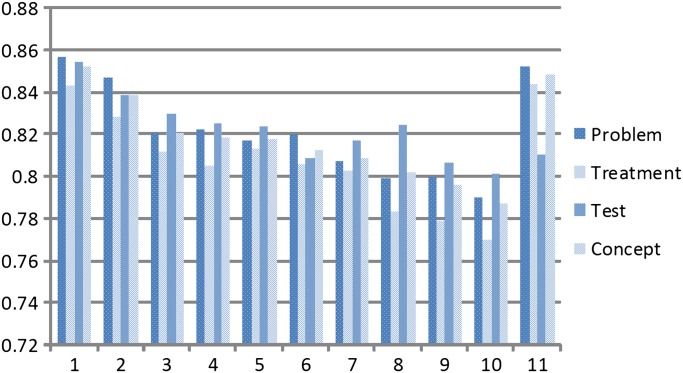
To compare our system with the systems submitted to the 2010 i2b2 challenge, figure 3 summarizes the top 10 systems39 and our system for concept extraction of medical problem, treatment, and test and the overall concept classes. Our system yields an F measure of 0.8524 for medical problem, 0.8441 for treatment, 0.8106 for test, and 0.8489 for all concepts (the sizes of the training data for medical problem, treatment, and test are 11 968, 8500, and 7369, respectively). Our system significantly outperforms (by a margin of 3.35%) the top-performing system on treatment. This indicates that our approach of dynamic switching of the two different models, one for telegraphic sentences and one for ordinary sentences, works very well.
Figure 3.
The results from the top 10 systems and our system for the concept task. On the abscissa, the numbers 1–10 denote the top 10 systems and the number 11 denotes our improved system.
Assertion task
Table 8 gives the results for the assertion task by seven different methods: rule-based classifier (RC), SVM-binary with the rule-based features (SVMB_RF), boosting with the rule-based features (BOOST_RF), logistic-regression-binary with the rule-based features (LRB_RF), SVM-multiclass with the rule-based features (SVMM_RF), voting by the four statistical classifiers (SVM-binary, boosting, logistic-regression-binary, and SVM-multiclass) without the rule-based features (VC_WRF), and voting by combining all the classifiers (SVM-binary, boosting, logistic-regression-binary, SVM-multiclass, and rule-based classifier) with the rule-based features (VC_RF).
Table 8.
Micro-averaged F measures for the assertion task from various methods
| Assertion category | Training set size | RC | SVMB_RF | BOOST_RF | LRB_RF | SVMM_RF | VC_WRF | VC_RF |
| Present | 13 025 | 0.9497 | 0.9510 | 0.9491 | 0.9513 | 0.9579 | 0.9386 | 0.9608 |
| Absent | 3609 | 0.9290 | 0.9322 | 0.9305 | 0.9315 | 0.9372 | 0.8858 | 0.9418 |
| Possible | 883 | 0.6800 | 0.7010 | 0.6291 | 0.6599 | 0.7001 | 0.4437 | 0.7019 |
| Conditional | 171 | 0.3371 | 0.3913 | 0.3372 | 0.3492 | 0.3472 | 0.17 | 0.3938 |
| Hypothetical | 717 | 0.7809 | 0.7870 | 0.7897 | 0.7988 | 0.9465 | 0.8122 | 0.9443 |
| Not_associated_with_the_patient | 145 | 0.8794 | 0.8786 | 0.8889 | 0.7607 | 0.9790 | 0.8075 | 0.9720 |
| Overall | 18 550 | 0.9220 | 0.9239 | 0.9214 | 0.9239 | 0.9374 | 0.9032 | 0.9392 |
BOOST_RF, boosting with the rule-based features; LRB_RF, logistic-regression-binary with the rule-based features; RC, rule-based classifier; SVMB_RF, SVM-binary with the rule-based features; SVMM_RF, SVM-multiclass with the rule-based features; VC_WRF, voting by the four statistical classifiers (SVM-binary, boosting, logistic-regression-binary, and SVM-multiclass) without rule-based features; VC_RF, voting by combining all classifiers (SVM-binary, boosting, logistic-regression-binary, SVM-multiclass, and rule-based classifier) with the rule-based features.
Table 8 shows that VC_RF is the best of the seven systems. RC outperforms VC_WRF by a margin of 1.88%. This illustrates the effectiveness of the rule-based classifier and the features produced by it. The statistical classifiers do not perform well without the rule-based features, as clearly shown when there is serious scarcity of data. For the small categories such as conditional and possible, the rule-based system outperforms the statistical classifiers by margins of 16.71% and 23.63%, respectively.
Relation task
Table 9 summarizes the performance improvement by the seven types of discriminating features. RF3 showed the most improvement. Together with the contributions of RF1 and RF2, this shows the importance of feature engineering in this task. The contribution of RF7, which is significant, shows that clustering of words alleviates the problem of data scarcity. Our system compared favorably with the state-of-the-art systems40 in the 2010 i2b2 challenge that did not rely on external web-based knowledge sources.
Table 9.
Micro-averaged results from feature contributions for the relation task
| Methods for the relation task | Precision | Recall | F measure | Increment |
| (b)+Baseline | 0.6984 | 0.6411 | 0.6685 | |
| (c)+RF1 | 0.7029 | 0.6567 | 0.6790 | 1.05% |
| (d)+RF2 | 0.7079 | 0.6603 | 0.6833 | 0.43% |
| (e)+RF3 | 0.7012 | 0.6935 | 0.6973 | 1.40% |
| (f)+RF4 | 0.7106 | 0.7015 | 0.7060 | 0.87% |
| (g)+Pairwise classifiers | 0.7226 | 0.7144 | 0.7185 | 1.25% |
| (h)+RF5 | 0.7249 | 0.7173 | 0.7212 | 0.27% |
| (i)+RF6 | 0.7403 | 0.7104 | 0.7251 | 0.39% |
| (j)+RF7 | 0.7400 | 0.7252 | 0.7326 | 0.75% |
RF1, concepts co-occurrence; RF2, verb-based rules; RF3, n-gram-sequential patterns; RF4, assertion knowledge; RF5, Stanford dependency path; RF6, tense information; RF7, word clustering.
Discussion
Our rule-based assertion system has the following advantages compared with the state-of-the-art methods. The dictionary we manually constructed is large and comprehensive, so that we can fully exploit external knowledge. We also use a voting mechanism for integrating the results of the different classifiers. Moreover, syntactic features are used in our classifiers. While syntactic features in previous works have not been used as widely as lexical features, the experiment results show that they improve the assertion task. This is one of the reasons why our system outperforms the other state-of-the-art systems.
Accurate marking of NPs and APs impacts on overall performance. Since only 88% of phrases are correctly marked for the training data even by the modified version of SharpNLP, 12% of concepts failed to be passed to the extraction stage. This shows that NLP tools need to be further improved to cope with the peculiarities of medical language. Medical records have many telegraphic sentences that may be deemed ungrammatical according to ordinary English grammar. Even non-telegraphic sentences contain many acronyms, local constructions that are not covered by ordinary English grammar, etc. These are challenges for NLP tools developed for the general domain.
Conclusion
This paper describes a state-of-the-art information extraction system based on extensive feature engineering combined with rule-based and machine learning methods. In order to treat telegraphic sentences interspersed among ordinary sentences, we proposed a method of dynamic switching of models. The method significantly improved the concept extractors. Since telegraphic sentences are interspersed among ordinary sentences in medical records, the same idea can be generalized and applied to other tasks that deal with medical records. In assertion classification, the result of a rule-based classifier is used as a feature in statistical classifiers, which successfully combine the rule-based approach with an ML-based classifier. This works well for small classes for which the training data are limited. In relation identification, the architecture of the pair-wise classifiers improves the performance significantly. A set of new discriminating features introduced in this paper are also shown to be very effective.
Acknowledgments
We would like to thank the organizers of the i2b2 NLP challenge, especially Dr Ozlem Uzuner and Dr Scott Duvall for their tremendous contribution.
Footnotes
Funding: This work was supported by Microsoft Research Asia (MSR Asia). The work was also supported by MSRA eHealth grant, Grant 61073077 from National Science Foundation of China and Grant SKLSDE-2011ZX-13 from State Key Laboratory of Software Development Environment in Beihang University.
Competing interests: None.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data sharing statement: The data are shared by the i2b2 Challenge organizer.
References
- 1.The Fourth i2b2/VA Challenge. https://www.i2b2.org/NLP/Relations/
- 2.CRF++. http://crfpp.sourceforge.net/
- 3.Statistical Classifiers. http://research.microsoft.com/en-us/downloads/19f63ff3-06c7-4fa9-8ee0-35abffe0e5be/default.aspx
- 4.SharpNLP Tools. http://sharpnlp.codeplex.com/
- 5.Link Grammar Parser. http://www.link.cs.cmu.edu/link/
- 6.Spyns P. Natural language processing in medicine: an overview. Methods Inf Med 1996;35:285–301 [PubMed] [Google Scholar]
- 7.Sager N, Lyman M, Bucknall C, et al. Natural language processing and the representation of clinical data. J Am Med Inform Assoc 1994;1:142–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Friedman C, Alderson PO, Austin JH, et al. A general natural-language text processor for clinical radiology. J Am Med Inform Assoc 1994;1:161–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meystre SM, Savova GK, Kipper-Schuler KC, et al. Extracting information from textual documents in the electronic health record: a review of recent research. Yearb Med Inform 2008;128:128–44 [PubMed] [Google Scholar]
- 10.Szolovits P. Adding a medical lexicon to an English parser. Proceedings of AMIA Annu Symp Proc. Washington DC, USA: AIMA, 2003:639–43 [PMC free article] [PubMed] [Google Scholar]
- 11.Ananiadou S, Pvvsalo S, Tsujii J, et al. Event extraction for systems biology by text mining the literature. Trends Biotechnol 2010;28:381–90 [DOI] [PubMed] [Google Scholar]
- 12.Sibanda TC. Was the Patient Cured? Understanding Semantic Categories and Their Relationships in Patient Records. M.S. Thesis. Cambridge, MA, USA: MIT, 2006 [Google Scholar]
- 13.Jagannathan V, Mullett CJ, Arbogast JG, et al. Assessment of commercial NLP engines for medication information extraction from dictated clinical notes. Int J Med Inform 2009;78:284–91 [DOI] [PubMed] [Google Scholar]
- 14.A Shared Task of Multi-Label Classification. http://computationalmedicine.org/challenge/previous
- 15.Uzuner O, Luo Y, Szolovits P. Evaluating the state-of-the-art in automatic de-identification. J Am Med Inform Assoc 2007;14:550–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Uzuner O, Goldstein I, Luo Y, et al. Identifying patient smoking status from medical discharge records. J Am Med Inform Assoc 2008;15:14–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Uzuner O. Recognizing obesity and co-morbidities in sparse data. J Am Med Inform Assoc 2009;16:561–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Uzuner O, Solti I, Cadag E. Extracting medication information from clinical text. J Am Med Inform Assoc 2010;17:514–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Uzuner O, Solti I, Xia F. Community annotation experiment for ground truth generation for the i2b2 medication challenge. J Am Med Inform Assoc 2010;17:519–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun Y, Nguyen A, Geva S, et al. Rule-based approach for identifying assertions in clinical free-text data. Proceedings of Australasian Document Computing Symposium (ADCS 2010). Melbourne, Australia: Australasian Document Computing Symposium, 2010:100–3 [Google Scholar]
- 21.Minard AL, Ligozat AL, Ben Abacha A, et al. Hybrid methods for improving information access in clinical documents: concept, assertion, and relation identification. J Am Med Inform Assoc 2011;18:588–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.NegEx. http://code.google.com/p/negex/
- 23.ConText. http://code.google.com/p/negex/
- 24.MESH Knowledge Base. http://www.ncbi.nlm.nih.gov/mesh
- 25.UMLS Knowledge Base. http://www.nlm.nih.gov/research/umls
- 26.Stemmer. http://tartarus.org/∼martin/PorterStemmer/
- 27.Head Noun Finder. http://nlp.cs.berkeley.edu/Main.html#Parsing
- 28.Lafferty J, McCallum A, Pereira F. Conditional Random Fields: Probabilistic models for segmenting and labeling sequence data. Proceedings of the 18th International Conference on Machine Learning (ICML 2001). Williamstown, MA, USA: International Machine Learning Society, 2001:282–9 [Google Scholar]
- 29.Duda RO, Hart PE, Stork DG. Pattern Classification. 2nd edn New York: John Wiley & Sons Inc, 2001 [Google Scholar]
- 30.Ratinov L, Roth D. Design challenges and misconceptions in named entity recognition. Proceedings of the 13th Conference on Computational Natural Language Learning (CoNLL 2009). Boulder, CO, USA: Association for Computational Linguistics, 2009:147–55 [Google Scholar]
- 31.Medical Suffixes. http://www.macroevolution.net/medical-suffixes.html
- 32.Deleger L, Grouin C, Zweigenbaum P. Extracting medical information from narrative patient records: the case of medication-related information. J Am Med Inform Assoc 2010;17:555–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clark A. Combining distributional and morphological information for part of speech induction. Proceedings of the 10th Conference of the European Chapter of the Association for Computational Linguistics (EACL 2003). Budapest, Hungary: Association for Computational Linguistics, 2003:59–66 [Google Scholar]
- 34.Bagley SC, White H, Golomb BA. Logistic regression in the medical literature: standards for use and reporting, with particular attention to one medical domain. J Clin Epidemiol 2001;54:979–85 [DOI] [PubMed] [Google Scholar]
- 35.Chan YS, Roth D. Exploring background knowledge for relation extraction. Proceedings of the 23rd International Conference on Computational Linguistics (COLING 2010). Beijing, China: Association for Computational Linguistics, 2010:152–60 [Google Scholar]
- 36.Miwa M, Pyysalo S, Hara T, et al. A comparative study of syntactic parsers for event extraction. Proceedings of Natural Language Processing in Biomedicine (BioNLP 2010). Uppsala, Sweden: Association for Computational Linguistics, 2010:37–45 [Google Scholar]
- 37.Miwa M, Saetre R, Kim JD, et al. Event extraction with complex event classification using rich features. J Bioinform Comput Biol 2010;8:131–46 [DOI] [PubMed] [Google Scholar]
- 38.Uzuner O, South BR, Shen S, et al. 2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text. J Am Med Inform Assoc 2011;18:552–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.The Results from Top 10 Systems for the First Task. https://www.i2b2.org/NLP/Relations/Download.php
- 40.Rink B, Harabagiu S, Roberts K. Automatic extraction of relations between medical concepts in clinical texts. J Am Med Inform Assoc 2011;18:594–600 [DOI] [PMC free article] [PubMed] [Google Scholar]