Abstract
Objective
Patient discharge summaries provide detailed medical information about hospitalized patients and are a rich resource of data for clinical record text mining. The textual expressions of this information are highly variable. In order to acquire a precise understanding of the patient, it is important to uncover the relationship between all instances in the text. In natural language processing (NLP), this task falls under the category of coreference resolution.
Design
A key contribution of this paper is the application of contextual-dependent rules that describe relationships between coreference pairs. To resolve phrases that refer to the same entity, the authors use these rules in three representative NLP systems: one rule-based, another based on the maximum entropy model, and the last a system built on the Markov logic network (MLN) model.
Results
The experimental results show that the proposed MLN-based system outperforms the baseline system (exact match) by average F-scores of 4.3% and 5.7% on the Beth and Partners datasets, respectively. Finally, the three systems were integrated into an ensemble system, further improving performance to 87.21%, which is 4.5% more than the official i2b2 Track 1C average (82.7%).
Conclusion
In this paper, the main challenges in the resolution of coreference relations in patient discharge summaries are described. Several rules are proposed to exploit contextual information, and three approaches presented. While single systems provided promising results, an ensemble approach combining the three systems produced a better performance than even the best single system.
Keywords: Coreference resolution, Markov logic network, clinical information extraction, natural language processing, ensemble classifier, text mining
Background and significance
Coreference resolution is the task of determining whether or not two noun phrases are used to refer to the same thing.1 The coreference resolution track in the 2011 i2b2/VA/Cincinnati Challenge2 focuses on the resolution of co-referential relations between concepts. The concepts in this task consist of entities related to a patient mentioned in the patient's discharge summary, including the patient's name and all pronouns referring to the patient, medications, and states describing the patient (eg, ‘back pain’). These concepts are common in clinical documents, and resolving their coreferences is essential to obtain a full view of the clinical situation.
In comparison with common coreference resolution tasks, identifying coreference relations in patient discharge summaries turns out to be more challenging. The first challenge is inconsistencies among the summary formats. Each summary may consist of different subsections, such as the ‘social history’ and ‘discharge medications’ shown in figure 1, which include patient information under different circumstances. The same concepts mentioned in different subsections may in fact be different entities. For example, the same medication, ‘clozapine’, mentioned in the ‘medications on admission’ and ‘discharge medications’ sections are considered non-coreferential, since each is taken under distinct conditions. Furthermore, the writing style is not specified, so several names can exist for a single entity. For example, names such as clinician, doctor, Dr and MD may all refer to the same person in a summary.
Figure 1.
A sample discharge summary from the i2b2 Beth test dataset.
Second, the coreferential concepts depend heavily on contextual information. For example, for concepts under the class, ‘problem’, ‘treatment’, and ‘test’, the time of occurrence strictly regulates the coreferential relationship among them. In addition to time, other restrictions such as dosage, administration, and quantities may further preclude these concepts from becoming coreference pairs. Specific prerequisites considering the individual and the location of the ‘problem’ (eg, internal organs or external body parts) also circumscribe the coreference pairs.
Finally, for convenience, clinicians often record certain types of information such as examination procedure, results, and medications as numbered lists with incomplete phrases (eg, ‘7. Clozapine 25 mg…’ in figure 1). Moreover, arbitrary acronyms and abbreviations of clinical terms are often found throughout most records, which may harm the performance of coreference resolution systems without domain knowledge. For instance, the abbreviation ‘PCP’ is often found in the discharge summaries as a person concept. Without clinical knowledge, the system will not be able to distinguish it as ‘primary care physician’, and thus leave this concept out of its appropriate chain.
In this paper, we propose three approaches to the clinical coreference resolution task: one is an unsupervised multistage rule-based (MR) system based on the sieve architecture3; another is based on the Markov logic network (MLN) model4; and the last is based on the maximum entropy (ME).5 In addition, we report the results of combining the above three systems. All systems exploit contextual information, such as subsection captions and quantity mentions, extracted from discharge summaries to improve coreference resolution performance. Several useful features are formally described in first-order logic (FOL) and evaluated on the i2b2 Beth and Partners datasets.
Methods
In the following subsections, we describe our three coreference resolution systems and the rules developed for the i2b2 coreference resolution challenge. These rules were developed by carefully examining the i2b2 training dataset and analyzing the main writing style and the layout of the discharge summaries.
System 1: MR coreference resolution system
The MR-based system is implemented as a series of rule-based models. Four stages are developed: string match, filtering, person concept resolution, and non-person concept resolution. The system applies stages of deterministic rule-based models one at a time from the highest to the lowest precision. In this work, we use the FOL formula to represent each rule.
In FOL, formulas consist of four types of symbol: constants, variables, functions, and predicates. ‘Constants’ represent objects in a specific domain. ‘Variables’ (eg, i, j) range over the objects. ‘Predicates’ represent relationships among objects or attributes of objects. An ‘atom’ is a predicate symbol applied to a list of arguments, which may be constants or variables. Our rules are recursively constructed from atoms using logical connectives and quantifiers. The Boolean operations of logical conjunction, disjunction, and negation are denoted by ‘∧’, ‘∨’, and ‘¬’, respectively. The symbol, ‘∃’, is an existential quantification, while ‘∃!’ is a uniqueness quantification. Note that ∃x.P(x) means that there is at least one x such that P(x) is true. But ∃x.P(x) means that there is exactly one x such that P(x) is true. Examples of FOL symbols and formulas are given in table 1.
Table 1.
Examples of first-order logic symbols used in this work
| Symbol type | Example | Description |
| Constant |
|
Treatment concept cluster |
|
Problem concept cluster | |
| Predicate | coreference (i,j) | i and j are variables referring to the ith and the jth concepts in a text. coreference (i,j) indicates that the two concepts are the same instance |
| Predicate | conceptCluster(i,c) | i indicates the ith concept and c indicates a concept cluster. conceptCluster(i,c) indicates that the concept cluster of the ith concept is c |
| exactMatch(i,j) | The string of the ith concept matches with the jth concept. | |
| Atom |
|
|
| Rule/formula |
|
If the string of the ith concept matches with the jth concept, and they belong to the same concept cluster, the two concepts are the same instance. |
A ‘ground atom’ is an atom whose arguments are all constants (eg, conceptCluster (‘liver transplantation’, ‘treatment’)). A ‘possible world’ is an assignment of truth values to a ground atom. In the MR system, each rule-based model considers the ‘body’ of a rule. If the possible world of the ‘body’ is true, the model could either link two concepts or remove linkages based on the definition of the ‘head’. For example, the rule defined in the last row of table 1 will link the two concepts i and j if the strings of the two concepts match with each other. Take the term ‘bipolar disorder’, which appears in figure 1 twice, as an example. The two instances must be linked together and form a coreference chain.
System 2: MLN-based coreference resolution system
In contrast with the MR system in which different stages are used to prioritize rules, there are no explicit stages in the MLN system. The MLN system associates rules with different weights to differentiate them. In order to highlight each rule's weighting factors, we use the ‘+’ notation. For example, consider the formula in Table 1, the ‘+’ before the variable ‘c’ indicates that the MLN model must learn a separate weight for each possible grounded value of c. Therefore, and are given two different weights after training.
In the MLN model, we learn the weights by constructing a Markov network from the predefined rule set and the given training data. In the network, a node is constructed for each possible grounded predicate. There is an edge between two nodes (grounded predicates) of the network if, and only if, the corresponding ground atoms appear together in a formula. The constructed network represents a joint distribution over possible worlds where z is the partition function, F is the set of all formulas in the MLN, gj is the set of groundings of the ith formula, and gj(x)=1 if the jth ground formula is true and gj (x)=0 otherwise. General algorithms for learning and inference in Markov logic are discussed in the work of Richardson and Domingos.4 We use 1-best Margin Infused Relax online learning Algorithm (MIRA)6 for learning weights and cutting plane inference7 with integer linear programming as the base solver for inference at test time as well as during the MIRA online learning process.
System 3: ME-based coreference resolution system
In the ME-based system, we formulated the filtering (cf stage 2) and the coreference resolution tasks (cf stages 3 and 4) as classification tasks and developed three models. Model1 used features equivalent to the formulas described in stage 2. Model2 and model3 used feature functions equivalent to the coreference resolution formulas described in stage 3 and stage 4, respectively. We transformed rule models defined in each stage into corresponding features. For example, the binary feature function defined for the formula
is listed as follows. where o refers to the outcome indicating whether the pair i and j is coreference or not, and h refers to the history (the two concepts' cluster and the matching result in this case).
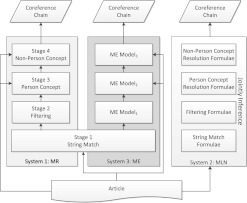
Figure 2 provides a summary of the three systems. As shown in figure 2, the MLN system integrates several stages using only one model. This is in contrast with the two separate-stage systems, MR and ME, where several models need to be trained and integrated by different strategies.
Figure 2.
An overview of the stage-based MR (multistage rule-based) and ME (maximum entropy) systems and the Markov logic network (MLN)-based system.
Predicate definition
We summarize the main predicates defined for the coreference resolution task in table 2. The first predicate, coreference(i,j), is referred to as ‘hidden’ because our systems need to determine it at test time. Others are considered ‘observed’, because they are known in advance. In the following subsections, we describe the rule-based models implemented in each stage of our MR system. These rules are used to construct a Markov network to train the MLN model, and are also transformed to the equivalent binary feature functions for ME training. The MLN/ME system then uses the models to infer hidden coreferences.
Table 2.
Predicate definitions
| Predicate | Description |
| coreference (i,j) | The two concepts i and j are the same instance |
| backwardNonPersonConcept(i,j,d) | The distance between the concept i and the non-person/pronoun concept j is d |
| backwardPersonConcept(i,j,d) | The distance between the concept i and the person concept j is d. The distance is calculated by counting the number of concepts in between them |
| conceptCluster(i,c) | The concept cluster of i is c |
| exactMatch(i,j) | The string of the ith concept matches with the jth concept |
| stemMatch(i,j) | The stem of the ith concept matches with the stem of the jth concept |
| gender(g) | The discharge summary contains the gender field g, for example ‘sex: M’ |
| hasDate(l) | The line l contains date descriptions such as 2013-03-26 |
| hasDosage(l,d) | The line l contains a description d related to the amount of medicine that the patient needs to take at one time |
| hasFollowingword(i,w) | The ith concept has the following word w |
| hasPrecedingWord(i,w) | The ith concept has the preceding word w |
| hasQuantity(i,q) | The concept i has a quantity description q, such as ‘10 mg’ |
| jaroWinkerDistance(i,j,s) | The Jaro–Winker distance between the concept i and j is s |
| line(i,l) | The concept i is on the line l in the discharge summary |
| overlap(i,j) | The two concepts i and j are overlapped |
| personalPronoun(i) | The concept i is a personal pronoun, such as ‘he’, ‘she’ and ‘you’ |
| personalRelativePronoun(i) | The concept i is a personal relative pronoun, such as who, whom, and whose |
| possessivePronoun(s1,s2) | The string s1 (eg, his) is the possessive pronoun of the string s2 (eg, he) |
| section(i,s) | The concept i is under the subsection s |
| string(i,s) | The string of the ith concept is s |
| wordPosition(i,s,e) | The first and the last word of the concept i is the sth and the eth words of the text, respectively |
| wordNetSimilarity(i,j,s) | The WordNet similarity score between the concept i and j is s |
Stage 1: string match
Previous work5 has reported that string matching is the feature that contributes most to coreference resolution. We use F.1 to ensure that the system links concepts if they are expressed by the same text string.
| F.1 |
Articles (a, an, the) and demonstrative pronouns (this, these, that, those) were removed from strings before comparison.
In addition to the exact matching method, we incorporate the Porter stemming algorithm8 to normalize each concept term, and add the following rule:
Stage 2: filtering
In this stage, our models use the contextual information extracted from the discharge summary to remove existent linkages between the three concept clusters: test, problem, and treatment.
| F.2.1 |
The first model removes two linked concepts if the following two cases are satisfied: (1) their concept cluster is not ‘person’ and ‘pronoun’; (2) one concept is located in a line containing date description, but the other does not co-occur with any date description.
| F.2.2 |
The second model removes the linkage of two treatment concepts if they do not have equivalent dosages.
| F.2.3 |
The third model is similar to the second model, but it considers the test concepts and checks whether or not they have the equivalent quantity. For instance, in the sentence ‘Temp noted to be low at 94 and she was placed on bear hugger which improved temp to 96.7’, both of the ‘temp’ concepts that appeared in this sentence are test concepts. Nevertheless, the number following each concept indicates that they are actually separate entities.
| F.2.4 |
The final model checks whether or not two exactly matching non-person/pronoun concepts belong to the same subsection. If they are under different sections, their linkage will be removed depending on S1 and S2 as well as the concept class ct. Continuing the example shown in figure 1, the two treatment concepts, ‘clozapine’, are originally linked as a coreference pair. But our final model in the filtering stage was aware of the fact that the two concepts was each under the section ‘medications on admission’ and ‘discharge medications’, respectively, and removed their linkage.
In practice, our systems only consider cases in which the text of the section S1 contains ‘hospital’ or ‘admission’ and the text of S2 contains ‘discharge’. For the MLN/ME systems, we associate the weight with the concept cluster ct and the section tag Sn. We transform all F.2 formulas to binary feature functions to train the ME model1.
Stage 3: person concept coreference resolution
In this stage, the MR system links two person concepts if any of the conditions below are satisfied.
Personal relative pronoun
The ith concept is a personal relative pronoun that modifies the antecedent person concept j. In the sentence ‘Ms. Franklin is a 34-year-old woman with Bipolar disorder and group home resident who by way of EMS for apparent alprazolam overdose.’ from figure 1, ‘who’ is a personal relative pronoun, and thus it is linked with its antecedent person concept, which is ‘Ms. Franklin’. For the MR system, the exact rule is: .
For the ME model2 and the MLN system, the rule is associated with different weights based on the string of the personal relative pronoun (eg, s is ‘who’ in the previous example) and the number of concepts d between concepts i and j (eg, d is ‘1’ in the example):
| F.3.1 |
Possessive pronoun
The concept i is a possessive pronoun of the concept j (eg, his and he). The rule is defined as follows.
| F.3.2 |
Note that, in this stage, the MR system only deals with person concepts (ie, the case that c1 = c2 = ‘person’).
Personal pronoun
We define two rules to resolve personal pronouns. The first rule links the personal pronoun i to the backward person concept j by considering the distance d between i and j.
| F.3.3.1 |
For the MR system, we only consider cases in which d is 1 (ie, the rule model directly links the personal pronoun i to the nearest person concept j).
The second rule links the concept i containing the word ‘patient’ to the most frequent personal pronoun concept j because, in the discharge summary, the most frequent personal pronoun usually refers to the described patient.
| F.3.3.2 |
Discharge summary information
Fields such as ‘sex’ provided by the discharge summary or patterns such as ‘<person concept>: <person concept>’ are exploited by our rule-based models.
| F.3.4.1 |
Unlike previous work9 10 that used alignment algorithms to collect syntactic patterns, we propose modeling the word sequence in the MLN model. After training the model, we select the syntactic patterns associated with positive weights. For example, the following formula can select patterns such as: ‘<person concept>: <person concept>’ and ‘<person> is <person>’.
| F.3.4.2 |
The collected patterns are used by the MR system and the ME model2. In contrast, the MLN system uses the weights determined by the word w to resolve the coreference.
Objective case
The concept i is an objective case of the concept j (eg, objective pronouns him and her). The rule is defined as follows.
| F.3.5 |
Finally, some rules that require domain-specific knowledge are used to remove linkages. For example:
| F.3.6 |
Concepts that contain ‘PCP’, which means the primary care physician, and ‘patient’ should not be considered as coreference pairs because they apparently indicate different people.
Stage 4: non-person concept coreference resolution
In this stage, the MR system and ME model3 resolve coreferences of non-person concepts, including treatment, problem, test, and pronoun.
String similarity
We used two algorithms to calculate the similarity of two concepts. The first is adapted from the work of Banerjee and Pedersen11 in which WordNet is used as a glossary to determine the semantic similarity between two concepts. The second is based on the Jaro–Winkler distance.12 Two predicates, wordNetSimilarity(i,j,s) and jaroWinkerDistance(i,j,s), are defined to indicate the similarity score s between the two concepts i and j. The MR system links two concepts together if their score exceeds a threshold λ. For the ME/MLN system, we put the calculated score into one of five bins as shown in the following rules.
| F.4.1.1 |
| F.4.1.2 |
Pronoun concept
For pronoun concepts, in this stage, the MR and ME systems remove chains containing only pronoun concepts through post-processing. In the MLN system, we add the following deterministic rule.
| F.4.2.1 |
The next rule links the pronoun concept i to the backward non-person/pronoun concept j, if the concept j is the only non-person/pronoun concept that precedes i.
| F.4.2.2 |
In F.4.2.3, the MR system links the pronoun concept i to the nearest non-person/pronoun concept j, but the ME/MLN system links them by the distance d and the string of the concept i.
| F.4.2.3 |
Finally, we applied F.4.3 to remove the chain that contains two overlapped concepts, such as ‘<person><person> her</person> mother</person>’ in figure 1, which may be added by F.4.1.1 and F.4.1.2. Since they obviously refer to different people, overlapping concepts such as these are removed from their coreference connection.
| F.4.3 |
Results
Dataset
The 2011 i2b2 coreference resolution challenge released three datasets annotated by three organizations: Beth Israel Deaconess Medical Center (the Beth dataset), Partners HealthCare (the Partners dataset), and the University of Pittsburgh. All records have been fully deidentified and are manually annotated according to the i2b2/VA annotation guidelines.13 Five concept classes—‘problems’, ‘treatments’, ‘tests’, ‘person’, and ‘pronoun’—are annotated. Generally speaking, ‘problems’ are the abnormalities of a person's body or mind, either observed or described by the clinician or the patient in the text. ‘Treatments’ are medical procedures and processes adopted to deal with the ‘problems’, including medicines, surgeries, and therapies. ‘Tests’ are medical procedures carried out on the patient or his/her body fluids or tissues to gain more information about his/her ‘problems’. The ‘persons’ class consists of all mentions regarding a person or a group of people. Words and phrases that contain proper names, personal pronouns, possessive pronouns, job titles, and groups are all part of this class. Finally, the ‘pronoun’ class accommodates all the other remaining pronouns not found in the ‘persons’ class. Owing to the late release of the Pittsburgh dataset and the difficulty of obtaining institutional review board approval, only the Beth and Partners datasets were downloaded for system development. In the following subsections, we first used the two datasets with concept annotations as the development sets and conducted 10-fold cross-validation (CV) to evaluate the proposed systems. We then reported their performance on the official Beth and Partners test sets.
CV results on the training set
In this section, we report the proposed systems' 10-fold CV results on the Beth and Partners dataset. For each dataset, we trained two supervised learning systems (ME and MLN): the first two systems (MLNBeth and MEBeth) were trained using the Beth dataset and the following systems (MLNPartners and MEPartners) used the Partners dataset. We report the performance of the proposed systems under the four concept clusters, ‘test’, ‘person’, ‘problem’, and ‘treatment’ in table 3. The results generated by the official i2B2 evaluation script are calculated by using the unweighted average of three metrics: MUC,14 BCUBED,15 and CEAF.16
Table 3.
Tenfold cross-validation results on the Beth/Partners dataset
| Dataset | Stage | Test | Person | Problem | Treatment | |||||
| Average F-score (%) | Diff | Average F-score (%) | Diff | Average F-score (%) | Diff | Average F-score (%) | Diff | |||
| Beth | 1 w/o F.0 | MR | 66.07 | – | 83.69 | – | 77.05 | – | 78.11 | – |
| MLN-1 | 66.07 | – | 83.69 | – | 77.05 | – | 78.11 | – | ||
| 1 | MR-1 | 66.23 | +0.16 | 83.90 | +0.21 | 77.07 | +0.02 | 78.19 | +0.08 | |
| MLN-1 | 66.23 | +0.16 | 83.90 | +0.21 | 77.07 | +0.02 | 78.19 | +0.08 | ||
| 1+2 | MR-2 | 71.34 | +5.11 | 83.90 | – | 76.73 | −0.34 | 76.49 | −1.70 | |
| MR-1+model1 | 65.48 | −0.75 | 83.90 | – | 77.07 | – | 78.20 | +0.01 | ||
| MLN-2 | 72.86 | +6.63 | 83.90 | – | 77.05 | −0.02 | 77.64 | −0.55 | ||
| 1+2+3 | MR-3 | 71.34 | – | 90.44 | +6.54 | 76.73 | – | 76.49 | – | |
| MR-1+model1+2 | 65.48 | – | 87.87 | +3.97 | 77.07 | – | 78.20 | – | ||
| MLN-3 | 73.04 | +0.18 | 90.73 | +6.83 | 76.70 | −0.35 | 78.48 | +0.84 | ||
| 1+2+3+4 | MR-4 | 77.21 | +5.87 | 90.50 | +0.06 | 77.96 | +1.23 | 76.44 | −0.05 | |
| MR-1+model1+2+3 | 69.36 | +3.88 | 87.87 | – | 77.38 | +0.31 | 77.58 | −0.63 | ||
| MLN-4 | 77.04 | +4.00 | 90.72 | −0.01 | 79.48 | +2.78 | 78.91 | +0.43 | ||
| Partners | 1 w/o F.0 | MR | 70.83 | – | 76.55 | – | 77.96 | – | 78.41 | – |
| MLN-1 | 70.83 | – | 76.55 | – | 77.96 | – | 78.41 | – | ||
| 1 | MR-1 | 71.12 | +0.29 | 76.60 | +0.05 | 77.99 | +0.03 | 78.64 | +0.23 | |
| MLN-1 | 71.12 | +0.29 | 76.60 | +0.05 | 77.99 | +0.03 | 78.64 | +0.23 | ||
| 1+2 | MR-2 | 72.80 | +1.68 | 76.60 | – | 77.89 | −0.10 | 74.38 | −4.26 | |
| MR-1+model1 | 67.64 | −3.48 | 76.60 | – | 78.04 | +0.05 | 78.64 | – | ||
| MLN-2 | 72.24 | +1.12 | 76.60 | – | 77.20 | −0.79 | 77.72 | −0.96 | ||
| 1+2+3 | MR-3 | 72.80 | – | 84.16 | +7.56 | 77.89 | – | 74.38 | – | |
| MR-1+model1+2 | 67.64 | – | 82.48 | +5.88 | 78.08 | +0.04 | 78.64 | – | ||
| MLN-3 | 72.21 | −0.03 | 85.24 | +8.64 | 77.91 | +0.71 | 78.13 | +0.41 | ||
| 1+2+3+4 | MR-4 | 80.30 | +7.50 | 84.42 | +0.26 | 79.60 | +1.71 | 76.58 | +2.20 | |
| MR-1+model1+2+3 | 73.19 | +5.55 | 82.52 | +0.04 | 79.35 | +1.27 | 80.63 | +1.99 | ||
| MLN-4 | 80.69 | +8.48 | 85.66 | +0.42 | 80 | +2.09 | 81.79 | +3.66 | ||
MLN-n is the MLNBeth/Partners system with all corresponding rules defined in stage 1 to n. For example, MLN-1 is the MLN system with all formulas defined in the ‘Stage 1: string match’ section. MLN-2 adds all formulas defined in the ‘Stage 2: filtering’ section on MLN-1. The column ‘Diff’ shows the improvement over the previous stage.
MLN, Markov logic network; MR, multistage rule-based; w/o, without.
In Table 3, for each dataset, the first row (stage 1 w/o F.0) shows the performance of the MR and the MLN systems when only the rule defined in the ‘Stage 1: exact match’ section was used. In the second row (stage 1), we compare the performance improvements that the two systems made after adding F.0. The results show that considering the concept cluster can reduce the likelihood of errors.
| F.0 |
The third row (stage 1+2) shows the achievement of the three systems after the addition of the proposed filtering stage. As you can see, the filtering stage significantly boosts both MR and MLN systems' performance on test concepts. However, both systems perform worse on the problem and treatment concept clusters.
After stage 3 (the fourth row), all systems achieve a satisfactory F-score on the person concept. Compared with MR-3 and the ME system, MLN-3 makes a larger performance gain on the person concept, and its performance on treatment and test concepts is better than MLN-2.
The last row shows the performance of our systems with all rules used. We find that MLN-4 outperforms the other two systems in three out of four concepts. Furthermore, performance in all concepts improved in MLN-4.
In a nutshell, we can see that the proposed rules generally improve coreference resolution performance on both datasets. However, on the treatment concept, the rules actually degrade performance of the MR system. After the four stages, MR performance on the treatment concept is worse than that of the baseline system (stage 1 without F.0). In contrast, the MLN system improves the baseline system's performance on all concept clusters in stage 4 for both datasets.
Results on the test set
Table 4 shows the results on the test dataset. By looking at tables 3 and 4, we can conclude that the proposed stages and formulas consistently improved coreference resolution performance on both training and test sets. In comparison with the other three concepts, the improvement of the treatment concept is not significant. The MLN system achieves the highest overall performance on the Beth and Partners datasets. However, in contrast with the training set, the MLN system did not dominate all concept clusters on the test set; the other two systems achieved a higher F-score on certain concept clusters.
Table 4.
Results on the test dataset
| Stage | Test | Person | Problem | Treatment | Overall | |||||
| Average F-score (%) | Diff | Average F-score (%) | Diff | Average F-score (%) | Diff | Average F-score (%) | Diff | Average F-score (%) | Diff | |
| Beth | ||||||||||
| MR-1/MLN-1 | 63.80 | – | 81.20 | – | 77.37 | – | 80.23 | – | 80.30 | – |
| MR-2 | 71.40 | +7.60 | 81.20 | – | 76.53 | −0.84 | 75.10 | −5.13 | 82.20 | +1.90 |
| MR-1+model1 | 65.07 | +1.27 | 81.23 | +0.03 | 77.40 | +0.03 | 80.27 | +0.04 | 83.60 | +3.30 |
| MLN-2 | 70.37 | +6.57 | 81.20 | – | 77.40 | +0.03 | 79.63 | −0.60 | 83.70 | +3.40 |
| MR-3 | 71.40 | – | 88.77 | +7.57 | 76.53 | – | 75.10 | – | 83.90 | +1.70 |
| MR-1+model1+2 | 65.07 | – | 85.13 | +3.90 | 77.40 | – | 80.27 | – | 84.50 | +0.90 |
| MLN-3 | 69.73 | −0.64 | 87.83 | +6.63 | 77.43 | +0.03 | 78.50 | −1.13 | 84.90 | +1.20 |
| MR-4 | 75.57 | +4.17 | 88.90 | +0.13 | 77.73 | +1.20 | 75.33 | +0.23 | 85.57 | +1.67 |
| MR-1+model1+2+3 | 67.60 | +2.53 | 85.30 | +0.17 | 79.50 | +2.90 | 80.77 | +0.50 | 85.20 | +0.70 |
| MLN-4 | 72.50 | +2.77 | 88.37 | +0.54 | 79.67 | +2.24 | 80.10 | +1.60 | 86.00 | +1.10 |
| Partners | ||||||||||
| MR-1/MLN-1 | 73.47 | – | 76.27 | – | 77.63 | – | 81.57 | – | 82.90 | – |
| MR-2 | 72.53 | −0.94 | 76.27 | – | 77.47 | −0.16 | 75.03 | −6.54 | 82.50 | −0.40 |
| MR-1+model1 | 73.53 | +0.06 | 76.27 | – | 77.63 | – | 81.57 | – | 84.70 | +1.80 |
| MLN-2 | 73.80 | +0.33 | 76.27 | – | 76.67 | −0.96 | 77.87 | −3.70 | 84.10 | +1.20 |
| MR-3 | 72.53 | – | 81.77 | +5.50 | 77.47 | – | 75.03 | – | 84.10 | −0.60 |
| MR-1+model1+2 | 73.53 | – | 80.83 | +4.56 | 77.63 | – | 81.57 | – | 85.80 | +1.70 |
| MLN-3 | 73.47 | −0.33 | 82.37 | +6.10 | 77.70 | +1.03 | 77.67 | -0.20 | 86.10 | +2.00 |
| MR-4 | 79.67 | +7.14 | 82.03 | +0.26 | 78.93 | +1.46 | 75.30 | +0.27 | 86.10 | +2.00 |
| MR-1+model1+2+3 | 75.27 | +1.74 | 80.90 | +0.07 | 80.50 | +2.87 | 81.93 | +0.36 | 86.50 | +0.70 |
| MLN-4 | 77.83 | +4.36 | 84.03 | +1.66 | 79.70 | +2.00 | 77.67 | – | 87.20 | +1.10 |
MLN-n is the MLNBeth/Partners system with all corresponding rules defined in stage 1 to n. For example, MLN-1 is the MLN system with all formulas defined in the ‘Stage 1: string match’ section. MLN-2 adds all formulas defined in the ‘Stage 2: filtering’ section on MLN-1. The column ‘Diff’ shows the improvement over the previous stage.
MLN, Markov logic network; MR, multistage rule-based.
Discussion
The hardest concept type—treatment
As shown in tables 3 and 4, our formulas encountered some difficulty in resolving the treatment concept coreferences. We believe that the inconsistent annotation of treatment concepts found in the training set may harm the effectiveness of the proposed rules. Following the official coreference guidelines,13 treatment concepts are only paired when they are of the same episode and dosage. However, some of the coreference pairs that do not completely follow this standard were found in the training/test corpus. For example, at times, a treatment concept can first appear without a known dosage and then be followed by another appearance with its dosage specified. Annotators would intuitively think of them as the same entity, but our filtering model removes the linkage. An example can be found in the Beth training set: ‘She was continued on Protonix over the course of her hospital stay …’ and ‘5. Protonix 40 mg p.o. q.d.’. In the dataset, the two Protonix mentions are annotated as a coreference pair.
Another case is related to the filtering rule for different subsections (F.2.5). Based on the annotation guidelines, treatments under the ‘admission’ and the ‘discharge’ subsections should be considered separate. However, there are also some instances where the same treatments under both subsections are annotated as coreference pairs. In the file ‘clinical-777.txt’ of the Beth dataset, ‘amiodarone’ is found in both the ‘medications on admission’ and ‘discharge medications’ subsections, and the two mentions are regarded as a coreference pair.
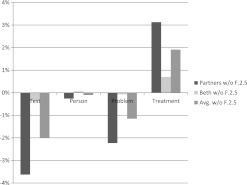
Figure 3 shows the performance gap of the MLN-4 system when we remove F.2.5 from the rule set. As you can see, performance on both the test and problem concepts fell significantly on both datasets. However, the treatment concept performance improved by 1.92% on average. Based on these results, we believe that if interannotator agreement of treatment concepts were improved, treatment concept resolution performance would improve.
Figure 3.
Performance gap when F.2.5 is removed from the MLN-4 system on the test set. w/o, without.
Furthermore, as described in the previous section, our systems relied on rough keywords and simple rules to detect subsection tags. It is legitimate to speculate that, if the discharge summaries were neatly categorized into sections, more efficient coreference resolution results could be achieved.
Ensemble results
We have so far focused on individual ‘best system’ results. Even though our CV results (table 3) show that the MLN-based system outperforms the MR system by as much as 5.21% and dominates in almost all concept types, the results on the test set (table 4) show that each system has its strengths on certain concept types. For example, on the Beth dataset, MR-4 achieves the highest F-score on the test/person concepts, but MR-1+model1+2+3 outperforms the others on the treatment concept. Several studies have demonstrated that an ‘ensemble system,’ which combines several systems' outputs, generally outperforms even the best single system.17 These results encouraged us to build an ensemble of the three systems.
We ran an experiment to create an ensemble system made up of the best test results generated by the three systems, MR-4, MLN-4 and MR-1+model1,2,3.. The ensemble chains were produced by combining the output of the three systems into a single ranked list. Rather than comparing similar components between several possible chains and selecting the best one, we break down each chain into coreference pairs to decide which chain a concept belongs to. Please refer to our online supplementary data for the details.
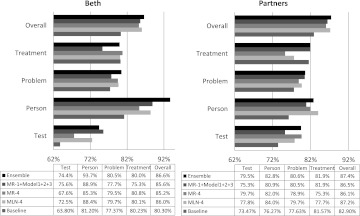
Figure 4 shows the ensemble results on the test set. As you can see, the proposed ensemble method achieves the highest overall F-score—86.6% and 87.4% on the Beth and Partners datasets, respectively. The average F-score of the ensemble system on both datasets is 87.21%, which beats the official mean F-score of 82.7% in the i2b2 coreference resolution track task 1C by 4.51%.
Figure 4.
Ensemble results on the Beth and Partners datasets. MLN, Markov logic network; MR, multistage rule-based.
Significance tests for the four coreference resolution systems
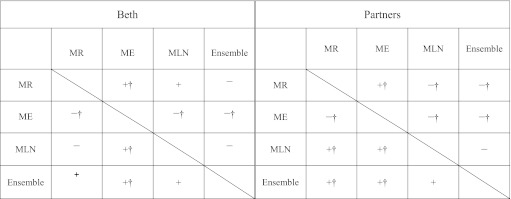
To confirm whether one configuration's performance is better than the other with statistical significance, we applied two-sample t tests by using the datasets DBeth and DPartners, which consists of the training and test sets of Beth and Partners, respectively. A total of 30 unique training/test sets were compiled. The training set of each unique set was compiled by randomly selecting 90% of the datasets (DBeth or DPartners), and the remaining 10% was used as the test set. We summed up the scores for the 30 sets, and calculated the averages for performance comparison. Table 5 shows the results.
Table 5.
Significance tests on both datasets
|
‘+’ or ‘–’ indicates that the performance of the system in this row is better/worse than the system in the intersecting column; ‘†’ refers to the significant difference (p<0.05). For example, in the Beth dataset, the symbol +† is assigned to the cell at the intersection of the ‘Ensemble’ row and the ‘ME’ column, which indicates that the ensemble system significantly outperforms the ME-based system.
ME, maximum entropy; MLN, Markov logic network; MR, multistage rule-based.
The results show that the performance of the ensemble system is superior to any single system in both datasets. Moreover, both the ensemble and the MLN-based systems significantly outperform the MR- and ME-based approaches on the Partners dataset. However, the significant performance improvement was not observed in the Beth dataset.
Conclusion
In this paper, we have described the main challenges in the resolution of coreference relations in patient discharge summaries, including the inconsistencies among summary formats, the analysis of contextual information, such as dosage, quantities, and section titles, as well as clinical domain knowledge. We have proposed several rules to exploit contextual information and presented three approaches to clinical concept coreference resolution. On the basis of our evaluation results on the i2b2 test dataset, our best single system achieves average F-scores of 86% and 87.2% on the Beth and Partners datasets, respectively. Using an ensemble approach combining our three systems, we have demonstrated that performance can be further improved by ∼0.6%, outperforming our baseline system (exact match) and the mean value of the official i2b2 results by ∼4%.
Supplementary Material
Footnotes
Contributors: HJD designed all the experiments and wrote most of this paper. CYC and CYW discussed and refined the paper. CYC wrote the MR-based and the MLN-based coreference resolution systems and conducted all experiments. PTL wrote the ME-based and the ensemble systems. CYW, the biologist in our laboratory, analyzed the coreference resolution results. RTHT and WLH led the whole project.
Funding: The 2011 i2b2/VA challenge and the workshop are funded in part by grant number 2U54LM008748 on Informatics for Integrating Biology to the Bedside from the National Library of Medicine. This challenge and workshop are also supported by resources and facilities of the VA Salt Lake City Health Care System with funding support from the Consortium for Healthcare Informatics Research (CHIR), VA HSR HIR 08-374 and the VA Informatics and Computing Infrastructure (VINCI), VA HSR HIR 08-204 and the National Institutes of Health, National Library of Medicine under grant number R13LM010743-01. MedQuist, the largest transcription technology and services vendor, co-sponsored the 2011 i2b2/VA challenge meeting at AMIA. This work was supported by the National Core Facility Program for Biotechnology, Taiwan (Bioinformatics Consortium of Taiwan, NSC100-2319-B-010-002) and NSC98-2221-E-155-060-MY3.
Competing interests: None.
Patient consent: The data are provided by Partners HealthCare, Beth Israel Deaconess Medical Center (MIMIC II Database), University of Pittsburgh, and the Mayo Clinic. These data are fully deidentified and manually annotated. More information along with the organizing committee and other important information can be found at http://i2b2.org/NLP/Coreference.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data sharing statement: Deidentified clinical records used in this research were provided by the i2b2 National Center for Biomedical Computing funded by U54LM008748 and were originally prepared for the Shared Tasks for Challenges in NLP for Clinical Data organized by Dr Ozlem Uzuner, i2b2 and SUNY.
References
- 1.Morton TS. Coreference for NLP Applications. Proceedings of the 38th Annual Meeting on Association for Computational Linguistics, Hong Kong, China, 1-8 October, 2000 [Google Scholar]
- 2.Uzuner O, Shen S, Forbush T, et al. Evaluating the state of the art in coreference resolution for electronic medical records. J Am Med Inform Assoc 2012;19:786–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Raghunathan K, Lee H, Rangarajan S, et al. A multi-Pass sieve for Coreference Resolution. Proceedings of the 2010 Conference On Empirical Methods In Natural Language Processing. Cambridge, Massachusetts: Association for Computational Linguistics, 2010:492–501 [Google Scholar]
- 4.Richardson M, Domingos P. Markov logic networks. Machine Learn 2006;62:107–36 [Google Scholar]
- 5.Soon WM, Ng HT, Lim DCY. A machine learning approach to coreference resolution of noun phrases. Comput Ling 2001;27:521–44 [Google Scholar]
- 6.Crammer K, Singer Y. Ultraconservative online algorithms for multiclass problems. J Machine Learn Res 2003;3:951–91 [Google Scholar]
- 7.Riedel S. Improving the Accuracy and Efficiency of MAP Inference for Markov logic. Proceedings of the 24th Conference on Uncertainty in Artificial Intelligence (UAI 2008). Helsinki, Finland: AUAI Press, 2008:468–78 [Google Scholar]
- 8.Porter MF. An algorithm for suffix stripping. Program 1980;14:130–7 [Google Scholar]
- 9.Huang M, Zhu X, Hao Y, et al. Discovering patterns to extract protein-protein interactions from full texts. Bioinformatics 2004;20:3604–12 [DOI] [PubMed] [Google Scholar]
- 10.Sung CL, Lee CW, Yen HC, et al. Alignment-based surface patterns for factoid question answering systems. Integrated Computer-Aided Eng 2009;16:259–69 [Google Scholar]
- 11.Banerjee S, Pedersen T. Extended Gloss Overlaps as a Measure of Semantic Relatedness. Proceedings of the 18th International Joint Conference on Artificial Intelligence (IJCAI). Acapulco, Mexico: Morgan Kaufmann Publishers Inc., 2003:805–10 [Google Scholar]
- 12.Winkler WE. String comparator metrics and enhanced decision rules in the Fellegi-Sunter model of record linkage. Proceedings of the Section on Survey Research Methods. SRMS 1990:354–9 [Google Scholar]
- 13.Uzuner O, Forbush T, Shen S, et al. 2011 i2b2/VA co-reference annotation guidelines for the clinical domain. J Am Med Inform Assoc 2011;18:552–621685143 [Google Scholar]
- 14.Vilain M, Burger J, Aberdeen J, et al. A Model-Theoretic Coreference Scoring Scheme. Sixth Message Understanding Conference (MUC-6). Columbia, Maryland, 6-8 November 1995 [Google Scholar]
- 15.Bagga A, Baldwin B. Entity-based Cross-document Coreferencing Using the Vector Space Model. Proceedings of the 17th International Conference on Computational Linguistics—Volume 1. Montreal, Quebec, Canada: Association for Computational Linguistics, 1998:79–85 [Google Scholar]
- 16.Human Language Technology Conference and Conference on Empirical Methods in Natural Language Processing. Proceedings of the Conference, 6-8 October 2005, Vancouver, British Columbia, Canada. The Association for Computational Linguistics, 2005. [Google Scholar]
- 17.Multiple Classifier Systems. First International Workshop, MCS 2000, Cagliari, Italy, 21-23 June 2000. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.