Abstract
Objective
To develop a system to extract follow-up information from radiology reports. The method may be used as a component in a system which automatically generates follow-up information in a timely fashion.
Methods
A novel method of combining an LSP (labeled sequential pattern) classifier with a CRF (conditional random field) recognizer was devised. The LSP classifier filters out irrelevant sentences, while the CRF recognizer extracts follow-up and time phrases from candidate sentences presented by the LSP classifier.
Measurements
The standard performance metrics of precision (P), recall (R), and F measure (F) in the exact and inexact matching settings were used for evaluation.
Results
Four experiments conducted using 20 000 radiology reports showed that the CRF recognizer achieved high performance without time-consuming feature engineering and that the LSP classifier further improved the performance of the CRF recognizer. The performance of the current system is P=0.90, R=0.86, F=0.88 in the exact matching setting and P=0.98, R=0.93, F=0.95 in the inexact matching setting.
Conclusion
The experiments demonstrate that the system performs far better than a baseline rule-based system and is worth considering for deployment trials in an alert generation system. The LSP classifier successfully compensated for the inherent weakness of CRF, that is, its inability to use global information.
Keywords: Text processing, natural language processing, medical records
Introduction
Electronic medical records contain a large reservoir of information describing patients' conditions. This can help physicians make accurate clinical decisions, provide information for medical research, and assist insurance billing. Electronic medical records are in the form of unstructured text, but research in medical information extraction has grown in recent years1–5 and includes the detection of named entities (NEs) and relationships among them6–11 to generate structured information representation from medical records.
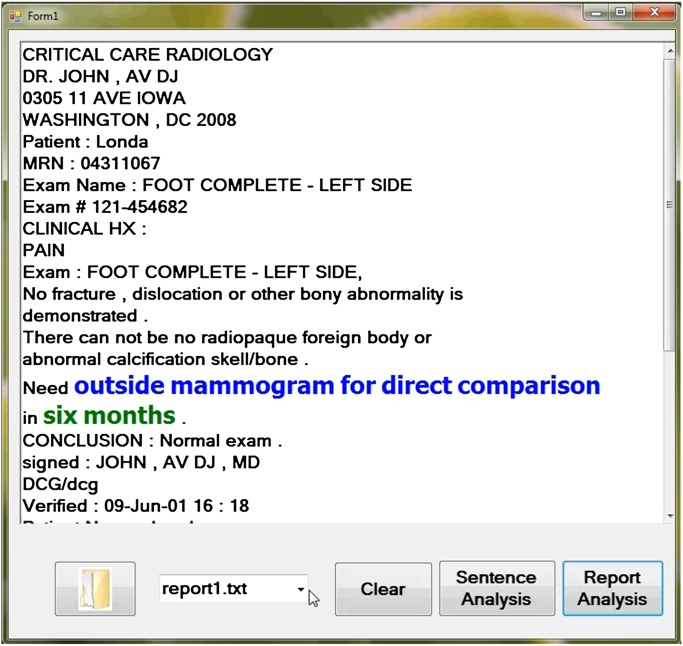
Although previously afforded little attention, a system which automatically generates an alert based on extracted information would be another promising application of medical records processing. In this paper, we focus on a system which automatically recognizes the positions of follow-up and time information in records and could be used as a component of an alert system. We define follow-up information as the information embedded in text that tells patients what they should do next. Such follow-up information includes further examination using a specific detection method such as MRI, CT or biopsy, suggesting comparison with other studies, and advising patients to make follow-up appointments with physicians. An example of follow-up information from a radiology report is shown in figure 1, where the text ‘outside mammogram for direct comparison’ is follow-up information and the phrase ‘six months’ is time information.
Figure 1.
An example of an alert to remind physicians and patients about follow-up information in a timely fashion (‘outside mammogram for direct comparison’ is follow-up information; ‘six months’ is time).
For this study, sentences with follow-up information which do not contain time information are assumed to have the time set to ‘at once,’ while sentences with time information but no follow-up information are disregarded. For simplicity, we call sentences with follow-up information ‘S_FU_I.’ A radiology report typically contains very few S_FU_I. This implies that both physicians and patients take a long time to find follow-up information in a record. It would be very useful if a system could remind patients in a timely fashion of what they have to do, and would significantly reduce the mental burden of physicians and patients.
Such an alert system can be constructed using an information extraction system which detects text fragments or phrases containing information on follow-up and time constraints. Since follow-up phrases usually contain clue words such as specific medical examinations to be conducted (eg, CT, MRI, etc), it might be presumed that a simple system of clue word matching with specialized lexicons would suffice. However, as our experiment in this paper shows, such a simple system produces very noisy results. Three factors make such a system noisy and unfeasible. The first is the presence of ambiguous words. For example, the acronym ‘CT’ is used for computed tomography and also for ‘cerebral tumor’ in radiology reports. The second reason, which is more common, is that the same clue words appear in contexts different from those of follow-up information. For example, while ‘CT’ in ‘the CT scan is recommended in 3 days’ signals follow-up, ‘CT’ in ‘CT scan was performed following angiodynamic bolus of 140cc’ obviously does not. The third is that such domain lexicons require tedious manual efforts to construct and would not be comprehensive in a given medical domain.
To resolve these difficulties, we have to adopt more sophisticated information extraction techniques, which can be developed using two methods. One is to refine a system by adding rules with comprehensive dictionary resources.12 The other is to prepare annotated texts and use machine learning techniques. We adopt the latter approach in this work. Since follow-up information is usually expressed in text as noun phrases (NP) with certain semantic characteristics, we adopt a common technique used in named entity recognizers, namely CRF (conditional random field). We trained CRF by using a large body of training data (20 000 radiology reports) made available by the Microsoft Medical Media Lab. The large size of the training data actually had a significant effect on performance. We further improved the performance of CRF13 14 by using a labeled sequential pattern15–18 (LSP)-based classifier to filter out sentences unlikely to contain follow-up information. In the test stage, while the CRF recognizer uses local contexts to recognize a follow-up phrase and its time constraint in a sentence, the LSP classifier uses global patterns in a sentence to narrow down a set of candidate S_FU_I sentences. More importantly, in the training phase, the LSP classifier disregards a large number of negative examples from the training dataset and thereby improves the consistency of local contexts of positive examples. This process of cleaning-up the training data significantly improved the performance of the CRF recognizer and thereby the performance of the whole system.
The contributions of this work are twofold. First, our work shows that an automatic alert generation system is possible and that such a system can be developed by using machine learning techniques with minimum cost. A large dataset (20 000 radiology reports) was used to test the feasibility of the system in a realistic setting. Our second contribution is a novel and generic method of named entity recognition (NER) which combines an LSP classifier with a CRF recognizer. The method is general enough to be applied to other tasks. In our method, LSP captures global patterns to choose candidate sentences before CRF identifies NEs or relevant phrases. The experiment shows that filtering out a large number of negative examples from the training set by an LSP classifier can significantly improve the performance of a CRF recognizer.
Related work
There have been only a few works on detecting follow-up information in medical records. However, since we formulate the undertaking as an information extraction task, in particular an NER task, we first summarize previous works on NER in the biomedical domain. On the other hand, although we treat the identification of time and temporal information in this paper as a special type of NER, the topic has attracted the interest of researchers and has been studied independently of NER, both in the general and the clinical domains. Time expressions have been intensively studied in terms of their semantic interpretation and inference based on this interpretation. Although interpretation and inference are crucial to our ultimate goal of an automatic alert system, we focus in this paper on the identification of time expressions. We discuss in detail several studies which are directly relevant to our work.
Most NER methods in the general and biology domains have used a CRF framework and provided it with features based on gazetteers or lexicons of the semantic classes (eg, person, location, company, gene, protein, etc) of interest. Li et al19 compared CRF with another common framework, SVM, for various NER tasks in medical texts and demonstrated that CRF outperformed SVM. Using CRF, Settles20 recognized biomedical NEs such as protein, DNA, RNA, cell-line, and cell-type. Their feature sets were composed of orthographic and semantic features. Interestingly, their experiment showed that orthographic features alone achieved performance comparable with that of a semantic features system when the training dataset was large enough. This is a promising result which implies that CRF with a standard set of features can achieve a reasonable performance level.
The i2b2 challenge tasks of concept extraction in 200921 and 201022 were typical of NER in the medical domain. The 2009 challenge21 only treated NEs of a medication class. The winning team23 used a CRF model to identify the boundaries of entities to achieve a performance of 0.8835 (F measure). Another team24 combined a CRF model with a rule-based method to recognize medication names, which showed similar performance. On the other hand, the 2010 challenge22 defined three classes (test, problem, and treatment). Most of participants used CRF as the framework together with feature engineering specific to the types of NEs in the challenge. Nine of the 10 teams in 201022 used CRF models to extract concepts of the three classes. The performance of the best system25 was 0.852 (F measure).
While the type of follow-up in this paper roughly corresponds to a class consisting of the test and treatment classes in the 2010 i2b2 challenge, some follow-up phrases are not necessarily NEs in the conventional sense. For instance, it includes phrases such as ‘outside mammogram for direct comparison.’ Furthermore, although they follow certain rules, time expressions are more productive than conventional classes of NEs. Despite such differences, our work shows that CRF can be applied to the extraction of both follow-up and time information with performance comparable with state-of-the-art systems.
However, our main contribution to NER is a novel technique of combining LSP with CRF to avoid the adverse effects of a large number of negative examples. Although CRF-based recognizers have delivered good performance in diverse tasks, CRF relies heavily on local contexts surrounding NEs and assumes that similar local contexts lead to the same judgments. However, this assumption does not necessarily hold in general. In the current task, a phrase in the ‘conclusion’ section of a report is more likely to be judged as follow-up than the same phrase in a similar local context in a different section such as ‘patient information.’ The training data which contain such seemingly contradictory judgments on the same phrases and contexts confuse a CRF recognizer. The effects of the global context on judgment are observed even among sentences in the same section. The same phrases (ie, phrases which can be follow-ups) in similar local contexts receive different judgments when they appear in sentences with different global characteristics.
In order to remove such contradictory data from the training set, we use an LSP classifier to filter out sentences unlikely to contain follow-up information. LSP has been widely applied to medical information to capture patterns that are somewhat global in nature. Jay et al26 discerned, categorized, and envisaged frequent patterns among patient paths using LSP. Their experiment showed that LSP could mine temporal symbolic data to discover global temporal patterns of patient behavior. Yang et al27 used LSP to detect healthcare fraud and abuse. Two groups of datasets, normal clinical instances and fraudulent clinical instances, were given to a classifier which used features generated by the labeled sequential patterns that LSP produced. Hanauer et al28 gathered time-motion data before and after a computer physician order entry (CPOE) implementation and used LSP to assist and inspect workflow changes associated with CPOE usage. Zheng et al29 automatically extracted hidden user interface navigational patterns from recorded electronic health record data. These LSP applications show that, unlike CRF, LSP is capable of detecting global patterns which are inherent to certain classes.
As for time expressions, their importance in the medical domain has been increasingly recognized. It is important to extract such information when a patient has a medical condition and the length of time they have had it. Mani et al30 and Schilder et al31 shows that time and temporal information has also been extensively studied in the general domain. However, since time expressions are deemed to follow certain linguistic rules which can be captured by simple rules, they have focused more on their semantic interpretation and temporal inferences than on the identification of time expressions. Identification of time expressions has been usually treated by explicit rules. Zhou et al32 and Hripcsak et al,33 for example, proposed a system architecture for a pipeline integrated approach to perform temporal information extraction, representation, and reasoning in medical narrative reports. Although the system is comprehensive regarding the treatment of temporal expressions and inferences in the medical domain, the identification of temporal expressions was carried out by a rule-based system or regular expression. Jung et al34 proposed an end-to-end system to build timelines from unstructured narrative medical records. Their core procedure is also based on explicit rules for logical form pattern-based extraction, concept extraction, temporal expression extraction, event extraction, and timelines creation. Their system relies on deep natural language understanding.
In contrast to these systems, our work pursues the possibility of applying NER techniques to the identification of time expressions. Since time expressions are annotated together with follow-up information, it is natural to leverage these annotations for the identification of time expressions. Bramsen et al35 used a supervised machine-learning framework based on linguistic and contextual features to derive the temporal order for discharge summaries. Their experiment results demonstrate that machine-learning methods can also achieve promising performance for temporal information extraction. Although, in contrast to their study, we do not treat temporal information such as temporal ordering explicitly, our work shows that NER techniques using CRF can achieve reasonable performance with minimum manual intervention.
Among the few systems in the medical domain which treat time expressions, the study by Denny et al12 is most relevant to our work. They proposed timing and status descriptors colonoscopy testing. While they used the KnowledgeMap concept identifier to extract colonoscopy concepts, they developed a rule-based method with regular expressions to extract time descriptors and normalized them. Among their six types of status indicator (‘Scheduling,’ ‘Considering,’ ‘Discussion,’ ‘In need of,’ ‘Receipt,’ and ‘Refusal’), ‘Scheduling’ and ‘In need of’ are very similar to ‘follow-up’ in our task. The two systems differ in the sense that, while we use machine learning intensively, they rely on meticulous manual rule writing. Since our ultimate goal of an alert system needs to have more explicit understanding of temporal information than the current system, we have to integrate our approach in the future with their type of approach.
The proposed method
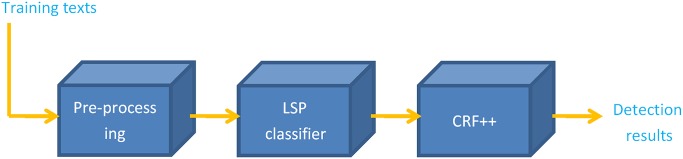
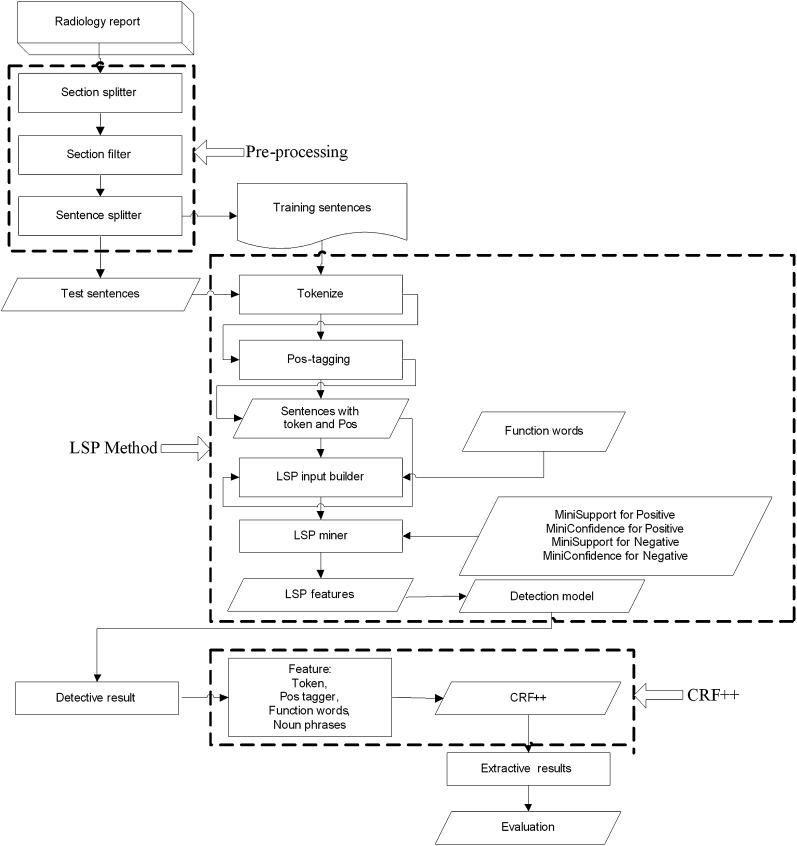
It is well known that the larger the training dataset, the better the performance of a CRF-based NER. However, this is the case only when the judgments in the training data are consistent in terms of local contexts. The training dataset must be kept clean by removing training data which give different interpretations to the same local contexts. In our case, since we have a disproportionally large number of negative examples of S-FU-I which always give a negative interpretation to any local context, it is important to disregard them from the training data for CRF. All sentences in sections such as ‘patient information’ should be removed. Furthermore, we use an LSP classifier to filter out irrelevant sentences in the sections which may contain S_FU_I as well. The proposed system consists of: (1) preprocessing unstructured medical reports and filtering out irrelevant sentences in certain pre-defined sections; (2) finding a set of S_FU_I candidates by using an LSP classifier; and (3) identifying follow-up phrases and their time constraints in candidate S_FU_I by using a CRF recognizer. Figures 2 and 3 show the general and detailed flow diagrams of the overall system. The same filtering steps (ie, by sections and by the LSP classifier) are used to create the training data for the CRF recognizer.
Figure 2.
Flow diagram of the proposed method.
Figure 3.
Detailed flow chart of the overall method.
Preprocessing radiology reports
The preprocessing steps are similar to those found in previously published literature.36 The overall preprocessing steps consist of section splitter, section filter, sentence splitter, sentence tokenizer, and part-of-speech (POS) tagger and NP finder.
Section splitter and section filter
A radiology report consist of several sections: ‘patient information,’ ‘exam,’ ‘clinical diagnosis,’ ‘impression,’ ‘conclusion,’ ‘radiologist signature,’ and so on. Among these sections, follow-up information appears in ‘exam,’ ‘impression,’ and ‘conclusion,’ so only these sections are passed to the next processing stages.
Processing by standard NLP tools
All sections are split into units of sentences, followed by tokenization (recognition of word boundaries), POS tagging, and NP recognition. POS tagging assigns POS information to each word in a sentence. We use a POS tagger developed by Schmid.37 An NP chunker recognizes NPs in a sentence based on POS tags assigned by the previous step. We use an NP chunker developed by Microsoft Research Redmond. While we use these specific NLP tools for POS tagging and NP chunking, any equivalent tools, which nowadays are readily available, would do the same job. The POS tags and NP chunk markers are to be used as features by the following major processing steps carried out by LSP and CRF.
LSP classifier to generate a set of S_FU_I candidates
The LSP classifier divides the dataset into two categories. One is a set of sentences which are likely to be S_FU_I, while the other is a set of the remaining sentences. In the training phase of CRF, only the former set of sentences is used. In the recognition phase, LSP filters out irrelevant sentences and passes only the former set to the CRF recognizer.
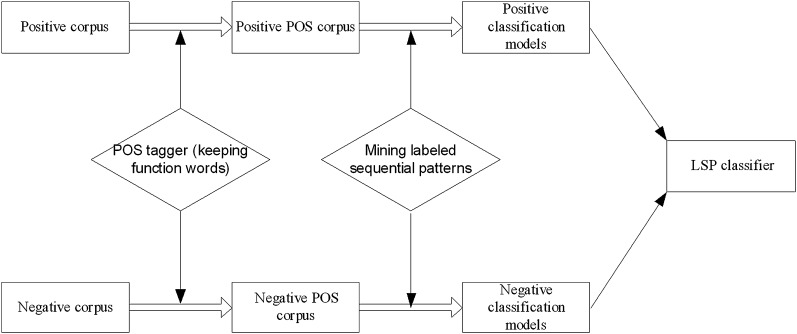
The LSP classifier we constructed has an architecture similar to that in Cong et al18 (see figure 4). The classifier itself is an SVM classifier which uses two set of features (ie, positive and negative sets), each of which corresponds to a set of patterns of POS tags generated by LSP. The training data set created by the previous stage (ie, sentences with POS tags in the three sections, ‘exam,’ ‘impression,’ and ‘conclusion’) is divided into two sets, one of which is a set of positive sentences (ie, S_FU_I) and the other a set of the rest. LSP is applied to each of these two sets to mine the characteristic POS patterns of each set. The existence of a pattern is used as a binary feature for the SVM classifier.
Figure 4.
Follow-up information LSP classifier.
The LSP classifier is an SVM classifier which uses binary features, each of which corresponds to a pattern mined by two LSPs. A set of patterns is mined for the positive set of S_FU_I and another for the negative set. A binary feature is set to 1 when the corresponding pattern exists in input. The number of patterns mined is controlled by the four parameters of MiniSupport for Positive (MPS), MiniConfidence for Positive (MPC), MiniSupport for Negative (MNS), and MiniConfidence for Negative (MNC) given to the two LSPs. MPS and MPC are threshold parameters for mining the positive set and are the minimum percentage of the known support and the minimum percentage of the known confidence, respectively. MNS and MNC are the corresponding parameters for mining the negative set (see Cong et al18 for details).
CRF recognizer for extracting follow-up and time information in sentences
We use a standard set of features with a set of clue words specific to this task (see the Experiment section for details). As CRF, we use the package CRF++ (available from http://crfpp.sourceforge.net/).
For the purpose of comparison, two baseline systems are constructed. Since we are interested in the performance of the combined system of LSP and CRF, both baseline systems use the same preprocessing stage as the proposed system.
Baseline system I: simple rule-based system
Simple observation of the dataset reveals that most S_FU_I contain one of a restricted set of verbs. The set consists of ‘recommend,’ ‘suggest,’ ‘correlate,’ ‘evaluate,’ ‘advise,’ ‘plan,’ ‘follow-up,’ and ‘follow’ and 2972 (74.36%) of 3997 follow-up sentences contain these verbs. In these sentences, the follow-up phrases appear as the object of these verbs, that is, when a verb is active, the NP following the verb is the follow-up phrase (eg, ‘Recommend CT’). On the other hand, when a verb is passive, the NP preceding the verb is the follow-up phrase (eg, ‘CT is recommended’). An NP is recognized by the SharpNLP parser.38 We prepared a set of regular expressions to identify time constraints.
Baseline system II: CRF system
The CRF system is the same as the proposed system, except that this baseline system does not use the LSP classifier. The same CRF recognizer was trained by the entire set of 121 748 sentences instead of a set chosen by the LSP classifier.
Experiment
Four experiments were designed to evaluate the feasibility of the proposed system for extracting follow-up and time information, to analyze performance compared with two baseline systems, to assess the impact of the size of the training set, and to determine the effect of different feature sets on the CRF recognizer.
Dataset
A large dataset containing 20 000 clinical radiology reports was made available by the Microsoft Medical Media Lab. These reports were generated from April 1, 2000 to May 1, 2000. A total of 121 748 sentences were collected from the three sections, ‘exam,’ ‘impression’ and ‘conclusion.’ There were 1 239 994 word tokens in the 121 748 sentences. Only 3997 of the 121 748 sentences contained follow-up information, manually judged as such by a medical doctor and double-checked by one of the authors of this paper. Annotation is performed by following a simple guideline on NE boundaries. For example, we decided not to include a preposition preceding a time expression such as ‘after’ in ‘after six months.’ Regarding the semantics of the follow-up class, we did not provide any formal guideline. Instead, the two annotators shared examples during the course of annotation and resolved disagreements. To check the quality of annotation, we randomly selected 50 new radiology reports containing follow-up information, and asked the same two annotators to annotate them independently. Inter-annotation agreement (κ) between the two annotators was 0.9578, which indicates that annotation was reliable. Unlike other tasks such as coreferences (where the inter-annotator κ is 0.7202 in the Mayo dataset and 0.4072 in the UPMC dataset39) and sentiment analyses (where the inter-annotator κ is 0.54640) in the 2011 i2b2 challenge, annotation of follow-up information was easily judged and stable across the two annotators.
Evaluation method
System performances were evaluated using the three standard performance metrics: precision (P), recall (R), and F measure (F). Since both follow-up and time information are usually expressed by phrases, two criteria for correct recognition are defined. One evaluation setting is based on exact matching which requires the recognized span of expression to be exactly equal to the manually labeled span in a sentence. Another setting is based on inexact matching which require the two spans, the automatically recognized span and the manually recognized span, to overlap.
For training and testing, we followed the standard leave-one-out method and the cross-validation method.41 We used sixfold cross-validation in all experiments. The averaged metrics of evaluation were computed.
Experiment results
The proposed method of combining an LSP classifier with a CRF recognizer has been quantitatively evaluated on 20 000 radiology reports. As shown in table 1, all three metrics in the two evaluation settings exceed 0.85 and 0.90 in the exact matching and inexact matching settings, respectively. The F measure in the inexact matching setting is 0.95. The performance of two types of follow-up information and time are also listed in table 1. For the time type, the F measure is 0.98 in our method compared with an F measure of 0.93 in the work by Denny et al.12
Table 1.
Performance metrics with precision (P), recall (R), and F measure (F) using the baseline method (Baseline I), the CRF method (Baseline II) and the proposed method
| Method | Exact matching | Inexact matching | ||||
| P | R | F | P | R | F | |
| Baseline I | 0.81 | 0.58 | 0.68 | 0.89 | 0.63 | 0.74 |
| Baseline II | 0.90 | 0.80 | 0.85 | 0.98 | 0.87 | 0.92 |
| Proposed | 0.90 | 0.86 | 0.88 | 0.98 | 0.93 | 0.95 |
| Follow-up | 0.89 | 0.85 | 0.87 | 0.97 | 0.92 | 0.94 |
| Time | 0.98 | 0.98 | 0.98 | 0.98 | 0.98 | 0.98 |
Bold values indicate the best performance.
For the purposes of comparison, we list three methods in table 1: our method, baseline system I, and baseline system II.
Baseline system I: simple rule-based system
The performance of such a simple system is low (see table 1) with an upper-bound of recall of 74.36%. In order to exceed this upper-bound, we have to provide rules which can identify S_FU_I such as ‘If avascular necrosis is suspected, MRI of both hips would be more sensitive for this,’ etc. To write the many rules required for recognizing such an implicit S_FU_I would be difficult. The fact that our system exceeds this upper-bound by a large margin indicates that the machine-learning approach processes implicit S_FU_I very well.
Baseline system II: CRF system
As shown in table 1, the CRF system underperformed in comparison with the proposed system, by 0.03 (F measure) in both the exact and inexact settings. In other words, the system with the LSP classifier reduces errors in the inexact matching setting by 6%. Before discussing why such a significant error reduction was achieved by the LSP classifier, we first examine the performance of the LSP classifier itself.
LSP classifier
Table 2 demonstrates how the performance of the LSP classifier changes depending on the parameters. Since the CRF works only on the set of sentences generated by the LSP classifier, the recall of the LSP classifier should be high. We set MPS, MPC, MNS, and MNC to 0.01, 0.65, 0.01, and 0.65, respectively. The recall of the LSP classifier is 0.95 with these parameters.
Table 2.
Performance metrics for the LSP experiment
| LSP performance | LSP parameters | Whole system performance | |||||||
| P | R | F | MPS | MPC | MNS | MNC | F_Exact | F_Inexact | |
| Test 1 | 0.83 | 0.90 | 0.86 | 0.01 | 0.85 | 0.01 | 0.85 | 0.85 | 0.92 |
| Test 2 | 0.83 | 0.90 | 0.86 | 0.02 | 0.85 | 0.02 | 0.85 | 0.85 | 0.92 |
| Test 3 | 0.84 | 0.89 | 0.86 | 0.03 | 0.85 | 0.03 | 0.85 | 0.84 | 0.91 |
| … | |||||||||
| … | |||||||||
| Test 36 | 0.80 | 0.91 | 0.85 | 0.06 | 0.70 | 0.06 | 0.70 | 0.87 | 0.94 |
| Test 37 | 0.80 | 0.91 | 0.85 | 0.07 | 0.70 | 0.07 | 0.70 | 0.87 | 0.94 |
| Test 38 | 0.81 | 0.91 | 0.86 | 0.08 | 0.70 | 0.08 | 0.70 | 0.87 | 0.94 |
| Test 39 | 0.81 | 0.90 | 0.85 | 0.09 | 0.70 | 0.09 | 0.70 | 0.86 | 0.93 |
| Test 40 | 0.81 | 0.90 | 0.85 | 0.10 | 0.70 | 0.10 | 0.70 | 0.86 | 0.93 |
| Test 41 | 0.75 | 0.94 | 0.83 | 0.01 | 0.65 | 0.01 | 0.65 | 0.88 | 0.95 |
| Test 42 | 0.75 | 0.94 | 0.83 | 0.02 | 0.65 | 0.02 | 0.65 | 0.88 | 0.95 |
| Test 43 | 0.75 | 0.93 | 0.83 | 0.03 | 0.65 | 0.03 | 0.65 | 0.88 | 0.95 |
| Test 44 | 0.76 | 0.93 | 0.84 | 0.04 | 0.65 | 0.04 | 0.65 | 0.88 | 0.95 |
| … | |||||||||
| Test 50 | 0.76 | 0.93 | 0.84 | 0.10 | 0.60 | 0.10 | 0.60 | 0.87 | 0.94 |
Bold values indicate the best performance.
LSP, labeled sequential pattern classifier; F_Exact: F measure in the exact matching; F_Inexact: F measure in the inexact matching; MNC, MiniConfidence for Negative; MNS, MiniSupport for Negative; MPC, MiniConfidence for Positive; MPS, MiniSupport for Positive.
Effect of dataset size on performance
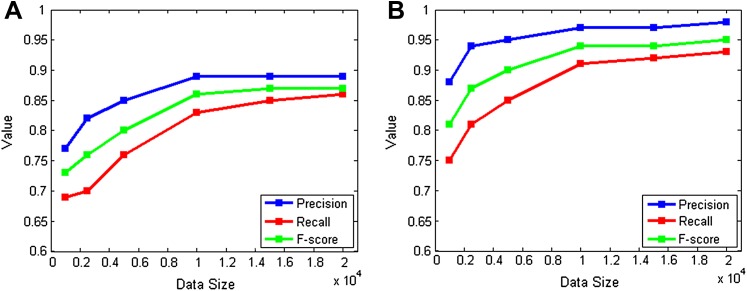
We used datasets of six different sizes: 1000, 2500, 5000, 10 000, 15 000, and 20 000. The results are given in table 3 and figure 5. This experiment shows that, when a larger dataset is used, a system based on machine learning techniques outperforms the simple rule-based system.
Table 3.
Precision (P), recall (R), and F measure (F) with various training datasets
| Dataset size | Exact matching | Inexact matching | ||||
| P | R | F | P | R | F | |
| 1000 | 0.77 | 0.69 | 0.73 | 0.88 | 0.75 | 0.81 |
| 2500 | 0.82 | 0.70 | 0.76 | 0.94 | 0.81 | 0.87 |
| 5000 | 0.85 | 0.76 | 0.80 | 0.95 | 0.85 | 0.90 |
| 10 000 | 0.90 | 0.84 | 0.87 | 0.97 | 0.91 | 0.94 |
| 15 000 | 0.90 | 0.85 | 0.87 | 0.97 | 0.92 | 0.94 |
| 20 000 | 0.90 | 0.86 | 0.88 | 0.98 | 0.93 | 0.95 |
Figure 5.
(A) Results with the exact matching settings; (B) results with the inexact matching settings.
Features used in the CRF recognizer
In all experiments, we use a set of features which can be easily derived (see table 4). The features we use are ‘tokens’ (ie, actual words in a sentence), ‘POS-tags,’ ‘clue-words’ (ie, the same set of verbs such as ‘recommend,’ ‘suggest,’ etc), and ‘noun phrases.’ We avoided sophisticated feature engineering which involves time-consuming trial-and-error processes.
Table 4.
Precision (P), recall (R), and F measure (F) with different feature combinations
| Exact matching | Inexact matching | |||||
| P | R | F | P | R | F | |
| token | 0.82 | 0.80 | 0.81 | 0.89 | 0.88 | 0.88 |
| token+POS_tag | 0.85 | 0.84 | 0.84 | 0.93 | 0.91 | 0.92 |
| token+clue_words | 0.86 | 0.83 | 0.84 | 0.94 | 0.89 | 0.91 |
| token+noun_phrases | 0.86 | 0.80 | 0.83 | 0.94 | 0.87 | 0.90 |
| token+POS_tag+clue_words | 0.89 | 0.86 | 0.87 | 0.97 | 0.91 | 0.94 |
| token+POS_tag+ noun_phrases | 0.88 | 0.85 | 0.86 | 0.96 | 0.91 | 0.93 |
| token+clue_words+noun_phrases | 0.88 | 0.85 | 0.86 | 0.96 | 0.91 | 0.93 |
| token+POS_tag+clue_words+ noun_phrases | 0.90 | 0.86 | 0.88 | 0.98 | 0.93 | 0.95 |
Discussion
Effects of the LSP classifier
The current version of the LSP classifier disregarded an average of 19 448 (116 685) sentences in the sixfold validation of 20 291 (121 748) sentences in the three sections. The LSP classifier wrongly filters out an average of 34 (200) sentences in the sixfold validation (recall is 0.95). Compared with baseline system II, despite 34 (200) sentences being wrongly filtered out, the recall of the whole system improves from 0.80 to 0.86 in the exact matching setting and from 0.87 to 0.93 in the inexact matching setting. This is because the LSP classifier correctly disregards 19 414 (116 485) negative sentences from the training dataset. This filtering improves the local consistency of positive examples, and as a result, the CRF recognizer achieves better generalization, which leads to higher recall.
The filtering improves the efficiency as well. The entire set of 20 000 reports can be processed in 349 452 ms by our system (processor: Intel Core Quad CPU Q9400 at 2.66 GHz, RAM: 2.00 GB). A large portion of the processing time (326 305 ms) is used by the LSP classifier. However, the number of sentences to be processed by the CRF recognizer is greatly reduced, from 121 748 to 5063. As a result, although baseline system II (ie, the CRF recognizer) takes 713 488 ms to process the entire set of sentences, the processing time of the CRF recognizer in our system is only 23 147 ms. The average time (17.47 ms) to process a report by our system is around half of that of the baseline II system (35.67 ms).
Feature engineering
It is worth pointing out that we minimized feature engineering in this research. The features that we used for the CRF recognizer are all independent of the specific domain and specific text type (radiology reports) and the specific task (extraction of follow-up information), except for the set of eight clue words such as ‘recommend,’ ‘suggest,’ etc. The features used by SVM in the LSP classifier were automatically mined by LSP, without human intervention. Avoiding trial-and-errors in feature engineering contributed to reducing development costs, which were less than one person's monthly salary.
Size of training data
The fourth experiment clearly shows the effect of the size of training data. Proper generalization is achieved with at least 10 000 reports, with performance then reaching a plateau. It is also worth noting that human judgment in this task is stable and as a result, the quality of training data is much better than for other tasks,39 40 which helps the machine learning approach which we took in this study. However, to reach the same level of performance, a rule-based approach would require much more human involvement in inspecting manual annotation.
Conclusion
This paper describes a system to extract follow-up information whose performance is sufficiently good for it to be embedded in an alert generation system for actual use. System performance was tested using a large set of real radiology reports. The relatively low cost of development and the domain/task independent design makes the proposed framework attractive. One can readily apply it to the extraction of follow-up from reports in different domains.
The method of using an LSP classifier to filter out negative examples from training data worked well in this task and significantly improved the performance of a CRF recognizer. Since the method is general, it can be applied to other tasks.
From the practical point of view, we will focus on further improving the performance of the system, in particular recall since much higher recall would be required for an automatic alert generation system.
Acknowledgments
We gratefully acknowledge the assistance of Dr Michael Gillam, Dr Hank Rappaport, Mr P D Singh and Mr Dobroslav Kolev in carrying out this work.
Footnotes
Funding: This work was supported by Microsoft Research Asia (MSR Asia). It was also supported by MSRA eHealth grant, Grant 61073077 from the National Science Foundation of China and Grant SKLSDE-2011ZX-13 from the State Key Laboratory of Software Development Environment of Beihang University in China.
Competing interests: None.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Spyns P. Natural language processing in medicine: an overview. Methods Inf Med 1996;35:285–301 [PubMed] [Google Scholar]
- 2.Friedman C, Shagina L, Lussier Y, et al. Automated encoding of clinical documents based on natural language processing. J Am Med Inform Assoc 2004;11:392–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meystre SM, Haug PJ. Comparing natural language processing tools to extract medical problems from narrative text. AMIA Annu Symp Proc 2005:525–9 [PMC free article] [PubMed] [Google Scholar]
- 4.Meystre SM, Haug PJ. Natural language processing to extract medical problems from electronic clinical documents: performance evaluation. J Biomed Inform 2006;39:589–99 [DOI] [PubMed] [Google Scholar]
- 5.Sager N, Lyman M, Bucknall C, et al. Natural language processing and the representation of clinical data. J Am Med Inform Assoc 1994;2:142–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Friedman C, Alderson PO, Austin JH, et al. A general natural-language text processor for clinical radiology. J Am Med Inform Assoc 1994;1:161–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hripcsak G, Austin JH, Alderson PO, et al. Use of natural language processing to translate clinical information from a database of 889,921 chest radiographic reports. Radiology 2002;224:157–63 [DOI] [PubMed] [Google Scholar]
- 8.Huang Y, Lowe HJ, Klein D, et al. Improved identification of noun phrases in clinical radiology reports using a high-performance statistical natural language parser augmented with the UMLS specialist lexicon. J Am Med Inform Assoc 2005;12:275–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bashyam V, Taira RK. A study of lexical behavior of sentences in chest radiology reports. AMIA Annu Symp Proc 2005:891. [PMC free article] [PubMed] [Google Scholar]
- 10.Friedlin J, McDonald CJ. A natural language processing system to extract and code concepts relating to congestive heart failure from chest radiology reports. AMIA Annu Symp Proc 2006:269–73 [PMC free article] [PubMed] [Google Scholar]
- 11.Elkin PL, Froehling D, Wahner-Roedler D, et al. NLP-based Identification of Pneumonia Cases from Free-text Radiological Reports. AMIA Annu Symp Proc 2005:172–6 [PMC free article] [PubMed] [Google Scholar]
- 12.Denny JC, Peterson JF, Choma NN, et al. Extracting timing and status descriptors for colonoscopy testing from electronic medical records. J Am Med Inform Assoc 2010;17:383–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lafferty J, McCallum A, Pereira F. Conditional Random Fields: Probabilistic Models for Segmenting and Labeling Sequence Data. Proceedings of the 18th International Conference on Machine Learning (ICML 2001). Williamstown, MA, USA: International Machine Learning Society, 2001:282–9 [Google Scholar]
- 14.Li X, Wang YY, Acero A. Extracting structured information from user queries with semi-supervised conditional random fields. Proceedings of the 32nd ACM Special Interest Group on Information Retrieval (SIGIR 2009). Boston, MA, USA ACM Special Interest Group on Information Retrieval, 2009:572–9 [Google Scholar]
- 15.Agrawal R, Srikant R. Mining sequential patterns. Proceedings of the 11th International Conference on Data Engineering (ICDE 1995). Taipei, Taiwan: International Conference on Data Engineering, 1995:3–14 [Google Scholar]
- 16.Ren JD, Zhou XL. A new incremental updating algorithm for mining sequential patterns. J Comput Sci 2006;2:318–21 [Google Scholar]
- 17.Sun G, Cong G, Liu X, et al. Mining sequential patterns and tree patterns to detect erroneous sentences. Proceedings of the 22nd Conference on Artificial Intelligence (AAAI 2007). Vancouver, British Columbia, Canada: Association for the Advancement of Artificial Intelligence, 2007:925–30 [Google Scholar]
- 18.Cong G, Wang L, Lin CY, et al. Finding question-answer pairs from online forums. Proceedings of the 31st ACM Special Interest Group on Information Retrieval (SIGIR 2008). Singapore: ACM Special Interest Group on Information Retrieval, 2008:467–74 [Google Scholar]
- 19.Li D, Kipper-Schuler K, Savova G, et al. Conditional random fields and support vector machines for disorder named entity recognition in clinical texts. Proceedings of Natural Language Processing in Biomedicine (BioNLP 2008). Columbus, OH, USA: Association for Computational Linguistics, 2008:94–5 [Google Scholar]
- 20.Settles B. Biomedical named entity recognition using conditional random fields and rich feature sets. Proceedings of Natural Language Processing in Biomedical Applications (NLPBA 2004). Barcelona, Spain: Association for Natural Language Processing in Biomedical Applications, 2004:104–7 [Google Scholar]
- 21.Uzuner O, Solti I, Cadag E. Extracting medication information from clinical text. J Am Med Inform Assoc 2010;17:514–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Uzuner O, South BR, Shen SY, et al. 2010 I2B2/VA challenge on concepts, assertions, and relations in clinical text. J Am Med Inform Assoc 2011;18:552–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Partrick J, Li M. High accuracy information extraction of medication information from clinical notes: 2009 I2B2 medication extraction challenge. J Am Med Inform Assoc 2010;17:524–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Z, Cao Y, Antieau L. Extracting medication information from patient discharge summaries. Proceedings of the Third I2B2 Workshop on Challenges in Natural Language Processing for Clinical Data 2009. Washington DC, USA, 2006 [Google Scholar]
- 25.De Bruiji B, Cherry C, Kiritchenko S, et al. Machine-learned solutions for three stages of clinical information extraction: the state of the art at I2B2 2010. J Am Med Inform Assoc 2011;18:557–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jay N, Herengt G, Albuisson E, et al. Sequential pattern mining and classification of patient path. MEDINFO 2004:1667 [Google Scholar]
- 27.Yang WS, Hwang SY. A process-mining framework for the detection of healthcare fraud and abuse. Expert Syst Appl 2006;31:56–68 [Google Scholar]
- 28.Hanauer DA, Zheng K. Detecting workflow changes after a CPOE implementation: a sequential pattern analysis approach. AMIA Annu Symp Proc 2008:963. [PubMed] [Google Scholar]
- 29.Zheng K, Padman R, Johnson MP, et al. An interface-driven analysis of user interactions with an electronic health records system. J Am Med Inform Assoc 2009;16:228–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mani I, Verhagen M, Wellner B, et al. Machine Learning of Temporal Relations. Proceedings of the 21st International Conference on Computational Linguistics and the 44th Annual Meeting of the Association for Computational Linguistics (ACL 2006). Sydney, Australia, 2006:753–60 [Google Scholar]
- 31.Schilder F, Katz G, Pustejovsky J. Annotating, extracting and reasoning about time and events. Lecture Notes Comput Sci 2007;4795:1–6 [Google Scholar]
- 32.Zhou L, Friedman C, Parsons S, et al. System architecture for temporal information extraction, representation and reasoning in clinical narrative reports. AMIA Annu Symp Proc 2005:869. [PMC free article] [PubMed] [Google Scholar]
- 33.Hripcsak G, Elhadad N, Chen YH, et al. Using empiric semantic correlation to interpret temporal assertions in clinical texts. J Am Med Inform Assoc 2009;16:220–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jung H, Allen J, Blaylock N, et al. Building timelines from narrative clinical records: initial results based-on deep natural language understanding. Proceedings of the 49th Annual Meeting of the Association for Computational Linguistics: Human Language Technologies (ACL 2011). Portland, Oregon, USA, 2011:146 [Google Scholar]
- 35.Bramsen P, Deshpande P, Lee YK, et al. Finding temporal order in discharge summaries. AMIA Annu Symp Proc 2006:81. [PMC free article] [PubMed] [Google Scholar]
- 36.Zeng QT, Goryachev S, Weiss S, et al. Extracting principal diagnosis, co-morbidity and smoking status for asthma research: evaluation of a natural language processing system. BMC Med Inform Decis Mak 2006;6:30–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmid H. Probabilistic part-of-speech tagging using decision trees. Proceedings of the International Conference on New methods in Language Processing (NeMLaP 1994). Manchester, 1994:44–9 [Google Scholar]
- 38.SharpNLP tools. http://sharpnlp.codeplex.com/
- 39.Savova GK, Chapman WW, Zheng J, et al. Anaphoric relations in the clinical narrative: corpus creation. J Am Med Inform Assoc 2011;18:459–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pestian J. Emotional Assignment. I2B2vacincinnati-2011-challenge-I2B2va-track-2@googlegroups.com
- 41.Duda RO, Hart PE, Stork DG. Pattern Classification. 2nd edn New York: John Wiley & Sons, Inc, 2003:389–90 [Google Scholar]