Abstract
Our understanding of both the nature and diversity of Staphylococcal immune evasion proteins has increased tremendously throughout the last several years. Among this group of molecules, members of the SCIN and Efb families of complement inhibitors have been the subject of particularly intense study. This work has demonstrated that both types of proteins exert their primary function by inhibiting C3 convertases, which lie at the heart of the complement-mediated immune response. Despite this similarity, however, significant differences in structure/function relationships and mechanisms of action exist between these bacterial proteins. Furthermore, divergent secondary effects on host immune responses have also been described for these two protein families. This review summarizes recent advances toward understanding the structure, function, and mechanism of the SCIN and Efb families, and suggests potential directions for the field over the coming years.
1 Complement-directed Immune Evasion by Staphylococcus aureus
Few human pathogens have kept the world in suspense over the past few decades as much as Staphylococcus aureus. Originally considered an opportunistic bacterium associated with superficial skin infections, S. aureus has evolved into a major medical problem in hospital environments and, increasingly, in otherwise healthy communities (DeLeo et al. 2010). It has been reported that about one third of healthy individuals in the United States are colonized with S. aureus, mostly in the nostrils and skin, without showing symptoms and the bacterium may therefore be often considered part of the normal flora (Gorwitz et al. 2008). Yet in more and more cases, S. aureus strains cause severe infections, including life-threatening septicemia, which can poorly be controlled by today’s therapeutic arsenal due to the bacterium’s extraordinary ability to acquire resistance to antibiotics (Chambers et al. 2009). In 2005, the number of deaths associated with methicillin-resistant S. aureus (MRSA) infections in the U.S.A. was estimated to be around 19,000, thereby exceeding those associated with HIV (Klevens et al. 2007; Kluytmans et al. 2009). Although MRSA strains have been described as early as 1960 (e.g. COL strain), the current community-associated MRSA strains (e.g. USA300) appear to be much more virulent (Chambers et al. 2009). Although the exact driving forces of this increased virulence are not clearly identified, immune evasion strategies are considered potential contributors to this phenomenon F (Chambers et al. 2009). Indeed, S. aureus has been described to interfere at various levels of the immune system and more than 50 expressed or secreted proteins are currently considered part of its immune evasion arsenal (Chavakis et al. 2007). Interestingly, many of those molecules appear to target the human complement system, therefore moving this ancient branch of innate immunity into the spotlight of research efforts (Foster 2005; Haspel et al. 2008).
Although complement is involved in many physiological functions ranging from cell homeostasis to tissue development and metabolism, the rapid recognition and elimination of microbial intruders defines a key role of this network of plasma proteins and cell surface receptors (Ricklin et al. 2010). After recognition of bacterial surfaces by specialized pattern recognition proteins, complement can be triggered by various routes (historically termed as “classical”, “lectin”, and “alternative” pathways) that all result in the activation of complement component C3 to its C3b fragment, which is deposited as an opsonin on the foreign surface. Together with two enzymes, i.e. factor B (fB) and factor D (fD), C3b forms a transient yet potent C3 convertase complex (C3bBb). This convertase activates even more C3 into C3b and thereby amplifies the complement response. In addition, C3b and its degradation products (iC3b, C3d) act as ligands for complement receptors (CR) that mediate phagocytosis of opsonized particles (i.e. CR1, CR3, CR4, and CRIg) or lower the threshold for B cell activation (CR2) and thereby bridge to adaptive immune responses. Finally, ongoing complement activation produces the potent proinflammatory anaphylatoxins C3a (as a byproduct of C3 cleavage) and C5a (through C5 convertases such as C3b3bBb), which attract and activate immune cells via binding to anaphylatoxin receptors (C3aR, C5aR, C5L2).
Whereas human cells are protected by expressing cell surface-bound or acquiring soluble complement regulators, microbial cells normally lack these regulators and are therefore susceptible to complement attack (Fig. 1a) (Ricklin et al. 2010). However, an impressive body of research in recent years revealed that S. aureus produces a fascinating panel of several inhibitors that block complement activity at all stages from initiation and amplification to phagocytosis and inflammatory/immune signaling (Fig. 1b, c) (Laarman et al. 2010; Lambris et al. 2008; Serruto et al. 2010). Owing to its central role in the cascade, the C3 convertase appears to be a particularly attractive target for S. aureus, and at least three different groups of proteins, i.e. the extracellular fibrinogen-binding protein (Efb) and staphylococcal complement inhibitor (SCIN) families as well as the staphylococcal binder of immunoglobulins (Sbi) have been identified and characterized. Of those, SCIN (i.e. SCIN-A/B/C) and Efb (and its homologue Ehp; also referred to as Ecb) have proven to be particularly strong inhibitors of convertase activity. These properties not only highlight their potential roles in the overall virulence of the bacterium, they may also render these rather small, secreted proteins (~10 kDa) attractive templates for therapeutic intervention in the ever-growing list of diseases where complement-mediated inflammation is known to play a central role (Ricklin et al. 2007). For this purpose, a detailed knowledge about these proteins’ exact mechanisms of action as well as key structures and residues that impart these unique activities is essential.
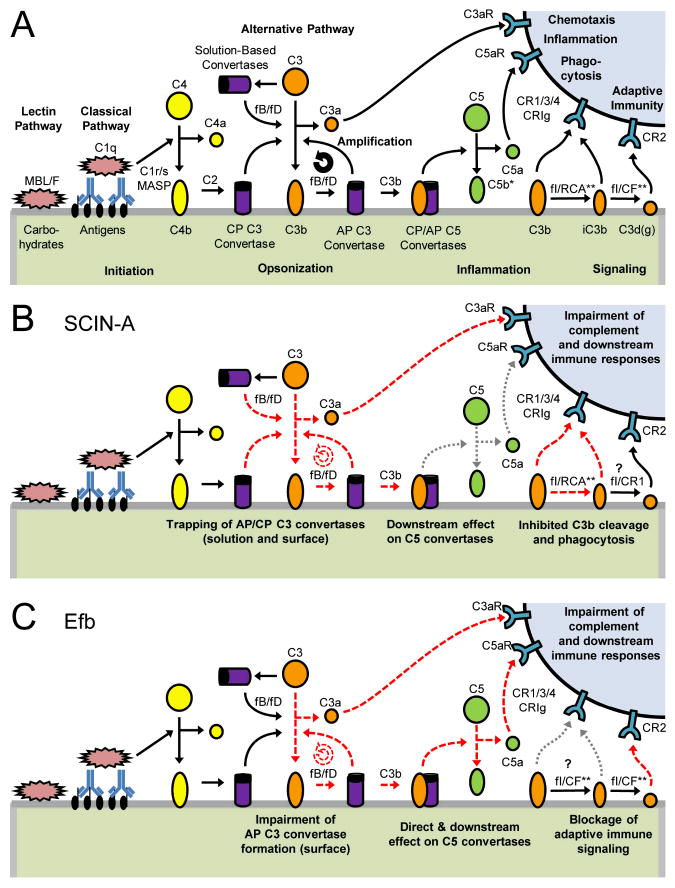
Figure 1.
Pathways of Complement Activation and Amplification in the Vicinity of an S. aureus cell. (A) A normal, uninhibited complement response that arises from either the classical, lectin, or alternative pathways, and which leads to changes in various immune effector cells (top right). (B) Disruption of the complement response by SCIN-A. (C) Disruption of the complement response by Efb. Note that red dashed lines signify processes affected directed by either of these secreted inhibitors, while dotted grey lines indicate secondary, indirect effects. Formation of the terminal complement complex is not shown, since the bacterium is resistant to its function owing to the peptidoglycan-rich structure of its gram positive cell wall (Frank 2001).
In view of their similar structures (trihelical bundle), their common binding partners (C3 and its activation fragments), and their identical functional targets (C3 convertases) in the complement cascade, it would seem obvious that the SCIN and Efb protein families share similar mechanisms of action. However, detailed molecular, structural, and functional studies in the past few years have surprisingly revealed that their binding modes and inhibitory mechanisms are fascinatingly distinct and include both directly competitive and allosteric strategies (Chen et al. 2010; Ricklin et al. 2009; Rooijakkers et al. 2009). There are even first indications that small differences within the same family (e.g. between Efb and Ehp) may result in slightly modified structural properties that could allow S. aureus to fine-tune its evasion response. As a consequence, this review discusses the structural and functional properties of these convertase-targeting evasion protein families, their exciting secondary effects on host immunity, and their potential implications for overall immune evasion by S. aureus.
2 Recent Developments Regarding the Staphylococcal Complement Inhibitor Family
2.1. Background
The founding member of the SCIN family (i.e., SCIN-A) was discovered through genomic analysis of an uncharacterized gene on the SaPI5 immune evasion cluster (Rooijakkers et al. 2005). Further work demonstrated that the SCIN family consists of three active members (SCIN-A, SCIN-B, and SCIN-C) (Rooijakkers et al. 2007), all of which share between 46–48% sequence identity with one another. A putative fourth SCIN family member (typically referred to as ORF-D, but herein as SCIN-D) is also expressed by S. aureus, and likewise shares relatively high amino acid sequence identity (~30%) to the other SCIN family members (Rooijakkers et al. 2007). However, SCIN-D does not appear to bind directly to any C3 fragments and thus exhibits no inhibition of complement activity (Rooijakkers et al. 2007). SCIN proteins are potent complement inhibitors and were shown to target both the classical and alternative pathway C3 convertases (i.e. C4b2b and C3bBb, respectively) (Jongerius et al. 2007; Rooijakkers et al. 2005), the latter of which is the central enzymatic component of complement amplification. Whereas many classes of complement modulators (in particular the host-derived ‘regulators of complement activation’ and their viral homologues (reviewed in (Zipfel et al. 2009))) possess decay accelerating activity and dissociate assembled C3 convertases (Wu et al. 2009), the active SCIN family members actually stabilize C3 convertases both in solution and at the bacterial surface (Rooijakkers et al. 2005; Rooijakkers et al. 2009). Recent work has elucidated many details about the molecular mechanism of SCIN proteins, and revealed how these bacterial inhibitors trap C3 convertases in a catalytically inactive state (Ricklin et al. 2009; Rooijakkers et al. 2009).
2.2. Structure and Active Sites of SCIN-A
A crucial first step toward understanding the complement inhibition properties of SCIN proteins was provided by the crystal structure of SCIN-A (Rooijakkers et al. 2007) (Fig. 2a). The SCIN-A structure revealed a three alpha helix bundle arranged in a compact coiled-coil, and which was most topologically similar to that described previously for the IgG-binding staphylococcal protein A (SpA) module (Gouda et al. 1992). In conjunction with multiple sequence alignment, the SCIN-A structure was used to guide construction of protein chimeras between SCIN-A and the reportedly inactive member of the SCIN family, SCIN-D (Rooijakkers et al. 2007). It was this transposition of amino acid sequences from SCIN-D into SCIN-A which subsequently identified a contiguous stretch of 18 amino acids (residues 31–48) that were critical for complement inhibition by SCIN-A, as judged by both inhibition and stabilization of the alternative pathway C3 convertase. As will be discussed shortly in Section 2.4. below, these peculiar stabilizing properties would prove invaluable to providing the first high-resolution structural data on the alternative pathway C3 convertase (C3bBb) (Rooijakkers et al. 2009).
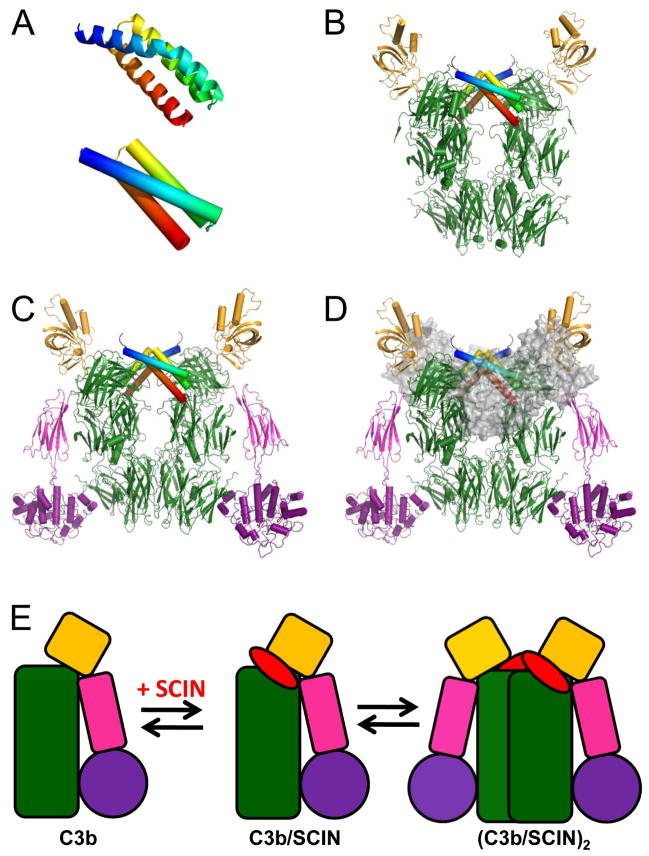
Figure 2.
Structural Properties of the SCIN-A Protein both Free and Bound to Complement Components. (A) Crystal structure of the unbound form of SCIN-A, shown in both ribbon (top) and tube (bottom) convention (Rooijakkers et al. 2007). The N terminus of the protein is colored blue in both images. (B) Crystal structure of SCIN-A bound to C3c, which adopts a (C3c/SCIN-A)2 pseudo-dimer (Garcia et al. 2010). (C) Crystal structure of SCIN-A bound to C3b (Garcia et al. 2010). A (C3b/SCIN-A)2 pseudo-dimer is found through the application of crystallographic symmetry operators. (D) Crystal structure of a dimeric, SCIN-A inhibited convertase (C3b/Bb/SCIN-A)2 (Rooijakkers et al. 2009). The Bb proteins are drawn as partially-transparent grey surfaces for clarity. (E) Schematic representation of C3b-based pseudo-dimers that form upon SCIN-A binding. Such structures can form either in the presence or absence of the convertase component Bb.
2.3. Structure/Function Analysis of SCIN-A Binding to C3c/C3b
Whereas initial interaction studies suggested that SCIN proteins could only bind within the context of fully assembled C3 convertases (Rooijakkers et al. 2005), an improved surface plasmon resonance (SPR)-based approach designed to mimic the physiological orientation of covalently deposited C3b provided the first evidence for direct binding between these proteins and C3b (Ricklin et al. 2009). In this same study, direct binding of SCIN-A to C3b was also characterized by solution-based isothermal titration calorimetry (ITC) (Ricklin et al. 2009). Even though these two analytical platforms differ considerably, the results obtained were consistent with one another, yielding apparent KD values within the range of 170–265 nM. Subsequent biochemical mapping of the SCIN-A binding site on C3b localized this interaction to the C3c fragment of the complement component, since SCIN-A failed to bind to recombinant C3d, and the C3b/SCIN-A interaction was almost completely blocked by a monoclonal anti-C3c antibody, i.e. mAb C3-9, which has previously been shown to affect the binding of key ligands of C3b, such as fH, CR1, and fB (Becherer 1992). As a consequence, identification of a direct, high-affinity interaction between SCIN-A and the C3c fragment of C3b provided an incentive for pursuing crystal structures of these binary complexes.
Indeed, structures derived from four different crystal forms of C3c/SCIN-A and C3b/SCIN-A were recently reported (Garcia et al. 2010) (Fig. 2b, c). Superposition of these structures’ refined coordinates indicated that they displayed a large degree of identity with one another, and were therefore directly comparable. For example, the relative orientations of the SCIN polypeptide and nearly all C3c-derived domains were practically indistinguishable among these structures. Furthermore, this primary contact site between SCIN-A and its complement target largely agreed with the predictions made previously through the functional analysis of SCIN protein chimeras (Rooijakkers et al. 2007). It was also significant that this SCIN-A contact site localized to the seventh a2-macroglobulin-like domain (MG7) of C3c/C3b. This feature was consistent with the previously mentioned biochemical mapping studies using mAb C3-9 (Ricklin et al. 2009), and also with negatively stained electron microscopy (EM) imaging that had mapped the C3-9 epitope to this region of C3b (Nishida et al. 2006).
The significant functional differences that accompany structural transitions between C3 and its downstream activation products are a fascinating feature of complement biology that has been well described for over two decades (Alsenz 1989; Hack et al. 1988; Lachmann 1982; Lambris 1988; Lambris 1985; Nilsson 1987; Nilsson 1982; Tamerius 1985), and one that has become much more readily interpretable with the many recent advances in understanding C3 structure and dynamics (Gros et al. 2008; Janssen et al. 2006; Janssen et al. 2005; Ricklin et al. 2010; Schuster et al. 2008). In this context, the biochemical nature of the SCIN-A binding site on C3c/C3b provided an excellent illustration of how a functionally-important neoantigen/neoepitope arises upon activation of C3 to C3b. In its native state, C3 is a large 185 kDa protein that is comprised of two polypeptide chains (α, β), which themselves are held together by two interchain disulfide bonds and an extensive non-covalent interface (Janssen et al. 2005). Proteolytic conversion to C3b causes a large number of residues that line the α/β interface to undergo structural rearrangement, many of which become surface exposed in the process (Janssen et al. 2006; Schuster et al. 2008). Comparison of the C3b/SCIN-A and C3c/SCIN-A structures to that of native C3 revealed that 15 of 23 residues that constitute the primary SCIN-A contact site were previously buried in the large α/β interface of the unactivated complement protein (Garcia et al. 2010). Consequently, SCIN-A cannot bind to native C3, yet specifically recognizes C3c and C3b with high affinity.
Aside from this, several important insights followed from correlating these new structures with various functional outcomes of SCIN-A binding to C3b. To begin, SCIN-A was shown to slightly decelerate the initial rate of convertase formation, even though it primarily stabilizes the assembled convertase complex (Ricklin et al. 2009); consistent with this, SCIN-A binding to surface-immobilized C3b partially inhibited fB binding. In addition, SCIN competitively blocked the binding the major soluble complement regulator factor H (fH), which exerts both decay acceleration of the convertase and cofactor activity for the factor I-mediated degradation of C3b to iC3b. Indeed, functional assays with a recombinant fH fragment (i.e. fH(1-4)) showed that the SCIN-stabilized convertase was essentially resistant for decay acceleration by fH, and that the presence of SCIN interfered with the cofactor function of fH(1-4). Altogether, these data strongly suggest an overlapping C3b binding site for SCIN-A and the host complement proteins fB and fH. Comparative analysis of the C3c/SCIN-A and C3b/SCIN-A structures with the existing structures of both C3b/fH(1-4) (Wu et al. 2009) and cobra venom factor bound to fB (CVF/B) (Janssen et al. 2009) provided a satisfying structural explanation for these observations: the primary SCIN-A binding site on C3c/C3b partially overlaps with that of fB, but more significantly occludes that of fH(1-4). Thus, by binding at a key functional region on C3b, SCIN-A protects inactivated forms of the convertase against host complement regulator activity (Fig. 1b).
2.4. Structural/Functional Basis for SCIN-A Inhibition of C3 Convertase Activity
In order to direct and restrict complement amplification to target surfaces, the cascade relies on a highly concerted and well-timed activation mechanism of its C3 convertase, the details of which have only been revealed very recently (Forneris et al. 2010; Rooijakkers et al. 2009; Wu et al. 2009). While it has long been shown that SCIN impairs the conversion of C3 to C3b by the convertase, the exact mode of intervention has been difficult to pinpoint and could not be readily explained by the binary structures of SCIN-A with C3c and C3b. In principle, SCIN could impair either the initial binding of native C3 to the convertase or the cleavage/activation process itself. However, previous interaction studies had shown that native C3 bound readily to surface-based, SCIN-A-inhibited convertases that had been assembled in vitro on biosensor surfaces, yet that bound C3 was not activated to C3b (Ricklin et al. 2009). This observation thereby excluded the possibility that SCIN-A blocks the initial binding of the convertase’s native C3 substrate, and instead suggested that the bacterial inhibitor must somehow “lock” C3bBb in an inactive state by preventing access of Bb to the target scissile bond in C3 (Ricklin et al. 2009; Rooijakkers et al. 2009).
The activity of decay-accelerating regulatory proteins notwithstanding, C3 convertases are themselves intrinsically transient complexes that dissociate irreversibly with half-lives on the order of 1–2 min (Pangburn et al. 1986). While this property is vitally important in tightly regulating the overall extent of complement activation, it also had long precluded detailed structural analysis of C3 convertases. With this in mind, the unique stabilizing properties of SCIN-A were innovatively used to obtain the first crystal structure of a fully assembled C3 convertase (C3bBb/SCIN-A) (Fig. 2d) (Rooijakkers et al. 2009). In this structure SCIN-A formed an essentially identical interface with the convertase C3b scaffold to that seen in the binary complexes described above (Garcia et al. 2010). However, the C-terminally oriented residues of alpha helices 1 and 3 in SCIN-A also formed a distinct interface with the proteolytic complement component, i.e. fragment Bb (Rooijakkers et al. 2009). Whereas this additional interface did not appear to induce any conformational changes in Bb (as judged by comparison to the unbound Bb structure (Ponnuraj et al. 2004)), it nevertheless appeared to fix the orientation of Bb relative to the remainder of the C3b molecule.
In the absence of its interactions with SCIN-A, the proteolytic Bb fragment is bound by the C345C domain of C3b and thereby remains loosely associated with its convertase scaffold. The C345C domain itself is conformationally flexible, and in many crystal structures has adopted slightly, but noticeably different positions when compared to the remaining C3c/C3b macroglobulin-like core (later referred to as “C3b core”) (Garcia et al. 2010; Rooijakkers et al. 2009). The most current models of convertase activity have therefore proposed that the C345C domain might function as a swinging platform, which allows proper orientation of the Bb active site relative to its C3 substrate (Rooijakkers et al. 2009). Taken together, all of these data have suggested that the SCIN-A-dependent loss of convertase activity lies in the ability of this bacterial protein to sterically restrict critical reorientations of C345C domain, which itself harbors the Bb protease (Garcia et al. 2010; Rooijakkers et al. 2009). In this manner, SCIN-A not only slows the rate of convertase formation as described above, but once it does assemble, it holds the essential convertase complex in an inactive conformation that itself is resistant to decay by host regulators (Fig. 1b).
2.5. Structural Basis and Functional Consequences of SCIN-induced Dimerization of C3b
A distinctive feature of all SCIN-bound crystal structures of C3c/C3b determined was the presence of crystallographic or non-crystallographic symmetry-related pseudo-dimers that formed along the long axis of the complement component (Garcia et al. 2010; Rooijakkers et al. 2009) (Fig. 2b–e). In all of these cases, the pseudo-dimer inducing site consisted almost exclusively of the first alpha helix of SCIN-A in contact with residues donated by the symmetry-related C3b molecule’s a′-chain. While this additional interaction may have arisen as an artifact of crystal packing forces, several lines of evidence argued against this possibility. First, the dimerization-promoting interface buried comparable surface area to that of the primary SCIN contact site described above (750 versus 900 Å2, as judged by the structure of C3b/SCIN-A) (Garcia et al. 2010). Second, direct C3b/SCIN-A binding studies using either solution or surface-based methods revealed a slight deviation from 1:1 Langmuir binding that was most pronounced at higher concentrations (Ricklin et al. 2009). Furthermore, small angle X-ray scattering (SAXS) analysis of C3b/SCIN-A samples were also consistent with an equilibrium mixture between 1:1 (C3b/SCIN-A) and 2:2 (C3b/SCIN-A)2 species in solution. Finally, the presence of SCIN-A in reactions that generate the alternative pathway C3 convertase (C3bBb) resulted in formation of stably inhibited convertase dimers (C3bBb/SCIN-A)2 when compared to controls (Rooijakkers et al. 2009). All of these observations together predicted that SCIN-A-induced dimerization of C3b or convertases may have important functional consequences.
In this regard, a recent report established a correlation between SCIN-A mediated convertase dimerization and the inhibition of phagocytosis (Jongerius et al. 2010b). In this study, a mutant form of SCIN-A was constructed that blocked convertase activity equally as well as its wild type counterpart, yet was unable to form the dimers seen in the convertase crystal structure (Rooijakkers et al. 2009). Intriguingly, while the mutant SCIN-A-inhibited convertases had no effect on the recognition of phagocytic receptors, the wild-type protein disrupted both complement receptor 1 (CR1) and CR of the Ig-superfamily (CRIg)-dependent recognition and phagocytosis of opsonized S. aureus cells (Jongerius et al. 2010b). While this result demonstrated a dimerization-dependent inhibition of phagocytosis, it did not address whether C3b/SCIN-A dimerization in the absence of Bb might elicit a similar effect (Garcia et al. 2010). Furthermore, though not demonstrated directly in this study, it was also proposed that SCIN-A may also interfere with CR3-dependent phagocytic responses by blocking the generation of iC3b (Jongerius et al. 2010b). This is because SCIN-A effectively competes for the C3b-binding site of fH and, in doing so, inhibits the cofactor activity needed to stimulate fI-mediated generation of iC3b (Ricklin et al. 2009).
Examining the crystal structures of both C3b/SCIN-A (Garcia et al. 2010) and C3bBb/SCIN-A (Rooijakkers et al. 2009) provided a physical explanation for the anti-phagocytic properties of SCIN-A (Jongerius et al. 2010b). When SCIN-A-dependent dimers of C3c/C3b or the C3bBb convertase assemble, a large contact interface is formed. This interface subsequently masks a large number of C3b residues, among which are those that comprise the binding site of CRIg, a receptor is predominantly found on phagocytic Kupffer cells involved in pathogen clearance by the liver (Helmy et al. 2006; Wiesmann et al. 2006). Whereas SCIN-induced dimerization also inhibited binding of CR1, the absence of a co-crystal structure for this receptor with C3b restricts a similarly detailed analysis for this observation. However, the high structural/functional similarity with fH suggests a similar competitive mechanism as observed for this regulator (see above). Consequently, it seems quite likely that the anti-phagocytic properties of SCIN follow from steric hindrance effects that prevent efficient binding of C3b by certain classes of CR proteins (Jongerius et al. 2010b). As a result, it now appears that a secondary function of SCIN-A is to facilitate the formation of pseudo-dimeric structures of C3b at the bacterial surface. These structures not only serve to attenuate complement activation (in the case of the convertase), but also to protect S. aureus cells from phagocytic uptake and destruction.
3 Recent Developments Regarding the Extracellular Fibrinogen-Binding Protein Family
3.1. Background
Efb was originally identified as a 16 kDa protein present in S. aureus conditioned culture medium capable of binding fibrinogen (Fg) (Boden et al. 1992). The gene encoding Efb lies within a second immune-evasion cluster within S. aureus genome (Jongerius et al. 2007), which also harbors the coding regions for SCIN-B and SCIN-C. Biochemical analyses of the Efb protein suggested that it was comprised of a modular architecture consisting of two distinct structural regions (Hammel et al. 2007b; Lee et al. 2004b). While several reports ultimately concluded that the N terminal region of Efb was responsible for its eponymous Fg-binding activity (Lee et al. 2004b; Palma 2001; Palma 1998), the function of its proteolytically-stable C terminal domain (residues 101 to 165, referred to in the literature and hereafter as Efb-C) remained inconclusive. Several years ago, the outcomes of several lines of investigation revealed that Efb-C binds directly to C3/C3b/C3d (Hammel et al. 2007b; Lee et al. 2004a; Lee et al. 2004b). Interestingly, a nearby gene on the same immune-evasion cluster was found to encode a 9.8 kDa protein denoted Ehp (Hammel et al. 2007a) (for Efb homologous protein; alternatively known as Ecb (Jongerius et al. 2007)) that shares 44% sequence identity with Efb-C. Although similar C3/C3b/C3d binding activities were subsequently reported for Ehp (Hammel et al. 2007a; Jongerius et al. 2007), this smaller molecule also possessed a unique ability to form ternary 1:2 complexes with C3d-containing proteins (Hammel et al. 2007a). As a result, Ehp was identified as a more potent complement inhibitor than Efb-C on a molar basis (Hammel et al. 2007a).
The C3 binding properties of both Efb and Ehp strongly suggested a virulence-promoting role during S. aureus pathogenesis. To this end, studies revealed that S. aureus strains lacking Efb were less infectious than wild-type clinical isolates in a mouse mastitis model (Mamo 1994), and functional Efb was essential for prolonged S. aureus survival in a rat model of wound healing (Palma 1996). Separately, inclusion of Ehp in a model of immune complex-mediated inflammation blocked the influx of neutrophils in the peritoneal cavity of experimental mice (Jongerius et al. 2007). In the latter case specifically, these in vivo effects arise directly from potent inhibition of C3 and C5 activity by the bacterial proteins. Whereas their inhibition at the level of C3 appears largely restricted to the alternative pathway, Efb and Ehp inhibited the turnover of C5 through all pathways (Hammel et al. 2007b; Jongerius et al. 2007). This observation pinpointed the activity of Efb-C and Ehp to the level of the C3b-containing C3 and C5 convertases (i.e. C3bBb, C3b3bBb, and C3b4b2b) (Jongerius et al. 2007). In the few brief years since these original discoveries, convertase inhibition has become a well-established mechanism of complement evasion. However, the structural and mechanistic basis for the C3 convertase-blocking activity of Efb-C has only recently been elucidated (Chen et al. 2010). Furthermore, additional and unexpected activities of Efb and its homolog Ehp have been identified that highlight the elaborate nature of the staphylococcal immune evasion arsenal.
3.2. Structure/Function Analysis of Efb-C Binding to C3, C3b, and C3d
The crystal structure of the Efb complement inhibitor domain (Efb-C) has been determined both free and bound to its cognate C3d fragment of C3/C3b (Hammel et al. 2007b). In these structures Efb-C adopts a compact three helix bundle fold that is related to, but topologically-distinct from that of either SCIN-A or SpA modules (Fig. 3a, b). Superposition analysis revealed that the Efb-C structure is essentially identical in both the free and C3d-bound states, which suggested that its C3d binding surface is preformed and requires little structural rearrangement. Further characterization of the C3d/Efb-C complex has provided a wealth of biochemical information on the nature of this interaction. First, the interaction is highly favorable thermodynamically with a KD on the order of 1–2 nM, as judged by both solution ITC and surface-based SPR studies. Second, the C3d/Efb-C complex is also exceedingly stable kinetically, and displays a mean half-life of approximately 60 minutes (Hammel et al. 2007b). Finally, Efb-C binds near an acidic cleft on C3d, and the C3d/Efb-C complex is mediated by an extensive array of largely ionic interactions. These occur almost exclusively between polar residues donated by the second alpha helix Efb-C and the loops which connect helices H2-H3, H4-H5, and H6-H7 of C3d.
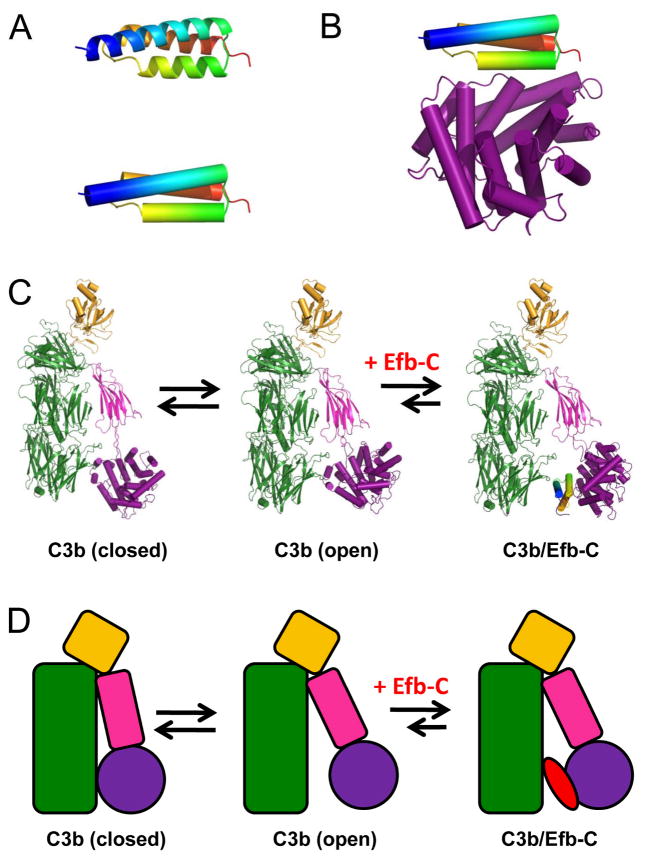
Figure 3.
Structural Properties of the Efb-C Protein both Free and Bound to Complement Components. (A) Crystal structure of the Efb-C protein, shown in both ribbon (top) and tube (bottom) convention (Hammel et al. 2007a). The N terminus of the protein is colored blue in both images. (B) Crystal structure of Efb-C bound to C3d (Hammel et al. 2007a). (C) Whereas solution structural transitions occur freely between the closed and open forms of C3b, Efb-C acts as a wedge to stabilize the open form (Chen et al. 2010). (D) Schematic representation of structural transitions in C3b, and the wedge effect upon Efb-C binding.
While the contributions of specific positions in C3d have not yet been examined by mutagenesis, this type of detailed structure/function analysis has been carried out for Efb-C (Hammel et al. 2007b; Haspel et al. 2010; Haspel et al. 2008). This work has demonstrated that positions R131 and N138 of Efb-C are particularly critical determinants for both the affinity and stability of the C3d complex. Studies on both single and pairwise substitutions at positions 131 and 138 revealed that interactions between Efb-C and C3d are dependent upon both the charge and polar nature of these side chains. Whereas the observed KD rose to 26 and 19 nM, respectively, when either residue alone was mutated to alanine (Haspel et al. 2010; Haspel et al. 2008), the corresponding double mutant had no residual affinity (Hammel et al. 2007b; Haspel et al. 2008). Furthermore, while loss of the side chain functionality had only minor effects on the ionic strength-dependent association phase, the rate of complex dissociation was greatly accelerated in each mutant (Haspel et al. 2008). In addition to these two residues, the results of molecular dynamics (MD) simulations and computational estimations of free energy release upon binding have suggested a supporting role for interactions involving residues K106, K110, V127, F142, and R165 of Efb-C (Haspel et al. 2008). However, this possibility has not been evaluated experimentally as of this time.
3.3. Allosteric Inhibition of C3 Convertase Function by Efb-C
Although the studies described above provided a detailed knowledge of Efb-C’s structure and the molecular basis for its interactions with C3/C3b/C3d, the mechanism through which Efb-C caused specific blockade of the alternative pathway remained unclear (Hammel et al. 2007b). However, several lines of evidence demonstrated that Efb-C binding either caused or stabilized an altered structure in C3b that caused disruption of complement activity. To begin, wild-type Efb-C, but not a nonfunctional mutant, rendered C3b increasingly sensitive to mild digestion by proteases in vitro. Furthermore, binding of mAb C3-9, which is specific for activated forms of C3 (e.g. C3b, iC3b, and C3c), was significantly enhanced by the presence of saturating levels of Efb-C when compared to C3b alone. This observation was most unexpected, because the Efb-C binding site on the TED/C3d domain of C3b lies nearly 100 Å distant from the MG7/MG8 domains, which themselves harbor the mAb C3-9 epitope (Nishida et al. 2006). Finally, the C3b/Efb-C complex displayed a noticeably lower formation rate constant when compared to those of native C3, hydrolyzed C3, and C3d (Hammel et al. 2007b). Since Efb-C binds with similar affinity to each of these C3 derivatives, this suggested that access of Efb-C to its TED/C3d domain binding site on C3b might somehow be restricted. Indeed, analysis of the C3d/Efb-C complex in this light revealed that Efb-C binding to C3b would result in a large steric clash with the MG1 domain of the C3b core (Hammel et al. 2007b; Janssen et al. 2006; Janssen et al. 2005); as a consequence, formation of a stable C3b/Efb-C complex could only be possible upon structural rearrangements within the C3b protein. All of these observations, together with other data which showed that Efb-C acts at the level of C3b-containing C3 and C5 convertases (Jongerius et al. 2007), suggested that Efb-C binding either resulted in or selected for an altered C3b conformation that subsequently disrupted alternative pathway convertase formation and/or function.
An investigation into the solution structural nature and functional consequences of these conformational changes was recently reported, which resulted in identification of Efb-C as the first known allosteric complement inhibitor (Chen et al. 2010). In this study, conformational differences upon Efb-C binding were detected through both hydrogen-deuterium exchange mass-spectrometry (HDX-MS) and SAXS-MD-based approaches. Efb-C binding caused the greatest changes in the HDX-MS profiles of the CUB domain of C3b, which tethers C3d/TED to the remainder of the α-chain of the core. Independent analysis of SAXS data via MD methods likewise revealed that the larger-scale conformational changes in C3b/Efb-C relative to unbound C3b could largely be explained through quaternary alterations in the CUB-TED region. As a consequence, Efb-C was proposed to act as a “wedge” upon binding its C3b target that effectively separates both the CUB and TED domains from their positions relative to unbound C3b (Fig. 3c). It is interesting to note that this “open-like” state is clearly distinct from the “closed” conformations observed in crystal structures of C3b (Garcia et al. 2010; Janssen et al. 2006; Rooijakkers et al. 2009; Wiesmann et al. 2006; Wu et al. 2009). Importantly, though, previous negatively-stained EM studies implied the physiological existence of C3b molecules with even more pronounced separation of TED from the C3b (Nishida et al. 2006), and the SAXS-MD data mentioned above indicate an equlibrium between open and closed forms of C3b in absence of Efb-C (Chen et al. 2010). Together, these results strongly suggest that a dynamic fluctuation exists between these opened and closed forms of the C3b molecule, although the physiological implication of this conformational distribution is not yet clear. In keeping with the most recent models of protein allostery (Tsai et al. 2009), this reveals that Efb-C binds to and stabilizes the “open” form of C3b (Fig. 3d), rather than “inducing” a new C3b conformation per se
Aside from these changes in the CUB-TED region, additional alterations were seen throughout other portions of C3b that are even more distant from the Efb-C binding site (e.g. in the MG 6–8 domains). Based on available co-crystal structures between C3b and major ligands, it was expected that these changes would modulate the affinity of C3b for at least some of its binding partners, as a large number of these structural changes mapped to functionally important areas of C3b. Most significantly among these, Efb-C binding to C3b was found to greatlyreduce binding of fB and to critically impair formation of the alternative pathway C3 convertase in vitro. Efb-C primarily affects the fast initial binding phase of fB and impairs binding of recombinant fragment Ba (Chen et al. 2010), which indicates that Efb-C-dependent inhibition results from its effects on the Ba-mediated loading step during convertase assembly (Forneris et al. 2010; Rooijakkers et al. 2009). This observation was supported by contact site analyses, which revealed that several contact residues for Ba were close to or within segments of altered HDX; in contrast, no HDX differences overlapped with the Bb binding site (Chen et al. 2010). At the time of this report, a high-resolution structure was only available for the complex of fB with CVF, but not with C3b; since then, however, the structures of C3bB and C3bBD have been described (Forneris et al. 2010). Importantly, a good correlation is still found between available crystallographic data, HDX, and SPR analyses, when these most recent structures for the basis for contact site comparison. All of this indicates that these independent yet complementary techniques are observing the same structure/function/mechanism relationship between Efb-C, C3b, and fB.
Aside from its effects on convertase formation, it is also noteworthy that Efb-C was found to modulate fH binding to C3b (Chen et al. 2010). Yet in this case, the molecular details of this phenomenon appear to be more confounding. Whereas Efb-C blocked binding of the regulatory fH(1-4) fragment to C3b, Efb-C clearly enhanced a second binding event that involves the fH C-terminus, i.e. fH(19-20). Overall, this leads to a net increase of full-length fH binding in the presence of Efb-C (Chen et al. 2010). Consistent with this, very recent structural studies have lent support to the possibility of Efb-C-mediated enhancement of the C3b/fH interaction (Kajander et al. 2011; Morgan et al. 2011). Specifically, two separate structures of fH(19-20) bound to C3d show that the Efb-C binding site on C3d appears to be largely available for forming a ternary complex with this fragment of fH. Nevertheless, this raises questions on how the bacterium could benefit by tethering fH to C3b in a presumably inactive conformation, for Efb-C potently blocks binding of fH(1-4) to C3b. Given these seemingly divergent conclusions, it is clear that the functional consequences of this peculiar modulatory effect will need to be more fully investigated over the coming years.
Considering these data as a whole, the ability of Efb-C to alter the conformational/dynamic properties of regions so far distant (~ 100 Å) from its actual binding site is startling. Yet this “action-at-a-distance” phenomenon is highly selective as only certain ligand-binding sites are affected. Though the inhibition of convertase assembly was not complete under the conditions used in the published assay, even the observed reduction by ~80% may drastically impair the ability of complement to initiate and amplify its response on target S. aureus cells. The specific nature of these effects argues strongly that disruption of certain C3b activities has been selected for and optimized during the course of bacterial evolution in the face of the innate immune response. In this regard, the necessity to block the function of C3b-containing convertases appears to be paramount.
3.4. Structure/Function Studies of the Efb Homologous Protein, Ehp/Ecb
A defining feature of the Staphylococcal immune evasion arsenal is the existence of multiple, structurally related proteins that appear to have similar, and sometimes overlapping functions (Chavakis et al. 2007; Foster 2005; Geisbrecht 2008; Lambris et al. 2008). In the case of Efb and its homolog Ehp (Hammel et al. 2007a), the situation more closely mirrors the latter of these scenarios. Whereas Efb has the ability to bind both C3 and Fg (Lee et al. 2004b), Ehp appears to lack this latter activity as it lacks the disordered, Fg-binding N-terminal region of Efb (Hammel et al. 2007b). In addition, quantitative studies of complement inhibition have demonstrated that Ehp is a nearly three-fold more potent inhibitor of the alternative pathway than is either Efb or its isolated complement inhibitor domain, Efb-C. Given its relatively high sequence identity to Efb-C, Ehp assumes a nearly identical fold when bound to C3d, as judged by a C3d/Ehp co-crystal structure. However, examination of the Ehp amino acid sequence identified a second C3/C3b/C3d-binding site that resides on the first alpha helix of Ehp. A combination of mutational, interaction, and functional analyses subsequently revealed that while this second site was of lower affinity than of those found on the second alpha helices of both Ehp and Efb-C, it was required for the enhanced inhibitory potency of Ehp (Reviewed in (Geisbrecht 2008)).
While the structure/function relationships and precise mechanism of Ehp have not been fully investigated to the same extent as Efb-C, several informative conclusions can still be drawn from the available literature. In much the same way as for Efb-C (Hammel et al. 2007b), studies using mAb C3-9 strongly suggested that Ehp binding instigates a conformational change in the C3 protein (Hammel et al. 2007a). To manifest consequences at a functional level, such changes would presumably need to occur at the level of C3b as well. So far, however, neither the monomeric C3b/Ehp nor dimeric C3b3b/Ehp complex (Hammel et al. 2007a) has been examined structurally by the SAXS-MD or HDX-MS methods described above (Chen et al. 2010). The targets of Ehp activity have been identified as the surface-bound, C3b-containing C3 and C5 convertases (i.e. C3bBb, C3b3bBb, and C3b4b2b) (Jongerius et al. 2007). Although this is consistent with its overall similarity to Efb-C, a recent study has suggested a slightly different means to this end (Jongerius et al. 2010a). While both Efb-C and Ehp inhibit formation of active solid-phase convertases, Ehp was shown to stabilize fB binding to the convertase C3b scaffold and block its fD-dependent activation to Bb. This activity stands in contrast to Efb-C, which instead prevents initial fB binding to immobilized C3b (Chen et al. 2010). It is worth noting that this stabilizing activity is vaguely reminiscent to that described for SCIN-A (Ricklin et al. 2009; Rooijakkers et al. 2005). Thus, it is tempting to speculate that the bivalent nature of Ehp may somehow contribute to this effect, much in a way that SCIN-A forms C3b-containing pseudo-dimers (Jongerius et al. 2010b; Ricklin et al. 2009). In any case, the previously described differences in their C3/C3b/C3d-binding properties, along with these more recently discovered routes to convertase inhibition represent an intriguing starting point for future studies on Efb-C and Ehp.
3.5. Disruption of Adaptive Immune Engagement by Efb and Ehp
In addition to its roles in recognizing and marking pathogens for elimination, the complement system plays a pivotal role in stimulating the downstream mechanisms of adaptive immunity. Central to this feature of innate-adaptive crosstalk is the CR2 receptor, which is expressed by and localized to the surface of B cells, follicular dendritic cells, and immature T-cells (Carroll 2004). As part of the B cell coreceptor complex (in conjunction with CD19 and CD81), CR2 lowers the threshold of activation and affects the maturation of B cells upon binding of C3d-opsonized particles. As a consequence, the C3d/CR2 interaction is pivotal to properly engaging the adaptive immune system following complement opsonization of pathogen-derived biomaterial. Examination of the C3d/Efb-C (Hammel et al. 2007b) and C3d/Ehp (Hammel et al. 2007a) crystal structures, along with structure/function data regarding C3d/CR2 binding (Clemenza et al. 2000) raised the possibility that these staphylococcal proteins might also disrupt the C3d/CR2 interaction (Ricklin et al. 2008). Consistent with this, SPR-based competition assays demonstrated that only a slight molar excess of either Efb-C or Ehp was needed to completely inhibit CR2 binding to C3d (Ricklin et al. 2008). Furthermore the presence of wild-type Efb-C, but not a non-functional double mutant, was also shown to block binding of crosslinked C3 to a CR2-expressing B cell lymphoma line (Raji cells). Importantly, an inhibitory effect was also seen in mouse splenocytes, where inclusion of wild-type Efb-C, but not the mutant, impaired the C3d-mediated B-cell co-stimulatory response as judged by release of intracellular Ca2+ (Henson et al. 2001). Thus, even though their primary role appears to lie in disrupting formation of alternative pathway-derived convertases, both Efb-C and Ehp also inhibit CR2-mediated engagement of the adaptive immune system. Such versatility within a single protein family underscores the powerful evasion mechanisms that S. aureus has co-evolved in the presence of the host immune system.
4 Conclusions and Future Directions
The exquisitely tailored, yet non-redundant convertase-blocking capabilities of the SCIN and Efb protein families represent only a small fraction of the overall S. aureus immune evasion arsenal. Yet in just a few short years, intense study of these molecules has profoundly expanded our molecular-level understanding of complement inhibition and regulation. Given these significant changes in only two segments of what has become a surprisingly large field, it is almost certain that additional and unexpected discoveries still lie in waiting. For while diverse anti-complement effects for these two protein families have been identified already, the literature also clearly underscores that these advances have been achieved primarily through study of the structure, function, and mechanism of the SCIN-A and Efb-C proteins alone. In this regard, a more detailed examination of related proteins may be more relevant now, and this may very well open new doors for further inquiry. For example, study of the remaining SCIN proteins may aid in delineating the unique interactions described here and elsewhere that are required for the two distinct, but related SCIN functions of convertase inhibition and disruption of phagocytosis. Similarly, continued investigation into the functional significance of the second C3/C3b/C3d binding site in Ehp may highlight heretofore unappreciated differences between this protein and Efb-C.
Finally, it is important to mention that nearly all studies on S. aureus complement inhibitors reported so far have been carried out from a reductionist perspective, where only a single proteins’ function is examined at a time. While experimentally tractable, this dramatically oversimplifies the physiological situation where multiple immune evasion proteins are expressed simultaneously. Given that several of these proteins actually target the same structures (e.g. convertases), the possibility for functional synergy cannot be discounted. Regardless of the precise approaches taken, further analyses of these unique proteins will advance our understanding of S. aureus immune evasion and human complement regulation. Not only that, it has the potential to provide a valuable template for the design of complement-directed therapeutics as well.
References
- Alsenz J, Becherer JD, Nilsson B, Lambris JD. Structural and functional analysis of C3 using monoclonal antibodies. Curr Top Microbiol Immunol. 1989;153:235–248. doi: 10.1007/978-3-642-74977-3_13. [DOI] [PubMed] [Google Scholar]
- Becherer JD, Alsenz J, Esparza I, Hack CE, Lambris JD. A segment spanning residues 727–768 of the complement C3 sequence contains a neoantigenic site and accomodates the binding of CR1, factor H, and factor B. Biochemistry. 1992;31:1787–1794. doi: 10.1021/bi00121a029. [DOI] [PubMed] [Google Scholar]
- Boden MK, Flock JI. Evidence for three different fibrinogen-binding proteins with unique properties from Staphylococcus aureus strain Newman. Microb Pathog. 1992;12:289–298. doi: 10.1016/0882-4010(92)90047-r. [DOI] [PubMed] [Google Scholar]
- Carroll MC. The complement system in regulation of adaptive immunity. Nat Immunol. 2004;5:981–986. doi: 10.1038/ni1113. [DOI] [PubMed] [Google Scholar]
- Chambers HF, Deleo FR. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat Rev Microbiol. 2009;7:629–641. doi: 10.1038/nrmicro2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavakis T, Preissner KT, Herrmann M. The anti-inflammatory activities of Staphylococcus aureus. Trends Immunol. 2007;28:408–418. doi: 10.1016/j.it.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Chen H, Ricklin D, Hammel M, Garcia BL, McWhorter WJ, Sfyroera G, Wu YQ, Tzekou A, Li S, Geisbrecht BV, Woods VL, Jr, Lambris JD. Allosteric inhibition of complement function by a staphylococcal immune evasion protein. Proc Natl Acad Sci U S A. 2010;107:17621–17626. doi: 10.1073/pnas.1003750107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemenza L, Isenman DE. Structure-guided identification of C3d residues essential for its binding to complement receptor 2 (CD21) J Immunol. 2000;165:3839–3848. doi: 10.4049/jimmunol.165.7.3839. [DOI] [PubMed] [Google Scholar]
- Deleo FR, Otto M, Kreiswirth BN, Chambers HF. Community-associated meticillin-resistant Staphylococcus aureus. Lancet. 2010;375:1557–1568. doi: 10.1016/S0140-6736(09)61999-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forneris F, Ricklin D, Wu J, Tzekou A, Wallace RS, Lambris JD, Gros P. Structures of C3b in complex with factors B and D give insight into complement convertase formation. Science. 2010;330:1816–1820. doi: 10.1126/science.1195821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster TJ. Immune evasion by Staphylococci. Nat Rev Microbiol. 2005;3:948–958. doi: 10.1038/nrmicro1289. [DOI] [PubMed] [Google Scholar]
- Frank MM. Annihilating host defense. Nat Med. 2001;7:1285–1286. doi: 10.1038/nm1201-1285. [DOI] [PubMed] [Google Scholar]
- Garcia BL, Ramyar KX, Tzekou A, Ricklin D, McWhorter WJ, Lambris JD, Geisbrecht BV. Molecular basis for complement recognition and inhibition determined by crystallographic studies of the staphylococcal complement inhibitor (SCIN) bound to C3c and C3b. J Mol Biol. 2010;402:17–29. doi: 10.1016/j.jmb.2010.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisbrecht BV. Staphylococcal complement inhibitors: biological functions, recognition of complement components, and potential therapeutic implications. Adv Exp Med Biol. 2008;632:221–236. [PubMed] [Google Scholar]
- Gorwitz RJ, Kruszon-Moran D, McAllister SK, McQuillan G, McDougal LK, Fosheim GE, Jensen BJ, Killgore G, Tenover FC, Kuehnert MJ. Changes in the prevalence of nasal colonization with Staphylococcus aureus in the United States, 2001–2004. J Infect Dis. 2008;197:1226–1234. doi: 10.1086/533494. [DOI] [PubMed] [Google Scholar]
- Gouda H, Torigoe H, Saito A, Sato M, Arata Y, Shimada I. Three-dimensional solution structure of the B domain of staphylococcal protein A: comparisons of the solution and crystal structures. Biochemistry. 1992;31:9665–9672. doi: 10.1021/bi00155a020. [DOI] [PubMed] [Google Scholar]
- Gros P, Milder FJ, Janssen BJC. Complement driven by conformational changes. Nat Rev Immunol. 2008;8:48–58. doi: 10.1038/nri2231. [DOI] [PubMed] [Google Scholar]
- Hack CE, Paardekooper J, Smeenk RJT, Abbink J, Eerenber AJM, Nuijens JH. Disruption of the internal thioester bond in the third component of complement, (C3) which results in the exposure of neodeterminants also present on activation products of C3. An analysis with monoclonal antibodies. J Immunol. 1988;141:1602–1609. [PubMed] [Google Scholar]
- Hammel M, Sfyroera G, Pyrpassopoulos S, Ricklin D, Ramyar KX, Pop M, Jin Z, Lambris JD, Geisbrecht BV. Characterization of Ehp: a secreted complement inhibitory protein from Staphylococcus aureus. J Biol Chem. 2007a;202:30051–30061. doi: 10.1074/jbc.M704247200. [DOI] [PubMed] [Google Scholar]
- Hammel M, Sfyroera G, Ricklin D, Magotti P, Lambris JD, Geisbrecht BV. A structural basis for complement inhibition by Staphylococcus aureus. Nat Immunol. 2007b;8:430–437. doi: 10.1038/ni1450. [DOI] [PubMed] [Google Scholar]
- Haspel N, Geisbrecht BV, Lambris JD, Kavraki LE. Multi-scale characterization of the energy landscape of proteins with application to the C3D/Efb-C complex. Proteins. 2010;78:1004–1014. doi: 10.1002/prot.22624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haspel N, Ricklin D, Geisbrecht BV, Kavraki LE, Geisbrecht BV, Lambris JD. Electrostatic contributions drive the interaction between Staphylococcus aureus protein Efb-C and its complement target C3d. Protein Sci. 2008;17:1894–1906. doi: 10.1110/ps.036624.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmy KY, Katschke KJ, Gorgani NN, Kljavin NM, Elliott JM, Diehl L, Scales SJ, Ghilardi N, van Lookeren Campagne M. CRIg: a macrophage complement receptor required for phagocytosis of circulating pathogens. Cell. 2006;124:915–927. doi: 10.1016/j.cell.2005.12.039. [DOI] [PubMed] [Google Scholar]
- Henson SE, Smith D, Boackle SA, Holers VM, Karp DR. Generation of recombinant human C3dg tetramers for the analysis of CD21 binding and function. J Immunol Methods. 2001;258:97–109. doi: 10.1016/s0022-1759(01)00471-9. [DOI] [PubMed] [Google Scholar]
- Janssen BJ, Gomes L, Koning RI, Svergun DI, Koster AJ, Fritzinger DC, Vogel CW, Gros P. Insights into complement convertase formation based on the structure of the factor B-cobra venom factor complex. EMBO J. 2009;28:2469–2478. doi: 10.1038/emboj.2009.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen BJC, Christodoulidou A, McCarthy A, Lambris JD, Gros P. Structure of C3b reveals conformational changes underlying complement activity. Nature. 2006;444:213–216. doi: 10.1038/nature05172. [DOI] [PubMed] [Google Scholar]
- Janssen BJC, Huizinga EG, Raaijmakers HCA, Roos A, Daha MR, Ekdahl-Nilsson K, Nilsson B, Gros P. Structures of complement component C3 provide insights into the function and evolution of immunity. Nature. 2005;437:505–511. doi: 10.1038/nature04005. [DOI] [PubMed] [Google Scholar]
- Jongerius I, Garcia BL, Geisbrecht BV, van Strijp JA, Rooijakkers SH. Convertase inhibitory properties of staphylococcal extracellular complement-binding protein. J Biol Chem. 2010a;285:14973–14979. doi: 10.1074/jbc.M109.091975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongerius I, Köhl J, Pandey MK, Ruyken M, van Kessel KP, van Strijp JA, Rooijakkers SH. Staphylococcal complement evasion by various convertase-blocking molecules. J Exp Med. 2007;204:2461–2471. doi: 10.1084/jem.20070818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongerius I, Puister M, Wu J, Ruyken M, van Strijp JA, Rooijakkers SH. Staphylococcal complement inhibitor modulates phagocyte responses by dimerization of convertases. J Immunol. 2010b;184:420–425. doi: 10.4049/jimmunol.0902865. [DOI] [PubMed] [Google Scholar]
- Kajander T, Lehtinen MJ, Hyvarinen S, Bhattacharjee A, Leung E, Isenman DE, Meri S, Goldman A, Jokiranta TS. Dual interaction of factor H with C3d and glycosaminoglycans in host-nonhost discrimination by complement. Proc Natl Acad Sci USA. 2011;108:2897–2902. doi: 10.1073/pnas.1017087108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klevens RM, Morrison MA, Nadle J, Petit S, Gershman K, Ray S, Harrison LH, Lynfield R, Dumyati G, Townes JM, Craig AS, Zell ER, Fosheim GE, McDougal LK, Carey RB, Fridkin SK Active Bacterial Core surveillance (ABCs) MRSA Investigators. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA. 2007;298:1763–1771. doi: 10.1001/jama.298.15.1763. [DOI] [PubMed] [Google Scholar]
- Kluytmans J, Struelens M. Meticillin resistant Staphylococcus aureus in the hospital. BMJ. 2009;338:b364. doi: 10.1136/bmj.b364. [DOI] [PubMed] [Google Scholar]
- Laarman A, Milder F, van Strijp J, Rooijakkers S. Complement inhibition by gram-positive pathogens: molecular mechanisms and therapeutic implications. J Mol Med. 2010;88:115–120. doi: 10.1007/s00109-009-0572-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachmann PJ, Pangburn MK, Oldroyd RG. Breakdown of C3 after complement activation. Identification of a new fragment, C3g, using monoclonal antibodies. J Exp Med. 1982;156:205–216. doi: 10.1084/jem.156.1.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambris JD. The multifunctional role of C3, the third component of complement. Immunol Today. 1988;9:387–393. doi: 10.1016/0167-5699(88)91240-6. [DOI] [PubMed] [Google Scholar]
- Lambris JD, Ganu VS, Hirani S, Muller-Eberhard HJ. Mapping of the C3d receptor (CR2)-binding site and a neoantigenic site in the C3d domain of the third component of complement. Proc Natl Acad Sci USA. 1985;82:4235–4239. doi: 10.1073/pnas.82.12.4235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambris JD, Ricklin D, Geisbrecht BV. Complement evasion by human pathogens. Nat Rev Microbiol. 2008;6:132–142. doi: 10.1038/nrmicro1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee LYL, Hook M, Haviland D, Wetsel RA, Yonter EO, Syribeys P, Vernachio J, Brown EL. Inhibition of complement activation by a secreted Staphylococcus aureus protein. J Infect Dis. 2004a;190:571–579. doi: 10.1086/422259. [DOI] [PubMed] [Google Scholar]
- Lee LYL, Liang X, Hook M, Brown EL. Identification and characterization of the C3 binding domain of the Staphylococcus aureus extracellular fibrinogen-binding protein (Efb) J Biol Chem. 2004b;279:50710–50716. doi: 10.1074/jbc.M408570200. [DOI] [PubMed] [Google Scholar]
- Mamo W, Boden M, Flock JI. Vaccination with Staphylococcus aureus fibrinogen-binding proteins (FgBPs) reduces colonisation of S. aureus in a mouse mastitis model. FEMS Immunol Med Microbiol. 1994;10:47–53. doi: 10.1111/j.1574-695X.1994.tb00010.x. [DOI] [PubMed] [Google Scholar]
- Morgan HP, Schmidt CQ, Guariento M, Blaum BS, Gillsepie D, Herbert AP, Ka-vanagh D, Mertens HD, Svergun DI, Johansson CM, Uhrin D, Barlow PN, Hannan JP. Structural basis for engagement by complement factor H of C3b on a self surface. Nat Struct Mol Biol. 2011 doi: 10.1038/nsmb.2018. E-pub Ahead of Print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson B, Svensson KE, Borwell P, Nilsson UR. Production of mouse monoclonal antibodies that detect distinct neoantigenic epitopes on bound C3b and iC3b but not on the corresponding soluble fragments. Mol Immunol. 1987;24:487–494. doi: 10.1016/0161-5890(87)90023-x. [DOI] [PubMed] [Google Scholar]
- Nilsson UR, Nilsson B. Analogous antigenic alterations elicited in C3 by physiologic binding and by denaturation in the presence of sodium dodecylsulfate. J Immunol. 1982;129:2594–2597. [PubMed] [Google Scholar]
- Nishida N, Walz T, Springer TA. Structural transitions of complement component C3 and its activation products. Proc Nat Acad Sci USA. 2006;103:19737–19742. doi: 10.1073/pnas.0609791104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palma M, Nozohoor S, Schennings T, Heimdahl A, Flock JI. Lack of the extracellular 19-kilodalton fibrinogen-binding protein from Staphylococcus aureus decreases virulence in experimental wound infection. Infect Immun. 1996;64:5284–5289. doi: 10.1128/iai.64.12.5284-5289.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palma M, Shannon O, Quezada HC, Berg A, Flock JI. Extracellular fibrinogen-binding protein, Efb, from Staphylococcus aureus blocks platelet aggregation due to its binding the alpha-chain. J Biol Chem. 2001;276:31691–31697. doi: 10.1074/jbc.M104554200. [DOI] [PubMed] [Google Scholar]
- Palma M, Wade D, Flock M, Flock JI. Multiple binding sites in the interaction between an extracellular fibrinogen-binding protein from Staphylococcus aureus and fibrinogen. J Biol Chem. 1998;273:13177–13181. doi: 10.1074/jbc.273.21.13177. [DOI] [PubMed] [Google Scholar]
- Pangburn MK, Muller-Eberhard HJ. The C3 convertase of the alternative pathway of human complement. Enzymic properties of the biomolecular proteinase. Biochem J. 1986;235:723–730. doi: 10.1042/bj2350723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponnuraj K, Xu Y, Macon K, Moore D, Volanakis JE, Narayana SV. Structural analysis of engineered Bb fragment of complement factor B: insights into the activation mechanism of the alternative pathway C3-convertase. Mol Cell. 2004;14:17–28. doi: 10.1016/s1097-2765(04)00160-1. [DOI] [PubMed] [Google Scholar]
- Ricklin D, Hajishengallis G, Yang K, Lambris JD. Complement: a key system for immune surveillance and homeostasis. Nat Immunol. 2010;11:785–797. doi: 10.1038/ni.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricklin D, Lambris JD. Complement-targeted therapeutics. Nat Biotechnol. 2007;25:1265–1275. doi: 10.1038/nbt1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricklin D, Ricklin-Lichtsteiner SK, Markiewski MM, Geisbrecht BV, Lambris JD. Cutting edge: members of the Staphylococcus aureus extracellular fibrinogen-binding protein family inhibit the interaction of C3d with complement receptor 2. J Immunol. 2008;181:7463–7467. doi: 10.4049/jimmunol.181.11.7463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricklin D, Tzekou A, Garcia BL, Hammel M, McWhorter WJ, Sfyroera G, Wu YQ, Holers VM, Herbert AP, Barlow PN, Geisbrecht BV, Lambris JD. A molecular insight into complement evasion by the staphylococcal complement inhibitor protein family. J Immunol. 2009;183:2565–2574. doi: 10.4049/jimmunol.0901443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooijakkers SH, Ruyken M, Roos A, Daha MR, Presanis JS, Sim RB, van Wamel WJ, van Kessel KP, van Strijp JA. Immune evasion by a staphylococcal complement inhibitor that acts on C3 convertases. Nat Immunol. 2005;6:920–927. doi: 10.1038/ni1235. [DOI] [PubMed] [Google Scholar]
- Rooijakkers SH, Wu J, Ruyken M, van Domselaar R, Planken KL, Tzekou A, Ricklin D, Lambris JD, Janssen BJ, van Strijp JA, Gros P. Structural and functional implications of the alternative complement pathway C3 convertase stabilized by a staphylococcal inhibitor. Nat Immunol. 2009;10:721–727. doi: 10.1038/ni.1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooijakkers SHM, Milder FJ, Bardoel BW, Ruyken M, van Strijp JAG, Gros P. Staphylococcal complement inhibitor: structure and active sites. J Immunol. 2007;179:2989–2998. doi: 10.4049/jimmunol.179.5.2989. [DOI] [PubMed] [Google Scholar]
- Schuster MC, Ricklin D, Papp K, Molnar KS, Coales SJ, Hamuro Y, Sfyroera G, Chen H, Winters MS, Lambris JD. Dynamic structural changes during complement C3 activation analyzed by Hydrogen/Deuterium exchange mass spectrometry. Mol Immunol. 2008;45:3142–3151. doi: 10.1016/j.molimm.2008.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serruto D, Rappuoli R, Scarselli M, Gros P, van Strijp JA. Molecular mechanisms of complement evasion: learning from staphylococci and meningococci. Nat Rev Microbiol. 2010;8:393–399. doi: 10.1038/nrmicro2366. [DOI] [PubMed] [Google Scholar]
- Tamerius JD, Pangburn MK, Muller-Eberhard HJ. Detection of a neoantigen on human C3bi and C3d by monoclonal antibody. J Immunol. 1985;135:2015–2019. [PubMed] [Google Scholar]
- Tsai CJ, Del Sol A, Nussinov R. Protein allostery, signal transmission, and dynamics: a classification scheme of allosteric mechanisms. Mol Biosyst. 2009;5:207–216. doi: 10.1039/b819720b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesmann C, Katschke KJ, Yin J, Helmy KY, Steffek M, Fairbrother WJ, McCallum SA, Embuscado L, DeForge L, Hass PE, van Lookeren Campagne M. Structure of C3b in complex with CRIg gives insights into regulation of complement activation. Nature. 2006;444:217–220. doi: 10.1038/nature05263. [DOI] [PubMed] [Google Scholar]
- Wu J, Wu YQ, Ricklin D, Janssen BJ, Lambris JD, Gros P. Structure of complement fragment C3b-factor H and implications for host protection by complement regulators. Nat Immunol. 2009;10:728–733. doi: 10.1038/ni.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel PF, Skerka C. Complement regulators and inhibitory proteins. Nat Rev Immunol. 2009;9:729–740. doi: 10.1038/nri2620. [DOI] [PubMed] [Google Scholar]