Abstract
Objective
We describe a novel, crowdsourcing method for generating a knowledge base of problem–medication pairs that takes advantage of manually asserted links between medications and problems.
Methods
Through iterative review, we developed metrics to estimate the appropriateness of manually entered problem–medication links for inclusion in a knowledge base that can be used to infer previously unasserted links between problems and medications.
Results
Clinicians manually linked 231 223 medications (55.30% of prescribed medications) to problems within the electronic health record, generating 41 203 distinct problem–medication pairs, although not all were accurate. We developed methods to evaluate the accuracy of the pairs, and after limiting the pairs to those meeting an estimated 95% appropriateness threshold, 11 166 pairs remained. The pairs in the knowledge base accounted for 183 127 total links asserted (76.47% of all links). Retrospective application of the knowledge base linked 68 316 medications not previously linked by a clinician to an indicated problem (36.53% of unlinked medications). Expert review of the combined knowledge base, including inferred and manually linked problem–medication pairs, found a sensitivity of 65.8% and a specificity of 97.9%.
Conclusion
Crowdsourcing is an effective, inexpensive method for generating a knowledge base of problem–medication pairs that is automatically mapped to local terminologies, up-to-date, and reflective of local prescribing practices and trends.
Keywords: Electronic health records, decision support systems, clinical, knowledge bases, medication systems, hospital, data collection, clinical decision support, error management and prevention, evaluation, monitoring and surveillance, ADEs, developing/using computerized provider order entry, knowledge representations, classical experimental and quasi-experimental study methods (lab and field), designing usable (responsive) resources and systems, statistical analysis of large datasets, electronic health records, clinical summarization, user interface, patient preferences, patient-centered, heart failure, psychological, nursing, clinical information systems
Background and significance
Typical electronic health records (EHRs) contain a variety of patient data elements almost universally organized by content type, including medications, laboratory results, problems, allergies, notes, visits, health maintenance items, and many others.1 2 While these elements are necessary, clinical care and medical decision making are most often organized around clinical conditions. For example, a clinician may want to know about all laboratory results, medications, and notes related to a particular patient's diabetes or hypertension. There is significant evidence to suggest that suboptimal presentations of clinical data in clinical information systems can impair medical decision making, contribute to medical errors, and reduce quality.3–6 With the enormous quantity of data housed within the EHR, this can also lead to frustration and inefficiency, often causing important clinical data to be overlooked.7 8 Efforts to simplify the way EHRs provide information to the end-user comprise an important area of investigation.2
Problem-oriented summaries of patients' EHRs may provide a means for optimizing the efficiency, quality, and safety of patient care by clinicians, especially in the setting of large quantities of electronically available data.9–12 The development of such summaries requires knowledge about the inter-relationships between data elements in the record. The most common and salient link type is the ‘treats’ or ‘is treated by’ link between medications and problems. Linking medications and problems enables additional functionality in EHRs, such as problem-oriented presentations of the medication list for clinicians and patients, indication-based prescribing, problem inference, various types of error detection, and more specific clinical decision support.
However, various difficulties exist with current procedures for linking medications and problems. Some standard terminologies and commercial and publicly available knowledge bases are available that contain selected information on medication–problem links.13 14 Development of these knowledge bases is difficult and expensive, and they require considerable maintenance. Similarly, knowledge bases linking other types of necessary clinical data, such as problems linked to laboratory results (ie, ‘diagnoses,’ ‘is diagnosed by,’ ‘is monitored by’), are not currently available. Data mining techniques for inferring such relationships have been proposed, including using association-rule mining to find frequency-based links15–17 or literature-mining approaches to extract links from free-text sources,18–20 but these approaches have important limitations and are generally biased toward more common links, often omitting infrequent links. Clinicians can also be asked to manually link medications or other data elements to problems. However, because such linking by clinicians is frequently optional in clinical systems and can be burdensome, it is most often underutilized, and linking can be incomplete or incorrect.21
We propose a novel crowdsourcing method for inferring medication–problem inter-relationships that takes advantage of manually asserted links between medications and problems. Crowdsourcing, which is defined as outsourcing a task to a group or community of people,22 23 can facilitate rapid generation of a large knowledge base. For example, Wikipedia, a free internet encyclopedia, depends on contributions from the public, and it was found to have accuracy comparable to the Encyclopedia Britannica.24 Crowdsourcing has also been utilized in pharmaceutical research to develop drug discovery resources25 and among patients to share treatment, symptom, progression, and outcome data.26 27 One preliminary report proposes the use of crowdsourcing to create SNOMED CT subsets.28 However, no prior study has utilized crowdsourcing to generate clinical knowledge within an existing clinician workflow.
We utilized crowdsourcing to generate a problem–medication knowledge base. In the described scenario, clinician EHR users represent the community or crowd, and generating problem–medication pairs represents the outsourced task. In this paper, we describe a method for identifying accurate problem–medication pairs obtained through crowdsourcing, which we validated through expert review.
Methods
Study setting
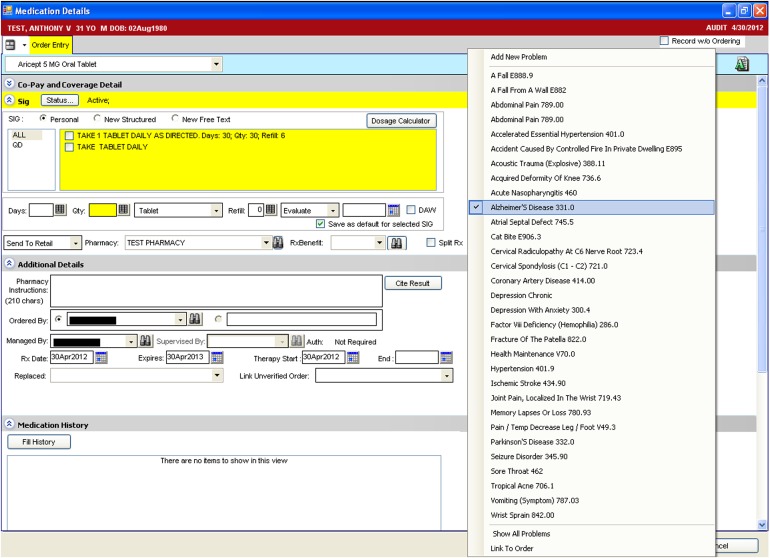
We conducted the study at a large, multi-specialty, ambulatory academic practice that provides medical care for adults, adolescents, and children throughout the Houston community. Clinicians utilized Allscripts Enterprise Electronic Health Record (v11.1.7; Chicago, Illinois, USA) to maintain patient notes and problem lists, order and view the results of laboratory tests, and prescribe medications. Clinicians are required to manually link medications to an indication within the patient's clinical problem list for all medications ordered through e-prescribing (figure 1); however, medications listed in the EHR not added through e-prescribing do not require selection of an indicated problem. Between June 1, 2010 and May 31, 2011, clinicians entered 418 221 medications and 1 222 308 problems for 53 108 patients.
Figure 1.
Sample screen for linking a medication to an indicated problem.
Method development
Identification of problem–medication pairs
We first retrieved medications from the clinical data warehouse that had been entered and linked to one or more problems during the study period. We excluded problem entries with an ICD-9 V code (eg, V70.0—Normal Routine History and Physical), as these concepts are for supplementary classification of factors and are not clinical problems, although they are frequently added to the problem list for billing purposes.
During the study period, 867 clinicians linked 231 223 medications (55.30%) to a problem (239 469 total links). The links included 41 203 distinct problem–medication pairs, comprised of 4903 distinct medications (46.91% of medications ordered at least once) and 4676 distinct problems (20.96% of problems entered at least once). Of the links, 25 434 (61.73%) were asserted more than once, 12 517 (30.38%) were asserted for more than one patient, and 12 996 (31.54%) were asserted by more than one clinician.
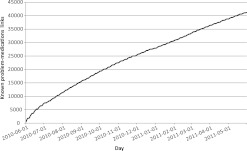
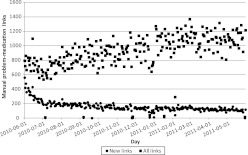
Clinicians asserted a mean of 879.30 links each weekday (656.24 including weekends). The mean number of clinicians asserting a link each day was 172.61 (125.70 including weekends) with an average of 5.09 daily links per clinician (3.63 including weekends). When only counting new links (ie, the first time any clinician linked ‘Metformin 500MG Oral Tablet’ to ‘Type II Diabetes Mellitus’ for any patient), the mean daily rate was 156.45 new links each weekday (112.97 including weekends). Figure 2 illustrates the total number of known problem–medication links each day during the study period. Figure 3 illustrates the number of links asserted by clinicians each day, separately including all links, and those links asserted for the first time.
Figure 2.
Number of known problem–medication links by day during the study period.
Figure 3.
Number of links asserted by clinicians each day during the study period.
Identification of correct links
During preliminary review of randomly selected problem–medication pairs, we identified a number of incorrect links (eg, simvastatin linked to hypertension). Many of these may have been entered in error during e-prescribing, while others may have been clinically justifiable with the linked problem as a secondary, related problem to the prescribed medication. However, our intended use of the knowledge base to summarize problems with indicated medications required the identification of more directly related links. We first explored the use of absolute frequency of manual links as a threshold. Including in the knowledge base only those links that had been asserted for more than one patient resulted in 12 517 distinct problem–medication pairs (30.38% of distinct pairs), accounting for 189 073 total links (78.96% of all links); however, informal review still revealed many erroneous links. Limiting inclusion to only those links asserted for more than 10 patients increased the number of correct links but reduced the number of included pairs to 1756 (4.26% of distinct links), accounting for 121 341 total links (50.67% of all links).
During the development process, we also noted that inclusion of pairs based on absolute frequency did not take into account the baseline probability of various medication–problem pairs occurring. For example, commonly co-occurring but unrelated pairs (eg, simvastatin and hypertension) could be erroneously linked sufficiently often to exceed a fixed threshold, and rare but strongly linked pairs (eg, magnesium and Gitelman syndrome) might not exceed the threshold even though they are correct. Therefore, we supplemented our fixed threshold with a relative threshold; for each problem–medication pair, we divided the number of patients for whom that link had been asserted by the number of patients with both the medication and problem in their record without regard to link. The ratio can be interpreted as the proportion of patients receiving a particular drug and with a particular problem for which a link between the drug and problem has been manually asserted; a ratio of 1.0 indicates that all patients with the specific drug and problem had that link asserted.
We stratified problem–medication pairs into threshold groups using patient link frequency into 1, 2, 3–4, 5–9, or ≥10 and link ratio values into <0.1, 0.1–0.19, 0.2–0.29, 0.3–0.49, or ≥0.5. We then evaluated 100 randomly selected problem–medication pairs from each group to determine a threshold for which links could be considered appropriate to use for inference of new links. An investigator (JAM), blinded to the threshold values, determined for each pair whether each medication was appropriate for use in the treatment of the manually linked problem according to the Lexi-Comp drug database (Wolters Kluwer, Hudson, Ohio, USA), a free-text resource, curated by pharmacists and pharmacology experts, that is commonly utilized by clinicians at our institution to look up drug information. The proportions of appropriate links in each group are shown in table 1. An increase in the number of patient links and the link ratio corresponded positively with link appropriateness.
Table 1.
Appropriateness thresholds for problem–medication link inference
| Link ratio | |||||
| <0.1 | 0.1–0.19 | 0.2–0.29 | 0.3–0.49 | ≥0.5 | |
| Patient link frequency | |||||
| 1 | 57% | 78% | 89% | 87% | 92% |
| 2 | 77% | 94% | 96%* | 96%* | 100%* |
| 3–4 | 77% | 99%* | 96%* | 96%* | 99%* |
| 5–9 | 76% | 98%* | 97%* | 100%* | 99%* |
| ≥10 | 96.8%* | 99%* | 99%* | 100%* | 100%* |
The link ratio is the proportion of patients receiving a particular drug and with a particular problem for whom a link between the drug and problem has been manually asserted.
Groups meeting the 95% or greater appropriateness threshold.
We selected 95% estimated appropriateness (cells indicated with an asterisk in table 1) as the threshold for pairs included in the knowledge base to be used to infer new problem–medication links for medications that were not linked by clinicians. Thus, we included those problem–medication pairs with either a link ratio ≥0.2 and having at least two patient links, a link ratio ≥0.1 and having at least three patient links, or having at least 10 patient links. The resulting knowledge base included 11 166 distinct problem–medication pairs (27.10% of distinct links), which were comprised of 2537 distinct medications and 1575 problems (see supplementary online data). The pairs in the knowledge base accounted for 183 127 total manual links asserted by clinicians (76.47% of all links).
Method validation
Application of the knowledge base
Using the knowledge base of manually linked problem–medication pairs that met the selected threshold, we retrospectively inferred links between co-occurring medications and problems for patients during the study period. For each problem–medication pair, we recorded separately whether a clinician manually linked the pair and whether application of the knowledge base inferred a link between the pair. We determined the number of new links inferred and the number of previously unlinked medications that were linked to a problem with the knowledge base.
Evaluation of the knowledge base
To evaluate the utility of the knowledge base, we randomly selected 100 patients with at least two medications to determine whether the retrospectively inferred links were appropriate and whether unlinked medications should have been linked. Two investigators (MJO, DB) independently reviewed 55 patient charts to identify true medication indications, 10 of which overlapped with the other reviewer (100 total reviewed patients). For each potential problem–medication pair (ie, the Cartesian product of each patient's problem list and medication list), the reviewers indicated whether a link would be appropriate or inappropriate according to a documented indication in the patient's chart or the Lexi-Comp gold standard. Therefore, a medication could have more than one indicated problem, and a problem could have more than one medication involved in its treatment. We calculated inter-rater reliability for the overlapping charts using the κ statistic. A third investigator (JAM) reviewed each case to determine the appropriateness of the link when the reviewers disagreed. With these results, we estimated the sensitivity and specificity of the knowledge base. This study was approved by The University of Texas Health Science Center at Houston's Committee for the Protection of Human Subjects (HSC-SHIS-10-0238).
Results
Inference of problem–medication links
After retrospective analysis using the knowledge base of only those links meeting the threshold criteria, we linked 68 316 previously unlinked medications to problems (36.53% of 186 998 unlinked medications), inferring 229 251 previously unasserted links. Manually linked and automatically linked medications totaled 299 539 (71.62%) linked medications, a difference increase of 16.32% compared to manual links alone.
Of the 118 682 medication instances that could not be linked using the knowledge base, 27 554 (23.22%) corresponded to links that had been previously asserted by a clinician but did not meet the threshold criteria for inclusion in the knowledge base. Of the 91 128 remaining unlinked medication instances, 23 006 (25.25%) were for 5550 distinct medications that had never been linked to a problem by a clinician and were therefore not included in the knowledge base. Also among the remaining unlinked medication instances were 18 452 medications (20.25%) occurring for 4671 patients with no problem list entries; 17 008 of these (4580 patients) were for medications that had been linked previously and were included in the knowledge base.
Accuracy of inferred problem–medication links
The 100 randomly selected patients for evaluation included 11 029 potential problem–medication pairs (669 medications and 1200 problems) with 356 pairs manually linked by clinicians. The knowledge base generated 611 linked pairs; manually linked pairs and knowledge base linked pairs totaled 698 problem–medication links.
Reviewers agreed on the appropriateness of problem–medication pairs for 94.5% of pairs (κ=0.68). After resolving disagreements, the reviewers determined that 726 potential pairs were appropriate links. Compared to expert review, manual links by clinicians achieved a sensitivity of 42.8% and specificity of 99.6%, and links inferred by the knowledge base achieved a sensitivity of 56.2% and specificity of 98.0%. Evaluation of links either asserted manually by clinicians or inferred from the knowledge base found a sensitivity of 65.8% and specificity of 97.9% (table 2).
Table 2.
Comparison of manual links and crowdsourced knowledge base to expert problem–medication link review
| Expert review | Total | |||
| + | – | |||
| Manual links | + | 311 | 45 | 356 |
| – | 415 | 10 258 | 10 673 | |
| Total | 726 | 10 303 | 11 029 | |
| Knowledge base | + | 408 | 203 | 611 |
| Inferred links | – | 318 | 10 100 | 10 418 |
| Total | 726 | 10 303 | 11 029 | |
| Manual links and knowledge base | + | 478 | 220 | 698 |
| Inferred links combined | – | 248 | 10 083 | 10 331 |
| Total | 726 | 10 303 | 11 029 | |
Discussion
We developed a knowledge base for problem–medication links by applying crowdsourcing techniques, collecting manual links asserted by clinicians between medications and clinical problems during e-prescribing. The methods for generating the knowledge base may be easily adopted by other institutions that have implemented EHRs allowing clinicians to link medications to clinical problems, requiring only that the institution extract the links and determine the local threshold for link inclusion in the knowledge base (ie, which patient frequency and link ratio groups had sufficient appropriateness).
The resulting knowledge base of problem–medication pairs has a number of potential uses. First, the knowledge base can assist semi-automation of problem-oriented clinical summaries, which may allow clinicians to provide more efficient, comprehensive care.11 Using only those links with an estimated appropriateness greater than 95% in the retrospective analysis, we increased the number of linked medications at the end of the study period from 55% to 72%, which would result in a more complete summary of patient medications compared to one generated only by manual links. The knowledge base can also be used to suggest links during e-prescribing, reducing clinician workload and potentially increasing the accuracy of linked problems and medications. Finally, the knowledge base can be used to identify undocumented patient problems for those medications that cannot be linked to an entry in a patient's problem list, contributing to the Stage 1 Meaningful Use goal of maintaining an up-to-date problem list of current and active diagnoses.29 Among medications not linked using the knowledge base, 20% were ordered for patients with no problem list entries, and 92% of these medications existed in the knowledge base from which a problem may have been inferred and added to the patient's problem list.
These findings also have significant implications beyond application of the problem–medication knowledge base; the use of crowdsourcing to generate knowledge has revolutionized the creation and use of encyclopedias,24 and it could have similar results in the EHR knowledge domain. One alternative to generating such knowledge is the use of existing knowledge bases, whether open standards-based or proprietary. However, this approach often requires the substantial time-consuming effort of mapping local data elements to the source elements, as EHRs frequently implement coding schemas that differ from standardized terminologies, and the results are still frequently incomplete or inaccurate.30–32 Data mining techniques represent another approach to generating knowledge bases, providing a localized solution for generating problem–medication links. Still, these methods can require large amounts of data and significant computational resources, and may result in a number of spurious associations due to commonly co-occurring but unrelated medications and problems.15–17 Knowledge base development through crowdsourcing is advantageous compared to these existing methods. Like data mining techniques, crowdsourcing allows the development of a knowledge base that uses the system's existing coding schema, eliminating the need to map data elements, a step which alone can be a source of errors. Crowdsourcing also allows the generation of clinical content without the need for external resources, using methods already existing in clinical workflows for a short period of time. Because the knowledge is generated using links asserted by clinicians, accuracy is likely to be high, and methods can be applied to only retain the data most likely to be correct. Further, as clinicians continue to assert links, the appropriateness metrics can be reassessed to include an increased number of links with higher accuracy; thus, the knowledge base improves over time. Finally, clinicians can continuously generate new knowledge as new clinical knowledge is available, such as new medications or new uses for existing medications. This allows an institution to always maintain an up-to-date knowledge base without spending considerable resources for updating an external knowledge base or repeating alternate methods for generating the knowledge.
Limitations
The crowdsourcing method for generating problem–medication links has some limitations. Like data mining methods, resulting knowledge bases may not be easily generalized for adoption in other settings with different underlying clinical terminologies. However, the methods may be more easily reproduced with less computational effort required by implementers. Generation of shared knowledge bases using links asserted by clinicians at multiple institutions may improve the generalizability.
Another limitation is the potential for clinicians to make incorrect links. Our proposed methods for calculating the appropriateness of links at various patient frequency and link ratio thresholds and selection of only those links in threshold groups estimated to be accurate for inclusion in the resulting knowledge base can help overcome this limitation. Further, although we selected an appropriateness cut-off of 95% appropriateness for inclusion of our resulting knowledge base, other knowledge base use cases may require a higher sensitivity or specificity and therefore may require the developer to select an alternate cut-off to meet the specific needs for that application.
Finally, because clinicians are only required to link e-prescribed medications to problems, medications that are added to the patient's chart but never e-prescribed, such as over-the-counter medications or supplements, are likely to be missing from our knowledge base. Combining the crowdsourcing approach with data mining, ontology-based, or other knowledge acquisition methods may contribute to the development of a more complete knowledge base.
Future work
Future work includes improving methods for evaluating the appropriateness of the problem–medication pairs in the knowledge base. An initial approach to improving the appropriateness involves increasing the size of the dataset to include links from multiple institutions, allowing higher frequency restrictions in the evaluation process. In addition to evaluating the pairs based on the patient link frequency and link ratio, we can also evaluate the pairs based on a reputation score of the clinician asserting the link, a metric that has previously been employed in e-commerce and online forum settings. This could be adapted by calculating the percentage of links asserted by a clinician that are shared by other clinicians to predict which clinicians have a percentage of appropriate links meeting the inclusion cut-off, and links asserted by clinicians meeting the cut-off would be included in the knowledge base in addition to those meeting the patient link frequency and link ratio threshold cut-offs.
We can also further this work by expanding the knowledge base to include additional clinical data elements that would also be beneficial in a clinical summary. For example, clinicians are also frequently required to link laboratory test orders to problems within the EHR, and the same methods could be applied to generate a problem–laboratory result pair knowledge base.
Finally, our findings that some manually asserted links are incorrect implicate a variety of scenarios meriting further research. Some incorrect links may persist as juxtaposition errors, where users unknowingly make a wrong selection by selecting the incorrect item that is adjacent to the correct item, which frequently results from poor system design.33 In one example, simvastatin is linked to hypertension instead of hypercholesterolemia when the two are adjacent in a long list of patient problems. Future work can identify the proportion of incorrect links that are related to juxtaposition errors, missing problem list entries, or other entry error, indicating a need for improved user interfaces, interventions to populate the problem list, and clinician education, respectively.
Conclusion
Crowdsourcing is an effective, inexpensive method for generating an accurate, up-to-date problem–medication knowledge base, which healthcare information systems can employ to generate problem-oriented summaries or infer missing problem list items to improve patient safety. Further research may improve the sensitivity of the knowledge base and expand the use of crowdsourcing to other EHR data types.
Supplementary Material
Footnotes
Contributors: ABM had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. ABM, AW, AL, and DFS: study concept and design; ABM, MJO, JAM, and DB: acquisition of data; ABM, AW, AL, MJO, JAM, DB, and DFS: analysis and interpretation of data; ABM, AW, and DFS: drafting of the manuscript; ABM, AW, AL, MJO, JAM, DB, and DFS: critical revision of the manuscript for important intellectual content and final approval.
Funding: This project was supported in part by Grant No. 10510592 for Patient-Centered Cognitive Support under the Strategic Health IT Advanced Research Projects Program (SHARP) from the Office of the National Coordinator for Health Information Technology. The UTHealth Clinical Data Warehouse is supported by NCRR Grant 3UL1RR024148.
Competing interests: None.
Ethics approval: Ethics approval was provided by the Committee for the Protection of Human Subjects at the University of Texas Health Science Center at Houston.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Laxmisan A, McCoy AB, Wright A, et al. Clinical Summarization Capabilities of Commercially-Available and Internally-developed Electronic Health Records. Appl Clin Inform 2012;3:80–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Health IT and Patient Safety: Building Safer systems for Better care - Institute of medicine [Internet]. http://iom.edu/Reports/2011/Health-IT-and-Patient-Safety-Building-Safer-Systems-for-Better-Care.aspx (accessed 9 Nov 2011). [PubMed]
- 3.Han YY, Carcillo JA, Venkataraman ST, et al. Unexpected increased mortality after implementation of a commercially sold computerized physician order entry system. Pediatrics 2005;116:1506–12 [DOI] [PubMed] [Google Scholar]
- 4.Horsky J, Kuperman GJ, Patel VL. Comprehensive analysis of a medication dosing error related to CPOE. J Am Med Inform Assoc 2005;12:377–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koppel R, Metlay JP, Cohen A, et al. Role of computerized physician order entry systems in facilitating medication errors. JAMA 2005;293:1197–203 [DOI] [PubMed] [Google Scholar]
- 6.McCoy AB, Waitman LR, Lewis JB, et al. A framework for evaluating the appropriateness of clinical decision support alerts and responses. J Am Med Inform Assoc 2012;19:346–52 http://www.ncbi.nlm.nih.gov/pubmed/21849334 (accessed 29 Aug 2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ash JS, Berg M, Coiera E. Some unintended consequences of information technology in health care: the nature of patient care information system-related errors. J Am Med Inform Assoc 2004;11:104–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ash JS, Sittig DF, Poon EG, et al. The extent and importance of unintended consequences related to computerized provider order entry. J Am Med Inform Assoc 2007;14:415–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sittig DF, Singh H. Legal, ethical, and financial dilemmas in electronic health record adoption and use. Pediatrics 2011;127:e1042–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sittig DF, Teich JM, Osheroff JA, et al. Improving clinical quality indicators through electronic health records: it takes more than just a reminder. Pediatrics 2009;124:375–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feblowitz JC, Wright A, Singh H, et al. Summarization of clinical information: a conceptual model. J Biomed Inform 2011;44:688–99 [DOI] [PubMed] [Google Scholar]
- 12.Sittig DF, Singh H. Rights and responsibilities of EHR users. CMAJ. Published Online First: 13 February 2012. doi:10.1503/cmaj.111599 [Google Scholar]
- 13.Carter JS, Brown SH, Erlbaum MS, et al. Initializing the VA medication reference terminology using UMLS metathesaurus co-occurrences. Proc AMIA Symp 2002:116–20 [PMC free article] [PubMed] [Google Scholar]
- 14.Elkin PL, Carter JS, Nabar M, et al. Drug knowledge expressed as computable semantic triples. Stud Health Technol Inform 2011;166:38–47 [PubMed] [Google Scholar]
- 15.Wright A, Chen ES, Maloney FL. An automated technique for identifying associations between medications, laboratory results and problems. J Biomed Inform 2010;43:891–901 [DOI] [PubMed] [Google Scholar]
- 16.Brown SH, Miller RA, Camp HN, et al. Empirical derivation of an electronic clinically useful problem statement system. Ann Intern Med 1999;131:117–26 [DOI] [PubMed] [Google Scholar]
- 17.Zeng Q, Cimino JJ, Zou KH. Providing concept-oriented views for clinical data using a knowledge-based system: an evaluation. J Am Med Inform Assoc 2002;9:294–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen ES, Hripcsak G, Xu H, et al. Automated acquisition of disease drug knowledge from biomedical and clinical documents: an initial study. J Am Med Inform Assoc 2008;15:87–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kilicoglu H, Fiszman M, Rodriguez A, et al. Semantic MEDLINE: A Web Application for Managing the Results of PubMed Searches. Turku, Finland: Proceedings of the Third International Symposium for Semantic Mining in Biomedicine (SMBM), 2008:69–76 [Google Scholar]
- 20.Duke JD, Friedlin J. ADESSA: a real-time decision support service for delivery of semantically coded adverse drug event data. AMIA Annu Symp Proc 2010;2010:177–81 [PMC free article] [PubMed] [Google Scholar]
- 21.Sweidan M, Williamson M, Reeve JF, et al. Evaluation of features to support safety and quality in general practice clinical software. BMC Med Inform Decis Mak 2011;11:27 [Google Scholar]
- 22.Tapscott D. Wikinomics: How Mass Collaboration Changes Everything. New York: Portfolio, 2006 [Google Scholar]
- 23.Howe J. The rise of crowdsourcing. Wired magazine 2006;14:1–4 [Google Scholar]
- 24.Giles J. Internet encyclopaedias go head to head. Nature 2005;438:900–1 [DOI] [PubMed] [Google Scholar]
- 25.Ekins S, Williams AJ. Reaching out to collaborators: crowdsourcing for pharmaceutical research. Pharm Res 2010;27:393–5 [DOI] [PubMed] [Google Scholar]
- 26.Hughes S, Cohen D. Can online consumers contribute to drug knowledge? a mixed-methods comparison of consumer-generated and professionally controlled psychotropic medication information on the internet. J Med Internet Res 2011;13:e53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brownstein CA, Brownstein JS, Williams DS, 3rd, et al. The power of social networking in medicine. Nat Biotechnol 2009;27:888–90 [DOI] [PubMed] [Google Scholar]
- 28.Parry DT, Tsung-Chun T. Crowdsourcing Techniques to Create a Fuzzy Subset of SNOMED CT for Semantic Tagging of Medical Documents. 2010 IEEE International Conference on Fuzzy systems (FUZZ). Barcelona: IEEE, 2010:1–8 doi:10.1109/FUZZY.2010.5584055 [Google Scholar]
- 29.Blumenthal D, Tavenner M. The “meaningful use” regulation for electronic health records. N Engl J Med 2010;363:501–4 [DOI] [PubMed] [Google Scholar]
- 30.Elkin PL, Brown SH, Husser CS, et al. Evaluation of the content coverage of SNOMED CT: ability of SNOMED clinical terms to represent clinical problem lists. Mayo Clin Proc 2006;81:741–8 [DOI] [PubMed] [Google Scholar]
- 31.Burton MM, Simonaitis L, Schadow G. Medication and indication linkage: a practical therapy for the problem list? AMIA Annu Symp Proc 2008:86–90 [PMC free article] [PubMed] [Google Scholar]
- 32.McCoy AB, Wright A, Laxmisan A, et al. A prototype knowledge base and SMART app to facilitate organization of patient medications by clinical problems. AMIA Annu Symp Proc 2011;2011:888–94 [PMC free article] [PubMed] [Google Scholar]
- 33.Campbell EM, Sittig DF, Ash JS, et al. Types of unintended consequences related to computerized provider order entry. J Am Med Inform Assoc 2006;13:547–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.