Abstract
The problem of simultaneously clustering columns and rows (co-clustering) arises in important applications, such as text data mining, microarray analysis, and recommendation system analysis. Compared with the classical clustering algorithms, co-clustering algorithms have been shown to be more effective in discovering hidden clustering structures in the data matrix. The complexity of previous co-clustering algorithms is usually O(m × n), where m and n are the numbers of rows and columns in the data matrix respectively. This limits their applicability to data matrices involving a large number of columns and rows. Moreover, some huge datasets can not be entirely held in main memory during co-clustering which violates the assumption made by the previous algorithms. In this paper, we propose a general framework for fast co-clustering large datasets, CRD. By utilizing recently developed sampling-based matrix decomposition methods, CRD achieves an execution time linear in m and n. Also, CRD does not require the whole data matrix be in the main memory. We conducted extensive experiments on both real and synthetic data. Compared with previous co-clustering algorithms, CRD achieves competitive accuracy but with much less computational cost.
Keywords: co-clustering, matrix decomposition
1. INTRODUCTION
Clustering is a fundamental data mining problem with a wide variety of applications. It seeks good partitioning of the data points such that points in the same cluster are similar to each other and the points in different clusters are dissimilar. Many real-life applications involve large data matrices. For example, in text and web log analysis, the term-document data can be represented as contingency table. In biology domain, the gene expression data are organized in matrices with rows representing genes and columns representing experimental conditions. Recently there has been a growing research interest in developing co-clustering algorithms that simultaneously cluster both columns and rows of the data matrix. Co-clustering takes advantage of the duality between rows and columns to effectively deal with the high dimensional data. It has successful applications in gene expression data analysis [3] and text mining [5].
Many formulations of the co-clustering problem have been proposed, such as hierarchical model [12], bi-clustering model [3], pattern-based model [20] and so on. The partitioning-based model, which was first introduced in [12], has attracted much interest, because of the simplicity of the formalization and its close relationships to other well studied problems, such as spectral clustering and matrix decomposition [1, 5, 6, 8, 14, 15]. In this paper, we focus on the partitioning-based co-clustering formulation. Suppose that the data matrix D consists of m rows and n columns. Given input parameters k and l, the partitioning-based co-clustering algorithms aim to partition the rows of data matrix into k clusters and columns into l clusters to optimize certain objective functions measuring the quality of clustering results. Please refer to Section 2 for detailed discussion on recent development of co-clustering algorithms.
Although theoretically well studied and widely applied, existing co-clustering algorithms usually have time complexity in the order of m × n. For general data matrices, the information-theoretic co-clustering algorithm introduced in [5] takes O(t(k + l)mn) time to find the clustering results, where t is the number of iterations. Matrix-decomposition (such as nonnegative matrix factorization (NMF) [13]) based co-clustering methods [8, 15] have similar time complexity. In real-life applications, however, the numbers of rows and columns of the data matrices are usually large. For example, the term-document datasets may contain at least tens of thousands of articles and thousands of words [17]. The high throughput microarray techniques can monitor the expression values of tens of thousands of genes under hundreds to thousands of experimental conditions [16]. Such high time complexity limits the applicability of existing algorithms to these large datasets. Furthermore, these algorithms implicitly make the assumption that the whole data matrix is held in the main memory, since the original data matrix needs to be accessed constantly during the execution of these algorithms.
To address these limitations of existing work, in this paper, we propose a general co-clustering framework, CRD1, for large datasets. This framework is based on recently developed sampling-based matrix decomposition method CUR [9, 18]. Unlike NMF based algorithms, the complexity of CRD algorithms is linear in m and n. Moreover, most of the operations in CRD involve only the sampled columns and rows. Therefore, we do not require the whole data matrix be in main memory. This is crucial for large datasets. CRD can be implemented using different algorithms such as k-means or information-theoretic co-clustering methods. We conduct extensive experiments on both synthetic and several well-known real-life datasets. The experimental results show that CRD can be orders of magnitude faster than previous information-theoretic methods and NMF based methods. At the same time, it achieves comparable accuracy to other methods.
2. RELATED WORK
Co-clustering has attracted much research interest in recent years. It simultaneously clusters columns and rows in the data matrix. In each iteration of the co-clustering procedure, rows(columns) are clustered based on their similarities in the column(row) clusters.
A notable early work is presented in [12] which adopts a splitting procedure to perform hierarchical row and column clustering on the data matrix. Recently, an information-theoretic algorithm specifically designed for contingency table is presented in [5]. The algorithm monotonically increases the preserved mutual information by intertwining row and column clusterings at successive stages. In [1], the authors further propose a more general co-clustering framework based on Bregman divergence, which includes squared Euclidean distance, KL-divergence, Itakura-Saito distance, etc., as special cases. Thus it is applicable to a wide range of data matrices. Given a general data matrix with n rows and m columns, the complexity of the information-theoretic co-clustering approaches is O(t(k + l)mn), where t is the number of iterations. This is because, in each iteration, the algorithm needs to compare each column or row with respect to the representative point of each cluster in the current clustering result. For sparse matrices, the algorithm can have O(t(k + l)|E|) complexity by efficient implementation, where |E| is the number of non-zero elements in the matrices.
Co-clustering is closely related to matrix decomposition. Singular value decomposition (SVD) [11] is perhaps the most well-known matrix decomposition method. The lower rank singular vector or eigenvector space provides a compact representation of the original space. Some research work [4, 7] clusters data points in the transformed space. However, since the factor matrices may contain negative values, it is hard to interpret the co-clustering results directly from SVD. Nonnegative matrix factorization (NMF) [13] imposes the constraint that the factor matrices contain only nonnegative values and has been successfully applied in document clustering [19]. In [15], the authors propose block value decomposition (BVD) for co-clustering. BVD generalizes the idea of NMF to factorize the original matrix into three nonnegative matrices, which provides intuitive interpretation of the co-clustering structure in the data matrix. The theoretic relationship between NMF and co-clustering is studied in [8]. It has been shown that the objective of k-means co-clustering and nonnegative matrix tri-factorizations with the orthogonal constraint are the same. A similar idea is also exploited in [14]. Similar to information-theoretic co-clustering, the time complexity of matrix decomposition based co-clustering methods usually is also in the order of m × n. For example, the complexity of BVD is O(t(k + l)mn) with t being the number of iterations. This is because updating the entries of the factor matrices usually involves multiplications of the data matrix.
Unlike PCA/SVD and NMF which generate new bases for the data space, recently developed matrix decomposition methods CUR [9] and CMD [18] sample columns and rows from the original data matrix to form factor matrices. Because of the random sampling procedure, these methods are non-deterministic. However, they can guarantee a provable bound on reconstruction error in probabilistic sense. CUR has been successfully applied in recommendation system [10].
3. PRELIMINARIES
In this section, we present notations that will be used in the paper and we provide formal definitions.
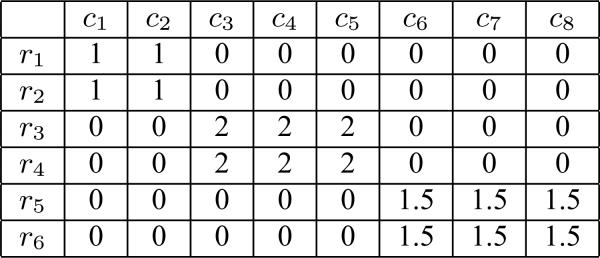
We denote the two-dimensional matrix(table) as . R = {r1, r2, ..., rm} represents the set of row vectors of M, where ri is the ith row vector. C = {c1, c2, ..., cn} represents the set of column vectors, where cj is the jth column vector. Thus, we have M = (c1...cn) = (r1...rm)T. The element at the ith row and jth column is denoted by mij. Note that in this paper, we use bold font, such as ci, to represent vector and use normal font, such as mij, to represent scalar. An example matrix consisting of 6 rows and 8 columns is shown in Figure 1.
Figure 1.
Example Matrix
A co-clustering of matrix M consists of a set of row clusters and column clusters. Let denote the row cluster set and denote the column cluster set. Each row of M belongs to one of the row clusters. This relationship can be considered as a many-to-one mapping from rows to row clusters, i.e., . The relationship between columns and column clusters is similar, i.e., .
Our CRD framework consists of two components.
Low rank matrix decomposition: The data matrix is decomposed using a subset of its rows and columns. The decomposition procedure must be fast and accurate.
Co-clustering on the subset of rows and columns: The selected rows and columns are co-clustered first. The cluster labels for the rest rows and columns are assigned based on those selected ones. In general, any co-clustering algorithm that optimizes its objective function by alternating the clustering of rows and columns can be used in CRD.
There may be many low rank decompositions for a given data matrix. However, not all of them are appropriate for co-clustering. Therefore, we provide definitions for qualified low rank row/column decompositions that can be used in CRD.
Definition 3.1. Low rank row decomposition: A low rank row decomposition of M approximates the original matrix using a subset of rows.
| (1) |
WR · MR is a qualified low rank row decomposition of M if it satisfies the following constraints,
, where ε < 1, is a user-specified approximation rate. ∥ · ∥F represents the Frobenius matrix norm [11].
, where .
Given , the corresponding rows in WR have , for all i ∈ {1, ..., m′} and j ∈ {1, ..., m′} − {i}.
Constraint 1 requires that matrix has small approximation error bounded by a user-specified threshold ε. The number of rows, m′, that need to be included in MR can be calculated from ε and the rank of M. Constraint 2 ensures that each row vector in MR is from M. And Constraint 3 ensures the correct mapping of the row cluster labels between selected rows and the rest rows. Details about the decomposition and its usage will be discussed in Sections 4 and 5 respectively.
Let WR be represented by its row vectors, i.e., WR = ( w1 w2 ... wm )T. Since , for each row vector in the approximation matrix , we have,
Therefore, for each row , its corresponding row in WR can be considered as a projection of onto the sub-space formed by the rows in MR.
The definition of low rank column decomposition of M is similar.
Definition 3.2 Low rank column decomposition: A low rank column decomposition of M approximates the original matrix using a subset of columns.
| (2) |
MC · WC is a qualified low rank column decomposition of M if it satisfies the following constraints,
, where ε < 1.
, where .
Given , the corresponding columns in WC have , for all i ∈ {1, ..., n′} and j ∈ {1, ..., n′} − {i}.
For example, we can find a low rank row/column decomposition for the matrix in Figure 1. They are
For both decompositions, we have . Note that the rows in MR and the columns in MC are all selected from M. And both WR and WC satisfy Constraint 3 in our definition.
Using the 4 matrices, {MR, WR, MC, WC}, we want to find a co-clustering of M which is as good as the result obtained by running previous co-clustering algorithms on M directly.
4. LOW RANK ROW/COLUMN DECOMPOSITION
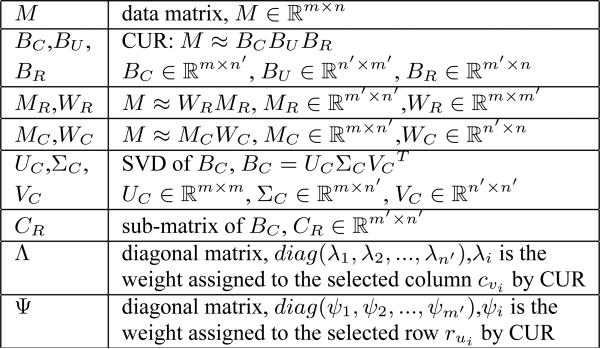
In our CRD framework, the row/column decomposition serves as a pre-processing step. Once M is decomposed into qualified matrices defined in Definitions 3.1 and 3.2, co-clustering algorithms will be applied on the decomposition matrices, {MC, WC, MR, WR}. In this section, we explain the details of the low rank row/column decomposition of the original matrix M. Before we start, we list a set of matrix notations that will be frequently used in this section in Figure 2.
Figure 2.
Notations for frequently used matrices
As defined in Definition 3.1, in low rank row decomposition, the matrix M is approximated by two decomposition matrices, MR and WR. MR consists of a set of row vectors selected from M which establish a row sub-space. And WR can be considered as a projection of all the row vectors in M onto the sub-space formed by rows in MR.
We can use SVD to calculate WR from MR. First, we construct the orthogonal basis of MR using SVD, i.e., MR = URΣRVRT. Without loss of generality, we can assume that m′ < n, and let be a sub-matrix of VR consisting of the first m′ row vectors in VR(right eigenvectors). Then we can get a low rank approximation of M as
Since we know that ,
Thus, .
In order to improve the efficiency of the entire algorithm, the approximate decomposition of M need to be calculated very efficiently. Most of the previous co-clustering algorithms have runtime complexity equal to or higher than O(t(k + l)mn) where k and l are the number of row/column clusters and t is the number of iterations. Therefore, the decomposition method we use need to have complexity lower than O(t(k + l)mn). When using SVD to calculate WR, its time complexity is O(mnm′) where m′ is the number of selected rows. Though m′ << m, m′ is greater than or equal to k. Thus, it is inefficient to use SVD directly to calculate WR.
In this paper, we use the CUR decomposition method proposed in [9, 18] to find qualified low rank row/column decompositions. The runtime complexity of CUR is mn + O((m + n)m′n′), in which mn is used to calculate the norm of each row/column vector. In fact, the norm of row and column vectors only need to be calculated once and can be stored with the matrix for further use. Besides mn, CUR has runtime O((m + n)m′n′) which is much smaller than O(t(k + l)mn).
The CUR method approximately decomposes M into three matrices, i.e.,
where is a set of weighted columns selected from is a set of weighted rows selected from M and . Rows and columns in BC and BR are randomly selected with probability in proportional to their norm. And BU is calculated based on BC and BR.
If matrix M has rank equal to k, it is proven in [9] that
holds with probability (1 − δ) if the numbers of selected columns and rows are at least
Thus, if we use a small δ, with high probability, the CUR decomposition satisfies the first constraint on the approximation error between M and in Definitions 3.1 and 3.2. Details about the decomposition algorithm can be found in [9, 18].
Before we can use CUR to generate our low rank row/column decomposition, we have to make some changes in the method.
BC and BR consist of weighted column and row vectors of M. While in our CRD algorithm, we need MC and MR consist of original row and column vectors of M.
We only need to decompose M into two matrices while CUR gives us three. A simple multiplication of two of the matrices in {BC, BU, BR} seems to be able to get WR and WC, i.e., WR = BC · BU and WC = BU · BR. However, we need to select rows and columns that do not violate Constraint 3 in Definitions 3.1 and 3.2. We will discuss it in the next section.
4.1 Generate Low Rank Row Decomposition Using CUR
First, we generate MR from BR. To approximate M, the three matrices used by CUR are,
| (3) |
where and .
By multiplying each row vector in BR with , we get
| (4) |
Obviously, MR in Equation 4 satisfies Constraint 2 in Definition 3.1.
Next, we generate a qualified WR. By replacing BR in Equation 3 with Ψ · MR, we get
| (5) |
In CUR, given BC and BR,
| (6) |
where VC and ΣC are singular vectors and singular values of BC, i.e., consists of the {u1, ..., um′}th rows of BC (the set of selected rows in BR) and each row in is weighted by the corresponding ψi. To remove the weight ψi from each row of , let
| (7) |
Using Equations 6 and 7, we have
| (8) |
Without loss of generality, we assume that n′ < m. Let be a sub-matrix of UC which consists of the first n′ column vectors in UC. We have
| (9) |
By combing Equations 8 and 9, we have
| (10) |
where consists of the {u1, ..., um′}th rows of which correspond to the selected rows in BR.
Let WR = BC · BU · Ψ2, we have
| (11) |
Constraint 3 in Definition 3.1 only involves those rows in WR which correspond to the selected rows in MR(BR). Let this sub-matrix of WR be , we have
| (12) |
Let satisfies Constraint 3 if and only if
| (13) |
is generated from according to the set of rows selected in BR. In order to generate a qualified WR, Equation 13 must be verified every time a new row is selected to be included in BR.
The algorithm to generate low rank row decomposition using CUR is shown in Figure 3.
Figure 3.
Generate Low Rank Row Decomposition Using CUR
In Figure 3, the steps labelled with CUR are the steps in the original CUR decomposition method. The remaining steps in Figure 3 are added to enforce the constraints defined on WR and MR. The qualification of WR is ensured by the test in Step 8. A new row is added to BR if and only if it does not violate Equation 13. The qualification of MR is ensured in Step 12.
Note that the SVD operation in Step 2 is calculated by eigenvalue decomposition on which is a small n′ × n′ matrix. is not calculated until Step 3.
4.2 Generate Low Rank Column Decomposition Using CUR
The algorithm in Figure 3 only ensures the qualification of WR and MR. Since the columns are randomly selected without any restriction in Step 1 in Figure 3, we cannot guarantee qualified MC and WC by simply letting MC = BCΛ−1 and WC = ΛBUBR.
However, if we transpose the original matrix M, exchange m′ and n′, and use them as input to the algorithm in Figure 3, we can get qualified MC and WC by letting
| (14) |
Therefore, in order to get a qualified low rank row/column decomposition, we need to run the algorithm in Figure 3 twice. In general, if the norms of row and column vectors of a data matrix have been calculated, our decomposition method will take O((m + n)m′n′) time to find a set of qualified matrices {MR, WR, MC, WC} where m′ and n′ are the numbers of selected rows and columns respectively. As we will show in the experiment section, the time used on finding the decomposition is minimal and can be neglected.
5. A GENERAL FRAMEWORK FOR CO-CLUSTERING USING ROW/COLUMN DECOMPOSITION: CRD
In order to perform co-clustering on large data matrix efficiently, in our CRD framework, the co-clustering is performed on the decomposition matrices instead of the original matrix M. It provides significant improvements in both space and time utilization.
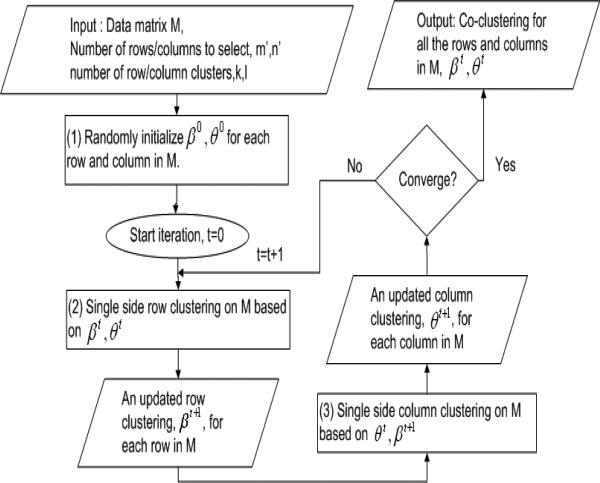
As we discussed in Section 2, one type of co-clustering algorithms cluster the rows and columns of a data matrix alternatively in successive iterations. These algorithms include the Information Theoretic Co-clustering algorithm [5], the Bregman Co-clustering algorithm [1] and the Fully Automatic Cross-association algorithm in [2]. In each iteration, they either keep the row side clusters fixed and re-cluster the columns or keep the column side clusters fixed and re-cluster the rows. It is proved that this single side clustering (row or column) can guarantee the objective function on the co-clustering structure converge to a local minimum (maximum). We call it the Iterative Single Side Clustering approach.
Suppose that there are k row clusters, , and l column clusters, . The co-clustering structure after the tth iteration is represented by βt and θt which map each row/column in the data matrix to a row/column cluster.
An illustration of the Iterative Single Side Clustering approach is shown in Figure 4.
Figure 4.
Co-clustering using Iterative Single Side Clustering Approach
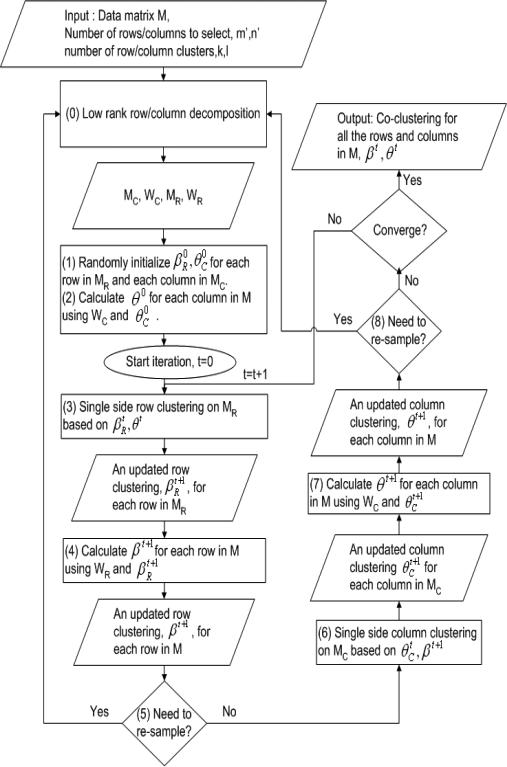
In this paper, we use this general approach as the co-clustering component in our CRD framework. In each iteration, instead of re-clustering all rows (or columns), we only re-cluster the selected rows in MR or the selected columns in MC. Then we assign cluster label to each row (or column) in M based on WR (or WC) and the cluster labels of the selected rows (columns).
Let the single-side clustering structure on the rows in MR and the columns in MC after the tth iteration be
A detailed illustration of CRD is shown in Figure 5.
Figure 5.
Framework for Co-clustering using Decomposition: CRD
As we can see from Figures 5 and 4, Steps (1,2) in Figure 5 correspond to Step (1) in Figure 4, Steps (3,4) in Figure 5 correspond to Step (2) in Figure 4 and Steps (6,7) in Figure 5 correspond to Step (3) in Figure 4. Also, for those steps in the iteration loop, the steps in Figure 5 have much smaller time complexity than the corresponding steps in Figure 4. The comparison is shown in Table 1.
Table 1.
Complexity of Corresponding Steps in Co-clustering
In general, co-clustering using Iterative Single Side Clustering approach can have runtime complexity equal to (or larger than) O(t(k + l)mn) where t is the number of iterations. While CRD only uses O(t(km′n + ln′m + m′m + n′n)) plus the time used in matrix decomposition, O((m + n)m′n′), which is a one-time cost at the beginning. Since usually we have m′ << m and n′ << n, CRD is much faster.
The single side clustering in Steps (3) and (6) of CRD is exactly the same as the one in the Iterative Single Side Clustering approach. Details about the steps can be found in [5, 1]. We will discuss the two new steps, Steps (4,7) and Steps (5,8) in the following sections respectively.
5.1 Mapping Cluster Labels from Selected Rows /Columns to All Rows/Columns
In Steps (2), (4) and (7) of CRD, we calculate β (θ) for each row (column) in M using WR (WC) and βR (θC). Since the procedure for calculating θ on columns is the same as that of calculating β on rows, we will only explain the calculation of β.
Using low rank row decomposition, M is approximated by WR and MR. Each row of WR can be considered as a projection of the corresponding row of M onto the sub-space formed by the rows in MR, i.e., there is a one-to-one relationship between a row of M and the corresponding row of WR. Therefore, when we get the row cluster label for each row of MR, we can use WR to calculate the cluster label for each row of M.
Suppose that for each row of MR, we have . For each row ri of M, we calculate its label vector si = [si1, si2, ..., sik], in which
| (15) |
Then we have,
| (16) |
Intuitively, sij is the correlation between row ri and row cluster . In Equation 16, ri is assigned to the row cluster having the highest correlation.
Now we prove the following property.
Property 5.1. For each row of MR and its corresponding row ri of M, if we use Equations 15 and 16 to calculate β, we have
| (17) |
Proof: According to Constraint 3 in Definition 3.1, we have |wij| > |wiz|, z ∈ {1, ..., m′} − {j}. Assume that . When calculating si, we get
According to Equation 15, we know sit ∈ {|wi1|, ..., |wim′|}, t ∈ {1, ..., k}. Thus we get
Applying Equation 16, we get
According to Property 5.1, when calculating β from βR, the selected rows of M will have the same cluster label as their corresponding rows in MR.
The calculation of θ is the same as that of β. Moreover, θ also holds a property similar to Property 5.1, which can be derived from Constraint 3 in Definition 3.2. We thus omit the formal discussion due to space limitations.
5.2 Re-sample Data Matrix
Although we add several constraints to the low rank row/column decomposition, the rows/columns are still randomly sampled from the original data matrix. It is possible that some row/column clusters are not sampled, i.e., none of the rows/columns in those clusters are selected in MR/MC. Though this problem does not happen frequently, its occurrence may impact the accuracy of our method. Therefore, we introduce a re-sampling step in CRD to solve the problem.
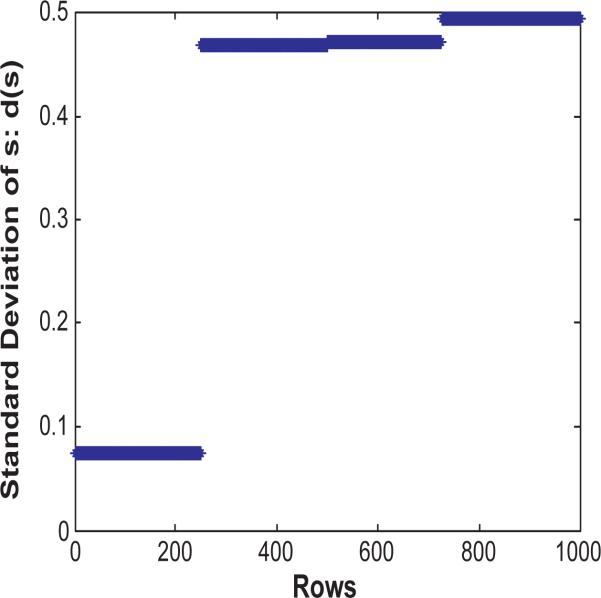
Consider the scenario where an existing row cluster is not sampled in MR, then for each row vector , all of the m′ row vectors in MR are almost equally irrelevant to rji. Thus if we calculate the label vectors of rji according to Equation 15, it is expected that the standard deviation of the elements in s is very low.
Let d(s) denote the standard deviation of the elements in s. An example is shown in Figure 6. The figure plots d(s) for each row in one iteration of the co-clustering. The first 250 rows form a cluster in the data. However, none of them is selected in MR. As we can see, for those rows, d(s) is much lower than other rows which belong to properly sampled clusters.
Figure 6.
Standard deviation of label vectors
In CRD, we use Chebyshev's Inequality to detect those rows and columns. Consider d(s) of all rows(columns) as a distribution with expected value μ and standard deviation σ, Chebyshev's inequality entails that, for any real number q > 0 we have the conservative bound:
| (18) |
We set q = 2 in CRD, i.e., if there are more than of the rows (columns) having |d(s) − μ| ≥ 2σ, the algorithm will quit the iteration and re-sample the rows (columns).
In general, larger τ induces lower re-sampling frequency and shorter runtime while smaller τ induces higher re-sampling frequency and longer runtime. Since Chebyshev's Inequality holds for any distribution, is a rather loose bound. In fact, we found in the experiments that the bound is too loose and does not induce re-sampling sufficiently. Therefore we test various values of τ to obtain the optimal frequency of re-sampling. Based on our experiments on both real and synthetic data, instead of 25%, τ = 12.5% has the best tradeoff between runtime and cluster purity (as defined in Section 6).
In Steps (5,8) of Figure 5, we calculate d(s) for each row(column). If the percentage of rows (columns) having |d(s) − μ| ≥ 2σ is larger than τ, the re-sampling works in the following way.
All the rows(columns) that are already in MR (MC) will be kept.
If the number of rows(columns) having d(s) ≤ μ − 2σ is t, then rows ( columns) will be randomly re-sampled only from this set of rows (columns).
When adding new rows/columns into MR and MC, they must not violate Constraints 1 ~ 3 in Definitions 3.1 and 3.2.
Note that in Step (5) of Figure 5, only rows are re-sampled, and in Step (8), only columns are re-sampled.
The re-sampling only involves the calculation of a new BU which is very fast. BC and BR only need to be expanded to include the new columns and rows.
After the re-sampling, the iteration will start over again. In Step (1), those rows (columns) that are already in MR (MC) will retain their cluster labels obtained before the re-sampling. All the new rows (columns) will start with the same new cluster label, e.g, βR(r) = k + 1 (θR(c) = l + 1).
In general, CRD has runtime complexity O(t(km′n + ln′m + m′m + n′n)) plus the time used in matrix decomposition, O((m + n)m′n′). For large dataset, it's easy to get m′ << m and n′ << n, and CRD can be orders of magnitude faster than the previous co-clustering algorithms that have runtime complexity at least O(t(k + l)mn).
6. EXPERIMENTS
In this section, we present results on both synthetic data and real data. In order to demonstrate the effectiveness and efficiency of our CRD algorithms, all the datasets have large number of rows and columns. Several other co-clustering algorithms were also implemented for comparison.
Datasets
We use two real datasets and one synthetic dataset in our experiments.
Multiple Features Dataset2: This dataset consists of features of handwritten numerals (‘0’–‘9’) extracted from a collection of Dutch utility maps. It contains 2000 numerals (rows) and 240 features. There are 10 classes in the dataset (from 0 to 9) and each of them has 200 numerals.
20 Newsgroup Dataset3: The package consists of 20000 documents from 20 major newsgroups. Each document is labelled by the major newsgroup in which it is involved. We preprocess the 20 Newsgroup dataset to build the corresponding two dimensional contingency table. Each document is represented by a row in the table and 2000 distinct words are selected to form 2000 columns. Words are selected using the same method as that in [17].
Synthetic dataset: We generated a set of synthetic data with different sizes and noise levels. Each synthetic data is an m × m square matrix consisting of 0 and 1. Co-clusters are embedded by setting different blocks in the matrix to be 1. And we use parameter e to control the noise level, i.e., the value of each element can be randomly flipped with probability e.
Algorithms
We implemented two versions of our CRD framework using different iterative single side clustering approaches.
CRD-ITC: CRD using information-theoretic co-clustering [5].
CRD-kmeans: CRD using Euclidian distance (k-means) co-clustering [1].
Unless otherwise noted, we set q = 2 and use bound τ = 12.5% for re-sampling in the CRD algorithms.
In order to show the efficiency of our CRD framework, we also implemented three recent co-clustering algorithms.
Performance Measurement
In the experiments, we compare the runtime and purity of co-clustering solution of the algorithms.
The purity measurement was used in [8]. The purity of a clustering solution is calculated as the weighted sum of the purity of each cluster and is given by,
where m is the total number of rows(columns), mi is the size of cluster i and is the number of rows(columns) in cluster i that has (real) class label j. The larger the value of purity, the better the clustering is.
In the real datasets, since we only know the true class labels of the rows, the purity is only calculated and compared on row clusters. While in the synthetic data, we compare the purity of both row and column clusters found by co-clustering.
All algorithms were implemented in MATLAB. All experiments were conducted on a PC with CPU P4 3GHz, 1G RAM and 80G HDD.
6.1 Synthetic Data
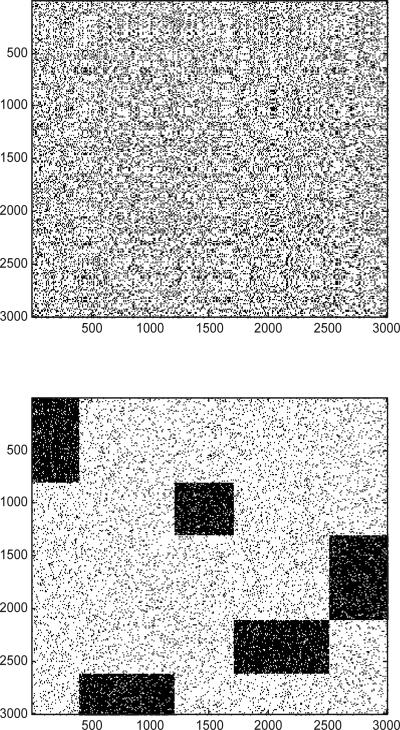
In this section, we compare the performance of the algorithms on a set of synthetic data. For each synthetic dataset of size m × m, we embed 5 row/column clusters into the matrix. An example dataset of size 3000 × 3000 is shown in Figure 7. The figure at the top plots the original data matrix: the dark point represents the ’1’ elements in the matrix. And the figure at the bottom shows the co-cluster structure after the orders of the columns/rows are rearranged.
Figure 7.
Example of a synthetic data matrix of size 3000×3000
We vary the following parameters separately in the experiments:
m: size of synthetic data matrix (for both row and column). Default value, 5000.
e: noise level of synthetic data. Default value, 0.1.
m′: number of selected rows (columns) in low rank row/column decomposition when using CRD. Default value, 20.
When varying one of the parameters, the rest use their default values.
Varying m
By varying m, the number of rows and columns in the data matrix are changed simultaneously, e.g., when m is doubled, the data matrix will have size 2m × 2m. We vary m from 3000 to 11000. The runtime and row/column cluster purity of the algorithms are plotted in Figures 8 and 9.
Figure 8.
Runtime performance when varying m
Figure 9.
Purity performance when varying m
As we can see, CRD algorithms are orders of magnitude faster than the other three algorithms. ONMF is the slowest algorithm because it uses more iterations to converge. When m becomes larger than 9000, the entire matrix is too large to be loaded into the MATLAB environment. Therefore, we did not get any results for ITC, Kmeans and ONMF for m ≥ 9000. While for our CRD algorithm, there is no problem for such large matrices, since it only needs to load {MC, WC, MR, WR} which are much smaller.
The runtime of calculating the low rank row/column decomposition is shown in Figures 8 at the bottom. The decomposition is very efficient. It only takes a small portion of the total runtime of CRD algorithms. When m ≥ 9000, since the data matrix cannot be entirely loaded, our decomposition algorithm have to load the matrix piece by piece when calculating the norm of each column and row. That's the reason for the steep change in runtime when m passes 9000.
The purity of all algorithms decreases when we increase the number of rows/columns in the matrix. One reason is that the increasing number of rows/columns (with noise) brings more local optimums into the data, which makes it more difficult for the algorithms to find the embedded co-clustering structure. The purity of CRD algorithms is comparable to that of the other algorithms as shown in Figure 9. Thus, the dramatic saving in its running time makes CRD the winner among all algorithms.
Varying e
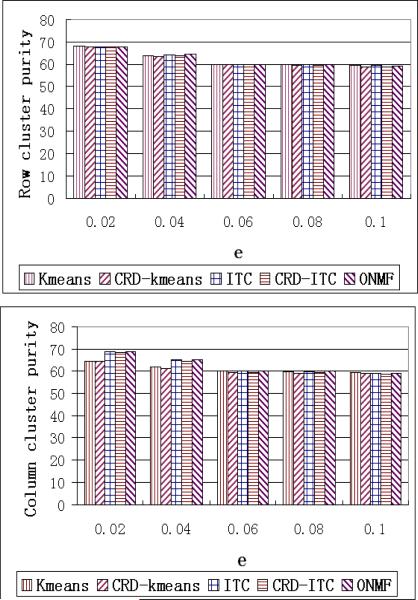
When we increase the value of e, the synthetic data have more noise. The largest row/column cluster embedded in the data covers only 20% of the rows/columns. When e exceeds 0.2, the embedded structure begins to be overwhelmed by noise. Therefore, in this set of experiments, we vary e from 0.02 to 0.1.
The runtime of the algorithms does not change so much with increasing e. So we only report the purity performance in Figure 10.
Figure 10.
Purity performance when varying e
The purity of CRD algorithms is comparable to others but is slightly lower when e becomes larger. One reason is that, the effectiveness (accuracy) of the low rank decomposition is sensitive to the increase in noise. Though we have the re-sampling procedure in CRD, the computation of the updated weight matrices, WR and WC, is still affected by the noise.
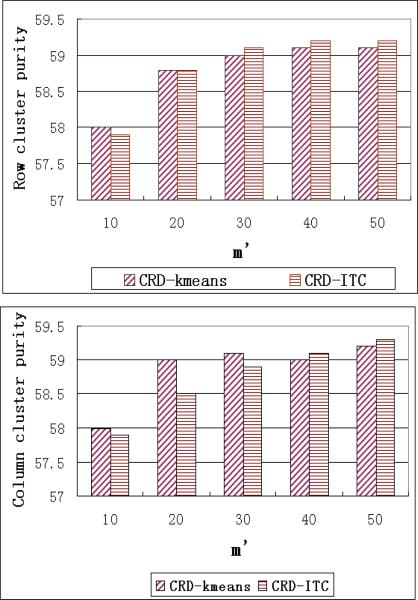
Varying m′
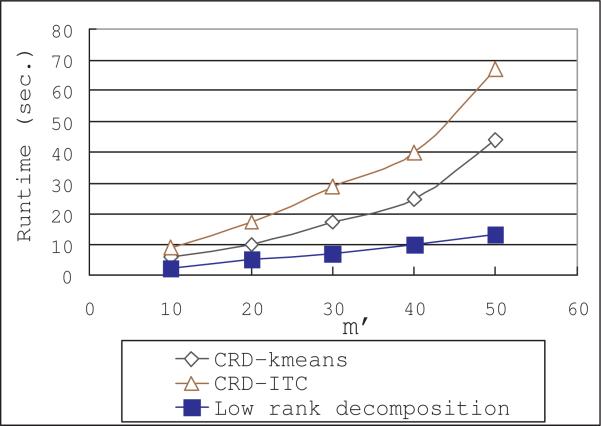
In this set of experiments, we vary the number of selected rows/columns, m′, from 10 to 50. This parameter does not affect the ITC, Kmeans and ONMF algorithms, therefore we only plot the cluster purity and runtime of our CRD algorithms in Figures 11 and 12 respectively.
Figure 11.
Purity performance when varying m′
Figure 12.
Runtime performance when varying m′
As we can see, the CRD algorithms and their pre-processing procedure, the decomposition algorithm, both have runtime increase linearly with respect to m′. Increasing the number of selected rows/columns can also improve the purity of the co-clustering solutions. However, when there are enough rows/columns in MC and MR, adding more rows/columns can only affect the purity slightly. As shown in Figure 11, when there are more than 30 rows/columns, the improvement of purity becomes very small.
Efficiency of Re-sampling
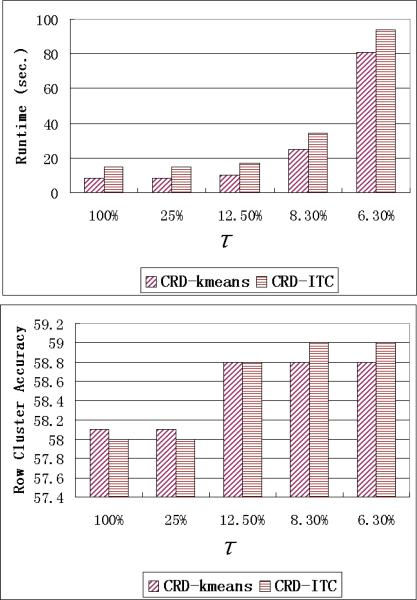
As we discussed in Section 5.2, the random sampling of matrix decomposition does not guarantee to sufficiently sample all cluster structures and re-sampling is used in CRD to make up for the missing clusters in random sampling. We use Equation 18 to detect those rows/columns that are not properly sampled. For all the previous experiments we set q = 2 and use bound τ = 12.5%, i.e., if there are more than 12.5% of the rows (columns) having |d(s) − μ| ≥ 2σ, the CRD algorithm will re-sample the rows (columns).
In order to show the effects of τ on re-sampling frequency and clustering performance, we vary τ from 100% to 6.3% in this set of experiments. Note that when the τ is equal to 100%, the CRD algorithm does not re-sample rows/colums in any case. The results on the 5000 × 5000 synthetic data are shown in Figure 13. Column cluster purity is omitted because it is similar to the row cluster purity.
Figure 13.
Runtime and Row cluster purity performance with different re-sampling bound τ
Considering runtime and clustering performance together, τ = 12.5% has the best performance. When τ = 25%, there is almost no re-sampling in CRD. When τ is set to 6.3%, the algorithm consumes much longer time but has no significant improvement in clustering performance. There are two reasons for the longer runtime when bound= 6.3%
More frequent matrix decomposition computation.
Larger size of {WR, MR, WC, MC}.
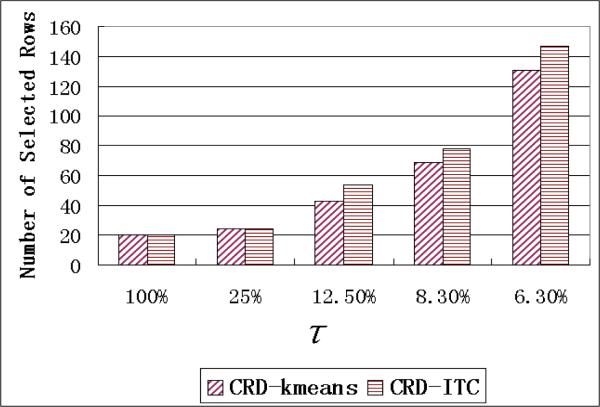
The numbers of selected rows for these bounds are shown in Figure 14. As we can see, when τ = 25%, only a few rows are re-sampled. And when τ = 6.3%, there are too many rows selected in the MR matrix.
Figure 14.
Number of selected rows with different re-sampling bound τ
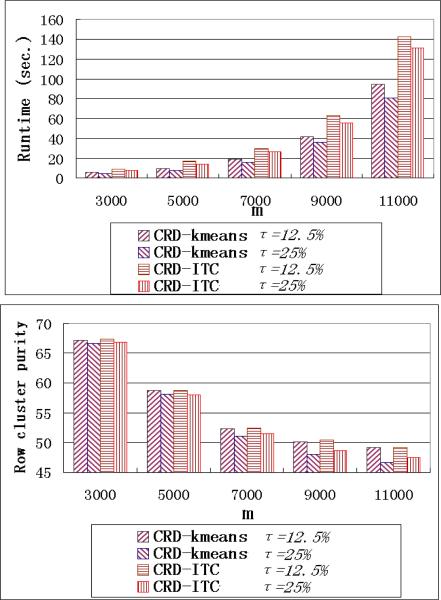
We also use τ = 25% and τ = 12.5% on datasets of different sizes and compare the performance. The results are shown in Figure 15. As expected, the runtime of the CRD algorithms with τ = 25% is always slightly faster. However, its purity of row clustering is lower than that of the CRD algorithms with τ = 12.5% especially when there are a large number of rows and columns in the data matrix, e.g., m ≥ 9000. In those cases, the initial low rank decomposition may have a larger risk to miss a cluster or sample insufficiently in a cluster with a mixed number of selected rows (columns), and the re-sampling steps becomes more important.
Figure 15.
Runtime and Row cluster purity performance of τ = 25% and τ = 12.5% on data of different sizes
6.2 Multiple Feature Dataset
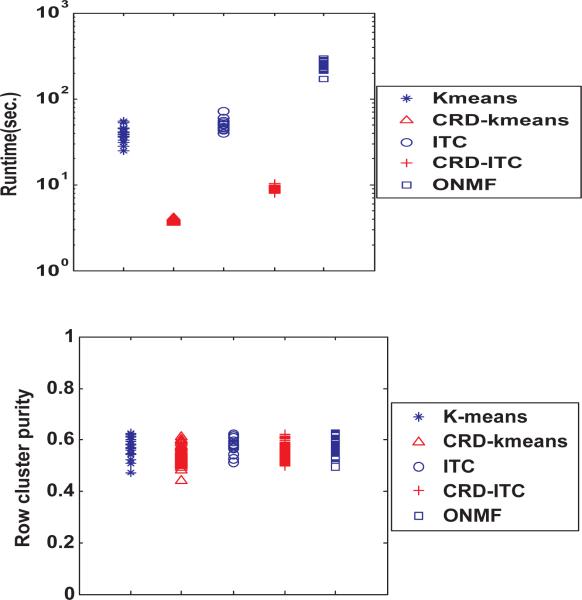
The multiple feature dataset contains 2000 rows and 240 columns. And the 2000 rows are equally divided into 10 classes. We run each algorithm 40 times on the dataset, and plot the distribution of their runtime and row cluster purity in Figure 16. Since the columns do not have class label, the column cluster purity cannot be calculated here.
Figure 16.
Runtime and Row cluster purity performance on Multiple Feature Dataset
Each point in Figure 16 represents the runtime or the purity of a single run of one of the algorithms. As we can see, our CRD algorithms are about 10 times faster than ITC and Kmeans and more than 20 times faster than ONMF. The performance advantage of the CRD algorithms is not as much as the one on the synthetic data because the multiple feature dataset has much less columns than rows, and thus most of the speedup comes from the row-side sampling only.
And if we compare the distributions of cluster purity in Figure 16, we can find that CRD algorithms performs as good as the other three algorithms.
6.3 20 Newsgroup Dataset
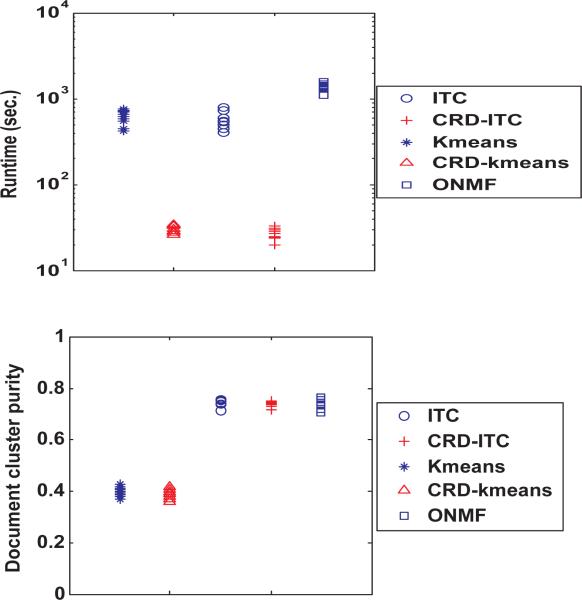
In this section, we use the 20 newsgroup dataset to compare the performance. The entire data matrix is too large to be loaded into MATLAB environment, therefore, we use a subset of the dataset which contains 5000 documents from 5 newsgroups. And 2000 words are selected as features using the same method as in [17]. We run each algorithms 10 times on the dataset and plot the distribution of runtime and row (document) cluster purity in Figure 17. Words do not have class label so that the purity of word clusters cannot be calculated.
Figure 17.
Runtime and Document cluster purity performance on 20 Newsgroups Dataset
CRD algorithms are still orders of magnitude faster than the other three algorithms. The Euclidian distance based algorithm, Kmeans and CRD-kmeans, cannot cluster the documents correctly and have much lower purity than others. ITC, CRD-ITC and ONMF has similar performance on document cluster purity.
The low rank decomposition on these two real datasets is fast and the runtime can be neglected.
Based on the experiment results we presented in this section, we can see that our CRD framework can be used to design co-clustering algorithms which are much faster and can achieve comparable co-clustering accuracy.
7. CONCLUSIONS
In this paper, we proposed a general framework for fast co-clustering on large data, CRD. CRD has two components. It first decomposes the data matrix into low rank row/column approximation matrices. Then co-clustering algorithms using iterative single-side clustering are used to cluster the approximation matrices. Several constraints are added in the matrix decomposition in order to ensure the performance of co-clustering. Because of the small size of the approximation matrices, CRD has runtime complexity O(t(km′n + ln′m + m′m + n′n)) which is orders of magnitude faster than O(t(k + l)mn), which is the runtime complexity of the previous co-clustering algorithms. We conducted experiments extensively on real datasets and synthetic datasets. The results show that our framework is both efficient and effective.
8. REPEATABILITY ASSESSMENT RESULT
Figure 8 (when m = 3000 and 5000), Figure 16 and Figure 17(top) have been verified by the SIGMOD repeatability committee.
Footnotes
Categories and Subject Descriptors
H.2.8 [Database Management]: Database Applications—Data mining
General Terms
Algorithms
Permission to make digital or hard copies of all or part of this work for personal or classroom use is granted without fee provided that copies are not made or distributed for profit or commercial advantage and that copies bear this notice and the full citation on the first page. To copy otherwise, to republish, to post on servers or to redistribute to lists, requires prior specific permission and/or a fee.
CRD stands for Co-clustering based on Column and Row Decomposition
9. REFERENCES
- 1.Banerjee A, Dhillon I, Ghosh J, Merugu S, Modha DS. A generalized maximum entropy approach to bregman co-clustering and matrix approximation. KDD. 2004 [Google Scholar]
- 2.Chakrabarti D, Papadimitriou S, Modha D, Faloutsos C. Fully automatic cross-associations.. ACM SIGKDD’04 Conference Proceedings; 2004. [Google Scholar]
- 3.Cheng Y, Church G. Biclustering of experssion data. Proc. of the Eighth Int. Conf. on Intelligent Systems for Molecular Biology. 2000 [PubMed] [Google Scholar]
- 4.Deerwester S, Dumais ST, Furnas GW, Landauer TK, Harshman R. Indexing by latent semantic analysis. Journal of the American Society of Information Science. 1990;41(6):391–407. [Google Scholar]
- 5.Dhillon IS, Mallela S, Modha DS. Information-theoretic co-clustering. KDD. 2003 [Google Scholar]
- 6.Ding C, He X, Simon H. On the equivalence of nonnegative matrix factorization and. spectral clustering. SDM. 2005 [Google Scholar]
- 7.Ding C, He X, Zha H, Gu M, Simon H. A min-max cut algorithm for graph partitioning and data clustering. ICDM. 2001 [Google Scholar]
- 8.Ding C, Li T, Peng W, Park H. Orthogonal nonnegative matrix tri-factorizations for clustering. KDD. 2006 [Google Scholar]
- 9.Drineas P, Kannan R, Mahoney MW. Fast monte carlo algorithms for matrices iii: Computing a compressed approximate matrix decomposition. SIAM Journal of Computing. 2005 [Google Scholar]
- 10.Drineas P, Kerenidis I, Raghavan P. Competitive recommendation systems. STOC. 2002 [Google Scholar]
- 11.Golub G, Loan A. Matrix computations. Johns Hopkins University Press; Baltimore, Maryland: 1996. [Google Scholar]
- 12.Hartigan JA. Direct clustering of a data matrix. Journal of the American Statistical Association. 1972;67(337):123–129. [Google Scholar]
- 13.Lee DD, Seung HS. Learning the parts of objects by non-negative matrix factorization. Nature. 1999;401:788–791. doi: 10.1038/44565. [DOI] [PubMed] [Google Scholar]
- 14.Li T. A general model for clustering binary data. KDD. 2005 [Google Scholar]
- 15.Long B, Zhang Z, Yu P. Co-clustering by block value decomposition. KDD. 2005 [Google Scholar]
- 16.Natsoulis G, et al. Classification of a large microarray data set: Algorithm comparison and analysis of drug signatures. Genome Res. 2005;15:724–736. doi: 10.1101/gr.2807605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Slonim N, Tishby N. Document clustering using word clusters via the information bottleneck method. SIGIR. 2000 [Google Scholar]
- 18.Sun J, Xie Y, Zhang H, Faloutsos C. Less is more: Compact matrix decomposition for large sparse graphs. SDM. 2007 [Google Scholar]
- 19.Xu W, Liu X, Gong Y. Document clustering based on non-negative matrix factorization. SIGIR. 2003 [Google Scholar]
- 20.Xu X, Lu Y, Tung A, Wang W. Mining shifting-and-scaling co-regulation patterns on gene expression profiles. ICDE. 2006 [Google Scholar]