Abstract
Objective
The classification of complex or rare patterns in clinical and genomic data requires the availability of a large, labeled patient set. While methods that operate on large, centralized data sources have been extensively used, little attention has been paid to understanding whether models such as binary logistic regression (LR) can be developed in a distributed manner, allowing researchers to share models without necessarily sharing patient data.
Material and methods
Instead of bringing data to a central repository for computation, we bring computation to the data. The Grid Binary LOgistic REgression (GLORE) model integrates decomposable partial elements or non-privacy sensitive prediction values to obtain model coefficients, the variance-covariance matrix, the goodness-of-fit test statistic, and the area under the receiver operating characteristic (ROC) curve.
Results
We conducted experiments on both simulated and clinically relevant data, and compared the computational costs of GLORE with those of a traditional LR model estimated using the combined data. We showed that our results are the same as those of LR to a 10−15 precision. In addition, GLORE is computationally efficient.
Limitation
In GLORE, the calculation of coefficient gradients must be synchronized at different sites, which involves some effort to ensure the integrity of communication. Ensuring that the predictors have the same format and meaning across the data sets is necessary.
Conclusion
The results suggest that GLORE performs as well as LR and allows data to remain protected at their original sites.
Keywords: Logistic regression, grid computing, H-L test, Area under the ROC curve, data mining, machine learning, statistical learning, predictive modeling, statistical learning, privacy technology
Introduction
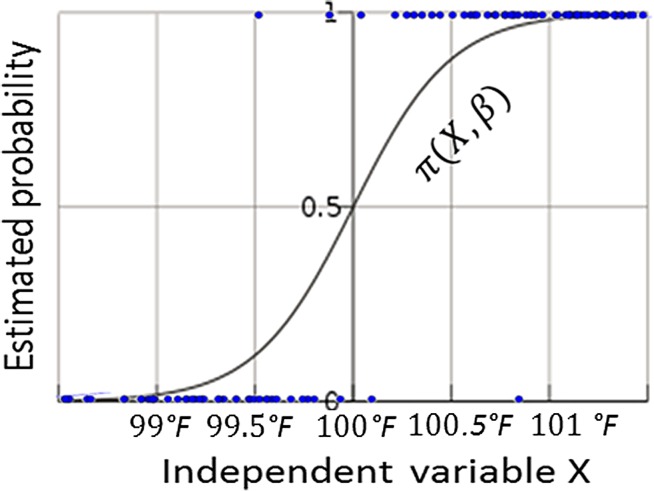
In biomedical, translational, and clinical research, it is important to share data to obtain sample sizes that are meaningful and potentially accelerate discoveries.1 This is necessary to expedite pattern recognition related to relatively rare events or conditions, such as complications from invasive procedures, adverse events associated with new medications, association of disease with a rare gene variant, and many others. Although electronic data networks have been established for this purpose, in the form of disease registries, clinical data warehouses for quality improvement and cohort discovery related to clinical trial recruitment, etc, many of these initiatives are based on federated models in which the actual data never leave the institution of origin, for example, as in the model used at the Clinical Evaluative Sciences in Ontario (ICES), Manitoba Centre for Health Policy (MCHP).2 However, the statistics and predictive models that can be developed in these distributed networks have been very limited, often consisting of simple counts (which still need to be somewhat obfuscated to preserve the privacy of individuals).3 4 Many clinical pattern recognition tasks5–8 are highly complex, involving multiple factors. To support human decision making in complex situations, numerous prediction models9–16 have been developed and applied in a clinical context. Recently, various systems were developed for assisting with tasks as diverse as automatically discovering drug treatment patterns in electronic health records,17 improving patient safety via automated laboratory-based adverse event grading,18 predicting the outcome of renal transplantation,19 guiding the treatment of hypercholesterolemia,11 making prognoses for patients undergoing surgical procedures,20 21 and estimating the success of assisted reproduction techniques.22 Multiple risk calculators for cardiovascular disease prediction are based on the Framingham study.13 Among the most popular prediction models, the logistic regression (LR)23 model is widely adopted in biomedical research, such as the Model for End-stage Liver Disease (MELD)24 and many other clinical applications,25–27 owing largely to its simplicity and the interpretability of the estimated parameters. In an LR model, the independent variables constitute a vector X of several variables that help classify a case into positive or negative as represented by the dependent binary variable Y. In order to do this, the LR model estimates coefficients for each of the dependent variables. For example, the classification of temperature (independent variable X) into ‘fever’ (dependent variable Y) may be done by an LR model and sufficient examples, such that the model ‘discovers’ that temperatures above 38°C (100.4°F) are associated with ‘fever’. The LR is based on a simple sigmoid function (see figure 1) and is backed by information theory,28 which provides a theoretical justification for its power.
Figure 1.
Illustration of a logistic regression model using one-dimensional data (for example, X=body temperature. is a sigmoid function relating temperature and the probability that a particular record contained the word ‘fever’. Dots on the upper and lower horizontal lines correspond to positive (‘fever’) and negative (absence of ‘fever’) observations, respectively. Beta is the estimated parameter.
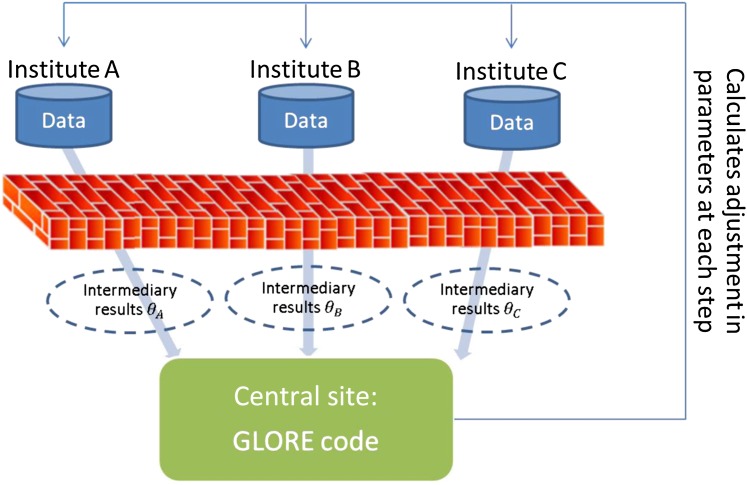
The classic LR model, however, has limitations in operating on federated data sets, or distributed data, since the training phase (ie, learning of parameters) involves looking at all the data, which are usually located at a central repository. Data that are distributed across institutions have to be combined for the classic LR algorithm to work. Although sharing and dissemination can largely improve the power of the analysis,29 it is often not possible to combine distributed data due to concerns related to the privacy of individuals30 31 or the privacy of institutions.32–34 Such a scenario brings new challenges to the classic learning problem of the LR model. Although we, among others, have shown that certain machine learning models like boosting35 and support vector machines36 can be trained in a distributed fashion37–40 and produce accurate models, extending this advantage to LR requires a specialized strategy. A recent work to compute LR with Map-reduce41 is most relevant to our work, but its focus lies in parallelization for computational speed rather than for privacy-preserving data analysis. Furthermore, it does not elaborate on how to provide evaluation indices for these models (eg, Hosmer-Lemeshow goodness-of-fit test or areas under the receiver operating characteristic (ROC) curve (AUCs)) in a distributed fashion. Researchers often refine their models for inclusion or exclusion of some predictors, variable transformations, and other pre-processing steps. Without evaluation strategies that can be privacy-preserving, the value of performing LR in a distributed fashion is very limited. Previous work by other authors in privacy-preserving distributed linear regression was based on vertical partitions of data: multiple data owners each had different attributes for the same observation.42 Our previous work in distributed support vector machines is also related to vertical partitions.40 In this manuscript, we propose a new algorithm, Grid Binary LOgistic REgression (GLORE), to fit a LR model in a distributed fashion using information from locally hosted databases containing different observations that share the same attributes (ie, horizontal partitions of data—stackable sets of patient records), without sharing the sensitive original patient data from these databases, as shown in figure 2. This is not a trivial task: distributed linear regression is much easier to implement than distributed logistic regression, since there is a closed form solution for the former but not for the latter.
Figure 2.
Pipeline for the Grid Binary LOgistic REgression (GLORE) model. Data sets hosted in different institutions (ie, A, B, and C) are processed locally through the same virtual engine (ie, GLORE code) to compute non-sensitive intermediary results, which are exchanged and combined to obtain the final global model parameters at the central site. A similar distributed process is executed for evaluation of the model.
Specifically, GLORE estimates the coefficients of an ordinary LR model by integrating decomposable intermediary results (and not the actual patient data) to calculate model parameters and test statistics. The resulting model is calculated in a privacy-preserving manner and performs as well as classic LR. In the Methodology section, we will discuss details for estimating model coefficients, estimating the variance-covariance matrix of coefficients, performing a goodness-of-fit test, and calculating the AUC in a distributed fashion. Commonly reported statistics for LR, including CIs, Z test statistics and their p values, and ORs can be obtained by using these estimated coefficients and their variance-covariance matrix.
Methodology
The LR model is an instance of a generalized linear model with a logit (ie, ) link function (illustrated in figure 1).
where , and denote probabilities of an event Y to be 1 (ie, ‘fever’) or 0 (ie, absence of ‘fever’), conditioned on observation of a feature vector X (eg, 101°F), respectively. The parameter vector β corresponds to the set of weights or coefficients that need to be estimated and that will be multiplied by the values for the features X (ie, Xβ) to make predictions.
Estimating model coefficients
In order to explain how GLORE works, it is important to remind readers how traditional LR works. To estimate the final value for the parameter vector β, an LR model iteratively maximizes the likelihood of obtaining X given an initial β. At each iteration, the algorithm produces , which is based on the previous . The initial β can start with all coefficients set to zero, eg, the estimated probability of is based on observations of a binary response Y and a feature vector X (ie, a set of predictors) from each of the data sites. To compute the maximum likelihood of getting the observed data, the LR estimation algorithm first finds the derivative of the log likelihood function, then applies the Newton-Raphson method43 to find its maximum, that is, the value of β for which the derivative function equals zero. For details of the log likelihood function, please refer to section 1 of the online supplementary appendix. We also explain how the Newton-Raphson method works in section 4 of the online supplementary appendix.
In each Newton-Raphson iteration for the LR parameter estimation problem, the first and second derivatives of log likelihood function are both decomposable, that is, they can be calculated separately for a subset of observations, and then combined, with the same result as if they were calculated on the complete set. Hence, a GLORE update can be finished by combining intermediary results from across all local sites. Please refer to the online appendix (sections 1 and 2) for technical details on model coefficient estimation.
Because intermediary results from individual sites do not lose any information, GLORE guarantees accurate estimation of parameters through summation. Note that the information exchanged consists of partially aggregated intermediary results rather than the raw data, hence they are more privacy-preserving than would be the case if we transmitted all patient data to a central site.
Furthermore, since the calculations can be done in parallel, each step takes only as long as the maximum time for the sites (ie, the slowest site will determine how long each step takes). The time to transmit the intermediary results is usually negligible, as only one vector of coefficient adjustments needs to be sent. After setting the same initial values, at each iteration, GLORE uses the summation of partial intermediary results from multiple sites to update the coefficients and sends them back to the sites for another iteration.
A central engine is efficient in terms of computation, but the process could occur via peer-to-peer transmittal of intermediary results. Assuming that there are m features (ie, variables) available over multiple sites, at each iteration, intermediary results of a (m+1)×(m+1) matrix (ie, the variance-covariance matrix of coefficients) and a (m+1)-dimensional vector (ie, the gradients for coefficient adjustment) from each site must be transmitted to the central engine or to all other sites, depending on the design choice. The GLORE framework with a central computing node can reduce the probability of network delays when compared to the GLORE framework based on a peer-to-peer architecture.
Estimating the variance-covariance matrix
Besides model coefficients, variance-covariance matrix estimation is also important, as it is necessary for the computation of the CIs of individual predictions.9 Like the model coefficient estimation, it can be done by integrating decomposable partial elements. Please refer to the online supplementary appendix (section 2) for technical details.
Estimating goodness-of-fit test statistics
The Hosmer and Lemeshow (H-L) test23 44 45 is commonly used to check model fit for LR. This section discusses how to perform an H-L test to check for GLORE's fit, without sharing patient data. The null and alternative hypotheses of the H-L test are that (1) ‘the model provides an adequate fit,’ and (2) ‘the model does not fit the data,’ respectively. That is, when the p values for this test are below 0.05, we can reject the hypothesis that the model fits the data well, meaning that the model is not well calibrated. To calculate the H-L statistic, we have to sort cases by their predictions and create groups from which we establish a proxy for the ‘true probability’ (ie, the fraction of positive cases in the group). Let us denote Oj as the number of positive observations in the jth group and Ej as the sum of predictions in the jth group, respectively. The parameter nj refers to the number of records in the jth group. In the box ‘Algorithm 1’ below, we introduce a procedure to obtain the H-L test statistic for GLORE (ie, the C version of this test, that is based on percentiles to determine the groups, which is more robust than the H version of the test that is based on fixed thresholds to determine the groups46) without sharing patient data (ie, individual patient features or individual patient outcomes). In the following algorithm, class labels are not shared with all other sites or the central engine. Instead, only the total number of labeled records with outcome ‘1’ per group from a site needs to be transmitted to the central engine.
Algorithm 1. Computing the H-L statistic for GLORE (C version).
Step 1. Each site transmits probability estimates, that is, 's for their observations to the central engine.
Step 2. The central engine sorts all 's and evenly groups the sorted 's into deciles*, and computes the estimated probability for each decile as the sum of predictions in that decile.
Step 3. The central engine sends the indices of predictions in each decile to their original sites.
Step 4. Each site finds the number of records labeled ‘1’ in each decile among its own records based on the indices from the central engine, and transmits this number to the central engine.
Step 5. The central engine combines the numbers of records labeled ‘1’ from all sites to obtain the total number of records labeled ‘1’ in each category, and, together with the results from step 2, computes the H-L statistic.
*Deciles are commonly used in the H-L C test, but other types of percentiles can be used depending on the size of the data set.
Estimating the area under the ROC curve (AUC)
The AUC47 is widely used to evaluate the discrimination performance of predictive models. To calculate the AUC, it is necessary to estimate the total number of pairs for which positive observations (ie, ‘one’-labeled records) rank higher than negative observations (ie, ‘zero’-labeled records). The box ‘Algorithm 2’ below shows the details of how AUCs are estimated without sharing individual patient features or patient outcomes. Besides transmissions between the central engine and all sites, this algorithm requires peer-to-peer communication to keep patient outcomes protected.
Algorithm 2. Computing the AUC using GLORE.
Step 1. Each site transmits probability estimates, that is, ’s of their observations to all other sites.
Step 2. Each site finds the ranking* of each predicted probability transmitted from all other sites among the zero-labeled records in this site, and sends the rankings of these probabilities back to their original sites.
Step 3. Each site calculates the rank sum for each of its one-labeled records using the ranks sent back from all other sites.
Step 4. Each site finds the ranking* of each of its one-labeled records among the zero-labeled records in this site.
Step 5. Each site computes the sums of the ranks regarding its own one-labeled records using the intermediary results from steps 3 and 4.
Step 6. Each site sends rank sums from step 5 and counts of their own one-labeled and zero labeled records to the central engine.
Step 7. The central engine computes the AUC as the summation of all rank sums from step 6 divided by the product of the total one-labeled and zero-labeled records.
*Ranking is the sum of the number of zero-labeled records with smaller predictions and half the number of zero-labeled records with equal predictions.
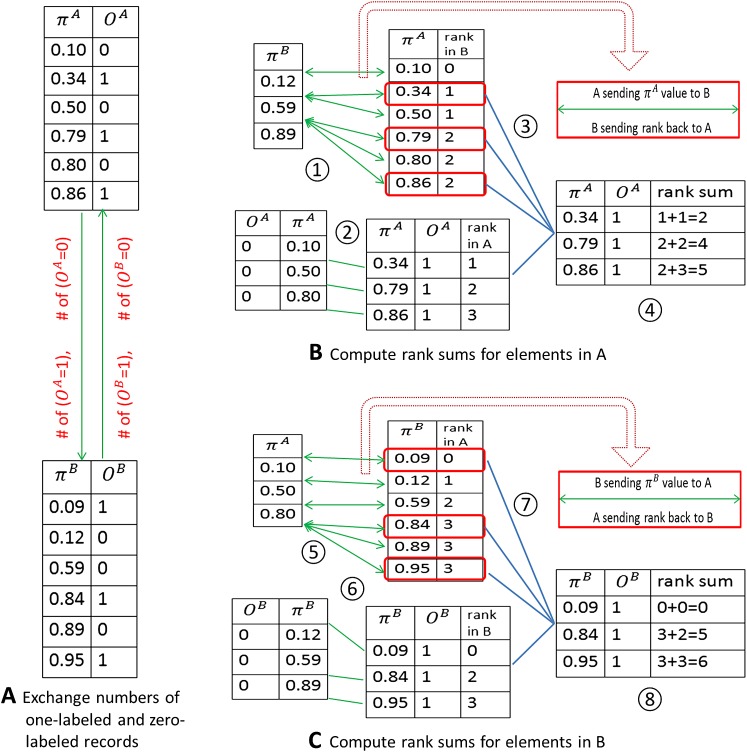
In figure 3 we use a simple artificial example with only two sites (A and B) to explain how algorithm 2 works. and OA indicate predicted probabilities and class labels from site A, and and OB are predicted probabilities and class labels from site B. Note that in procedures 1 and 5, only predicted probabilities (ie, no class labels) are sent to the other site, as in our previous strategy for H-L tests. The AUC, which is equivalent to the c-index,48 can be calculated in three steps: (1) count all one-labeled records that have predictions that are larger than the predictions across all zero-labeled records; (2) count all one-labeled records that have predictions that are equal to the predictions across all zero-labeled records; and (3) sum the counts of step (1) and half the counts of step (2) and divide this number by the total number of pairs formed by zero-labeled and one-labeled records.47
Figure 3.
Calculating the area under the curve (AUC) using GLORE. (A) Exchange numbers of one-labeled and zero-labeled records between site A and site B. (B) Compute rank sums for records in A. 1: Calculate the rank of each probability in A among zero-labeled records in B. 2: Calculate the rank of each one-labeled probability in A among zero-labeled records in A. 3: Find the one-labeled records from step 1 (ie, bounded in red boxes). 4: Combine ranks for one-labeled records from procedures 2 and 3 to get the rank sums for A. (c) Compute rank sums for records in site B. 5: Calculate the rank of each probability in B among zero-labeled records in A. 6: Calculate the rank of each one-labeled probability in B among zero-labeled records in B. 7: Find the one-labeled records from step 5 (ie, bounded in red boxes). 8: Combine ranks for one-labeled records from procedures 6 and 7 to get rank sums for A.
Sometimes, we might also want to display an entire ROC curve instead of calculating a single AUC score. In this case, using as threshold each prediction , a full contingency table (ie, true positive, false positive, true negative, and false negative) can be calculated for each threshold. The ROC curve results from the points (1−specificity, sensitivity) that are calculated from each of these contingency tables. Please refer to Zou et al49 and a review article by Lasko et al50 for details on ROC curves. In GLORE, one site needs to send all predictions and their corresponding contingency tables to the central engine. The central engine then needs to merge the information to compute the sensitivity and specificity at all thresholds. It is worth noting that, although this procedure is straightforward, it may lead to more privacy leak than algorithm 1, since class labels (ie, patient outcomes) associated with predicted probabilities can be recovered by the central or peer site, depending on the strategy selected.
Remark
We verified in both simulated and clinical data experiments that the proposed GLORE will produce the same estimated coefficients, that is, with precision 0 (10−15), together with accurate variance-covariance matrix estimation, goodness-of-fit statistics, and AUC, when compared to the classic LR trained in a centralized manner. It is also worth noting that, although the GLORE coefficient estimation process needs to transmit intermediary results in each Newton-Raphson iteration, usually a small number (<15) of iterations is necessary to achieve high precision such as O(10−6). After the parameter estimation is done, only a one-time data transmission is needed for estimating the variance-covariance matrix, computing the model fit statistic, and computing the AUC.
Experiments
We used the statistic computing language R to conduct our experiments with simulated data in which the true generating model is known, and also on clinical data to validate GLORE.
Simulation study
In a simulation study, we compared two-site GLORE (assuming data are evenly partitioned between two sites) and ordinary LR (combining all data for computation). We used a total sample size of 1000 (500 for each site) and nine features (ie, variables). First, we simulated all features from a standard normal distribution, then simulated the response from a binomial distribution assuming that the log odds of the response being 1 was a linear function of features (ie, all coefficients were set to 1). We conducted the study on 100 runs to compare coefficient estimation difference between GLORE and LR for the same simulated data. This simple study shows that the number of Newton-Raphson iterations to convergence is always six when 10−6 precision is set for the iteration stop criterion.
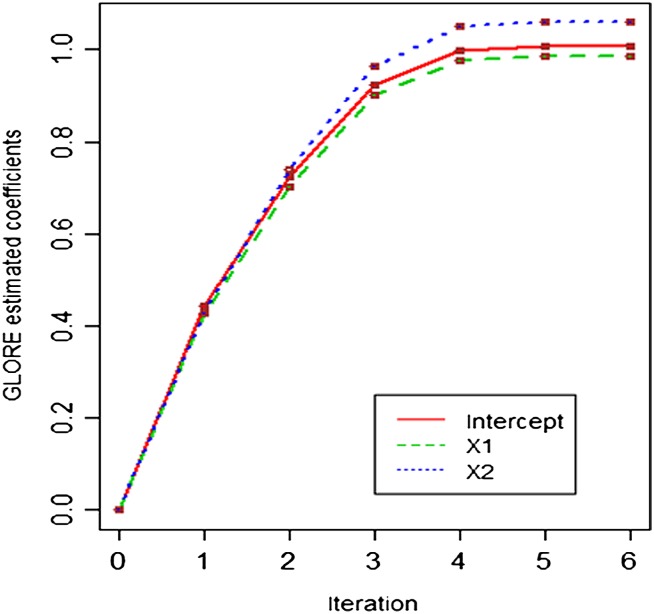
Table 1 shows the mean absolute difference between two-site GLORE and LR estimations for all 10 coefficients (nine features plus one intercept) at each iteration, where the mean is calculated for 100 runs. There are no substantial differences between estimations from GLORE and LR for all coefficients at all iterations. We also pick one of the 100 runs to graphically present the convergence paths of the estimations for three coefficients (intercept, X1, and X2) in figures 3 and 4, showing that there is no difference between two convergence paths for these three coefficients.
Table 1.
Mean absolute difference between two-site GLORE and LR estimations
| Iteration | 1st | 2nd | 3rd | 4th | 5th | 6th |
| Intercept | 7.33E−17 | 3.05E−16 | 2.09E−16 | 1.11E−16 | 8.55E−17 | 8.88E−17 |
| X1 | 4.42E−16 | 3.34E−16 | 2.52E−16 | 1.02E−16 | 8.77E−17 | 8.88E−17 |
| X2 | 5.30E−16 | 3.15E−16 | 2.35E−16 | 1.02E−16 | 8.44E−17 | 8.66E−17 |
| X3 | 4.46E−16 | 2.60E−16 | 2.28E−16 | 1.21E−16 | 7.99E−17 | 9.21E−17 |
| X4 | 3.87E−16 | 3.12E−16 | 2.29E−16 | 1.19E−16 | 7.55E−17 | 8.10E−17 |
| X5 | 4.88E−16 | 3.19E−16 | 2.11E−16 | 1.18E−16 | 8.66E−17 | 8.55E−17 |
| X6 | 4.62E−16 | 3.09E−16 | 2.45E−16 | 1.30E−16 | 8.10E−17 | 7.55E−17 |
| X7 | 4.25E−16 | 2.96E−16 | 2.45E−16 | 1.29E−16 | 9.21E−17 | 8.88E−17 |
| X8 | 4.61E−16 | 3.06E−16 | 2.43E−16 | 1.24E−16 | 6.44E−17 | 7.77E−17 |
| X9 | 4.45E−16 | 3.26E−16 | 2.48E−16 | 1.17E−16 | 7.77E−17 | 8.55E−17 |
Figure 4.
The convergence paths of the two-site GLORE estimations for intercept, X1, and X2. The estimation difference between GLORE and classic LR is smaller than 10−15 for all iterations, as shown in table 1.
Experiments using a clinical data set
Our clinical data set is related to myocardial infarction at Edinburgh, UK,51 which has one binary outcome, 48 features and 1253 records, and was used to illustrate GLORE with two sites. Records are evenly partitioned (627 vs 626) between the two sites. We picked nine non-redundant features in this data set using methods described in Kennedy et al51 for GLORE fitting. We also used another data set,49 which contains two cancer biomarkers (CA-19 and CA-125). This data set has 141 records, one binary outcome denoting the presence or absence of cancer, and two features denoting the two cancer markers. The 141 records were split into 71 and 70 for two sites. Tables 2 and 3 show coefficient estimates and their standard errors, Z test statistics and p values for the Edinburgh data and for the CA-19 and CA-125 cancer marker data, respectively. Using algorithm 1, the H-L test statistic equals 8.036 with a p value 0.430 for the Edinburgh data, and the H-L test statistic equals 3.510 with a p value 0.898 for the CA-19 and CA-125 data, which are no different from the results obtained from traditional centralized LR models. Seven and 12 Newton-Raphson iterations were needed for convergence with 10−6 precision for the Edinburgh data and the CA-19/CA-125 data, respectively. In addition, using algorithm 2, we found that the AUCs were 0.965 and 0.891 for the Edinburgh data and for the CA-19 and CA-125 data, respectively, which are no different from the results obtained from traditional centralized LR models.
Table 2.
Coefficient estimation for the Edinburgh data using two-site GLORE
| Estimation | Standard error | Z value | Pr(>|z|) | |
| Intercept | −4.3485 | 0.2968 | −14.6508 | 0.00E+00 |
| Pain in left arm | 0.1816 | 0.2680 | 0.6777 | 4.98E−01 |
| Pain in right arm | 0.1764 | 0.3061 | 0.5763 | 5.64E−01 |
| Nausea | 0.1323 | 0.3862 | 0.3426 | 7.32E−01 |
| Hypoperfusion | 2.2511 | 0.6590 | 3.4160 | 6.36E−04 |
| ST elevation | 5.5556 | 0.4404 | 12.6150 | 0.00E+00 |
| New Q waves | 4.1453 | 0.6747 | 6.1435 | 8.07E−10 |
| ST depression | 3.4173 | 0.2815 | 12.1392 | 0.00E+00 |
| T wave inversion | 1.2030 | 0.2635 | 4.5649 | 5.00E−06 |
| Sweating | 0.2721 | 0.2510 | 1.0837 | 2.79E−01 |
Table 3.
Coefficient estimation for CA-19 and CA-125 data using 2-site GLORE
| Estimation | Standard error | Z value | Pr(>|z|) | |
| Intercept | −1.4645 | 0.3881 | −3.7739 | 1.61E−04 |
| CA19 | 0.0274 | 0.0085 | 3.2063 | 1.34E−03 |
| CA125 | 0.0163 | 0.0077 | 2.1008 | 3.57E−02 |
Discussion
Our study shows that the proposed GLORE framework can use intermediary results that do not contain patient data from multiple local sites to build an accurate prediction model without increasing the computation cost. This is important in situations in which data cannot leave a particular institution, but the institution still wants to contribute to the development of a predictive model. For the study of rare diseases and many other situations in which polling data from multiple sources has the potential to improve the power of statistical analyses and generalizability of predictive models, developing techniques that allow shared analyses without necessarily allowing collaborating parties to see each other's data is very important. Many authors have reported on federated queries across distributed clinical data warehouses,52 53 and how the results of some of these queries need to be transformed to protect the privacy of the individuals in these data sets.3 Sharing intermediary results calculated at each site is less prone to privacy compromise, although further studies of the privacy risk involved with the use of this strategy are certainly warranted.
In any model-building task, significant pre-processing of data needs to be performed. GLORE should follow the same general guidelines as those recommended for ordinary LR. For example, redundant predictors should be removed; categorical predictors need to be converted to a set of dummy variables based on the number of categorical values (this can be done by statistical software or manually). Furthermore, pre-processing operations done at all sites need to ensure that the resulting data are comparable across the sites.
The data need to be harmonized (at the syntactic and semantic levels) before GLORE can be successfully applied. Data harmonization can be challenging even within a single institution. Creating new data models to suit the purposes of a particular application may be tempting, but researchers should consider the mapping of concepts to an information model and standardized terminology to make the model extensible and the effort in data harmonization reusable. The rate of missing values in a particular site has to stay below an agreed level, and data imputation techniques need to be jointly discussed. Researchers should agree on a minimal set of variables that can be easily mapped and are expected to have sufficient predictive ability or usefulness in adjusting for confounders (as per automated multivariate feature selection procedures and/or expert opinion). It is advisable that the investigators exchange descriptive statistics and test out their environments using artificial data. Expert opinion should be sought as to whether it makes sense to combine data if the data from different institutions were collected under different circumstances and employed different assumptions. A trivial case, for example, occurs when one institution had a binary variable named ‘out-of-control diabetes,’ and another has actual continuous HbA1C values: it is important to determine how ‘out-of-control’ was determined before trying to turn the continuous value for HbA1C into a binary variable. It may not make sense to combine the data if the determination of ‘out-of-control’ was not clear. The security settings need to be agreed upon. Firewalls, authentication protocols, and other security aspects need to be extensively tested before implementation. The robustness of network connections and intermediary result transmission needs to be tested for real-time computation, or an asynchronous process needs to be established.
The combination of data from different sites is often desirable in surveillance systems, in which low signals need to be detected from noisy data with the help of a large number of samples. The proposed system is applicable when the institutions are collaborating in such systems but cannot share individual level data. As noted above, the method is not applicable when it is not possible to harmonize the data across institutions, when the data have too many missing values, when the descriptive statistics suggest strong site-specific patterns that cannot be adjusted for, or when the security and network infrastructures do not allow for reliable real-time computation (although an asynchronous version of the proposed infrastructure could be used in this case).
Limitations
We have not compared the added value of models derived from the combination of data from different sites with models derived from a single site. The combination will only add value if the data from the additional site are representative of the population on which the model will be applied. Additionally, there are communication costs at each iteration, and unreliable connections may lead to interrupted analyses that can only be resumed when connections are restored. Furthermore, the privacy preserving qualities of GLORE have not been fully investigated in a rigorous framework such as differential privacy.54 It is possible that, for very small data sets at a particular institution, privacy can be compromised by releasing partial elements or prediction values obtained at those institutions. This problem would be more exacerbated in the distributed calculation of the H-L test, and even worse for ROC curve plotting, as discussed before.
Conclusion
We showed that a LR model performed in a distributed fashion provides the same results as a conventional LR model performed centrally. This has implications in terms of preservation of individual privacy and may facilitate construction of predictive models across institutions that have limited ability to actually share patient data. GLORE is not a panacea for multiple obstacles that exist for researchers to collaborate, but provides a reliable solution for the problem of having too few cases to construct and evaluate a predictive model at a single institution.
Supplementary Material
Acknowledgments
We thank Dr. Hamish Fraser and Dr. Kelly Zou for providing the clinical data, and Mr. Kiltesh Patel for helpful discussions. We thank two anonymous reviewers for their feedback, which helped us improve the original manuscript.
Footnotes
Contributors: YW and LOM contributed equally to the writing of this article. The other authors are ranked according to their contributions.
Funding: The authors were funded in part by NIH grants R01LM009520, U54HL108460, R01HS019913, and UL1RR031980.
Competing interests: None.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Ohno-Machado L, Bafna V, Boxwala AA, et al. iDASH: integrating data for analysis, anonymization, and sharing. J Am Med Inform Assoc 2012;19:196–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Willison DJ. Use of data from the electronic health record for health research: current governance challenges and potential approaches. In: Johnston S, Ranford J, eds. OPC Guidance Documents, Annual Reports to Parliament. Ottawa, Ont: Office of the Privacy Commissioner of Canada, 2009:1–32 [Google Scholar]
- 3.Murphy SN, Gainer V, Mendis M, et al. Strategies for maintaining patient privacy in i2b2. J Am Med Inform Assoc 2011;18(Suppl 1):103–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vinterbo SA, Sarwate AD, Boxwala A. Protecting count queries in study design. J Am Med Inform Assoc 2012;19:750–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Denekamp Y, Boxwala AA, Kuperman G, et al. A meta-data model for knowledge in decision support systems. AMIA Annu Symp Proc 2003:826. [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang X. Predictions for Biomedical Decision Support. 2010. http://reports-archive.adm.cs.cmu.edu [Google Scholar]
- 7.Katz MS, Efstathiou JA, D'Amico AV, et al. The 'CaP Calculator': an online decision support tool for clinically localized prostate cancer. BJU Int 2010;105:1417–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ohno-Machado L, Wang SJ, Mar P, et al. Decision support for clinical trial eligibility determination in breast cancer. Proc AMIA Symp 1999:340–4 [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang X, Boxwala A, El-Kareh R, et al. A patient-Driven Adaptive Prediction Technique (ADAPT) to improve personalized risk estimation for clinical decision support. J Am Med Inform Assoc 2012;19:e137–e44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang X, El-Kareh R, Ohno-Machado L. Improving Predictions in Imbalanced Data Using Pairwise Expanded Logistic Regression. Washington, DC: AMIA Annual Symposium Proceedings, 2011:625–34 [PMC free article] [PubMed] [Google Scholar]
- 11.Karp I, Abrahamowicz M, Bartlett G, et al. Updated risk factor values and the ability of the multivariable risk score to predict coronary heart disease. Am J Epidemiol 2004;160:707–16 [DOI] [PubMed] [Google Scholar]
- 12.Leslie WD, Lix LM, Johansson H, et al. Independent clinical validation of a Canadian FRAX tool: fracture prediction and model calibration. J Bone Mineral Res 2010;25:2350–8 [DOI] [PubMed] [Google Scholar]
- 13.Wilson PW, D'Agostino RB, Levy D, et al. Prediction of coronary heart disease using risk factor categories. Circulation 1998;97:1837–47 [DOI] [PubMed] [Google Scholar]
- 14.Matheny M, Ohno-Machado L, Resnic F. Discrimination and calibration of mortality risk prediction models in interventional cardiology. J Biomed Inform 2005;38:367–75 [DOI] [PubMed] [Google Scholar]
- 15.Ohno-Machado L. Modeling medical prognosis: survival analysis techniques. J Biomed Informat 2001;34:428–39 [DOI] [PubMed] [Google Scholar]
- 16.Ohno-Machado L, Resnic FS, Matheny ME. Prognosis in critical care. Annu Rev Biomed Eng 2006;8:567–99 [DOI] [PubMed] [Google Scholar]
- 17.Savova GK, Olson JE, Murphy SP, et al. Automated discovery of drug treatment patterns for endocrine therapy of breast cancer within an electronic medical record. J Am Med Inform Assoc 2012;19:e83–e9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Niland JC, Stiller T, Neat J, et al. Improving patient safety via automated laboratory-based adverse event grading. J Am Med Inform Assoc 2011;19:111–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lasserre J, Arnold S, Vingron M, et al. Predicting the outcome of renal transplantation. J Am Med Inform Assoc 2012;19:255–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Talos I, Zou K, Ohno-Machado L, et al. Supratentorial low-grade glioma resectability: statistical predictive analysis based on anatomic MR features and tumor characteristics. Radiology 2006;239:506–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Resnic F, Ohno-Machado L, Selwyn A, et al. Simplified risk score models accurately predict the risk of major in-hospital complications following percutaneous coronary intervention. Am J Cardiol 2001;88:5–9 [DOI] [PubMed] [Google Scholar]
- 22.Racowsky C, Ohno-Machado L, Kim J, et al. Is there an advantage in scoring early embryos on more than one day? Hum Reprod 2009;24:2104–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hosmer DW, Lemeshow S. Applied Logistic Regression. New York: Wiley-Interscience, 2000 [Google Scholar]
- 24.Kamath PS, Kim W. The model for end-stage liver disease (MELD). Hepatology 2007;45:797–805 [DOI] [PubMed] [Google Scholar]
- 25.Boxwala AA, Kim J, Grillo JM, et al. Using statistical and machine learning to help institutions detect suspicious access to electronic health records. J Am Med Inform Assoc 2011;18:498–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frisse ME, Johnson KB, Nian H, et al. The financial impact of health information exchange on emergency department care. J Am Med Inform Assoc 2012;19:328–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seidling HM, Phansalkar S, Seger DL, et al. Factors influencing alert acceptance: a novel approach for predicting the success of clinical decision support. J Am Med Inform Assoc 2011;18:479–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shtatland ES, Barton MB. Information theory makes logistic regression special. Annual Conference of NorthEast SAS Users' Group (neseg). Pittsburgh, PA: NESEG, 1998:190–93 [Google Scholar]
- 29.Osl M, Dreiseitl S, Kim J, et al. Effect of data combination on predictive modeling: a study using gene expression data. AMIA Annu Symp Proc 2010:567–71 [PMC free article] [PubMed] [Google Scholar]
- 30.El Emam K, Hu J, Mercer J, et al. A secure protocol for protecting the identity of providers when disclosing data for disease surveillance. J Am Med Inform Assoc 2011;18:212–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loukides G, Denny JC, Malin B. The disclosure of diagnosis codes can breach research participants' privacy. J Am Med Inform Assoc 2010;17:322–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vaszar LT, Cho MK, Raffin TA. Privacy issues in personalized medicine. Pharmacogenomics 2003;4:107–12 [DOI] [PubMed] [Google Scholar]
- 33.Sweeney L. Privacy and medical-records research. N Engl J Med 1998;338:1077 author reply 77–8. [PubMed] [Google Scholar]
- 34.Calloway SD, Venegas LM. The new HIPAA law on privacy and confidentiality. Nurs Adm Q 2002;26:40–54 [DOI] [PubMed] [Google Scholar]
- 35.Freund Y, Schapire RE. A decision-theoretic generalization of on-line learning and an application to boosting. J Comput Syst Sci 1997;55:119–39 [Google Scholar]
- 36.Vapnik NV. The Nature of Statistical Learning Theory. New York: Springer-Verlag, 2000 [Google Scholar]
- 37.Gambs S, Kégl B, Aïmeur E. Privacy-preserving boosting. Data Min Knowl Discov 2007;14:131–70 [Google Scholar]
- 38.Vaidya J, Yu H, Jiang X. Privacy-preserving SVM classification. Knowl Inform Syst 2008;14:161–78 [Google Scholar]
- 39.Yu H, Jiang X, Vaidya J. Privacy-preserving SVM using nonlinear kernels on horizontally partitioned data. Symposium on Applied Computing (SAC). Dijon, France: ACM, 2006:603–10 [Google Scholar]
- 40.Yu H, Vaidya J, Jiang X. Privacy-preserving svm classification on vertically partitioned data. Advances in Knowledge Discovery and Data Mining. 2006;3918:647–56 [Google Scholar]
- 41.Chu CT, Kim SK, Lin YA, et al. Map-reduce for machine learning on multicore. Adv Neural Inform Process Syst 2007;19:281–88 [Google Scholar]
- 42.Sanil AP, Karr AF, Lin X, et al. Privacy Preserving Regression Modelling via Distributed Computation. Proceedings of the Tenth ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. Seattle, WA: ACM, 2004:677–82 [Google Scholar]
- 43.Minka T. A Comparison of Numerical Optimizers for Logistic Regression. Pittsburgh, PA: Carnegie Mellon University, Technical Report, 2003 [Google Scholar]
- 44.Hosmer DW, Hosmer T, Le Cessie S, et al. A comparison of goodness-of-fit tests for the logistic regression model. Stat Med 1997;16:965–80 [DOI] [PubMed] [Google Scholar]
- 45.Kramer AA, Zimmerman JE. Assessing the calibration of mortality benchmarks in critical care: the Hosmer-Lemeshow test revisited. Crit Care Med 2007;35:2052–56 [DOI] [PubMed] [Google Scholar]
- 46.Lemeshow S, Hosmer DW. A review of goodness of fit statistics for use in the development of logistic regression models. Am J Epidemiol 1982;115:92–106 [DOI] [PubMed] [Google Scholar]
- 47.Hanley J, McNeil B. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology 1982;143:29–36 [DOI] [PubMed] [Google Scholar]
- 48.Harrell F, Califf R, Pryor D, et al. Evaluating the yield of medical tests. JAMA 1982;247:2543–46 [PubMed] [Google Scholar]
- 49.Zou KH, Liu AI, Bandos AI, et al. Statistical Evaluation Of Diagnostic Performance: Topics in ROC Analysis. Boca Raton, FL: Chapman & Hall/CRC Biostatistics Series, 2011 [Google Scholar]
- 50.Lasko TA, Bhagwat JG, Zou KH, et al. The use of receiver operating characteristic curves in biomedical informatics. J Biomed Inform 2005;38:404–15 [DOI] [PubMed] [Google Scholar]
- 51.Kennedy RL, Burton AM, Fraser HS, et al. Early diagnosis of acute myocardial infarction using clinical and electrocardiographic data at presentation: derivation and evaluation of logistic regression models. Eur Heart J 1996;17:1181–91 [DOI] [PubMed] [Google Scholar]
- 52.Weber GM, Murphy SN, McMurry AJ, et al. The Shared Health Research Information Network (SHRINE): a prototype federated query tool for clinical data repositories. J Am Med Inform Assoc 2009;16:624–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stolba N, Nguyen TM, Tjoa AM. Data Warehouse Facilitating Evidence-Based Medicine. In: Nguyen TM, ed. Complex Data Warehousing and Knowledge Discovery for Advanced Retrieval Development: Innovative Methods and Applications. Hershey, PA: IGI Global, 2010:174–207 [Google Scholar]
- 54.Dwork C. Differential privacy. Proceedings of the 33rd International Colloquium on Automata, Languages and Programming (ICALP) (2); 10-14 July 2006, Venice, Italy. Germany: Springer, 2006. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.